Abstract
Maintenance of “gut health” is considered a priority in commercial chicken farms, although a precise definition of what constitutes gut health and how to evaluate it is still lacking. In research settings, monitoring of gut microbiota has gained great attention as shifts in microbial community composition have been associated with gut health and productive performance. However, microbial signatures associated with productivity remain elusive because of the high variability of the microbiota of individual birds resulting in multiple and sometimes contradictory profiles associated with poor or high performance. The high costs associated with the testing and the need for the terminal sampling of a large number of birds for the collection of gut contents also make this tool of limited use in commercial settings. This review highlights the existing literature on the chicken digestive system and associated microbiota; factors affecting the gut microbiota and emergence of the major chicken enteric diseases coccidiosis and necrotic enteritis; methods to evaluate gut health and their association with performance; main issues in investigating chicken microbial populations; and the relationship of microbial profiles and production outcomes. Emphasis is given to emerging noninvasive and easy-to-collect sampling methods that could be used to monitor gut health and microbiological changes in commercial flocks.
Key words: biomarkers, gut health, flock monitoring, noninvasive sampling, 16S rRNA sequencing
INTRODUCTION
Improving flock productive performance is of major concern for the poultry industry. Although improvements in genetic selection and feed formulation have increased production parameters such as feed efficiency and body weight of broilers, nonuniformity on performance within and between successive flocks is still an issue (Stanley et al., 2014; Van Limbergen et al., 2020). Because of that, many nutritional interventions have been developed and their efficacy is usually measured by observing improvements in productive performance (Yadav and Jha, 2019). However, many factors related to health, housing, and management can influence bird performance such as the source of birds, light regime, ventilation, stocking density, feed, water quality, and diseases (Van Limbergen et al., 2020), making it difficult to disentangle the effect of each factor on the observed performance.
Among the factors affecting performance, overall gut health, which can be defined as the “state of symbiotic equilibrium between the microbiota and intestinal tract where animal health and welfare remain unaltered” is considered of major importance (Celi et al., 2019). Specifically, enteric diseases such as coccidiosis and necrotic enteritis (NE) have become a major issue in commercial broilers after strict regulations on the use of in-feed antimicrobials (M'Sadeq et al., 2015). While a presumptive diagnosis of coccidiosis and NE can usually be made when the clinical disease is present, subclinical forms are often difficult to detect because of the lack of practical and reliable diagnostic tools (M'Sadeq et al., 2015). Gut health score systems have been used to determine the severity of intestinal damage associated with coccidiosis (Johnson and Reid, 1970), NE (Keyburn et al., 2006), and general intestinal inflammatory conditions such as dysbiosis (De Gussem, 2010). The routine scoring of a representative number of birds in commercial flocks is often not practical and not consistently performed. Ideal biomarkers of gut health should be easily detected in noninvasive and easy-to-collect samples such as litter and excreta; however, despite the large number of research groups dedicated to finding ideal biomarkers, the use of these tools outside research settings has been limited (Ducatelle et al., 2018).
The main objective of this review is to highlight the existing literature on the chicken digestive microbiota and its function, factors affecting the gut microbiota and emergence of enteric diseases, and methods to evaluate gut health and their association with performance. The focus of this literature review is on broilers, although some of the information presented is relevant for other poultry industries.
THE AVIAN DIGESTIVE SYSTEM
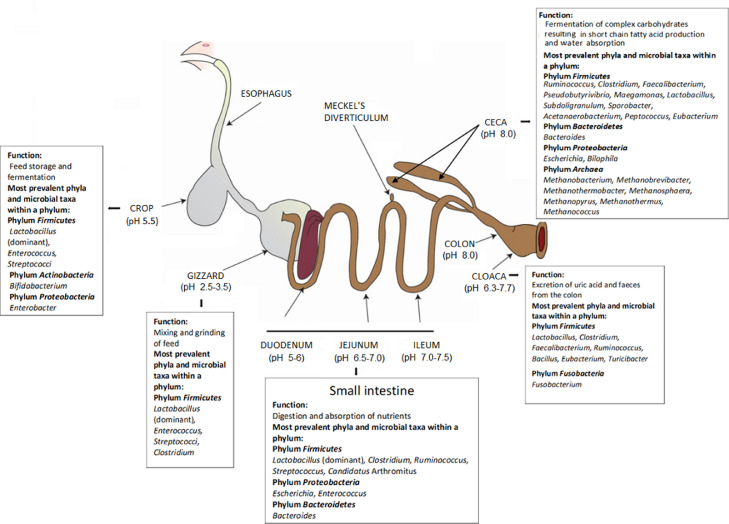
The gastrointestinal tract (GIT) of the chicken is short compared to mammals so that the passage rate of the digesta in the poultry GIT is faster. Although the type of feed given to the chickens determines the retention time of the digesta, the transit of feed in the GIT is in general between 3 and 4 h (Hughes, 2008). The major organs of the GIT and the main microbiota in each organ are presented in Figure 1.
Figure 1.
Major organs of the gastrointestinal tract and the major microbiota of each organ. Modified from Yeoman et al. (2012). The information included the figure were extracted from Rehman et al. (2007), Wielen et al. (2002), Yadav and Jha (2019), Sekelja et al. (2012), Engberg et al. (2004), Jin et al. (1997), Munyaka et al. (2016), Wang et al. (2016), Lu et al. (2003), Danzeisen et al. (2011), Gong et al. (2007), Saengkerdsub et al. (2007a), Saengkerdsub et al. (2007b), Qu et al. (2008), Stanley et al. (2014), Lim et al. (2015), Ohh (2011), Ravindran (2013).
The digestive tract of birds starts with the beak which is used for prehension. Salivary glands in the mouth then secrete mucus which provides lubrication, antiviral and antibacterial protection to the mucosa, and the mucus also facilitates the transport of feed from the mouth to the esophagus (Scanes and Pierzchala-Koziec, 2014). Primary peristalsis in the esophagus moves the feed into the crop or directly into the stomach (Rodrigues and Choct, 2018). The chicken stomach contains a glandular part, named proventriculus, and a muscular part, named gizzard. The contraction and relaxation of the gizzard determine the feed flux which will bypass the crop when the gizzard is empty and will enter the crop when the gizzard is fully or partially filled (Denbow, 2015). The crop is a distended pouch that plays a major role in fermentation (Fuller and Brooker, 1974; Scanes and Pierzchala-Koziec, 2014). Contraction of the crop wall results in the evacuation of feed into the gizzard via proventriculus (Denbow, 2015). Protein digestion is initiated in the proventriculus aided by the secretion of hydrochloric acid and pepsinogen in this organ (Scanes and Pierzchala-Koziec, 2014). The volume of the proventriculus is small so the feed rapidly transits into the gizzard; however, the feed can reflux back into the proventriculus several times for enzymatic digestion (Rodrigues and Choct, 2018). The gizzard is responsible for mixing and grinding the feed until particles are smaller than 0.1 mm when it is then passed into the small intestine (Hetland et al., 2002; Jacob et al., 2011).
The small intestine, which is composed of the duodenum, jejunum, and ileum, is a major site of chemical digestion and absorption after which the feed is passed into the 2 large ceca which are responsible for fermentation and digestion of starch, cellulose, and indigestible carbohydrates (Mead, 1989; Stanley et al., 2014, 2015). The lower passage rate of digesta in the ceca compared to other sections of the GIT allows for increased fermentation of the digesta resulting in increased concentration of short-chain fatty acids (SCFAs) production by the resident microbiota and water absorption (Clench and Mathias, 1995; Rehman et al., 2007; Rodrigues and Choct, 2018). Emptying of the cecum takes place every 24 to 48 h (Clench and Mathias, 1995). Digesta move through the ileo-cecal junction into the colon, from which the digesta can move to the ceca by retrograde antiperistalsis or to the cloaca by peristaltic contraction (Duke, 1989). The colon is very small and little to no digestion or absorption takes place in it. From the colon, the digesta is passed into the cloaca and mixed with uric acid before being expelled out.
THE MICROBIOTA OF THE CHICKEN INTESTINAL TRACT
The microbiota of the gastrointestinal tract is a complex ecosystem predominantly comprised of bacteria, but also contains viruses, archaea, fungi, and protozoa (Smulikowska, 2006; Wei et al., 2013). This section focuses on the bacterial communities that are part of the intestinal microbiota and for simplicity; the term “microbiota” will refer to the bacterial microbiota hereafter.
The Role of the Intestinal Microbiota
The importance of intestinal microbiota in the nutrition, health, physiology, and immunity of the chicken has become evident in recent years (Apajalahti et al., 2004; Sekirov et al., 2010; Pan and Yu, 2014; Kers et al., 2018). The commensal microbiota is attached to the epithelium of the intestine and plays an important role in maintaining homeostasis and in the protection against colonization by pathogens (Yegani and Korver, 2008; Diaz Carrasco et al., 2019). The microbiota interacts with the chicken immune system assisting the training and maturation of the immune cells to ensure that commensal microorganisms are tolerated, pathogens are recognized and curbed and those potentially pathogenic commensal microorganisms such as Clostridium perfringens and Escherichia coli are kept in check (Adil and Magray, 2012; Klindworth et al., 2013). It also helps in the development of the lamina propria, mucus layer, and epithelial monolayer through the production of metabolites such as SCFAs, indoles, vitamins, and antimicrobial compounds, increasing the epithelial absorptive surface (Dibner and Richards, 2005; Yegani and Korver, 2008; Shakouri et al., 2009; Oakley et al., 2014). An in-depth review of microbiota and chicken immune interactions can be found at Kogut et al. (2020).
Microbiota of the cecum also plays a role in host nitrogen metabolism which is highly associated with the efficiency of chickens to extract energy from feed (Oakley et al., 2014). Uric acid and feces mixed in the cloaca may go to the cecum due to the retrograde peristalsis where bacteria convert uric acid to ammonia which is utilized by the host to synthesize amino acids such as glutamine (Vispo and Karasov, 1997). However, the microbiota may also compete for energy and protein and reduce the availability of nutrients for the host (Pan and Yu, 2014). Some of the intestinal microbiota can catabolize bile acids which retards fat digestion and absorption (Furuse and Yokota, 1985; Engberg et al., 2000). When protein is not fully digested and absorbed in the small intestine, it undergoes further fermentation in the large intestine with the potential production of toxic amines which can decrease bird growth and performance (Gaskins et al., 2002). Detailed reviews of the interaction between gut microbiota and diet have been recently published (Pan and Yu, 2014; Yadav and Jha, 2019).
Microbiota in the Different Sections of the Gut
Initial Colonization of the Gut
After chicken eggs are laid they are usually sent to hatcheries that adhere to strict hygienic conditions. Bacteria can penetrate through the eggshell while it is moist (Berrang et al., 1999) and although transmission of bacteria such as Salmonella enterica serovar Enteritidis from hens to chicks has been described, there is somewhat limited evidence of transovarial transmission of bacteria (Tankson et al., 2002; Cortés et al., 2004; Kubasova et al., 2019; Akinyemi et al., 2020). Therefore the majority of colonization of the chick gut with bacteria begins right after hatching via contact with environmental sources at the hatchery, mainly people handling the eggs, transportation boxes, and any feed and water or bedding material available (Kizerwetter-Świda and Binek, 2008; Stanley et al., 2014). Young chickens at 1 and 2 d of age are delivered to commercial farms where they are usually placed in bedding material, although rearing of chicks in plastic nets is also practiced in some countries such as China (Chen et al., 2020; Yan et al., 2021). Several microbiota taxa from new or reused bedding material and other environmental sources such as feed continue the initial colonization of the gut (Wang et al., 2016). As the chickens grow, the microbiota undergoes temporal successions and become more complex and diverse (Wei et al., 2013).
Microbiota in Different Parts of the Gastrointestinal Tract
Overall, the 5 most dominant bacterial phyla found in the chicken GIT are Firmicutes, Tenericutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Wei et al., 2013) (Figure 1). Lactobacillus is the primary bacterial taxa found in the crop, gizzard, duodenum, and ileum. Bifidobacterium and Enterobacter are also commonly detected in the crop (Yeoman et al., 2012; Saxena et al., 2016). The acidic nature of the stomach provides an unfavorable environment for most bacteria so it harbors fewer bacteria than other regions of the GIT (Rehman et al., 2007). The gizzard has primarily lactobacilli with few enterococci, coliforms, and lactose-negative enterobacteria (Zoetendal et al., 2004; Sekelja et al., 2012; Yadav and Jha, 2019). Clostridium perfringens, a commensal but potentially pathogenic bacterium, has also been found in gizzard at a very low level (103 bacteria/g of contents) (Engberg et al., 2004).
The most dominant phylum in the small intestine of chickens is Firmicutes (Yeoman et al., 2012), and this phylum is largely represented by the genus Lactobacillus and to a lesser extent by Clostridium, Ruminococcus, Streptococcus, and Candidatus Arthromitus (Jin et al., 1997; Lu et al., 2003; Yeoman et al., 2012; Munyaka et al., 2016; Wang et al., 2016). Bacterial members of the phylum Proteobacteria, mainly Escherichia and Enterococcus have also been reported in the ileum (Jin et al., 1997; Lu et al., 2003; Yeoman et al., 2012; Wang et al., 2016).
The highest bacterial density and diversity in the chicken intestinal tract are in the cecum (Dibner and Richards, 2005; Shakouri et al., 2009; Oakley et al., 2014). The most abundant phyla found in the cecum are Firmicutes followed by Bacteroidetes while Proteobacteria and Archaea are present in lesser amounts (Figure 1). The most dominant genera in the cecum within the phylum Firmicutes were found to be Lactobacillus, Ruminococcus, Faecalibacterium, and Clostridium (Jin et al., 1997; Gong et al., 2002; Qu et al., 2008; Wang et al., 2016) (Figure 1).
Lactobacillus, Clostridium, Faecalibacterium, Ruminococcus, Bacillus, and Eubacterium (phylum Firmicutes) are the predominant genera in cloaca and excreta (Sekelja et al., 2012; Stanley et al., 2014; Lim et al., 2015); however, the microbiota found in cloacal mucosa and excreta are not stable and abrupt temporal fluctuations occur due to the cyclic emptying of different regions of the GIT (Deusch et al., 2015).
Factors Affecting the Intestinal Microbiota Composition
This section covers some of the major factors that influence intestinal microbial composition such as age, sex, and breed, diet, and litter conditions (Kers et al., 2018).
