Abstract
Replication of the eukaryotic genome is a highly regulated process and stringent control is required to maintain genome integrity. In this review, we will discuss the many aspects of the chromatin and nuclear environment that play key roles in the regulation of both unperturbed and stressed replication. Firstly, the higher order organisation of the genome into A and B compartments, topologically associated domains (TADs) and sub-nuclear compartments has major implications in the control of replication timing. In addition, the local chromatin environment defined by non-canonical histone variants, histone post-translational modifications (PTMs) and enrichment of factors such as heterochromatin protein 1 (HP1) plays multiple roles in normal S phase progression and during the repair of replicative damage. Lastly, we will cover how the spatial organisation of stalled replication forks facilitates the resolution of replication stress.
Keywords: DNA replication, chromatin, DNA repair, nucleus, DNA damage
1 Introduction
Faithful replication of the genome is important to allow successful inheritance of genetic material from parental to daughter cells in all organisms. Eukaryotic DNA replication is preceded in G1 phase with the licencing of pre-replicative complexes (pre-RC), containing head-to-head dimers of minichromosome maintenance (MCM) helicase, to replicative origins throughout the genome in an origin recognition complex (ORC)-dependent manner (Parker et al., 2017). After the G1 to S phase transition, these pre-RCs are activated in a process called origin firing, which is highly dependent on Dbf4-dependent kinase (DDK) cyclin-dependent kinase (CDK) activity (Boos and Ferreira, 2019). Origin firing does not happen simultaneously across the genome; instead, different regions are replicated at different stages throughout S phase and this process is coordinated by various factors such as the local chromatin environment and sub-nuclear localisation. Control of origin firing is vital to ensure coordinated replication of the entire genome (Boos and Ferreira, 2019). Upon origin firing, Cdc45, and GINS are recruited to MCM to form the active CMG helicase which unwinds DNA into single stranded DNA (ssDNA) in a bidirectional manner, forming two replication forks (Burgers and Kunkel, 2017). Replication then occurs from these forks and DNA in synthesised in a semi-conservative manner. Due to the unidirectionality of the CMG helicase and replicative polymerases, the leading strand is synthesised continuously by polymerase epsilon (Polε) and the lagging strand is synthesised in discontinuous Okazaki fragments by polymerases alpha (Polα) and delta (Polδ) (Burgers and Kunkel, 2017).
Throughout S phase, replication forks may be challenged by various sources of replication stress that stall DNA synthesis through a variety of mechanisms. Some endogenous sources include challenging secondary DNA structures within repetitive sequences, barriers to replisome movement such as torsional stress or DNA-protein crosslinks (DPCs) and collisions with the transcription machinery (Zeman and Cimprich, 2014). Importantly, the high levels of endogenous replication stress observed in cancer cells are exploited to develop novel anti-tumour drugs (Ubhi and Brown, 2019). In addition, several exogenous agents can be used experimentally to induce replication stress; for example, hydroxyurea (HU) stalls replication by depleting the cellular supply of deoxyribonucleotides (Bianchi et al., 1986). In the event of replication stress, the cell coordinates a variety of pathways to maintain genome stability and ensure the completion of replication.
These processes occur in a highly organised genome. The genome is spatially arranged in a hierarchical manner from nuclear compartments and chromosome territories (CTs) to topologically associated domains (TADs) and nucleosomes with a range of chromatin states (Gibcus and Dekker, 2013). Genome-wide chromosome conformation capture methods (such as Hi-C) are important tools to delineate how different regions of chromatin interact with each other, allowing the identification of TADs: loops of chromatin with borders that restrict the activity of regulatory elements between different TADs (Dixon et al., 2012; Li et al., 2018). In addition, genomic regions are identified according to their transcriptional status as either A or B compartments. A compartments are transcriptionally active, open and gene-rich whereas B compartments are transcriptionally silent, compact and gene-poor (Gibcus and Dekker, 2013). At the smaller scale, genetic material is packaged into units called nucleosomes comprised of DNA wrapped around a histone octamer of two H2A-H2B dimers and two histone H3-H4 dimers which are connected by H1-bound linker DNA (Luger et al., 2012). The local chromatin environment defined by composition of these nucleosomes, histone post-translational modifications (PTMs) and enrichment of non-histone proteins can affect the levels of transcription and chromatin compaction. In this sense, chromatin is categorised into relaxed, transcriptionally active euchromatin and compact, transcriptionally inactive heterochromatin. For example, histone marks enriched within active chromatin include methylated H3K4 and acetylated H3K9 and inactive marks include trimethylated H3K9 and H3K27 (Kouzarides, 2007). Regions of chromatin can also be localised within specific sub-nuclear compartments; for example, the nuclear/nucleolar periphery, nuclear pores, within nucleoli and in the nuclear interior.
Other than transcriptional regulation, genome organisation has major regulatory roles in the repair of DNA damage and in faithful DNA replication. In this Review, we will outline how the organisation of the genome, ranging from higher order spatial arrangement to the local chromatin environment, impacts the complex process of replication and the response to replication stress.
2 How Chromatin Affects Unperturbed Replication
2.1 Replication Timing
2.1.1 Higher Order Compartments and Nuclear Location
The spatial positioning of specific genomic regions greatly influences their replication timing (RT). This region-specific replication timing program is established during early G1 phase during a period called the timing decision point (TDP), coinciding with the restoration of genome organisation following cell division (Dimitrova and Gilbert, 1999; Dileep et al., 2015). Furthermore, TADs identified through mapping of the entire genome in several cell types correlate with the basic units of replication timing known as replication domains (RD); therefore, the global organisation of the genome contributes to RT at a greater level than the activation of individual replication origins (Pope et al., 2014). Notably, early and late replicating regions correlate with transcriptionally active A and silent B compartments, respectively, and this correlation can be followed even with resolution of nuclear sub-compartments (Ryba et al., 2010; Yaffe et al., 2010; Rao et al., 2014). Histone modifications enriched at specific nuclear compartments involved in transcriptional activation/repression also contribute to this control of replication timing (see below). Higher order reorganisation of the genome during mouse stem cell differentiation, which is coordinated with changes in transcription and cell fate determination, is also associated with changes in the RT program (Hiratani et al., 2008, 2010). Recently it was shown that reorganisation of the TADs is important in modulating origin firing efficiency (Li et al., 2018). During G1 phase, TADs can be rearranged by displacement of CTCF and chromatin decompaction mediated by transcription, which results in the relocation of replication origins to the TAD boundaries. At the spatial boundary of TADs, origin firing is more efficient due to the presence of proliferating cell nuclear antigen (PCNA) clusters that facilitate replication (Li et al., 2021). This describes a mechanism by which transcription plays a role in the regulation of replication initiation, a phenomenon described in multiple studies (Blin et al., 2019; Chen et al., 2019; Li et al., 2021).
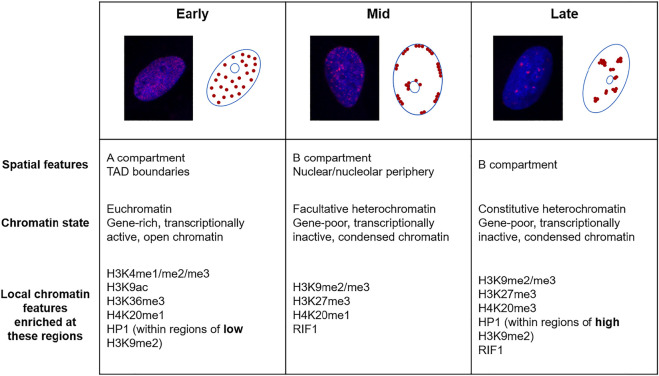
In mammalian cells, the progression of S phase follows a distinct pattern, where euchromatin is replicated in early S phase, facultative heterochromatin at the nuclear/nucleolar periphery in mid S phase and constitutive heterochromatin in late S phase (Heinz et al., 2018) (Figure 1). Some heterochromatic regions of the genome, known as lamina-associated domains (LADs) are associated with the nuclear periphery and bind to the nuclear envelope via lamin proteins. These regions are generally replicated during mid-S phase, gene-poor and categorised under the transcriptionally repressed B compartment. In support of the nuclear periphery environment playing roles in replication timing, artificial targeting of mouse chromocentres containing constitutive heterochromatin to the nuclear periphery advances their replication timing from late to mid-S phase (Heinz et al., 2018). It has been hypothesised that the nuclear periphery contains the replication factors required for origin firing during mid S phase and therefore provides an environment where artificially tethered chromatin is replicated during this time. This advance in replication timing is not due to de novo deposition of facultative heterochromatin marks following artificial tethering, although constitutive histone marks are progressively lost at these regions following subsequent divisions (Heinz et al., 2018).
FIGURE 1.
The replication timing of the eukaryotic genome. Fluorescence microscopy images of U2OS cells labelled with 5-ethynyl-2′-deoxyuridine (EdU, red) and DAPI (blue) and schematics showing the three distinguishable sub-phases of replication; early, mid and late. Below are descriptions of the chromatin replicated in each of these sub-phases.
An important factor that connects genome organisation with the replication timing program is RIF1, a protein mainly involved in maintaining the late replication status of specific genomic regions (Hayano et al., 2012; Yamazaki et al., 2012; Peace et al., 2014; Foti et al., 2016; Hafner et al., 2018; Gnan et al., 2021). Specifically, RIF1 interacts with the nuclear lamina and plays a key role in regulating the replication timing of the LADs (Roux et al., 2012; Foti et al., 2016). Recruitment of RIF1 to RIF1 associated domains (RADs) leads to chromatin reorganisation of these regions and delays the replication timing through regulating origin firing (Yamazaki et al., 2012; Davé et al., 2014; Hiraga et al., 2014; Hiraga et al., 2017; Foti et al., 2016; Alver et al., 2017). This control of replication origin firing by RIF1 is highly dependent on its conserved interaction with protein phosphatase 1 (PP1) (Davé et al., 2014; Hiraga et al., 2014; Hiraga et al., 2017; Mattarocci et al., 2014; Alver et al., 2017; Gnan et al., 2021). Mechanistically, recruitment of PP1 to replication origins by RIF1 reverses the phosphorylation of the MCM complex by DDK, a modification that is required for initiation of replication, and therefore prevents premature replication at specific genomic loci (Davé et al., 2014; Hiraga et al., 2014).
2.1.2 Link Between Histone Modifications and Replication Timing
In both mouse and human cell types, there is a broad correlation between early replicating regions with “active” chromatin marks such as methylated H3K4, H3K36me3, H4K20me1 and acetylated H3K9 but not “repressive” marks including di- and trimethylated H3K9 and H3K27me3 (Hiratani et al., 2008; Yokochi et al., 2009; Ryba et al., 2010; Picard et al., 2014) (Figure 1). In one study in human cells, the strongest correlation between later replication and repressive histone marks was detected with H3K9me2, which is commonly enriched at the nuclear periphery (Ryba et al., 2010).
In mouse embryonic stem cells (mESCs), the degree of DNA methylation affects replication timing through affecting histone modifications particularly at pericentric heterochromatin (Jørgensen et al., 2007a; Takebayashi et al., 2021). Partial loss of DNA methylation (Dnmt1 single knockout), as well as loss of other repressive chromatin modifiers, results in earlier replication of pericentric major satellite repeats (Jørgensen et al., 2007a). Interestingly, complete abolishment (Dnmt1, Dnmt3a, and Dnmt3b triple knockout) results in abnormal enrichment of H3K27me3 at some loci of pericentric heterochromatin, where it instead causes a delay in replication timing (Takebayashi et al., 2021). However, this redistribution of H3K27me3 does not cause RT changes in other regions of the genome and replication timing at these loci appears to be more dependent on transcriptional changes caused by loss of DNA methylation. In another study, it was demonstrated that H3K27me3 enrichment broadly correlates with mid-S phase replicating origins (Picard et al., 2014).
Monomethylation and dimethylation of lysine 20 of histone H4 (H4K20me1 and me2) are found throughout the genome and coincide with chromatin compaction and transcriptional inactivation, whereas trimethylation (H4K20me3) is exclusively enriched at pericentric heterochromatin and imprinting control regions (Schotta et al., 2004; Karachentsev et al., 2005; Delaval et al., 2007). The methyltransferase PR-Set7 deposits H4K20me1 at replication origins and facilitates loading of the pre-RC complex during licencing. This activity is regulated by ubiquitin-mediated degradation of PR-Set7 during S phase to prevent re-replication (Tardat et al., 2010). In contrast, SET8-dependent methylation of H4K20 has been implicated in repressing replication origin licencing through facilitating chromatin compaction (Shoaib et al., 2018). H4K20me1 is present in approximately half of origins and enriched in early- and mid-replicating regions (Picard et al., 2014). The control of the proportion of nucleosomes with each methylation state of H4K20 is critical to regulate origin selection to ensure no re-replication of DNA, allowing faithful replication of the genome (Schotta et al., 2008; Beck et al., 2012). In addition, H4K20me2 is enriched at replication origins, where it acts as a binding site for ORC1, and H4K20me3 is vital for ensuring the late replication of pericentric heterochromatin (Kuo et al., 2012; Brustel et al., 2017).
2.1.3 Modulation of Replication Timing by Heterochromatin Protein 1
Heterochromatin protein 1 (HP1) is a non-histone protein that is recruited to regions of heterochromatin through binding to methylated H3K9 (Lachner et al., 2001). This factor is an identifier of heterochromatin in several organisms and plays a major role in maintaining the local chromatin environment of heterochromatin. Therefore, the different isoforms of HP1 (HP1α, β and γ in humans) play varying roles in regulating processes including transcription, chromatin compaction (Eissenberg et al., 1990; Kwon and Workman, 2011), cell differentiation (Mattout et al., 2015; Casale et al., 2019) and DNA repair (Dinant and Luijsterburg, 2009; Bártová et al., 2017). In Drosophila, HP1 has contrasting roles in replication timing depending on nuclear localisation and H3K9 methylation status. HP1 has been implicated in promoting the early replication timing of some regions with low H3K9me2 enrichment and in mediating later replication of regions with greater H3K9 methylation, such as pericentric heterochromatin (Schwaiger et al., 2010) (Figure 1). Additionally, HP1 is sufficient to induce the late replicating characteristic normally seen in heterochromatic regions when artificially tethered to earlier replicating loci (Pokholkova et al., 2015). Interestingly, HP1 interacts with ORC, a complex that promotes origin firing, and this association is important in HP1α localisation and heterochromatin formation (Pak et al., 1997; Prasanth et al., 2010).
