Summary
c-MYC controls global gene expression and regulates cell proliferation, cell differentiation, cell cycle, metabolism and apoptosis. According to some estimates, MYC is dysregulated in ≈70% of human cancers and strong evidence implicates aberrantly expressed MYC in both tumor initiation and maintenance. In vivo studies show that MYC inhibition elicits a prominent anti-proliferative effect and sustained tumor regression while any alteration on healthy tissue remains reversible. This opens an exploitable window for treatment that makes MYC one of the most appealing therapeutic targets for cancer drug development. This review describes the main functional and structural features of the protein structure of MYC and provides a general overview of the most relevant or recently identified interactors that modulate MYC oncogenic activity. This review also summarizes the different approaches aiming to abrogate MYC oncogenic function, with a particular focus on the prototype inhibitors designed for the direct and indirect targeting of MYC.
Keywords: MYC, Therapeutic strategies, Cancer, Small-molecule inhibitors, MYC protein interactors
Introduction
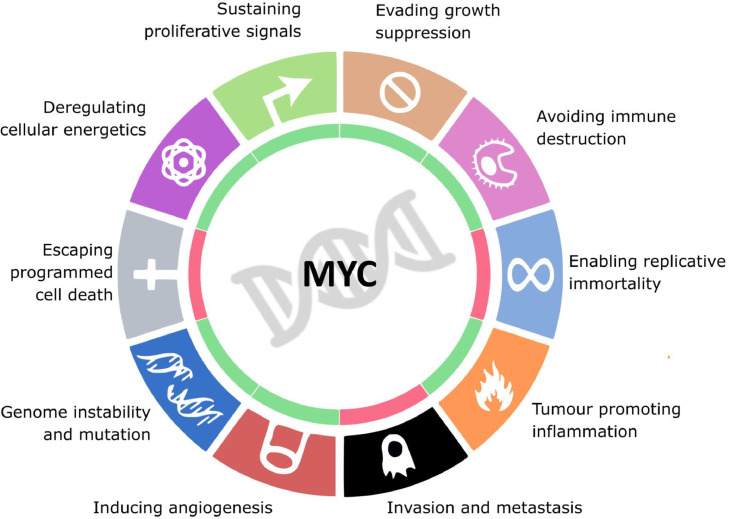
The oncoprotein MYC is a pleiotropic transcription factor that modulates global gene expression and regulates critical cellular processes including proliferation, differentiation, cell cycle, metabolism and apoptosis. Strong evidence supports aberrant MYC expression as a driver of both tumor initiation and maintenance1 and is associated with all the “hallmark” features of cancer2,3 (Figure 1). The role of MYC as a general or specific transcription factor has been traditionally controversial, although recent studies further support the notion that MYC is a general amplifier of highly expressed genes.4, 5, 6 The MYC oncogene family includes c-MYC, MYCN (N-MYC) and MYCL (L-MYC). All three paralogs have a similar function but show distinct expression timings and tissue specificities during development. c-MYC is ubiquitously expressed during tissue development and in a broad variety of tumors. N-MYC is expressed in neural tissue and early hematopoietic development and is deregulated in different cancer types including neuroblastoma, rhabdomyosarcoma, medulloblastoma, Wilms tumor or retinoblastoma although it can functionally replace c-MYC in some contexts.7 L-MYC is expressed in lungs and particularly overexpressed in small cell lung carcinomas although it is considered to have lower transforming activity.7,8 MYC activity is usually tightly controlled at the transcriptional and protein level, but is estimated to be aberrantly expressed in up to 70% of human cancers, many of which are highly aggressive (e.g. acute leukemia and high-grade lymphoma) and/or show poor response to treatment (e.g., small-cell lung carcinoma and neuroblastoma).1 In support of the oncogene addiction model, it has been demonstrated that transient MYC inhibition in vivo results in sustained tumor regression by promoting proliferative arrest, differentiation, apoptosis and cellular senescence in cancer cells. Notably, the anti-proliferative effects on normal tissue remain minimal and reversible.9,10 Furthermore, MYC has an unforeseen major role in enabling tumors to escape immunosurveillance through various mechanisms including decrease of MHCI expression or upregulation of inhibitory cytokines and immune check-point proteins such as PD-L1 and CD47, providing a compelling rational for combining MYC inhibition and immune checkpoint blockade.11, 12, 13 Together, these data suggest small molecules targeting MYC have the potential to exploit a clinically meaningful therapeutic window making MYC one of the most enticing targets for cancer drug development.
Figure 1.
MYC regulates all hallmarks of cancer. Oncogenic levels of MYC promote proliferative signalling, inhibition of growth suppression, escaping immune response, tumour inflammation, angiogenesis, genome instability and changes in cellular metabolism; and inhibits cell replicative immortality, metastasis and the escape of programmed cell death.
Several approaches have attempted to inhibit MYC directly or indirectly at all levels of its regulation. Some strategies that have shown promising results in vivo have focused on preventing MYC gene expression using antisense oligonucleotides (ASOs) that target MYC mRNA14 or G4-quadruplex binders that stabilize these DNA secondary structures. Other strategies are based on inhibiting MYC synthetic lethal genes required for tumorigenesis or tumor maintenance such as BUD31,15 NUAK1 (ARK5)16 or eIF4F.17 Other approaches receiving interest aim to reduce MYC stability and function at the protein level.18,19 However, despite massive efforts, targeting MYC with clinical grade small molecules still represents an intractable challenge, particularly at the protein level. The fact that important functional domains of MYC are intrinsically disordered (Figure 2a) and lack an enzymatically-active site, has precluded structure-guided drug design. In addition, the high affinity interaction between MYC and its obligate partner MAX along with the partial functional redundancy of the different MYC family members and their nuclear localization pose an enduring obstacle for the design of effective MYC inhibitors.20
Figure 2.
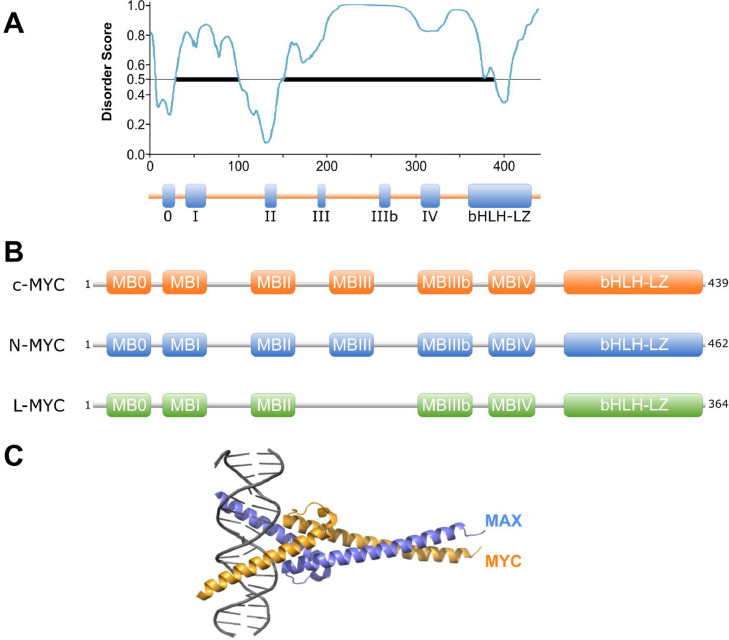
Structure of MYC protein. A) Intrinsically disordered region prediction for c-MYC (predictor tool PONDR). B) Organization of the main protein domains and conservation across different MYC super-family members. C) X-ray structure of MYC:MAX heterodimer binding DNA (PDB ID: 1NKP).
This review describes the main functional and structural features of the MYC protein, delineates the most relevant or recently identified MYC cofactors that modulate its oncogenic activity and summarizes recent advances on targeting MYC using small molecule inhibitors.
