Abstract
Fluorophore-assisted carbohydrate electrophoresis (FACE) is a straightforward, sensitive method for determining the presence and relative abundance of individual oligomannosyl residues in Candida mannoprotein, the major antigenic determinant located on the outer surface of the yeast cell wall. The single terminal aldehydes of oligomannosyl residues released by hydrolysis were tagged with the charged fluorophore 8-aminonaphthalene-1,3,6-trisulfonate (ANTS) and separated with high resolution on the basis of size by polyacrylamide gel electrophoresis. ANTS fluorescence labeling was not biased by oligomannoside length; therefore, band fluorescence intensity was directly related to the relative abundance of individual oligomannoside moieties in heterogeneous samples. FACE analysis revealed the major oligomannosides released by acid hydrolysis and β-elimination of Fehling-precipitated mannan from Candida albicans, which were the same as those previously reported in studies based on mass and nuclear magnetic spectroscopic analysis. FACE was also amenable to the analysis of samples obtained by direct hydrolysis of whole yeast cells. Whole-cell acid hydrolysis and whole-cell β-elimination of two isolates each of C. albicans, C. glabrata, C. krusei, C. lusitaniae, C. parapsilosis, C. rugosa, C. stellatoidea, and C. tropicalis resulted in oligomannoside gel banding patterns that were species and strain specific for the 16 isolates surveyed. Whereas some bands were specific for an individual isolate or species, other bands were shared by two or three species in various groupings. Differences in the mannoprotein composition of C. albicans A9 and four spontaneous cell surface mutants were also detected. Mannan “fingerprints,” or banding pattern profiles, derived from the electrophoretic mobilities of individual bands relative to the migration of acid-hydrolyzed dextran (relative migration index) yielded profiles characteristic of individual isolates not revealed by standard assimilation and biochemical profiles. FACE represents an accessible, sensitive, and quantitative analytical tool enabling the characterization of yeast mannan complexity.
Candida albicans is an opportunistic fungal pathogen with host interaction abilities that range from normal commensal through life-threatening, disseminated disease. The interplay between Candida and host defenses is paramount in determining infection outcome. The major antigenic determinant in the outermost region of the C. albicans cell wall is mannoprotein (MP). This structure makes initial contact with the host (35) and participates in immunomodulation (1, 3, 8, 20) and adherence (2, 10, 28–30, 37, 40, 52), and antimannan antibodies may be protective (17, 18). Its major constituent is d-mannose, presented as N-linked (90%) and O-linked (10%) oligomannosides (5, 19, 39, 54). The complex N-linked mannan consists of an extended α-1,6-oligomannosyl backbone with α-1,2- and α-1,3-linked oligomannoside side chains that may in turn branch through a phosphodiester linkage to β-1,2-oligomannosides. The O-linked mannan consists of short, linear α-1,2- and α1,3-linked oligomannosides. The complexity and function of mannan are dynamic and are influenced by the nutritional environment and cell morphology (4, 6, 7, 12, 13, 35). The structural specifics and subtleties of the MP may be ascertained by the use of sophisticated analytical tools, such as multidimensional nuclear magnetic resonance (NMR) (19, 32, 33, 43, 44, 46, 48). Unfortunately, the difficulty of data interpretation and the scarcity of polysaccharide NMR expertise limit rapid and widespread use of this analytical approach. A simple method for determining the presence and relative abundance of oligomannoside species on the Candida yeast cell wall will contribute to our understanding of the role of these moieties in the pathogenesis of candidiasis.
Fluorophore-assisted carbohydrate electrophoresis (FACE) is a high-resolution polyacrylamide gel electrophoretic procedure that separates oligosaccharides on the basis of size (23, 24). Individual carbohydrate moieties are tagged at the terminal aldehyde with the highly charged fluorophore 8-aminonaphthalene-1,3,6-trisulfonate (ANTS), which imparts a uniformly strong negative charge to each oligosaccharide or monomeric reducing sugar and enables the polyacrylamide gel electrophoretic size separation. The relative abundance of each saccharide residue present in the starting mixture is represented by the fluorescence intensity of the resulting band on the gel (22, 23, 47).
In this report, we describe the application of FACE for determining the relative abundances of mannoside residues in the Candida cell wall. We demonstrate that the products obtained by conventional MP extraction and fractionation methods, acid hydrolysis and β-elimination, are amenable to FACE analysis. We also report on the development of a rapid whole-cell hydrolysis procedure that does not require prior mannan extraction or carbohydrate purification. FACE analysis of the whole-cell fractionation products revealed species- and strain-specific oligomannoside banding patterns.
MATERIALS AND METHODS
Organisms and culture conditions.
The identification of all strains, C. lusitaniae ATCC 64125, C. parapsilosis ATCC 22019, C. stellatoidea ATCC 11006, C. stellatoidea ATCC 36232, C. glabrata ATCC 2001, C. krusei ATCC 6258 (American Type Culture Collection, Manassas, Va.), C. albicans 3153A, serotype A (a gift from A. Cassone), C. albicans A9, serotype B, and its spontaneous cell surface mutants, C. albicans A9-V2, A9-V4, A9-V8, and A9-V10 (a gift from W. L. Whelan) (50), C. lusitaniae Y1.190, C. parapsilosis Y1.797, C. glabrata M33568, C. tropicalis Y1.787, C. tropicalis Y1.802, C. rugosa 8.47 and C. rugosa 8.48 (a gift from K. Hazen), and C. krusei 01 (a gift from D. Brawner), was verified with the API 20 assimilation profile index (Bio Mérieux Vitek, Inc., Hazelwood, Mo.). The source cultures were grown from glycerol (50%) stocks as previously described (30).
Mannan extraction and fractionation.
The C. albicans A9 yeast cells were grown in glucose (2%, wt/vol)-yeast extract (0.3%, wt/vol)-peptone (1%, wt/vol) (GYEP) (Difco, Becton Dickinson, San Jose, Calif.) broth at 37°C under constant aeration until stationary phase was obtained (24 h). The stationary-phase cells were transferred to fresh GYEP, and 24-h cells were harvested by centrifugation at 3,000 × g for 10 min and washed extensively with cold (4°C) deionized water (dH2O). The cells were kept in a tube submerged in an ice-water slurry to minimize modification of the yeast cell wall during processing. Short-term (fractionation completed within 2 h) Fehling precipitation was performed as described previously (32, 34). Dialysis was carried out by using Spectra/Por 2 Molecularporous Dialysis Membranes (molecular weight cutoff, 12,000 to 14,000; Spectrum Medical Industries, Inc., Houston, Tex.). Aqueous stock solutions of the MP were prepared at 10 mg/ml and stored at −20°C. The protein and carbohydrate contents of the extract were determined with the bicinchoninic acid (BCA) Protein Assay reagent (Pierce, Rockford, Ill.) and by the phenol-sulfuric acid assay (9), respectively.
