Graphical abstract
Keywords: Human airway metagenome, Rare species, Graph kernels, Random forest, Microbiome development
Highlights
-
•
Core and rare airway bacteria form a stable background network in healthy children.
-
•
Protein sequence usage is complementary between core and rare taxa.
-
•
Rare taxa are the key components to differentiate a healthy from a CF metagenome.
-
•
In the early CF airways, the bacterial background network is underdeveloped.
-
•
CF network robustness increases by inserting a key combination of healthy taxa.
Abstract
The default removal of low-abundance (rare) taxa from microbial community analyses may lead to an incomplete picture of the taxonomic and functional microbial potential within the human habitat. Publicly available shotgun metagenomics data of healthy children and children with cystic fibrosis (CF) were reanalysed to study the development of the rare species biosphere, which was here defined by either the 15th, 25th or 35th species abundance percentile. We found that healthy children contained an age-independent network of abundant (core) and rare species with both entities being essential in maintaining the network structure. The protein sequence usage for more than 100 bacterial metabolic pathways differed between the core and rare species biosphere. In CF children, the background structure was underdeveloped and random forest bootstrapping based on all constituents of the early airway metagenome and host-associated factors indicated that rare taxa were the most important variables in deciding whether a child was healthy or suffered from the life-limiting CF disease. Attempts failed to make the age-independent CF network as robust as the healthy structure when an increasing number of bacterial taxa from the healthy network was incorporated into the CF structure by computer-based model simulations. However, the transfer of a key combination of taxa from the healthy to the CF network structure with high species diversity and low species dominance, correlated with a more robust CF network and a topological approximation of CF and healthy graph structures. Rothia mucilaginosa, Streptococci and rare species were essential in improving the underdeveloped CF network.
1. Introduction
In the last two decades and with the fast development of high-throughput sequencing technologies, a large number of studies gave evidence that both the human and microbial gene repertoires are essential in shaping human health and disease [1], [2], [3], [4]. But despite the enormous growth of knowledge in this research area, it remains challenging to define the microbial features that universally characterise a healthy human microbiome to date [5], [6]. One reason for this lasting uncertainty is that no two humans harbour the same microbial community signature in terms of species composition and abundance pattern. The large inter-subject variability is, among other factors, influenced by age, environment, genetics, lifestyle, socioeconomic status, ethnicity, and geography [7], [8], [9]. Another reason is that during the culture-independent taxonomic classification of microbes, publicly available reference-based alignment pipelines count the number of reads mapping to a reference sequence without considering the distribution of reads across the genome [10], [11], [12], [13], [14], [15]. As a consequence, the low-abundance taxa (rare species) are filtered out by default during the read alignment procedure or downstream data analysis. They cannot be distinguished from the high DNA background noise that is routinely observed with high-throughput sequencing applications [16], [17], [18]. By focusing on the abundant (core) species for which an acceptable genome coverage was obtained, pipelines can circumvent the low methodological-based signal-to-noise ratio and ensure a robust analysis of the core microbial communities inhabiting the environments of interest. However, the removal of rare taxa leads to an incomplete picture of the microbiome, because bacterial species within complex microbial communities follow a power law distribution with a higher number of rare species than core species [19], [20], [21]. Thus, neglecting the rare species biosphere leads to a loss of potentially disease- or health-associated variables before the community analysis has been initiated. Thereafter, data analyses and interpretations are limited to the small subsection of the core species biosphere.
In the past, a key role of rare species in providing the microbial community with functional flexibility and ecological stability has been described for various ecological habitats [20], [21], [22], [23], [24], [25]. We therefore assumed that rare species of healthy and diseased human airways in the early years of life occupy a similar pivotal role in their microbial community. With the objective to investigate the development of the rare species biosphere and unravel the yet unknown contribution of rare species to healthy or chronically diseased airway habitats, we re-analysed publicly available cross-sectional metagenome data obtained from deep cough swabs of 46 healthy children, 41 children with cystic fibrosis (CF) and negative controls [26]. Our recent software publication raspir in combination with gapseq, a tool introduced by Zimmermann et al. (2021) facilitated the taxonomic and functional identification of core and rare species from shotgun metagenomic sequencing data and reference genomes, respectively, with reduced false discovery and omission rates [27], [28]. Since previous reports have demonstrated that metagenome investigations are affected by the reference database of choice [29] and the normalisation strategy of count data for addressing the compositional behaviour of microbiome sequencing data [30], [31], [32], [33], we tested our model simulations, random forest bootstrapping aggregations, ecological network analysis and kernel-based machine learning applications on infant metagenome datasets, generated from read alignments towards either a pan-genome or a one-strain-per-species reference database. Moreover, we generated datasets based on three different read count normalisation strategies, namely variance-stabilising transformations (VST), relative log expression (RLE) and bacterial to human cell ratios (BCPHC) and worked with three distinct rarity thresholds (15th, 25th and 35th species abundance percentile) to define the core and rare species biosphere. This approach was essential for extracting the method-independent biological effects of the rare species biosphere on CF or healthy airway microbial communities in infancy, considering that the type of data normalisation may affect species abundance estimations and hence distort which species are classified as “core” or “rare” according to the 15th, 25th or 35th species abundance percentile.
Independent of the normalisation method, reference database of choice, and rarity threshold, we found that healthy children harboured an age-independent background network of core and rare bacteria, which were equally important in maintaining the network structure. In CF children, the airway metagenome was defined by non-persisting core and rare species that were only detected at a particular developmental stage. The presence or absence of rare species was found to be the key variable in differentiating between the healthy and CF airway metagenome in the early years of life.
2. Materials & methods
2.1. Data acquisition
We re-analysed our previously published shotgun metagenomic sequencing data obtained from deep cough swabs of 41 CF and 46 healthy children between zero and six years of age (ENA PRJEB38221) [26]. As previously reported, the clinical study was approved by the ethics committee of MHH (No. 7674) and the parents or legal guardians gave written consent prior to sample collection. Sampling was accompanied by an obligate cough of the participant. The samples were immediately stored at −80 °C until further processed with negative controls and sequenced on the Illumina NextSeq 500/550 platform to generate short (75 bp) single-end reads [26].
2.2. Data preparation and normalisation
The whole metagenome sequencing alignment pipeline (version 1.8.0) of Davenport and colleagues [34] was utilised for the removal of read duplicates and low-complexity reads as well as for read trimming and alignments towards a pan-genome or a one-strain-per-species reference database with the Burrows-Wheeler Aligner [35]. For each sample, the rare species identifier tool (raspir, version 1.0.2) [27] was run to filter out microbial species with non-uniform read distributions towards the reference genome. Raw read counts were normalised to human reads (bacterial cell per human cell, BCPHC), in which case the length of the diploid human genome was divided by a million to account for the bacterial count scale. The quotient was multiplied by the normalised bacterial read count (normalised to a million reference base pairs) and the final product was divided by the human read count [36], [37]. For the same dataset of raw read counts, a variance-stabilising transformation (VST) was calculated from fitted dispersion-mean relations to generate approximate homoscedastic data [38]. As previously described by McMurdie and Holmes (2014), negative normalised values were set to zero and a pseudo count of one was applied to raw count data [39]. Thirdly, raw reads counts were normalised by relative log expression (RLE) [38], in which case the geometric mean across all samples was calculated, the median ratio of each sample to the median library was taken and read counts per million were computed. The samples were then grouped according to their disease state (healthy versus CF) and based on the following age groups: the first year of life (0 years), the toddlers (1–3 years of age) and the pre-school children (4–6 years of age). While the 25th species abundance percentile is commonly used as the threshold to define the rare species biosphere in ecological applications [25], we performed parts of the data analysis with three different cut-off values (15th, 25th and 35th species abundance percentile) aiming to avoid working with one rigid threshold definition.