However, mouse HP1 was shown to be dispensable for the maintenance of late replication within pericentric heterochromatin, with the methylation status of H3K9 being more important in establishing the RT of these regions (Wu et al., 2006). Therefore, it is still unclear whether the roles of HP1 in controlling the late replication of pericentric heterochromatin is conserved across organisms or whether H3K9 methylation, a repressive mark which recruits HP1, is more important.
2.2 Restoration of Chromatin Following Replication
2.2.1 Formation of Nucleosomes Behind the Fork
During replication, histones must be dissociated from the DNA to allow progression of the replication fork and it is important that they are accurately restored on the parental strand and duplicated onto the daughter strand. Movement of the replisome destabilises chromatin, leading to decondensation and increased mobility of linker histone H1 (Gasser et al., 1996; Contreras et al., 2003; Kuipers et al., 2011). Indeed, mobility of H1 regulated by its phosphorylation is involved in controlling replication timing, presumably by allowing chromatin relaxation (Contreras et al., 2003; Alexandrow and Hamlin, 2005; Katsuno et al., 2009; Thiriet and Hayes, 2009).
Histone deposition is not identical across the leading and lagging strands of replication, and therefore strand-specific mechanisms of chromatin restoration have been identified which are dependent on several components of the replisome. For example, MCM2 and polymerase α directly bind to parental H3-H4 and recycle it onto the lagging strand, and Dpb3 and Dpb4 (accessory subunits of polymerase ε) facilitate the deposition of H3-H4 onto the leading strand (Gan et al., 2018; Petryk et al., 2018; Yu et al., 2018; Li et al., 2020). It is important that these strand-specific mechanisms of histone deposition are highly regulated to allow symmetric inheritance of histones, alongside their post-translational modifications, during duplication of the genome.
As the replication fork progresses, parental histones are recycled onto the nascent DNA and newly synthesised histones are added in any gaps with the help of histone chaperones (Alabert and Groth, 2012; Stewart-Morgan et al., 2020). For example, chromatin assembly factor 1 (CAF-1) is an important chaperone which interacts with the replication factor PCNA to deposit histones as replication occurs and has roles in maintaining heterochromatin and transcriptional regulation (Yu et al., 2015). The anti-silencing function 1 (ASF1) chaperone interacts with Mcm2-7 and deposits parental and new H3-H4 dimers directly behind the replication fork in a process that is disrupted during replication stress (Groth et al., 2007; Jasencakova et al., 2010). Another important histone chaperone is called facilitates chromatin transcription (FACT), which also interacts with the replication machinery and has roles in both the eviction and deposition of histones during replication (Formosa, 2012). In order to maintain the epigenomic features of the parental DNA, specific histone variants must also be deposited onto the nascent DNA using specialised histone chaperones. This restoration of chromatin does not always occur in a replication-dependent manner; for example, centromere protein A (CENP-A, a H3 variant) deposition by Holliday junction recognition protein (HJURP) at centromeres and H3.3 deposition by death-associated protein 6 (DAXX) at pericentric and telomeric heterochromatin occur after replication in the following G1 phase (Ahmad and Henikoff, 2002; Jansen et al., 2007; Foltz et al., 2009; Goldberg et al., 2010; Lewis et al., 2010).
2.2.2 Maintenance of Histone Modifications
Following the deposition of nucleosomes onto nascent DNA, it is important that histone PTMs from the parental chromatin are inherited so that the newly synthesised chromatin maintains the same transcriptional regulation, conformation, and replication timing. Methylation and acetylation marks are maintained on parental histones, but these are only present on half of the chromatin following replication due to the incorporation of new unmodified histones, so further processes are required to ensure full restoration of PTMs (Scharf et al., 2009; Xu et al., 2012a; Alabert et al., 2015). If a residue is mono-/dimethylated (e.g., H3K9me1/me2, H3K27me1/me2 and H4K20me1/me2), this mark is diluted following replication and newly incorporated histones obtain these PTMs de novo within one cell cycle to become identical to the parental histones (Alabert et al., 2015). On the other hand, trimethylation (e.g., H3K9me3 and H3K27me3) is established slowly and continuously, independent of replication, across several cell divisions (Alabert et al., 2015; Reverón-Gómez et al., 2018). Consequently, oscillations in the levels of histone marks can be observed throughout the cell cycle, and these are not equal at all loci and for all types of histone modification. For example, the H3K4me3 mark enriched at active promoters recovers much quicker than repressive marks such as H3K9me3 and H3K27me3 (Alabert et al., 2015; Reverón-Gómez et al., 2018; Stewart-Morgan, Petryk and Groth, 2020).
2.2.3 Roles of Heterochromatin Protein 1 in Maintaining Heterochromatin
In mouse cells, the interaction of HP1 with the histone chaperone CAF-1, particularly the large p150 subunit, is essential for S phase progression (Quivy et al., 2008). Disruption of this interaction impedes replication of pericentric heterochromatin in a manner that is independent of the canonical histone deposition roles for CAF-1. Intriguingly, p150 is essential for mouse embryo viability during the period of development where HP1-enriched domains are formed (the 8–16 cell stage), suggesting major importance for the p150-HP1 interaction in preserving the survival of cells that are rapidly proliferating and therefore undergoing frequent DNA replication (Houlard et al., 2006).
Additionally, the p150-HP1 complex colocalises with the histone methyltransferase SETDB1 to promote monomethylation of non-nucleosomal histone H3.1 (Loyola et al., 2009). This is important to allow formation of H3K9me1 at newly incorporated histones immediately following DNA replication, which then nucleates formation of trimethylated H3K9 catalysed by Suv39H1/H2. Additionally, the HP1-CAF-1-SETDB1 complex has roles in depositing HP1 at sites of pericentric regions already enriched in H3K9me3. Therefore, CAF-1 has numerous roles in preserving repressive H3K9me3 and HP1 enrichment at pericentric heterochromatin following replication (Loyola et al., 2009).
3 Role of the Local Chromatin Environment During Replication Stress
Replication stress (RS) is defined as the slowing or blocking of replication fork progression by a range of endogenous and exogenous sources. Mild RS only results in the slowing of the replication fork velocity and the activation of dormant origins in order to complete replication (Técher et al., 2017). However, as RS gets more severe, the cellular response becomes more intricate. Prolonged RS may lead to uncoupling of the replisome, resulting in production of stretches of replication protein A (RPA)-bound ssDNA which activate the ataxia telangiectasia and Rad3-related (ATR) kinase (Byun et al., 2005). Downstream effectors of ATR such as checkpoint kinase 1 (CHK1) activate the intra-S phase checkpoint which protects the genome from further instability by inhibiting late origin firing and cell cycle progression and promoting DNA repair pathways (Iyer and Rhind, 2017). In a process called fork reversal, the stalled replication fork is remodelled to form a four-way junction to provide protection against excessive degradation (Neelsen and Lopes, 2015). Although this fork reversal, in combination with the recruitment of multiple factors including RAD51 and FANCD2, prevents degradation by nucleases such as MRE11, DNA2 and MUS81, some controlled resection is required to allow rescue of replication (Bryant et al., 2009; Schlacher et al., 2012; Thangavel et al., 2015; Lemaçon et al., 2017). Replication restart following this remodelling of the fork requires the recruitment of several HR proteins, importantly RAD51, which facilitate homology-directed restart (Ait Saada et al., 2018). Therefore, components of homologous recombination, a pathway canonically associated with DSB repair, play various roles in fork remodelling, protection and restart in the event of replication stress. In this section, we will discuss how several aspects of the local chromatin environment are involved in the regulation of these pathways that play central roles in the resolution of replication stress (Table 1).
TABLE 1.
The roles of histone variants and modifications in replication stress.
| Histone variant/modification | Chaperone/enzyme | Functions in replication stress | Reference(s) |
|---|---|---|---|
| H2A.X | — | Phosphorylation at serine-139 (γH2AX) by ATM/ATR is an early marker of damage and involved in repairing replicative lesions in checkpoint-blind yeast | Ward and Chen (2001), Redon et al. (2003), Sirbu et al. (2011), Lyu et al. (2019), Saldivar et al. (2018) |
| γH2AX maintains normal S/G2 phase transition during unperturbed replication | |||
| macroH2A | FACT | Promotes HR factor recruitment to stressed forks and persists at fragile sites after replication stress resolution to protect from future replication stress | Kim et al. (2018) |
| LSH | Promotes HR factor recruitment to stressed forks | Xu et al. (2021) | |
| Depletion causes increased H4K20me2, which supresses fork protection | |||
| — | Protects the inactive X chromosome from replication stress | Sebastian et al. (2020) | |
| H2A.Z (Htz1) | SWR-C | Prevents misincorporation on dNTPs and collapse of stalled replication forks | Van et al. (2015), Srivatsan et al. (2018) |
| CENP-Α | HJURP | Suppresses formation of centromeric R-loops to prevent TRCs | Giunta et al. (2021) |
| H2A K13/K15 Ub | RNF168 | Facilitates normal S phase progression and promotes fork restart | Schmid et al. (2018); Nakamura et al. (2021) |
| H2B K123Ub | Bre1 (yeast) | Stabilises nucleosomes on newly replicated DNA an facilitates fork progression | Trujillo and Osley (2012), Lin et al. (2014), Northam and Trujillo (2016), Hung et al. (2017) |
| Activates the intra-S phase checkpoint | |||
| Maintains error-free DNA damage tolerance | |||
| H3K4me1/me3 | MLL3/4 | Promotes MRE11-depedendent fork degradation in BRCA-deficient cells | Ray Chaudhuri et al. (2016) |
| SETD1A | Promotes RAD51-mediated fork protection through H3.1 chaperoning | Higgs et al. (2018) | |
| Set1 (yeast) | Prevents TRCs in active regions | Chong et al. (2020) | |
| H3K9me2/me3 | Met-2, set-25 (C. elegans) | Represses genes in repetitive regions to prevent R-loop formation | Zeller et al. (2016) |
| H3K27me3 | EZH2 | Recruits MUS81 to facilitate fork restart | Rondinelli et al. (2017) |
| H4K20me1/me3 | PR-Set7 | Prevents replication stress by controlling fork number and velocity | Jørgensen et al. (2007a), Tardat et al. (2007), Saredi et al. (2016) |
| H4K20me0 on new histones provides binding site for TONSL-MMS22L to facilitate post-replicative repair | |||
| H4 K5/K12 ac | HAT1 | Prevents replication stress and protects stressed forks from MRE11-dependent degradation | Nagarajan et al. (2013), Agudelo Garcia et al. (2020) |
| H4K8ac | PCAF | Promotes MRE11- and EXO1-dependent degradation of stalled forks in BRCA2-deficient cells | Kim et al. (2020) |
3.1 Histone Variants
In humans, there are several variations of histones H2A, H2B, H3, and H4 which have different sequences to their canonical histone counterparts and may be localised to specific genomic regions to influence the structure and function of the chromatin (Martire and Banaszynski, 2020). Here, we will describe the importance of some of these histone variants during replication and particularly in the event of replication stress.
3.1.1 Histone H2AX
The histone H2A variant H2A.X has major implications in genome stability: specifically, H2A.X phosphorylated at serine-139 rapidly after DSB induction, creating a mark known as γH2AX (Rogakou et al., 1998). γH2AX formation catalysed by the ATR signalling cascade also occurs following replication stalling before the collapse of forks in DSBs, although inhibition of Chk1, a downstream factor of ATR, has been shown to induce γH2AX formation (Ward and Chen, 2001; Sirbu et al., 2011). This association of γH2AX presumably facilitates recruitment of DNA repair proteins and studies in yeast have revealed a role in repairing replicative damage in cells without intra-S phase checkpoint activation (Redon et al., 2003). Interestingly, γH2AX distribution following replication stress is not equal across the genome and greater association is seen in commonly fragile regions containing compact chromatin which are depleted of transcription start sites and CpG islands (Lyu et al., 2019). Therefore, this modification could be primarily important to promote certain pathways involved in the resolution of replicative damage in specific chromatin contexts, or this enrichment could reflect persistent replication stress particularly in fragile regions of the genome that are difficult to repair. In addition, ATR kinase activity and H2AX phosphorylation also occur during unperturbed S phase to facilitate the correctly timed transition into G2 phase (Saldivar et al., 2018). Consequently, γH2AX not only has direct roles during stressed replication but also is essential to maintain genome integrity by preventing premature entry into mitosis and under-replication of the genome.
3.1.2 Macro H2A
MacroH2A is a subfamily of histone H2A variants comprised of three isoforms called macroH2A1.1, macroH2A1.2 (which are splice variants from the same gene) and macroH2A2 (Rasmussen et al., 1999; Costanzi and Pehrson, 2001). Structurally, these proteins are composed of a H2A domain with an N-terminal non-histone macro domain and are approximately three times the size of other H2A variants (Pehrson and Fried, 1992). Some functions of this histone variant have been linked to X chromosome inactivation (Costanzi and Pehrson, 1998; Mermoud et al., 1999; Rasmussen et al., 2000), transcriptional regulation (Ouararhni et al., 2006; Gamble et al., 2010; Creppe et al., 2012) and nucleosome organisation (Angelov et al., 2003; Abbott et al., 2004; Chakravarthy and Luger, 2006; Muthurajan et al., 2011; Chakravarthy et al., 2012). Additionally, macroH2A is enriched at heterochromatin regions, particularly those marked by H3K9me3, and has roles in higher genome organisation (Douet et al., 2017; Kozlowski et al., 2018).