MYC structure
c-MYC (hereafter MYC) is a 439 amino acid oncoprotein that contains a well characterized C-terminal DNA-binding and an N-terminal transactivation domain (TAD). The C-terminal region ∼100 residues, comprises a basic helix-loop-helix leucine zipper (bHLH-LZ) segment that regulates the heterodimerization between MYC and its bHLH-LZ partner MAX, mediating its binding to gene promoters.21 In contrast, the TAD is comprised of residues 1-143, conforming an intrinsically disordered domain that regulates MYC's biological activity and is essential for MYC-mediated transcriptional activation.18,22 This intrinsically disordered nature of MYC structure has been linked to the formation of transcriptional condensates at super-enhancer sites and may represent, if confirmed, a previously overlooked opportunity to tackling MYC with certain antineoplastic agents that can be preferentially partitioned into such phase-separated condensates.23,24
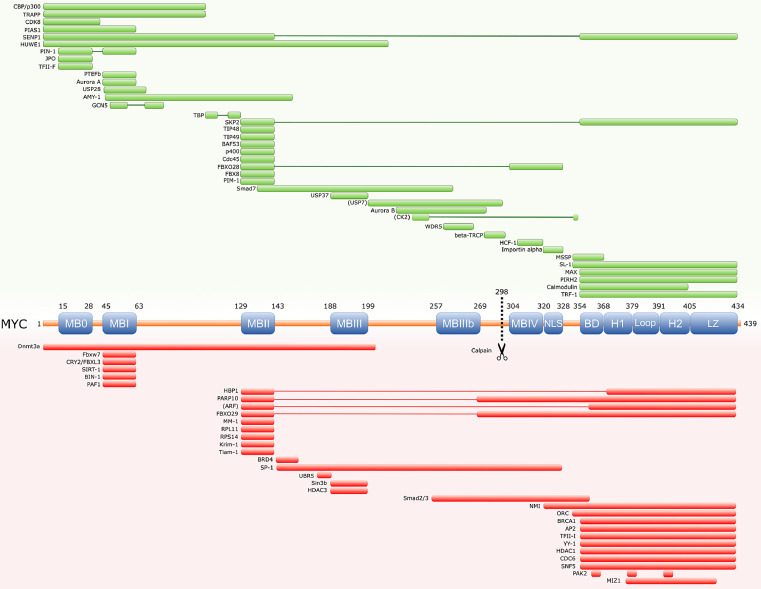
The protein sequence of MYC contains several evolutionary-conserved segments including the so called MYC homology boxes (MB), that modulate MYC function and are mostly shared amongst all MYC paralogs, L-MYC and N-MYC (Figure 2b).25,26 Each MB shows distinct, sometimes cell dependent, contributions to MYC activity although functional redundancies have been described. MBs are often involved in highly dynamic protein-protein interactions with a wide number of cofactors that contribute to modulate MYC's function as a master regulator (reviewed in27) (Figure 3). These interactions can be multivalent and occasionally involve distal binding sites outside these conserved domains. In fact, on many occasions MBs do not function as isolated regulatory entities and instead establish dynamic interactions that define complex patterns of cooperation modulating important aspects of the protein.
Figure 3.
Schematic of MYC protein interactors. The minimal interaction region between both proteins is mapped. Green boxes represent activators of MYC function. Red boxes represent repressors of MYC activity.
N-terminal Transactivation Domain
The N-terminal TAD of MYC comprises MB0, MBI and MBII subdomains involved in the regulation of the transcriptional activity and stability of the protein.26,28, 29, 30, 31
MBI contains the canonical phosphodegron composed by residues S62 and T58 that regulates MYC protein stability through a sequential and hierarchical phosphorylation process. The residue S62 is first phosphorylated by RAS/MEK/ERK/CDK2 priming the subsequent phosphorylation by GSK3β at T58, followed by the dephosphorylation of S62 mediated by PIN-1 and PP2A (reviewed in32). Recent findings demonstrated the phosphorylation of MYC at S67, which is proximal to the S62 residue, by Aurora B Kinase counteracts GSK3β and promotes protein stability.33 These series of post-translational modifications govern the SCFFBXW7/Ub-proteasome-mediated degradation of MYC and ultimately has a prominent impact on its function. The deregulation of this pathway has strong effects on MYC stability and is frequently linked to malignant cell proliferation. For example, in Burkitt's lymphoma, in which the MYC gene is translocated (t(8:14)), T58 in MBI is recurrently mutated, leading to protein stabilization.34 In addition, SCFFBXW7 is one of the most commonly mutated proteins of the ubiquitin-proteasome system in human cancer.35
MBII (residues 129-143) represents the most widely studied segment within the MYC TAD.26 This domain acts as a binding hub for a myriad of critical interactors (Figure 3) and is considered fundamental for MYC biological activity.31 In vivo, MBII is also indispensable for full MYC oncogenicity.30
Central region
The MBIII regulates protein stability and contributes to MYC cell transformation.26,36 MBIIIb encompasses a 10-amino-acid segment that is part of a PEST domain rich in proline, glutamic acid, threonine and serine residues that expands from positions 226-270 and is involved in the regulation of MYC stability but not in ubiquitination.37 MBIV domain may only be required for transformation in some biological contexts although there is a high in vitro assay variability amongst different cellular models. The main nuclear localization signal (NLS) sequence in MYC lies on residues 320-328 and, although it regulates the transport of MYC into the nucleus through the association with Importin α, it is not indispensable for MYC nuclear localization suggesting that other subdomains might be marginally involved in the process.38,39
C-terminal bHLH-LZ domain
The C-terminal region of the MYC protein represents the most widely studied domain. It contains the basic helix-loop-helix leucine zipper (bHLH-LZ) spanning residues 357-439 and plays a key role in DNA binding. At physiological levels, MYC preferentially binds at the canonical E box sequence 5’-CACGTG-3’ of target gene promoters whereas at deregulated levels it also binds the open chromatin containing the more prevalent non-canonical 5’-CANNTG-3’ E-boxes.40 The bHLH-LZ domain also stabilizes the interaction between MYC and MAX forming an obligate heterodimer that is essential for MYC binding to DNA and regulation of gene expression.41,42
The structural model of the MYC-MAX heterodimer in the apo form has been recently resolved providing important structural information of the bHLH-LZ and, in contrast to older models, avoiding the conformational changes induced by DNA binding.41,43 This new model facilitates structure-based design of drugs that could target the formation of the MYC-MAX dimer, or impede its binding to DNA, providing at the same time relevant structural information related to its interactions with other dimer-associated cofactors.
Not surprisingly, the bHLH-LZ domain is considered an attractive therapeutic target and significant efforts have been made over the last decades aiming to develop small molecule compounds that target the MYC-MAX association. Although clinical grade has not yet been achieved for such compounds, these efforts have provided important insights into MYC biology and represent the basis for the generation of improved therapeutic agents.
Targeting MYC
Direct inhibition of the MYC-MAX heterodimer and targeted degradation of MYC
The role of MAX as an obligate partner of MYC has led to significant efforts towards the development of MYC-MAX heterodimer inhibitors (Figure 2c; Table 1). Over the last decade, new chemically refined compounds such as KJ-Pyr-944 or SaJM589 have been identified with improved overall in vivo properties in comparison to the prototypic small molecules 10058-F4 and 10074-G5. SaJM589 suppressed cell proliferation in diverse cancer cell lines by disrupting MYC-MAX heterodimerization and potentially by promoting proteasome-mediated MYC degradation.45 However, to fully realize the therapeutic potential of SaJM589, in vivo studies in different cancer models are still required. Another recently identified small compound, MYCMI-6, also inhibits MYC-MAX heterodimerization by binding the bHLH-LZ domain of MYC and abrogates MYC-mediated transcription 46. In vitro, MYCMI-6 supresses MYC-dependent cell growth and this effect correlates with the level of MYC expression in tumor cells. The administration of MYCMI-6 in mouse bearing xenografts of SK-N-DZ neuroblastoma cells resulted in the reduction of tumor cell proliferation and microvasculature density, as well as the induction of apoptosis. In contrast to the previous small molecules that inhibit heterodimerization, the compound KSI-3716 blocks MYC-MAX binding to DNA.47 KSI-3716 is able to inhibit tumor growth in bladder murine xenograft models and has proven effective even in xenografts resistant to Gemcitabine chemotherapy.48,49
Table 1.
Pre-clinical studies and clinical trials evaluating the use of compounds for the direct or indirect inhibition of MYC protein.