Fractionations of the MP by acid hydrolysis to release the β-1,2-oligomannosides (acid-labile moieties) and by β-elimination to release the O-linked α-1,2- and α-1,3-linked oligomannosides were carried out as described previously (45). Briefly, for acid hydrolysis, 500 μl of the 10-mg/ml stock MP solution was added to 500 μl of 20 mM HCl and incubated at 100°C for 1 h. For β-elimination, 500 μl of the stock MP solution was added to 500 μl of 200 mM NaOH and incubated for 18 h at 20 to 23°C. Following hydrolysis, the samples were neutralized with either 100 mM NaOH or 1 N HCl. A 200-μl aliquot of each fraction was filtered with an ultrafiltration unit (Pierce Kwik Spin; molecular weight cutoff, 100,000) by centrifugation at 1,000 × g for 20 min. The filtered aliquots were freeze dried, suspended in dH2O at 10 mg/ml, and stored at −20°C. The bulk unfiltered fractions were freeze dried for shelf storage.
Whole-cell hydrolysis.
The stationary-phase yeast cells were serially transferred to GYEP agar (20%, wt/vol) plates twice, at 24-h intervals, prior to harvesting for mannan extraction and fractionation. In the rapid whole-cell hydrolysis, approximately 109 cells were lifted from the surface of GYEP agar plates, placed into 1 ml of 100 mM NaOH, suspended by use of a vortex mixer and agitated continuously on a Labquake Shaker (Barnstead/Thermolyne, Dubuque, Iowa) at 26°C. The supernatant materials containing the whole-cell β-elimination (WC-β) products were harvested by centrifugation at 14,000 × g for 15 min at 20 to 23°C. The cells were washed twice with 1 ml of cold dH2O (4°C) and suspended into 1 ml of 10 mM HCl for acid hydrolysis in a boiling-water bath for 60 min. The vortex mixer was used to suspend the cells at 30 min during hydrolysis and after cooling. The supernatant fluids containing the whole-cell acid (WC-A) products were harvested by centrifugation as stated above. Prior to storage at −20°C, the protein and carbohydrate contents of the whole-cell extracts were determined as for Fehling-precipitated mannan.
Carbohydrate standards.
Carbohydrate standards were prepared from an enzyme-catalyzed hydrolysis of wheat starch (24) and from acid-hydrolyzed dextran (51). Fifty microliters of a 10-mg/ml solution of wheat starch (S-2760; Sigma Chemical Co., St. Louis, Mo.) in 100 mM ammonium acetate buffer (AAB), pH 5.5, was hydrolyzed with α-amylase (A6380; Sigma) at 37°C for 30 min (25). Hydrolysis was stopped by the addition of 1 ml of ice-cold 100% ethanol (EtOH; McCormick Distillation Co., Inc., Weston, Mo.), and the solution was incubated at −70°C for 30 min. The EtOH was evaporated to dryness under CO2 at 20 to 23°C, and the hydrolyzed wheat starch was suspended into AAB, pH 5.5, at 2 mg/ml. One hundred milligrams of dextran (D-7265; molecular weight, 298,000; Sigma) in 10 ml of 0.1 N HCl was heated at 100°C for 4 h and then freeze dried, and a stock solution was prepared at 10 mg/ml of dH2O. Aqueous 10 mM carbohydrate standard solutions were also prepared from d-glucose (G-8270; Sigma), d-mannose (P545; Baker Chemical Co., Sanford, Maine), d-galactose (G-0750; Sigma), d-arabinose (A-3131; Sigma), maltotetraose (M-8253; Sigma), and maltooligosaccharide (M-3639; estimated average molecular weight, 1260; Sigma). All carbohydrate stock solutions were stored at −20°C.
ANTS labeling of oligosaccharides.
One hundred nanomolar aliquots of the aqueous carbohydrate solutions were freeze dried and tagged with ANTS as described elsewhere (25). Others have shown that under the conditions described below, ANTS labeling of the terminal aldehydes is complete (23). Each dried carbohydrate sample was suspended in 5.0 μl each of 0.2 M ANTS (Molecular Probes, Eugene, Oreg.) in acetic acid-water (3:17, vol/vol) and freshly made 1.0 M sodium cyanoborohydride (Aldrich Chemical Co., Milwaukee, Wis.) in dimethyl sulfoxide (D-5879; Sigma) and incubated at 37°C for 16 h. The samples were dried under nitrogen at 45°C, suspended in 50 μl of loading buffer (62.5 mM Tris-HCl, pH 6.8, containing 20% glycerol), and stored at −70°C.
Electrophoresis of ANTS-labeled oligosaccharides.
The electrophoretic method used was an adaptation of those previously reported (23, 25, 26, 47). The resolving gel was 32% acrylamide–2.4% bisacrylamide (PlusOne Ready Sol IEF 40; Pharmacia Biotech, Piscataway, N.J.) in a 140- by 160- by 0.75-mm glass cassette. For every 35 ml of resolving gel, 150 μl of 10% ammonium persulfate (A-6761; Sigma) and 15 μl of N,N,N′,N′,-tetramethylethylenediamine (TEMED [17-1312-01; Pharmacia Biotech]) were added. The stacking gel was 8% acrylamide–0.6% bisacrylamide containing 50 and 5 μl of ammonium persulfate and TEMED, respectively, for every 6 ml of stacking gel. The running buffer and the gel buffer were 0.025 M Tris base–0.192 M glycine (pH 8.4) and 0.42 M Tris base (pH 8.5), respectively. Electrophoresis was run at a constant voltage of 620 V for 3 h in a cooled buffer system (model SE600; Hoefer Scientific Instruments, San Francisco, Calif.). Trace amounts of trypan blue (molecular weight 960), bromophenol blue (molecular weight 670), and phenol red (molecular weight 376) in loading buffer were included as a visual reference in any unused well.
Visualization, photography, and image analysis.
For visualization of the ANTS-labeled oligosaccharides, the gel was removed from the glass cassette and placed onto the surface of a light box with UV illumination (300 nm; Ultra-Violet Products, Inc., San Gabriel, Calif.). The gels were photographed through a no. 12 Kodak Wratten gelatin filter with Polaroid type 57 film, at a film speed of ISO 3000/36°, at f11 and an exposure time of 3 to 10 s.