2.3. Data analysis
2.3.1. Investigations of the age-dependent and age-independent bacteria
Taxonomic overlaps of the taxa in the core and rare species biosphere per age group (0 years, 1–3 years, 4–6 years) in CF and healthy children were studied with Venn Diagrams [40] and paired line plots. The statistical comparison of species numbers between healthy and CF children in different age groups was based on the Fisher's Exact test for count data. Disease state-specific ‘background species’ as well as ‘non-persisting species’ were defined to differentiate between the early colonisers present in all age groups, starting from the first year of life (0 years) up to the preschool age (4–6 years) and the fluctuating colonisers that were solely detected at certain developmental stages of the children, respectively.
2.3.2. Bootstrapping aggregation
A random forest analysis based on bootstrapping aggregations was applied to identify the key determinants distinguishing healthy from CF airway metagenomes in the early years of life [41], [42]. All non-random contributing variables with mean decrease accuracy above zero were extracted and classified in terms of ‘rare species’ or ‘core species’ as defined by the 15th, the 25th or the 35th abundance percentile or ‘host-associated’ factors. Host-associated variables included age, body mass index and gender. The classification performance was validated with the out-of-bag (OOB) estimate of error rate, class errors and the Boruta algorithm [43]. The Boruta algorithm generates shadow attributes by randomly shuffling the original input variables and iteratively comparing random with the corresponding original variables. Consequently, non-random features of importance for the CF versus healthy classification could be distinguished from random feature assignments. Random variables were removed from the downstream data analysis. Random forest and Boruta wrapper application runs were repeated 100 times with different seeds set for the classification and for the feature selection procedure with the objective to avoid a selection-based bias.
2.3.3. Functional investigations of age-independent bacteria
The functional gene profile of background core and rare species (25th abundance percentile) was investigated by running gapseq (version 1.1) [28]. Here, only those species were included which were confirmed as ‘background species’ by both read alignments to the one-strain-per-species (DB1) or pan-genome (DB2) reference database. As explained by the authors of the tool, metabolic pathways are often made of several sub-reactions and key enzymes that together execute a biological process. For the MetaCyc [44] metabolic pathway analysis, gapseq approached a customised protein database that stored sequence data of proteins and enzymes involved in the underlying sub-reactions of 1779 bacterial metabolic pathways. The protein reference sequences and the corresponding metadata used by gapseq were obtained from UniProt [45], BRENDA [46], and the Enzyme Nomenclature Committee [28]. The metabolic pathway analysis of gapseq was then based on a homology search by tblastn [47], in which the DNA sequence of the reference genome was screened for matching protein sequences in the reference sequence pools of sub-reactions with the following cut-off values: Bitscore ≥ 200, coverage ≥ 75% [28]. Finally, a metabolic pathway was considered to be present in a reference genome if sequence evidence was found for at least 80% (‘completeness threshold’) of the reactions per pathway [28]. Since we link the absence of sequence evidence based on a manually curated protein database with the absence of known sequences with sufficient homology in the corresponding reference genome and not with the absence of the metabolic pathway within the bacterial gene repertoire itself, we exchanged the term ‘completeness threshold’ with ‘functional matching (FM) score’. In other words, the FM score quantifies the percentage of reactions that were detected per pathway in the same way as the completeness threshold [28] but the final interpretation of the output differed. We then extracted all those metabolic pathways with at least 10% difference in mean FM score between the healthy background core and rare species biosphere. FM scores of the extracted metabolic pathways were centred and scaled to perform a principal component analysis and to evaluate the functional cluster behaviour of taxa in the core or rare species biosphere obtained from VST-, RLE-, or BCPHC-normalised data. Afterwards, all variables were extracted that contributed to the data separation in the first dimension. MetaCyc pathways were converted into MetaCyc superclasses [44].
Moreover, we statistically compared the average FM scores of bacteria in the healthy and CF core and rare species biosphere with the FM score per pathway of the CF hallmark pathogen Pseudomonas aeruginosa PAO1 reference sequence by applying the non-parametric Mann-Whitney U test. The effect size r was calculated, which is the Mann–Whitney U test statistics divided by the square-rooted sample size.
We also searched the reference genomes of background species for known adhesin protein sequences. The reference protein sequences were obtained from NCBI with the following command line: adhesin[All Fields] AND (“Bacteria”[Organism] AND swissprot[filter]. A blastx search was performed with default settings. Significant matches between bacterial reference genomes and the protein database were extracted (BLAST expectation value < 0.01). The adhesin profile of background species was compared between healthy and CF airways in the early years of life to get a first impression on the potential long-term training of the immune system based on the early airway microbial communities in healthy and CF infants. The reference database of known adhesins is publicly available (see data availability statement).
2.3.4. Ecological species co-occurrence network analysis with graph kernels
Undirected ecological network analysis was based on the best practice guidelines for species co-occurrence network construction [26], [48]. Spearman’s rank correlation matrices were obtained for healthy and CF children from BCPHC-, RLE- or VST-normalised count data of background core and rare species as defined by the 25th abundance percentile and confirmed by both reference databases. All significant positive correlations (p-values < 0.01, Spearman’s rank correlation coefficient > 0.20) were extracted and the continuous graph layout algorithm Fruchterman-Reingold was applied to CF and healthy co-occurrence data of background taxa [49], [50]. Network robustness and vulnerability were evaluated by removing network species based on descending degree centrality of contributing nodes (targeted attack) or by random node removal (random attack) while tracking the overall network connectivity [51]. For network simulation runs, an increasing number of healthy background taxa (n = 1 – 20) was randomly selected with replacement and transferred into the CF network. All simulation runs were repeated 100 times with different seeds set for the random node selector. Fréchet-distances were obtained based on Alt und Godau’s (1995) algorithm with runtime O(pq log(pq)) [52] to measure the similarity of CF and healthy network attack chains based on their corresponding edges p and q, respectively. The orientation-preserving classical Fréchet-distance algorithm was selected instead of the more commonly used Euclidean or Hausdorff-distance, because not only the distance between pairs of points, but also the ordering of points can be biologically relevant when comparing two-dimensional network attack curves [51]. The topological similarity between CF and healthy network graph structures was evaluated with a neighbourhood aggregation graph kernel. The 1-dimensional Weisfeiler-Lehman subtree kernel matched neighbourhoods of discretely labelled nodes in the network structures with five iterations (h = 5) and was computed on a pair of graphs with m edges and h iterations in time O(hm) [53]. The kernel computations were based on the efficient C++ implementations of the R software package graphkernels (version 1.6) [54]. Null models were used to validate whether the observed effect of modulated CF networks was significantly different from the observed effect obtained from 100 randomly generated networks per modulated CF network. The corresponding p-values and z-scores (observed − mean(null)/sd(null)) were obtained. All non-random modulated CF networks were grouped into the categories “stabilisation effect” or “destabilisation effect”, when a non-random CF graph structure was more or less similar in its robustness to a healthy network structure compared to the original CF network structure. A subset of non-random simulation runs was selected (70%, training dataset) for running a binominal regression analysis and for extracting the taxonomic features causing either a stabilisation or destabilisation event of the original CF network structure. The model performance was then validated with the test dataset (30%) by plotting the true positive rate against the false positive rate and evaluating the area under the curve (AUC). Data analysis was performed in R [55].
