Abstract
Objective
Adenosine deaminase acting on RNA-1 (ADAR1) enzyme is a type I interferon (IFN)-stimulated gene (ISG) catalyzing the deamination of adenosine-to-inosine, a process called A-to-I RNA editing. A-to-I RNA editing takes place mainly in Alu elements comprising a primate-specific level of post-transcriptional gene regulation. Whether RNA editing is involved in type I IFN responses in systemic sclerosis (SSc) patients remains unknown.
Methods
ISG expression was quantified in skin biopsies and peripheral blood mononuclear cells derived from SSc patients and healthy subjects. A-to-I RNA editing was examined in the ADAR1-target cathepsin S (CTSS) by an RNA editing assay. The effect of ADAR1 on interferon-α/β-induced CTSS expression was assessed in human endothelial cells in vitro.
Results
Increased expression levels of the RNA editor ADAR1, and specifically the long ADAR1p150 isoform, and its target CTSS are strongly associated with type I IFN signature in skin biopsies and peripheral blood derived from SSc patients. Notably, IFN-α/β-treated human endothelial cells show 8-10-fold increased ADAR1p150 and 23-35-fold increased CTSS expression, while silencing of ADAR1 reduces CTSS expression by 60-70%. In SSc patients, increased RNA editing rate of individual adenosines located in CTSS 3′ UTR Alu elements is associated with higher CTSS expression (r = 0.36–0.6, P < 0.05 for all). Similar findings were obtained in subjects with activated type I IFN responses including SLE patients or healthy subjects after influenza vaccination.
Conclusion
ADAR1p150-mediated A-to-I RNA editing is critically involved in type I IFN responses highlighting the importance of post-transcriptional regulation of proinflammatory gene expression in systemic autoimmunity, including SSc.
Keywords: Autoimmunity, Type I interferon, Gene expression, A-to-I RNA editing, Systemic sclerosis
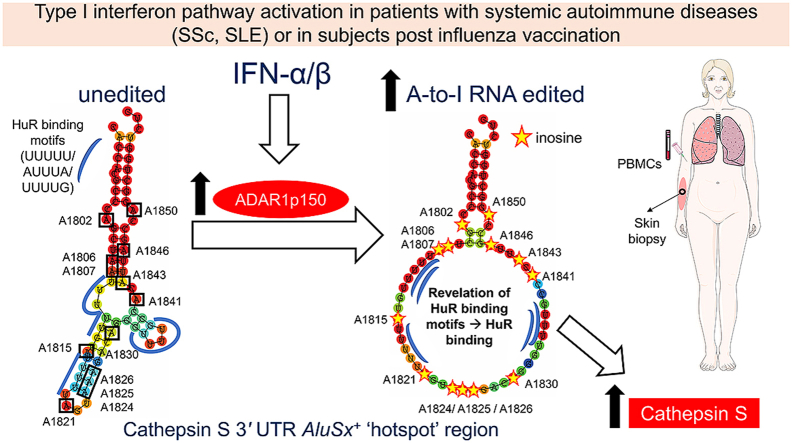
Graphical abstract
Increased type I interferon (IFN-α/β) in patients with systemic sclerosis (SSc), systemic lupus erythematosus (SLE) or healthy subjects after influenza vaccination leads to increased expression of the ADAR1p150 enzyme. Increased ADAR1p150 in turn leads to increased adenosine-to-inosine (A-to-I) RNA editing of Alu elements. We were able to detect a ‘hotspot’ region within AluSx+ in cathepsin S 3′ untranslated region, characterized by the presence of 13 A-to-I RNA editing events within 50 bps and of 5 binding motifs of the stabilizing single-stranded RNA-binding protein HuR. A-to-I RNA editing unwinds the local double-stranded RNA structure into a more single-stranded conformation, thus revealing HuR binding motifs. Increased A-to-I RNA editing rate of the ‘hotspot’ region in SSc and SLE patients, or after acute induction of a systemic type I IFN response, may thus facilitate recruitment of the stabilizing single stranded RNA-binding protein HuR to its target motifs, ultimately increasing CTSS transcript stability and expression.
Highlights
-
•
Adenosine deaminase acting on RNA-1 (ADAR1) is an interferon-stimulated gene.
-
•
ADAR1p150 enzyme isoform is the main RNA editor in systemic sclerosis (SSc).
-
•
Increased ADAR1p150 and cathepsin S expression levels are associated with type I interferon in patients with SSc.
-
•
Increased adenosine-to-inosine RNA editing contributes to cathepsin S expression in patients with SSc.
-
•
Similar findings are observed also in SLE or after acute induction of type IFN response by influenza vaccination.
1. Introduction
Systemic sclerosis (SSc) or Scleroderma is a detrimental autoimmune disease characterized by immune activation, vascular dysfunction and fibrosis, as principal features of its pathogenesis [1]. A prominent activation of the type I interferon (IFN) pathway has been consistently reported in peripheral blood cells and target-tissues (skin, lung) of patients with SSc [[2], [3], [4], [5], [6], [7], [8]], even from the earliest phases of disease before overt fibrosis [9]. Moreover, GWAS studies have shown an association between polymorphisms in interferon regulatory factors (IRFs) and SSc (reviewed in Ref. [10]). Treatment of SSc patients with IFN-α led to deterioration of skin and lung function during 1-year follow-up further supporting the detrimental role of type I IFNs in SSc progression [11]. On the other hand, treatment of SSc patients with a monoclonal antibody against the common type I IFN receptor (IFNAR) showed favorable safety profile associated with decreased IFN- and TGF-β-induced gene expression in patients’ skin [12,13]. Despite extensive evidence supporting the pivotal role of type I IFN in SSc pathogenesis, it remains largely unclear how type I IFN may contribute to disease development and progression [10,14].
Adenosine deaminase acting on RNA-1 (ADAR1) is an IFN-inducible gene that catalyzes the deamination of adenosine to inosine, a process called A-to-I RNA editing [[15], [16], [17]]. A-to-I RNA editing controls multiple aspects of RNA metabolism and can thus fine-tune gene expression at the post-transcriptional level [16,17]. However, the role of A-to-I RNA editing in human diseases characterized by type I IFN activation remains poorly understood [18,19]. RNA editing takes place mainly in Alu elements, which are dispersed throughout the human transcriptome and can form local double-stranded RNA (dsRNA) structures due to high sequence complementarity [20]. RNA editing of Alu elements may alter this local dsRNA architecture into a more single-stranded conformation [21], due to weak pairing of inosine with uracil. This allows the recruitment of single-strand RNA binding proteins (RBPs), such as the stabilizing Human antigen R (HuR; ELAVL1) protein, which may greatly affect transcript stability [21]. Importantly, Alu elements are conserved only among primates [22], making Alu A-to-I RNA editing a primate-specific level of post-transcriptional gene regulation. A-to-I RNA editing is increased in chronic inflammatory diseases, such as atherosclerosis [21] and rheumatoid arthritis [23], but returns to normal levels after successful anti-inflammatory treatment [23]. Herein, we tested the hypothesis that increased type I IFN may drive ADAR1-induced RNA editing in SSc, and other disease contexts characterized by chronic or acute type I IFN pathway activation, serving as an additional post-transcriptional layer of proinflammatory gene regulation shared among systemic autoimmune diseases.
2. Materials and methods
2.1. Demographics and clinical characteristics of the study PBMC cohort
Peripheral blood was collected in EDTA tubes (BD Vacutainer) from 31 patients with SSc (2013 EULAR/ACR classification criteria [24]), 25 patients with systemic lupus erythematosus (SLE) (2012 SLICC criteria [25]) and 23 healthy controls (HC). Exclusion criteria for the whole study cohort included viral or bacterial infection during the past month and severe co-morbidities (cancer, heart or kidney failure). Demographics, clinical and laboratory features [interstitial lung disease, skin involvement, Raynaud's phenomenon; puffy fingers; digital ulcers; erythrocyte sedimentation rate (ESR; mm/1st h.)] and disease-specific auto-antibody status were recorded at baseline (Supplementary Table 1). All participants gave informed consent in compliance with the Declaration of Helsinki, which had been previously approved by the Ethics Committee of Laiko Hospital, Athens, Greece (Protocol Nr.:1368/17-11-2016).
2.2. Influenza vaccination study
In order to examine the effect of acute in vivo type I IFN induction, peripheral blood was collected from 9 healthy individuals [mean±SD age: 32 ± 6 years, 7 (77.8%) women] before- and 24 h after-vaccination against influenza. The selection of this time-point was based on a previous study examining the kinetics of type I IFN response in human peripheral blood cells after influenza vaccination up to 48 h post-vaccination [26].
2.3. Isolation of peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMCs) were isolated by standard methods (Ficoll density gradient centrifugation, Ficoll-Paque PLUS, GE Healthcare) within 2 h from venipuncture, as previously described [23]. Isolated PBMCs were washed twice with phosphate-buffered saline (PBS) without calcium/magnesium and subsequently lysed in Trizol (Invitrogen, ThermoFisher Scientific) and stored at −80 °C, as previously described [23].
2.4. RNA isolation and reverse transcription
Total RNA was extracted from patient and control PBMCs using Direct-Zol RNA Miniprep kit (Zymo research), including an additional DNase digestion step, as previously described [23]. One (1) μg of total RNA was reverse transcribed into complementary DNA (cDNA) using the M-MLV reverse transcriptase kit (Invitrogen, ThermoFisher Scientific), as previously described [23].
2.5. Quantitative polymerase chain reaction
The expression of the two ADAR1 isoforms, ADAR1p110 and ADAR1p150, was quantified by Taqman assays (ADAR1p110: Hs01017596; ADAR1p150: Hs01020780, Applied Biosystems), as previously described [23]. TATA-Box binding protein (TBP) (Hs00427621, Applied Biosystems) served as the housekeeping gene.