The photographs were scanned by using a Hewlett-Packard ScanJet 6200C at a resolution of 300 dpi and the images were inverted (inverse pixels) using Adobe Photoshop 4.0. The tagged-image format file (TIFF)-based images were analyzed using SigmaGel gel analysis software (SPSS Science Inc., Chicago, Ill.). The oligosaccharide concentration in the individual bands, defined as regions exhibiting intensities of >10% of background, was calculated based on band fluorescence intensity (pixel number). Relative migration indices (RMI) of the oligosaccharides (x) were calculated based on the migration of each oligosaccharide relative to a mixture of maltooligosaccharides of known structure (47) derived from the acid-hydrolyzed dextran, by the equation RMIx = [(dx − dn)/(dn + 1 − dn)] + n, where d is the distance migrated and n is the number of glucose residues in the oligosaccharide. For example, a band 0.5 of the unit distance between glucose (RMI 1.00) and maltose (RMI 2.00) has an RMI of 1.50. As a measure of the unit distance between two reference points, the RMI for any single oligosaccharide will remain constant, independent of image size. The RMI of bands appearing between the ANTS front, RMI 0.00, and glucose, RMI 1.00, were calculated in the same manner.
RESULTS
Electrophoretic band intensity correlates with carbohydrate concentration.
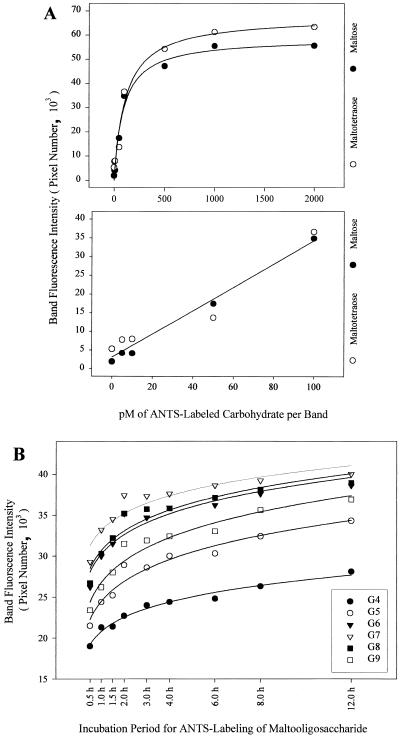
The sensitivity and quantitative limits of the methodology were determined by electrophoretic analysis of serial dilutions of the maltose and maltotetraose standards. At replicate concentrations less than 5 pM, considerable variation in fluorescence intensities was recorded, although as little as 2 pM/band could be seen visually. The linear relationship between band fluorescence intensity and carbohydrate concentration in the range of 5 to 100 pM was used to calculate relative abundance (Fig. 1A).
FIG. 1.
Band fluorescence intensity as a function of carbohydrate concentration and oligomannoside length. (A) The relationship between band intensity and carbohydrate concentration was determined. Band fluorescence intensities of serial dilutions of maltose and maltotetraose were calculated and related to carbohydrate concentration for triplicate samples (r2 = 0.96140). (B) Nonpreferential ANTS labeling of maltooligosaccharides of various lengths was demonstrated. Aliquots of the maltooligosaccharide were derivatized under identical conditions for 0.5 to 12 h. The oligosaccharide concentration of the maltotetraose (G4) through maltonanose (G9) demonstrated an ANTS-labeling rate independent of oligomer length.
A time course derivatization of the maltooligosaccharide standard for 0.5 to 12 h indicated that ANTS labeling of the single terminal aldehyde per oligosaccharide chain occurred without bias to oligosaccharide length (Fig. 1B). That is, no one chain length was derivatized more readily than any other chain length. The relative abundance of all ANTS-labeled maltooligosaccharides, as indicated by band fluorescence intensity, remained constant at all time points tested throughout the incubation period.
Electrophoretic resolution of ANTS-labeled oligosaccharides.
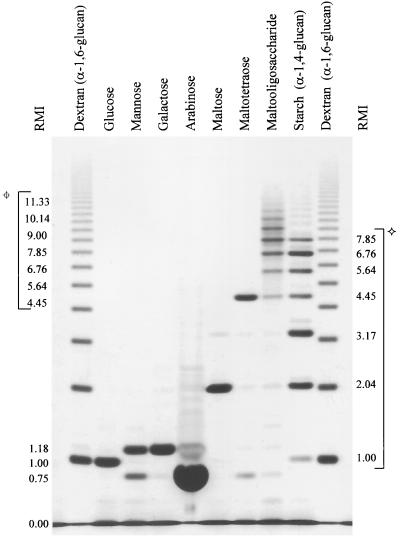
The enzymatic hydrolysis of the heterogeneous wheat starch yielded a preponderance of biose through heptaose polymers. Conversely, the graded diminished intensity of the dextran bands with an increase in oligomer length is indicative of true random hydrolysis of this homogeneous substrate. The electrophoretic resolution of the ANTS-labeled oligosaccharides derived from the acid-hydrolyzed dextran was sufficient to easily distinguish monomeric glucose through a 20-unit oligomer of glucose (Fig. 2).
FIG. 2.
FACE results for the ANTS-labeled standard oligosaccharides and definition of the RMI. The RMI of monomeric glucose and the oligosaccharides derived from dextran were assigned unit values based on the number of hexose residues, i.e., glucose RMI 1.00, maltose RMI 2.00, etc. Every other migration position is expressed as a fraction of the linear distance between two adjacent reference points. Migration positions between the ANTS front, RMI 0.00, and glucose, RMI 1.00, were calculated in the same manner. The RMI of monomeric glucose (1.00), mannose (1.18), galactose (1.18), and arabinose (0.75) and of the α-1,4-oligoglucosides (✧) and maltooligosaccharide (⌖) are indicated.
Monomeric mannose gave an RMI of 1.18 as compared to glucose at an RMI of 1.00 and maltose at an RMI of 2.00. These RMI differences show the influence of hydroxyl positions on migration rate. Additionally, the distances migrated by oligosaccharides of equal length derived from the hydrolysis of dextran and wheat starch illustrate the subtle effect of glycosidic linkage type on electrophoretic movement through the gel. The α-1,6-oligoglucosides of dextran migrated at a slightly accelerated rate compared to the α-1,4-oligoglucosides of equal length derived from wheat starch.
Finally, the sensitivity of FACE can be used to evaluate the purity of commercial carbohydrate products. For example, a putative pentose monomer with a migration rate identical to that of arabinose, RMI 0.75, was present to varying degrees in reagent grade mannose, galactose, maltotetraose, and the maltooligosaccharide (Fig. 2).