Age, Sex, and Breed
The maturation of the microbiota which is defined as the age at which microbial community matures and does not change over time has been shown to vary between studies. In a study in commercial broiler farms, Kers et al. (2020) found that cecum phylogenetic diversity stabilized after 21 d of chicken age. However, a study by Lu et al. (2003) found that microbial communities in the cecum were similar at d 14 and 28 and different compared to d 49 suggesting that the microbiota stabilized after 28 d of chicken age under experimental conditions. Torok et al. (2009) also showed large differences in cecal microbial communities between d 14 and 28 in broilers under experimental conditions. Despite those differences, it is accepted that the GIT microbial richness increases and the microbial composition changes with age, that the microbiota becomes more stable as age increases, and that the environment plays a role in when the stabilization will occur (Feye et al., 2020).
At the beginning of the production cycle, on d 0, the family Clostridiaceae was dominant in cecal contents in broilers (Kers et al., 2019a; Kers et al., 2020). Lu et al. (2003) showed that at 3 d of age Lactobacillus represented 25% of the total bacteria in the cecal contents. The concentration of Lactobacillus in cecal contents at d 3 was 100 times higher compared to d 42 (Gong et al., 2008). Among the Lactobacillus, Lactobacillus delbrueckii and Lactobacillus acidophilus were the dominant species in the cecum (Lu et al., 2003). In the same study, the cecal contents were dominated by Clostridium saccharolyticum, Clostridium oroticum, and Clostridium orbiscindens at 7 d of age; Ruminococcus schinkii and Clostridium indolis at d 14 to 28; and Eubacterium at d 49 (Lu et al., 2003). In a study by Ranjitkar et al. (2016), the relative abundance of Enterococcus in ileal contents decreased from 25% at d 8 to 1% at d 15 and remained unchanged till d 36 while the level of Clostridium increased from 1 to 18% and Streptococcus from 5 to 15% from d 8 to 36.
Although the effect of sex is more pronounced when chickens undergo sexual maturity, differences have also been observed in younger broilers and seem to be correlated with sex-based differences in glycan and lipid metabolism by the cecal microbiota (Lumpkins et al., 2008; Lee et al., 2017; Cui et al., 2021). In the cecum of 35-day-old Ross 308 broilers, Oscilospira and Tenericutes were higher in females and Bacteroides were higher in males (Lee et al., 2017). In a study by Cui et al. (2021), the abundance of Bacteroides, Megamonas, Megasphaera, and Phascolarctobacterium were higher in males while Akkermansia was higher in females in the cecum of 35-day-old chickens. Torok et al. (2013) also found that sex influences total eubacterial numbers with males having an increased abundance of eubacteria compared to females in cecal contents at d 22 and 42. Lumpkins et al. (2008) showed that microbial communities of the ileum also differed between male and female broilers at d 3. While the overall effect of sex on the chicken GIT microbiota is unclear, whole-genome profiling of prepubertal mice has shown that even in the absence of high levels of circulating sex hormones, mice with identical microbiota have intrinsic sex-specific gene regulation in the GIT and that sex differences in the microbiota after puberty are correlated with sex differences in GIT expression of multiple genes (Vemuri et al., 2019).
Chicken genotype affects the microbiota (Pandit et al., 2018; Ji et al., 2020; Tumova et al., 2021), although this effect appears to be limited to specific taxa (Chintoan-Uta et al., 2020; Wen et al., 2021). Kers et al. (2018) compiled data from 12 studies reporting 16S rRNA sequencing of cecal samples of Cobb and Ross breeds. Actinobacteria was present in Cobb chickens in all 4 studies of this breed, and in 3 out of 8 studies reporting data on Ross chickens. Similarly, Bacteroidetes were present in Cobb chickens in all 4 studies and 6 out of 8 studies in Ross chickens. Large differences in the cecal microbial composition between chickens of Hubbard and Ross breeds at an early age have also been reported (Richards et al., 2019). On d 0, Hubbard cecal microbiota was dominated by Enterobacteriaceae while Cobb cecal microbiota was dominated by Enterococcaceae and Clostridiaceae. On d 3, a higher abundance of Bifidobacteriaceae but a lesser level of Enterobacteriaceae was seen in Hubbard compared to Ross chickens but from d 7, no differences were observed in the microbial composition between breeds (Richards et al., 2019).
From the above findings, it is evident that age, sex, and breed can influence the microbiota of the poultry, and detailed information should be provided in microbiota studies regarding these parameters.
Diet
Diet has a major influence on the gut microbiota composition and both the composition of the diet and the physical form of feed (pellet or mash) affect digestibility and nutrient absorption in the gut (Apajalahti et al., 2001, 2004). When broilers were fed a wheat-based diet in pellet form there was an increased concentration of coliforms and enterococci in the ileum and C. perfringens in the cecum compared to the same feed in mash form (Engberg et al., 2002). Compared to grounded wheat in pellet form, whole wheat feeding resulted in the reduction of C. perfringens and lactose negative enterobacteria and increased numbers of Bifidobacterium and bacterial diversity in ceca (Apajalahti et al., 2001; Engberg et al., 2004). Higher levels of Lactobacillus spp were found throughout the intestinal tract of chickens fed whole wheat compared to chickens fed with pellets of grounded wheat (Engberg et al., 2004).
Chickens fed a rye-based diet had increased levels of coliforms in the duodenum and ileum and increased levels of lactic acid bacteria in the cecum, duodenum, and ileum compared to chickens fed a corn-based diet (Tellez et al., 2014). Diets rich in nonstarch polysaccharides (NSPs), fish meal, and bone meal have been associated with the increased proliferation of C. perfringens and predispose chickens to NE (Williams et al., 2003; Williams, 2005; M'Sadeq et al., 2015). NSPs increase the viscosity in the intestinal lumen, decrease the passage rate and enzymatic activities, and reduce the feed conversion efficiency (Choct and Annison, 1992). The increased retention time of the digesta, especially in diets of high protein or with unbalanced amino acid profiles, provides substrates and facilitates colonization of pathogenic bacteria in the small intestine, including C. perfringens (Waldenstedt et al., 2000; Annett et al., 2002; Timbermont et al., 2011; Loh and Blaut, 2012). Fish meal also contains higher amount of zinc and glycine and a positive correlation between the C. perfringens abundance and glycine concentration has been shown (Wilkie et al., 2005; Dahiya et al., 2007). In addition to feed ingredients and physical feed form, feed deprivation and withdrawal also influence gut health and microbiota composition leading to increased Salmonella colonization (Burkholder et al., 2008; Thompson et al., 2008; Lamot et al., 2014).
Torok et al. (2008) investigated the difference in gut microbial communities in broilers fed barley or barley supplemented with exogenous enzymes and found that the bacterial community in cecum and ileum was different between treatments. The addition of exogenous enzymes increased the butyrate and lactic acid-producing bacteria in the ceca (Munyaka et al., 2016). Different feed additives such as enzymes, probiotics, prebiotics, and symbiotics have been used to modulate gut microbiota and improve the immune system of poultry (Jha and Berrocoso, 2015; Yadav and Jha, 2019).
Litter Type and Conditions
Litter, which is a mixture of excreta, bedding material, feather, dander, feed, and various microorganisms, can have a direct influence on gut microbiota composition. Studies describing the microbial composition of litter are scarce and usually focus on the detection of pathogenic microorganisms such as Salmonella, E. coli, Campylobacter, and C. perfringens (Bennett et al., 2005). In some countries such as the United States and Brazil, it is common to reuse litter in broiler flocks up to 6 cycles of grow-out (Coufal et al., 2006; Roll et al., 2011) while in Canada and Australia, fresh bedding materials is usually used in every grow out period (Cockerill et al., 2020; Feye et al., 2020). As chickens consume up to 4% of their diet as litter during the first 2 wk of age (Malone et al., 1983), litter type and microbial composition directly influence the colonization and development of gut microbiota in chickens (Torok et al., 2009).
Using culture-based techniques, Terzich et al. (2000) investigated total bacteria, coliforms, Staphylococcus, Gram-positive, and Gram-negative bacterial counts in samples of litter that had been used for at least 3 chicken grow-out cycles in the United States. The average total number of bacteria was 2.54 × 1011 colony forming units (CFU)/g while the average count of Gram-negative or Gram-positive was 1.60 × 1011 CFU/g; Staphylococcus and coliforms were commonly detected (Terzich et al., 2000). Reused wood shaving litter was found to enable Campylobacter jejuni and Campylobacter coli to survive for at least 20 d while fresh litter inhibited the growth of these pathogens (Kassem et al., 2010).
Using a sequencing-based technique, Wang et al. (2016) found that reused pine shaving litter contained mostly halotolerant alkaliphiles and Faecalibacterium prausnitzii while fresh litter was dominated by Devosia, Yaniella, Acinetobacter, Trichococcus, and Luteimonas. In the same study, ileal mucosa of broilers raised on reused litter had a higher abundance of Enterococcus at d 10 while Lactobacillus was higher at d 35 compared to birds raised on fresh litter. Similarly, in cecal contents, Blautia, Anaerotruncus, and Faecalibacterium were at higher levels in birds raised on the re-used litter while Lactobacillus, Escherichia, Subdoligranulum, Bacteroides, and Clostridium XIVb were higher in cecal contents of birds raised on fresh litter at d 10 (Wang et al., 2016). On d 35, chickens raised on reused litter had a higher abundance of Faecalibaterium and Oscillibacter in cecal contents while the abundance of Subdoligranulum was higher in cecal contents of birds raised on fresh litter (Wang et al., 2016). Cressman et al. (2010) described that 7-day-old broilers raised on reused litter had unclassified Clostridiales group of bacteria as the dominating feature in the ileal mucosa while the Lactobacillus were dominant in chickens raised on fresh litter.
The microbial communities also vary depending on the bedding material (Torok et al., 2009). The microbial communities in the cecum of 28-day-old broilers raised on chopped straw were significantly different from broilers raised in sawdust; similarly, there were differences in the cecal microbial communities of broilers raised on rice hulls, chopped straw, or shredded paper (Torok et al., 2009). Total aerobic mesophilic bacteria, enterococci, Enterobacteriaceae, and Staphylococcus aureus counts were found to be higher in pine wood shavings compared to dried rose dreg at 42 d (Aktan and Sagdic, 2004).
The management of litter may also influence chicken gut microbiota as the Shannon diversity and richness of microbiota has been shown to vary between dry and wet litter with higher diversity found in wet litter compared to dry litter (Dumas et al., 2011; Oakley et al., 2013). Although not consistently, wet litter is associated with poor gut health in poultry (Collett, 2012). These results show that litter quality, type, and management have a direct impact on gut microbiota composition, although the relationship of early microbial colonization of the gut in various litter management conditions and chicken productive performance is unknown.
Some Methodological Aspects of Studying Microbiota
Although 16S rRNA gene sequencing has become a popular approach to study microbiota in humans and animals, direct comparison between microbiota detected in different studies could be hampered by differences in sample processing. Some of the technical aspects that could bring potential bias to microbiota studies are discussed below. In-depth reviews have been recently published covering sample size calculation (Casals-Pascual et al., 2020), laboratory methodological aspects (Pollock et al., 2018), and bioinformatics (Bharti and Grimm, 2021).
Sample Size
Because of the high bird-to-bird microbiota variability, selecting the appropriate sample size is crucial to detect differences between groups in microbiota studies (Stanley et al., 2014; Bharti and Grimm, 2021). In humans, Falony et al. (2016) described that over 500 samples per group are required to accurately identify microbial shifts and sample sizes of 100 individuals per group are considered relatively small. In contrast, chicken microbiota studies have often less than 10 samples per treatment at each time point (Kers et al., 2020). Appropriate sample sizes should be calculated considering a high population variance and weak treatment effect sizes to exclude bias in the results (Bharti and Grimm, 2021). Different statistical models have been developed for power test and sample size calculation in microbiome studies (Xia et al., 2018), ideally using data from preliminary experiments to calculate the expected variances and treatment effect sizes (Qian et al., 2020).
Sample Collection
In a given GIT region, samples can be collected directly from digesta, mucosa, or a swab of digesta and/or mucosa. Although the impact of the choice of sample type has not been fully studied in chickens, a comparison between the microbiota of cecal luminal contents and cecal mucosa in broilers showed a higher abundance of Bacteroides and a lower level of Lachnospiraceae in luminal contents compared to the mucosa (Richards et al., 2019), suggesting that the sample type may have an impact on microbial communities.
Storage Condition of Samples
Ideally, gut contents or excreta should be stored at -80°C immediately after collection to prevent changes in the microbial community composition, but the maintenance of cold chain from sample collection until transference to a laboratory is not always possible (Gorzelak et al., 2015). It has been established that the microbial composition of human stools stored at 4°C for up to 72 h was similar to the control samples stored at -80°C (Choo et al., 2015; Tedjo et al., 2015). However, in stool samples stored at room temperature for 3 d, there was a decrease in abundance of the phylum Firmicutes, mainly genus Anaerostipes, Ruminococcus, Faecalibacterium, and Lachnospiraceae; and an increase in the abundance of phylum Actinobacteria mainly the genus Bifidobacterium compared to samples stored at -80°C (Roesch et al., 2009; Choo et al., 2015). Preservatives such as RNAlater have been shown to decrease DNA purity and lower microbial diversity compared to samples stored at −80 °C for 3 d (Dominianni et al., 2014). Similarly, buffers containing EDTA have been shown to increase the abundance of Proteobacteria and reduce the abundance of Firmicutes and Actinobacteria in stool samples stored in this buffer for 3 d compared to samples stored at −80°C (Choo et al., 2015). Other preservatives such as OMNIgene.GUT (DNA Genotek, OMR-200, Ontario, Canada) decreased the relative abundance of Lachnospiraceae and increased the abundance of Sutterella and Faecalibacterium when samples were stored for 3 d compared to storage at -80°C without buffer (Chen et al., 2019). Consequently, all samples in an experiment should undergo to the same cryopreservation protocol to avoid the introduction of biases in the analysis.
DNA Extraction Methods
The methods of DNA extraction employed have a direct impact on the microbiota composition and diversity (Scupham et al., 2007; Costea et al., 2017; Abundo et al., 2021). Different commercially available extraction kits use different protocols for bacterial cell lysis such as enzymatic and chemical digestion, and/or mechanical disruption of bacterial cell walls such as bead-beating (Videnska et al., 2019). Mechanical disruption, particularly bead-beating following chemical digestion is recommended for lysis of the cell walls of Gram-positive bacteria as it increases the DNA yield and microbial diversity (Lu et al., 2015; Fidler et al., 2020). Complete absence of the Gram-positive Bifidobacterium spp was observed upon extracting the DNA without a bead-beating step while the same genus was well represented after a bead-beating step was added (Walker et al., 2015). However, bead beating for a long time results in the shearing of DNA into small fragments and the protocol needs to be optimized depending upon the sample matrix (von Wintzingerode et al., 1997; Dilhari et al., 2017). Once the extraction method is optimized, the same protocol should be consistently followed to allow for direct comparison of the microbiota (Boers et al., 2019). The use of mock bacterial communities of known composition is a recommended technical control in every study to account for the bias introduced by the DNA extraction protocol (Salter et al., 2014).