Expression of CTSS was quantified with SYBR Premix Ex Taq (Takara)/PowerUP SYBR Green Master Mix (Applied Biosystems) on ViiA7/QuantStudio 7 Flex system. RPLP0 served as housekeeping gene (for primer sequence see Supplementary Table 2). For the calculation of type I IFN score, we measured the expression of 3 genes preferentially induced by type I IFN (IFIT1, MX1, IFI44), as previously described [[27], [28], [29]]. The relative expression of each gene was determined according to formula 2−ΔCt, where ΔCt = Ct (gene)-Ct (housekeeping gene).
2.6. Transcript-specific RNA editing analysis
A-to-I RNA editing of individual adenosines was examined, as we have previously described [21,23,30], in AluJo- and AluSx+ elements of CTSS 3′ untranslated region (UTR). Primer sequences are available in Supplementary Table 2.
2.7. Expression analysis in skin biopsies of SSc patients
2.7.1. RNA sequencing analysis
Expression of interferon-stimulated genes (ISGs), ADAR1 and CTSS was examined in skin biopsies derived from 47 SSc patients and 33 controls [31,32]. The .sra files were downloaded from NCBI GEO project GSE130955 and subsequently transformed to .fastq files using the SRA Toolkit (http://ncbi.github.io/sra-tools/) provided by the NCBI. The paired-end reads were trimmed for adapters and quality using Trim Galore version 0.6.6 [33] and subsequently passed an overall quality control using FASTQC version 0.11.9 [34] and MultiQC [35] version 1.9 tools. The trimmed and quality checked reads were then aligned to human genome GRCh38.p13 with default options for paired-end read mapping from Ensembl [36] using STAR [37] aligner version 2.7.0e-foss-2018b and Ensembl [36] GRCh38.99.gtf annotation file. The generated .bam files were then used as input for htseq-count [38], with parameters -f bam -a 0 -s no -t exon, to produce tab delimited tables of read counts for each gene.
2.7.2. Differential gene expression analysis (DGEA)
DGEA was performed using DESeq2 [39] R package from Bioconductor version 3.11 using Rstudio version February 1, 5033 and R language version 3.6.3. The count tables were read into R using DESeqDataSetFromHTSeqCount function (default settings) and the appropriate design of the DESeq data set was applied to take into account all samples from all conditions hence enabling the inter-sample comparison of the same gene. Finally, a table of normalized counts per gene was extracted.
2.8. In silico RNA folding analysis
Using the UCSC browser for the human genome 38, we identified the 3′ UTR of CTSS including the 3 Alu elements (AluSz6-, AluJo-, AluSx+). Subsequently, we used the RNAfold webtool from the Vienna RNA Websuite [40] to predict the RNA folding structure, as we have previously described [30]. Folding analysis was performed using the default parameters and the algorithm ‘minimum free energy (MFE) and partition function’. The same parameters were used for all folding analyses.
2.9. Cell culture and transfection
Human umbilical vein endothelial cells (HUVECs; Lonza) were cultured in EBM medium (Lonza) supplemented with 10% FBS (Gibco) and growth factors (EGM-Bulletkit-no ascorbic acid). Cells were cultured until passage 5 (P5) and all experiments were performed between P3–P5.
For gene silencing studies, the endothelial cells were grown until the fourth passage and then seeded in 60 mm dishes (Greiner). As HUVECs reached 70–80% confluence, cells were then transfected with an siRNA targeting specifically ADAR1 or a non-targeting (scrambled) siRNA using Lipofectamine RNAiMAX (Invitrogen) at 55 nM. Transfection was carried out in a reduced-serum medium (OptiMEM, Gibco). After transfection, cells were incubated in growth medium for further 48 h.
For the stimulation studies, we used human IFN-α-2α (11100-1, R&D) at 1000 U/ml (=2.2 ng/ml) and human IFN-α A/D (11200-1, R&D) at 1000 U/ml (=3.39 ng/ml) concentration, as had been previously described for the induction of ADAR1p150 [41,42]. Moreover, we used human IFN-β (8499-IF-010, R&D) at 2800 U/ml (=10 ng/ml) and human IFN-α-2α at 4560 U/ml (=10 ng/ml) concentration. All treatments were performed in full EBM medium supplemented with 10% FBS, after a 2-h serum starvation of the cells in OptiMEM.
2.10. Statistical analysis
Statistical analysis was conducted with SPSS v26.0 and GraphPad Prism 7. Normality of continuous variables was assessed by D'Agostino-Pearson, Kolmogorov-Smirnov and Shapiro-Wilk tests. Pairwise differences were evaluated with independent samples Student's t-test (with Welch's correction when variances of the groups were unequal) or with the non-parametric Mann-Whitney U test for continuous variables, and chi-squared test for nominal variables. Correlations between continuous variables were explored by Pearson's correlation coefficient test. Results were considered statistically significant when P < 0.05.
3. Results
3.1. Increased ADAR1p150 expression is associated with type I IFN signature in SSc skin and peripheral blood
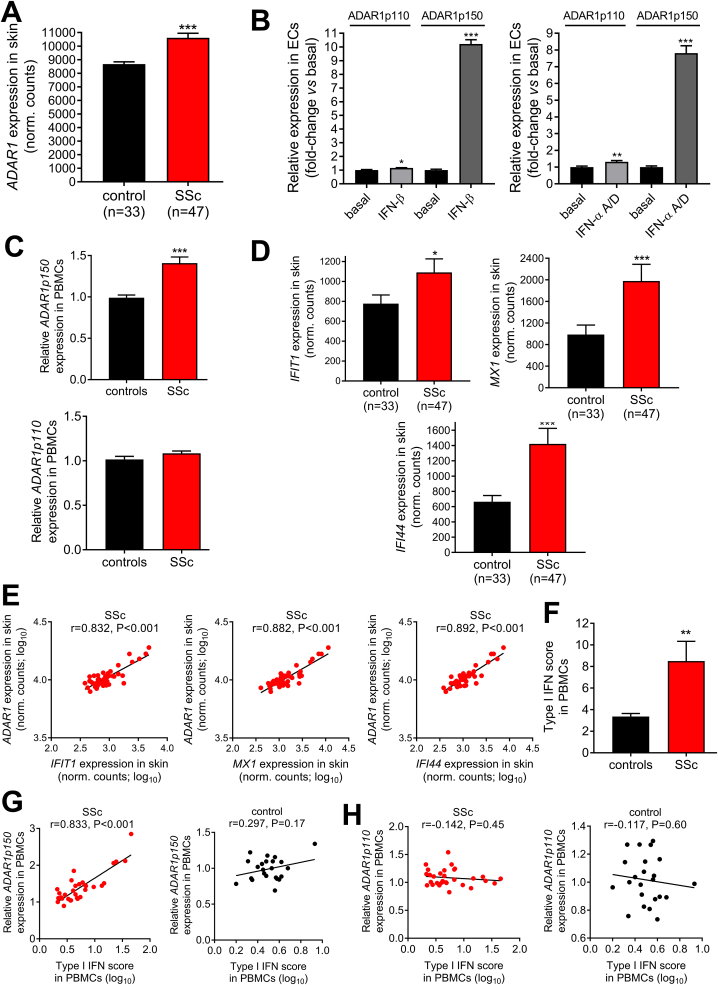
Expression analysis of an RNA-sequencing dataset of skin biopsies derived from 47 SSc patients and 33 controls showed a significant upregulation of ADAR1 in SSc (P < 0.001; Fig. 1A). ADAR1 has two isoforms, a shorter 110 kDa-long isoform (ADAR1p110), which is constitutively expressed, and a longer 150 kDa-long isoform (ADAR1p150), which is transcribed by an interferon-responsive promoter [41,42]. Treatment of human endothelial cells with IFN-α or IFN-β led to a 8-10-fold increase in ADAR1p150 isoform expression, while ADAR1p110 was only slightly affected (1.1-1.2-fold increase) (Fig. 1B and Supplementary Figure 1). The type I IFN-stimulated genes IFIT1, MX1 and IFI44 were also measured as control (Supplementary Figure 2), validating successful induction of a type I IFN response. In line with these results, isoform specific expression analysis in PBMCs showed increased ADAR1p150 expression in SSc (P < 0.001; Fig. 1C, upper panel), while ADAR1p110 was comparable between patients and controls (Fig. 1C, lower panel).
Fig. 1.
Increased ADAR1 expression in SSc skin and peripheral blood mononuclear cells is associated with type I IFN scores.A. Bar graph shows the levels (normalized counts) of ADAR1 in SSc (n = 47) vs control (n = 33) skin biopsies. Expression levels were generated by the publicly available RNA-seq. dataset GSE130955 with the use of STAR aligner and R package DeSeq2. B. ADAR1p110 and ADAR1p150 expression levels in endothelial cells treated with 2,800U (=10 ng/ml) IFN-β (left panel; n = 5) or 1000 U/ml (=3.4 ng/ml) human IFN-α A/D (right panel; n = 7) for 24 h compared to vehicle (PBS for IFN-β and PBS-BSA 0.1% for IFN-α A/D). C. ADAR1p150 (upper panel) and ADAR1p110 (lower panel) expression levels in PBMCs of healthy individuals (n = 23) and SSc patients (n = 31) as quantified by TaqMan qRT-PCR. D. Bar graphs depict the expression levels (normalized counts) of the 3 type I IFN-induced genes used to calculate type I IFN score in skin biopsies of SSc patients (n = 47) vs control (n = 33) skin biopsies. Expression levels were generated by the publicly available RNA-seq. dataset GSE130955 with the use of the R package DeSeq2. E. Scatter plots show the correlation between ADAR1 and ISGs (IFIT1, MX1, IFI44) in skin biopsies of SSc patients (n = 47). F. Type I IFN score in PBMCs of healthy individuals (n = 23) and SSc patients (n = 31). For the calculation of type I IFN score the relative mRNA expression of 3 genes selectively induced by type I IFN (IFIT1, MX1 and IFI44) was measured with RT-qPCR. G,H. Scatter plots depict the correlation between individual type I IFN scores and ADAR1p150 expression levels (G) or ADAR1p110 expression levels (H) in PBMCS of SSc patients (red, left panels) and healthy individuals (black, right panels). Individual expression levels in PBMCs in all graphs are plotted as fold-change vs HC median. Bar graphs represent mean and SEM. Pairwise comparisons were performed by Mann-Whitney U test or independent samples t-test. Correlation coefficients r were derived by Pearson's correlation coefficient test. *P < 0.05, **P < 0.01, ***P < 0.001.