Alongside functions in homology-directed DSB repair (Xu et al., 2012b; Khurana et al., 2014), macroH2A1.2 also has roles during replication stress, where it is deposited onto chromatin by the FACT histone chaperone (Kim et al., 2018). This activity is particularly important at common fragile sites (CFS), regions that are more prone to replication stress induced damage. MacroH2A deposition is also assisted by the LSH chromatin remodeller and promotes BRCA1 and RAD51 recruitment to stalled forks to promote fork protection and facilitate repair (Kim et al., 2018; Xu et al., 2021). Recently, it was shown that loss of this macroH2A deposition is associated with an increase of H4K20me2 at stalled forks which then favours 53BP1 recruitment to stalled forks rather than BRCA1, causing a detrimental effect on fork protection (Xu et al., 2021). MacroH2A1.1 and 1.2 are enriched on the mammalian female inactive X (Xi) chromosome, a highly condensed genomic region with increased susceptibility to replication stress (Costanzi and Pehrson, 1998; Koren and McCarroll, 2014). The macroH2A1.2 variant is involved in supressing replication stress at the Xi, whereas the splice variant macroH2A1.1 activates the alternative end joining DSB repair pathway, leading to Xi anaphase defects in the absence of H2A1.2 (Sebastian et al., 2020). Overall, these studies suggest an importance for macroH2A in facilitating the proper repair of difficult-to-replicate genomic loci such as CFS’s and the inactive X chromosome. The replication stress-dependent recruitment of macroH2A to fragile sites is mainly transient but some remains after resolution of replication stress and protects these difficult to replicate regions from future replicative damage (Kim et al., 2018). Since macroH2A has roles in repressing gene expression, X inactivation and nucleosome organisation, it would be important to see whether this continued enrichment alters transcription and genome organisation at these sites.
3.1.3 H2A.Z
H2A.Z is a variant of histone H2A that is incorporated throughout the cell cycle and has the ability to alter the physical properties of the nucleosome and create specialised chromatin structures (Fan et al., 2002, 2004). This histone variant has conserved functions in the regulation of transcription in euchromatic genes (Zhang et al., 2005; Hardy et al., 2009). Nucleosomes associated with H2A.Z also enhance replication origin firing efficiency through promoting H4K20 dimethylation and ORC1 recruitment (Long et al., 2020). In yeast, incorporation of the H2A.Z homologue Htz1 by Swi2/Snf2-related chromatin remodelling complex (SWR-C) is important in maintaining genome stability following both DSBs (Kalocsay et al., 2009; Horigome et al., 2014) and replication stress (Van et al., 2015; Srivatsan et al., 2018). Specifically, Hitz1 has roles in preventing the misincorporation of nucleotides during replication and in promoting the repair of replicative damage (Van et al., 2015; Srivatsan et al., 2018). Incorporation of Htz1 during replication stress has been hypothesised to prevent collapse of forks into DSBs by two mechanisms: either it stabilises the fork to prevent replisome dissociation, or it is incorporated after replisome dissociation to prevent further degradation of the fork by nucleases (Srivatsan et al., 2018). Furthermore, the balance of H2A.Z on chromatin is vital: as removal by the INO80 chromatin remodeller is also important in maintaining replication fork stability and to allow HR to occur (Papamichos-Chronakis et al., 2011; Lademann et al., 2017).
3.1.4 CENP-Α at Centromeres
Despite being located between pericentromeric regions expressing features of heterochromatin, mammalian centromeres display active epigenetic marks (e.g., H3K4me2 and H3K36me2) combined with production of long non-coding RNA transcripts (Sullivan and Karpen, 2004). In human cells, centromeres are composed of tandem alpha satellite repeats and enriched in CENP-A, a histone H3 variant specific for centromeres which has key roles in formation of kinetochores to allow proper chromosome segregation (Barra and Fachinetti, 2018). Because of their involvement in chromosome segregation, the maintenance of centromere integrity during replication is vital to ensure proper cell division.
Unlike many canonical histone variants, CENP-Α is not deposited onto newly replicated DNA during S-phase but is instead incorporated in the following G1-phase by its dedicated histone chaperone, HJURP (Jansen et al., 2007). Contrary to this, CENP-A has recently been shown to possess key roles in maintaining centromere integrity during DNA replication (Giunta et al., 2021). CENP-A deposition prevents the formation of centromeric R-loops formed as a consequence of transcription-replication conflicts (TRCs) during late S phase, thereby promoting fork progression. Centromeres depleted of CENP-A display multiple replication-associated defects such as error-prone mitotic DNA synthesis (MiDAS) and chromosomal translocations, breakages, and fragmentation. These chromosomal aberrations are the result of recombination between alpha satellite repeats in an R-loop dependent manner rather than being due to defects in chromosomal segregation during mitosis (Giunta et al., 2021). The specialised function for CENP-A in removing R-loops is important because some alpha-satellite RNA transcripts remain associated with the centromere and could therefore disrupt the progression of incoming replication forks (McNulty et al., 2017).
3.2 Role of Linker Histone H1
In human cells, eviction of the linker histone H1 by the histone chaperone SET results in sensitivity to DNA damage (Mandemaker et al., 2020). Although depletion of SET does not alter normal S phase progression, it does cause resistance to the replication stress-inducing agent HU; suggesting important roles for chromatin-bound H1 during perturbed replication (Mandemaker et al., 2020). In agreement, overexpression of SET causes slower S phase progression upon treatment of HU (Kalousi et al., 2015). SET also promotes retention of the heterochromatin factors KAP1 and HP1 on chromatin which causes chromatin compaction and inhibition of HR repair of collapsed forks (Kalousi et al., 2015). Potentially, loss of SET and subsequent H1 retention and chromatin decompaction could allow for greater access of DNA repair factors to replicative lesions, thereby promoting proper repair and cell survival.
In addition, the linker histone dH1 in Drosophila has been implicated in preventing the accumulation of R-loops, a structure which can induce replication stress (Bayona-Feliu et al., 2017). Upon depletion of dH1, transcriptional repression is relieved within heterochromatin regions, leading to the accumulation of R-loops. Interestingly, R-loop accumulation was not seen upon depletion of HP1a, suggesting that this effect is only seen upon loss of H1 and is not a general effect of transcriptional derepression in heterochromatin (Bayona-Feliu et al., 2017), although loss of repression in repetitive regions has been associated with R-loop prevention in C. elegans (Zeller et al., 2016). Notably, in mouse cells depleted of histone H1, fork stalling as a result of transcription-replication conflicts has also been observed (Almeida et al., 2018). H1 loss leads to chromatin decompaction which causes acceleration of both transcription initiation and replication leading to pathogenic accumulation of R-loops and collisions between the transcription and replication machineries (Almeida et al., 2018).
The linker histone H1 has been shown to have roles in both preventing the onset of replication stress and in facilitating the repair of replicative damage. Currently, it is unclear how H1 elicits these effects: whether it is due to its roles in organising nucleosome particles into stable higher order structures and maintaining proper spacing between nucleosomes or in the formation of heterochromatin and maintaining transcriptional repression (Happel and Doenecke, 2009).
3.3 Histone Modifications and Replication Stress
3.3.1 Histone Ubiquitination
Ubiquitination of histone H2AK13 and K15 adjacent to DNA lesions plays a central role in the DNA damage response, where these marks facilitate the recruitment of several DNA repair factors (Smeenk and Mailand, 2016). During unchallenged S phase, H2A ubiquitination by RNF168 and activation of the DNA damage response (DDR) is required for normal replication progression and to prevent fork stalling (Schmid et al., 2018). In addition, RNF168 plays a role in preventing accumulation of reversed forks through restarting stalled replication, particularly at difficult to replicate repetitive regions of the genome (Schmid et al., 2018). Formation of γH2AX by ATR and ATM kinases occurs upstream of H2A ubiquitination and is involved in coordinating this DDR activation during S phase (Schmid et al., 2018; Nakamura et al., 2021). Interestingly, ATM inhibition increases activation of the histone ubiquitin response upon fork breakage by camptothecin. This is possibly because ATM is involved in a negative feedback loop alongside PLK1 where end resection at single-ended DBSs deactivates the histone ubiquitination pathway (Nakamura et al., 2021).
Monoubiquitinated histone H2B is a dynamic histone mark first associated with transcription and repair (Kao, 2004; Fleming et al., 2008) also possessing other roles in controlling chromatin compaction during DSB repair (Moyal et al., 2011; Nakamura et al., 2011). In yeast, the ubiquitin ligase responsible for H2B monoubiquitination, Bre1, is maintained on replicating DNA and is further enriched at stressed replication forks (Trujillo and Osley, 2012; Lin et al., 2014; Hung et al., 2017). During replication, H2B ubiquitination plays a role in regulating nucleosome assembly onto replicating DNA, which facilitates normal fork progression and replisome stability during replication stalling by HU (Trujillo and Osley, 2012). In contrast, H2Bub has also been shown to restrict fork progression upon HU-induced stress through coordinating chromatin assembly and activation of the Rad53-dependent intra-S checkpoint (Lin et al., 2014). Intriguingly, other roles for H2Bub during replication stress are linked to its involvement in regulating DNA damage tolerance (DDT) pathways (Northam and Trujillo, 2016; Hung et al., 2017). This mark promotes polymerase eta (Polη)-dependent translesion synthesis (TLS) and suppresses the more mutagenic polymerase zeta (Polζ)-dependent pathway following fork stalling by HU and UV treatment (Northam and Trujillo, 2016). In another study, monoubiquitinated H2B was shown to promote recombination-dependent lesion bypass following treatment with alkylating agents, specifically by altering the chromatin dynamics and allowing RAD51 recruitment (Hung et al., 2017). This mark then facilitates repair of these bypassed lesions post-replication, possibly by promoting activation of the G2/M checkpoint. Therefore, this histone modification has cell cycle-specific roles for maintaining genome replication following replicative damage checkpoint (Hung et al., 2017). Whilst these studies have elucidated the importance of H2B monoubiquitination and chromatin dynamics during replication stress, it remains unclear how this modification has seemingly contradictory roles in both stalling of replication through checkpoint activation and coordinating DDT pathways to allow DNA synthesis beyond lesions and subsequent post-replicative repair.
3.3.2 Histone H3 Modifications
Methylation of H3K4 has been implicated to have contrasting roles in the protection of stalled replication forks in mammalian cells depending on which methyltransferase deposits this mark (Ray Chaudhuri et al., 2016; Higgs et al., 2018). In mouse cells, MLL3/4 is responsible for depositing H3K4me1 and H3K4me3 at stressed forks, where it promotes fork degradation by MRE11 in BRCA-deficient mouse cells with existing defects in fork protection (Ray Chaudhuri et al., 2016). Therefore, loss of H3K4 methylation by MLL3/4 rescues fork protection in BRCA-deficient cells, leading to chemoresistance (Ray Chaudhuri et al., 2016). In contrast, monomethylation of H3K4 at stressed forks by human SETD1A promotes RAD51-dependent replication fork protection from degradation by DNA2 and CHD4 (Higgs et al., 2018). This fork protection is mediated through chaperoning of histone H3.1 by the Fanconi anaemia factor FANCD2 (Higgs et al., 2018), a factor involved in interstrand crosslink repair, RAD51-mediated replication fork protection and the replication of fragile sites (Schlacher et al., 2012; Sato et al., 2012; Madireddy et al., 2016; Higgs et al., 2018). It is interesting that methylation of H3K4 by different methyltransferases leads to opposite effects on fork protection and this could mean that these enzymes are either active at different times during the replication stress response, are recruited differentially to certain genomic loci or are responsible for methylation within a specific local chromatin context (e.g., presence of H2BUb) (Wu et al., 2008; Higgs et al., 2018). Additionally, H3K4me is commonly found in active regions, so it would be important to investigate whether these mechanisms are mostly important at these regions or whether the mark is added de novo throughout the genome in response to replication stress. Notably, fork protection upon loss of MLL3/4 was observed only in BRCA-deficient cells whereas fork degradation by SETD1A loss was observed in BRCA-WT cells (Ray Chaudhuri et al., 2016; Higgs et al., 2018). Therefore, the promotion of fork degradation by MLL3/4 mediated methylation of H3K4 may specifically occur when HR-mediated fork protection is lost.
In yeast, H3K4 methylation plays a role in slowing down fork progression through highly transcribed regions to prevent transcription-replication conflicts, thereby ensuring faithful replication (Chong et al., 2020). Indeed, transcriptionally active genes are commonly decorated with H3K4me3 (Santos-Rosa et al., 2002; Guenther et al., 2007). Another histone H3 modification with roles in preventing TRCs is methylated H3K9 (Zeller et al., 2016), a mark commonly enriched at heterochromatin that plays a role in HP1 recruitment and transcriptional repression (Bannister et al., 2001; Lachner et al., 2001; Nakayama, 2001). In C. elegans, H3K9me2/me3 function to repress genes in repetitive elements, thereby suppressing R-loop formation and preventing replication stress (Zeller et al., 2016).