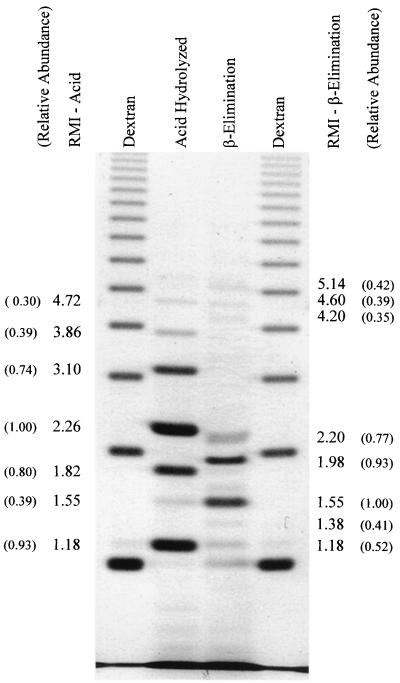
ANTS-labeled oligomannosides obtained by short-term Fehling precipitation.
The major phosphodiester-linked oligomannosides released by acid hydrolysis of the C. albicans A9 Fehling-precipitated mannan gave RMIs of 1.18, 1.82, 2.26, and 3.10 (Fig. 3). Minor bands appeared at RMI of 3.86, 4.45, and 5.76. The O-linked oligomannosides released following β-elimination presented a unique banding pattern, with the exception of mannose at RMI 1.18, with additional bands at RMI 1.55, 1.98, 2.20, 4.31, and 5.14 (Fig. 3).
FIG. 3.
Relative abundances of oligosaccharides obtained by acid hydrolysis and β-elimination of Fehling-precipitated mannan from C. albicans A9. The RMI values and the relative abundances of the individual moieties are indicated. Relative abundance was calculated using the linear regression (r2 = 0.96140) derived from known concentrations of maltotetraose (Fig. 1B).
Whole-cell hydrolysis of various wild-type Candida spp.
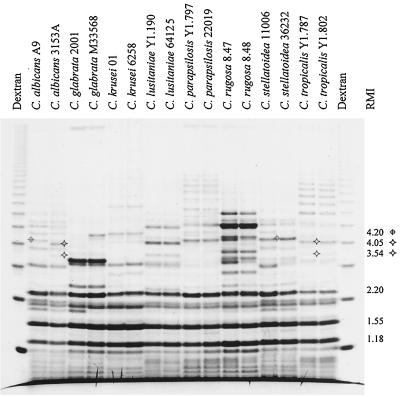
The array of oligomannosides released by whole-cell hydrolysis revealed relative abundance patterns that were specific for each of the eight Candida species surveyed (Fig. 4). Most paired Candida species presented the same bands, but the strains exhibited differences in individual band intensities. For example, the bands at RMI 2.26 in C. lusitaniae 64125 and C. stellatoidea 11006 are more intense than the corresponding bands in C. lusitaniae Y1.190 and C. stellatoidea 36232. These differences in band intensity represent true differences in relative abundance and do not reflect incomplete hydrolysis; the differences occur independently of fluorescence intensity in other bands. In some instances, oligomannosides were present in one strain but not in another, as seen by the fact that C. glabrata 2001, but not C. glabrata M33568, showed a band at RMI 1.82. Although some of the species and strain differences may be determined by simple inspection, the RMI of the major bands were calculated and tabulated for convenience in demonstrating intra- and interstrain band relatedness (Table 1).
FIG. 4.
FACE profile of oligosaccharides released by whole-cell acid hydrolysis of 16 Candida strains. The oligosaccharide banding patterns were specific for each Candida species. The RMI of selected bands illustrating band intensity differences, RMI 2.26 and RMI 3.10 (⌖) in C. stellatoidea, or band occurrence differences, RMI 1.82 (✧) in C. glabrata 2001, are indicated. The RMI and relative abundances for all major bands (bands with intensities 10% greater than background) were calculated and tabulated (Table 1).
TABLE 1.
RMIa of oligosaccharides released by whole-cell acid hydrolysis of 16 Candida strains
| RMI | Band fluorescence intensity scoreb for:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. albicans A9 | C. albicans 3153A | C. glabrata 2001 | C. glabrata M33568 | C. krusei 01 | C. krusei 6258 | C. lusitaniae Y1.190 | C. lusitaniae 64125 | C. parapsilosis Y1.797 | C. parapsilosis 22016 | C. rugosa 8.47 | C. rugosa 8.48 | C. stellatoidea 11006 | C. stellatoidea 36232 | C. tropicalis Y1.787 | C. tropicalis Y1.802 | |
| 1.00 | 3 | 3 | 4 | 4 | 2 | 2 | 3 | 3 | 2 | 2 | 4 | 4 | 2 | 2 | 3 | 3 |
| 1.18 | 4 | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 1.82 | 2 | 2 | 4 | 1 | 1 | 2 | 2 | 1 | 1 | |||||||
| 2.05 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| 2.15 | 1 | 1 | 1 | 1 | ||||||||||||
| 2.26 | 2 | 2 | 1 | 2 | 3 | 1 | 1 | 2 | ||||||||
| 3.10 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | ||||||||
| 3.48 | 1 | 1 | ||||||||||||||
| 3.52 | 1 | 1 | ||||||||||||||
| 3.86 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| 4.45 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 4.72 | 1 | 1 | ||||||||||||||
| 5.76 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 6.85 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 7.85 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 9.20 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Migration distance relative to that of maltooligosaccharides derived from dextran; e.g., a band 0.5 of the unit distance between glucose (RMI 1.00) and maltose (RMI 2.00) has an RMI of 1.50.
Band fluorescence intensity was measured using SigmaGel analytical software as described in the text and rated as a percentage of pixel number ranging from background (0%) through saturation (100%): 1 = 0 to 25%; 2 = 26 to 50%; 3 = 51 to 75%; 4 = 76 to 100% saturation.
The oligomannosides released by whole-cell β-elimination presented banding patterns (Fig. 5) that were distinctly different for the eight species tested, and differences were noted even between the paired strains of C. albicans, C. glabrata, and C. stellatoidea. For example, C. albicans 3153A, serotype A, exhibits bands at RMI 3.54 and RMI 4.05 that are not seen in C. albicans A9, serotype B. Again, for ease of comparison, the RMI of the major bands were calculated and tabulated (Table 2).
FIG. 5.
FACE profile of oligosaccharides released by whole-cell β-elimination of 16 Candida strains. The oligosaccharide banding patterns were specific for each Candida species. The RMI of bands that are specific for C. albicans 3153A and C. tropicalis (✧) or more prominent in C. albicans A9 and C. stellatoidea (⌖) are indicated. The RMI for all major bands (bands with intensities 10% greater than background) were calculated and tabulated (Table 2).