Choice of Primers
16S rRNA gene amplicon sequencing is based on the amplification of a hypervariable region of the bacterial 16S rRNA gene by PCR (Adhikari, 2019). The 16S rRNA gene is around 1550 nucleotides long and contains 9 hypervariable regions (V1-V9) that allow the differentiation between bacterial species (Van de Peer et al., 1996), and different studies target different hypervariable regions for amplification and different primers have been used to amplify the same hypervariable region (Bergmann et al., 2011; Walker et al., 2015; Barb et al., 2016; Pollock et al., 2018). Studies have shown that some of the commonly used universal primers may underestimate the abundance level of certain bacterial taxa or may not amplify some biologically relevant bacterial taxa (Bergmann et al., 2011; Walker et al., 2015). Barb et al. (2016) compared the efficacy of 6 universal primers designed to target V2, V3, V4, V6-7, V8, and V9 regions of 16S rRNA gene on the identification of the bacteria present in mock samples and found that the highest divergence from the original mock microbial community was found for primer sets targeting the V9 region, and the least divergence was found for primer sets targeting V2, V4, and V6-7. Fouhy et al. (2016) also found differences in genus and species detected using 3 different universal primer sets illustrating that the choice of primer could impact microbiota studies. The choice of primers should allow high phylogenetic resolution and low bias (Klindworth et al., 2013; Fuks et al., 2018).
PCR Cycles and DNA Polymerase Types
Further biases could also be introduced in the sequencing results because of the number of PCR cycles and the type of DNA polymerase used. Increased PCR cycles (15–30) lead to the formation of artifacts and a large number of chimera sequences which could lead to misinterpretation of results (Suzuki and Giovannoni, 1996; von Wintzingerode et al., 1997; Ishii and Fukui, 2001; Hongoh et al., 2003). This effect is much more pronounced when using high-fidelity DNA polymerase with proofreading activity compared with normal Taq DNA polymerase (Ahn et al., 2012). In a study by Hongoh et al. (2003), 2 high-fidelity polymerases, namely PfuUltra II Fusion HS DNA Polymerase (Stratagene, La Jolla, CA) and Ex Taq (Takara, Ontario, Canada) produced significantly different microbial community structures. It is recommended to optimize the cycling conditions and to evaluate the effect of different polymerase enzymes in samples of mock bacterial communities before the commencement of the study and keep the same conditions throughout the same study (Pollock et al., 2018).
MICROBIOTA PROFILES AND PRODUCTIVE PERFORMANCE
Identifying and understanding the role of specific microbiota profiles associated with efficient feed conversion is of utmost importance to develop and test dietary interventions (Stanley et al., 2016). Numerous studies have been performed to identify GIT microbiota associated with high and low-performance in chickens with often nonreproducible or contradictory results among trials as demonstrated by Stanley et al. (2016), with additional literature recently reviewed by Diaz Carrasco et al. (2019).
Stanley et al. (2016) investigated the differences in cecal microbiota of high- and low-performance birds across 3 experimental trials using the same experimental facility, feed batch, and source of birds. In that study, specific microbiota was differentially abundant in high and low performing birds in each trial but some of the microbiota present in high performing birds in one trial was absent in other trials and none of the microbial taxa was consistently present in high or low performing birds across the 3 trials (Stanley et al., 2016). Although specific microbial taxa could not be conclusively associated with a production outcome, high performance, as measured by feed conversion ratio (FCR), were in general enriched with bacterial communities known to degrade cellulose and resistant starch, mainly members of the families Lachnospiraceae (e.g., Clostridium lactatifermentans), Ruminococcaceae (e.g., Clostridium islandicum, Faecalibacterium prausnitzii) Erysipelotrichaceae (e.g., Clostridium spp), Bacteroides fragalis and Clostridium cellulosi while bacterial communities negatively correlated with performance belonged to uncultured members of an unknown class of Firmicutes and species of clostridium belonging to the family Clostridiaceae (Stanley et al., 2013a; Stanley et al., 2016).
Those findings were similar to previous studies in experimental chickens in which Ruminococcus torques and Clostridium lactatifermentans isolated in ceca were associated with high performance (Torok et al., 2013) while Lactobacillus salivarius, L. aviarius, and L. crispatus in ileum were associated with decreased performance (Torok et al., 2011). As evidenced by the findings above, although some Lactobacillus strains can be a major energy source for the host via their ability to produce SCFAs not all species or strains are beneficial. Lactobacillus gasseri SBT2055 has been reported to reduce body weight in obese humans (Kadooka et al., 2010) while L. salivarius has been associated with bile deconjugating activity which is associated with a decrease in the growth rate of chickens (Knarreborg et al., 2002). Streptococcus faecium and C. perfringens are also strong bile deconjugating bacteria (Stutz and Lawton, 1984; Knarreborg et al., 2002). In a study in commercial broiler flocks, Johnson et al. (2018) found that the microbiota in the cecum and ileum positively correlated with bird weight was Bacteroides, Bilophila, and Butyricimonas while the taxa negatively correlated with bird weight included Anaerotruncus, Bacteroides, Blautia, Clostridium, Coprobacillus, Coprococcus, Eggerthella, Enterococcus, Lactobacillus, and Oscillospira.
In a study by Singh et al. (2012) in experimental broilers, excreta of low-performance birds, as measured by FCR, were enriched with Acinetobacter, Anaerosporobacter, Arcobacter, Dysgonomonas, Ignatzschineria, Megamonas, Myroides, Providencia, and Wohlfahrtiimonas and excreta of high performing birds were enriched with Alistipes, Barnesiella, Cloacibacillus, Escherichia/Shigella, Faecalibacterium, Helicobacter, Oscillibacter, and Phascolarctobacterium while the abundance of Lactobacillus, Enterococcus and Bacteroides were found to be similar in both low- and high-FCR birds.
As revised above, the majority of the studies have focused on the association of cecum and ileum microbiota with production. The development of practical and noninvasive tools to monitor microbiota associated with production would greatly enhance this field of research (Diaz Carrasco et al., 2019). Monitoring the microbiota at flock-level using samples such as pooled excreta, litter or poultry dust would be perhaps an alternative to individual bird sampling as a way to reduce the noise generated by the within bird variation observed in GIT microbiota studies. Another approach for overcoming the variability of the GIT microbiota is to further interrogate the functionality of the observed microbial population, either by doing predictive functional bioinformatics analysis or by directly measuring microbial metabolites of interest in the cecum using high-performance liquid chromatography, which is particularly useful for measuring microbial fatty acids and lactic acid outputs that are related to gut health, or using liquid-chromatography-tandem mass spectroscopy, which captures the microbial metabolomic profile (Kers et al., 2019b; Niu et al., 2020; Park et al., 2020; Slizewska et al., 2020).
DISEASES OF THE ENTERIC TRACT: COCCIDIOSIS AND NECROTIC ENTERITIS
Coccidiosis
Coccidiosis is an enteric disease that can be caused by any of 7 species of Eimeria in chickens. From those, Eimeria mitis and E. praecox are less pathogenic compared to E. brunetti, E. maxima, E. acervulina, E. necatrix, and E. tenella (Williams, 2005). Globally the annual cost of coccidiosis was estimated to be US$3 billion (Williams, 1999) but this figure is largely outdated. Coccidiosis can occur in clinical or subclinical forms. In the clinical form, it is associated with mortality, bloody diarrhea, and weight loss whereas subclinical coccidiosis is manifested by reduced feed efficiency and poor weight gain (Williams, 1999). The infection with Eimeria usually affects the growth and feed conversion for 2 to 3 wk, after which a period of compensatory growth occurs (Voeten et al., 1988). Subclinical coccidiosis may have no effect on FCR and chicken growth when birds are infected in the first week of life because of compensatory growth, or when birds are infected on the last week of grow-out because in this time-frame there is not enough damage to the intestinal mucosa (Voeten et al., 1988; Kipper et al., 2013).
Traditional diagnosis of coccidiosis is based on counts of oocysts in excreta or litter, which is costly, labor-intensive, and requires skilled personnel to differentiate the oocysts based on the morphology (De Gussem, 2007). Based on this method, an Eimeria oocyst count of more than 50,000/g of pooled excreta has been associated with loss of performance in chicken flocks when more than 50% of oocysts are of medium size (corresponding to infections with E. necatrix, E. tenella, or E. praecox) or oocysts of large size (E. brunetti or E. maxima); while the presence of small oocysts (E. acervulina) was not associated to significant production losses (Haug et al., 2008). Molecular based approaches, mainly using PCR, have been effective to subtype and quantifying Eimeria species in excreta and litter in both experimental and commercial flocks (Morris and Gasser, 2006; Morris et al., 2007; Morgan et al., 2009; Vrba et al., 2010; Godwin and Morgan, 2015). More recently, DNA of C. perfringens and Eimeria species have been detected in poultry house dust of experimental and commercial flocks (Ahaduzzaman et al., 2021a; Bindari et al., 2021a).
In commercial broiler flocks, monitoring of coccidiosis is usually performed by scoring several intestinal sections based on the predisposition of the Eimeria species; a score of 0 is given for the absence of gross pathology in an intestinal segment and a score of +4 is given for severe gross lesions (Johnson and Reid, 1970). This method can be subjective and requires a high level of expertise to correctly score lesions as other diseases with similar pathologies may co-occur, for example, NE (Shirley et al., 2005). Coccidiosis is an important predisposing factor for NE and different Eimeria spp administration regimens have been used to induce NE experimentally (Van Immerseel et al., 2009; Dierick et al., 2021).
Coccidiosis also alters gut microbiota. Ceca of healthy birds were found to be dominated by the genera Subdoligranulum, Coprococcus, Alistipes, Lactobacillus, and Faecalibacterium while ceca from Eimeria spp (E. acervulina, E. maxima, and E. tenella) infected birds were dominated by Escherichia/Shigella and Bacteroides (Kley et al., 2012). In a study by Macdonald et al. (2017), Bacillales and Lactobacillales were decreased while Enterobacteriaceae was increased following E. tenella infection in the cecum; however, the overall diversity of the microbial taxa was not changed. Wu et al. (2014) reported a reduction in the number of Ruminococcaceae groups and an increase in 3 unknown Clostridium species in birds challenged with E. acervulina, E. maxima, and E. brunetti compared to unchallenged groups. Hume et al. (2006) also observed a clear shift in the microbiota of duodenal, ileum, and cecal contents following E. acervulina, E. maxima, and E. tenella infection compared to unchallenged birds.
Necrotic Enteritis
Necrotic enteritis (NE) is an economically important enteric disease affecting broilers (M'Sadeq et al., 2015). Strains of C. perfringens type A carrying necrotic enteritis B-like toxin (netB) plasmid are associated with increased risk of NE, although the presence of netB does not seem sufficient nor necessary to cause clinical disease (Rood et al., 2016). Similar to coccidiosis, NE can manifest clinically or subclinically. The clinical presentation includes diarrhea and associated wet litter, decreased appetite, ruffled feathers, anorexia, depression, and death (Al-Sheikhly and Al-Saieg, 1980; Williams, 2005). In NE-affected flocks, the mortality rate is about 1% per day and birds usually die within a few hours of displaying clinical signs (Helmboldt and Bryant, 1971). The subclinical form may result in reduced feed intake and poor FCR (Lovland and Kaldhusdal, 2001; Morris and Gasser, 2006) which may lead to great economic losses (Dahiya et al., 2006).
Clostridium perfringens is found in the intestinal tract of healthy chickens usually in lower amounts (102–104 CFU/g digesta) without causing disease (Kondo, 1988; Caly et al., 2015). Changes in the GIT environment via feed, disease, stress, and immunosuppression can facilitate C. perfringens proliferation to concentrations of 107 to 109 cfu/g digesta in NE affected chickens (Kondo, 1988; McDevitt et al., 2006). Birds that survive NE outbreaks have impaired digestion and absorptive capacity of nutrients (Paiva and McElroy, 2014); however, microbiota shifts in those birds remain to be investigated.
The onset of NE is associated with a clear shift of microbiota but it is unclear if the microbial shift was a predisposing factor or a consequence of the disease (Antonissen et al., 2016). Stanley et al. (2012) found that birds with NE induced by a wheat-based diet with 50% fish meal starting on d 15, followed by a C. perfringens in-feed challenge on d 19 to 22 had decreased abundance of the butyrate-producing bacteria Weissella confusa and an increase in the abundance of unclassified Mollicutes compared to healthy birds examined at d 23. Feng et al. (2010) tested the difference in abundance of Enterococcus, Enterobacteriaceae, Clostridium, and Lactobacillus between birds fed with and without C. perfringens contaminated feed and found that only lactobacilli were reduced 2 d postinfection compared to the controls. Antonissen et al. (2016) conducted an extensive review on microbial shifts associated with NE and concluded that different predisposing factors for NE reduced the abundance of segmented filamentous bacteria, butyric acid and lactic acid producing bacteria.
Diagnosis of NE is usually performed by the scoring of intestinal lesions (Keyburn et al., 2006; Vidanarachchi et al., 2013), while the culture of digesta for detection of C. perfringens and toxins and histopathological techniques are also available (Smyth, 2016). One of the major issues with the scoring of intestinal lesions in the field is misidentifying autolytic changes or coccidiosis lesions, especially from E. necatrix, as of NE (Smyth, 2016). Monitoring of both Eimeria species and C. perfringens using noninvasive easy to collect population-level samples would perhaps provide useful research or on-farm tools to evaluate the management interventions of both diseases at flock-level (Ahaduzzaman et al., 2020; Bindari et al., 2021a).
METHODS FOR MONITORING SHIFTS IN CHICKEN INTESTINAL STATUS
A wide range of tools have been evaluated in their ability to accurately evaluate healthy or diseased enteric states (Ducatelle et al., 2018; Celi et al., 2019) but to date, no standardized, reliable and practical method is available. Currently, the gold standard to measure gut health consists of microscopic examination of small intestine sections for evaluation of the villus height and crypt depth ratio (de Verdal et al., 2010). It is, however, time-consuming, and requires the collection of samples and a specialized laboratory, which is not readily available outside of research settings. In commercial settings, scoring of different intestinal sections is usually performed when an enteric disease is observed or anticipated but this method is time-consuming, requires skilled staff and the diagnosis can be confounded as NE, coccidiosis and dysbiosis produce similar gross lesions and these conditions may overlap.