Trimethylated H327 is also a mark of transcriptionally inactive chromatin and is deposited by enhancer of zeste homologue 2 (EZH2), a component of the polycomb repressive complex 2 (PRC2), during both G1 and S phases (Hansen et al., 2008; Morey and Helin, 2010). Alongside its important roles in transcription and cell identity (Wiles and Selker, 2017), H3K27me3 also plays a role during replication stress (Rondinelli et al., 2017). Methylated H3K27 is involved in the recruitment of MUS81 to stalled forks, an endonuclease which creates DSBs to facilitate HR-mediated fork restart (Hanada et al., 2007; Rondinelli et al., 2017). Whilst activation of this pathway is required for replication restart in BRCA2-deficient cells, excessive fork degradation by MUS81 can be toxic. Therefore, in a similar manner to with the MLL3/4/H3K4me/MRE11 axis (Ray Chaudhuri et al., 2016), loss of EZH2 and subsequent fork protection from MUS81 endonuclease activity leads to chemoresistance in BRCA-deficient cells (Rondinelli et al., 2017). These roles of epigenetic modifications in protecting the replication fork, thereby allowing cancer cells to become resistant to therapies, have major clinical implications. For example, expression levels of EZH2 could be used as a biomarker to predict resistance to drugs such as poly (ADP-ribose) polymerase (PARP) inhibitors and allow more stratified treatment for BRCA-deficient cancers (Rondinelli et al., 2017).
3.3.3 Histone H4 Modifications
In addition to its roles in replication timing, the methyltransferase PR-Set7 responsible for H4K20 methylation has roles in facilitating S phase progression and during replication stress, where it interacts with the key replication factor PCNA (Jørgensen et al., 2007b). In addition, depletion of this factor affects the number and velocity of replication forks and causes activation of p53 and Chk1 dependent checkpoints. Loss of H4K20 methylation in this manner increases the frequency of replicative DNA breaks which then recruit repair factors such as RPA, RAD51 and 53BP1 (Jørgensen et al., 2007b; Tardat et al., 2007). Importantly, unmethylated H4K20 which marks newly incorporated histones plays a role in regulating the post-replicative repair of DNA lesions obtained during S phase (Saredi et al., 2016). H4K20me0 provides a binding site for TONSL-MMS22L in G2/M phases, and this complex then promotes HR repair (Duro et al., 2010; Saredi et al., 2016). Overall, these studies show roles for H4K20 methylation status in the regulation of replication fork progression and, in addition, the repair of replicative lesions after S phase completion.
Histone acetyltransferase 1 (HAT1) catalyses the acetylation of histone H4 at lysine 5 and lysine 12 and this activity is important in the processing of newly deposited histones during replication (Nagarajan et al., 2013). Depletion of HAT1 results in replication stalling and sensitivity of cells to replication stress induced by HU (Nagarajan et al., 2013; Agudelo Garcia et al., 2020). The importance of HAT1 in maintaining genome stability during replication stress is due to its roles in protecting stalled forks from degradation by MRE11 (Agudelo Garcia et al., 2020). In contrast, acetylation of H4K8 by p300/CBP-associated factor (PCAF) promotes nucleolytic degradation of stalled forks in BRCA-deficient cells, suggesting differential roles for histone acetylation on replication fork protection depending on which residue is modified and by which acetyltransferase (Kim et al., 2020). Acetylation of H4K8 by PCAF provides a binding site for MRE11 and EXO1 nucleases, and this activity is suppressed by ATR-mediated phosphorylation of PCAF to maintain tight control of stalled fork degradation. In BRCA2-deficient tumours where fork protection is compromised, low levels of PCAF activity is associated with resistance to PARP inhibition through restoring fork protection (Kim et al., 2020).
3.4 HP1 During Replication Stress
In a more recent study, HP1β has been shown to have some important roles during normal replication progression and in the presence replication stress in mammalian cells. Knockout of HP1 in mouse embryonic fibroblasts (MEFs) led to reduced cell growth and fork speeds in combination with enhanced formation of DNA damage foci (Charaka et al., 2020). Upon depletion of deoxyribonucleotides, both mouse and human (Hela) cells depleted of HP1 displayed increased fork stalling and defective fork restart. In support for key roles of HP1 during replication stress, depletion also sensitises human and mouse cells to HU and cisplatin, resulting in increased levels of chromosomal aberrations (Charaka et al., 2020). It is currently unknown exactly how HP1 plays a role in the cellular response to replication stress, however it could share some similarities with its roles in DSB repair. For example, HP1 depletion leads to reduced recruitment of BRCA1 to DSBs, so perhaps the importance of HP1 in the resolution of replication stress is dependent on an ability to recruit BRCA1, which then promotes RAD51-dependent fork protection (Schlacher et al., 2012; Lee et al., 2013). On the other hand, HP1 retention on heterochromatin has been implicated in preventing homology-directed repair of heterochromatic DSBs and mobilisation of HP1β plays a role in the activation of DDR signalling (Ayoub et al., 2008; Kalousi et al., 2015). Currently, it is unclear which specific pathways HP1 plays a role in to resolve replication stress and whether it is strictly important for RS in heterochromatin or whether it has global roles.
4 Movement of Stressed Replication Forks
The mobility of DNA lesions, namely DSBs, has been well documented and has been shown to be vital for proper repair in specific circumstances. In mammalian cells for example, whilst heterochromatic DSBs are positionally stable in G1 and are repaired by non-homologous end joining (NHEJ), they are relocated to the periphery of heterochromatin domains during S and G2 phases to allow recruitment of HR factors (Tsouroula et al., 2016). The mobility of DSBs is a conserved response that allows formation of the RAD51 filament and repair of DSBs, especially within heterochromatin, and SUMOylation is an important post translational modification which controls this process (Oza et al., 2009; Ryba et al., 2010). Therefore, certain nuclear compartments provide a protective environment for DNA repair, and this is also important in the event of replication stress. In this section we will discuss how some stressed forks do not remain positionally stable and are relocated to specific nuclear compartments in a similar manner to DSBs.
4.1 The Nuclear Pore Complex and Replication Stress
4.1.1 Importance in Repetitive vs. Non-repetitive Sequences
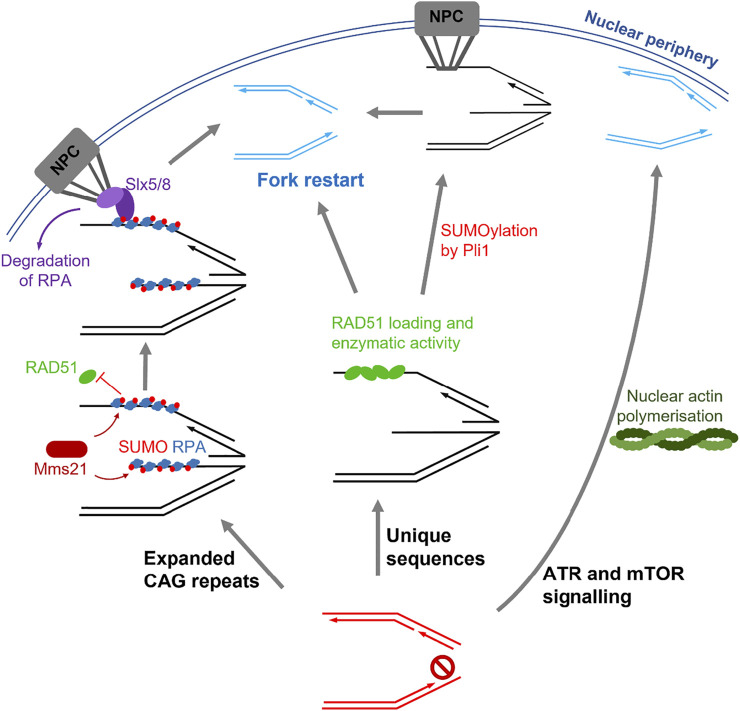
During S phase in yeast, replication stress prone expanded CAG repeats relocate to the nuclear periphery where they interact with components of the nuclear pore complex (NPC) to prevent chromosomal breakages (Su et al., 2015). For this movement to occur, the repair proteins RPA, Rad59 and Rad52 are SUMOylated by Mms21 SUMO E3 ligase which permits their interaction with the SUMO interacting motif (SIM) of Slx5 (Whalen et al., 2020). SUMOylation of the ssDNA-binding protein RPA inhibits Rad51 binding at the stalled fork and this inhibition is lost when collapsed forks associate with NPCs, possibly by degradation of SUMOylated proteins promoted by the SUMO-targeted ubiquitin ligase (STUbL) Slx5/8 (Su et al., 2015; Whalen et al., 2020). Therefore, homology-directed fork restart is suppressed until movement of these repetitive sequences to the nuclear pore. Intriguingly, in a study where replication stress was induced at a unique sequence in yeast, RAD51 binding and activity were able to occur before anchoring to the NPC (Kramarz et al., 2020). Here, SUMOylation by Pli1 was shown to promote fork mobility, and anchoring of the fork to the NPC caused removal of the SUMO chains and allowed recombination-dependent restart. Alternatively, recombination-dependent restart could still even occur without NPC anchoring when SUMOylation by Pli1 was selectively inhibited (Kramarz et al., 2020).
Overall, these studies suggest that different mechanisms of replication stress resolution occur depending on the nature of the DNA sequence (Figure 2). In repetitive sequences (i.e., expanded CAG repeats), RAD51 binding is limited until tethering to the NPC, which is essential to allow replication restart and prevention of DSBs (Su et al., 2015; Whalen et al., 2020). Conversely, within non-repetitive sequences, RAD51 activity occurs at stalled forks before relocation to the NPC, and restart can occur with or without NPC tethering (Kramarz et al., 2020). Therefore, mobility of stalled forks and tethering to the NPC may only be necessary for repetitive sequences, which are likely to undergo detrimental recombination events if in close proximity to other repetitive sequences. The involvement of SUMO is similar in both repetitive and unique sequences, where it promotes mobility of stalled forks but is removed upon tethering to the NPC for replication restart to occur (Su et al., 2015; Kramarz et al., 2020; Whalen et al., 2020).
FIGURE 2.
Summary of the pathways involved in stressed fork mobility. Left: fork stalling induced by expanded CAG repeats leads to SUMOylation of repair proteins such as RPA by Mms21, which block RAD51 binding. Tethering of the fork to the nuclear pore complex (NPC) by Slx5/8 binding releases this inhibition and allows fork restart. Centre: Replication fork stalling within unique sequences results in loading of RAD51 in the nucleoplasm followed by SUMOylation events by Pli1, which allow NPC tethering and fork restart. Alternatively, fork restart can also occur in the absence of SUMOylation and NPC tethering. Right: ATR and mTOR signalling during replication stress leads to polymerisation of nuclear actin. Stalled forks then move along the actin filaments to the nuclear periphery, where fork restart can occur.
4.1.2 Anchoring of Stressed Telomeres to the Nuclear Pore Complex
Telomeres in yeast and human cells are relocated to the nuclear pore complex in response to genomic stress (Khadaroo et al., 2009; Churikov et al., 2016; Pinzaru et al., 2020). In budding yeast, this relocalisation of eroded telomeres is SUMO- and STUbL-dependent and promotes type II recombination [the mechanism of alternative lengthening of telomeres (ALT) in yeast] (Churikov et al., 2016). In human cancers, replication stress at telomeres can be arise due to dysfunction of the telomere protection factor POT1 (Pinzaru et al., 2016; Pinzaru et al., 2020). This type of replicative stress causes increased MiDaS at telomeres and relocation of a small fraction of them to nuclear pore. This movement is promoted by polymerisation of nuclear actin and is required to maintain telomere integrity in POT1-deficient cells. Indeed, disruption of the nuclear pore complex in cells harbouring a DNA binding deficient POT1 mutant exacerbates telomere repeat instability and detrimental telomeric recombination events (Pinzaru et al., 2020).
4.1.3 Processing of R-Loops at the Nuclear Pore Complex
Several studies have implicated roles for the NPC in the resolution of transcription-replication conflicts. It has been hypothesised that transcribed genes are transiently localised to the nuclear pores to aid in nuclear export of transcription products in a process called gene gating (Blobel, 1985). In yeast, localisation of transcribed genes to the NPC is important in preventing pathological R-loop formation, suggesting that gene gating and the subsequent nuclear transport of nascent RNA is important in preventing TRCs (García-Benítez et al., 2017). The human NPC component Tpr has also been implicated in the processing of DNA-RNA hybrids (Kosar et al., 2021). Interestingly, activation of the ATR-dependent checkpoint during replication stress releases transcribed genes from the nuclear pore to facilitate fork restart (Bermejo et al., 2011). These results suggest that proximity to the nuclear pore is important in preventing R-loop-dependent replication stress within transcribed genes, but upon exposure to other sources of replication stress, these genes are released from the NPC possibly to allow R-loop formation which consequently promotes DDR signalling and fork restart (García-Benítez et al., 2017). In addition, topoisomerases have been shown to suppress RNA-DNA hybrid formation during replication through modulating gene topology (Tuduri et al., 2009; Achar et al., 2020). Specifically, Top2 maintains negative supercoiling at gene boundaries, which facilitates normal transcription and suppresses hybrid formation specifically during S phase (Achar et al., 2020).
4.2 Mobility of Stressed Replication Forks by Nuclear Actin
Whilst it remains unclear whether stressed fork mobility is caused by passive diffusion or involves an active mechanism, recent studies have shown that the contractile fibre actin has roles during replication stress, suggesting fork movement is an active process. Transient polymerisation of actin, forming filamentous actin (F)-actin, occurs in the nucleus and is involved in the relocation of DSBs, particularly within heterochromatin, to facilitate HR repair (Belin et al., 2015; Caridi et al., 2018; Schrank et al., 2018). During unperturbed replication, nuclear actin polymerisation is important to allow recruitment of replication factors and S phase progression (Parisis et al., 2017). Additionally, upon replication stress, F-actin formation at stressed replication foci reversibly increases in an ATR and mTOR signalling-dependent manner (Lamm et al., 2020). Stalled forks then move along the F-actin, facilitated by myosin motor proteins, to localise with the nuclear periphery. This localisation allows replication restart, and therefore resolution of replication stress before collapse into a single ended DSB, which distinguishes this pathway from the movement of DSBs to allow HR (Schrank et al., 2018; Lamm et al., 2020) (Figure 2). In addition, the nuclear volume increases as a result of F-actin formation, possibly due to chromatin decompaction, and this response is not seen during DSB repair (Lamm et al., 2020). The movement of stalled forks to the nuclear lamina may allow direct interactions with the nuclear envelope and be important in the recruitment of repair factors; indeed, the laminA/C proteins has been implicated in promoting the recruitment of RPA, RAD51 and FANCD2 (Singh et al., 2013). On the other hand, DSBs located at the nuclear periphery within LADs suppress HR by inhibiting the recruitment of RAD51 (Lemaître et al., 2014). In the future, it will be therefore important to understand the roles of the nuclear lamina in the regulation of homologous-directed repair of DSBs and stalled replication forks.