TABLE 2.
RMIa of oligosaccharides released by whole-cell β-elimination of 16 Candida strains
| RMI | Band fluorescence intensity scoreb for:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. albicans A9 | C. albicans 3153A | C. glabrata 2001 | C. glabrata M33568 | C. krusei 01 | C. krusei 6258 | C. lusitaniae Y1.190 | C. lusitaniae 64125 | C. parapsilosis Y1.797 | C. parapsilosis 22016 | C. rugosa 8.47 | C. rugosa 8.48 | C. stellatoidea 11006 | C. stellatoidea 36232 | C. tropicalis Y1.787 | C. tropicalis Y1.802 | |
| 1.00 | 1 | 1 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 1 | 1 |
| 1.18 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 1.55 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 1.82 | 2 | 1 | 3 | 2 | 2 | |||||||||||
| 1.92 | 3 | 2 | 3 | 3 | 4 | 4 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 2 | 2 |
| 1.98 | 3 | 2 | 3 | 3 | 4 | 4 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 2 | 2 |
| 2.20 | 4 | 3 | 4 | 4 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 3 | 2 | 2 |
| 2.36 | 1 | 1 | ||||||||||||||
| 2.44 | 2 | 2 | 1 | 1 | 1 | 1 | ||||||||||
| 2.85 | 1 | 1 | 2 | 2 | ||||||||||||
| 3.05 | 2 | 1 | 1 | 1 | 2 | 1 | ||||||||||
| 3.15 | 3 | 3 | 2 | 2 | ||||||||||||
| 3.25 | 4 | 4 | 3 | 2 | ||||||||||||
| 3.40 | 2 | 2 | ||||||||||||||
| 3.54 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| 3.66 | 2 | 2 | ||||||||||||||
| 3.80 | 1 | 1 | 1 | |||||||||||||
| 4.05 | 1 | 2 | 2 | 1 | 1 | |||||||||||
| 4.20 | 1 | 2 | 2 | 3 | 2 | 2 | ||||||||||
| 4.48 | 1 | |||||||||||||||
| 4.60 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| 5.14 | 1 | 1 | 4 | 4 | 1 | 1 | ||||||||||
| 5.95 | 2 | 2 | 1 | 1 | ||||||||||||
| 6.40 | 1 | 1 | ||||||||||||||
| 6.65 | 1 | 1 | 1 | 1 | ||||||||||||
Migration distance relative to that of maltooligosaccharides derived from dextran; e.g., a band 0.5 of the unit distance between glucose (RMI 1.00) and maltose (RMI 2.00) has an RMI of 1.50.
Band fluorescence intensity was measured using SigmaGel analytical software as described in the text and rated as a percentage of pixel number ranging from background (0%) through saturation (100%): 1 = 0 to 25%; 2 = 26 to 50%; 3 = 51 to 75%; 4 = 76 to 100% saturation.
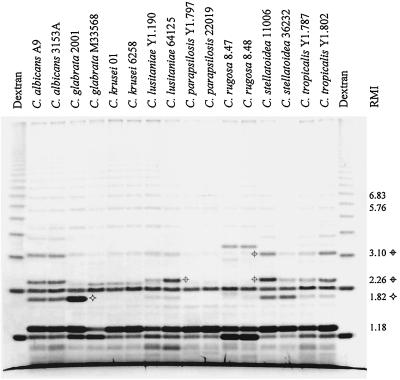
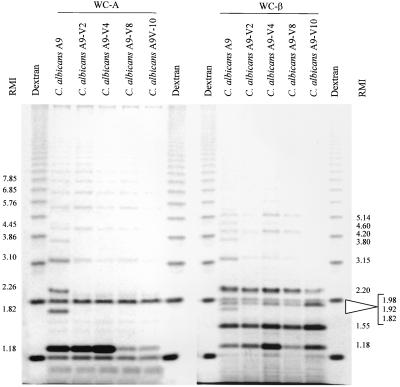
Whole-cell hydrolysis of C. albicans A9 and four cell wall mutants.
All of the C. albicans A9 adherence variants demonstrated losses of oligomannoside residues, as was expected for these mutants. No variant displayed the band at RMI 1.82 (WC-A and WC-β), and A9-V10 did not display any WC-β bands with RMI greater than 2.20 (Figure 6). Relative-abundance differences were also demonstrated for the bands at RMI 1.18 (WC-A and WC-β), RMI 3.10 (WC-A), and RMI 1.98, 2.20, 3.15, 4.20, 4.60, and 5.14 (WC-β).
FIG. 6.
FACE profile of oligosaccharides released by whole-cell acid hydrolysis and β-elimination of C. albicans A9 and four cell wall mutants, C. albicans A9-V2, C. albicans A9-V4, C. albicans A9-V8, and C. albicans A9-V10. The RMI of the fractionated products are indicated.
DISCUSSION
The carbohydrate part of the Candida cell wall is a dynamic and complex cellular component. Because the Candida cell wall is a constituent of adherence, immunomodulation, and host defense reactions in the host, characterization of its structure is paramount in understanding the course of infection. FACE presents a straightforward method for determining the presence and relative abundance of the oligomannoside species in the yeast cell wall.
The detection and imaging system used herein is readily available in most laboratories and is facilitated by the large Stokes shift of ANTS fluorescence (λex, 365 nm; λem, 515 nm). The film response was easily manipulated by varying the exposure times, but the relationship between fluorescence band intensity and carbohydrate concentration remained constant (data not shown). That is, the relationship remained linear in the range of 5 to 100 pM, with a decrease in sensitivity at higher carbohydrate concentrations. ANTS fluorescence labeling is restricted to the single terminal aldehyde of the carbohydrate, and the reductive deamination is not biased by oligosaccharide length. Therefore, fluorescence band intensity is a direct measure of the relative abundance of individual oligomannoside moieties in a heterogeneous sample.
Excellent resolution of the major oligomannoside bands was achieved using the described parameters. Oligosaccharides of the same length but with different glycosidic linkage types demonstrated a progressive difference in migration rate related to increased length (Fig. 2). The α-1,4-oligosaccharides derived from starch, described as flexible helices, show a slightly retarded migration rate compared to the α-1,6-oligosaccharides derived from dextran, which are described as flexible coils (31). The RMI of mannose and galactose (1.18) relative to that of glucose (1.00) may be due to an electrophoretic affinity phenomenon involving the gel matrix and position of the carbohydrate hydroxyl groups (22). The calculated RMI are not intended as “absolute” values for any particular oligosaccharide but rather as a reference for the comparison of samples analyzed under identical conditions. Reducing the gel acrylamide concentration and/or increasing the voltage improved the resolution of oligomeric saccharides greater than 20 units in length and compressed the migration zone of the smaller moieties (data not shown). Therefore, manipulation of electrophoretic parameters confers flexibility on FACE, enabling the procedure to be tailored for a particular sample.