3. Results
3.1. The taxonomic overlap of core and rare species in different developmental stages of the early human life
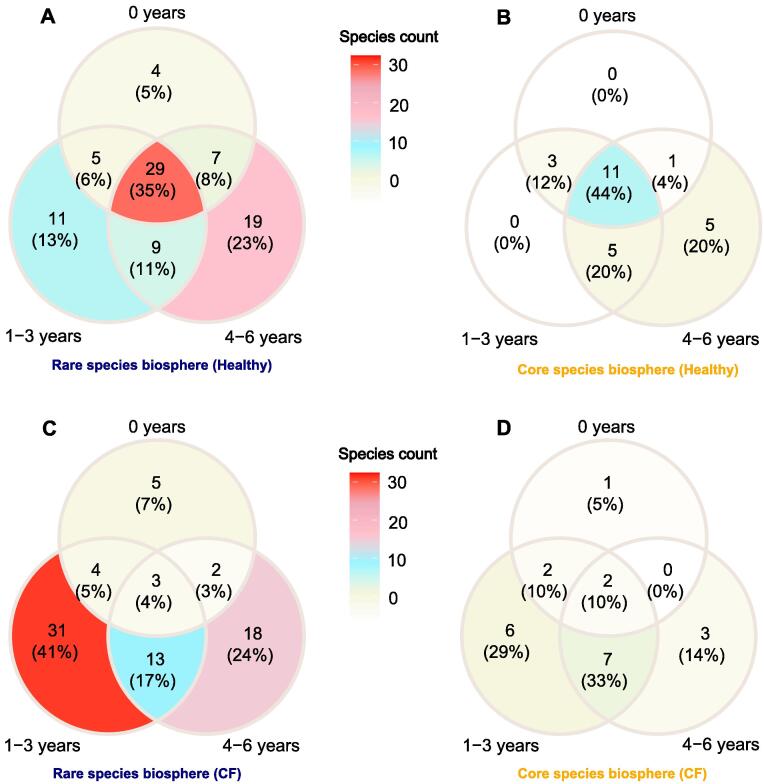
In healthy airways, the majority of rare (35%, Fig. 1A) and core species (44%, Fig. 1B) were detected across all age groups (‘background species’). Some rare taxa (13%) were only found in toddlers (1–3 years, Fig. 1A) and another 23% were solely detected in pre-school children (4–6 years, Fig. 1B). Core and rare species numbers increased with age (Fig. 1B, Supplementary Fig. S1A-B). In CF airways however, a minority of rare (4%, Fig. 1C) and of core species (10%, Fig. 1D) were found from early on and up to the pre-school age. Most of the rare species (41%) were solely recovered in the toddler’s age group (1–3 years). Higher core and rare species numbers were observed in the CF toddlers compared to the CF infants in the first year of life and CF pre-school children. The age-independent background community of bacterial taxa was hence underdeveloped in CF compared to healthy children (Fig. 1, Supplementary Fig. S1, Supplementary Tables S1 and S2).
Fig. 1.
Venn Diagrams reveal the taxonomic overlap of bacterial species in the core or rare species biosphere as defined by the 25th abundance percentile of healthy and CF children. (A) Representation of the least abundant (rare) species in healthy airways per age group (0 years, 1–3 years and 4–6 years). The majority of rare species (35%) were shared between all age groups and are hence ‘age-independent’ because they were present from early on (0 years) until pre-school age (4–6 years). (B) Visualisation of the most abundant (core) species in healthy airways per age group. The majority of healthy core species (44%) was found in all age groups. (C) The taxonomic overlap of rare species in CF airways between all age groups. Most of the rare species were found in the toddler age group (1–3 years). (D) Representation of the core species in CF airways per age group. Most bacterial taxa were only transiently present in one age group of CF children. Note: In the CF cohort, there were 5 infants below the age of one, 20 between 1 and 3 years of age and 16 children between 4 and 6 years of age. In the healthy cohort, there were 25 infants below the age of one, 7 children between 1 and 3 years of age and 14 preschool children were between 4 and 6 years of age. The Venn Diagrams were constructed based on read alignments towards the pan-genome database. The findings of all three normalisation strategies (BCPHC, RLE, VST) were merged to generate a global overview of species distribution with age. The background species can be extracted from Supplementary Table S1 and raw count data is available from Supplementary Table S2.
3.2. Rare species as key determinants of a healthy airway metagenome
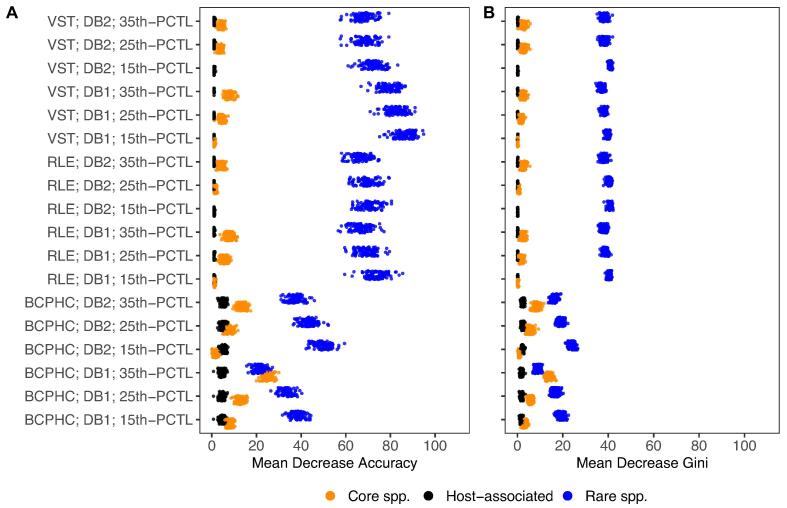
The classification of children into the groups ‘CF disease’ or ‘healthy’ based on their airway metagenome and host-associated factors revealed that most of the features contributing to the random forest decision were associated with the rare taxa. In general, this observation was made irrespective of whether a one-strain-per-species (DB1) or pan-genome (DB2) reference database was selected for the read alignment procedure or read counts were normalised by BCPHC, VST or RLE (Fig. 2A-B). However, for BCPHC-normalised read counts based on read alignments towards a pan-genome database and with the following rarity threshold: 35th species abundance percentile, core species explained most of the difference between CF and healthy children. The database and threshold settings caused a distinct situation in which more taxa were detected in the core species than the rare species biosphere, which explains this strategy-dependent observation of core species becoming more important than rare species in the classification process. For RLE and VST-normalised data, logarithmic-like transformations compressed the data and hence boosted the effect of rare taxa, causing the significantly higher mean decrease accuracy of rare species compared to core species and host-associated factors for all rarity thresholds and both reference databases (Fig. 2A). A similar observation was obtained for the mean decrease Gini (Fig. 2B, Supplementary Table S3). The importance of host-associated variables, including age, body mass index and gender was negligible during the classification process.
Fig. 2.
Random forest classification [41], [42] of children into the pre-defined categories ‘CF disease’ and ‘healthy’ by constituents of the airway microbial metagenome and host variables including age, body mass index and gender. (A) Representation of the classification outcome based on the mean decrease accuracy obtained with the following three normalisation strategies: BCPHC (Bacterial cell per human cell), RLE (Relative log expression), and VST (Variance-stabilising transformation), with two reference databases, namely the one-strain-per-species (DB1) and the pan-genome (DB2) reference database and with the 15th, the 25th and the 35th species abundance percentile (PCTL) to define the core and rare species biosphere. (B) Representation of the classification results based on the mean decrease Gini. Note: The mean out-of-bag (OOB) estimate of error rate for the random forest classification was 0.11 (standard deviation = 0.06), the mean CF class error was 0.09 (standard deviation = 0.08) and the mean healthy class error was 0.12 (standard deviation = 0.07). For host-associated, core species and rare species columns, only non-random features are shown with a mean decrease accuracy above zero.