Expression of type I IFN-stimulated genes was also increased in SSc skin (all P < 0.05, Fig. 1D) showing a strong positive correlation with ADAR1 levels (r = 0.832–0.892, all P < 0.001; Fig. 1E). Similarly, 3-fold increased ISG expression was detected in PBMCs of SSc patients (P = 0.004; Fig. 1F). Of interest, ADAR1p150 expression levels were strongly associated with type I IFN scores in SSc PBMCs (r = 0.833, P < 0.001; Fig. 1G, left panel), but not in controls (r = 0.297, P = 0.17; Fig. 1G, right panel), while ADAR1p110 did not correlate with type I IFN score in either patients or controls (Fig. 1H). Taken together, these results suggested that ADAR1, and specifically the long ADAR1p150 isoform, is part of type I IFN responses in both SSc skin and blood cells.
3.2. Type I IFN-driven post-transcriptional regulation of cathepsin S in SSc
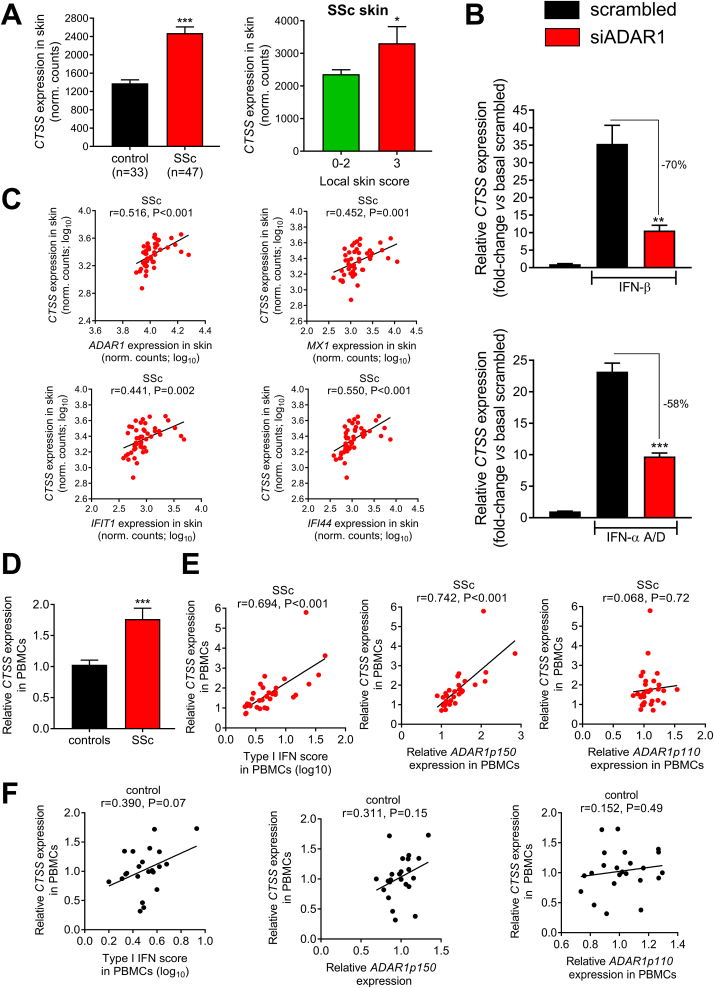
Next, we examined whether cathepsin S (CTSS), a well-established target of ADAR1 [21,23], was also increased in SSc. CTSS is involved in antigen presentation [43] and extracellular matrix remodeling and has been previously shown to correlate with extent of skin involvement in SSc [6]. We observed 2-fold increased expression of CTSS in SSc skin (Fig. 2A, left panel) and especially in those patients with high local skin score (Fig. 2A, right panel). Of interest, treatment of ECs with either IFN-α A/D or IFN-β led to a striking 25-35-fold increase in CTSS expression, which was dampened by 58-70% when ADAR1 was silenced (Fig. 2B) suggesting a predominant effect of ADAR1 on CTSS expression under IFN-α/β conditions. Similarly, a dose-dependent increase in CTSS was observed when endothelial cells were treated with 1000 U/ml and 4560 U/ml IFN-α-2α (Supplementary Figure 3). Individual CTSS expression levels in SSc skin correlated with ADAR1 (r = 0.516, P < 0.001) and ISGs (r = 0.441–0.550, all P < 0.01) expression (Fig. 2C).
Fig. 2.
Increased expression of the ADAR1-targetCTSSin SSc skin and peripheral blood mononuclear cells.A. Bar graph shows the levels (normalized counts) of CTSS in SSc (n = 47) vs control (n = 33) skin biopsies (left) or in SSc patients with highest local skin score vs the rest SSc patients (right). Expression levels were generated by the publicly available RNA-seq. dataset GSE130955 with the use of the R package DeSeq2. B. HUVECs were transfected with an siRNA targeting ADAR1 or control (scrambled) non-targeting siRNA and incubated for 48 h. Subsequently, cells were treated with 2800 U/ml (=10 ng/ml) human IFN-β (n = 5) or 1000 U/ml (=3.4 ng/ml) human IFN-α A/D (n = 7) or vehicle (PBS for IFN-β and PBS-BSA 0.1% for IFN-α A/D) for additional 24 h. Bar graphs show relative CTSS expression levels as fold-change vs basal scrambled. C. Scatter plots show the correlation between CTSS and ADAR1 or ISGs (IFIT1, MX1, IFI44) in skin biopsies of SSc patients (n = 47). D. Bar graph shows the relative CTSS expression levels in PBMCs of SSc patients (n = 31) vs control subjects (n = 23) as quantified by SYBR-Green RT-qPCR. E. Scatter plots show the correlation between CTSS expression levels and individual type I IFN scores (left), ADAR1p150 (middle) or ADAR1p110 (right) in SSc patients (n = 31). F. Scatter plots show the correlation between CTSS expression levels and individual type I IFN scores (left), ADAR1p150 (middle) or ADAR1p110 (right) in PBMCs of healthy controls (n = 23). Individual expression levels in PBMCs in all graphs are plotted as fold-change vs HC median. Bar graphs represent mean and SEM. Pairwise comparisons were performed by Mann-Whitney U test or independent samples t-test. Correlation coefficients r were derived by Pearson's correlation coefficient test. *P < 0.05, **P < 0.01, ***P < 0.001.
Similarly, we observed a 2-fold increase of CTSS expression in PBMCs derived from SSc patients (Fig. 2D). Expression levels of CTSS were associated with modified Rodnan skin score and were increased in patients with digital ulcers (Supplementary Figure 4). There was a strong correlation between CTSS and type I IFN score (r = 0.694, P < 0.001; Fig. 2E, left panel) or ADAR1p150 expression (r = 0.742, P < 0.001; Fig. 2E, middle panel) in SSc PBMCs, but not in controls (Fig. 2F- left and middle panel), suggesting that this may be a disease-specific mechanism of gene regulation. CTSS expression was not associated with ADAR1p110 in either SSc or control PBMCs (Fig. 2E and F - right panel). Finally, a regression analysis showed that type I IFN score in PBMCs of SSc patients lost its association with CTSS after adjusting for ADAR1p150 expression, suggesting that the effect of IFN on CTSS was mainly mediated through ADAR1p150.
3.3. Increased Alu adenosine-to-inosine RNA editing in patients with SSc
Having established that expression of ADAR1p150 and its target CTSS is increased in patients with SSc, we asked whether this was accompanied by an increase in A-to-I RNA editing. We next examined the A-to-I RNA editing of Alu elements in the 3′ UTR of CTSS using a well-established in-house transcript-specific RNA editing assay [21,23] to determine the editing rate of individual adenosine residues in PBMCs of healthy individuals and SSc patients. According to UCSC Genome Browser (Hg38), CTSS 3′ UTR has 3 Alu elements, two on the negative strand (AluSz6- and AluJo-) and one on the positive strand (AluSx+) (Fig. 3A). RNA foldback analysis revealed that AluJo- and the opposite-oriented AluSx+ form a long dsRNA structure (Fig. 3B), which is a perfect substrate for ADAR1 binding and editing.
Fig. 3.
Mapping of A-to-I RNA editing events inCTSS AluJo-andAluSx+.A. Graphical representation of CTSS 3′ UTR depicting the 3 Alu elements (AluSz6-, AluJo-, AluSx+). B. RNA foldback analysis of the secondary structure formed by AluJo- and the opposite-oriented AluSx+. C. Mapping of the adenosine residues that undergo RNA editing throughout the two Alu regions of our interest, AluSx+ and AluJo-. A-to-I RNA editing rate of individual adenosine residues was detected by an RNA editing assay in PBMCs of healthy individuals (black) and SSc patients (red) in AluJo- (HC n = 13, SSc n = 9) and AluSx+ (HC n = 23, SSc n = 31). *P < 0.05, #P < 0.10 by Mann-Whitney U test.
First, we mapped RNA editing events in both AluJo- and AluSx+ in a pilot cohort of 9 SSc patients and 13 HC. We were able to detect RNA editing in 23 out of 85 adenosines located in AluJo- (Fig. 3C) and in 39 out of 62 adenosines located in AluSx+. Therefore, we then focused on RNA editing of AluSx+ (Fig. 3C). In line with the increased expression of ADAR1p150, we observed an increase in Alu A-to-I RNA editing in 31 SSc patients compared to 23 controls (P < 0.05 for 15 detected editing sites in AluSx+; Fig. 3C), similar to what we had previously observed in rheumatoid arthritis [23].