To overcome these limitations, biomarkers that are indicative of gut inflammation or disruption of the gut barrier permeability have been widely investigated. Gut permeability, a term describing the capacity of the intestinal epithelium to allow the passage of molecules through nonmediated diffusion, is affected by several factors including infectious and noninfectious diseases, stress, hormones, diet, and others (Travis and Menzies, 1992). Biomarkers for gut health can be grouped into 3 broad categories, 1) Host protein biomarkers that are indicative of gut barrier damage such as plasma protein detection in gut contents (e.g., hemoglobin subunit beta); or detection of gut epithelium proteins in the blood (e.g., enzyme diaminoxidase); or detection of acute-phase proteins (e.g., C-reactive protein, ovotransferrin, alpha-1 antitrypsin) in blood or excreta produced in response to inflammation or as a result of translocation of bacteria to the liver; 2) Microbial metabolites (e.g., D-lactate) and macromolecules (e.g., fatty acid-binding protein) in blood as evidence of gut barrier failure, or the profile of microbiota metabolites in the cecum (e.g., fatty acids or lactic acid); 3) Biomarkers related to microbiota patterns.
This section will focus on the third category, while biomarkers from categories 1 and 2 above that have been used in poultry are summarized in Table 1 and recent reviews by Ducatelle et al. (2018) and Celi et al. (2017). Among those biomarkers, reference values indicative of gut health are available for microscopic examination of villus height and depth and villus/crypt ratio in broilers at d 23 (de Verdal et al., 2010); while reference values for other biomarkers have not been established.
Table 1.
Potential biomarkers for monitoring gut health in poultry.
| Biomarkers | Biological samples for examination | Tools used to measure biomarker | Pros | Cons | References |
|---|---|---|---|---|---|
| Biomarkers in the intestinal wall | |||||
| Microscopic examination of the intestine (measurement of villus length/crypt depth ratio) | Direct microscopic evaluation of intestinal wall | Histology | The gold standard method to evaluate intestine health. Reference values are available. |
Requires skilled personnel to collect samples and a specialized laboratory for sample processing. For diagnostic purposes, additional tests are required increasing costs | (de Verdal et al., 2010; Chen et al., 2015; Ducatelle et al., 2018) |
| Intestinal inflammation and gut wall appearance | Direct macroscopic evaluation of intestine | Gross evaluation of tissues | Direct measurement of gut appearance, scores are available to determine the extent of intestinal inflammation for necrotic enteritis, coccidiosis, and dysbiosis | It is subjective, requires a skilled veterinarian for lesion scoring and terminal sampling of birds | (Johnson and Reid, 1970; Keyburn et al., 2006; De Gussem, 2010). |
| Acute-phase proteins | |||||
| Ovotransferrin | Blood and excreta | Immunoassay | Cost-effective, no need to sacrifice chickens for sample collection | The biomarker is susceptible to proteolysis and samples should be tested from fresh excreta. Has only been reported to be increased in experimental meat chickens in a necrotic enteritis model, E. maxima, E. tenella, and E. coli infection | (Rath et al., 2009; Goossens et al., 2018) |
| Alpha-1 antitrypsin | Blood and excreta | Immunoassay | The enzyme is resistant to proteolysis, cost-effective technique, no need to sacrifice chickens for sample collection | The suitability of this biomarker in experimental meat chickens is debatable as the concentration of this protein in excreta or blood remained unchanged between control birds and birds with damaged intestinal integrity induced by fasting or by administration of dexamethasone and dextran sodium sulfate | (Gilani et al., 2017; Barekatain et al., 2020) |
| α1-acid glycoprotein | Blood | Immunoassay | Cost-effective, no need to sacrifice chickens for sample collection | No report of detection in excreta, it has been reported to be induced in blood after administration of dexamethasone in broilers | (Chen et al., 2015; Barekatain et al., 2020) |
| Host protein biomarkers | |||||
| Citrulline | Blood | Immunoassay | Cost-effective, no need to sacrifice chickens, and has the potential to be used as a marker of small intestine barrier in poultry | Decreased level of plasma citrulline was correlated with reduced enterocyte mass in chickens fed a rye-based diet compared to chickens fed a corn-based diet after 20 d. Need to be tested in other conditions of intestinal disturbance. | (Baxter et al., 2019) |
| Fibronectin | Blood and excreta | Immunoassay | Cost-effective, no need to sacrifice chickens, and has the potential to be used as a marker of intestinal inflammation in poultry | Increased levels on gut leakage model induced by administration of dexamethasone and E. maxima and E. acervulina. Need to be tested in other conditions of intestinal disturbance | (Owen et al., 2008; De Meyer et al., 2019; Barekatain et al., 2020) |
| Diaminoxidase | Blood | Immunoassay | Cost-effective, no need to sacrifice chickens for sample collection | Only studied in laying hens as a marker of gut barrier failure after feed withdrawal for 12 h. Not reported in broilers, needs to be tested in other conditions of intestinal disturbance | (Lei et al., 2013) |
| Fatty acid-binding protein | Blood and excreta | Immunoassay, PCR | Cost-effective, no need to sacrifice chickens for sample collection | Due to the higher molecular weight of fatty acid-binding protein (15000 Da), severe damage of intestinal epithelium is required for its passage from the intestine to the blood. In excreta, one study showed no difference between gut barrier damage induced by dexamethasone using ELISA. However, in another study, the DNA of this biomarker was increased in blood after birds were administered twice the dose of a coccidiosis vaccine containing a mixture of E. acervulina, E. maxima, and E. tenella. Needs further investigation. |
(Iizuka and Konno, 2011; Chen et al., 2015; Barekatain et al., 2020) |
| Myeloid protein-1, Alpha-actinin-4, Apolipoprotein A-1, Hemoglobin subunit beta, Nucleophosmin, Ovoinhibitor, Transthyretin | Colon contents | PCR | Cost-effective, colon contents can be collected without sacrificing chickens | Only tested in gut leakage model induced by coccidia administration (E. maxima and E. acervulina). Needs testing in other conditions of intestinal disturbance | (De Meyer et al., 2019) |
| Metabolome profiling | Blood | Ultra-high performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) | No need to sacrifice chickens for sample collection. Characterizes biochemical and metabolic changes in the host, can be used for biomarker discovery and characterization of metabolites and metabolic changes of the host and microbiota-associated products in broilers after inoculation with Eimeria spp., eating restriction patterns, and supplementation with enzymes, antibiotics, prebiotics, and probiotics. | Expensive, time-consuming, requires specialized equipment and intensive bioinformatics | (Aggrey et al., 2019; Cao et al., 2020; Gonzalez-Uarquin et al., 2020; Wang et al., 2021; Zhang et al., 2021) |
| Microbial shift or detection of bacterial metabolites as biomarkers | |||||
| Detection of microbial shift such as a decreased abundance of family Enterobacteriaceae as a marker of poor gut health |
Gut contents and excreta | Sequencing based approach/PCR | Identification of microbial shifts could be used for testing of management interventions or occurrence of antibiotic resistance genes | Expensive, time-consuming, and difficult to find consistent microbial taxa associated with health and disease due to extensive bird to bird variation | (Ducatelle et al., 2018; Hernandez-Patlan et al., 2019a) |
| Bacterial metabolite ‘D-lactate’ in blood | Blood | Immunoassay | No need to sacrifice chickens for sample collection | Only studied in laying hens after feed restriction for 12 h. Needs testing in meat chickens and other conditions of intestinal disturbance. | (Lei et al., 2013; Gilani et al., 2016) |
| Bacterial count in liver | Liver | Culture | A more accurate approach to evaluate translocation of bacteria as a result of gut barrier failure | Need to sacrifice birds, time-consuming, and expensive. Has been reported to be increased in birds fed with the rye-based diet compared to the corn-based diet, not studied in intestinal disease models. | (Tellez et al., 2014; Ducatelle et al., 2018) |
| Genes encoding bacterial enzymes in the butyrate production pathway | Excreta and gut contents | PCR | Cost-effective, no need to sacrifice chickens for sample collection | Has not been studied in leaky gut models but reported to be increased amounts in improved feed conversion efficient birds fed with xylooligosaccharides and wheat-rye based diet compared to only wheat-rye based diet | (De Maesschalck et al., 2015; Ducatelle et al., 2018) |
| Endotoxin production by gram-negative bacteria | Blood | PCR | Cost-effective, no need to sacrifice chickens for sample collection | Has only been reported to increase in chickens administered twice the recommended dose of coccidiosis vaccine containing a mixture of E. acervulina, E. maxima, and E. tenella | (Chen et al., 2015) |
| Fatty acids and lactic acid profiles, other bacterial metabolites | Gut contents (cecum) | High-performance liquid chromatography (HPLC), Liquid chromatography-tandem mass spectroscopy (LC-MS/MS) | Monitor real-time, dynamic changes in bacterial metabolites, providing direct analysis of bacterial metabolic activity. | It is expensive and requires specialized equipment. If LC-MS/MS is used intensive bioinformatics is needed. Requires the sacrifice of chickens for sample collection, maintenance of cold chain after sample collection, and rapid processing of samples. | (Kers et al., 2019b; Niu et al., 2020; Park et al., 2020; Slizewska et al., 2020) |
| Others | |||||
| Fluorescein isothiocyanate–dextran (FITC-d) | Blood | Immunoassay | Measures epithelial permeability | Limited to research settings since the compounds need to be gavaged and monitored in blood as a tool to determine gut barrier failure | (Niewold, 2014; Immerseel, 2019; Barekatain et al., 2020; Vuong et al., 2021) |
Biomarkers Related to the Microbiota Patterns
In poultry, microbial shifts have been mainly studied in NE models (Antonissen et al., 2016). Some of the important microbial signatures of dysbiosis identified in both humans and poultry are described herein.
Depleted level of family Ruminococcacaea and Lachnospiracaea, mainly butyrate-producing Clostridiales, has been reported in stool samples of humans with Crohn's disease (Duvallet et al., 2017). The same family was seen to be depleted in experimental chickens following the feeding of fish meal and Eimeria inoculation (Wu et al., 2014). Most of the bacterial species under the family Ruminococcacaea and Lachnospiracaea are butyrate producers that activate glucagon-like peptide 2 (GLP-2) secretion which maintains intestinal epithelial cells integrity. Butyrate supplementation in feed has also been shown to reduce the incidence and severity of NE (Timbermont et al., 2010; Antonissen et al., 2016). Therefore, the absence of members of these families could be an indication of gut barrier damage. However, excessive butyrate could also lead to poor gut health. A study on rats showed that increasing the butyrate concentration from 5 mM to 100 mM was toxic to colonic goblet cells and decreased colon mucin secretion (Barcelo et al., 2000). In broilers, dietary sodium butyrate supplementation at 2,000 mg/kg decreases feed efficiency (Hu and Guo, 2007). A detailed dose-response study is warranted to know the appropriate dose of butyrate in the diet of chickens.
Roseburia hominis (family Lachnospiracaea), Faecalibacterium prausnitzii (family Ruminococcacaea), and Butyricicoccus pullicaecorum (family Clostridiaceae) were found to be under-represented in stool samples of humans with ulcerative colitis (Machiels et al., 2014; Tilg and Danese, 2014). A decrease in Faecalibacterium in excreta has also been shown in E. tenella infection in poultry (Huang et al., 2018). Roseburia hominis, F. prausnitzii, and B. pullicaecorum are butyrate producers, have anti-inflammatory properties, and regulate innate immunity (Eeckhaut et al., 2016; Lopez-Siles et al., 2017; Patterson et al., 2017). Butyricicoccus pullicaecorum is also commonly used in probiotics and has been shown to increase feed efficiency and aid in the protection against NE in challenged broilers (Eeckhaut et al., 2016; Keerqin et al., 2021).
The abundance of the phylum Proteobacteria has been regarded as a signature of dysbiosis and disease in humans (Shin et al., 2015). Many opportunistic bacteria are under the phylum Proteobacteria such as Escherichia, Salmonella, Campylobacter, and Proteus. Therefore, an increase in the abundance of Proteobacteria could be a useful indicator of dysbiosis. More specifically family Enterobacteriaceae was found to increase in intestinal epithelial dysfunction as a result of noncommunicable disease (cardiac disease, colorectal cancer, metabolic diseases) and diarrhea in humans, and the same family was found to be depleted in the ileal contents of birds with NE induced by supplementation of Salmonella Typhimurium, E. maxima and C. perfringens (Hernandez-Patlan et al., 2019b).
While a clear shift in microbiota has been shown in clinical NE and coccidiosis, it is often difficult to detect changes in subtle gut health conditions (Shin et al., 2015; Duvallet et al., 2017; Ducatelle et al., 2018). In poultry research, several attempts have been made to define the microbiota associated with gut health and productivity but the microbiota consistently associated with high or low-performing flocks has not been identified (see Section 4). Considerable bird to bird and flock to flock variations in microbiota resulted in the lack of reproducibility of bacterial profiles related to production across trials (Stanley et al., 2013b; Stanley et al., 2016). Although a sequence-based approach has been useful to characterize the GIT microbiota, a more comprehensive analysis of microbial composition combined with the microbial and host metabolites (metabolomics) using liquid chromatography and mass spectroscopy would be more insightful on defining host-gut microbiome interactions (Aggrey et al., 2019; Kers et al., 2019b; Cao et al., 2020; Gonzalez-Uarquin et al., 2020; Niu et al., 2020; Park et al., 2020; Slizewska et al., 2020; Wang et al., 2021; Zhang et al., 2021). Although metabolomics offers great promise for biomarker discovery and mechanistic investigation on in-feed additives, they are time-consuming and expensive which limits their application in commercial settings.
The Use of Noninvasive Sampling as a Proxy for Gut Microbiota
In human gut microbiota research, stools are considered a reference sample type despite their limitations compared to the detection of bacteria in the intestinal mucosa (Tang et al., 2020). In poultry, the majority of studies are conducted using cecum microbiota although some of the research has been performed to identify suitable alternatives. The nonterminal sampling method most commonly used is excreta or cloacal swabs (Stanley et al., 2014). However, the microbiota of excreta varies at different time points due to the periodic emptying of different GIT sections (Sekelja et al., 2012). Although excreta has been shown to contain microbial taxa present in the cecum, there are quantitative differences in the microbial composition between sample types (Stanley et al., 2015). Similarly, microbial communities in excreta are more similar to litter compared to cecum or ileum (Johnson et al., 2018). Therefore, using excreta as a proxy of gut contents would likely miss some important core gut microbial taxa (Stanley et al., 2014; Videvall et al., 2018).