| Target | Compound | Malignancy | Phase | Trial number | Main adverse effects |
|---|---|---|---|---|---|
| MYC-MAX | Omomyc | Advanced solid tumors. | Phase I/II | NCT04808362 | - |
| KJ-Pyr-9 | Breast cancer xenografts. | Pre-clinical | |||
| SaJM589 | B cell line P493-6, Ramos (Burkitt's lymphoma cell line), HL-60 and KG1a (acute myeloid leukemia cell lines). | Pre-clinical | |||
| MYCMI-6 | Neuroblastoma | Pre-clinical | |||
| KSI-3716 | Bladder cancer xenograft. | Pre-clinical | |||
| MAX:MAX | KI-MS2-008 | T-cell acute lymphoblastic leukemia and hepatocellular carcinoma mouse models. | Pre-clinical | ||
| Aurora A kinase | MLN8237/ Alisertib | Relapsed/Refractory peripheral T-cell lymphoma, non-Hodgkin lymphoma, advanced-non hematological malignancies, lung, breast, head and neck, gastroesophageal malignancies, and advanced or metastatic sarcoma. | Phase I/II/III |
NCT01482962 NCT00807495 NCT01045421 NCT01653028 |
Neutropenia, anemia or gastrointestinal disorders. |
| CD532 | Neuroblastoma xenografts. | Pre-clinical | |||
| PLK1 | BI6727 (Volasertib) | Ovarian cancer. | Phase II | NCT01121406 | Anemia, neutropenia and thrombocytopenia. |
| PP2A | DT-061/(AZD6244) | KRAS-driven lung cancer mouse models, lung adenocarcinoma xenografts, non-small cell lung cancer xenografts. | Pre-clinical | ||
| FTY720 | leukemia, colon cancer, non-small cell lung cancer, breast cancer, hepatocellular carcinoma, prostate cancer. | Pre-clinical | |||
| OP449 | AML, breast cancer, and pancreatic cancer. | Pre-clinical | |||
| Perphenazine | Brain, breast, colon, pancreatic, liver, skin and lung cancer, ovarian and oral carcinoma and leukemia and lymphoma cell lines. T-ALL and melanoma xenografts. | Pre-clinical | |||
| LB-100 | Myelodysplastic syndromes, extensive stage lung small-cell carcinoma and astrocytoma, glioblastoma multiforme, giant cell glioblastoma, glioma and oligodendrogliomas. | Phase I/II |
NCT03886662 NCT04560972 NCT03027388 |
- | |
| Pin-1 | KPT-6566 | Pancreatic, lung, prostate, and breast cancer cell lines. Lung cancer xenografts. | Pre-clinical | ||
| ATRA | Advanced adenoid cystic carcinoma, pancreatic ductal adenocarcinoma, breast cancer. | Phase I/II |
NCT03999684 NCT03307148 NCT04113863 |
-** | |
| BJP-06-005-3 | Pancreatic ductal adenocarcinoma. | Pre-clinical | |||
| Sulfopin | Neuroblastoma and pancreatic mouse model, and neuroblastoma zebrafish model. | Pre-clinical | |||
| PIM1 | AZD1208 | Acute myelogeneous leukemia and advanced solid malignancies including malignant lymphoma. | Phase I |
NCT01489722 NCT01588548 |
Guillain–Barré syndrome, increased blood creatinine, neutropenia, anemia and gastrointestinal disorders. |
| TP-3654 | Advanced solid tumors. | Phase I | NCT03715504 | - | |
| PIM447 | Myelofibrosis, acute myeloid leukemia, high risk myelodysplastic syndrome, multiple myeloma. | Phase I |
NCT02370706 NCT02078609 NCT01456689 NCT02160951 |
Thrombocytopenia, anemia or fatigue. | |
| SGI-1776 | Refractory prostate cancer and relapsed/refractory non Hodgkin's lymphoma, relapsed/refractory leukemias, | Phase I/II |
NCT00848601 NCT01239108 |
Cardiac toxicity. | |
| SEL24/MEN1703 | Acute myeloid leukemia. | Phase I/II | NCT03008187 | - | |
| SKP2 | SZL-P1-41 | Prostate and long tumor xenografts. | Pre-clinical | ||
| Dioscin | Colorectal adenocarcinoma xenografts. | Pre-clinical | |||
| FKA | Synovial sarcoma, osteosarcoma xenografts and prostate cancer in vivo models. | Pre-clinical | |||
| SKPin C1 | Uveal melanoma xenografts. | Pre-clinical | |||
| BRD4 (and other BET proteins) | OTX015/MK-8628 | Acute leukemia, solid tumors. | Phase I/II |
NCT02698189 NCT02698176 NCT02296476 NCT02259114 NCT01713582 |
Thrombocytopenia, anemia or gastrointestinal disorders. |
| GSK2820151 | Solid tumors. | Phase I | NCT02630251 | -* | |
| ZEN-3694 | Metastatic castration-prostate cancer and triple-negative breast cancer. | Phase I/II |
NCT02705469 NCT02711956 NCT03901469 |
Eye disorders, nausea or fatigue. | |
| CPI-0610 | Lymphoma, multiple myeloma, myelofibrosis leukemia, myelocytic, acute myelodysplastic/myeloproliferative neoplasm myelodysplastic syndrome (MDS). | Phase I/II |
NCT02158858 NCT01949883 NCT02157636 |
Gastrointestinal disorders, fatigue, anemia or thrombocytopenia. | |
| GSK525762/I-BET762 | Hematological malignancies. | Phase I/II |
NCT01943851 NCT01587703 |
Thrombocytopenia and gastrointestinal events. | |
| INCB057643 | Any advanced malignancy. | Phase I/II | NCT02711137 | Nausea, thrombocytopenia or fatigue. | |
| JQ1 | Multiple myeloma, T-ALL, B-ALL, acute myeloid leukemia cell lines. Medulloblastoma, glioblastoma, neuroblastoma, colorectal carcinoma and breast cancer cells. | Pre-clinical | |||
| USP7 | P22077 | Neuroblastoma and hepatocellular carcinoma xenografts. | Pre-clinical | ||
| XL177A | Ewing Sarcoma, acute myeloid leukemia, and multiple myeloma cell lines. | Pre-clinical | |||
| GNE-6640 | Acute myeloid leukemia cell lines and eosinophilic leukemia xenografts. | Pre-clinical | |||
| GNE6776 | Eosinophilic leukemia xenografts. | Pre-clinical | |||
| FT671 | Multiple myeloma xenografts. | Pre-clinical | |||
| HUWE1 | BI8622/ BI8626 | Multiple myeloma cell-lines and in vivo models. | Pre-clinical | ||
| Aurora B kinase | Acute myeloid leukaemia and advanced solid malignancies. |
NCT00497731 NCT00497991 |
Neutropenia, leukopenia, gastrointestinal disorders or stomatitis. | ||
| HUWE1 | BI8622/ BI8626 | Multiple myeloma cell-lines and in vivo models. | Pre-clinical | ||
| HDAC-(PI3K) | CUDC-907 | multiple myeloma, lymphoma, thyroid cancer and in solid tumors. | Phase I/II |
NCT02674750 NCT01742988 NCT02909777 NCT03002623 |
Gastrointestinal events or thrombocytopenia, lymphopenia, leukopenia or anemia. |
Only1/2 patients per group.
The small molecule MYCi975 was recently identified following a screen of 16 million compounds using an in silico five-point pharmacophore model.11 MYCi975 directly binds to the HLH domain of MYC, enhancing T58 phosphorylation, in turn reducing MYC stability, transcriptional activity and cancer cell viability. In vivo, MYCi975 showed reasonable pharmacokinetic properties, with the capacity to reduce tumor growth, at the same time potentiating immune cell infiltration at the site of the tumor. Most notably, when combined with anti-PD-1 immunotherapy, MYCi975 significantly reduced tumor growth suggesting combining MYC inhibition and immune checkpoint blockade is an enticing potential therapeutic strategy to test in future trials.
Omomyc is a 90 amino acid MYC mini-mutant that comprises the bHLH-LZ domain and competes with c-MYC, N-MYC and L-MYC for binding to DNA displacing the MYC/MAX heterodimers and inhibiting transcription of target genes.50, 51, 52 The role of Omomyc as an effective dominant negative form able to inhibit MYC has been evidenced in several cellular models showing anti-proliferative and pro-apoptotic effects.52,53 In vivo, Omomyc-mediated MYC inhibition elicited a potent anti-proliferative effect and sustained tumor regression with no detrimental effect on healthy tissue, and denoted, for the first time, MYC inhibition as a feasible anti-cancer therapeutic strategy.10 Recently, Omomyc mini-protein showed cell-penetrating activity and partial localization to the nucleus reverting the expression of MYC-regulated genes,54 although the MYC/Omomyc interaction is reported to occur primarily in the cytoplasm where Omomyc can bind excess MYC monomers and prevent it entering the nucleus.51 Co-treatment with Omomyc and Paclitaxel abrogated tumor growth of human lung cancer xenografts in mice.54 Despite its short effective half-life,51 a dose-escalation phase I/II clinical trial started in 2021 making OMO-103 (Omomyc) the first direct MYC inhibitor to reach clinical phase studies in patients with advanced solid tumors including non-small cell lung, colorectal and triple-negative breast cancer.
Omomyc inhibits MYC in part by promoting its proteasomal degradation.51 In recent years, the notion of directly targeting undruggable oncoproteins like MYC using Proteolysis-targeting chimeras (PROTACs) has grown in interest and the idea of enhancing the affinity of MYC-specific ubiquitin ligases through hetero-bifunctional small molecules to promote its degradation seems plausible but remains to be shown. Some of the high-affinity molecules that target MYC could potentially be an excellent initial platform for the development of future anti-MYC PROTAC strategies. The PROTAC-mediated degradation of MYC partners, in contrast, has proven successful in pre-clinical models. The compound ARV-771 robustly degrades BET proteins in castration-resistant prostate cancer subsequently depleting the expression of c-MYC.55 Similarly, in neuroblastoma, ARV-825 targets BET proteins resulting in inhibition of both c-MYC and N-MYC.56 The bacterial protease Lon has also recently been described to directly degrade c-MYC in bladder and colon cancer murine models, presenting an additional potential tool to treat MYC-dependent tumors based on MYC degradation.57
Indirect inhibition of MYC
To date, most efforts for developing clinically applicable small molecule inhibitors of MYC have focused on targeting the MYC/MAX heterodimer, but the majority of compounds developed thus far show low cellular or in vivo potency, suboptimal pharmacological properties or unacceptable off-target effects.1,11 Due to the intrinsically disordered nature of its protein structure, MYC has been traditionally considered “undruggable”. However, the MYC protein presents transient secondary structure elements that become conformationally stable and can be structurally resolved when bound to other cofactors. These heterodimeric crystal models, such as those obtained for MYC-WDR558 or MYC-Aurora A,59 provide a platform for the structure-based design of MYC-inhibitors potentially with a greater specificity compared to compounds that recognize disordered polypeptides.28,60 Notably, some of these cofactors are functionally essential for MYC oncogenicity and can be potentially targeted for the indirect inhibition of MYC (Table 1; Figure 3).