Moreover, we observed that there was a ‘hotspot’ region in CTSS AluSx+ (A1802-A1850), which contained 13 A-to-I RNA editing sites (Fig. 4A) and 5 binding motifs for the stabilizing RNA-binding protein HuR (ELAVL1). Inosine residues are in turn recognized as guanosine by the cellular machinery ultimately leading to an A-to-G base substitution (Fig. 4B–I, upper panels), which allows its quantification by sequencing. Increased A-to-I RNA editing in SSc PBMCs was observed in 11 out of these 13 adenosines (Fig. 4B–I, middle panels), leading to an increase in average RNA editing rate as well (Supplementary Figure 5A). Of note, A-to-I RNA editing rate of individual adenosine residues was positively correlated with ADAR1p150 levels in SSc patients (r = 0.460–0.737, P < 0.05; Fig. 4B–I, lower panels), while only 1 site reached significance in controls (data not shown). Similar results were obtained when examining the average editing rate of all 13 adenosine residues (SSc: r = 0.695, P < 0.001; HC: r = 0.302, P = 0.17; Supplementary Fig. 5B and C).
Fig. 4.
Association between RNA editing of individual adenosine residuesin CTSSAluSx+‘hotspot’ regionand ADAR1p150 expression in PBMCs of SSc patients.A. RNA fold-back analysis of the ‘hotspot’ region within AluSx+, characterized by the presence of 13 A-to-I RNA editing events within 50 bps and of 5 binding motifs of the stabilizing single-stranded RNA-binding protein HuR. The scheme shows predicted local secondary RNA structure in the absence of A-to-I RNA editing. Colors represent probability of base pairing with blue indicating low and red indicating high pairing probability. Blue lines denote HuR binding sites (UUUUU/AUUUA/UUUUG). B–I. A-to-I RNA editing of individual adenosines within this ‘hotspot’ region. Chromatopherograms (upper panel) show adenosine-to-inosine RNA editing events. Inosines are recognized as guanosines as shown. Tukey box plots (middle) show A-to-I RNA editing levels in PBMCs of SSc patients (n = 31) vs control subjects (n = 23). Scatter plots (lower) show the correlation of individual adesonine RNA editing rates with ADAR1p150 expression levels in PBMCs of SSc patients (n = 31). RNA editing rates of individual adenosines were determined following analysis of chromatopherograms. Individual expression levels in all graphs are plotted as fold-change vs HC median. Correlation coefficients r were derived by Pearson's correlation coefficient test.
We have previously shown that ADAR1-induced A-to-I RNA editing of CTSS Alu elements regulates transcript stability by facilitating binding of the stabilizing RNA-binding protein HuR [21]; and this process was increased in patients with chronic inflammatory diseases such as atherosclerosis [21] and active rheumatoid arthritis [23]. Indeed, using an in silico RNA foldback analysis, we observed that the introduction of the detected A-to-I editing events in this ‘hotspot’ region within AluSx+ revealed an alternative predicted RNA secondary structure, which had as a result the exposure of the HuR biding sites (UUUUU, AUUUA, UUUUG) from double-stranded to single-stranded RNA (Fig. 5A). This disruption of dsRNA into single-stranded RNA (ssRNA) in 3′ UTR is essential for binding of HuR and controls RNA stability [21]. Importantly, RNA editing rate of individual adenosine residues located in close proximity with HuR binding sites strongly correlated with CTSS expression in patients with SSc (r = 0.357–0.600, all P < 0.05; Fig. 5B–I) while it showed no correlation in controls (Fig. 5B–I). Moreover, editing rate of the adenosine residues located within this 50-bp region showed a much stronger correlation with CTSS transcript levels compared to the editing rate of adenosine residues located elsewhere in AluSx+ (Supplementary Figure 6). Similar results were obtained when examining the average editing rate of all 13 adenosine residues of the 'hotspot' (SSc: r = 0.562, P = 0.001; HC: r = -0.190, P = 0.40; Supplementary Fig. 5E,F), indicating that the bulk conformational changes of this ‘hotspot’ are important for the regulation of transcript levels. Taken together, these results suggest that type I IFN-induced increase in A-to-I RNA editing rate of CTSS 3′ UTR is incremental for CTSS transcript expression in SSc.
Fig. 5.
Association between RNA editing of individual adenosine residues in close proximity with HuR binding motifs andCTSSexpression in PBMCs of SSc patients.A. RNA fold-back analysis of the ‘hotspot’ region within AluSx+, characterized by the presence of 13 A-to-I RNA editing events within 50 bps and of 5 binding motifs of the stabilizing single-stranded RNA-binding protein HuR. The scheme shows predicted local secondary RNA structure after being A-to-I RNA edited. Colors represent probability of base pairing with blue indicating low and red indicating high pairing probability. Blue lines denote HuR binding sites (UUUUU/AUUUA/UUUUG). B–I. Scatter plots show correlation of individual adenosine RNA editing rates with CTSS expression levels in PBMCs of SSc patients (red; n = 31) or healthy individuals (black; n = 23). RNA editing rates of individual adenosines were determined following analysis of chromatopherograms. Individual expression levels in all graphs are plotted as fold-change vs HC median. Correlation coefficients r were derived by Pearson's correlation coefficient test.
3.4. Increased ADAR1p150-induced A-to-I RNA editing in systemic lupus erythematosus and in healthy controls after influenza vaccination
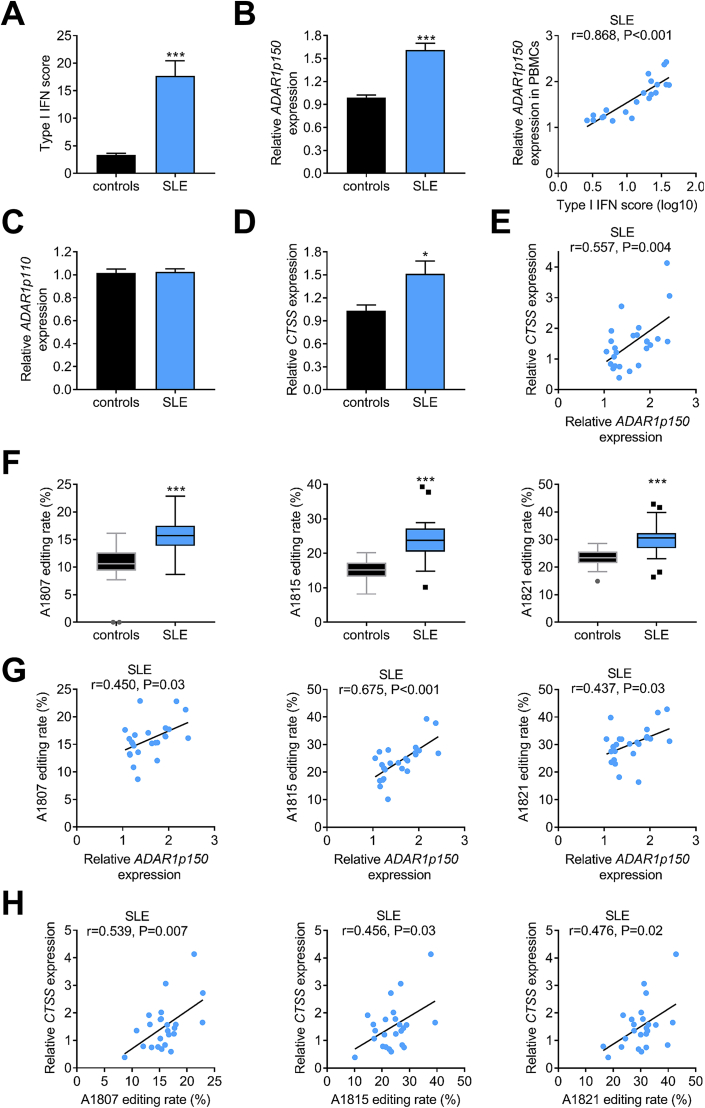
Next, we asked whether the observed increase of ADAR1p150-induced A-to-I RNA editing in SSc was also evident in another disease characterized by chronic type I IFN activation, i.e. in SLE [2]. Indeed, SLE patients also showed increased type I IFN score (Fig. 6A) and ADAR1p150 expression (Fig. 6B, left panel), which strongly correlated with each other (r = 0.868, P < 0.001; Fig. 6B, right panel). On the other hand, ADAR1p110 expression did not differ between SLE patients and controls (Fig. 6C). The A-to-I RNA editing rate of 10 out of the 13 edited individual adenosine residues in the ‘hotspot’ region of AluSx+ was increased in SLE compared to control subjects (data not shown). Interestingly, the average RNA editing rate of the adenosines located in the CTSS AluSx+ ‘hotspot’ region was significantly increased in SLE patients compared to control subjects (Supplementary Figure 5A). No statistical significant difference was observed between average RNA editing rate of the ‘hotspot’ region between SLE and SSc patients (Supplementary Figure 5A). Finally, CTSS expression was also increased in SLE patients compared to controls (Fig. 6D) showing positive correlation with ADAR1p150 expression (Fig. 6E). We identified 3 adenosines (A1807, A1815, A1821) within the AluSx+ ‘hotspot’ region, which presented with increased RNA editing rate in SLE patients compared to controls and their RNA editing rate was associated with both ADAR1p150 mRNA (Fig. 6G) and CTSS mRNA expression levels (Fig. 6H). Similarly, a trend association was observed between average RNA editing rate of the ‘hotspot’ region of the CTSS 3′ UTR with CTSS (r = 0.353, P = 0.098; Supplementary Figure 5G).
Fig. 6.