Cecal droppings and boot sock samples have been suggested as suitable alternatives to cecal contents for studying longitudinal microbiota composition (Pauwels et al., 2015; Kers et al., 2018). However, the collection of cecal droppings is difficult as chickens produce one cecal dropping after the production of 7-8 excreta droppings (Williams, 1995), and the use of boot sock sampling needs to be further evaluated (Kers et al., 2019a).
A potential noninvasive sample is poultry house dust. Previous research has shown that the poultry dust is composed mostly of dried excreta, and to a lesser extent from feed and feather dander (Ahaduzzaman et al., 2021b), and nucleic acid of viruses, bacteria, and protozoa can be readily detected in the dust of commercial chicken flocks (Walkden-Brown et al., 2013; Ahaduzzaman et al., 2020; Assen et al., 2020; Bindari et al., 2021a). More recently the use of poultry dust has been proposed as an alternative sample type to study microbiota at flock-level but further investigation is needed (Bindari et al., 2021b, c). Combining a sequencing approach with quantitative PCR for absolute measurements of microbial communities would assist in more accurately analyzing changes in microbial taxa and further investigating the microbiota function (Barlow et al., 2020; Jian et al., 2020). This approach may be more suitable for use on routine monitoring of commercial broiler farms.
CONCLUSIONS
Phasing out of in-feed antibiotics in many countries has been associated with the re-emergence of enteric diseases such as coccidiosis, necrotic enteritis, and dysbiosis. The establishment of practical tools to monitor gut health has the potential to provide timely information which would help to identify, implement, and evaluate targeted interventions quickly. Monitoring of the chicken gut microbiota has been the most widely used research tool to infer gut health; however, microbial signatures related to production remain elusive despite great research efforts in this area. Many host and environmental factors have been shown to influence the microbial community structure, including nutritional and husbandry management, chicken breed, and age, in addition to various methodological factors such as sample storage and data analysis. Microbiota studies must provide details on these variables for accurate interpretation of the results. Efforts should be made by the research community to standardize the methodology used in chicken microbiota studies to allow for a direct comparison of data sets among studies, which ideally should include functional analysis of the microbiota.
The use of noninvasive and easy-to-collect population-level samples, such as boot socks and poultry dust, do not require terminal sampling and offer promise to provide flock-level metrics on microbial composition and burden of enteric pathogens using a reduced number of samples. This would therefore reduce costs and potentially facilitate on-farm studies which are crucial to fully assess management interventions including testing of in-feed additives as the outcomes observed in experimental studies under controlled conditions do not always translate to findings in commercial settings. Further research on the usefulness of population-level sampling approaches to monitor microbiota composition and their association with flock production is warranted.
ACKNOWLEDGMENTS
We thank PoultryHub Australia for kindly providing the chicken digestive tract schematic figure used in this review.
DISCLOSURES
The authors declare no conflict of interest.
REFERENCES
- Abundo M.E., Ngunjiri J.M., Taylor K.J., Ji H., Ghorbani A., KC M., Weber B.P., Johnson T.J., Lee C.-W. Assessment of two DNA extraction kits for profiling poultry respiratory microbiota from multiple sample types. PLoS One. 2021;16 doi: 10.1371/journal.pone.0241732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, B. 2019. Investigation of microbiota in health and disease of poultry. University of Arkansas, Fayetteville. Thesis and Dissertations. 3371.
- Adil S., Magray S. Impact and manipulation of gut microflora in poultry: a review. J. Anim. Vet. Adv. 2012;11:873–877. [Google Scholar]
- Aggrey S.E., Milfort M.C., Fuller A.L., Yuan J., Rekaya R. Effect of host genotype and Eimeria acervulina infection on the metabolome of meat-type chickens. PLoS One. 2019;14 doi: 10.1371/journal.pone.0223417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahaduzzaman M., Keerqin C., Kumar A., Musigwa S., Morgan N., Kheravii S.K., Sharpe S., Williamson S., Wu S.-B., Walkden-Brown S.W. Detection and quantification of Clostridium perfringens and Eimeria spp. in poultry dust using real-time PCR under experimental and field conditions. Avian Dis. 2021;65:77–85. doi: 10.1637/aviandiseases-D-20-00084. [DOI] [PubMed] [Google Scholar]
- Ahaduzzaman M., Milan L., Morton C.L., Gerber P.F., Walkden-Brown S.W. Characterization of poultry house dust using chemometrics and scanning electron microscopy imaging. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahaduzzaman M., Walkden-Brown S.W., Gerber P.F., Keerqin C., Kumar A., Musigwa S., Morgan N., Kheravii S.K., Sharpe S., Williamson S. Detection and quantification of Clostridium perfringens and Eimeria spp. in poultry dust using real-time PCR under experimental and field conditions. Avian Dis. 2020;65:77–85. doi: 10.1637/aviandiseases-D-20-00084. [DOI] [PubMed] [Google Scholar]
- Ahn J.-H., Kim B.-Y., Song J., Weon H.-Y. Effects of PCR cycle number and DNA polymerase type on the 16S rRNA gene pyrosequencing analysis of bacterial communities. J. Microbiol. 2012;50:1071–1074. doi: 10.1007/s12275-012-2642-z. [DOI] [PubMed] [Google Scholar]
- Akinyemi F.T., Ding J., Zhou H., Xu K., He C., Han C., Zheng Y., Luo H., Yang K., Gu C., Huang Q., Meng H. Dynamic distribution of gut microbiota during embryonic development in chicken. Poult. Sci. 2020;99:5079–5090. doi: 10.1016/j.psj.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktan S., Sagdic O. Dried rose (Rosa damascena Mill.) dreg: an alternative litter material in broiler production. South Afr. J. Anim. Sci. 2004;34:75–79. [Google Scholar]
- Al-Sheikhly F., Al-Saieg A. Role of coccidia in the occurrence of necrotic enteritis of chickens. Avian Dis. 1980;24:324–333. [PubMed] [Google Scholar]
- Annett C., Viste J., Chirino-Trejo M., Classen H., Middleton D., Simko E. Necrotic enteritis: effect of barley, wheat and corn diets on proliferation of Clostridium perfringens type A. Avian Pathol. 2002;31:598–601. doi: 10.1080/0307945021000024544. [DOI] [PubMed] [Google Scholar]
- Antonissen G., Eeckhaut V., Van Driessche K., Onrust L., Haesebrouck F., Ducatelle R., Moore R.J., Van Immerseel F. Microbial shifts associated with necrotic enteritis. Avian Pathol. 2016;45:308–312. doi: 10.1080/03079457.2016.1152625. [DOI] [PubMed] [Google Scholar]
- Apajalahti J., Kettunen A., Graham H. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World's Poult. Sci. J. 2004;60:223–232. [Google Scholar]
- Apajalahti J.H., Kettunen A., Bedford M.R., Holben W.E. Percent G+ C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 2001;67:5656–5667. doi: 10.1128/AEM.67.12.5656-5667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assen A.M., Stillman M., Alfirevich S., Gerber P.F., Groves P.J., Walkden-Brown S.W. Assessment of A20 infectious laryngotracheitis vaccine take in meat chickens using swab and dust samples following mass vaccination in drinking water. Vet. Microbiol. 2020;251 doi: 10.1016/j.vetmic.2020.108903. [DOI] [PubMed] [Google Scholar]
- Barb J.J., Oler A.J., Kim H.-S., Chalmers N., Wallen G.R., Cashion A., Munson P.J., Ames N.J. Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA using mock samples. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo A., Claustre J., Moro F., Chayvialle J., Cuber J., Plaisancié P. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut. 2000;46:218–224. doi: 10.1136/gut.46.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barekatain R., Howarth G.S., Willson N.-L., Cadogan D., Wilkinson S. Excreta biomarkers in response to different gut barrier dysfunction models and probiotic supplementation in broiler chickens. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.T., Bogatyrev S.R., Ismagilov R.F. A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat. Commun. 2020;11:2590. doi: 10.1038/s41467-020-16224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter M.F.A., Latorre J.D., Dridi S., Merino-Guzman R., Hernandez-Velasco X., Hargis B.M., Tellez-Isaias G. Identification of serum biomarkers for intestinal integrity in a broiler chicken malabsorption model. Front. Vet. Sci. 2019;6:144. doi: 10.3389/fvets.2019.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.S., Beltran R., Higgins S.E., Hargis B.M., Moore R., Byrd J.A., II, Corsiglia C., Caldwell D. Effect of addition of hydrated lime to litter on recovery of selected bacteria and poult performance. J. Appl. Poult. Res. 2005;14:721–727. [Google Scholar]
- Bergmann G.T., Bates S.T., Eilers K.G., Lauber C.L., Caporaso J.G., Walters W.A., Knight R., Fierer N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrang M.E., Frank J.F., Buhr R.J., Bailey J.S., Cox N.A. Eggshell membrane structure and penetration by Salmonella Typhimurium. J. Food Prot. 1999;62:73–76. doi: 10.4315/0362-028x-62.1.73. [DOI] [PubMed] [Google Scholar]
- Bharti R., Grimm D.G. Current challenges and best-practice protocols for microbiome analysis. Briefings Bioinf. 2021;22:178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindari Y.R., Kheravii S.K., Morton C.L., Wu S.-B., Walkden-Brown S.W., Gerber P.F. Molecular detection of Eimeria species and Clostridium perfringens in poultry dust and pooled excreta of commercial broiler chicken flocks differing in productive performance. Vet. Parasitol. 2021;291 doi: 10.1016/j.vetpar.2021.109361. [DOI] [PubMed] [Google Scholar]
- Bindari Y.R., Moore R.J., Van T.T.H., Hilliar M., Wu S.-B., Walkden-Brown S.W., Gerber P.F. Microbial communities of poultry house dust, excreta and litter are partially representative of microbiota of chicken caecum and ileum. PLoS One. 2021;16 doi: 10.1371/journal.pone.0255633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindari Y.R., Moore R.J., Van T.T.H., Walkden‑Brown S., Gerber P.F. Microbial taxa in dust and excreta associated with the productive performance of commercial meat chicken flocks. Anim. Microbiome. 2021;3:66. doi: 10.1186/s42523-021-00127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boers S.A., Jansen R., Hays J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:1059–1070. doi: 10.1007/s10096-019-03520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder K., Thompson K., Einstein M., Applegate T., Patterson J. Influence of stressors on normal intestinal microbiota, intestinal morphology, and susceptibility to Salmonella enteritidis colonization in broilers. Poult. Sci. 2008;87:1734–1741. doi: 10.3382/ps.2008-00107. [DOI] [PubMed] [Google Scholar]
- Caly D.L., D'Inca R., Auclair E., Drider D. Alternatives to antibiotics to prevent necrotic enteritis in broiler chickens: a microbiologist's perspective. Front. Microbiol. 2015;6:1336. doi: 10.3389/fmicb.2015.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Zeng X., Liu J., Yan F., Xiang Z., Wang Y., Tao F., Yang C. Change of serum metabolome and cecal microflora in broiler chickens supplemented with grape seed extracts. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.610934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals-Pascual C., González A., Vázquez-Baeza Y., Song S.J., Jiang L., Knight R. Microbial diversity in clinical microbiome studies: sample size and statistical power considerations. Gastroenterology. 2020;158:1524–1528. doi: 10.1053/j.gastro.2019.11.305. [DOI] [PubMed] [Google Scholar]
- Celi P., Cowieson A., Fru-Nji F., Steinert R., Kluenter A.-M., Verlhac V. Gastrointestinal functionality in animal nutrition and health: new opportunities for sustainable animal production. Anim. Feed Sci. Technol. 2017;234:88–100. [Google Scholar]
- Celi P., Verlhac V., Calvo E.P., Schmeisser J., Kluenter A.-M. Biomarkers of gastrointestinal functionality in animal nutrition and health. Anim. Feed Sci. Technol. 2019;250:9–31. [Google Scholar]
- Chen J., Tellez G., Richards J.D., Escobar J. Identification of potential biomarkers for gut barrier failure in broiler chickens. Front. Vet. Sci. 2015;2:14. doi: 10.3389/fvets.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., W Saatkamp H., Cortenbach J., Jin W. Comparison of Chinese broiler production systems in economic performance and animal welfare. Animals. 2020;10:491. doi: 10.3390/ani10030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hui P.C., Hui M., Yeoh Y.K., Wong P.Y., Chan M.C., Wong M.C., Ng S.C., Chan F.K., Chan P.K. Impact of preservation method and 16S rRNA hypervariable region on gut microbiota profiling. Msystems. 2019;4:1. doi: 10.1128/mSystems.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintoan-Uta C., Wisedchanwet T., Glendinning L., Bremner A., Psifidi A., Vervelde L., Watson K., Watson M., Stevens M.P., Dozois C.M. Role of cecal microbiota in the differential resistance of inbred chicken lines to colonization by Campylobacter jejuni. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02607-19. e02607-02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choct M., Annison G. Anti-nutritive effect of wheat pentosans in broiler chickens: roles of viscosity and gut microflora. Br. Poult. Sci. 1992;33:821–834. doi: 10.1080/00071669208417524. [DOI] [PubMed] [Google Scholar]
- Choo J.M., Leong L.E., Rogers G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clench M.H., Mathias J.R. The avian cecum: a review. Wilson Bull. 1995;107:93–121. [Google Scholar]
- Cockerill S.A., Gerber P., Walkden-Brown S., Dunlop M.W. Suitability of litter amendments for the Australian chicken meat industry. Anim. Prod. Sci. 2020;60:1469–1481. [Google Scholar]
- Collett S.R. Nutrition and wet litter problems in poultry. Anim. Feed Sci. Technol. 2012;173:65–75. [Google Scholar]
- Cortés C.R., Isaías G.T., Cuello C.L., Flores J.M.V., Anderson R.C., Campos C.E. Bacterial isolation rate from fertile eggs, hatching eggs, and neonatal broilers with yolk sac infection. Revista latinoamericana de Microbiologia. 2004;46:12–16. [PubMed] [Google Scholar]