Targeting MAX monomers and MAX-MAX homodimers
Apart from heterodimerizing with MYC, MAX can form homodimers or bind the proteins MGA and MXD.61 This MAX extended network antagonizes MYC function by sequestering MAX from MYC. The compound NSC13728 stabilizes MAX/MAX homodimers, thereby decreasing MYC-mediated transcription and oncogenic transformation in chicken embryonic fibroblasts.62 KI-MS2-008 is another compound that has been recently reported to stabilize the MAX-MAX homodimers in vitro.63 KI-MS2-008 reduces the expression of MYC target genes and cancer cell growth in MYC-dependent cell lines and suppresses tumor growth in mouse models of T-cell acute lymphoblastic leukemia (T-ALL) and hepatocellular carcinoma (Table 1). These encouraging results obtained with MAX stabilizers supports the feasibility of modulating MAX dimerization as an alternative to targeting the MYC-MAX heterodimer. In addition, other interactors of the extended MAX network may provide the foundations for the development of similar therapeutic strategies.
Targeting MYC post-translational modifiers
MYC protein stability is tightly controlled by the Ub-proteasome system, a process in which the MBI phosphodegron is critically but not exclusively involved. Some of the proteins implicated in the regulation of MYC protein stability are considered as potentially exploitable therapeutic targets (Table 1).
Targeting of MYC/SCFFBXW7 axis through Aurora-A kinase or PLK1
The compound oridonin activates SCFFBXW7 leading to the degradation of MYC, inducing apoptosis in leukemia and lymphoma cells.64 Aurora A kinase prevents SCFFBXW7-mediated MYC proteolysis by binding MBI through an interaction that relies on the phosphorylation status of T58 and S62.60,65 The inhibition of Aurora A kinase using small molecule inhibitors has been explored as a strategy to target MYC function indirectly. Compounds such as MLN8237/alisertib and CD532 render the Aurora A-c-MYC/N-MYC complex dysfunctional and promote tumor regression in neuroblastoma and human hepatocarcinoma cells.65,66 CD532 promotes N-MYC protein degradation in neuroblastoma xenografts,67 while MLN8237/alisertib showed a compelling antitumor effect in various Phase I/II clinical trials including patients with non-Hodgkin lymphoma, advanced non-hematological malignancies and metastatic sarcomas, although it is associated with significant side effects, particularly hematological.68 Unfortunately, most of the small molecule inhibitors that target this complex have not reached Phase III clinical trials, and for those that have, such as alisertib (tested in relapsed/refractory peripheral T-cell lymphoma), have not shown encouraging results.69
The Polo-Like Kinase-1 (PLK1) directly binds and phosphorylates FBXW7 promoting its autopolyubiquitination and degradation resulting in stabilization of N-MYC. In a feedforward loop, stabilized N-MYC activates PLK-1 transcription further supporting N-MYC oncogenicity. The selective inhibition of PLK-1 using BI6727 in combination with the BCL2 inhibitor ABT199 reduces neuroblastoma tumor volume in vivo supporting the clinical potential of combined PLK1/BCL2 inhibitors for MYC-over expressing malignancies.70 In a phase II clinical trial, single agent BI6727 (Volasertib) has shown anti-tumor activity in patients with ovarian cancer.71
PP2A activation and Pin-1 abrogation
PP2A is frequently inactivated in human cancers.72 The important role played by PP2A in the SCFFBXW7-mediated degradation of MYC prompted the study of either the direct or indirect therapeutic activation of PP2A to potentially enhance the dephosphorylation of MYC pS62 and its subsequent proteasomal degradation. Small molecule activators of PP2A (SMAPS) have emerged as potential therapeutic drugs for the indirect inhibition of MYC specially in kinase-inhibition resistant tumors.72, 73, 74, 75 In a recent study, the treatment of non-small cell lung cancer xenografts with the SMAP DT-061 suppressed tumor growth accompanied by abrogation of c-MYC protein.76 The combined treatment of DT-061 with the MEK inhibitor AZD6244, or compounds such as FTY720 and FTY720 analogues OP449 or perphenazine, have proven effective in multiple pre-clinical models and are considered promising for cancer therapeutics.72,77, 78, 79 Despite its role as a tumor suppressor, inhibition of PP2A-C by the small molecule inhibitor LB-100 has a potent sensitization effect for chemotherapy and radiotherapy that is currently being tested alone or in combined regimes in Phase I clinical trials including patients with myelodysplastic syndromes, small-cell lung cancer and recurrent glioblastoma.77
The proline isomerase PIN1 is essential in regulating MYC proteolysis via the Ub-proteasome system. PIN1 expression is increased in most cancers and is associated with poor outcome.80 KPT-6566 is a recently described small molecule that binds and inhibits the PIN1 active site and targets it for degradation.81 At the same time, remnant chemical species that promote DNA damage originate from the interaction between PIN1 and KPT-6566. Through this dual mechanism of action, KPT-6566 induces cell death specifically in cancer cells in vitro and limits lung metastasis growth in vivo. The retinoid ATRA, was identified following a mechanism-based screen as a new specific binder to the PIN1 active site that inhibits PIN1 function and induces its degradation in acute promyelocytic leukemia and breast cancer cells.82 Moreover, ATRA was recently reported to suppress Tamoxifen-resistant breast cancer cell growth in vitro and in tumor xenografts.83 Recently, the highly selective compound BJP-06-005-3 was designed to probe Cys113 in the PIN1 active site and suppressed cell viability in pancreatic ductal adenocarcinoma.84 However, despite being a promising tool, further chemical optimization may be needed to improve BJP-06-005-3 bioavailability and in vivo performance.85 Similarly, the compound Sulfopin has recently been reported to target Cys113 downregulating MYC-target genes and reducing tumor progression in N-MYC driven neuroblastoma in vivo, as well as pancreatic cancer models. Despite the moderate effect on tumor viability in vitro, Sulfopin shows activity and very low toxicity in vivo suggesting that higher doses or combination with other treatments might be suitable for future treatments in MYC-driven malignancies.86
Inhibition of PIM1 kinase
PIM1 promotes T58 dephosphorylation and S62/S329 phosphorylation, stabilizing the MYC protein and increasing both its oncogenic transcriptional activity and transforming capacity.87 PIM1 directly interacts with MBII and is recruited to MYC target genes where it phosphorylates nucleosomes at H3S10 contributing to MYC-dependent transcriptional activation and cellular transformation.88 PIM1, along with other PIM kinases, is aberrantly expressed in several malignancies and is considered an attractive target for therapy. Accordingly, a number of small molecule PIM inhibitors have been tested in clinical trials suggesting a potential therapeutic role.89 The small molecule AZD1208 is a pan-PIM kinase inhibitor that inhibits cell growth, induces apoptosis and sensitizes to radiation.90 AZD1208 has been tested in phase I clinical trials including acute myeloid leukemia (AML) and advanced solid tumors;91 however, despite showing on-target engagement, it lacked clinical efficacy as a monotherapy, with a relatively high toxicity, particularly gastrointestinal and hematological.92 However, the combination of AZD1208 with other agents such as immunotherapeutics, immune checkpoint blockade or synthetic lethal targets might have potential to improve functional outcomes.93 Toxicity has also proven an issue with the PIM kinase inhibitor SGI-1776, where QT prolongation was observed on a phase I clinical trial, attributed to inhibition of the cardiac potassium channel hERG, leading to trial termination (NCT00848601). TP-3654 (SGI-9481), a second-generation pan-PIM kinase inhibitor that lacks off-target activity against hERG, has shown higher potency and lower toxicity in clinical trials for solid tumors.94 Other examples of PIM kinase inhibitors under evaluation are PIM447 which has activity in multiple myeloma, and SEL24/MEN1703, currently tested in a Phase I/II clinical trial in patients with AML.91,95 Targeting PIM kinases using small molecule inhibitors remains a promising anti-tumor strategy, and the outcome of ongoing clinical trials in terms of efficacy and toxicity are eagerly awaited.
Targeting SKP2
The E3 ligase SKP2 binds MBII along with the HLH-LZ domain, and targets MYC for ubiquitination and proteasome degradation. Distinct from SCFFBXW7, SKP2 increases MYC turnover activating transcription of some MYC target genes.96,97 High expression of SKP2 correlates with poor outcome in multiple malignancies98 and its inactivation is considered a potential therapeutic strategy. Several SKP2 inhibitors, including compounds such as SZL-P1-41,99 FKA100,101 or Dioscin,102 have been developed and show suppression of SKP2 oncogenic function in pre-clinical models, providing proof-of-principle for therapeutic targeting of SKP2 in human cancer therapy with associated high efficacy and safety. Further improvement on metabolic and pharmacokinetic properties of these compounds may eventually boost the development of candidates with clinical potential.