A striking feature of FACE analysis of the Fehling-precipitated mannan from C. albicans A9 is the disparate migration rates of the ANTS-labeled oligomannosides released by the two fractionation procedures, acid hydrolysis and β-elimination. The apparent disparity in migration rates may be attributed to differences in conformation. The β-1,2-oligomannosides released by acid hydrolysis are described as crumpled ribbons, whereas the α-1,2- and α,1-3-linked oligomannosides released by β-elimination are described as flexible helixes (31). The migration rates of these oligomannosides are also different from those of the α-1,4- or α-1,6-oligoglucosides of equal length derived from starch or dextran, respectively. Others have shown by size exclusion column chromatography, mass spectrometry, and NMR analysis (27, 42, 45, 46, 49) that the major moieties released by acid hydrolysis are mannotriose and mannotetraose and that the major moieties released by β-elimination are mannobiose and mannotriose. Relative abundance determined by band fluorescence intensity indicates that these moieties correspond to the acid products at RMI 2.26 and 3.10 and β-elimination products at RMI 1.55 and 1.98, respectively (Fig. 3). The putative identities of the faint bands at RMI 1.55, 3.86, and 4.72 (acid hydrolyzed) and RMI 1.38, 2.20, 4.20, 4.60, and 5.14 (β-elimination) could not be assessed by relative abundance comparison, as the literature reports are not corroborative. The low-abundance moieties may also represent nonspecifically released oligosaccharides that were trapped by the Fehling mannan precipitate and not removed during washing. The sensitivity of FACE analysis, therefore, renders it a valuable tool in evaluating the product uniformity of individual mannan preparations.
The major oligomannoside moieties detected from fractionated Fehling-precipitated mannan were also evident in the whole-cell preparations despite the small sample size and no prerequisite for carbohydrate purification. The Fehling-precipitated mannan contained approximately 20 mg of carbohydrate per mg of protein, whereas the whole-cell preparations ranged from a high of 1 mg of carbohydrate (C. rugosa 8.47 and C. rugosa 8.48) to a low of 0.3 mg of carbohydrate (C. albicans A9-V10) per mg of protein (data not shown). The bands unique to the whole-cell preparations may represent oligomannosides that were not precipitated as a copper complex during Fehling fractionation. Alternatively, they may be nonmannan carbohydrates released from other parts of the cell wall or from the cytosol during direct whole-cell hydrolysis. For example, the carbohydrate moieties at RMI 1.00, 1.18, and 1.82 were obtained from both the acid (WC-A) and alkaline (WC-β) extractions of whole cells of all 16 strains. The moieties migrating between the ANTS front and glucose may represent metabolic intermediates released from the cell cytoplasm during extraction.
Under the prescribed conditions, FACE banding patterns were predictable and were species and strain specific. The resulting oligomannoside “fingerprints” revealed interspecies differences both in the occurrence of specific moieties, e.g., the WC-β band at RMI 4.48 in C. glabrata, and in the relative abundances of specific moieties, e.g., the WC-A band at RMI 3.10 in C. stellatoidea. Intraspecies “fingerprints” may also be indicative of isolate serotypes. It has been observed that C. albicans, serotype A, is antigenically identical to C. tropicalis spp. whereas C. albicans, serotype B, is antigenically identical to C. stellatoidea spp. (19). C. albicans 3153A (serotype A) shares bands at RMI 3.54 and RMI 4.05 with C. tropicalis that are not seen in C. albicans A9 (serotype B). On the other hand, C. stellatoidea exhibits a major band at RMI 4.20 that is notably more prominent in C. albicans A9 than in C. albicans 3153A. FACE analysis was also useful in demonstrating differences among C. albicans A9, isolated from the oral cavity of an AIDS patient, and its cell surface mutants (50). NMR analysis indicated that all variant strains lacked the acid-labile β-1,2-oligomannosides attached via α1→PO4 linkages (50). The WC-A banding pattern illustrated that all mutants lost the moieties at RMI 1.82 and RMI 2.26 that may correspond to the putative β-1,2-mannobiose and β-1,2-mannotriose in the Fehling-precipitated mannan extract of wild-type C. albicans A9. Two of the variants, A9-V8 and A9-V10, were also deficient in the band at RMI 1.18 (mannose), a result that supports the NMR findings. However, additional banding pattern differences were also seen in the WC-β fraction, indicative of changes in the O-linked moieties of the variants. Any or all of these observed differences might be associated with the variants' decreased adherence to buccal epithelial cells and complement factors (14, 39, 50).
FACE provides a straightforward method for determining the presence and relative abundance of oligosaccharide species on the Candida yeast cell wall and will contribute to our understanding of their role in the pathogenesis of candidiasis. We are continuing to define the application of FACE to the in vivo expression of phosphomannan on the cell surface and to carbohydrate epitope identification.
ACKNOWLEDGMENTS
This work was supported by grants 5RO1 AI24912, RO1 AI31769, and 1 PO1 AI37194 from the National Institutes of Health.