- Costea P.I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., Tramontano M., Driessen M., Hercog R., Jung F.-E. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- Coufal C., Chavez C., Niemeyer P., Carey J. Measurement of broiler litter production rates and nutrient content using recycled litter. Poult. Sci. 2006;85:398–403. doi: 10.1093/ps/85.3.398. [DOI] [PubMed] [Google Scholar]
- Cressman M.D., Yu Z., Nelson M.C., Moeller S.J., Lilburn M.S., Zerby H.N. Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl. Environ. Microbiol. 2010;76:6572–6582. doi: 10.1128/AEM.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Zhang X., Cheng R., Ansari A.R., Elokil A.A., Hu Y., Chen Y., Nafady A.A., Liu H. Sex differences in growth performance are related to cecal microbiota in chicken. Microb. Pathog. 2021;150 doi: 10.1016/j.micpath.2020.104710. [DOI] [PubMed] [Google Scholar]
- Dahiya J., Hoehler D., Van Kessel A.G., Drew M.D. Dietary encapsulated glycine influences Clostridium perfringens and Lactobacilli growth in the gastrointestinal tract of broiler chickens. J. Nutr. 2007;137:1408–1414. doi: 10.1093/jn/137.6.1408. [DOI] [PubMed] [Google Scholar]
- Dahiya J., Wilkie D., Van Kessel A., Drew M. Potential strategies for controlling necrotic enteritis in broiler chickens in post-antibiotic era. Anim. Feed Sci. Technol. 2006;129:60–88. [Google Scholar]
- Danzeisen J.L., Kim H.B., Isaacson R.E., Tu Z.J., Johnson T.J. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS One. 2011;6:e27949. doi: 10.1371/journal.pone.0027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gussem M. Proceedings of the 16th European Symposium on Poultry Nutrition. 2007. Coccidiosis in poultry: review on diagnosis, control, prevention and interaction with overall gut health; pp. 253–261. [Google Scholar]
- De Gussem M. Macroscopic scoring system for bacterialenteritis in broiler chickens and turkeys. World Veterinary Poultry Association Meeting; Merelbeke, Belgium; 2010. [Google Scholar]
- De Maesschalck C., Eeckhaut V., Maertens L., De Lange L., Marchal L., Nezer C., De Baere S., Croubels S., Daube G., Dewulf J. Effects of xylo-oligosaccharides on broiler chicken performance and microbiota. Appl. Environ. Microbiol. 2015;81:5880–5888. doi: 10.1128/AEM.01616-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer F., Eeckhaut V., Ducatelle R., Dhaenens M., Daled S., Dedeurwaerder A., De Gussem M., Haesebrouck F., Deforce D., Van Immerseel F. Host intestinal biomarker identification in a gut leakage model in broilers. Vet. Res. 2019;50:1–14. doi: 10.1186/s13567-019-0663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Verdal H., Mignon-Grasteau S., Jeulin C., Le Bihan-Duval E., Leconte M., Mallet S., Martin C., Narcy A. Digestive tract measurements and histological adaptation in broiler lines divergently selected for digestive efficiency. Poult. Sci. 2010;89:1955–1961. doi: 10.3382/ps.2010-813. [DOI] [PubMed] [Google Scholar]
- Denbow D.M. Sturkie’s Avian Physiology. Elsevier; New York, NY: 2015. Gastrointestinal anatomy and physiology; pp. 337–366. [Google Scholar]
- Deusch S., Tilocca B., Camarinha-Silva A., Seifert J. News in livestock research—use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Comput. Struct. Biotechnol. J. 2015;13:55–63. doi: 10.1016/j.csbj.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Carrasco J.M., Casanova N.A., Fernández Miyakawa M.E. Microbiota, gut health and chicken productivity: what is the connection? Microorganisms. 2019;7:374. doi: 10.3390/microorganisms7100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner J., Richards J. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 2005;84:634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- Dierick E., Ducatelle R., Van Immerseel F., Goossens E. Research Note: The administration schedule of coccidia is a major determinant in broiler necrotic enteritis models. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2020.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilhari A., Sampath A., Gunasekara C., Fernando N., Weerasekara D., Sissons C., McBain A., Weerasekera M. Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. Amb Express. 2017;7:1–11. doi: 10.1186/s13568-017-0477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominianni C., Wu J., Hayes R.B., Ahn J. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014;14:1–6. doi: 10.1186/1471-2180-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Goossens E., De Meyer F., Eeckhaut V., Antonissen G., Haesebrouck F., Van Immerseel F. Biomarkers for monitoring intestinal health in poultry: present status and future perspectives. Vet. Res. 2018;49:1–9. doi: 10.1186/s13567-018-0538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke G.E. Relationship of cecal and colonic motility to diet, habitat, and cecal anatomy in several avian species. J. Exp. Zool. 1989;252:38–47. doi: 10.1002/jez.1402520507. [DOI] [PubMed] [Google Scholar]
- Dumas M.D., Polson S.W., Ritter D., Ravel J., Gelb J., Jr, Morgan R., Wommack K.E. Impacts of poultry house environment on poultry litter bacterial community composition. PLoS One. 2011;6:e24785. doi: 10.1371/journal.pone.0024785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8:1–10. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhaut V., Wang J., Van Parys A., Haesebrouck F., Joossens M., Falony G., Raes J., Ducatelle R., Van Immerseel F. The probiotic Butyricicoccus pullicaecorum reduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front. Microbiol. 2016;7:1416. doi: 10.3389/fmicb.2016.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg R., Hedemann M., Jensen B. The influence of grinding and pelleting of feed on the microbial composition and activity in the digestive tract of broiler chickens. Br. Poult. Sci. 2002;43:569–579. doi: 10.1080/0007166022000004480. [DOI] [PubMed] [Google Scholar]
- Engberg R., Hedemann M., Leser T., Jensen B. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 2000;79:1311–1319. doi: 10.1093/ps/79.9.1311. [DOI] [PubMed] [Google Scholar]
- Engberg R., Hedemann M., Steenfeldt S., Jensen B. Influence of whole wheat and xylanase on broiler performance and microbial composition and activity in the digestive tract. Poult. Sci. 2004;83:925–938. doi: 10.1093/ps/83.6.925. [DOI] [PubMed] [Google Scholar]
- Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., Kurilshikov A., Bonder M.J., Valles-Colomer M., Vandeputte D. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- Feng Y., Gong J., Yu H., Jin Y., Zhu J., Han Y. Identification of changes in the composition of ileal bacterial microbiota of broiler chickens infected with Clostridium perfringens. Vet. Microbiol. 2010;140:116–121. doi: 10.1016/j.vetmic.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Feye K., Baxter M., Tellez-Isaias G., Kogut M., Ricke S. Influential factors on the composition of the conventionally raised broiler gastrointestinal microbiomes. Poult. Sci. 2020;99:653–659. doi: 10.1016/j.psj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler G., Tolnai E., Stagel A., Remenyik J., Stundl L., Gal F., Biro S., Paholcsek M. Tendentious effects of automated and manual metagenomic DNA purification protocols on broiler gut microbiome taxonomic profiling. Sci. Rep. 2020;10:1–16. doi: 10.1038/s41598-020-60304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy F., Clooney A.G., Stanton C., Claesson M.J., Cotter P.D. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:1–13. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks G., Elgart M., Amir A., Zeisel A., Turnbaugh P.J., Soen Y., Shental N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome. 2018;6:1–13. doi: 10.1186/s40168-017-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R., Brooker B. Lactobacilli which attach to the crop epithelium of the fowl. Am. J. Clin. Nutr. 1974;27:1305–1312. doi: 10.1093/ajcn/27.11.1305. [DOI] [PubMed] [Google Scholar]
- Furuse M., Yokota H. Effect of the gut microflora on chick growth and utilisation of protein and energy at different concentrations of dietary protein. Br. Poult. Sci. 1985;26:97–104. doi: 10.1080/00071668508416791. [DOI] [PubMed] [Google Scholar]
- Gaskins H., Collier C., Anderson D. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 2002;13:29–42. doi: 10.1081/ABIO-120005768. [DOI] [PubMed] [Google Scholar]
- Gilani S., Howarth G., Kitessa S., Tran C., Forder R., Hughes R. New biomarkers for increased intestinal permeability induced by dextran sodium sulphate and fasting in chickens. J. Anim. Physiol. Anim. Nutr. 2017;101:e237–e245. doi: 10.1111/jpn.12596. [DOI] [PubMed] [Google Scholar]
- Gilani S., Howarth G.S., Kitessa S.M., Forder R.E., Tran C.D., Hughes R.J. New biomarkers for intestinal permeability induced by lipopolysaccharide in chickens. Anim. Prod. Sci. 2016;56:1984–1997. [Google Scholar]
- Godwin R.M., Morgan J.A. A molecular survey of Eimeria in chickens across Australia. Vet. Parasitol. 2015;214:16–21. doi: 10.1016/j.vetpar.2015.09.030. [DOI] [PubMed] [Google Scholar]
- Gong J., Forster R.J., Yu H., Chambers J.R., Sabour P.M., Wheatcroft R., Chen S. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS MicroBiol. Lett. 2002;208:1–7. doi: 10.1111/j.1574-6968.2002.tb11051.x. [DOI] [PubMed] [Google Scholar]
- Gong J., Si W., Forster R.J., Huang R., Yu H., Yin Y., Yang C., Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 2007;59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Gong J., Yu H., Liu T., Gill J., Chambers J., Wheatcroft R., Sabour P. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 2008;104:1372–1382. doi: 10.1111/j.1365-2672.2007.03699.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Uarquin F., Kenez A., Rodehutscord M., Huber K. Dietary phytase and myo-inositol supplementation are associated with distinct plasma metabolome profile in broiler chickens. Animal. 2020;14:549–559. doi: 10.1017/S1751731119002337. [DOI] [PubMed] [Google Scholar]
- Goossens E., Debyser G., Callens C., De Gussem M., Dedeurwaerder A., Devreese B., Haesebrouck F., Flügel M., Pelzer S., Thiemann F. Elevated faecal ovotransferrin concentrations are indicative for intestinal barrier failure in broiler chickens. Vet. Res. 2018;49:1–8. doi: 10.1186/s13567-018-0548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzelak M.A., Gill S.K., Tasnim N., Ahmadi-Vand Z., Jay M., Gibson D.L. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug A., Gjevre A.-G., Skjerve E., Kaldhusdal M. A survey of the economic impact of subclinical Eimeria infections in broiler chickens in Norway. Avian Pathol. 2008;37:333–341. doi: 10.1080/03079450802050705. [DOI] [PubMed] [Google Scholar]
- Helmboldt C., Bryant E. The pathology of necrotic enteritis in domestic fowl. Avian Dis. 1971;15:775–780. [PubMed] [Google Scholar]
- Hernandez-Patlan D., Solis-Cruz B., Pontin K.P., Hernandez-Velasco X., Merino-Guzman R., Adhikari B., López-Arellano R., Kwon Y.M., Hargis B.M., Arreguin-Nava M.A. Impact of a Bacillus direct-fed microbial on growth performance, intestinal barrier integrity, necrotic enteritis lesions, and ileal microbiota in broiler chickens using a laboratory challenge model. Front. Vet. Sci. 2019;6:108. doi: 10.3389/fvets.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Patlan D., Solis-Cruz B., Pontin K.P., Hernandez-Velasco X., Merino-Guzman R., Adhikari B., López-Arellano R., Kwon Y.M., Hargis B.M., Arreguin-Nava M.A., Tellez-Isaias G., Latorre J.D. Impact of a bacillus direct-fed microbial on growth performance, intestinal barrier integrity, necrotic enteritis lesions, and ileal microbiota in broiler chickens using a laboratory challenge model. Front. Vet. Sci. 2019;6:108. doi: 10.3389/fvets.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetland H., Svihus B., Olaisen V. Effect of feeding whole cereals on performance, starch digestibility and duodenal particle size distribution in broiler chickens. British Poult. Sci. 2002;43:416–423. doi: 10.1080/00071660120103693. [DOI] [PubMed] [Google Scholar]
- Hongoh Y., Yuzawa H., Ohkuma M., Kudo T. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS MicroBiol. Lett. 2003;221:299–304. doi: 10.1016/S0378-1097(03)00218-0. [DOI] [PubMed] [Google Scholar]
- Hu Z., Guo Y. Effects of dietary sodium butyrate supplementation on the intestinal morphological structure, absorptive function and gut flora in chickens. Anim. Feed Sci. Technol. 2007;132:240–249. [Google Scholar]
- Huang G., Tang X., Bi F., Hao Z., Han Z., Suo J., Zhang S., Wang S., Duan C., Yu Z. Eimeria tenella infection perturbs the chicken gut microbiota from the onset of oocyst shedding. Vet. Parasitol. 2018;258:30–37. doi: 10.1016/j.vetpar.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Hughes R. Relationship between digesta transit time and apparent metabolisable energy value of wheat in chickens. Br. Poult. Sci. 2008;49:716–720. doi: 10.1080/00071660802449145. [DOI] [PubMed] [Google Scholar]
- Hume M., Clemente-Hernández S., Oviedo-Rondón E. Effects of feed additives and mixed Eimeria species infection on intestinal microbial ecology of broilers. Poult. Sci. 2006;85:2106–2111. doi: 10.1093/ps/85.12.2106. [DOI] [PubMed] [Google Scholar]
- Iizuka M., Konno S. Wound healing of intestinal epithelial cells. World J. Gastroenterol. 2011;17:2161. doi: 10.3748/wjg.v17.i17.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immerseel F.V. Proceedings of Australian Poultry Symposium. 2019. Host and microbial biomarkers for intestinal health and disease in broilers; pp. 93–98. [Google Scholar]
- Ishii K., Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Pescatore T., Cantor A. University of Kentucky; Lexington, KY: 2011. Avian Digestive System. [Google Scholar]
- Jha R., Berrocoso J. Dietary fiber utilization and its effects on physiological functions and gut health of swine. Animal. 2015;9:1441–1452. doi: 10.1017/S1751731115000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Xu Y., Luo C., He Y., Xu X., Yan X., Li Y., Shu D., Qu H. Effects of the DMRT1 genotype on the body weight and gut microbiota in the broiler chicken. Poult. Sci. 2020;99:4044–4051. doi: 10.1016/j.psj.2020.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian C., Luukkonen P., Yki-Jarvinen H., Salonen A., Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Ho Y., Abdullah N., Kudo H., Jalaludin S. Studies on the intestinal microflora of chicken under tropical condition. Asian-Austral. J. Anim. Sci. 1997;10:495–504. [Google Scholar]
- Johnson J., Reid W.M. Anticoccidial drugs: lesion scoring techniques in battery and floor-pen experiments with chickens. Exp. Parasitol. 1970;28:30–36. doi: 10.1016/0014-4894(70)90063-9. [DOI] [PubMed] [Google Scholar]