USP7 inhibition
USP7 is a multi-substrate deubiquitinating enzyme that plays a key role in proliferation and tumorigenesis, partly because it prevents the proteolysis of c-MYC and N-MYC.103 In neuroblastoma patients, USP7 overexpression is associated with a poor outcome and its inhibition in neuroblastoma xenografts by the small molecule P22077 prevents tumor growth via N-MYC destabilization suggesting the potential of USP7 inhibition as an exploitable therapy.104 In other malignancies such as hepatocellular carcinoma, USP7 overexpression correlates with disease progression and its inhibition by P22077 also suppresses tumor growth in vivo.105 Other small molecule USP7 inhibitors such as XL177A, GNE-6640, GNE6776 or FT671 suppress the growth of different cancer cell lines and inhibit tumor growth in xenograft models.106, 107, 108 It is also worth noting that USP7 has been shown to have an oncogenic and tumor suppressor role within different subtypes of T-ALL,109 indicating that careful patient selection and biomarker analyses will need to be considered when USP7 inhibitors are tested in clinical trials.
Aurora B kinase
Aurora B (AURKB), a kinase that plays an important role during cell division, is frequently overexpressed in different types of cancer, including ALL and AML, and is associated with poor prognosis in patients.110 A recent study in T-ALL shows that the direct interaction between MYC and AURKB promotes leukemogenesis and depicts a model by which AURKB increases MYC stability after the phosphorylation of S67 antagonizing the binding of GSK3β.33 In the same study, AURKB depletion reduced MYC-induced gene expression and MYC protein levels in both primary cells and cell lines. The treatment with the highly selective Aurora B inhibitor AZD1152 combined with Vincristine suppresses tumor growth in FBXW7WT T-ALL xenografts suggesting a new therapeutic strategy for T-ALL.33 In two Phase I/II clinical trials, treatment with the compound AZD1152 resulted in a hematologic response rate in 25% of patients with AML,111 but no responses were seen in advanced solid malignancies.112 Overall, AZD1152 showed good tolerability in both studies providing promising preliminary evidence of an exploitable therapeutic window for AURKB inhibition as a potential anticancer strategy in hematological malignancies (Table 1).
The MYC, HUWE1 and MIZ1 Trimer
The transcription factor MIZ1 binds MYC through the bHLH-LZ region and abrogates MYC transcriptional activity via two mechanisms.113 The first involves the trimerization of MYC, MIZ1 and DNMT3A followed by DNA methylation and subsequent inhibition of MYC target genes.114 The second mechanism is based on the competition for MYC binding between MIZ1 and the E2 ligase HUWE1.115,116 Depending on the cellular context, HUWE1 can act as a tumor suppressor or tumor promoter by regulating the turnover of MYC/MIZ1 and balancing the ratio of both proteins at gene promoters.115,117 Due to the profound functional implication of the MYC/MIZ1 interaction on MYC activity, fostering this association via small molecule inhibition of HUWE1 has been explored as a therapy.113,115,116,118,119 The small compounds BI8622 and BI8626 are specific inhibitors of HUWE1 that enhance MYC proteolysis and suppress MYC-dependent transactivation resulting in inhibition of cell growth in colon carcinoma and multiple myeloma.116,120 In vivo, HUWE1 knock-down clearly reduces tumor burden, but both BI8622 and BI8626 have suboptimal pharmacokinetics that prevents the assessment of their in vivo efficacy in MYC-driven tumor models. Nevertheless, these pre-clinical studies support the notion that it is possible to develop effective antitumor strategies by targeting HUWE1. It is likely that other mechanisms of regulation like MIZ1-HUWE1 are involved in the interaction between MYC and other transcription factors that act as functional repressors. Extensive characterization of these mechanisms would allow the development of novel therapeutic strategies for MYC-related malignancies.
Other MYC modifiers
Apart from SCFFBXW7, several other E3 ubiquitin ligases and proteases, such as CRY2-FBXL3, USP28, USP7 or USP37, SCFFBXO28, FBXO29 and RNF115 (reviewed in121), are involved in modulation of c-MYC and/or N-MYC stability. PIAS1 is involved in SUMOylation122 or ubiquitination of residues K51 and K52 via an SCFFBXW7-independent mechanism123,124 albeit the functional effect of these modifications on MYC are likely to be context dependent. All these MYC post-translational modifiers could be explored as potential therapeutic targets for MYC-driven malignancies.
Targeting of chromatin modifiers
BRD4
BRD4 is a transcriptional regulator involved in DNA replication and reparation that plays a key role in the maintenance of cell proliferation.125 Apart from being a well-known transcriptional modulator of the MYC and MYCN genes, BRD4 directly interacts, phosphorylates and destabilizes MYC at the protein level through the formation of a ternary complex with ERK1 creating a regulatory network that balances MYC activity.126 Simultaneously, MYC regulates BRD4-mediated acetylation of histones establishing a negative feedback loop that controls its own transcription. Due to its important implication in controlling MYC homeostasis, BRD4 has been intensively investigated as a therapeutic target using small compounds in MYC-driven cancers. The small molecule bromodomain inhibitor JQ1 binds BRD2/4 and blocks the association of BRD4 at acetylated histones within the MYC locus decreasing the expression of all MYC superfamily members.127,128 As a result, anti-tumor effects are observed in diverse hematological malignancies and solid tumors.127,129 JQ1 is the first-in-class compound that established proof-of-principle for BET inhibition as a cancer therapy, however, its poor in vivo pharmacokinetic properties preclude its use in clinical trials. Alternatively, numerous other improved BRD4 inhibitors such as OTX015/MK-8628, GSK2820151, ZEN-3694, CPI-0610, GSK525762/I-BET762 and INCB057643 have been developed and their safety, pharmacokinetics and activity are currently under assessment in Phase I and Phase I/II clinical trials including several cancers130,131 (Table 1). Unfortunately, none of these inhibitors have yet received regulatory approval. More studies are necessary to optimize optimal dosing schedules and to explore potential combinations with other compounds that might improve efficacy.
HDACs and TRRAP
A myriad of components of the pre-replicative complex, including CDC6,132 ORC1,133 SNF5,134,135 CBP/300 and HDACs,36,136,137 associate with MYC through the bHLH-LZ domain, regulating DNA replication origin in response to cellular replication stress and genomic instability.138,139 Apart from their role in gene regulation, the deacetylases HDAC1/2 also modulate MYC stability through direct Sin3b-mediated deacetylation of the MYC peptidic chain accompanied with a reduction of MYC transcriptional activity.140 Similarly, HDAC3 attenuates MYC-mediated gene expression and MYC-associated pro-apoptotic activity through a direct protein-protein interaction.36,137 Overexpression of HDACs is observed in different cancer types and reversing epigenetic anomalies using HDAC inhibitors is considered an enticing therapeutic strategy in cancer. Accordingly, several compounds are currently tested in clinical trials (Table 1).141 The compound CUDC-907 is a dual inhibitor of both HDACs and PI3K that suppresses growth in MYC-dependent cancer cells142 and demonstrates efficacy against prostate cancer mouse xenografts.143 Results of phase I/II clinical trials reported anti-tumor activity of CUDC-907 in multiple myeloma, lymphoma, thyroid cancer and in solid tumors with MYC alterations, suggesting the therapeutic potential of CUDC-907 for the treatment of diverse MYC-driven malignancies.75,144,145
Transformation-transactivation domain-associated protein (TRRAP) is one of the best characterized MYC cofactors. TRRAP is the integral subunit of the GCN5-containing and the TIP60-containing histone acetyltransferase (HAT) complexes and it is crucial for their recruitment to MYC target genes, a pivotal element in regulating transcriptional activation.146, 147, 148, 149, 150 TRRAP also directly acetylates MYC136,146,151 and the association of both proteins is indispensable for MYC oncogenic activity in most contexts.152 The recent in-depth structural characterization of the MYC-TRRAP interaction may form the basis for the development of novel MYC small molecule inhibitors.148
WDR5 and MYC
The WD40-repeat protein WDR5 promotes the assembly of multiple chromatin regulatory complexes responsible of the recruitment of MYC onto chromatin. The interaction between MYC and WDR5 depends on the hydrophobic residues of the highly conserved MBIIIb core EEIDVV and is important for transcription of protein synthesis-related genes, gene target recognition and for somatic cell reprogramming (Table 1).153 However, how WDR5-MYC interaction regulates target genes is still not completely understood and the implications of MBIIIb in cell transformation remains elusive due to contradictory results from in vitro transformation assays using different MYC deleterious mutants.26,153 In vivo, blocking the MYC-WDR5 interaction is emerging as a potential anti-tumor therapeutic strategy. In mice with stablished Burkitt's lymphoma tumors, exchanging wild type MYC with a WDR5-binding defective MYC mutant promotes tumor regression, offering a potentially exploitable therapeutic window.153 A series of high affinity small compounds that bind the MYC-site of WDR5 targeting the interaction between both proteins have been developed in vitro and are currently under optimization to improve their drug-like properties prior to cell or animal testing.154 It is not only the MYC-binding site of WDR5 that is an enticing therapeutic target; the small molecule C6 binds the WIN-site of WDR5 and displaces it from chromatin with the subsequent eviction of MYC and downregulation of genes associated with protein synthesis.153,155
Outstanding Questions
Over the last decades, MYC has moved from being considered “undruggable” into one of the most appealing therapeutic targets for cancer treatment. Despite the major challenges in directly inhibiting MYC, particularly in translating promising in vitro results to meaningful in vivo efficacy, the first direct MYC inhibitors are now starting clinical trial evaluation. Omomyc, for instance, has promising in vivo properties in pre-clinical testing and the results of early phase trials in patients with advanced solid tumors will provide valuable insights into both the efficacy and toxicity of this approach. The complex protein structure of MYC still poses a substantial challenge for development and optimization of direct MYC inhibitors. Recent advances in the in silico prediction of a protein structure based on the polypeptidic sequence may represent an important breakthrough towards the refinement of incomplete crystallographic data.156 Nevertheless, the higher demand on computational resources may present an important limitation for many research facilities and no in silico model is yet capable of fully deciphering the complexities of intrinsic disordered domains.