REFERENCES
- 1.Ataoglu H, Zueco J, Sentandreu R. Characterization of epitopes recognized by Candida factor 1 and 9 antisera by use of Saccharomyces cerevisiae mnn mutants. Infect Immun. 1993;61:3313–3317. doi: 10.1128/iai.61.8.3313-3317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buurman E T, Westwater C, Hube B, Brown A J P, Odds F C. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc Natl Acad Sci USA. 1998;95:7670–7675. doi: 10.1073/pnas.95.13.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A, Cassone A, Bistono F, Cutler J E, Magliani W, Murphy J W, Polonelli L, Romani L. Antibody and/or cell-mediated immunity, protective mechanisms in fungal disease: an ongoing dilemma or an unnecessary dispute? Med Mycol. 1998;36:95–105. [PubMed] [Google Scholar]
- 4.Casanova M, Lopez-Ribot J L, Martinez J P, Sentandreu R. Characterization of cell wall proteins from yeast and mycelial cells of Candida albicans by labeling with biotin: comparison with other techniques. Infect Immun. 1992;60:4898–4906. doi: 10.1128/iai.60.11.4898-4906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassone A. Cell wall of Candida albicans: its functions and its impact on the host. Curr Top Med Mycol. 1989;3:248–314. doi: 10.1007/978-1-4612-3624-5_10. [DOI] [PubMed] [Google Scholar]
- 6.Cassone A. Immunogenic and immunomodulatory properties of mannoproteins from Candida albicans. Can J Bot. 1994;73:S1192–S1198. [Google Scholar]
- 7.Chaffin W L, Skudlarek J, Morrow K J. Variable expression of a surface determinant during proliferation of Candida albicans. Infect Immun. 1988;56:302–309. doi: 10.1128/iai.56.2.302-309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domer J E. Candida cell wall mannan: a polysaccharide with diverse immunologic properties. Microbiology. 1998;17:33–51. doi: 10.3109/10408418909105721. [DOI] [PubMed] [Google Scholar]
- 9.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Biochem. 1955;28:350–356. [Google Scholar]
- 10.Edwards J E, Mayer C L, Filler S G, Wadsworth E, Calderone R A. Cell extracts of Candida albicans block adherence of the organisms to endothelial cells. Infect Immun. 1992;60:3087–3091. doi: 10.1128/iai.60.8.3087-3091.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elorza M V, Marcilla A, Sentandreu R. Wall mannoproteins of the yeast and mycelial cells of Candida albicans: nature of the glycosidic bonds and polydispersity of their mannan moieties. J Gen Microbiol. 1988;134:2393–2403. doi: 10.1099/00221287-134-8-2393. [DOI] [PubMed] [Google Scholar]
- 12.Elorza M V, Murgui A, Sentandreu R. Dimorphism in Candida albicans: contribution of mannoproteins to the architecture of yeast and mycelial cell walls. J Gen Microbiol. 1985;131:2209–2216. doi: 10.1099/00221287-131-9-2209. [DOI] [PubMed] [Google Scholar]
- 13.Ener B, Douglas L J. Correlation between cell-surface hydrophobicity of Candida albicans and adhesion to buccal epithelial cells. FEMS Microbiol Lett. 1992;99:37–42. doi: 10.1016/0378-1097(92)90284-u. [DOI] [PubMed] [Google Scholar]
- 14.Fukayama M, Calderone R A. Adherence of cell surface mutants of Candida albicans to buccal epithelial cells and analyses of the cell surface proteins of the mutants. Infect Immun. 1991;59:1341–1345. doi: 10.1128/iai.59.4.1341-1345.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukazawa Y, Kagaya K. Molecular bases of adhesion of Candida albicans. J Med Vet Mycol. 1997;35:87–99. doi: 10.1080/02681219780000971. [DOI] [PubMed] [Google Scholar]
- 16.Gyanchandani A, Khan Z K, Farooqui N, Goswami M, Ranade S A. Rapid analysis of Candida albicans strains recovered from different immunocompromised patients (ICP) reveals an apparently non-random infectivity of the strains. Biochem Mol Biol Int. 1998;44:19–27. doi: 10.1080/15216549800201022. [DOI] [PubMed] [Google Scholar]
- 17.Han Y, Kanbe T, Cherniak R, Cutler J E. Biochemical characterization of Candida albicans epitopes that can elicit protective and nonprotective antibodies. Infect Immun. 1997;65:4100–4107. doi: 10.1128/iai.65.10.4100-4107.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, Riesselman M H, Cutler J E. Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect Immun. 2000;68:1649–1654. doi: 10.1128/iai.68.3.1649-1654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasenclever H F, Mitchell W O, Loewe J. Antigenic studies of Candida. J Bacteriol. 1961;82:574–577. doi: 10.1128/jb.82.4.574-577.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayette M P, Strecker G, Gaille C, Dive D, Camus D, MacKenzie D W R, Poulain D. Presence of human antibodies reacting with Candida albicans O-linked oligomannosides revealed by using an enzyme-linked immunosorbent assay and neoglycolipids. J Clin Microbiol. 1992;30:411–417. doi: 10.1128/jcm.30.2.411-417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hearn V M, Cole G T, Susuki S. Fungal antigens. In: Van Regenmortel M H V, editor. Structure of antigens. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 211–260. [Google Scholar]
- 22.Hu G, Vallee B L. A gel retardation assay for the interaction of proteins and carbohydrates by fluorophore-assisted carbohydrate electrophoresis. Anal Biochem. 1994;218:185–191. doi: 10.1006/abio.1994.1158. [DOI] [PubMed] [Google Scholar]
- 23.Jackson P. The use of polyacrylamide gel electrophoresis for the high resolution of reducing saccharides labeled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Biochem J. 1990;270:705–713. doi: 10.1042/bj2700705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson P. High-resolution polyacrylamide gel electrophoresis of fluorophore-labeled reducing saccharides. Methods Enzymol. 1994;230:250–265. doi: 10.1016/0076-6879(94)30017-8. [DOI] [PubMed] [Google Scholar]
- 25.Jackson P. The analysis of fluorophore-labeled glycans by high-resolution polyacrylamide gel electrophoresis. Anal Biochem. 1994;216:243–252. doi: 10.1006/abio.1994.1038. [DOI] [PubMed] [Google Scholar]
- 26.Jackson P, Williams G R. Polyacrylamide gel electrophoresis of reducing saccharides labeled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid: application to the enzymological structural analysis of oligosaccharides. Electrophoresis. 1991;12:94–86. doi: 10.1002/elps.1150120118. [DOI] [PubMed] [Google Scholar]