- Johnson T.J., Youmans B.P., Noll S., Cardona C., Evans N.P., Karnezos T.P., Ngunjiri J.M., Abundo M.C., Lee C.-W. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.00362-18. e00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadooka Y., Sato M., Imaizumi K., Ogawa A., Ikuyama K., Akai Y., Okano M., Kagoshima M., Tsuchida T. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur. J. Clin. Nutr. 2010;64:636–643. doi: 10.1038/ejcn.2010.19. [DOI] [PubMed] [Google Scholar]
- Kassem I.I., Sanad Y., Gangaiah D., Lilburn M., Lejeune J., Rajashekara G. Use of bioluminescence imaging to monitor Campylobacter survival in chicken litter. J. Appl. Microbiol. 2010;109:1988–1997. doi: 10.1111/j.1365-2672.2010.04828.x. [DOI] [PubMed] [Google Scholar]
- Keerqin C., Rhayat L., Zhang Z.-H., Gharib-Naseri K., Kheravii S., Devillard E., Crowley T., Wu S.-B. Probiotic Bacillus subtilis 29,784 improved weight gain and enhanced gut health status of broilers under necrotic enteritis condition. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., de Oliveira J.E., Fischer E.A., Tersteeg-Zijderveld M.H., Konstanti P., Stegeman J.A., Smidt H., Velkers F.C. Associations between phenotypic characteristics and clinical parameters of broilers and intestinal microbial development throughout a production cycle: a field study. Microbiol. Open. 2020;9:e1114. doi: 10.1002/mbo3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., Fischer E.A., Stegeman J.A., Smidt H., Velkers F.C. Comparison of different invasive and non-invasive methods to characterize intestinal microbiota throughout a production cycle of broiler chickens. Microorganisms. 2019;7:431. doi: 10.3390/microorganisms7100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., Velkers F.C., Fischer E.A., Hermes G.D., Stegeman J.A., Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., Velkers F.C., Fischer E.A.J., Hermes G.D.A., Lamot D.M., Stegeman J.A., Smidt H. Take care of the environment: housing conditions affect the interplay of nutritional interventions and intestinal microbiota in broiler chickens. Anim. Microbiome. 2019;1:10. doi: 10.1186/s42523-019-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyburn A.L., Sheedy S.A., Ford M.E., Williamson M.M., Awad M.M., Rood J.I., Moore R.J. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 2006;74:6496–6500. doi: 10.1128/IAI.00806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipper M., Andretta I., Lehnen C.R., Lovatto P.A., Monteiro S.G. Meta-analysis of the performance variation in broilers experimentally challenged by Eimeria spp. Vet. Parasitol. 2013;196:77–84. doi: 10.1016/j.vetpar.2013.01.013. [DOI] [PubMed] [Google Scholar]
- Kizerwetter-Świda M., Binek M. Bacterial microflora of the chicken embryos and newly hatched chicken. J. Anim. Feed Sci. 2008;17:224–232. [Google Scholar]
- Kley M.-V., Oviedo-Rondón E., Dowd S., Hume M., Nalian A. Effect of Eimeria infection on cecal microbiome of broilers fed essential oils. Int. J. Poult. Sci. 2012;11:747–755. [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knarreborg A., Engberg R.M., Jensen S.K., Jensen B.B. Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl. Environ. Microbiol. 2002;68:6425–6428. doi: 10.1128/AEM.68.12.6425-6428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut M.H., Lee A., Santin E. Microbiome and pathogen interaction with the immune system. Poult. Sci. 2020;99:1906–1913. doi: 10.1016/j.psj.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo F. In vitro lecithinase activity and sensitivity to 22 antimicrobial agents of Clostridium perfringens isolated from necrotic enteritis of broiler chickens. Res. Vet. Sci. 1988;45:337–340. [PubMed] [Google Scholar]
- Kubasova T., Kollarcikova M., Crhanova M., Karasova D., Cejkova D., Sebkova A., Matiasovicova J., Faldynova M., Pokorna A., Cizek A., Rychlik I. Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamot D., Van De Linde I., Molenaar R., Van Der Pol C., Wijtten P., Kemp B., Van Den Brand H. Effects of moment of hatch and feed access on chicken development. Poult. Sci. 2014;93:2604–2614. doi: 10.3382/ps.2014-04123. [DOI] [PubMed] [Google Scholar]
- Lee K.-C., Kil D.Y., Sul W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017;55:939–945. doi: 10.1007/s12275-017-7202-0. [DOI] [PubMed] [Google Scholar]
- Lei K., Li Y., Yu D., Rajput I., Li W. Influence of dietary inclusion of Bacillus licheniformis on laying performance, egg quality, antioxidant enzyme activities, and intestinal barrier function of laying hens. Poult. Sci. 2013;92:2389–2395. doi: 10.3382/ps.2012-02686. [DOI] [PubMed] [Google Scholar]
- Lim S., Cho S., Caetano-Anolles K., Jeong S.G., Oh M.H., Park B.Y., Kim H.J., Cho S., Choi S.H., Ryu S. Developmental dynamic analysis of the excreted microbiome of chickens using next-generation sequencing. J. Mol. Microbiol. Biotechnol. 2015;25:262–268. doi: 10.1159/000430865. [DOI] [PubMed] [Google Scholar]
- Loh G., Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes. 2012;3:544–555. doi: 10.4161/gmic.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M., Duncan S.H., Garcia-Gil L.J., Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovland A., Kaldhusdal M. Severely impaired production performance in broiler flocks with high incidence of Clostridium perfringens-associated hepatitis. Avian Pathol. 2001;30:73–81. doi: 10.1080/03079450020023230. [DOI] [PubMed] [Google Scholar]
- Lu J., Idris U., Harmon B., Hofacre C., Maurer J.J., Lee M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003;69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hugenholtz P., Batstone D.J. Evaluating DNA extraction methods for community profiling of pig hindgut microbial community. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkins B., Batal A., Lee M. The effect of gender on the bacterial community in the gastrointestinal tract of broilers. Poult. Sci. 2008;87:964–967. doi: 10.3382/ps.2007-00287. [DOI] [PubMed] [Google Scholar]
- M'Sadeq S.A., Wu S., Swick R.A., Choct M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim. Nutr. 2015;1:1–11. doi: 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald S.E., Nolan M.J., Harman K., Boulton K., Hume D.A., Tomley F.M., Stabler R.A., Blake D.P. Effects of Eimeria tenella infection on chicken caecal microbiome diversity, exploring variation associated with severity of pathology. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., Ballet V., Claes K., Van Immerseel F., Verbeke K. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- Malone G., Chaloupka G., Saylor W. Influence of litter type and size on broiler performance: 1. Factors affecting litter consumption. Poult. Sci. 1983;62:1741–1746. [Google Scholar]
- McDevitt R., Brooker J., Acamovic T., Sparks N. Necrotic enteritis; a continuing challenge for the poultry industry. World's Poult. Sci. J. 2006;62:221–247. [Google Scholar]
- Mead G. Microbes of the avian cecum: types present and substrates utilized. J. Exp. Zool. 1989;252:48–54. doi: 10.1002/jez.1402520508. [DOI] [PubMed] [Google Scholar]
- Morgan J., Morris G., Wlodek B., Byrnes R., Jenner M., Constantinoiu C., Anderson G., Lew-Tabor A., Molloy J., Gasser R. Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Mol. Cell. Probes. 2009;23:83–89. doi: 10.1016/j.mcp.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Morris G., Gasser R. Biotechnological advances in the diagnosis of avian coccidiosis and the analysis of genetic variation in Eimeria. Biotechnol. Adv. 2006;24:590–603. doi: 10.1016/j.biotechadv.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Woods W.G., Richards D.G., Gasser R.B. The application of a polymerase chain reaction (PCR)-based capillary electrophoretic technique provides detailed insights into Eimeria populations in intensive poultry establishments. Mol. Cell. Probes. 2007;21:288–294. doi: 10.1016/j.mcp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Munyaka P., Nandha N., Kiarie E., Nyachoti C., Khafipour E. Impact of combined β-glucanase and xylanase enzymes on growth performance, nutrients utilization and gut microbiota in broiler chickens fed corn or wheat-based diets. Poult. Sci. 2016;95:528–540. doi: 10.3382/ps/pev333. [DOI] [PubMed] [Google Scholar]
- Niewold T. Wageningen Academic Publishers; Wageningen, The Netherlands: 2014. Intestinal Health: Key to Maximise Growth Performance in Livestock. [Google Scholar]
- Niu J., Zhang J., Wei L., Ma X., Zhang W., Nie C. Cottonseed meal fermented by Candida tropical reduces the fat deposition in white-feather broilers through cecum bacteria-host metabolic cross-talk. Appl. Microbiol. Biotechnol. 2020;104:4345–4357. doi: 10.1007/s00253-020-10538-7. [DOI] [PubMed] [Google Scholar]
- Oakley B.B., Lillehoj H.S., Kogut M.H., Kim W.K., Maurer J.J., Pedroso A., Lee M.D., Collett S.R., Johnson T.J., Cox N.A. The chicken gastrointestinal microbiome. FEMS MicroBiol. Lett. 2014;360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- Oakley B.B., Morales C.A., Line J., Berrang M.E., Meinersmann R.J., Tillman G.E., Wise M.G., Siragusa G.R., Hiett K.L., Seal B.S. The poultry-associated microbiome: network analysis and farm-to-fork characterizations. PLoS One. 2013;8:e57190. doi: 10.1371/journal.pone.0057190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh S.-J. Meta analysis to draw the appropriate regimen of enzyme and probiotic supplementation to pigs and chicken diets. Asian-Austral. J. Anim. Sci. 2011;24:573–586. [Google Scholar]
- Owen H.C., Roberts S.J., Ahmed S.F., Farquharson C. Dexamethasone-induced expression of the glucocorticoid response gene lipocalin 2 in chondrocytes. Am. J. Physiol. Endocrinol. Metab. 2008;294:E1023–E1034. doi: 10.1152/ajpendo.00586.2007. [DOI] [PubMed] [Google Scholar]
- Paiva D., McElroy A. Necrotic enteritis: applications for the poultry industry. J. Appl. Poult. Res. 2014;23:557–566. [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit R.J., Hinsu A.T., Patel N.V., Koringa P.G., Jakhesara S.J., Thakkar J.R., Shah T.M., Limon G., Psifidi A., Guitian J., Hume D.A., Tomley F.M., Rank D.N., Raman M., Tirumurugaan K.G., Blake D.P., Joshi C.G. Microbial diversity and community composition of caecal microbiota in commercial and indigenous Indian chickens determined using 16s rDNA amplicon sequencing. Microbiome. 2018;6:115. doi: 10.1186/s40168-018-0501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I., Zimmerman N.P., Smith A.H., Rehberger T.G., Lillehoj E.P., Lillehoj H.S. Dietary supplementation with Bacillus subtilis direct-fed microbials alters chicken intestinal metabolite levels. Front. Vet. Sci. 2020;7:123. doi: 10.3389/fvets.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson A.M., Mulder I.E., Travis A.J., Lan A., Cerf-Bensussan N., Gaboriau-Routhiau V., Garden K., Logan E., Delday M.I., Coutts A.G. Human gut symbiont Roseburia hominis promotes and regulates innate immunity. Front. Immunol. 2017;8:1166. doi: 10.3389/fimmu.2017.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels J., Taminiau B., Janssens G., De Beenhouwer M., Delhalle L., Daube G., Coopman F. Cecal drop reflects the chickens' cecal microbiome, fecal drop does not. J. Microbiol. Methods. 2015;117:164–170. doi: 10.1016/j.mimet.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Pollock J., Glendinning L., Wisedchanwet T., Watson M. The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02627-17. e02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.-B., Chen T., Xu Y.-P., Chen L., Sun F.-X., Lu M.-P., Liu Y.-X. A guide to human microbiome research: study design, sample collection, and bioinformatics analysis. Chin. Med. J. 2020;133:1844. doi: 10.1097/CM9.0000000000000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu A., Brulc J.M., Wilson M.K., Law B.F., Theoret J.R., Joens L.A., Konkel M.E., Angly F., Dinsdale E.A., Edwards R.A. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One. 2008;3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar S., Lawley B., Tannock G., Engberg R.M. Bacterial succession in the broiler gastrointestinal tract. Appl. Environ. Microbiol. 2016;82:2399–2410. doi: 10.1128/AEM.02549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath N., Anthony N., Kannan L., Huff W., Huff G., Chapman H., Erf G., Wakenell P. Serum ovotransferrin as a biomarker of inflammatory diseases in chickens. Poult. Sci. 2009;88:2069–2074. doi: 10.3382/ps.2009-00076. [DOI] [PubMed] [Google Scholar]
- Ravindran V. Feed enzymes: The science, practice, and metabolic realities. J. Appl. Poult. Res. 2013;22:628–636. [Google Scholar]
- Rehman H.U., Vahjen W., Awad W.A., Zentek J. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch. Anim. Nutr. 2007;61:319–335. doi: 10.1080/17450390701556817. [DOI] [PubMed] [Google Scholar]
- Richards P., Fothergill J., Bernardeau M., Wigley P. Development of the caecal microbiota in three broiler breeds. Front. Vet. Sci. 2019;6:201. doi: 10.3389/fvets.2019.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues I., Choct M. The foregut and its manipulation via feeding practices in the chicken. Poult. Sci. 2018;97:3188–3206. doi: 10.3382/ps/pey191. [DOI] [PubMed] [Google Scholar]
- Roesch L.F., Casella G., Simell O., Krischer J., Wasserfall C.H., Schatz D., Atkinson M.A., Neu J., Triplett E.W. Influence of fecal sample storage on bacterial community diversity. Open Microbiol. J. 2009;3:40. doi: 10.2174/1874285800903010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll V., Dai Prá M., Roll A. Research on Salmonella in broiler litter reused for up to 14 consecutive flocks. Poult. Sci. 2011;90:2257–2262. doi: 10.3382/ps.2011-01583. [DOI] [PubMed] [Google Scholar]
- Rood J.I., Keyburn A.L., Moore R.J. NetB and necrotic enteritis: the hole movable story. Avian Pathol. 2016;45:295–301. doi: 10.1080/03079457.2016.1158781. [DOI] [PubMed] [Google Scholar]
- Saengkerdsub S., Anderson R.C., Wilkinson H.H., Kim W.-K., Nisbet D.J., Ricke S.C. Identification and quantification of methanogenic archaea in adult chicken ceca. Appl. Environ. Microbiol. 2007;73:353–356. doi: 10.1128/AEM.01931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengkerdsub S., Herrera P., Woodward C., Anderson R., Nisbet D., Ricke S. Detection of methane and quantification of methanogenic archaea in faeces from young broiler chickens using real-time PCR. Lett. Appl. Microbiol. 2007;45:629–634. doi: 10.1111/j.1472-765X.2007.02243.x. [DOI] [PubMed] [Google Scholar]
- Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F., Turner P., Parkhill J., Loman N.J., Walker A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:1–12. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Saxena V., Tomar S., Sapcota D., Gonmei G. Characterisation of caecum and crop microbiota of Indian indigenous chicken targeting multiple hypervariable regions within 16S rRNA gene. Br. Poult. Sci. 2016;57:381–389. doi: 10.1080/00071668.2016.1161728. [DOI] [PubMed] [Google Scholar]
- Scanes C.G., Pierzchala-Koziec K. Biology of the gastrointestinal tract in poultry. Avian Biol. Res. 2014;7:193–222. [Google Scholar]
- Scupham A., Jones J., Wesley I. Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J. Appl. Microbiol. 2007;102:401–409. doi: 10.1111/j.1365-2672.2006.03094.x. [DOI] [PubMed] [Google Scholar]
- Sekelja M., Rud I., Knutsen S., Denstadli V., Westereng B., Naes T., Rudi K. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl. Environ. Microbiol. 2012;78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S.L., Antunes L.C.M., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shakouri M., Iji P., Mikkelsen L.L., Cowieson A. Intestinal function and gut microflora of broiler chickens as influenced by cereal grains and microbial enzyme supplementation. J. Anim. Physiol. Anim. Nutr. 2009;93:647–658. doi: 10.1111/j.1439-0396.2008.00852.x. [DOI] [PubMed] [Google Scholar]
- Shin N.-R., Whon T.W., Bae J.-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Shirley M.W., Smith A.L., Tomley F.M. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv. Parasitol. 2005;60:285–330. doi: 10.1016/S0065-308X(05)60005-X. [DOI] [PubMed] [Google Scholar]
- Singh K., Shah T., Deshpande S., Jakhesara S., Koringa P., Rank D., Joshi C. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol. Biol. Rep. 2012;39:10595–10602. doi: 10.1007/s11033-012-1947-7. [DOI] [PubMed] [Google Scholar]
- Slizewska K., Markowiak-Kopec P., Zbikowski A., Szeleszczuk P. The effect of synbiotic preparations on the intestinal microbiota and her metabolism in broiler chickens. Sci. Rep. 2020;10:4281. doi: 10.1038/s41598-020-61256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulikowska S. Biology of Growing Animals. Elsevier; London, UK: 2006. Manipulation of the poultry ecosystem through biotechnology; pp. 597–609. [Google Scholar]
- Smyth J.A. Pathology and diagnosis of necrotic enteritis: is it clear-cut? Avian Pathol. 2016;45:282–287. doi: 10.1080/03079457.2016.1158780. [DOI] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Chen H., Hughes R.J., Moore R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015;15:51. doi: 10.1186/s12866-015-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Denman S.E., Haring V.R., Crowley T.M., Hughes R.J., Moore R.J. Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet. Microbiol. 2013;164:85–92. doi: 10.1016/j.vetmic.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Hughes R.J., Denman S.E., Moore R.J. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One. 2013;8:e84290. doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Hughes R.J., Geier M.S., Moore R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016;7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014;98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- Stanley D., Keyburn A.L., Denman S.E., Moore R.J. Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet. Microbiol. 2012;159:155–162. doi: 10.1016/j.vetmic.2012.03.032. [DOI] [PubMed] [Google Scholar]
- Stutz M., Lawton G. Effects of diet and antimicrobials on growth, feed efficiency, intestinal Clostridium perfringens, and ileal weight of broiler chicks. Poult. Sci. 1984;63:2036–2042. doi: 10.3382/ps.0632036. [DOI] [PubMed] [Google Scholar]
- Suzuki M.T., Giovannoni S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Jin G., Wang G., Liu T., Liu X., Wang B., Cao H. Current sampling methods for gut microbiota: a call for more precise devices. Front. Cell. Infect. Microbiol. 2020;10:151. doi: 10.3389/fcimb.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tankson J., Thaxton J., Vizzier-Thaxton Y. Bacteria in heart and lungs of young chicks. J Appl. Microbiol. 2002;92:443–450. doi: 10.1046/j.1365-2672.2002.01546.x. [DOI] [PubMed] [Google Scholar]
- Tedjo D.I., Jonkers D.M., Savelkoul P.H., Masclee A.A., van Best N., Pierik M.J., Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez G., Latorre J.D., Kuttappan V.A., Kogut M.H., Wolfenden A., Hernandez-Velasco X., Hargis B.M., Bottje W.G., Bielke L.R., Faulkner O.B. Utilization of rye as energy source affects bacterial translocation, intestinal viscosity, microbiota composition, and bone mineralization in broiler chickens. Front. Genet. 2014;5:339. doi: 10.3389/fgene.2014.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzich M., Pope M.J., Cherry T.E., Hollinger J. Survey of pathogens in poultry litter in the United States. J. Appl. Poult. Res. 2000;9:287–291. [Google Scholar]
- Thompson K., Burkholder K., Patterson J., Applegate T. Microbial ecology shifts in the ileum of broilers during feed withdrawal and dietary manipulations. Poult. Sci. 2008;87:1624–1632. doi: 10.3382/ps.2007-00324. [DOI] [PubMed] [Google Scholar]
- Tilg H., Danese S. Roseburia hominis: a novel guilty player in ulcerative colitis pathogenesis? Gut. 2014;63:1204–1205. doi: 10.1136/gutjnl-2013-305799. [DOI] [PubMed] [Google Scholar]
- Timbermont L., Haesebrouck F., Ducatelle R., Van Immerseel F. Necrotic enteritis in broilers: an updated review on the pathogenesis. Avian Pathol. 2011;40:341–347. doi: 10.1080/03079457.2011.590967. [DOI] [PubMed] [Google Scholar]
- Timbermont L., Lanckriet A., Dewulf J., Nollet N., Schwarzer K., Haesebrouck F., Ducatelle R., Van Immerseel F. Control of Clostridium perfringens-induced necrotic enteritis in broilers by target-released butyric acid, fatty acids and essential oils. Avian Pathol. 2010;39:117–121. doi: 10.1080/03079451003610586. [DOI] [PubMed] [Google Scholar]
- Torok V., Dyson C., McKay A., Ophel-Keller K. Quantitative molecular assays for evaluating changes in broiler gut microbiota linked with diet and performance. Anim. Prod. Sci. 2013;53:1260–1268. [Google Scholar]
- Torok V., Hughes R., Ophel-Keller K., Ali M., MacAlpine R. Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult. Sci. 2009;88:2474–2481. doi: 10.3382/ps.2008-00381. [DOI] [PubMed] [Google Scholar]
- Torok V.A., Hughes R.J., Mikkelsen L.L., Perez-Maldonado R., Balding K., MacAlpine R., Percy N.J., Ophel-Keller K. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl. Environ. Microbiol. 2011;77:5868–5878. doi: 10.1128/AEM.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok V.A., Ophel-Keller K., Loo M., Hughes R.J. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 2008;74:783–791. doi: 10.1128/AEM.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis S., Menzies I. Intestinal permeability: functional assessment and significance. Clin. Sci. 1992;82:471–488. doi: 10.1042/cs0820471. [DOI] [PubMed] [Google Scholar]
- Tumova E., Chodova D., Skrivanova E., Lalouckova K., Subrtova-Salmonova H., Ketta M., Machander V., Cotozzolo E. Research Note: The effects of genotype, sex, and feeding regime on performance, carcasses characteristic, and microbiota in chickens. Poult. Sci. 2021;100:760–764. doi: 10.1016/j.psj.2020.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintzingerode F., Göbel U.B., Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Chapelle S., De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Immerseel F., Rood J.I., Moore R.J., Titball R.W. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–36. doi: 10.1016/j.tim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Van Limbergen T., Sarrazin S., Chantziaras I., Dewulf J., Ducatelle R., Kyriazakis I., McMullin P., Méndez J., Niemi J.K., Papasolomontos S. Risk factors for poor health and performance in European broiler production systems. BMC Vet. Res. 2020;16:1–13. doi: 10.1186/s12917-020-02484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri R., Sylvia K.E., Klein S.L., Forster S.C., Plebanski M., Eri R., Flanagan K.L. The microgenderome revealed: sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility. Semin. Immunopathol. 2019;41:265–275. doi: 10.1007/s00281-018-0716-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidanarachchi J.K., Mikkelsen L.L., Constantinoiu C., Choct M., Iji P. Natural plant extracts and prebiotic compounds as alternatives to antibiotics in broiler chicken diets in a necrotic enteritis challenge model. Anim. Prod. Sci. 2013;53:1247–1259. [Google Scholar]
- Videnska P., Smerkova K., Zwinsova B., Popovici V., Micenkova L., Sedlar K., Budinska E. Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-49520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videvall E., Strandh M., Engelbrecht A., Cloete S., Cornwallis C.K. Measuring the gut microbiome in birds: comparison of faecal and cloacal sampling. Mol. Ecol. Resour. 2018;18:424–434. doi: 10.1111/1755-0998.12744. [DOI] [PubMed] [Google Scholar]
- Vispo C., Karasov W.H. Gastrointestinal Microbiology. Springer; London, UK: 1997. The interaction of avian gut microbes and their host: an elusive symbiosis; pp. 116–155. [Google Scholar]
- Voeten A.C., Braunius W.W., Orthel F.W., van Rijen M.A.J. Influence of coccidiosis on growth rate and feed conversion in broilers after experimental infections with Eimeria acervulina and Eimeria maxima. Vet. Q. 1988;10:256–264. doi: 10.1080/01652176.1988.9694182. [DOI] [PubMed] [Google Scholar]
- Vrba V., Blake D.P., Poplstein M. Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Vet. Parasitol. 2010;174:183–190. doi: 10.1016/j.vetpar.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Vuong C.N., Mullenix G.J., Kidd M.T., Bottje W.G., Hargis B.M., Tellez-Isaias G. Research Note: Modified serum fluorescein isothiocyanate dextran (FITC-d) assay procedure to determine intestinal permeability in poultry fed diets high in natural or synthetic pigments. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenstedt L., Elwinger K., Lunden A., Thebo P., Bedford M., Uggla A. Intestinal digesta viscosity decreases during coccidial infection in broilers. Br. Poult. Sci. 2000;41:459–464. doi: 10.1080/713654959. [DOI] [PubMed] [Google Scholar]
- Walkden-Brown S.W., Islam A., Groves P.J., Rubite A., Sharpe S.M., Burgess S.K. Development, application, and results of routine monitoring of Marek's disease virus in broiler house dust using real-time quantitative PCR. Avian Dis. 2013;57:544–554. doi: 10.1637/10380-92112-REG.1. [DOI] [PubMed] [Google Scholar]
- Walker A.W., Martin J.C., Scott P., Parkhill J., Flint H.J., Scott K.P. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:1–11. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lilburn M., Yu Z. Intestinal microbiota of broiler chickens as affected by litter management regimens. Front. Microbiol. 2016;7:593. doi: 10.3389/fmicb.2016.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu Y., Wu Y., Mahmood T., Chen J., Guo X., Wu W., Wang B., Guo Y., Yuan J. Impact of different durations of fasting on intestinal autophagy and serum metabolome in broiler chicken. Animals (Basel) 2021;11:2183. doi: 10.3390/ani11082183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Wen C., Yan W., Mai C., Duan Z., Zheng J., Sun C., Yang N. Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome. 2021;9:126. doi: 10.1186/s40168-021-01040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielen P., Keuzenkamp D., Lipman L.v., Knapen F., Biesterveld S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 2002;44:286–293. doi: 10.1007/s00248-002-2015-y. [DOI] [PubMed] [Google Scholar]
- Wilkie D.C., Van Kessel A.G., White L.J., Laarveld B., Drew M.D. Dietary amino acids affect intestinal Clostridium perfringens populations in broiler chickens. Can. J. Anim. Sci. 2005;85:185–193. [Google Scholar]
- Williams R. Epidemiological studies of coccidiosis in the domesticated fowl (Gallus gallus): II. Physical condition and survival of Eimeria acervulina oocysts in poultry-house litter. Appl. Parasitol. 1995;36:90–96. [PubMed] [Google Scholar]
- Williams R. A compartmentalised model for the estimation of the cost of coccidiosis to the world's chicken production industry. Int. J. Parasitol. 1999;29:1209–1229. doi: 10.1016/s0020-7519(99)00086-7. [DOI] [PubMed] [Google Scholar]
- Williams R. Intercurrent coccidiosis and necrotic enteritis of chickens: rational, integrated disease management by maintenance of gut integrity. Avian Pathol. 2005;34:159–180. doi: 10.1080/03079450500112195. [DOI] [PubMed] [Google Scholar]
- Williams R., Marshall R., La Ragione R., Catchpole J. A new method for the experimental production of necrotic enteritis and its use for studies on the relationships between necrotic enteritis, coccidiosis and anticoccidial vaccination of chickens. Parasitol. Res. 2003;90:19–26. doi: 10.1007/s00436-002-0803-4. [DOI] [PubMed] [Google Scholar]
- Wu S.-B., Stanley D., Rodgers N., Swick R.A., Moore R.J. Two necrotic enteritis predisposing factors, dietary fishmeal and Eimeria infection, induce large changes in the caecal microbiota of broiler chickens. Vet. Microbiol. 2014;169:188–197. doi: 10.1016/j.vetmic.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Xia Y., Sun J., Chen D.-G. Statistical Analysis of Microbiome Data With R. Springer; Singapore: 2018. Power and sample size calculations for microbiome data; pp. 129–166. [Google Scholar]
- Yadav S., Jha R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J. Anim. Sci. Biotechnol. 2019;10:1–11. doi: 10.1186/s40104-018-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Lv Z., An S., Xing K., Wang Z., Lv M., Choct M., Guo Y., Zhou G. Effects of rearing system and narasin on growth performance, gastrointestinal development, and gut microbiota of broilers. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2020.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegani M., Korver D. Factors affecting intestinal health in poultry. Poult. Sci. 2008;87:2052–2063. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoman C.J., Chia N., Jeraldo P., Sipos M., Goldenfeld N.D., White B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 2012;13:89. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- Zhang T., Ding H., Chen L., Lin Y., Gong Y., Pan Z., Zhang G., Xie K., Dai G., Wang J. Antibiotic-induced dysbiosis of microbiota promotes chicken lipogenesis by altering metabolomics in the cecum. Metabolites. 2021;11:487. doi: 10.3390/metabo11080487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E.G., Collier C.T., Koike S., Mackie R.I., Gaskins H.R. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 2004;134:465–472. doi: 10.1093/jn/134.2.465. [DOI] [PubMed] [Google Scholar]