Increased ADAR1p150-induced A-to-I RNA editing in systemic lupus erythematosus.A. Bar graphs show the levels of type I IFN score in PBMCs of healthy individuals (n = 23) and SLE patients (n = 25). For the calculation of type I IFN score the relative mRNA expression of 3 genes selectively induced by type I IFN (IFIT1, MX1 and IFI44) was measured with RT-qPCR. B. Bar graph shows relative expression levels of ADAR1p150 (left panel) as quantified by TaqMan RT-qPCR in PBMCs of SLE patients vs control subjects, while the scatter plot (right panel) depicts the correlation between individual type I IFN scores and ADAR1p150 expression levels in PBMCs of SLE patients. C. Bar graph shows relative expression levels of ADAR1p110 as quantified by TaqMan RT-qPCR in PBMCs derived from SLE patients vs control subjects. D. Bar graph shows the levels of CTSS expression in PBMCs of healthy individuals and SLE patients as quantified by SYBR-Green RT-qPCR. E. Scatter plot depicts the correlation between ADAR1p150 and CTSS expression levels in PBMCS of SLE patients (n = 25). F. Tukey box plots show editing rate of 3 individual nucleotides located in the ‘hotspot’ region of CTSS AluSx+ in PBMCS derived from SLE patients vs control subjects. G,H. Scatter plots depict the relationship between ADAR1p150 (G) or CTSS (H) expression levels with editing rate of 3 individual adenosines located in CTSS AluSx+ ‘hotspot’ in PBMCS derived from SLE patients. Individual expression levels in PBMCs in all graphs are plotted as fold-change vs HC median. Bar graphs represent mean and SEM. Pairwise comparisons were performed by Mann-Whitney U test or independent samples t-test. Correlation coefficients r were derived by Pearson's correlation coefficient test.
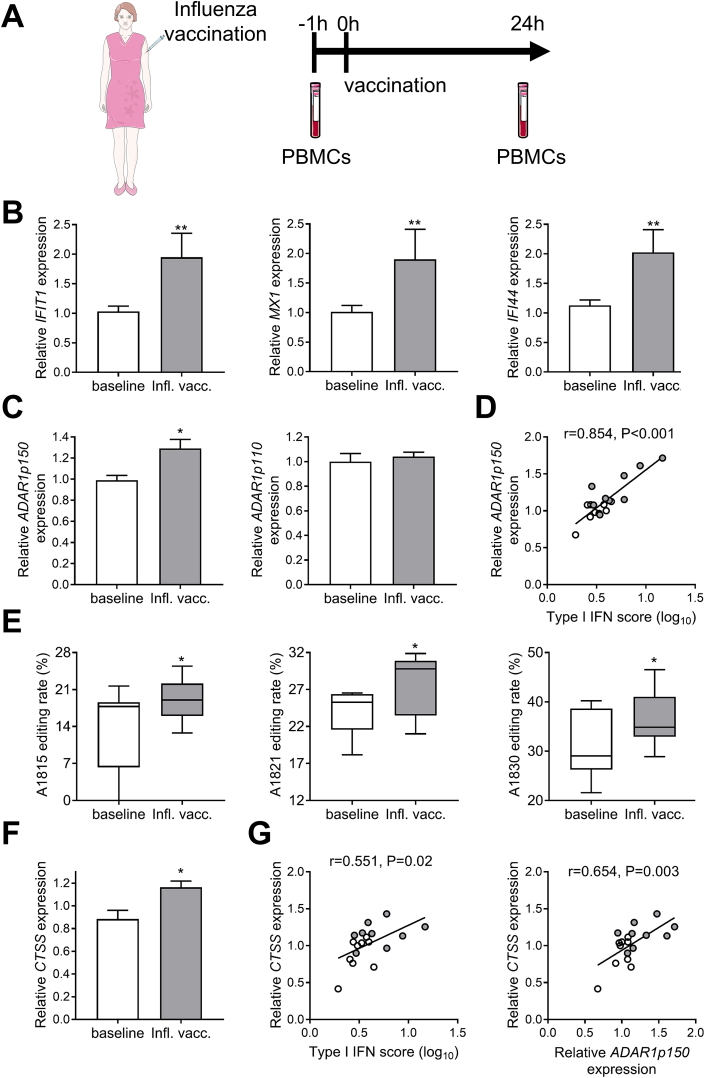
Finally, we examined healthy individuals who were vaccinated against influenza (Fig. 7A), as this has been previously shown to induce a specific and transient activation of the type I IFN system in human leukocytes [26]. Indeed, we validated the upregulation of 3 genes selectively induced by type I IFN 24 h post-vaccination (Fig. 7B). Of interest, we observed that this acute immune activation led to an increase in ADAR1p150 levels (Fig. 7C, left panel), while it did not affect ADAR1p110 (Fig. 7C, right panel). ADAR1p150 levels showed a strong positive correlation with individual type I IFN scores (r = 0.854, P < 0.001; Fig. 7D). The enzymatic activity of ADAR1, as measured by A-to-I RNA editing of adenosine residues in CTSS AluSx+ 'hotspot', was also significantly upregulated (Fig. 7E). Finally, CTSS expression was increased after influenza vaccination (Fig. 7F) and correlated with type I IFN score (r = 0.551, P = 0.02; Fig. 7G, left panel), ADAR1p150 (r = 0.654, P = 0.003; Fig. 7G, right panel) and RNA editing rate (after inluenza vaccination, A1825: r = 0.698, P = 0.037).
Fig. 7.
Induction of type I IFN by influenza vaccination recapitulates the observed ADAR1-mediated increase inCTSSexpression. A. Schematic representation of the study design. PBMCs were isolated from 9 healthy individuals before vaccination and 24 h after vaccination against influenza. B. Relative expression of 3 type I IFN-stimulated genes IFIT1 (left panel), MX1 (middle panel) and IFI44 (right panel) in PBMCs as quantified by RT-qPCR. C. Relative expression of the two ADAR1 isoforms ADAR1p150 (left panel) and ADAR1p110 (right panel) as quantified by TaqMan expression assays. D. Scatter plot showing the relationship between type I IFN score and ADAR1p150 expression levels. E. RNA editing rate of individual adenosine residues in CTSS AluSx+ ‘hotspot’ region. F. Relative expression levels of CTSS as quantified by RT-qPCR G. Scatter plots show the relationship between type I IFN score (left panel) or ADAR1p150 (right panel) and CTSS expression levels in PBMCs. Bar graphs represent mean and SEM. Individual expression levels in all graphs are plotted as fold-change vs HC median. In scatter plots shown in D and G, white points represent individuals before influenza vaccination and gray points the same individuals 24 h after influenza vaccination. Paired comparisons were performed by Wilcoxon signed-rank test or paired t-test. Correlation coefficients r were derived by Pearson's correlation coefficient test.
4. Discussion
The main findings of our study are that: 1) ADAR1 is increased in the skin and PBMCs of SSc patients in association with a prominent type I IFN signature; 2) the ADAR1-target CTSS, a protease involved in antigen presentation and extracellular matrix remodeling, is also increased in skin and peripheral blood of SSc patients; 3) increased A-to-I RNA editing rate of Alu elements in CTSS 3′ UTR is associated with increased target gene (CTSS) expression; 4) similar results are obtained in SLE patients suggesting a mechanism shared among autoimmune patients with chronic type I IFN pathway activation; 5) acute type I IFN pathway activation by influenza vaccination recapitulates the transcriptional alterations observed in patients with SSc and SLE; and 6) silencing of ADAR1 significantly dampens the type I IFN-induced CTSS upregulation, suggesting a predominant role of post-transcriptional gene regulation by RNA editing.
Deregulation of the type I IFN pathway has been consistently reported in peripheral blood cells and target-tissues of patients with SSc [[2], [3], [4], [5], [6], [7], [8], [9], [10]]. Moreover, activation of the type I IFN pathway has been associated with higher disease severity including extent of both skin and lung involvement, and presence of pulmonary hypertension [6,7,44,45], while baseline type I IFN levels were also recently shown to have a predictive value for response to immunosuppressive treatment (cyclophosphamide, mycophenolate mofetil) in SSc-related interstitial lung disease [46]. More importantly, SSc patients treated with IFN-α in an early study showed more rapid lung function decline [11], while a monoclonal antibody against the common type I IFN receptor, IFNAR, showed promising results regarding T cell activation and collagen accumulation in patients’ skin [13]. However, mechanistic links between increased type I IFN and inflammation/fibrosis in SSc, extending beyond its direct transcriptional effects of interferon-inducible genes, are very limited [10,47].
ADAR1-induced RNA editing is a widespread RNA modification, which can affect literally every aspect of RNA metabolism and thus comprises a critical post-transcriptional regulator of gene expression [16,17]. ADAR1 is indispensable for life as shown by the in utero lethality of ADAR1-knockout mice [[48], [49], [50], [51]], while in humans mutations in the catalytic region of ADAR1 have been causatively linked to Aicardi-Goutières syndrome [52]. While lack of ADAR1 induces an aberrant immune response partly mediated by type I IFN [50,51], overexpression of ADAR1 may also exert proinflammatory effects [21,23], underlying the need for a balanced ADAR1 expression and activity. ADAR1, and more specifically the long ADAR1p150 isoform, has been found increased in chronic inflammatory disorders such as cancer [53], atherosclerosis [21], SLE [18,54] and rheumatoid arthritis [23]. In line with these previous studies, we show herein that ADAR1 is increased in both the skin and peripheral blood of SSc patients, showing a strong association with individual type I IFN scores.
The enzymatic activity of ADAR1, namely A-to-I RNA editing, was increased in the Alu elements located in 3′ UTR of CTSS, a well-established target of ADAR1 [21] with central role in MHC-II antigen loading and production of autoantibodies [43]. In line with our results, a previous report by Roth and colleagues showed that Alu editing index, the weighted mean editing rate of millions adenosine residues located in Alu elements [55], was increased in the blood of patients with SLE, especially in those with high type I IFN score [18]. Similarly, we observed an association between type I IFN signature, ADAR1p150 and RNA editing of individual adenosines in CTSS AluSx+. Of note, vaccination of healthy individuals against influenza, which has been previously shown to induce an acute type I IFN upregulation [26], recapitulated the transcriptomic alterations observed in SSc and SLE patients. In our study both chronic activation of type I IFN responses, as observed in SSc and SLE patients, and acute induction of a systematic type I IFN response, as observed in healthy individuals 24 h after influenza vaccination, led to the upregulation of ADAR1p150-mediated A-to-I RNA editing and increased CTSS expression. However, we must acknowledge that an acute response can be different from chronic interferon signaling, as shown in the context of viral infection [56,57]. Future studies examining how A-to-I RNA editing contributes to regulation of host antiviral gene expression during acute and chronic induction of interferon responses are warranted.