5 Conclusion and Future Perspectives
The organisation of the genome plays roles in facilitating progression of DNA replication on multiple levels. At the higher levels of genome organisation, arrangement of genetic material into higher order structures and localisation within specific compartments play a major role in controlling the replication timing program. In addition, the composition of the nucleosome as defined by histone variants and PTMs, and the enrichment of non-histone proteins all modulate replication timing (Figure 1). In many cases, this replication timing is strongly linked to transcription where active regions correlate with early replication and inactive regions with late. The importance of this link between transcription and replication timing and whether there is any causality between these two processes are still unclear. Possibly, the compacted state of repressed chromatin may act as a barrier to early replication, localisation of inactive/active regions at specific loci may impact origin firing or it may be favourable to duplicate replication stress prone sequences (e.g., gene-poor repetitive regions) later in S phase.
In addition, the chromatin environment plays important roles in the resolution of replication stress. Particularly, the presence of histone variants and modifications at stressed loci play major roles in promoting or repressing specific repair pathways (Table 1). However, the question remains whether these variants/marks are deposited de novo at all genomic regions during replication stress and remain at these loci after the resolution of replication stress to protect against future challenges or whether they are present only at specific regions that are more difficult to repair.
The SUMO-directed movement of stressed replication forks to facilitate recombination-dependent repair shares some striking similarities with the mobility of DSBs to the nuclear periphery. This mobility of stalled forks is primarily important within repetitive regions such as expanded CAG repeats and telomeres, and the mobility of DSBs occurs when breaks are situated in heterochromatin (Figure 2). The movement of these specific regions to the nuclear periphery could be important for at least two reasons: movement may prevent detrimental recombination events between repeats and/or the environment of the nuclear periphery may provide a protective environment for the repair of these difficult sequences. Indeed, association of stressed forks with components of the nuclear pore complex is vital for repair of replicative lesions. Future studies linking the roles of nuclear actin and movement of stressed forks to the NPC would be beneficial to understand how these fork movements are regulated and propagated.
Tumours are highly prone to replication stress due to their uncontrolled proliferation and deregulated cellular signalling, and this feature is already being targeted in several therapies in clinic (Ubhi and Brown, 2019). Consequently, some of the features of the chromatin could be exploited to provide novel targeted therapies for cancer. Histone marks that regulate the protection of stalled forks form nucleolytic degradation such as methylated H3K4 and H3K27 and acetylated H4K8 have major implications in the treatment of cancers, specifically those harbouring BRCA1/2 mutations. Therefore, measuring the expression of the enzymes that deposit/remove these marks and the enrichment of the marks themselves could provide biomarkers that could be used in the future to predict resistance of tumours to certain therapies and allow greater stratification of treatment. In addition, drugs that target specific enzymes and modify the epigenome and sensitise cells to other therapies. For example, histone deacetylase (HDAC) inhibitors are already used in the clinic and combination of these with other anti-cancer drugs are being explored (Suraweera et al., 2018). Finally, the movement of stalled forks by nuclear actin is mediated through ATR and mTOR signalling, so these pathways provide attractive targets for anti-cancer drugs. Indeed, polymerisation of nuclear actin facilitates survival of cancer cells (Lamm et al., 2020). In summary, future studies elucidating the role of chromatin during replication stress are important in discovering new and more stratified cancer treatments.
Author Contributions
JW and ES wrote the review.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbott D. W., Laszczak M., Lewis J. D., Su H., Moore S. C., Hills M., et al. (2004). Structural Characterization of MacroH2A Containing Chromatin. Biochemistry 43 (5), 1352–1359. 10.1021/bi035859i [DOI] [PubMed] [Google Scholar]
- Achar Y. J., Adhil M., Choudhary R., Gilbert N., Foiani M. (2020). Negative Supercoil at Gene Boundaries Modulates Gene Topology. Nature 577 (7792), 701–705. 10.1038/s41586-020-1934-4 [DOI] [PubMed] [Google Scholar]
- Agudelo Garcia P. A., Lovejoy C. M., Nagarajan P., Park D., Popova L. v., Freitas M. A., et al. (2020). Histone Acetyltransferase 1 Is Required for DNA Replication fork Function and Stability. J. Biol. Chem. 295 (25), 8363–8373. 10.1074/JBC.RA120.013496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K., Henikoff S. (2002). The Histone Variant H3.3 Marks Active Chromatin by Replication-independent Nucleosome Assembly. Mol. Cel 9 (6), 1191–1200. 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- Ait Saada A., Lambert S. A. E., Carr A. M. (2018). Preserving Replication fork Integrity and Competence via the Homologous Recombination Pathway. DNA Repair 71, 135–147. 10.1016/J.DNAREP.2018.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C., Barth T. K., Reverón-Gómez N., Sidoli S., Schmidt A., Jensen O. N., et al. (2015). Two Distinct Modes for Propagation of Histone PTMs across the Cell Cycle. Genes Dev. 29 (6), 585–590. 10.1101/gad.256354.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C., Groth A. (2012). Chromatin Replication and Epigenome Maintenance. Nat. Rev. Mol. Cel Biol 13 (3), 153–167. 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Alexandrow M. G., Hamlin J. L. (2005). Chromatin Decondensation in S-phase Involves Recruitment of Cdk2 by Cdc45 and Histone H1 Phosphorylation. J. Cel Biol. 168 (6), 875–886. 10.1083/jcb.200409055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R., Fernández-Justel J. M., Santa-María C., Cadoret J.-C., Cano-Aroca L., Lombraña R., et al. (2018). Chromatin Conformation Regulates the Coordination between DNA Replication and Transcription. Nat. Commun. 9 (1). 10.1038/s41467-018-03539-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver R. C., Chadha G. S., Gillespie P. J., Blow J. J. (2017). Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cel Rep. 18 (10), 2508–2520. 10.1016/j.celrep.2017.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D., Molla A., Perche P. Y., Hans F., Côté J., Khochbin S., et al. (2003). The Histone Variant MacroH2A Interferes with Transcription Factor Binding and SWI/SNF Nucleosome Remodeling. Mol. Cel 11 (4), 1033–1041. 10.1016/S1097-2765(03)00100-X [DOI] [PubMed] [Google Scholar]
- Ayoub N., Jeyasekharan A. D., Bernal J. A., Venkitaraman A. R. (2008). HP1-β Mobilization Promotes Chromatin Changes that Initiate the DNA Damage Response. Nature 453 (7195), 682–686. 10.1038/nature06875 [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., et al. (2001). Selective Recognition of Methylated Lysine 9 on Histone H3 by the HP1 Chromo Domain. Nature 410 (6824), 120–124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Barra V., Fachinetti D. (2018). The Dark Side of Centromeres: Types, Causes and Consequences of Structural Abnormalities Implicating Centromeric DNA. Nat. Commun. 9 (1). 10.1038/s41467-018-06545-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártová E., Malyšková B., Komůrková D., Legartová S., Suchánková J., Krejčí J., et al. (2017). Function of Heterochromatin Protein 1 during DNA Repair. Protoplasma 254 (3), 1233–1240. 10.1007/s00709-017-1090-3 [DOI] [PubMed] [Google Scholar]
- Bayona-Feliu A., Casas-Lamesa A., Reina O., Bernués J., Azorín F. (2017). Linker Histone H1 Prevents R-Loop Accumulation and Genome Instability in Heterochromatin. Nat. Commun. 8 (1). 10.1038/s41467-017-00338-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D. B., Burton A., Oda H., Ziegler-Birling C., Torres-Padilla M.-E., Reinberg D. (2012). The Role of PR-Set7 in Replication Licensing Depends on Suv4-20h. Genes Dev. 26 (23), 2580–2589. 10.1101/gad.195636.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin B. J., Lee T., Mullins R. D. (2015). DNA Damage Induces Nuclear Actin Filament Assembly by Formin-2 and Spire-1/2 that Promotes Efficient DNA Repair. eLife 4. 10.7554/eLife.07735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., Carotenuto W., et al. (2011). The Replication Checkpoint Protects Fork Stability by Releasing Transcribed Genes from Nuclear Pores. Cell 146 (2), 233–246. 10.1016/j.cell.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V., Pontis E., Reichard P. (1986). Changes of Deoxyribonucleoside Triphosphate Pools Induced by Hydroxyurea and Their Relation to DNA Synthesis. J. Biol. Chem. 261 (34). 10.1016/S0021-9258(18)66672-4 [DOI] [PubMed] [Google Scholar]
- Blin M., le Tallec B., Nähse V., Schmidt M., Brossas C., Millot G. A., et al. (2019). Transcription-dependent Regulation of Replication Dynamics Modulates Genome Stability. Nat. Struct. Mol. Biol. 26 (1), 58–66. 10.1038/s41594-018-0170-1 [DOI] [PubMed] [Google Scholar]
- Blobel G. (1985). Gene Gating: a Hypothesis. Proc. Natl. Acad. Sci. 82 (24), 8527–8529. 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D., Ferreira P. (2019). Origin Firing Regulations to Control Genome Replication Timing. Genes 10 (3), 199. 10.3390/genes10030199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustel J., Kirstein N., Izard F., Grimaud C., Prorok P., Cayrou C., et al. (2017). Histone H4K20 Tri-Methylation at Late-Firing Origins Ensures Timely Heterochromatin Replication. EMBO J. 36 (18). 10.15252/embj.201796541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H. E., Petermann E., Schultz N., Jemth A.-S., Loseva O., Issaeva N., et al. (2009). PARP Is Activated at Stalled forks to Mediate Mre11-dependent Replication Restart and Recombination. Embo J. 28 (17), 2601–2615. 10.1038/emboj.2009.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M. J., Kunkel T. A. (2017). Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 86 (1), 417–438. 10.1146/annurev-biochem-061516-044709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T. S., Pacek M., Yee M.-c., Walter J. C., Cimprich K. A. (2005). Functional Uncoupling of MCM Helicase and DNA Polymerase Activities Activates the ATR-dependent Checkpoint. Genes Dev. 19 (9), 1040–1052. 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi C. P., D’Agostino C., Ryu T., Zapotoczny G., Delabaere L., Li X., et al. (2018). Nuclear F-Actin and Myosins Drive Relocalization of Heterochromatic Breaks. Nature 559 (7712), 54–60. 10.1038/s41586-018-0242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casale A. M., Cappucci U., Fanti L., Piacentini L. (2019). Heterochromatin Protein 1 (HP1) Is Intrinsically Required for post-transcriptional Regulation of Drosophila Germline Stem Cell (GSC) Maintenance. Sci. Rep. 9 (1). 10.1038/s41598-019-40152-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S., Luger K. (2006). The Histone Variant Macro-H2a Preferentially Forms "Hybrid Nucleosomes". J. Biol. Chem. 281 (35), 25522–25531. 10.1074/jbc.M602258200 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S., Patel A., Bowman G. D. (2012). The Basic Linker of macroH2A Stabilizes DNA at the Entry/exit Site of the Nucleosome. Nucleic Acids Res. 40 (17), 8285–8295. 10.1093/nar/gks645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charaka V., Tiwari A., Pandita R. K., Hunt C. R., Pandita T. K. (2020). Role of HP1β during Spermatogenesis and DNA Replication. Chromosoma 129 (3–4), 215–226. 10.1007/s00412-020-00739-4 [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Keegan S., Kahli M., Tonzi P., Fenyö D., Huang T. T., et al. (2019). Transcription Shapes DNA Replication Initiation and Termination in Human Cells. Nat. Struct. Mol. Biol. 26 (1), 67–77. 10.1038/s41594-018-0171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S. Y., Cutler S., Lin J.-J., Tsai C.-H., Tsai H.-K., Biggins S., et al. (2020). H3K4 Methylation at Active Genes Mitigates Transcription-Replication Conflicts during Replication Stress. Nat. Commun. 11 (1). 10.1038/s41467-020-14595-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D., Charifi F., Eckert-Boulet N., Silva S., Simon M.-N., Lisby M., et al. (2016). SUMO-dependent Relocalization of Eroded Telomeres to Nuclear Pore Complexes Controls Telomere Recombination. Cel Rep. 15 (6), 1242–1253. 10.1016/j.celrep.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Contreras A., Hale T. K., Stenoien D. L., Rosen J. M., Mancini M. A., Herrera R. E. (2003). The Dynamic Mobility of Histone H1 Is Regulated by Cyclin/CDK Phosphorylation. Mol. Cel Biol 23 (23), 8626–8636. 10.1128/MCB.23.23.8626-8636.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C., Pehrson J. R. (1998). Histone macroH2A1 Is Concentrated in the Inactive X Chromosome of Female Mammals. Nature 393 (6685), 599–601. 10.1038/31275 [DOI] [PubMed] [Google Scholar]
- Costanzi C., Pehrson J. R. (2001). MACROH2A2, a New Member of the MACROH2A Core Histone Family. J. Biol. Chem. 276 (24), 21776–21784. 10.1074/jbc.M010919200 [DOI] [PubMed] [Google Scholar]