3.3. Functional comparison of background bacteria
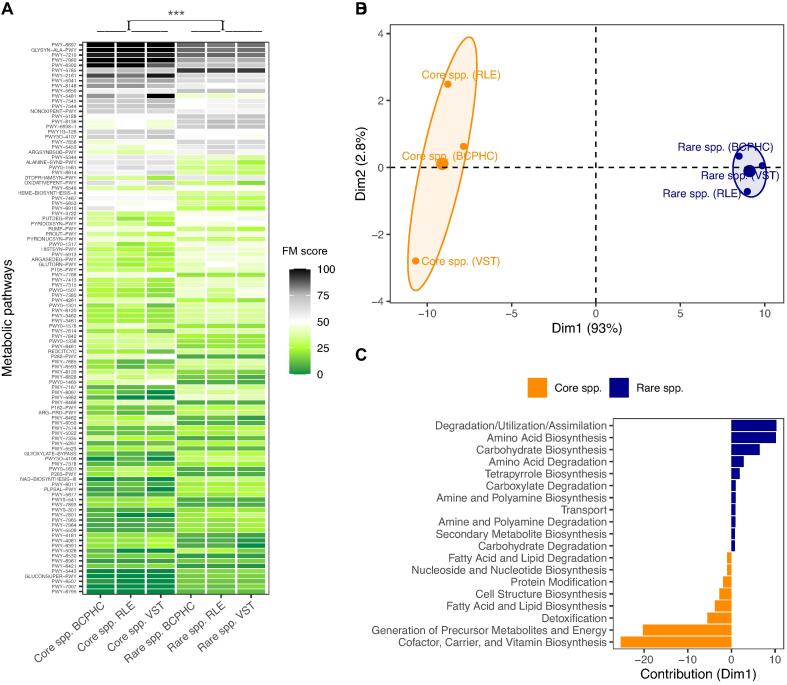
As described by Doolittle and Booth (2016), the taxonomic makeup of a microbial community may vary and fluctuate as a function of time, contrary to the biochemical function of the microbiome, which seems to be more conserved with time and across habitats [56]. Correspondingly, investigations of the functional capacities of the core and rare species biosphere are important to gain insights into the more stable part of community airway ecology in infancy. We thus investigated the functional capacity of core and rare background species based on the reference genomes for which uniform read distributions were obtained with raspir [30]. The tool gapseq utilised a pre-defined protein sequence reference pool for every sub-reaction involved in known core metabolic pathways of bacteria (n = 1779) [34]. Here, we defined a functional matching score (FM score) based on the percentage of reactions per metabolic pathway. While the overall intra-genus Canberra distance based on FM scores of core and rare species was generally more similar than the inter-genus distance (Supplementary Fig. S2), distinct differences between the core and rare microbial community of the healthy airway habitat were detected (Fig. 3A). These differences were based on the presence or absence of known DNA sequences associated with sub-reactions involved in alternative and central metabolic pathways (Fig. 3A, Supplementary Table S4). Indeed, a principal component analysis of the scaled and centred FM scores revealed a distinct clustering behaviour of core and rare species (Fig. 3B). The extraction of variables contributing to the separation in the first dimension revealed that the core species biosphere contained higher FM scores for cofactor, carrier and vitamin biosynthesis, the generation of precursor metabolites and energy, whereas the rare species covered degradation pathways, amino acid metabolism and carbohydrate biosynthesis (Fig. 3C).
Fig. 3.
Comparison of the protein sequence usage (FM score) between the core and rare background species in healthy children. (A) Heatmap visualisation of the mean differences in FM scores between the core (left) and rare (right) background taxa as defined by the 25th species abundance percentile and based on both reference databases. The metabolic pathway analysis used a homology search by tblastn (bitscore ≥ 200 and a coverage ≥ 75%) in which the DNA sequence of the corresponding reference genome was screened for matching protein reference sequences. Per reference metabolic pathway, a matching score (FM score) between 0 (green) and 100 (black) was assigned based on the percentage of the number of sequences per sub reaction that were detected in the corresponding reference genomes. The heatmap hence visualises the mean FM score obtained for the core species community (left) and the rare species community (right) for each normalisation strategy, separately. All the displayed pathways (n = 108) exhibited a mean group difference in their FM scores of at least 10%. The metabolic pathways with the corresponding MetaCyc superclasses [44] can be obtained from Supplementary Table S4. The statistical difference between the core and rare species biosphere was obtained with a rank-based Mann-Whitney U test (p-value < 0.001). (B) A principal component analysis (PCA) was performed based on the scaled and centred FM scores obtained from all 108 metabolic pathways defined in Fig. 3A. (C) Visualisation of the PCA variables in terms of Metacyc superclasses [44] contributing to the separation of the core and rare species biosphere in the first dimension (Dim1) of the PCA in Fig. 3B. The negative values (orange colour) on the x-axis reflect a positive contribution of the separation towards the negative coordinate space (core species) whereas positive values (blue colour) correspond to a positive contribution towards the positive coordinate space (rare species).
We also compared mean FM scores per metabolic pathway of background core and rare species isolated from healthy or CF airways with the FM scores obtained from the reference genome of the CF hallmark pathogen P. aeruginosa. The P. aeruginosa taxon was selected for these investigations because the DNA has been reported to be stochastically detected at low numbers in healthy and CF airways between 0 and 6 years of age [26], even though the opportunistic pathogen does not typically belong to the airway microbial community. P. aeruginosa is hence a verified taxonomic outlier to which all children are regularly exposed in the early years of life. However, CF in contrast to healthy children are at high risk of developing acute and chronic airway infections later in life with P. aeruginosa being the most dominant species of the lower airway community [37]. We found no significant difference between the mean FM scores per metabolic pathway of background core and rare species isolated from healthy airways and the FM score of the CF hallmark pathogen P. aeruginosa (Supplementary Fig. S3). However, for CF children, a small but significant deviation in FM scores between the background core species biosphere and the P. aeruginosa reference genome was unravelled, whereas the mean FM scores of the CF rare species biosphere and the P. aeruginosa genome remained similar (Supplementary Fig. S3).
Next, we performed a blast search of all background species against a reference database of known bacterial adhesin protein sequences. The reason why we focused on adhesins is that bacterial adhesive structures and specific or non-specific adhesion mechanisms determine the colonisation behaviour of bacteria in the host and provide the major interface of bacterial-host cell interaction and crosstalk [57]. While bacterial adhesins were extensively explored for invading pathogens in terms of virulence factors, the role of commensal adhesins in stimulating the immune system or manipulating the host physiology remains understudied [57]. Furthermore, the oropharyngeal barrier is often damaged in patients with CF from early on because viral and subsequent bacterial colonisation cannot be properly contained [58]. Hence, the spectrum of adhesins decreases in CF in contrast to the full repertoire of adhesins in the healthy oropharynx. The analysis revealed a distinct bacterial adhesin background pattern between healthy and CF airways, as well as between the core and the rare species biosphere in the early years of life (Supplementary Fig. S4).