Finally, we showed that A-to-I RNA editing rate of individual adenosines is strongly associated with the expression of the target gene (CTSS). However, CTSS expression seems to be lower in SLE patients compared to SSc patients despite higher IFN scores, ADAR1p150 and RNA editing rates. In line with this, the RNA editing rate of only 3 out of 13 adenosine residues in the CTSS AluSx+ ‘hotspot’ correlated with CTSS expression in SLE patients, in contrast with 9 out of 13 in SSc patients. As we have previously shown, adenosine-to-inosine RNA editing of AluSx+ in CTSS is integral for the recruitment of the stabilizing single-stranded RNA-binding protein HuR to its target motifs in CTSS AluSx+ [21]. However, a number of factors may affect final CTSS transcript levels including: 1) a potential competitive binding of other, destabilizing RBPs on AU-rich elements in SLE which could restrict HuR binding and transcript stabilization [58]; 2) a possible deregulation of other RBPs that may interfere with CTSS mRNA stability; and/or 3) the potential presence of other concomitant post-transcriptional regulatory mechanisms, such as SLE-induced microRNAs [59] that target CTSS could also account for the relatively low CTSS expression levels despite the significantly increased RNA editing rates of CTSS adenosine residues in SLE (10 out of 13 ‘hotspot’ adenosines were found to be significantly increased in SLE compared to controls).
More importantly, herein we showed that silencing of ADAR1 reduced the type IFN-induced upregulation of CTSS by 60-70%, suggesting that the contribution of RNA editing in regulation of CTSS expression outweighs the potential direct transcriptional effects. We have previously shown with a series of mechanistic studies that RNA editing of CTSS Alu elements results in the unwinding of local dsRNA structure into a more single-stranded conformation facilitating the binding of the stabilizing RNA-binding protein HuR [21]. In line with this, we observed herein a strong association between CTSS expression and editing rate of individual adenosines located in close proximity with HuR binding sites in CTSS 3′ UTR AluSx+. Of note, this association was present only in patients' cells, suggesting that RNA editing may act as a disease-specific mechanism of post-transcriptional gene regulation. CTSS has been linked with systemic autoimmune responses, as inhibition of CTSS has been shown to reduce autoantibody production and prevent tissue damage in animal models of Sjogren's syndrome and SLE/SLE nephritis [[60], [61], [62]]. On the other hand, CTSS overexpressing transgenic mice developed lupus-like phenotype associated with increased type I IFNs [63]. Increased expression and/or activity of CTSS has been detected in chronic inflammatory conditions, including obesity [64], atherosclerotic cardiovascular disease [21,65], psoriasis [66], Sjogren's Syndrome [67], idiopathic pulmonary fibrosis, pulmonary arterial hypertension and COPD [68]. In our study, we observed increased CTSS expression levels both in PBMCs and skin biopsies derived from patients with SSc as well as in PBMCs derived from SLE patients and from subjects after influenza vaccination. Further, we describe a positive association of CTSS expression levels with extent of skin fibrosis (mRSS and local skin score), as well as with digital ulcers. We must acknowledge that our study population included mainly patients at early stages of disease (median disease duration: 2 years) and mostly patients with limited SSc. Therefore, our findings may not essentially be extrapolated into patients with diffuse SSc. Indeed, a single previous study reported decreased cathepsin S serum levels in a small cohort of patients with diffuse SSc and normal kidney function [69]. Future studies are called to investigate the role of CTSS in SSc development as well as the involvement of RNA editing of CTSS and other ADAR1 targets in early vs. late stages of SSc, as well as in SSc disease subtypes (limited vs. diffuse).
Of interest, we have previously shown that A-to-I editing events were often located in close proximity to HuR binding motifs [21]. Whole transcriptome analysis revealed that A-to-I edited transcripts had a greater number of HuR binding motifs, especially in the neighboring up- and downstream 100 nucleotide-sequences of each edited site [21]. These results suggest that ADAR1-induced RNA editing may control the stability of several RNA transcripts through alteration of HuR binding [21]. Moreover, a recent study in THP-1 monocytes showed an enrichment of HuR binding in 3′ UTR sites of interferon responsive genes upon IRF3 stimulation compared to unstimulated cells [70]. Whether RNA editing could account for this shift to 3′ UTR binding of HuR through alteration of local structure remains to be elucidated. Future transcriptome-wide, but also gene-specific, studies are warranted to determine the contribution of ADAR1-induced RNA editing in shaping the transcriptome of patients with systemic autoimmune diseases.
5. Conclusions
In conclusion, our results support the role of A-to-I RNA editing as an additional, post-transcriptional level of proinflammatory gene regulation in SSc, which is shared among patients with systemic autoimmunity.
Credit author statement
Nikolaos I. Vlachogiannis: Methodology, Investigation, Formal analysis, Writing – original draft, Validation, Visualization; Simon Tual-Chalot: Investigation, Formal analysis, Validation; Eleftherios Zormpas: Software, Formal analysis; Francesca Bonini: Investigation, Formal analysis; Panagiotis A. Ntouros: Investigation; Maria Pappa: Investigation; Vasiliki-Kalliopi Bournia: Investigation; Maria G. Tektonidou: Resources; Vassilis L. Souliotis: Resources; Clio P. Mavragani: Resources; Kimon Stamatelopoulos: Validation; Aikaterini Gatsiou: Methodology, Validation, Formal analysis, Writing – review & editing, Supervision; Petros P. Sfikakis: Resources, Supervision, Writing – review & editing, Funding acquisition; Konstantinos Stellos: Conceptualization, Validation, Formal analysis, Resources, Supervision, Writing – review & editing, Project administration, Funding acquisition.
Declaration of competing interest
None.
Acknowledgements
This study was supported by the German Research Foundation DFG (SFB834 B12, project number 75732319) and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (MODVASC, grant agreement No 759248). NIV was supported with a scholarship from the Alexander S. Onassis Public Benefit Foundation during his doctoral studies, which covered part of the presented work. PPS is supported by educational grants (ELKE - National and Kapodistrian University of Athens, Greece; Nr 0974). The authors thank Dr Adrianos Nezos for technical assistance. Fig. 7 and Graphical abstract contain items that have been adapted from Servier Medical Art by Servier (https://smart.servier.com – licensed under Creative Commons Attribution 3.0 Unported License).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaut.2021.102755.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gabrielli A., Avvedimento E.V., Krieg T. Scleroderma. N. Engl. J. Med. 2009;360:1989–2003. doi: 10.1056/NEJMra0806188. [DOI] [PubMed] [Google Scholar]
- 2.Higgs B.W., Liu Z., White B., Zhu W., White W.I., Morehouse C., Brohawn P., Kiener P.A., Richman L., Fiorentino D., Greenberg S.A., Jallal B., Yao Y. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann. Rheum. Dis. 2011;70:2029–2036. doi: 10.1136/ard.2011.150326. [DOI] [PubMed] [Google Scholar]
- 3.Assassi S., Mayes M.D., Arnett F.C., Gourh P., Agarwal S.K., McNearney T.A., Chaussabel D., Oommen N., Fischbach M., Shah K.R., Charles J., Pascual V., Reveille J.D., Tan F.K. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum. 2010;62:589–598. doi: 10.1002/art.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barturen G., Babaei S., Català-Moll F., Martínez-Bueno M., Makowska Z., Martorell-Marugán J., Carmona-Sáez P., Toro-Domínguez D., Carnero-Montoro E., Teruel M., Kerick M., Acosta-Herrera M., Le Lann L., Jamin C., Rodríguez-Ubreva J., García-Gómez A., Kageyama J., Buttgereit A., Hayat S., Mueller J., Lesche R., Hernandez-Fuentes M., Juarez M., Rowley T., White I., Marañón C., Gomes Anjos T., Varela N., Aguilar-Quesada R., Garrancho F.J., López-Berrio A., Rodriguez Maresca M., Navarro-Linares H., Almeida I., Azevedo N., Brandão M., Campar A., Faria R., Farinha F., Marinho A., Neves E., Tavares A., Vasconcelos C., Trombetta E., Montanelli G., Vigone B., Alvarez-Errico D., Li T., Thiagaran D., Blanco Alonso R., Corrales Martínez A., Genre F., López Mejías R., Gonzalez-Gay M.A., Remuzgo S., Ubilla Garcia B., Cervera R., Espinosa G., Rodríguez-Pintó I., De Langhe E., Cremer J., Lories R., Belz D., Hunzelmann N., Baerlecken N., Kniesch K., Witte T., Lehner M., Stummvoll G., Zauner M., Aguirre-Zamorano M.A., Barbarroja N., Castro-Villegas M.C., Collantes-Estevez E., de Ramon E., Díaz Quintero I., Escudero-Contreras A., Fernández Roldán M.C., Jiménez Gómez Y., Jiménez Moleón I., Lopez-Pedrera R., Ortega-Castro R., Ortego N., Raya E., Artusi C., Gerosa M., Meroni P.L., Schioppo T., De Groof A., Ducreux J., Lauwerys B., Maudoux A.-L., Cornec D., Devauchelle-Pensec V., Jousse-Joulin S., Jouve P.-E., Rouvière B., Saraux A., Simon Q., Alvarez M., Chizzolini C., Dufour A., Wynar D., Balog A., Bocskai M., Deák M., Dulic S., Kádár G., Kovács L., Cheng Q., Gerl V., Hiepe F., Khodadadi L., Thiel S., de Rinaldis E., Rao S., Benschop R.J., Chamberlain C., Dow E.R., Ioannou Y., Laigle L., Marovac J., Wojcik J., Renaudineau Y., Borghi M.O., Frostegård J., Martín J., Beretta L., Ballestar E., McDonald F., Pers J.-O., Alarcón-Riquelme M.E. Integrative analysis reveals a molecular stratification of systemic autoimmune diseases. Arthritis Rheum. 2021;73:1073–1085. doi: 10.1002/art.41610. [DOI] [PubMed] [Google Scholar]