- Creppe C., Posavec M., Douet J., Buschbeck M. (2012). MacroH2A in Stem Cells: a story beyond Gene Repression. Epigenomics 4 (2), 221–227. 10.2217/epi.12.8 [DOI] [PubMed] [Google Scholar]
- Davé A., Cooley C., Garg M., Bianchi A. (2014). Protein Phosphatase 1 Recruitment by Rif1 Regulates DNA Replication Origin Firing by Counteracting DDK Activity. Cel Rep. 7 (1), 53–61. 10.1016/j.celrep.2014.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K., Govin J., Cerqueira F., Rousseaux S., Khochbin S., Feil R. (2007). Differential Histone Modifications Mark Mouse Imprinting Control Regions during Spermatogenesis. Embo J. 26 (3), 720–729. 10.1038/sj.emboj.7601513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V., Ay F., Sima J., Vera D. L., Noble W. S., Gilbert D. M. (2015). Topologically Associating Domains and Their Long-Range Contacts Are Established during Early G1 Coincident with the Establishment of the Replication-Timing Program. Genome Res. 25 (8), 1104–1113. 10.1101/gr.183699.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. S., Gilbert D. M. (1999). The Spatial Position and Replication Timing of Chromosomal Domains Are Both Established in Early G1 Phase. Mol. Cel 4 (6), 983–993. 10.1016/S1097-2765(00)80227-0 [DOI] [PubMed] [Google Scholar]
- Dinant C., Luijsterburg M. S. (2009). The Emerging Role of HP1 in the DNA Damage Response. Mol. Cel Biol 29 (24), 6335–6340. 10.1128/MCB.01048-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., et al. (2012). Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 485 (7398), 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet J., Corujo D., Malinverni R., Renauld J., Sansoni V., Marjanović M. P., et al. (2017). MacroH2A Histone Variants Maintain Nuclear Organization and Heterochromatin Architecture. J. Cel Sci. 10.1242/jcs.199216 [DOI] [PubMed] [Google Scholar]
- Duro E., Lundin C., Ask K., Sanchez-Pulido L., MacArtney T. J., Toth R., et al. (2010). Identification of the MMS22L-TONSL Complex that Promotes Homologous Recombination. Mol. Cel 40 (4), 632–644. 10.1016/j.molcel.2010.10.023 [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. (1990). Mutation in a Heterochromatin-specific Chromosomal Protein Is Associated with Suppression of Position-Effect Variegation in Drosophila melanogaster . Proc. Natl. Acad. Sci. 87 (24), 9923–9927. 10.1073/pnas.87.24.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. Y., Gordon F., Luger K., Hansen J. C., Tremethick D. J. (2002). The Essential Histone Variant H2A.Z Regulates the Equilibrium between Different Chromatin Conformational States. Nat. Struct. Biol. 10.1038/nsb767 [DOI] [PubMed] [Google Scholar]
- Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004). H2A.Z Alters the Nucleosome Surface to Promote HP1α-Mediated Chromatin Fiber Folding. Mol. Cel 16 (4), 655–661. 10.1016/j.molcel.2004.10.023 [DOI] [PubMed] [Google Scholar]
- Fleming A. B., Kao C.-F., Hillyer C., Pikaart M., Osley M. A. (2008). H2B Ubiquitylation Plays a Role in Nucleosome Dynamics during Transcription Elongation. Mol. Cel 31 (1), 57–66. 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., Bassett E. A., Wood S., et al. (2009). Centromere-Specific Assembly of CENP-A Nucleosomes is Mediated by HJURP. Cell 137 (3), 472–484. 10.1016/J.CELL.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. (2012). The Role of FACT in Making and Breaking Nucleosomes. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1819 (3–4), 247–255. 10.1016/j.bbagrm.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti R., Gnan S., Cornacchia D., Dileep V., Bulut-Karslioglu A., Diehl S., et al. (2016). Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Mol. Cel 61 (2), 260–273. 10.1016/j.molcel.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble M. J., Frizzell K. M., Yang C., Krishnakumar R., Kraus W. L. (2010). The Histone Variant macroH2A1 marks Repressed Autosomal Chromatin, but Protects a Subset of its Target Genes from Silencing. Genes Dev. 24 (1), 21–32. 10.1101/gad.1876110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H., Serra-Cardona A., Hua X., Zhou H., Labib K., Yu C., et al. (2018). The Mcm2-Ctf4-Polα Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cel 72 (1), 140–151. 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Benítez F., Gaillard H., Aguilera A. (2017). Physical Proximity of Chromatin to Nuclear Pores Prevents Harmful R Loop Accumulation Contributing to Maintain Genome Stability. Proc. Natl. Acad. Sci. USA 114 (41), 10942–10947. 10.1073/pnas.1707845114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R., Koller T., Sogo J. M. (1996). The Stability of Nucleosomes at the Replication Fork. J. Mol. Biol. 258 (2), 224–239. 10.1006/jmbi.1996.0245 [DOI] [PubMed] [Google Scholar]
- Gibcus J. H., Dekker J. (2013). The Hierarchy of the 3D Genome. Mol. Cel 49 (5), 773–782. 10.1016/J.MOLCEL.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Hervé S., White R. R., Wilhelm T., Dumont M., Scelfo A., et al. (2021). CENP-A Chromatin Prevents Replication Stress at Centromeres to Avoid Structural Aneuploidy. Proc. Natl. Acad. Sci. USA 118 (10), e2015634118. 10.1073/pnas.2015634118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnan S., Flyamer I. M., Klein K. N., Castelli E., Rapp A., Maiser A., et al. (2021). Nuclear Organisation and Replication Timing Are Coupled through RIF1-PP1 Interaction. Nat. Commun. 12 (1). 10.1038/s41467-021-22899-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. D., Banaszynski L. A., Noh K.-M., Lewis P. W., Elsaesser S. J., Stadler S., et al. (2010). Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell 140 (5), 678–691. 10.1016/J.CELL.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A., Corpet A., Cook A. J. L., Roche D., Bartek J., Lukas J., et al. (2007). Regulation of Replication Fork Progression through Histone Supply and Demand. Science 318 (5858), 1928–1931. 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007). A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 130 (1), 77–88. 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner L., Lezaja A., Zhang X., Lemmens L., Shyian M., Albert B., et al. (2018). Rif1 Binding and Control of Chromosome-Internal DNA Replication Origins Is Limited by Telomere Sequestration. Cel Rep. 23 (4), 983–992. 10.1016/j.celrep.2018.03.113 [DOI] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S. L., van Drunen E., Onizawa H., Beverloo H. B., et al. (2007). The Structure-specific Endonuclease Mus81 Contributes to Replication Restart by Generating Double-Strand DNA Breaks. Nat. Struct. Mol. Biol. 14 (11), 1096–1104. 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., et al. (2008). A Model for Transmission of the H3K27me3 Epigenetic Mark. Nat. Cel Biol 10 (11), 1291–1300. 10.1038/ncb1787 [DOI] [PubMed] [Google Scholar]
- Happel N., Doenecke D. (2009). Histone H1 and its Isoforms: Contribution to Chromatin Structure and Function. Gene 431 (1–2), 1–12. 10.1016/J.GENE.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Hardy S., Jacques P.-É., Gévry N., Forest A., Fortin M.-È., Laflamme L., et al. (2009). The Euchromatic and Heterochromatic Landscapes Are Shaped by Antagonizing Effects of Transcription on H2A.Z Deposition. Plos Genet. 5 (10), e1000687. 10.1371/journal.pgen.1000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H. (2012). Rif1 Is a Global Regulator of Timing of Replication Origin Firing in Fission Yeast. Genes Dev. 26 (2), 137–150. 10.1101/gad.178491.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz K. S., Casas-Delucchi C. S., Török T., Cmarko D., Rapp A., Raska I., et al. (2018). Peripheral Re-localization of Constitutive Heterochromatin Advances its Replication Timing and Impairs Maintenance of Silencing marks. Nucleic Acids Res. 46 (12), 6112–6128. 10.1093/nar/gky368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs M. R., Sato K., Reynolds J. J., Begum S., Bayley R., Goula A., et al. (2018). Histone Methylation by SETD1A Protects Nascent DNA through the Nucleosome Chaperone Activity of FANCD2. Mol. Cel 71 (1), 25–41. 10.1016/j.molcel.2018.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S.-i., Alvino G. M., Chang F., Lian H.-y., Sridhar A., Kubota T., et al. (2014). Rif1 Controls DNA Replication by Directing Protein Phosphatase 1 to Reverse Cdc7-Mediated Phosphorylation of the MCM Complex. Genes Dev. 28 (4), 372–383. 10.1101/gad.231258.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S. i., Ly T., Garzón J., Hořejší Z., Ohkubo Y. n., Endo A., et al. (2017). Human RIF 1 and Protein Phosphatase 1 Stimulate DNA Replication Origin Licensing but Suppress Origin Activation. EMBO Rep. 18 (3), 403–419. 10.15252/embr.201641983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., et al. (2010). Genome-wide Dynamics of Replication Timing Revealed by In Vitro Models of Mouse Embryogenesis. Genome Res. 20 (2), 155–169. 10.1101/gr.099796.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.-W., et al. (2008). Global Reorganization of Replication Domains during Embryonic Stem Cell Differentiation. Plos Biol. 6 (10), e245. 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C., Oma Y., Konishi T., Schmid R., Marcomini I., Hauer M. H., et al. (2014). SWR1 and INO80 Chromatin Remodelers Contribute to DNA Double-Strand Break Perinuclear Anchorage Site Choice. Mol. Cel 55 (4), 626–639. 10.1016/j.molcel.2014.06.027 [DOI] [PubMed] [Google Scholar]
- Houlard M., Berlivet S., Probst A. v., Quivy J.-P., Héry P., Almouzni G., et al. (2006). CAF-1 Is Essential for Heterochromatin Organization in Pluripotent Embryonic Cells. Plos Genet. 2 (11), e181. 10.1371/journal.pgen.0020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung S.-H., Wong R. P., Ulrich H. D., Kao C.-F. (2017). Monoubiquitylation of Histone H2B Contributes to the Bypass of DNA Damage during and after DNA Replication. Proc. Natl. Acad. Sci. USA 114 (11), E2205–E2214. 10.1073/pnas.1612633114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer D., Rhind N. (2017). The Intra-S Checkpoint Responses to DNA Damage. Genes 8 (2), 74. 10.3390/genes8020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L. E. T., Black B. E., Foltz D. R., Cleveland D. W. (2007). Propagation of Centromeric Chromatin Requires Exit from Mitosis. J. Cel Biol. 176 (6), 795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z., Scharf A. N. D., Ask K., Corpet A., Imhof A., Almouzni G., et al. (2010). Replication Stress Interferes with Histone Recycling and Predeposition Marking of New Histones. Mol. Cel 37 (5), 736–743. 10.1016/j.molcel.2010.01.033 [DOI] [PubMed] [Google Scholar]
- Jørgensen H. F., Azuara V., Amoils S., Spivakov M., Terry A., Nesterova T., et al. (2007a). The Impact of Chromatin Modifiers on the Timing of Locus Replication in Mouse Embryonic Stem Cells. Genome Biol. 8 (8), R169. 10.1186/gb-2007-8-8-r169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S., Elvers I., Trelle M. B., Menzel T., Eskildsen M., Jensen O. N., et al. (2007b). The Histone Methyltransferase SET8 Is Required for S-phase Progression. J. Cel Biol. 179 (7), 1337–1345. 10.1083/jcb.200706150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N. J., Jentsch S. (2009). Chromosome-wide Rad51 Spreading and SUMO-H2A.Z-dependent Chromosome Fixation in Response to a Persistent DNA Double-Strand Break. Mol. Cel 33 (3), 335–343. 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Kalousi A., Hoffbeck A.-S., Selemenakis P. N., Pinder J., Savage K. I., Khanna K. K., et al. (2015). The Nuclear Oncogene SET Controls DNA Repair by KAP1 and HP1 Retention to Chromatin. Cel Rep. 11 (1), 149–163. 10.1016/j.celrep.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Kao C.-F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M. A. (2004). Rad6 Plays a Role in Transcriptional Activation through Ubiquitylation of Histone H2B. Genes Dev. 18 (2), 184–195. 10.1101/gad.1149604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D., Sarma K., Reinberg D., Steward R. (2005). PR-Set7-dependent Methylation of Histone H4 Lys 20 Functions in Repression of Gene Expression and Is Essential for Mitosis. Genes Dev. 19 (4), 431–435. 10.1101/gad.1263005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y., Suzuki A., Sugimura K., Okumura K., Zineldeen D. H., Shimada M., et al. (2009). Cyclin A-Cdk1 Regulates the Origin Firing Program in Mammalian Cells. Proc. Natl. Acad. Sci. 106 (9), 3184–3189. 10.1073/pnas.0809350106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M. T., Luciano P., Eckert-Boulet N., Germann S. M., Simon M. N., et al. (2009). The DNA Damage Response at Eroded Telomeres and Tethering to the Nuclear Pore Complex. Nat. Cel Biol 11 (8), 980–987. 10.1038/ncb1910 [DOI] [PubMed] [Google Scholar]