3.4. Ecological network analysis of background commensals in the early years of life
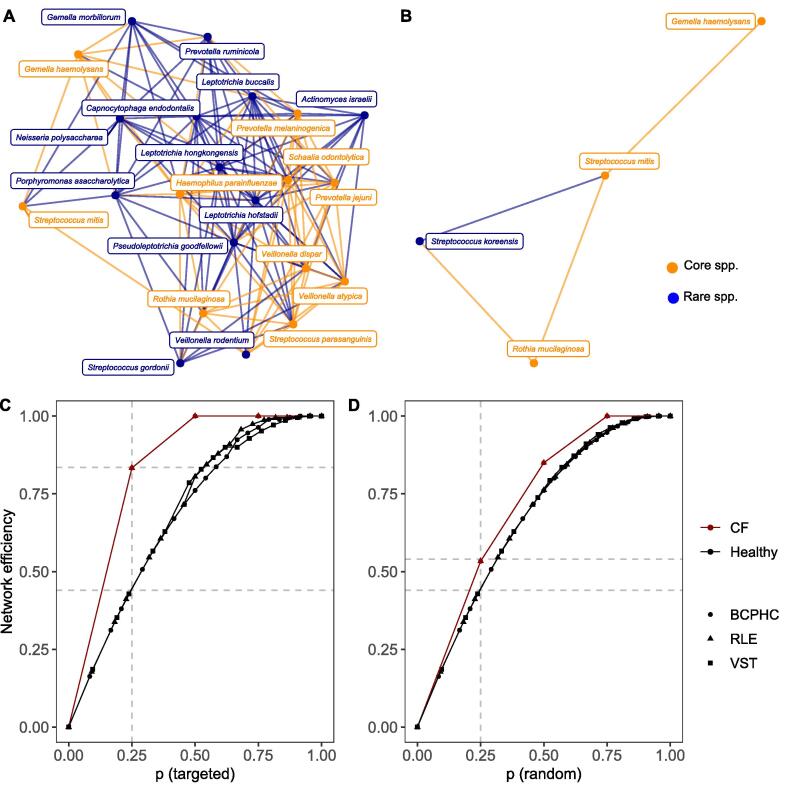
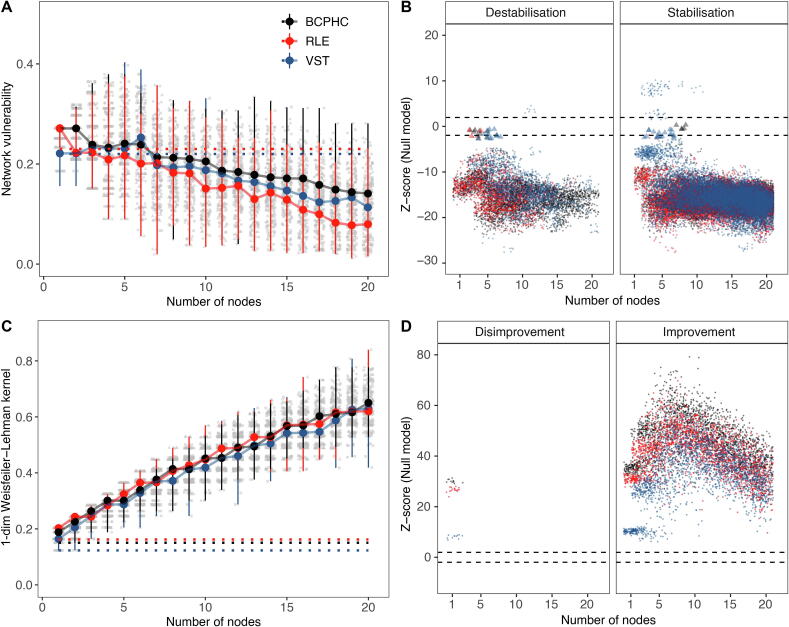
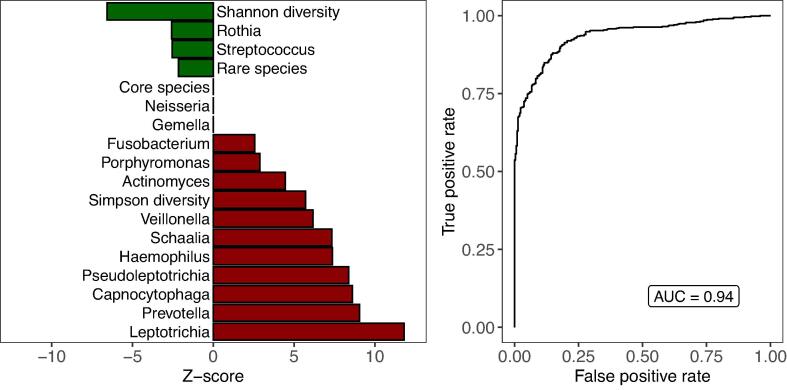
We performed an ecological species co-occurrence network analysis with graph kernels to explore and compare the age-independent background networks of microbial communities in healthy and CF infants. With computer simulations we modified the transient and labile CF background structure by inserting various combinations of healthy taxa, aiming to exploit the distinct characteristics that further destabilise the underdeveloped CF network or make it more robust by adopting healthy-like global features. Independent of the normalisation strategy of raw count data, the healthy background network of bacterial taxa exhibited strong positive correlations between core and rare species with high numbers of contributing species (Fig. 4A, Supplementary Fig. S5). The background network of bacterial commensals in children with CF contained only a few contributing species that were loosely connected (Fig. 4B, Supplementary Fig. S5). While a healthy background network under targeted attack broke down in a uniform manner independent of the normalisation strategy, the CF background network collapsed at high speed and in progressive stages (Fig. 4C). The CF and healthy networks were similarly affected by a random attack strategy (Fig. 4D). The vulnerability of CF networks to targeted attacks was decreased by inserting a key combination of health-associated background taxa (Fig. 5A). Overall, the more core or rare species were incorporated into the CF network, the smaller the distance between CF and healthy background networks in terms of network robustness and vulnerability. However, a large variability was observed between the simulation runs (Fig. 5A). Depending on the combination of inserted taxa, the CF network robustness became either more similar (stabilisation) or dissimilar (destabilisation) to the healthy network compared to the original CF network (Fig. 5A). In general, the modulated CF networks displayed destabilisation (Fig. 5B, left panel) or stabilisation (Fig. 5B, right) features to a significantly different extent than expected by chance under a null hypothesis (p-value < 0.05). In<0.2% of simulated structures, no significant difference was observed (Fig. 5B, enlarged triangular shapes). These random network structures were subsequently removed for the following part of the data analysis. The topological distance between CF and healthy network decreased in terms of the neighbourhood aggregation kernel when health-associated background taxa were incorporated into the CF network (Fig. 5C). We compared the observed graph structures of modulated CF networks with random network structures by null models and found that simulated structures were significantly different from random background structures (p-value < 0.05). We subsequently evaluated the selection features stabilising or destabilising the non-random CF network structures with a binominal regression analysis. The investigation revealed a stabilisation effect of the CF network structure whenever healthy taxa were incorporated with high species diversity (Shannon diversity index) and low species dominance (Simpson diversity index). Furthermore, the transfer of Rothia mucilaginosa and Streptococcus spp. (Fig. 6), as well as adding rare instead of core taxa was important in increasing the robustness of the CF network to targeted network attacks (Fig. 6).
Fig. 4.
Ecological network analysis of the background core (orange colour) and rare (blue colour) species in the early human airways. (A) The background co-occurrence network of bacterial taxa in healthy children between zero and six years of age obtained from RLE-normalised count data. (B) The background co-occurrence network of bacterial taxa in CF children between zero and six years of age obtained from RLE-normalised count data. Note forFig. 4A, 4B: Networks build from VST- and BCPHC-normalised data are provided in Supplementary Fig. S5. Plus, only those background species are depicted, which were detected by both reference databases (pan-genome and one-strain-per-species). (C) Network connectivity dynamics per fraction of nodes removed (p) by targeted network attacks. An increasing fraction of species was removed with descending degree centralities from all healthy (black) and CF (red) background networks with more than three contributing nodes. For BCPHC, RLE and VST-normalised data, Fréchet-distances of 0.23, 0.23 and 0.22 were obtained between CF and healthy attack curves, respectively. (D) Network connectivity dynamics per fraction of nodes removed (p) by random network attacks. To simulate random network attacks, species were randomly removed from all healthy (black) and CF (red) background networks. For BCPHC, RLE and VST-normalised data, Fréchet-distances of 0.06, 0.06 and 0.07 were obtained between CF and healthy attack curves, respectively.
Fig. 5.