- 5.Farina G., Lafyatis D., Lemaire R., Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2010;62:580–588. doi: 10.1002/art.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice L.M., Ziemek J., Stratton E.A., McLaughlin S.R., Padilla C.M., Mathes A.L., Christmann R.B., Stifano G., Browning J.L., Whitfield M.L., Spiera R.F., Gordon J.K., Simms R.W., Zhang Y., Lafyatis R. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2015;67:3004–3015. doi: 10.1002/art.39287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christmann R.B., Sampaio-Barros P., Stifano G., Borges C.L., de Carvalho C.R., Kairalla R., Parra E.R., Spira A., Simms R., Capellozzi V.L., Lafyatis R. Association of interferon- and transforming growth factor β-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis: gene expression in SSc-related progressive lung fibrosis. Arthritis Rheum. 2014;66:714–725. doi: 10.1002/art.38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eloranta M.-L., Franck-Larsson K., Lovgren T., Kalamajski S., Ronnblom A., Rubin K., Alm G.V., Ronnblom L. Type I interferon system activation and association with disease manifestations in systemic sclerosis. Ann. Rheum. Dis. 2010;69:1396–1402. doi: 10.1136/ard.2009.121400. [DOI] [PubMed] [Google Scholar]
- 9.Brkic Z., van Bon L., Cossu M., van Helden-Meeuwsen C.G., Vonk M.C., Knaapen H., van den Berg W., Dalm V.A., Van Daele P.L., Severino A., Maria N.I., Guillen S., Dik W.A., Beretta L., Versnel M.A., Radstake T. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann. Rheum. Dis. 2016;75:1567–1573. doi: 10.1136/annrheumdis-2015-207392. [DOI] [PubMed] [Google Scholar]
- 10.Skaug B., Assassi S. Type I interferon dysregulation in systemic sclerosis. Cytokine. 2020;132:154635. doi: 10.1016/j.cyto.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Black C.M., Silman A.J., Herrick A.I., Denton C.P., Wilson H., Newman J., Pompon L., Shi-Wen X. Interferon-alpha does not improve outcome at one year in patients with diffuse cutaneous scleroderma: results of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 1999;42:299–305. doi: 10.1002/1529-0131(199902)42:2<299::AID-ANR12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg A., Geppert T., Schiopu E., Frech T., Hsu V., Simms R.W., Peng S.L., Yao Y., Elgeioushi N., Chang L. others, Dose-escalation of human anti-interferon-alpha receptor monoclonal antibody MEDI-546 in subjects with systemic sclerosis: a phase 1, multicenter, open label study. Arthritis Res. Ther. 2014;16:R57. doi: 10.1186/ar4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X., Higgs B.W., Bay-Jensen A.C., Karsdal M.A., Yao Y., Roskos L.K., White W.I. Suppression of T Cell activation and collagen accumulation by an anti-IFNAR1 mAb, anifrolumab, in adult patients with systemic sclerosis. J. Invest. Dermatol. 2015;135:2402–2409. doi: 10.1038/jid.2015.188. [DOI] [PubMed] [Google Scholar]
- 14.Muskardin T.L.W., Niewold T.B. Type I interferon in rheumatic diseases. Nat. Rev. Rheumatol. 2018;14:214–228. doi: 10.1038/nrrheum.2018.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatsiou A., Vlachogiannis N., Lunella F.F., Sachse M., Stellos K. Adenosine-to-Inosine RNA editing in health and disease. Antioxid Redox Signal. 2018;29:846–863. doi: 10.1089/ars.2017.7295. [DOI] [PubMed] [Google Scholar]
- 18.Roth S.H., Danan-Gotthold M., Ben-Izhak M., Rechavi G., Cohen C.J., Louzoun Y., Levanon E.Y. Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus. Cell Rep. 2018;23:50–57. doi: 10.1016/j.celrep.2018.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shallev L., Kopel E., Feiglin A., Leichner G.S., Avni D., Sidi Y., Eisenberg E., Barzilai A., Levanon E.Y., Greenberger S. Decreased A-to-I RNA editing as a source of keratinocytes' dsRNA in psoriasis. RNA. 2018;24:828–840. doi: 10.1261/rna.064659.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jaé N., Rossbach O., Amrhein C., Sigala F., Boon R.A., Fürtig B., Manavski Y., You X., Uchida S., Keller T., Boeckel J.-N., Franco-Cereceda A., Maegdefessel L., Chen W., Schwalbe H., Bindereif A., Eriksson P., Hedin U., Zeiher A.M., Dimmeler S. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 22.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 23.Vlachogiannis N.I., Gatsiou A., Silvestris D.A., Stamatelopoulos K., Tektonidou M.G., Gallo A., Sfikakis P.P., Stellos K. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J. Autoimmun. 2020;106:102329. doi: 10.1016/j.jaut.2019.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Hoogen F., Khanna D., Fransen J., Johnson S.R., Baron M., Tyndall A., Matucci-Cerinic M., Naden R.P., Medsger T.A., Carreira P.E., Riemekasten G., Clements P.J., Denton C.P., Distler O., Allanore Y., Furst D.E., Gabrielli A., Mayes M.D., van Laar J.M., Seibold J.R., Czirjak L., Steen V.D., Inanc M., Kowal-Bielecka O., Müller-Ladner U., Valentini G., Veale D.J., Vonk M.C., Walker U.A., Chung L., Collier D.H., Ellen Csuka M., Fessler B.J., Guiducci S., Herrick A., Hsu V.M., Jimenez S., Kahaleh B., Merkel P.A., Sierakowski S., Silver R.M., Simms R.W., Varga J., Pope J.E. Classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 2013;72:1747–1755. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 25.Petri M., Orbai A.-M., Alarcón G.S., Gordon C., Merrill J.T., Fortin P.R., Bruce I.N., Isenberg D., Wallace D.J., Nived O., Sturfelt G., Ramsey-Goldman R., Bae S.-C., Hanly J.G., Sánchez-Guerrero J., Clarke A., Aranow C., Manzi S., Urowitz M., Gladman D., Kalunian K., Costner M., Werth V.P., Zoma A., Bernatsky S., Ruiz-Irastorza G., Khamashta M.A., Jacobsen S., Buyon J.P., Maddison P., Dooley M.A., van Vollenhoven R.F., Ginzler E., Stoll T., Peschken C., Jorizzo J.L., Callen J.P., Lim S.S., Fessler B.J., Inanc M., Kamen D.L., Rahman A., Steinsson K., Franks A.G., Sigler L., Hameed S., Fang H., Pham N., Brey R., Weisman M.H., McGwin G., Magder L.S. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–2686. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obermoser G., Presnell S., Domico K., Xu H., Wang Y., Anguiano E., Thompson-Snipes L., Ranganathan R., Zeitner B., Bjork A., Anderson D., Speake C., Ruchaud E., Skinner J., Alsina L., Sharma M., Dutartre H., Cepika A., Israelsson E., Nguyen P., Nguyen Q.-A., Harrod A.C., Zurawski S.M., Pascual V., Ueno H., Nepom G.T., Quinn C., Blankenship D., Palucka K., Banchereau J., Chaussabel D. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlachogiannis N.I., Nezos A., Tzioufas A.G., Koutsilieris M., Moutsopoulos H.M., Mavragani C.P. Increased frequency of the PTPN22W* variant in primary Sjogren's Syndrome: association with low type I IFN scores. Clin. Immunol. 2016;173:157–160. doi: 10.1016/j.clim.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Vlachogiannis N.I., Pappa M., Ntouros P.A., Nezos A., Mavragani C.P., Souliotis V.L., Sfikakis P.P. Association between DNA damage response, fibrosis and type I interferon signature in systemic sclerosis. Front. Immunol. 2020;11:582401. doi: 10.3389/fimmu.2020.582401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nezos A., Gravani F., Tassidou A., Kapsogeorgou E.K., Voulgarelis M., Koutsilieris M., Crow M.K., Mavragani C.P. Type I and II interferon signatures in Sjogren's syndrome pathogenesis: contributions in distinct clinical phenotypes and Sjogren's related lymphomagenesis. J. Autoimmun. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlachogiannis N.I., Sachse M., Georgiopoulos G., Zormpas E., Bampatsias D., Delialis D., Bonini F., Galyfos G., Sigala F., Stamatelopoulos K., Gatsiou A., Stellos K. Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease. J. Mol. Cell. Cardiol. 2021;160:111–120. doi: 10.1016/j.yjmcc.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skaug B., Khanna D., Swindell W.R., Hinchcliff M.E., Frech T.M., Steen V.D., Hant F.N., Gordon J.K., Shah A.A., Zhu L., Zheng W.J., Browning J.L., Barron A.M.S., Wu M., Visvanathan S., Baum P., Franks J.M., Whitfield M.L., Shanmugam V.K., Domsic R.T., Castelino F.V., Bernstein E.J., Wareing N., Lyons M.A., Ying J., Charles J., Mayes M.D., Assassi S. Global skin gene expression analysis of early diffuse cutaneous systemic sclerosis shows a prominent innate and adaptive inflammatory profile. Ann. Rheum. Dis. 2020;79:379–386. doi: 10.1136/annrheumdis-2019-215894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pachera E., Assassi S., Salazar G.A., Stellato M., Renoux F., Wunderlin A., Blyszczuk P., Lafyatis R., Kurreeman F., de Vries-Bouwstra J., Messemaker T., Feghali-Bostwick C.A., Rogler G., van Haaften W.T., Dijkstra G., Oakley F., Calcagni M., Schniering J., Maurer B., Distler J.H., Kania G., Frank-Bertoncelj M., Distler O. Long noncoding RNA H19X is a key mediator of TGF-β-driven fibrosis. J. Clin. Invest. 