- 27.Jouault T, Lepage G, Bernigaud A, Trinel P A, Fradin C, Wieruszeski J, Strecker G, Poulain D. β-1,2-Linked oligomannosides from Candida albicans act as signals for tumor necrosis factor alpha production. Infect Immun. 1995;63:2378–2381. doi: 10.1128/iai.63.6.2378-2381.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanbe T, Cutler J E. Evidence for adhesin activity in the acid-stable moiety of the phosphomannoprotein cell wall complex of Candida albicans. Infect Immun. 1994;62:1662–1668. doi: 10.1128/iai.62.5.1662-1668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanbe T, Cutler J E. Minimum chemical requirements for adhesin activity of the acid-stable part of Candida albicans cell wall phosphomannoprotein complex. Infect Immun. 1998;66:5812–5818. doi: 10.1128/iai.66.12.5812-5818.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanbe T, Han Y, Redgrave B, Riesselman M H, Cutler J E. Evidence that mannans of Candida albicans are responsible for adherence of yeast forms to spleen and lymph node tissue. Infect Immun. 1993;61:2578–2584. doi: 10.1128/iai.61.6.2578-2584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy J F, White C A. Bioactive carbohydrates: in chemistry, biochemistry and biology. Chichester, England: Ellis Horwood Limited; 1983. [Google Scholar]
- 32.Kobayashi H, Shibata N, Mitobe H, Ohkubo Y, Suzuki S. Structural study of phosphomannan of yeast-form cells of Candida albicans J-1012 strain with special reference to application of mild acetolysis. Arch Biochem Biophys. 1989;272:364–375. doi: 10.1016/0003-9861(89)90230-0. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi H, Shibata N, Nakada M, Chaki S, Mizugami K, Ohkubo Y, Suzuki S. Structural study of cell wall phosphomannan of Candida albicans NIH B-792 (serotype B) strain, with special reference to 1H and 13C NMR analysis of acid-labile oligomannosyl residues. Arch Biochem Biophys. 1990;278:195–204. doi: 10.1016/0003-9861(90)90248-w. [DOI] [PubMed] [Google Scholar]
- 34.Kocourek J, Ballou C E. Method for fingerprinting yeast cell wall mannan. J Bacteriol. 1969;100:1175–1181. doi: 10.1128/jb.100.3.1175-1181.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kukuruzinka M A, Bergh M L E, Jackson B J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Ballou C E. Preparation of mannobiose, mannotriose, and a new mannotetraose from Saccharomyces cerevisiae mannan. Biochemistry. 1965;4:257–264. [Google Scholar]
- 37.Li R K, Cutler J E. Chemical definition of an epitope/adhesin molecule on Candida albicans. J Biol Chem. 1993;268:18293–18299. [PubMed] [Google Scholar]
- 38.Macura A B, Voss A, Melchers W J G, Meis J F G M, Syslo J, Heczko P B. Characterization of pathogenetic determinants of Candida albicans strains. Zentbl Bakteriol. 1998;287:501–508. doi: 10.1016/s0934-8840(98)80191-6. [DOI] [PubMed] [Google Scholar]
- 39.Masuoka J, Hazen K C. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology. 1997;143:3015–3021. doi: 10.1099/00221287-143-9-3015. [DOI] [PubMed] [Google Scholar]
- 40.Miyakawa Y, Kuribayashi T, Kagaya K, Suzuki M, Nakase T, Fukazawa Y. Role of specific determinants in mannan of Candida albicans serotype A in adherence to human buccal epithelial cells. Infect Immun. 1992;60:2493–2499. doi: 10.1128/iai.60.6.2493-2499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molinari A, Gomez M J, Crateri P, Torosantucci A, Cassone A, Arancia G. Differential cell surface expression of mannoprotein epitopes in yeast and mycelial forms of Candida albicans. Eur J Cell Biol. 1993;60:146–153. [PubMed] [Google Scholar]
- 42.Shibata N, Kobayashi H, Tojo M, Suzuki S. Characterization of phosphomannan-protein complexes isolated from viable cells of yeast and mycelial forms of Candida albicans NIHB-792 strain by the action of Zymolyase-100T. Arch Biochem Biophys. 1986;251:697–708. doi: 10.1016/0003-9861(86)90379-6. [DOI] [PubMed] [Google Scholar]
- 43.Shibata N, Kobayashi H, Takahashi S, Okawa Y, Hisamichi K, Suzuki S. Structural study on a phosphorylated mannotetraose obtained from the phosphomannan of Candida albicans NIH B-792 strain by acetolysis. Arch Biochem Biophys. 1991;290:535–542. doi: 10.1016/0003-9861(91)90578-7. [DOI] [PubMed] [Google Scholar]
- 44.Shibata N, Ikuta K, Imai T, Satoh Y, Satoh R, Suzuki A, Kojima C, Kobayashi H, Hisamichi K, Suzuki S. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. J Biol Chem. 1995;270:1113–1122. doi: 10.1074/jbc.270.3.1113. [DOI] [PubMed] [Google Scholar]
- 45.Shibata N, Ichikawa T, Tojo M, Takahashi M, Okubo Y, Suzuki S. Immunochemical study on the mannans of Candida albicans NIH A-207, NIH B-792, and J-1012 strains prepared by fractional precipitation with cetyltrimethylammonium bromide. Arch Biochem Biophys. 1985;243:338–348. doi: 10.1016/0003-9861(85)90511-9. [DOI] [PubMed] [Google Scholar]
- 46.Shibata N, Fukasawa S, Kobayashi H, Tojo M, Yonezu T, Ambo A, Ohkubo Y, Suzuki S. Structural analysis of phospho-d-mannan-protein complexes isolated from yeast and mold form cells of Candida albicans NIH A-207 serotype A strain. Carbohydr Res. 1989;187:239–253. doi: 10.1016/0008-6215(89)80006-0. [DOI] [PubMed] [Google Scholar]
- 47.Stack R J, Sullivan M T. Electrophoretic resolution and fluorescence detection of N-linked glycoprotein oligosaccharides after reductive amination with 8-aminonaphthalene-1,3,6-trisulphonic acid. Glycobiology. 1992;2:85–92. doi: 10.1093/glycob/2.1.85. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki S, Shibata N, Kobayashi H. Immunochemistry of Candida mannans. In: Latge J P, Boucias D, editors. Fungal cell wall and immune response. Berlin, Germany: Springer-Verlag; 1991. pp. 111–121. [Google Scholar]
- 49.Trinel P A, Faille C, Jacquinot P M, Cailliez J-C, Poulain D. Mapping of Candida albicans oligomannosidic epitopes by using monoclonal antibodies. Infect Immun. 1992;60:3845–3851. doi: 10.1128/iai.60.9.3845-3851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan W L, Delga J M, Wadsworth E, Walsh T J, Kwon-Chung K J, Calderone R, Lipke P N. Isolation and characterization of cell surface mutants of Candida albicans. Infect Immun. 1990;58:1552–1557. doi: 10.1128/iai.58.6.1552-1557.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita K, Mizuochi T, Kobata A. Analysis of oligosaccharides by gel filtration. Methods Enzymol. 1982;83:105–126. doi: 10.1016/0076-6879(82)83008-5. [DOI] [PubMed] [Google Scholar]
- 52.Yan S, Rodrigues R G, Cahn-Hidalgo D, Walsh T J, Roberts D D. Hemoglobin induces binding of several extracellular matrix proteins to Candida albicans. J Biol Chem. 1998;273:5638–5644. doi: 10.1074/jbc.273.10.5638. [DOI] [PubMed] [Google Scholar]
- 53.Young K D. A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J Bacteriol. 1996;178:3962–3966. doi: 10.1128/jb.178.13.3962-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu L, Lee K K, Ens K, Doig P C, Carpenter M R, Staddon W, Hodges R S, Paranchych W, Irvin R T. Partial characterization of a Candida albicans fimbrial adhesin. Infect Immun. 1994;62:2834–2842. doi: 10.1128/iai.62.7.2834-2842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]