In silico network simulations to evaluate CF network topology and robustness in the presence of healthy background species. (A) In the presence of targeted attacks, network vulnerability of modulated CF networks was compared with the network vulnerability of the healthy network. A Fréchet distance of 0 indicated perfectly matching curves. Network vulnerability decreased with increasing number of species (Spearman’s rank correlation, BCPHC: p-value < 0.0001, coefficient = -0.51, CI = [-0.53; −0.49]; RLE: p-value < 0.0001, coefficient = -0.69, CI = [-0.71; −0.68]; VST: p-value < 0.0001, coefficient = -0.72, CI = [0.74; 0.71]). The robustness of the original CF background network is provided (dotted, horizontal lines). The grey dots represent the output of the simulation runs with 100 seeds set for the random node generator. The enlarged and coloured dots depict the median output per normalisation strategy. The error lines give information on the minimum and maximum final performance. (B) Representation of the outcome of the null model analysis to identify whether modulated CF networks display destabilisation (left) and stabilisation (right) characteristics to a significantly different extent than expected by chance under a null hypothesis. The colours black, red and blue correspond to simulations based on BCPHC-, RLE-, or VST-normalised count data, respectively. Points depict simulated CF networks which were significantly different from random background structures of the null model (p-value < 0.05). Enlarged triangular shapes, show simulated CF networks, which were not different from random structures (p-value > 0.05). (C) Based on the 1-dimensional (1-dim) Weisfeiler-Lehman graph kernel, modulated CF and healthy network similarity increased with two and more inserted species (Spearman’s rank correlation, BCPHC: p-value < 0.0001, coefficient = 0.95, CI = [0.95; 0.96]; RLE: p-value < 0.0001, coefficient = 0.94, CI = [0.93; 0.94]; VST: p-value < 0.0001, coefficient = 0.95, CI = [0.94; 0.95]). (D) Representation of the outcome of the null model analysis to identify whether modulated CF networks display disimprovement (left) and improvement (right) characteristics in terms of graph topology to a significantly different extent than expected by chance under a null hypothesis. In general, disimprovement was limited to simulation runs based on the incorporation of less than three bacterial taxa from the healthy network structure. All simulated networks were distinct from random background structures (no triangles). Note: We only worked with and display core and rare background species which were verified by both reference databases (pan-genome and one-strain-per-species).
Fig. 6.
A binominal regression analysis reveals the underlying transfer features of stabilisation (green) or destabilisation (red) events observed in non-random, modulated CF networks. The original dataset was randomly split into train (70%) and test (30%) datasets. (A) The outcome of the regression analysis based on the train dataset (70%) shows that CF network vulnerability was reduced (green) when healthy taxa were inserted with high species diversity (Shannon diversity). Plus, the transfer of Rothia mucilaginosa, Streptococcus spp. and rare species in general was associated with a stabilisation effect. CF network vulnerability was increased on the other hand (red), when species, e.g., of the genera Gemella, Fusobacteria, Veillonella, Prevotella and so on, were transferred with high species dominance (Simpson diversity). (B) The test dataset (30%) was now approached to validate the accuracy of the established model in Fig. 6A. The Area Under the Receiver Operating Characteristics (AUROC) curve, plotting the true positive rate (y-axis) against the false positive rate (x-axis) revealed an acceptable prediction performance of our model in terms of network stabilisation versus network destabilisation events (AUC = 0.94). Note: We only worked with and display core and rare background species which were verified by both reference databases (pan-genome and one-strain-per-species).
4. Discussion
The bacterial co-occurrence network of the lower airway habitat is a dynamic system, continuously shaped by newly arriving microbes from the upper respiratory tract and the loss of bacteria which were targeted by the host defence system [59], [60], [61]. In a previous study, we showed that the airways of healthy newborns and toddlers contain a strongly interconnected co-occurrence network of core taxa from early on [26]. This network was characterised by high species diversity and bacterial loads of all the contributing species. Here, we approached our recently published software tool that scans the within-species conservation of the global chromosomal organisation by evaluating the distribution of raw reads mapping towards circular reference genomes [27]. Since we re-analysed deep shotgun metagenomic sequencing data of single-end read runs, where the maximum number of genome positions was probed, insights were obtained into the contribution of rare species in maintaining and stabilising the bacterial co-occurrence network of human airways in the first six years of life. We performed the data analysis with three different normalisation methods to account for the compositional behaviour of microbiome data. The BCPHC normalisation has high ecological relevance considering that human read counts are used as natural spike-in control to obtain an estimation of the underlying microbial abundance pattern [36]. Since the human host forms an essential part of the ecological airway habitat, its quantitative information can be maintained by the BCPHC normalisation strategy. However, the method can be affected by free DNA in the sample or library preparation procedures [36]. We therefore also performed RLE and VST normalisations. These methods have been shown to perform well when applied to shotgun metagenomics and 16S rRNA gene sequencing data by obtaining unbiased p-values, controlling the false discovery rate in simulation scenarios and enabling robust interpretations of correlation and proportionality estimates [30], [39], [62].
The gastrointestinal human microbiome has been described as stable during adult life and unstable with regard to the overall taxonomy in the first few years of life [63]. In this study, we show that the healthy airway microbial community consists of both background and non-persisting bacteria. The background taxa are the early airway colonisers that remain to be detected until pre-school age, independently of the child’s developmental stage. These persistent background commensals form a stable network in which core and rare taxa are equally important from early on. Depending on the developmental stage of the child, non-persisting core and rare species enter the network for a limited period of time. So, the healthy airway metagenome was found to contain background and non-persisting bacteria that are stimulating the immune system on a long-term and short-term basis, respectively. In CF children, the background network was found to be underdeveloped and the majority of rare species in CF airways was associated with the toddler age group (1–3 years of age). Since a close-to-healthy network structure of high-abundant taxa was only detected in CF toddlers, a long-term immune system stimulation is missing in CF pre-school children for most of the time and the training of the immune system is defined by non-persisting bacteria or pathogens later in life [37], [64], [65]. Leitão et al. (2016) [23] utilised simulation studies of environmental ecosystems and thereby unravelled the disproportional influence of rare species on the functional structure of an ecosystem. They reported that rare species extinction causes a disturbance on the long-term supply of goods and services and thus destabilises the entire ecosystem [23]. The detection of the majority of rare species in CF toddlers with their subsequent loss in pre-schoolers may hence explain the known reduction in core species diversity and occurrence of network fragmentation as the disease progresses [37], [64], [65], [66]. Furthermore, the loss of rare species may create an open niche in the CF airway habitat that can be chronically filled by incoming CF hallmark pathogens such as P. aeruginosa. However, to confirm these results that were based on cross-sectional metagenome data, it is essential to perform follow-up longitudinal metagenome studies, tracking both the healthy and diseased early airway development in individuals over time.