The indirect inhibition of MYC through targeting binding proteins that are critical for its oncogenicity have emerged as a promising alternative. The identification of novel key cofactors for MYC function in promoting cancer opens new exploitable ways for the indirect inhibition of MYC (Figure 4). In addition, the identification of novel critical binding partners may facilitate our understanding of the molecular mechanisms by which MYC promotes oncogenesis and tumor maintenance providing at the same time useful information for the development of future successful therapies. However, MYC interacts with a plethora of different proteins and these interactions are usually context dependent thus the effects of indirect MYC inhibition are likely to be tumor specific.
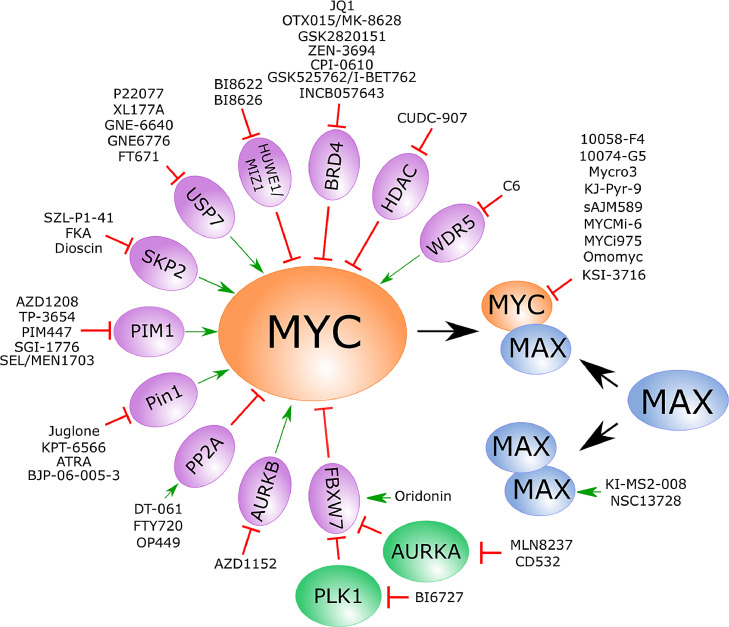
Figure 4.
Strategies for direct and indirect small-molecule inhibition of MYC protein.
Given the importance of MYC to tumor maintenance, it is possible that a variety of resistance mechanisms might be engaged by tumor cells to escape therapeutic targeting. For instance, mutations interfering with drug engagement might be selected for with certain small molecule inhibitors, or genomic alterations of the E3 ligase complex might occur on chronic exposure to PROTACs, as recently described for BET-PROTACs.157 Moreover, indirect targeting of MYC may induce highly adaptive signaling pathways, potentially associated with therapeutic resistance. In contrast, direct targeting of MYC with compounds such as Omomyc might be less likely to promote drug resistance since MYC acts as a central regulatory node of gene expression without obvious bypass pathways.158 The fact that c-MYC, N-MYC and L-MYC can functionally replace each other in some contexts adds an additional layer of complexity to the development of specific MYC-inhibitors, particularly as their interactomes vary. Although some small molecules have shown the ability to inhibit different MYC family members, the future development of new pan-MYC inhibitors will increase the chances of success when translating the results from bench-to-bedside. Finally, combination strategies with small molecule MYC inhibitors and immunomodulators or indirect MYC inhibitors along with other antitumoral agents should be included in future studies. However, the fact that MYC inactivation can induce cellular senescence159 needs to be considered when designing such combination trials, particularly those that include chemotherapeutic drugs whose efficacy is cell cycle dependent. Significant progress has been achieved to date, but further research is still required to better understand the regulation mechanism of several MYC interactors, and to bring MYC inhibitors to daily clinical practice.
Search strategy and selection criteria
Data for this Review were identified by searches of PubMed, and references from relevant articles using the search terms “MYC small molecule inhibitor”, “MYC binding protein OR MYC interactor” and “(MYC-binding protein) small molecule inhibitor”. All relevant information and reference code for clinical trials was retrieved from Clinicaltrials.gov. Abstracts and reports from meetings were excluded. Only articles published in English were included and original reports published after 2016 were prioritized highlighting more novel findings. Nevertheless, references back to 1988 have been included for contextualization.
Contributors
V. Llombart contributed on the conceptualization, data curation, funding acquisition, investigation, project administration, resources, writing of original draft and editing. M. R. Mansour contributed on the conceptualization, funding acquisition, resources, supervision, validation, and review of the manuscript.
Declaration of interests
No conflicts of interest.
Acknowledgments
This work was funded by Cancer Research UK to V.L and to M.M.
References
- 1.Carabet L.A., Rennie P.S., Cherkasov A. Therapeutic Inhibition of Myc in Cancer. Structural Bases and Computer-Aided Drug Discovery Approaches. Int J Mol Sci. 2018;20(1) doi: 10.3390/ijms20010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Gabay M., Li Y., Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6) doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nie Z.Q., Guo C.H., Das S.K., Chow C.C., Batchelor E., Simons S.S., et al. Dissecting transcriptional amplification by MYC. Elife. 2020;9 doi: 10.7554/eLife.52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeid R., Lawlor M.A., Poon E., Reyes J.M., Fulciniti M., Lopez M.A., et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nature Genetics. 2018;50(4):515. doi: 10.1038/s41588-018-0044-9. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., et al. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masso-Valles D., Beaulieu M.E., Soucek L.MYC, MYCL and. MYCN as therapeutic targets in lung cancer. Expert Opin Ther Targets. 2020;24(2):101–114. doi: 10.1080/14728222.2020.1723548. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z., Chen S.S., Clarke S., Veschi V., Thiele C.J. Targeting MYCN in Pediatric and Adult Cancers. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.623679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell-Intrinsic and Host-Dependent Mechanisms. Genes Cancer. 2010;1(6):597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soucek L., Whitfield J., Martins C.P., Finch A.J., Murphy D.J., Sodir N.M., et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han H., Jain A.D., Truica M.I., Izquierdo-Ferrer J., Anker J.F., Lysy B., et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell. 2019;36(5):483–497. doi: 10.1016/j.ccell.2019.10.001. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhanasekaran R., Deutzmann A., Mahauad-Fernandez W.D., Hansen A.S., Gouw A.M., Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2021 doi: 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrier P.I., Peltenburg LT. Relationship between myc oncogene activation and MHC class I expression. Adv Cancer Res. 1993;60:181–246. doi: 10.1016/s0065-230x(08)60826-x. [DOI] [PubMed] [Google Scholar]
- 14.Dhanasekaran R., Park J., Yevtodiyenko A., Bellovin D.I., Adam S.J., Kd A.R., et al. MYC ASO Impedes Tumorigenesis and Elicits Oncogene Addiction in Autochthonous Transgenic Mouse Models of HCC and RCC. Mol Ther Nucleic Acids. 2020;21:850–859. doi: 10.1016/j.omtn.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L., Ulbrich J., Muller J., Wustefeld T., Aeberhard L., Kress T.R., et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483(7391):608–612. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 17.Lin C.J., Nasr Z., Premsrirut P.K., Porco J.A., Jr., Hippo Y., Lowe S.W., et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 2012;1(4):325–333. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tansey Mammalian MYC Proteins and Cancer. New Journal of Science. 2014;Volume 2014 [Google Scholar]
- 19.Duffy M.J., O'Grady S., Tang M., Crown J. MYC as a target for cancer treatment. Cancer Treat Rev. 2021;94 doi: 10.1016/j.ctrv.2021.102154. [DOI] [PubMed] [Google Scholar]
- 20.Chen H., Liu H., Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amati B., Brooks M.W., Levy N., Littlewood T.D., Evan G.I., Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72(2):233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 22.Kato G.J., Barrett J., Villa-Garcia M., Dang CV. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boija A., Klein I.A., Sabari B.R., Dall'Agnese A., Coffey E.L., Zamudio A.V., et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175(7):1842–1855. doi: 10.1016/j.cell.2018.10.042. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein I.A., Boija A., Afeyan L.K., Hawken S.W., Fan M., Dall'Agnese A., et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368(6497):1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flinn E.M., Busch C.M., Wright AP. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol. 1998;18(10):5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J.M., Varmus H., et al. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu W.B., Helander S., Pilstal R., Hickman K.A., Lourenco C., Jurisica I., et al. Myc and its interactors take shape. Biochim Biophys Acta. 2015;1849(5):469–483. doi: 10.1016/j.bbagrm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Helander S., Montecchio M., Pilstal R., Su Y., Kuruvilla J., Elven M., et al. Pre-Anchoring of Pin1 to Unphosphorylated c-Myc in a Fuzzy Complex Regulates c-Myc Activity. Structure. 2015;23(12):2267–2279. doi: 10.1016/j.str.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amin C., Wagner A.J., Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993;13(1):383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalkat M., Resetca D., Lourenco C., Chan P.K., Wei Y., Shiah Y.J., et al. MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol Cell. 2018;72(5):836–848. doi: 10.1016/j.molcel.2018.09.031. e7. [DOI] [PubMed] [Google Scholar]