- Khurana S., Kruhlak M. J., Kim J., Tran A. D., Liu J., Nyswaner K., et al. (2014). A Macrohistone Variant Links Dynamic Chromatin Compaction to BRCA1-dependent Genome Maintenance. Cel Rep. 8 (4), 1049–1062. 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Lee S. Y., Choi J.-H., Woo H. G., Xhemalce B., Miller K. M. (2020). PCAF-mediated Histone Acetylation Promotes Replication Fork Degradation by MRE11 and EXO1 in BRCA-Deficient Cells. Mol. Cel 80 (2), 327–344. 10.1016/j.molcel.2020.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sturgill D., Sebastian R., Khurana S., Tran A. D., Edwards G. B., et al. (2018). Replication Stress Shapes a Protective Chromatin Environment across Fragile Genomic Regions. Mol. Cel 69 (1), 36–47. 10.1016/j.molcel.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A., McCarroll S. A. (2014). Random Replication of the Inactive X Chromosome. Genome Res. 24 (1), 64–69. 10.1101/gr.161828.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosar M., Giannattasio M., Piccini D., Maya-Mendoza A., García-Benítez F., Bartkova J., et al. (2021). The Human Nucleoporin Tpr Protects Cells from RNA-Mediated Replication Stress. Nat. Commun. 12 (1). 10.1038/s41467-021-24224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin Modifications and Their Function. Cell 128 (4), 693–705. 10.1016/J.CELL.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kozlowski M., Corujo D., Hothorn M., Guberovic I., Mandemaker I. K., Blessing C., et al. (2018). MacroH2A Histone Variants Limit Chromatin Plasticity through Two Distinct Mechanisms. EMBO Rep. 19 (10). 10.15252/embr.201744445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramarz K., Schirmeisen K., Boucherit V., Ait Saada A., Lovo C., Palancade B., et al. (2020). The Nuclear Pore Primes Recombination-dependent DNA Synthesis at Arrested forks by Promoting SUMO Removal. Nat. Commun. 11 (1). 10.1038/s41467-020-19516-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers M. A., Stasevich T. J., Sasaki T., Wilson K. A., Hazelwood K. L., McNally J. G., et al. (2011). Highly Stable Loading of Mcm Proteins onto Chromatin in Living Cells Requires Replication to Unload. J. Cel Biol. 192 (1), 29–41. 10.1083/jcb.201007111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A. J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J. K., et al. (2012). The BAH Domain of ORC1 Links H4K20me2 to DNA Replication Licensing and Meier-Gorlin Syndrome. Nature 484 (7392), 115–119. 10.1038/nature10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. H., Workman J. L. (2011). The Changing Faces of HP1: From Heterochromatin Formation and Gene Silencing to Euchromatic Gene Expression. BioEssays 33 (4), 280–289. 10.1002/bies.201000138 [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. (2001). Methylation of Histone H3 Lysine 9 Creates a Binding Site for HP1 Proteins. Nature 410 (6824), 116–120. 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- Lademann C. A., Renkawitz J., Pfander B., Jentsch S. (2017). The INO80 Complex Removes H2A.Z to Promote Presynaptic Filament Formation during Homologous Recombination. Cel Rep. 19 (7), 1294–1303. 10.1016/j.celrep.2017.04.051 [DOI] [PubMed] [Google Scholar]
- Lamm N., Read M. N., Nobis M., van Ly D., Page S. G., Masamsetti V. P., et al. (2020). Nuclear F-Actin Counteracts Nuclear Deformation and Promotes fork Repair during Replication Stress. Nat. Cel Biol 22 (12), 1460–1470. 10.1038/s41556-020-00605-6 [DOI] [PubMed] [Google Scholar]
- Lee Y.-H., Kuo C.-Y., Stark J. M., Shih H.-M., Ann D. K. (2013). HP1 Promotes Tumor Suppressor BRCA1 Functions during the DNA Damage Response. Nucleic Acids Res. 41 (11), 5784–5798. 10.1093/nar/gkt231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D., Jackson J., Quinet A., Brickner J. R., Li S., Yazinski S., et al. (2017). MRE11 and EXO1 Nucleases Degrade Reversed Forks and Elicit MUS81-Dependent Fork Rescue in BRCA2-Deficient Cells. Nat. Commun. 8 (1). 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., et al. (2014). Nuclear Position Dictates DNA Repair Pathway Choice. Genes Dev. 28 (22), 2450–2463. 10.1101/gad.248369.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Elsaesser S. J., Noh K.-M., Stadler S. C., Allis C. D. (2010). Daxx Is an H3.3-specific Histone Chaperone and Cooperates with ATRX in Replication-independent Chromatin Assembly at Telomeres. Proc. Natl. Acad. Sci. 107 (32), 14075–14080. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu M., Shen Y. (2018). Gene Regulation in the 3D Genome. Hum. Mol. Genet. 27 (R2), R228–R233. 10.1093/hmg/ddy164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xue B., Zhang M., Zhang L., Hou Y., Qin Y., et al. (2021). Transcription-coupled Structural Dynamics of Topologically Associating Domains Regulate Replication Origin Efficiency. Genome Biol. 22 (1). 10.1186/s13059-021-02424-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Hua X., Serra-Cardona A., Xu X., Gan S., Zhou H., et al. (2020). DNA Polymerase α Interacts with H3-H4 and Facilitates the Transfer of Parental Histones to Lagging Strands. Sci. Adv. 6 (35). 10.1126/sciadv.abb5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-Y., Wu M.-Y., Gay S., Marjavaara L., Lai M. S., Hsiao W.-C., et al. (2014). H2B Mono-Ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly. Plos Genet. 10 (10), e1004667. 10.1371/journal.pgen.1004667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Zhang L., Lv M., Wen Z., Zhang W., Chen X., et al. (2020). H2A.Z Facilitates Licensing and Activation of Early Replication Origins. Nature 577 (7791), 576–581. 10.1038/s41586-019-1877-9 [DOI] [PubMed] [Google Scholar]
- Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J. P., Imhof A., et al. (2009). The HP1α-CAF1-SetDB1‐containing Complex Provides H3K9me1 for Suv39‐mediated K9me3 in Pericentric Heterochromatin. EMBO Rep. 10 (7), 769–775. 10.1038/embor.2009.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Dechassa M. L., Tremethick D. J. (2012). New Insights into Nucleosome and Chromatin Structure: an Ordered State or a Disordered Affair? Nat. Rev. Mol. Cel Biol 13 (7), 436–447. 10.1038/nrm3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X., Chastain M., Chai W. (2019). Genome-wide Mapping and Profiling of γH2AX Binding Hotspots in Response to Different Replication Stress Inducers. BMC Genomics 20 (1). 10.1186/s12864-019-5934-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddy A., Kosiyatrakul S. T., Boisvert R. A., Herrera-Moyano E., García-Rubio M. L., Gerhardt J., et al. (2016). FANCD2 Facilitates Replication through Common Fragile Sites. Mol. Cel 64 (2), 388–404. 10.1016/j.molcel.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemaker I. K., Zhou D., Bruens S. T., Dekkers D. H., Verschure P. J., Edupuganti R. R., et al. (2020). Histone H1 Eviction by the Histone Chaperone SET Reduces Cell Survival Following DNA Damage. J. Cel Sci. 133 (9). 10.1242/jcs.235473 [DOI] [PubMed] [Google Scholar]
- Martire S., Banaszynski L. A. (2020). The Roles of Histone Variants in fine-tuning Chromatin Organization and Function. Nat. Rev. Mol. Cel Biol 21 (9), 522–541. 10.1038/s41580-020-0262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarocci S., Shyian M., Lemmens L., Damay P., Altintas D. M., Shi T., et al. (2014). Rif1 Controls DNA Replication Timing in Yeast through the PP1 Phosphatase Glc7. Cel Rep. 7 (1), 62–69. 10.1016/j.celrep.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Mattout A., Aaronson Y., Sailaja B. S., Raghu Ram E. v., Harikumar A., Mallm J.-P., et al. (2015). Heterochromatin Protein 1β (HP1β) Has Distinct Functions and Distinct Nuclear Distribution in Pluripotent versus Differentiated Cells. Genome Biol. 16 (1). 10.1186/s13059-015-0760-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty S. M., Sullivan L. L., Sullivan B. A. (2017). Human Centromeres Produce Chromosome-specific and Array-specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Developmental Cel 42 (3), 226–240. 10.1016/j.devcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J. E., Costanzi C., Pehrson J. R., Brockdorff N. (1999). Histone Macroh2a1.2 Relocates to the Inactive X Chromosome after Initiation and Propagation of X-Inactivation. J. Cel Biol. 147 (7), 1399–1408. 10.1083/jcb.147.7.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L., Helin K. (2010). Polycomb Group Protein-Mediated Repression of Transcription. Trends Biochem. Sci. 35 (6), 323–332. 10.1016/j.tibs.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.-Y., et al. (2011). Requirement of ATM-dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Mol. Cel 41 (5), 529–542. 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthurajan U. M., McBryant S. J., Lu X., Hansen J. C., Luger K. (2011). The Linker Region of MacroH2A Promotes Self-Association of Nucleosomal Arrays. J. Biol. Chem. 286 (27), 23852–23864. 10.1074/jbc.M111.244871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P., Ge Z., Sirbu B., Doughty C., Agudelo Garcia P. A., Schlederer M., et al. (2013). Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4. Plos Genet. 9 (6), e1003518. 10.1371/journal.pgen.1003518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D. V. N. P., et al. (2011). Regulation of Homologous Recombination by RNF20-dependent H2B Ubiquitination. Mol. Cel 41 (5), 515–528. 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kustatscher G., Alabert C., Hödl M., Forne I., Völker-Albert M., et al. (2021). Proteome Dynamics at Broken Replication forks Reveal a Distinct ATM-Directed Repair Response Suppressing DNA Double-Strand Break Ubiquitination. Mol. Cel 81 (5), 1084–1099. 10.1016/j.molcel.2020.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J.-i., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. S. (2001). Role of Histone H3 Lysine 9 Methylation in Epigenetic Control of Heterochromatin Assembly. Science 292 (5514), 110–113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- Neelsen K. J., Lopes M. (2015). Replication fork Reversal in Eukaryotes: from Dead End to Dynamic Response. Nat. Rev. Mol. Cel Biol 16 (4), 207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- Northam M. R., Trujillo K. M. (2016). Histone H2B Mono-Ubiquitylation Maintains Genomic Integrity at Stalled Replication forks. Nucleic Acids Res., gkw658. 10.1093/nar/gkw658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouararhni K., Hadj-Slimane R., Ait-Si-Ali S., Robin P., Mietton F., Harel-Bellan A., et al. (2006). The Histone Variant mH2A1.1 Interferes with Transcription by Down-Regulating PARP-1 Enzymatic Activity. Genes Dev. 20 (23), 3324–3336. 10.1101/gad.396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S. L., Miele A., Dekker J., Peterson C. L. (2009). Mechanisms That Regulate Localization of a DNA Double-Strand Break to the Nuclear Periphery. Genes Develop. 23 (8). 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D. T., Pflumm M., Chesnokov I., Huang D. W., Kellum R., Marr J., et al. (1997). Association of the Origin Recognition Complex with Heterochromatin and HP1 in Higher Eukaryotes. Cell 91 (3), 311–323. 10.1016/S0092-8674(00)80415-8 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L. (2011). Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell 144 (2), 200–213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisis N., Krasinska L., Harker B., Urbach S., Rossignol M., Camasses A., et al. (2017). Initiation of DNA Replication Requires Actin Dynamics and Formin Activity. Embo J. 36 (21), 3212–3231. 10.15252/embj.201796585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. W., Botchan M. R., Berger J. M. (2017). Mechanisms and Regulation of DNA Replication Initiation in Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 52 (2), 107–144. 10.1080/10409238.2016.1274717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace J. M., Ter-Zakarian A., Aparicio O. M. (2014). Rif1 Regulates Initiation Timing of Late Replication Origins throughout the S. cerevisiae Genome. PLoS ONE 9 (5), e98501. 10.1371/journal.pone.0098501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R., Fried V. A. (1992). MacroH2A, a Core Histone Containing a Large Nonhistone Region. Science 257 (5075), 1398–1400. 10.1126/science.1529340 [DOI] [PubMed] [Google Scholar]
- Petryk N., Dalby M., Wenger A., Stromme C. B., Strandsby A., Andersson R., et al. (2018). MCM2 Promotes Symmetric Inheritance of Modified Histones during DNA Replication. Science 361 (6409), 1389–1392. 10.1126/science.aau0294 [DOI] [PubMed] [Google Scholar]
- Picard F., Cadoret J.-C., Audit B., Arneodo A., Alberti A., Battail C., et al. (2014). The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells. Plos Genet. 10 (5), e1004282. 10.1371/journal.pgen.1004282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaru A. M., Hom R. A., Beal A., Phillips A. F., Ni E., Cardozo T., et al. (2016). Telomere Replication Stress Induced by POT1 Inactivation Accelerates Tumorigenesis. Cel Rep. 15 (10), 2170–2184. 10.1016/j.celrep.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaru A. M., Kareh M., Lamm N., Lazzerini-Denchi E., Cesare A. J., Sfeir A. (2020). Replication Stress Conferred by POT1 Dysfunction Promotes Telomere Relocalization to the Nuclear Pore. Genes Dev. 34 (23–24), 1619–1636. 10.1101/gad.337287.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholkova G. v., Koryakov D. E., Pindyurin A. v., Kozhevnikova E. N., Belyakin S. N., Andreyenkov O. v., et al. (2015). Tethering of SUUR and HP1 Proteins Results in Delayed Replication of Euchromatic Regions in Drosophila melanogaster Polytene Chromosomes. Chromosoma 124 (2), 209–220. 10.1007/s00412-014-0491-8 [DOI] [PubMed] [Google Scholar]
- Pope B. D., Ryba T., Dileep V., Yue F., Wu W., Denas O., et al. (2014). Topologically Associating Domains Are Stable Units of Replication-Timing Regulation. Nature 515 (7527), 402–405. 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Shen Z., Prasanth K. v., Stillman B. (2010). Human Origin Recognition Complex Is Essential for HP1 Binding to Chromatin and Heterochromatin Organization. Proc. Natl. Acad. Sci. 107 (34), 15093–15098. 10.1073/pnas.1009945107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J.-P., Gérard A., Cook A. J. L., Roche D., Almouzni G. (2008). The HP1-p150/CAF-1 Interaction Is Required for Pericentric Heterochromatin Replication and S-phase Progression in Mouse Cells. Nat. Struct. Mol. Biol. 15 (9), 972–979. 10.1038/nsmb.1470 [DOI] [PubMed] [Google Scholar]
- Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., et al. (2014). A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159 (7), 1665–1680. 10.1016/J.CELL.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. P., Huang T., Mastrangelo M.-A., Loring J., Panning B., Jaenisch R. (1999). Messenger RNAs Encoding Mouse Histone macroH2A1 Isoforms Are Expressed at Similar Levels in Male and Female Cells and Result from Alternative Splicing. Nucleic Acids Res. 27 (18), 3685–3689. 10.1093/nar/27.18.3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. P., Mastrangelo M.-A., Eden A., Pehrson J. R., Jaenisch R. (2000). Dynamic Relocalization of Histone Macroh2a1 from Centrosomes to Inactive X Chromosomes during X Inactivation. J. Cel Biol. 150 (5), 1189–1198. 10.1083/jcb.150.5.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A. A., Lee J.-E., et al. (2016). Replication fork Stability Confers Chemoresistance in BRCA-Deficient Cells. Nature 535 (7612), 382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C., Pilch D. R., Rogakou E. P., Orr A. H., Lowndes N. F., Bonner W. M. (2003). Yeast Histone 2A Serine 129 Is Essential for the Efficient Repair of Checkpoint‐blind DNA Damage. EMBO Rep. 4 (7), 678–684. 10.1038/sj.embor.embor871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverón-Gómez N., González-Aguilera C., Stewart-Morgan K. R., Petryk N., Flury V., Graziano S., et al. (2018). Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol. Cel 72 (2), 239–249. e5. 10.1016/J.MOLCEL.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 273 (10), 5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rondinelli B., Gogola E., Yücel H., Duarte A. A., van de Ven M., van der Sluijs R., et al. (2017). EZH2 Promotes Degradation of Stalled Replication forks by Recruiting MUS81 through Histone H3 Trimethylation. Nat. Cel Biol 19 (11), 1371–1378. 10.1038/ncb3626 [DOI] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M., Burke B. (2012). A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cel Biol. 196 (6), 801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., et al. (2010). Evolutionarily Conserved Replication Timing Profiles Predict Long-Range Chromatin Interactions and Distinguish Closely Related Cell Types. Genome Res. 20 (6), 761–770. 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar J. C., Hamperl S., Bocek M. J., Chung M., Bass T. E., Cisneros-Soberanis F., et al. (2018). An Intrinsic S/G 2 Checkpoint Enforced by ATR. Science 361 (6404), 806–810. 10.1126/science.aap9346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C. T., et al. (2002). Active Genes Are Tri-methylated at K4 of Histone H3. Nature 419 (6905), 407–411. 10.1038/nature01080 [DOI] [PubMed] [Google Scholar]
- Saredi G., Huang H., Hammond C. M., Alabert C., Bekker-Jensen S., Forne I., et al. (2016). H4K20me0 marks post-replicative Chromatin and Recruits the TONSL-Mms22l DNA Repair Complex. Nature 534 (7609), 714–718. 10.1038/nature18312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., Tachiwana H., et al. (2012). Histone Chaperone Activity of Fanconi Anemia Proteins, FANCD2 and FANCI, Is Required for DNA Crosslink Repair. EMBO J. 31 (17), 3524–3536. 10.1038/emboj.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf A. N. D., Meier K., Seitz V., Kremmer E., Brehm A., Imhof A. (2009). Monomethylation of Lysine 20 on Histone H4 Facilitates Chromatin Maturation. Mol. Cel Biol 29 (1), 57–67. 10.1128/MCB.00989-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., Jasin M. (2012). A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-Brca1/2. Cancer Cell 22 (1), 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J. A., Berti M., Walser F., Raso M. C., Schmid F., Krietsch J., et al. (2018). Histone Ubiquitination by the DNA Damage Response Is Required for Efficient DNA Replication in Unperturbed S Phase. Mol. Cel 71 (6), 897–910. 10.1016/j.molcel.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., et al. (2004). A Silencing Pathway to Induce H3-K9 and H4-K20 Trimethylation at Constitutive Heterochromatin. Genes Dev. 18 (11), 1251–1262. 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Sengupta R., Kubicek S., Malin S., Kauer M., Callén E., et al. (2008). A Chromatin-wide Transition to H4K20 Monomethylation Impairs Genome Integrity and Programmed DNA Rearrangements in the Mouse. Genes Dev. 22 (15), 2048–2061. 10.1101/gad.476008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B. R., Aparicio T., Li Y., Chang W., Chait B. T., Gundersen G. G., et al. (2018). Nuclear ARP2/3 Drives DNA Break Clustering for Homology-Directed Repair. Nature 559 (7712), 61–66. 10.1038/s41586-018-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M., Kohler H., Oakeley E. J., Stadler M. B., Schübeler D. (2010). Heterochromatin Protein 1 (HP1) Modulates Replication Timing of the Drosophila Genome. Genome Res. 20 (6), 771–780. 10.1101/gr.101790.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian R., Hosogane E. K., Sun E. G., Tran A. D., Reinhold W. C., Burkett S., et al. (2020). Epigenetic Regulation of DNA Repair Pathway Choice by MacroH2A1 Splice Variants Ensures Genome Stability. Mol. Cel 79 (5), 836–845. 10.1016/j.molcel.2020.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaib M., Walter D., Gillespie P. J., Izard F., Fahrenkrog B., Lleres D., et al. (2018). Histone H4K20 Methylation Mediated Chromatin Compaction Threshold Ensures Genome Integrity by Limiting DNA Replication Licensing. Nat. Commun. 9 (1). 10.1038/s41467-018-06066-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Hunt C. R., Pandita R. K., Kumar R., Yang C.-R., Horikoshi N., et al. (2013). Lamin A/C Depletion Enhances DNA Damage-Induced Stalled Replication Fork Arrest. Mol. Cel. Biol. 33 (6), 1210–1222. 10.1128/MCB.01676-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B. M., Couch F. B., Feigerle J. T., Bhaskara S., Hiebert S. W., Cortez D. (2011). Analysis of Protein Dynamics at Active, Stalled, and Collapsed Replication forks. Genes Dev. 25 (12), 1320–1327. 10.1101/gad.2053211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G., Mailand N. (2016). Writers, Readers, and Erasers of Histone Ubiquitylation in DNA Double-Strand Break Repair. Front. Genet. 7. 10.3389/fgene.2016.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A., Li B.-Z., Szakal B., Branzei D., Putnam C. D., Kolodner R. D. (2018). The Swr1 Chromatin-Remodeling Complex Prevents Genome Instability Induced by Replication fork Progression Defects. Nat. Commun. 9 (1). 10.1038/s41467-018-06131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Morgan K. R., Petryk N., Groth A. (2020). Chromatin Replication and Epigenetic Cell Memory. Nat. Cell Biol. 22 (4). 10.1038/s41556-020-0487-y [DOI] [PubMed] [Google Scholar]
- Su X. A., Dion V., Gasser S. M., Freudenreich C. H. (2015). Regulation of Recombination at Yeast Nuclear Pores Controls Repair and Triplet Repeat Stability. Genes Dev. 29 (10), 1006–1017. 10.1101/gad.256404.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Karpen G. H. (2004). Centromeric Chromatin Exhibits a Histone Modification Pattern that Is Distinct from Both Euchromatin and Heterochromatin. Nat. Struct. Mol. Biol. 11 (11), 1076–1083. 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A., O’Byrne K. J., Richard D. J. (2018). Combination Therapy with Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 8. 10.3389/fonc.2018.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S.-i., Ryba T., Wimbish K., Hayakawa T., Sakaue M., Kuriya K., et al. (2021). The Temporal Order of DNA Replication Shaped by Mammalian DNA Methyltransferases. Cells 10 (2), 266. 10.3390/cells10020266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., et al. (2010). The Histone H4 Lys 20 Methyltransferase PR-Set7 Regulates Replication Origins in Mammalian Cells. Nat. Cel Biol 12 (11), 1086–1093. 10.1038/ncb2113 [DOI] [PubMed] [Google Scholar]
- Tardat M., Murr R., Herceg Z., Sardet C., Julien E. (2007). PR-Set7-dependent Lysine Methylation Ensures Genome Replication and Stability through S Phase. J. Cel Biol. 179 (7), 1413–1426. 10.1083/jcb.200706179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Técher H., Koundrioukoff S., Nicolas A., Debatisse M. (2017). The Impact of Replication Stress on Replication Dynamics and DNA Damage in Vertebrate Cells. Nat. Rev. Genet. 18 (9), 535–550. 10.1038/nrg.2017.46 [DOI] [PubMed] [Google Scholar]
- Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., et al. (2015). DNA2 Drives Processing and Restart of Reversed Replication forks in Human Cells. J. Cel Biol. 208 (5), 545–562. 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet C., Hayes J. J. (2009). Linker Histone Phosphorylation Regulates Global Timing of Replication Origin Firing. J. Biol. Chem. 284 (5), 2823–2829. 10.1074/jbc.M805617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo K. M., Osley M. A. (2012). A Role for H2B Ubiquitylation in DNA Replication. Mol. Cel 48 (5), 734–746. 10.1016/j.molcel.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K., Furst A., Rogier M., Heyer V., Maglott-Roth A., Ferrand A., et al. (2016). Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cel 63 (2), 293–305. 10.1016/j.molcel.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Tuduri S., Crabbé L., Conti C., Tourrière H., Holtgreve-Grez H., Jauch A., et al. (2009). Topoisomerase I Suppresses Genomic Instability by Preventing Interference between Replication and Transcription. Nat. Cel Biol 11 (11), 1315–1324. 10.1038/ncb1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubhi T., Brown G. W. (2019). Exploiting DNA Replication Stress for Cancer Treatment. Cancer Res. 79 (8), 1730–1739. 10.1158/0008-5472.CAN-18-3631 [DOI] [PubMed] [Google Scholar]
- Van C., Williams J. S., Kunkel T. A., Peterson C. L. (2015). Deposition of Histone H2A.Z by the SWR-C Remodeling Enzyme Prevents Genome Instability. DNA Repair 25, 9–14. 10.1016/j.dnarep.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J. (2001). Histone H2AX Is Phosphorylated in an ATR-dependent Manner in Response to Replicational Stress. J. Biol. Chem. 276 (51), 47759–47762. 10.1074/jbc.C100569200 [DOI] [PubMed] [Google Scholar]
- Whalen J. M., Dhingra N., Wei L., Zhao X., Freudenreich C. H. (2020). Relocation of Collapsed Forks to the Nuclear Pore Complex Depends on Sumoylation of DNA Repair Proteins and Permits Rad51 Association. Cel Rep. 31 (6), 107635. 10.1016/j.celrep.2020.107635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles E. T., Selker E. U. (2017). H3K27 Methylation: a Promiscuous Repressive Chromatin Mark. Curr. Opin. Genet. Development 43, 31–37. 10.1016/j.gde.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Wang P. F., Lee J. S., Martin-Brown S., Florens L., Washburn M., et al. (2008). Molecular Regulation of H3K4 Trimethylation by Wdr82, a Component of Human Set1/COMPASS. Mol. Cel Biol 28 (24), 7337–7344. 10.1128/MCB.00976-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Singh P. B., Gilbert D. M. (2006). Uncoupling Global and fine-tuning Replication Timing Determinants for Mouse Pericentric Heterochromatin. J. Cel Biol. 174 (2), 185–194. 10.1083/jcb.200601113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Xu Y., Gursoy-Yuzugullu O., Price B. D. (2012a). The Histone Variant macroH2A1.1 Is Recruited to DSBs through a Mechanism Involving PARP1. FEBS Lett. 586 (21), 3920–3925. 10.1016/j.febslet.2012.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Wang W., Chen S., Zhu B. (2012b). A Model for Mitotic Inheritance of Histone Lysine Methylation. EMBO Rep. 13 (1), 60–67. 10.1038/embor.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Ni K., He Y., Ren J., Sun C., Liu Y., et al. (2021). The Epigenetic Regulator LSH Maintains fork protection and Genomic Stability via MacroH2A Deposition and RAD51 Filament Formation. Nat. Commun. 12 (1). 10.1038/s41467-021-23809-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E., Farkash-Amar S., Polten A., Yakhini Z., Tanay A., Simon I. (2010). Comparative Analysis of DNA Replication Timing Reveals Conserved Large-Scale Chromosomal Architecture. Plos Genet. 6 (7), e1001011. 10.1371/journal.pgen.1001011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Ishii A., Kanoh Y., Oda M., Nishito Y., Masai H. (2012). Rif1 Regulates the Replication Timing Domains on the Human Genome. EMBO J. 31 (18), 3667–3677. 10.1038/emboj.2012.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T., Poduch K., Ryba T., Lu J., Hiratani I., Tachibana M., et al. (2009). G9a Selectively Represses a Class of Late-Replicating Genes at the Nuclear Periphery. Proc. Natl. Acad. Sci. 106 (46), 19363–19368. 10.1073/pnas.0906142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Gan H., Serra-Cardona A., Zhang L., Gan S., Sharma S., et al. (2018). A Mechanism for Preventing Asymmetric Histone Segregation onto Replicating DNA Strands. Science 361 (6409), 1386–1389. 10.1126/science.aat8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Liu J., Deng W.-M., Jiao R. (2015). Histone Chaperone CAF-1: Essential Roles in Multi-Cellular Organism Development. Cell. Mol. Life Sci. 72 (2), 327–337. 10.1007/s00018-014-1748-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., Gasser S. M. (2016). Histone H3K9 Methylation Is Dispensable for Caenorhabditis elegans Development but Suppresses RNA:DNA Hybrid-Associated Repeat Instability. Nat. Genet. 48 (11), 1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
- Zeman M. K., Cimprich K. A. (2014). Causes and Consequences of Replication Stress. Nat. Cel Biol 16 (1), 2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Roberts D. N., Cairns B. R. (2005). Genome-Wide Dynamics of Htz1, a Histone H2A Variant that Poises Repressed/Basal Promoters for Activation through Histone Loss. Cell 123 (2), 219–231. 10.1016/j.cell.2005.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]