Investigations of the functional capacities of microbial communities are of high interest and can provide novel insights into the microbial airway inhabitants and the airway environment itself. However, functional annotation and the underlying gene orthologous databases are still in development. Despite the enormous number of annotations that are available in the public domain, a large number of entries remain classified as ‘hypothetical’ and wet-lab experiments are currently lacking to confirm the candidate proteins [67]. Also, short reads are often not discriminative enough to distinguish between functions and it is recommended to use paired-end data for generating and mapping the long contigs instead of short reads to the databases [68]. This strategy cannot be approached to gain insights into the functional capacity of rare species since genome coverages are low. Therefore, we performed the functional analysis based on the corresponding reference genomes instead of directly utilising DNA reads obtained from the biological samples. Major differences in FM scores were detected between the core and the rare microbial background communities in the human airways. These differences were not only associated with alternative metabolic pathways but also with central metabolic functions, e.g., usage of essential metabolites, bacterial respiration and energy generation. A high FM score in one group contrary to a low FM score in the other one (Fig. 3) does not imply the presence or absence of the corresponding core metabolic pathways, respectively. It does however indicate that in the latter case protein sequences are missing with sufficient homology towards the known sequences in pre-defined sequence reference pools of the sub-reactions per metabolic pathway. A different pool of still unknown protein sequences may be utilised by the group instead to accomplish the same functional task, suggesting a division of labour between the core and rare microbial community in terms of nucleotide, codon and motif usages. We assume that the observed sequence differences in exotic and core pathways enable the permanent co-existence of core and rare species in an otherwise highly competitive airway environment. As described in section 3.3, we also compared the mean FM scores of the core and rare species biosphere in healthy or CF airways with the FM scores obtained directly from the P. aeruginosa reference genome PAO1, because P. aeruginosa DNA has been reported to be stochastically detected at low numbers in healthy and CF airways between 0 and 6 years of age [26], even though the opportunistic pathogen does not typically belong to the airway microbial community. P. aeruginosa is hence a verified taxonomic outlier to which all children are regularly exposed in the early years of life. However, CF in contrast to healthy children are at high risk of developing acute and chronic airway infections later in life with P. aeruginosa being the most dominant species of the lower airway community [37]. Interestingly, we found no significant differences in the overall protein sequence usage between healthy core/rare species and PAO1, suggesting that a high competition for resources prevents the overgrowth of P. aeruginosa in the healthy airway habitat. While the mean FM scores of the rare species biosphere in the CF airway habitat was also similar to the P. aeruginosa genome scores, the CF core species biosphere differed slightly but significantly from the pathogen’s scores. We thus hypothesise that in the early CF airways, the rare species compete with the pathogen for resources by using a similar pool of known protein sequences. The subsequent loss of rare species in the CF pre-school age may hence again be assumed to facilitate the overgrowth of P. aeruginosa. However, our functional investigations have to be interpreted with caution. The typical core microbes may have been studied more extensively in the past than the often-unknown and unculturable rare species, so there is a literature bias in gene databases in favour of the more abundant species [69], [70]. On the other hand, the identification of moonlighting proteins, which are multifunctional molecules involved in various metabolic pathways is still at the beginning [71], [72]. So, known bacterial key enzymes may undertake a number of different tasks in unrelated biological processes and therefore functionally fill the gaps in case of missing protein sequences. Furthermore, the gapseq tool prioritises the mapping of sequences and reactions by EC number even though some reactions in the MetaCyc [44] database have more than one EC number annotation, which may introduce false FM scores [28]. Nonetheless, gapseq achieves higher prediction accuracies than other known functional pipelines and can hence be considered as state-of-the art software [28]. Ultimately, only comparative metatranscriptomics and metaproteomics will reveal what is actually expressed by core and rare airway species and hence give us more information on relevant microbe-microbe or host-microbe interactions [73].
In conclusion, we were able to show that rare species play the key role in differentiating between healthy and CF airway metagenomes in the early years of life. Rare species contribute to a stable and robust species co-occurrence airway network and seem to facilitate the metabolic integrity of the healthy human airway metagenome. The computer-based model simulations revealed a stabilisation effect of the CF network after the transfer of a key combination of bacterial taxa with high species number, high species diversity and low species dominance. Also, R. mucilaginosa and Streptococcus spp. were found to play a key role in reducing the vulnerability of labile CF background networks. With the here approached algorithms however, it has not been possible to make the networks identical. Nonetheless, rare species were particularly important in improving the underdeveloped CF background network. It is hence essential to investigate the currently uncharacterised taxonomic and functional potential of rare species in future studies to get a more comprehensive picture of the universal features characterising a healthy or diseased human microbiome.
Data availability
Coding scripts and reference databases are available from https://github.com/mmpust/airway-metagenome-simulations. Microbial sequencing data can be obtained from the European Nucleotide Archive (study accession number PRJEB38221).
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, SFB900/3 – 158989968 A2 and Z1) and the Volkswagenstiftung and the Niedersächsisches Ministerium für Wissenschaft und Kultur (Big Data initiative, ZN3432). M.-M.P. is a member of the PhD program Infection Biology coordinated by the Center of Infection Biology at MHH and a scholar of the Studienstiftung des deutschen Volkes.
CRediT authorship contribution statement
Marie-Madlen Pust: Conceptualization, Methodology, Data curation, Software, Formal analysis, Validation, Writing – original draft. Burkhard Tümmler: Conceptualization, Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Dr. Colin Davenport (Research Core Unit Genomics, Medical School Hannover) and Jannes Gleß (IT-Service, Medical School Hannover) for providing and maintaining the high-performance computing clusters and thereby enabling data generation and analysis with extensive parallel computing operations. The authors acknowledge the contribution of PD Dr. Anna-Maria Dittrich and Dr. Isa Rudolf who recruited and sampled study participants and thereby facilitated the generation of the primary sequencing data, which has been published in 2020. The graphical abstract was modified from ‘Cystic Fibrosis Airways’, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.12.008.
Contributor Information
Marie-Madlen Pust, Email: Pust.Marie-Madlen@mh-hannover.de.
Burkhard Tümmler, Email: Tuemmler.burkhard@mh-hannover.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wang B., Yao M., Lv L., Ling Z., Li L. The human microbiota in health and disease. Engineering. 2017;3(1):71–82. [Google Scholar]
- 2.Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu N.-N., Ma Q., Ge Y., Yi C.-X., Wei L.-Q., Tan J.-C., et al. Microbiome dysbiosis in lung cancer: from composition to therapy. NPJ Precis Oncol. 2020;4(1) doi: 10.1038/s41698-020-00138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanahan F., Ghosh T.S., O’Toole P.W. The healthy microbiome—What is the definition of a healthy gut microbiome? Gastroenterology. 2021;160(2):483–494. doi: 10.1053/j.gastro.2020.09.057. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F., Fraser C., Ringel Y., Sanders M., Sartor R.B., Sherman P., et al. Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin A.M., Sitarik A.R., Havstad S.L., Fujimura K.E., Wegienka G., Cassidy-Bushrow A.E., et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. 2016;6(1) doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta V.K., Paul S., Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye S.H., Siddle K.J., Park D.J., Sabeti P.C. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178(4):779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huson D.H., Auch A.F., Qi J.i., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D., Song L.i., Breitwieser F.P., Salzberg S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 14.Breitwieser F.P., Baker D.N., Salzberg S.L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19:198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 16.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F., et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12(1) doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weyrich L.S., Farrer A.G., Eisenhofer R., Arriola L.A., Young J., Selway C.A., et al. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019;19(4):982–996. doi: 10.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhofer R., Minich J.J., Marotz C., Cooper A., Knight R., Weyrich L.S. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019;27(2):105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Ma Z.S. Power law analysis of the human microbiome. Mol Ecol. 2015;24(21):5428–5445. doi: 10.1111/mec.13394. [DOI] [PubMed] [Google Scholar]
- 20.Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V., et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017;11(4):853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch M.D.J., Neufeld J.D. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13(4):217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 22.Karpinets T.V., Gopalakrishnan V., Wargo J., Futreal A.P., Schadt C.W., Zhang J. Linking associations of rare low-abundance species to their environments by association networks. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitão R.P., Zuanon J., Villéger S., Williams S.E., Baraloto C., Fortunel C., et al. Rare species contribute disproportionately to the functional structure of species assemblages. Proc R Soc B. 2016;283(1828):20160084. doi: 10.1098/rspb.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia Y., Leung M.H.Y., Tong X., Wilkins D., Lee P.K.H., Lozupone C. Rare Taxa Exhibit Disproportionate Cell-Level Metabolic Activity in Enriched Anaerobic Digestion Microbial Communities. mSystems. 2019;4(1) doi: 10.1128/mSystems.00208-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raphael M.G., Molina R. Island Press; Washington, DC: 2007. Conservation of rare or little-known species: Biological, social, and economic considerations; p. 44. [Google Scholar]
- 26.Pust M.-M., Wiehlmann L., Davenport C., Rudolf I., Dittrich A.-M., Tümmler B. The human respiratory tract microbial community structures in healthy and cystic fibrosis infants. NPJ Biofilms Microbiomes. 2020;6(1) doi: 10.1038/s41522-020-00171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pust M.M., Tümmler B. Identification of core and rare species in metagenome samples based on shotgun metagenomic sequencing, Fourier transforms and spectral comparisons. ISME Commun. 2021;1:2. doi: 10.1038/s43705-021-00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann J., Kaleta C., Waschina S. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 2021;22:81. doi: 10.1186/s13059-021-02295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valiente-Mullor C., Beamud B., Ansari I., Francés-Cuesta C., García-González N., Mejía L., et al. One is not enough: On the effects of reference genome for the mapping and subsequent analyses of short-reads. PLOS Comput Biol. 2021;17(1):e1008678. doi: 10.1371/journal.pcbi.1008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badri M., Kurtz Z.D., Bonneau R., Müller C.L. Shrinkage improves estimation of microbial associations under different normalization methods. NAR Genom Bioinform. 2021;2020:2. doi: 10.1093/nargab/lqaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gloor G.B., Macklaim J.M., Vu M., Fernandes A.D. Compositional uncertainty should not be ignored in high-throughput sequencing data analysis. Austrian J Stat. 2016;45(4):73–87. [Google Scholar]
- 32.Aitchison J. The statistical analysis of compositional data. J R Stat Soc Ser B. 1982;44(2):139–160. [Google Scholar]
- 33.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: And this is not optional. Front Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davenport C, Scheithauer T (2017) Wochenende – A whole genome/metagenome sequencing alignment pipeline (version 1.1). Github repository, https://github.com/MHH-RCUG/Wochenende.