- 31.Cowling V.H., Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16(4):242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Farrell A.S., Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4(3) doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J., Wang J., Yue M., Cai X., Wang T., Wu C., et al. Direct Phosphorylation and Stabilization of MYC by Aurora B Kinase Promote T-cell Leukemogenesis. Cancer Cell. 2020;37(2):200–215. doi: 10.1016/j.ccell.2020.01.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhatia K., Huppi K., Spangler G., Siwarski D., Iyer R., Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5(1):56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 35.Yeh C.H., Bellon M., Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17(1):115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbst A., Hemann M.T., Tworkowski K.A., Salghetti S.E., Lowe S.W., Tansey WP. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 2005;6(2):177–183. doi: 10.1038/sj.embor.7400333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregory M.A., Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20(7):2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dang C.V., Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conti E., Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8(3):329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 40.Blackwell T.K., Huang J., Ma A., Kretzner L., Alt F.W., Eisenman R.N., et al. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13(9):5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair S.K., Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 42.Blackwood E.M., Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 43.Sammak S., Hamdani N., Gorrec F., Allen M.D., Freund S.M.V., Bycroft M., et al. Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA. Biochemistry. 2019;58(29):3144–3154. doi: 10.1021/acs.biochem.9b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart J.R., Garner A.L., Yu J., Ito Y., Sun M., Ueno L., et al. Inhibitor of MYC identified in a Krohnke pyridine library. Proc Natl Acad Sci U S A. 2014;111(34):12556–12561. doi: 10.1073/pnas.1319488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi S.H., Mahankali M., Lee S.J., Hull M., Petrassi H.M., Chatterjee A.K., et al. Targeted Disruption of Myc-Max Oncoprotein Complex by a Small Molecule. ACS Chem Biol. 2017;12(11):2715–2719. doi: 10.1021/acschembio.7b00799. [DOI] [PubMed] [Google Scholar]
- 46.Castell A., Yan Q., Fawkner K., Hydbring P., Zhang F., Verschut V., et al. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Sci Rep. 2018;8(1):10064. doi: 10.1038/s41598-018-28107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong K.C., Ahn K.O., Yang CH. Small-molecule inhibitors of c-Myc transcriptional factor suppress proliferation and induce apoptosis of promyelocytic leukemia cell via cell cycle arrest. Mol Biosyst. 2010;6(8):1503–1509. doi: 10.1039/c002534h. [DOI] [PubMed] [Google Scholar]
- 48.Jeong K.C., Kim K.T., Seo H.H., Shin S.P., Ahn K.O., Ji M.J., et al. Intravesical instillation of c-MYC inhibitor KSI-3716 suppresses orthotopic bladder tumor growth. J Urol. 2014;191(2):510–518. doi: 10.1016/j.juro.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 49.Seo H.K., Ahn K.O., Jung N.R., Shin J.S., Park W.S., Lee K.H., et al. Antitumor activity of the c-Myc inhibitor KSI-3716 in gemcitabine-resistant bladder cancer. Oncotarget. 2014;5(2):326–337. doi: 10.18632/oncotarget.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soucek L., Helmer-Citterich M., Sacco A., Jucker R., Cesareni G., Nasi S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17(19):2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 51.Demma M.J., Mapelli C., Sun A., Bodea S., Ruprecht B., Javaid S., et al. Omomyc Reveals New Mechanisms To Inhibit the MYC Oncogene. Mol Cell Biol. 2019;39(22) doi: 10.1128/MCB.00248-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiorentino F.P., Tokgun E., Sole-Sanchez S., Giampaolo S., Tokgun O., Jauset T., et al. Growth suppression by MYC inhibition in small cell lung cancer cells with TP53 and RB1 inactivation. Oncotarget. 2016;7(21):31014–31028. doi: 10.18632/oncotarget.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Annibali D., Whitfield J.R., Favuzzi E., Jauset T., Serrano E., Cuartas I., et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beaulieu M.E., Jauset T., Masso-Valles D., Martinez-Martin S., Rahl P., Maltais L., et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci Transl Med. 2019;11(484) doi: 10.1126/scitranslmed.aar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raina K., Lu J., Qian Y., Altieri M., Gordon D., Rossi A.M., et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113(26):7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z., Lim S.L., Tao Y., Li X., Xie Y., Yang C., et al. PROTAC Bromodomain Inhibitor ARV-825 Displays Anti-Tumor Activity in Neuroblastoma by Repressing Expression of MYCN or c-Myc. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.574525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler D.S.C., Cafaro C., Putze J., Wan M.L.Y., Tran T.H., Ambite I., et al. A bacterial protease depletes c-MYC and increases survival in mouse models of bladder and colon cancer. Nat Biotechnol. 2021;39(6):754–764. doi: 10.1038/s41587-020-00805-3. [DOI] [PubMed] [Google Scholar]
- 58.Thomas L.R., Adams C.M., Fesik S.W., Eischen C.M., Tansey WP. Targeting MYC through WDR5. Mol Cell Oncol. 2020;7(2) doi: 10.1080/23723556.2019.1709388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richards M.W., Burgess S.G., Poon E., Carstensen A., Eilers M., Chesler L., et al. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc Natl Acad Sci U S A. 2016;113(48):13726–13731. doi: 10.1073/pnas.1610626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayliss R., Burgess S.G., Leen E., Richards MW. A moving target: structure and disorder in pursuit of Myc inhibitors. Biochem Soc Trans. 2017;45(3):709–717. doi: 10.1042/BST20160328. [DOI] [PubMed] [Google Scholar]
- 61.Conacci-Sorrell M., McFerrin L., Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4(1) doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang H., Bower K.E., AEt Beuscher, Zhou B., Bobkov A.A., Olson A.J., et al. Stabilizers of the Max homodimer identified in virtual ligand screening inhibit Myc function. Mol Pharmacol. 2009;76(3):491–502. doi: 10.1124/mol.109.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Struntz N.B., Chen A., Deutzmann A., Wilson R.M., Stefan E., Evans H.L., et al. Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem Biol. 2019;26(5):711–723. doi: 10.1016/j.chembiol.2019.02.009. e14. [DOI] [PubMed] [Google Scholar]
- 64.Huang H.L., Weng H.Y., Wang L.Q., Yu C.H., Huang Q.J., Zhao P.P., et al. Triggering Fbw7-mediated proteasomal degradation of c-Myc by oridonin induces cell growth inhibition and apoptosis. Mol Cancer Ther. 2012;11(5):1155–1165. doi: 10.1158/1535-7163.MCT-12-0066. [DOI] [PubMed] [Google Scholar]
- 65.Dauch D., Rudalska R., Cossa G., Nault J.C., Kang T.W., Wuestefeld T., et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med. 2016;22(7):744–753. doi: 10.1038/nm.4107. [DOI] [PubMed] [Google Scholar]
- 66.Brockmann M., Poon E., Berry T., Carstensen A., Deubzer H.E., Rycak L., et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24(1):75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gustafson W.C., Meyerowitz J.G., Nekritz E.A., Chen J., Benes C., Charron E., et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26(3):414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tayyar Y., Jubair L., Fallaha S., McMillan NAJ. Critical risk-benefit assessment of the novel anti-cancer aurora a kinase inhibitor alisertib (MLN8237): A comprehensive review of the clinical data. Crit Rev Oncol Hematol. 2017;119:59–65. doi: 10.1016/j.critrevonc.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 69.O'Connor O.A., Ozcan M., Jacobsen E.D., Roncero J.M., Trotman J., Demeter J., et al. Randomized Phase III Study of Alisertib or Investigator's Choice (Selected Single Agent) in Patients With Relapsed or Refractory Peripheral T-Cell Lymphoma. J Clin Oncol. 2019;37(8):613–623. doi: 10.1200/JCO.18.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao D., Yue M., Su H., Ren P., Jiang J., Li F., et al. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol Cell. 2016;64(3):493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 71.Pujade-Lauraine E., Selle F., Weber B., Ray-Coquard I.L., Vergote I., Sufliarsky J., et al. Volasertib Versus Chemotherapy in Platinum-Resistant or -Refractory Ovarian Cancer: A Randomized Phase II Groupe des Investigateurs Nationaux pour l'Etude des Cancers de l'Ovaire Study. J Clin Oncol. 2016;34(7):706–713. doi: 10.1200/JCO.2015.62.1474. [DOI] [PubMed] [Google Scholar]