2020;130:4888–4905. doi: 10.1172/JCI135439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krueger F. Trim Galore. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 34.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 35.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., Billis K., Boddu S., Marugán J.C., Cummins C., Davidson C., Dodiya K., Fatima R., Gall A., Giron C.G., Gil L., Grego T., Haggerty L., Haskell E., Hourlier T., Izuogu O.G., Janacek S.H., Juettemann T., Kay M., Lavidas I., Le T., Lemos D., Martinez J.G., Maurel T., McDowall M., McMahon A., Mohanan S., Moore B., Nuhn M., Oheh D.N., Parker A., Parton A., Patricio M., Sakthivel M.P., Abdul Salam A.I., Schmitt B.M., Schuilenburg H., Sheppard D., Sycheva M., Szuba M., Taylor K., Thormann A., Threadgold G., Vullo A., Walts B., Winterbottom A., Zadissa A., Chakiachvili M., Flint B., Frankish A., Hunt S.E., IIsley G., Kostadima M., Langridge N., Loveland J.E., Martin F.J., Morales J., Mudge J.M., Muffato M., Perry E., Ruffier M., Trevanion S.J., Cunningham F., Howe K.L., Zerbino D.R., Flicek P. Ensembl. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithm Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George C.X., Samuel C.E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson J.B., Samuel C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell Biol. 1995;15:5376–5388. doi: 10.1128/MCB.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riese R.J., Wolf P.R., Brömme D., Natkin L.R., Villadangos J.A., Ploegh H.L., Chapman H.A. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Mayes M.D., Tan F.K., Wu M., Reveille J.D., Harper B.E., Draeger H.T., Gonzalez E.B., Assassi S. Correlation of interferon-inducible chemokine plasma levels with disease severity in systemic sclerosis. Arthritis Rheum. 2013;65:226–235. doi: 10.1002/art.37742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christmann R.B., Hayes E., Pendergrass S., Padilla C., Farina G., Affandi A.J., Whitfield M.L., Farber H.W., Lafyatis R. Interferon and alternative activation of monocyte/macrophages in systemic sclerosis-associated pulmonary arterial hypertension. Arthritis Rheum. 2011;63:1718–1728. doi: 10.1002/art.30318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Assassi S., Li N., Volkmann E.R., Mayes M.D., Rünger D., Ying J., Roth M.D., Hinchcliff M., Khanna D., Frech T., Clements P.J., Furst D.E., Goldin J., Bernstein E.J., Castelino F.V., Domsic R.T., Gordon J.K., Hant F.N., Shah A.A., Shanmugam V.K., Steen V.D., Elashoff R.M., Tashkin D.P. Predictive significance of serum interferon-inducible protein score for response to treatment in systemic sclerosis-related interstitial lung disease. Arthritis Rheum. 2021;73:1005–1013. doi: 10.1002/art.41627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M., Skaug B., Bi X., Mills T., Salazar G., Zhou X., Reveille J., Agarwal S.K., Blackburn M.R., Mayes M.D., Assassi S. Interferon regulatory factor 7 (IRF7) represents a link between inflammation and fibrosis in the pathogenesis of systemic sclerosis. Ann. Rheum. Dis. 2019;78:1583–1591. doi: 10.1136/annrheumdis-2019-215208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2003;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 49.Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C.P., McLaughlin P.J., Jantsch M.F., Dorin J., Adams I.R., Scadden A.D.J., Ohman M., Keegan L.P., O'Connell M.A. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pestal K., Funk C.C., Snyder J.M., Price N.D., Treuting P.M., Stetson D.B. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43:933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice G.I., Kasher P.R., Forte G.M.A., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A., Jenkinson E.M., Bacino C.A., Battini R., Bertini E., Brogan P.A., Brueton L.A., Carpanelli M., De Laet C., de Lonlay P., del Toro M., Desguerre I., Fazzi E., Garcia-Cazorla À., Heiberg A., Kawaguchi M., Kumar R., Lin J.-P.S.-M., Lourenco C.M., Male A.M., Marques W., Mignot C., Olivieri I., Orcesi S., Prabhakar P., Rasmussen M., Robinson R.A., Rozenberg F., Schmidt J.L., Steindl K., Tan T.Y., van der Merwe W.G., Vanderver A., Vassallo G., Wakeling E.L., Wassmer E., Whittaker E., Livingston J.H., Lebon P., Suzuki T., McLaughlin P.J., Keegan L.P., O'Connell M.A., Lovell S.C., Crow Y.J. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., Levanon E.Y. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13:267–276. doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- 54.Quinones-Valdez G., Tran S.S., Jun H.-I., Bahn J.H., Yang E.-W., Zhan L., Brümmer A., Wei X., Van Nostrand E.L., Pratt G.A., Yeo G.W., Graveley B.R., Xiao X. Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2019;2:1–14. doi: 10.1038/s42003-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roth S.H., Levanon E.Y., Eisenberg E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods. 2019;16:1131–1138. doi: 10.1038/s41592-019-0610-9. [DOI] [PubMed] [Google Scholar]
- 56.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murira A., Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front. Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otsuka H., Fukao A., Funakami Y., Duncan K.E., Fujiwara T. Emerging evidence of translational control by AU-rich element-binding proteins. Front. Genet. 2019;10:332. doi: 10.3389/fgene.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srikantan S., Tominaga K., Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saegusa K., Ishimaru N., Yanagi K., Arakaki R., Ogawa K., Saito I., Katunuma N., Hayashi Y. Cathepsin S inhibitor prevents autoantigen presentation and autoimmunity. J. Clin. Invest. 2002;110:361–369. doi: 10.1172/JCI200214682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rupanagudi K.V., Kulkarni O.P., Lichtnekert J., Darisipudi M.N., Mulay S.R., Schott B., Gruner S., Haap W., Hartmann G., Anders H.-J. Cathepsin S inhibition suppresses systemic lupus erythematosus and lupus nephritis because cathepsin S is essential for MHC class II-mediated CD4 T cell and B cell priming. Ann. Rheum. Dis. 2015;74:452–463. doi: 10.1136/annrheumdis-2013-203717. [DOI] [PubMed] [Google Scholar]
- 62.Kim S.J., Schätzle S., Ahmed S.S., Haap W., Jang S.-H., Gregersen P.K., Georgiou G., Diamond B. Increased Cathepsin S in Prdm1−/− dendritic cells alters TFH repertoire and contributes to lupus. Nat. Immunol. 2017;18:1016–1024. doi: 10.1038/ni.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J., Jang S., Choi M., Kang M., Lim S.-G., Kim S.-Y., Jang S., Ko J., Kim E., Yi J., Choo Y., Kim M.O., Ryoo Z.Y. Overexpression of cathepsin S exacerbates lupus pathogenesis through upregulation TLR7 and IFN-α in transgenic mice. Sci. Rep. 2021;11:16348. doi: 10.1038/s41598-021-94855-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naour N., Rouault C., Fellahi S., Lavoie M.-E., Poitou C., Keophiphath M., Eberlé D., Shoelson S., Rizkalla S., Bastard J.-P., Rabasa-Lhoret R., Clément K., Guerre-Millo M. Cathepsins in human obesity: changes in energy balance predominantly affect cathepsin s in adipose tissue and in circulation. J. Clin. Endocrinol. Metab. 2010;95:1861–1868. doi: 10.1210/jc.2009-1894. [DOI] [PubMed] [Google Scholar]
- 65.Liu C.-L., Guo J., Zhang X., Sukhova G.K., Libby P., Shi G.-P. Cysteine protease cathepsins in cardiovascular disease: from basic research to clinical trials. Nat. Rev. Cardiol. 2018;15:351–370. doi: 10.1038/s41569-018-0002-3. [DOI] [PubMed] [Google Scholar]
- 66.Ainscough J.S., Macleod T., McGonagle D., Brakefield R., Baron J.M., Alase A., Wittmann M., Stacey M. Cathepsin S is the major activator of the psoriasis-associated proinflammatory cytokine IL-36γ. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E2748–E2757. doi: 10.1073/pnas.1620954114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamm-Alvarez S.F., Janga S.R., Edman M.C., Madrigal S., Shah M., Frousiakis S.E., Renduchintala K., Zhu J., Bricel S., Silka K., Bach D., Heur M., Christianakis S., Arkfeld D.G., Irvine J., Mack W.J., Stohl W. Tear cathepsin S as a candidate biomarker for sjögren’s syndrome: tear CTSS as a biomarker for SS. Arthritis Rheum. 2014;66:1872–1881. doi: 10.1002/art.38633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown R., Nath S., Lora A., Samaha G., Elgamal Z., Kaiser R., Taggart C., Weldon S., Geraghty P., Cathepsin S. Investigating an old player in lung disease pathogenesis, comorbidities, and potential therapeutics. Respir. Res. 2020;21:111. doi: 10.1186/s12931-020-01381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toyama S., Yamashita T., Saigusa R., Miura S., Nakamura K., Hirabayashi M., Miyagawa T., Fukui Y., Omatsu J., Yoshizaki A., Sato S., Asano Y. Decreased serum cathepsin S levels in patients with systemic sclerosis-associated interstitial lung disease. J. Dermatol. 2020;47:1027–1032. doi: 10.1111/1346-8138.15458. [DOI] [PubMed] [Google Scholar]
- 70.Rothamel K., Arcos S., Kim B., Reasoner C., Lisy S., Mukherjee N., Ascano M. ELAVL1 primarily couples mRNA stability with the 3’ UTRs of interferon-stimulated genes. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.