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chouvarine P., Wiehlmann L., Moran Losada P., DeLuca D.S., Tümmler B., Dalby A.R. Filtration and normalization of sequencing read data in whole-metagenome shotgun samples. PLoS ONE. 2016;11(10):e0165015. doi: 10.1371/journal.pone.0165015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losada P.M., Chouvarine P., Dorda M., Hedtfeld S., Mielke S., et al. The cystic fibrosis lower airways microbial metagenome. ERJ Open Res. 2016;2:00096–2015. doi: 10.1183/23120541.00096-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:1–12. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurdie P.J., Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLOS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao C.H., Yu G., Cai P. ggVennDiagram: An intuitive, easy-to-use, and highly customizable R package to generate Venn Diagram. Front Genet. 2021;12:1598. doi: 10.3389/fgene.2021.706907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liaw A., Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 42.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 43.Kursa M.B., Rudnicki W.R. Feature selection with the Boruta package. J Stat Softw. 2010;36 [Google Scholar]
- 44.Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E., et al. The MetaCyc database of metabolic pathways and enzymes-a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–D453. doi: 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeske L., Placzek S., Schomburg I., Chang A., Schomburg D. BRENDA in 2019: A European ELIXIR core data resource. Nucleic Acids Res. 2019;47(D1):D542–D549. doi: 10.1093/nar/gky1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. BLAST+: Architecture and applications. BMC Bioinform. 2009;10(1) doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry D., Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst 1695; 2006.
- 50.Fruchterman T.M.J., Reingold E.M. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21(11):1129–1164. [Google Scholar]
- 51.Albert R., Jeong H., Barabási A.-L. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 52.Fréchet M.M. Sur quelques points du calcul fonctionnel. Rend Circ Mat Palermo. 1906;22(1):1–72. [Google Scholar]
- 53.Shervashidze N., Schweitzer P., van Leeuwen E.J., Mehlhorn K., Borgwardt K.M. Weisfeiler-Lehman Graph Kernels. J Mach Learn Res. 2011;12:2539–2561. [Google Scholar]
- 54.Sugiyama M., Ghisu M.E., Llinares-López F., Borgwardt K., Wren J. graphkernels: R and Python packages for graph comparison. Bioinformatics. 2018;34(3):530–532. doi: 10.1093/bioinformatics/btx602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC. R: A Language and Environment for Statistical Computing; 2020.
- 56.Doolittle W.F., Booth A. It’s the song, not the singer: an exploration of holobiosis and evolutionary theory. Biol Philos. 2016;321:5–24. [Google Scholar]
- 57.Pizarro-Cerdá J., Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124(4):715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Lingner M., Herrmann S., Tümmler B. Adherence of Pseudomonas aeruginosa to cystic fibrosis buccal epithelial cells. ERJ Open Res. 2017;3(1):00095-2016. doi: 10.1183/23120541.00095-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Falkowski N.R., Huffnagle G.B., et al. Bacterial topography of the healthy human lower respiratory tract. MBio. 2017;8(1) doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10(2):299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Beck J.M., Huffnagle G.B., et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira M.B., Wallroth M., Jonsson V., Kristiansson E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genomics. 2018;19:274. doi: 10.1186/s12864-018-4637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhr G.T., Dohnalová L., Thaiss C.A. The Dimension of Time in Host-Microbiome Interactions. mSystems. 2019;4(1) doi: 10.1128/mSystems.00216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coburn B., Wang P.W., Diaz Caballero J., Clark S.T., Brahma V., Donaldson S., et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5(1) doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khanolkar R.A., Clark S.T., Wang P.W., Hwang D.M., Yau Y.C.W., Waters V.J., et al. Ecological Succession of Polymicrobial Communities in the Cystic Fibrosis Airways. mSystems. 2020;5(6) doi: 10.1128/mSystems.00809-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinn R.A., Whiteson K., Lim Y.W., Zhao J., Conrad D., LiPuma J.J., et al. Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes. 2016;2(1) doi: 10.1038/s41522-016-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ijaq J., Chandrasekharan M., Poddar R., Bethi N., Sundararajan V.S. Annotation and curation of uncharacterized proteins- challenges. Front Genet. 2015;6:119. doi: 10.3389/fgene.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamames J., Cobo-Simón M., Puente-Sánchez F. Assessing the performance of different approaches for functional and taxonomic annotation of metagenomes. BMC Genomics. 2019;20:960. doi: 10.1186/s12864-019-6289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haynes W.A., Tomczak A., Khatri P. Gene annotation bias impedes biomedical research. Sci Rep. 2018;8:1–7. doi: 10.1038/s41598-018-19333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schnoes A.M., Ream D.C., Thorman A.W., Babbitt P.C., Friedberg I., Orengo C.A. Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLoS Comput Biol. 2013;9(5):e1003063. doi: 10.1371/journal.pcbi.1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeffery C.J. Protein moonlighting: What is it, and why is it important? Philos Trans R Soc B. 2018;373(1738):20160523. doi: 10.1098/rstb.2016.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hernández S., Franco L., Calvo A., Ferragut G., Hermoso A., Amela I., et al. Bioinformatics and moonlighting proteins. Front Bioeng Biotechnol. 2015;3 doi: 10.3389/fbioe.2015.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Starr A.E., Deeke S.A., Li L., Zhang X.u., Daoud R., Ryan J., et al. Proteomic and metaproteomic approaches to understand host-microbe interactions. Anal Chem. 2018;90(1):86–109. doi: 10.1021/acs.analchem.7b04340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coding scripts and reference databases are available from https://github.com/mmpust/airway-metagenome-simulations. Microbial sequencing data can be obtained from the European Nucleotide Archive (study accession number PRJEB38221).