- 72.Kauko O., O'Connor C.M., Kulesskiy E., Sangodkar J., Aakula A., Izadmehr S., et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci Transl Med. 2018;10(450) doi: 10.1126/scitranslmed.aaq1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farrington C.C., Yuan E., Mazhar S., Izadmehr S., Hurst L., Allen-Petersen B.L., et al. Protein phosphatase 2A activation as a therapeutic strategy for managing MYC-driven cancers. J Biol Chem. 2020;295(3):757–770. doi: 10.1074/jbc.RA119.011443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sangodkar J., Perl A., Tohme R., Kiselar J., Kastrinsky D.B., Zaware N., et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J Clin Invest. 2017;127(6):2081–2090. doi: 10.1172/JCI89548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allen-Petersen B.L., Sears RC. Mission Possible: Advances in MYC Therapeutic Targeting in Cancer. BioDrugs. 2019;33(5):539–553. doi: 10.1007/s40259-019-00370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leonard D., Huang W., Izadmehr S., O'Connor C.M., Wiredja D.D., Wang Z., et al. Selective PP2A Enhancement through Biased Heterotrimer Stabilization. Cell. 2020;181(3):688–701. doi: 10.1016/j.cell.2020.03.038. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O'Connor C.M., Perl A., Leonard D., Sangodkar J., Narla G. Therapeutic targeting of PP2A. Int J Biochem Cell Biol. 2018;96:182–193. doi: 10.1016/j.biocel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tohme R., Izadmehr S., Gandhe S., Tabaro G., Vallabhaneni S., Thomas A., et al. Direct activation of PP2A for the treatment of tyrosine kinase inhibitor-resistant lung adenocarcinoma. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.125693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otreba M., Kosmider L. In vitro anticancer activity of fluphenazine, perphenazine and prochlorperazine. A review. J Appl Toxicol. 2021;41(1):82–94. doi: 10.1002/jat.4046. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y., Wu Y.R., Yang H.Y., Li X.Z., Jie M.M., Hu C.J., et al. Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 2018;9(9):883. doi: 10.1038/s41419-018-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campaner E., Rustighi A., Zannini A., Cristiani A., Piazza S., Ciani Y., et al. A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat Commun. 2017;8:15772. doi: 10.1038/ncomms15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei S., Kozono S., Kats L., Nechama M., Li W., Guarnerio J., et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. 2015;21(5):457–466. doi: 10.1038/nm.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang S., Chen Y., Liang Z.M., Li N.N., Liu Y., Zhu Y., et al. Targeting Pin1 by All-Trans Retinoic Acid (ATRA) Overcomes Tamoxifen Resistance in Breast Cancer via Multifactorial Mechanisms. Front Cell Dev Biol. 2019;7:322. doi: 10.3389/fcell.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pinch B.J., Doctor Z.M., Nabet B., Browne C.M., Seo H.S., Mohardt M.L., et al. Identification of a potent and selective covalent Pin1 inhibitor. Nat Chem Biol. 2020;16(9):979–987. doi: 10.1038/s41589-020-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pu W., Li J., Peng Y. Cysteine-113 covalency inspires the development of Pin1 inhibitor. Signal Transduct Target Ther. 2020;5(1):225. doi: 10.1038/s41392-020-00339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dubiella C., Pinch B.J., Koikawa K., Zaidman D., Poon E., Manz T.D., et al. Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo. Nature Chemical Biology. 2021;17(9):954. doi: 10.1038/s41589-021-00786-7. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y., Wang Z., Li X., Magnuson N.S. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27(35):4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 88.Zippo A., De Robertis A., Serafini R., Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9(8):932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X., Song M., Kundu J.K., Lee M.H., Liu ZZ. PIM Kinase as an Executional Target in Cancer. J Cancer Prev. 2018;23(3):109–116. doi: 10.15430/JCP.2018.23.3.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirschner A.N., Wang J., van der Meer R., Anderson P.D., Franco-Coronel O.E., Kushner M.H., et al. PIM kinase inhibitor AZD1208 for treatment of MYC-driven prostate cancer. J Natl Cancer Inst. 2015;107(2) doi: 10.1093/jnci/dju407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luszczak S., Kumar C., Sathyadevan V.K., Simpson B.S., Gately K.A., Whitaker H.C., et al. PIM kinase inhibition: co-targeted therapeutic approaches in prostate cancer. Signal Transduct Target Ther. 2020;5(1):7. doi: 10.1038/s41392-020-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cortes J., Tamura K., DeAngelo D.J., de Bono J., Lorente D., Minden M., et al. Phase I studies of AZD1208, a proviral integration Moloney virus kinase inhibitor in solid and haematological cancers. Br J Cancer. 2018;118(11):1425–1433. doi: 10.1038/s41416-018-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chatterjee S., Chakraborty P., Daenthanasanmak A., Iamsawat S., Andrejeva G., Luevano L.A., et al. Targeting PIM Kinase with PD1 Inhibition Improves Immunotherapeutic Antitumor T-cell Response. Clin Cancer Res. 2019;25(3):1036–1049. doi: 10.1158/1078-0432.CCR-18-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Foulks J.M., Carpenter K.J., Luo B., Xu Y., Senina A., Nix R., et al. A small-molecule inhibitor of PIM kinases as a potential treatment for urothelial carcinomas. Neoplasia. 2014;16(5):403–412. doi: 10.1016/j.neo.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raab M.S., Thomas S.K., Ocio E.M., Guenther A., Goh Y.T., Talpaz M., et al. The first-in-human study of the pan-PIM kinase inhibitor PIM447 in patients with relapsed and/or refractory multiple myeloma. Leukemia. 2019;33(12):2924–2933. doi: 10.1038/s41375-019-0482-0. [DOI] [PubMed] [Google Scholar]
- 96.Kim S.Y., Herbst A., Tworkowski K.A., Salghetti S.E., Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 97.von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11(5):1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14(4):233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan C.H., Morrow J.K., Li C.F., Gao Y., Jin G., Moten A., et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154(3):556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J., Sato K., O'Donnell E., Singla A., Yaguare S., Aldahamsheh O., et al. Skp2 depletion reduces tumor-initiating properties and promotes apoptosis in synovial sarcoma. Transl Oncol. 2020;13(10) doi: 10.1016/j.tranon.2020.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y., Zvi Y.S., Batko B., Zaphiros N., O'Donnell E.F., Wang J., et al. Down-regulation of Skp2 expression inhibits invasion and lung metastasis in osteosarcoma. Sci Rep. 2018;8(1):14294. doi: 10.1038/s41598-018-32428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou L., Yu X., Li M., Gong G., Liu W., Li T., et al. Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhattacharya S., Ghosh MK. HAUSP regulates c-MYC expression via de-ubiquitination of TRRAP. Cell Oncol (Dordr) 2015;38(4):265–277. doi: 10.1007/s13402-015-0228-6. [DOI] [PubMed] [Google Scholar]
- 104.Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med. 2016;22(10):1180–1186. doi: 10.1038/nm.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang W., Zhang J.X., Xu C.Z., Zhang S.Q., Bian S.Y., Jiang F., et al. Ubiquitin-specific protease 7 is a drug-able target that promotes hepatocellular carcinoma and chemoresistance. Cancer Cell Int. 2020;20(1) doi: 10.1186/s12935-020-1109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schauer N.J., Liu X., Magin R.S., Doherty L.M., Chan W.C., Ficarro S.B., et al. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci Rep. 2020;10(1):5324. doi: 10.1038/s41598-020-62076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kategaya L., Di Lello P., Rouge L., Pastor R., Clark K.R., Drummond J., et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550(7677):534–538. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 108.Turnbull A.P., Ioannidis S., Krajewski W.W., Pinto-Fernandez A., Heride C., Martin A.C.L., et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550(7677):481–486. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jin Q., Martinez C.A., Arcipowski K.M., Zhu Y.X., Gutierrez-Diaz B.T., Wang K.K., et al. USP7 Cooperates with NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clinical Cancer Research. 2019;25(1):222–239. doi: 10.1158/1078-0432.CCR-18-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chieffi P., Aurora B. A new promising therapeutic target in cancer. Intractable Rare Dis Res. 2018;7(2):141–144. doi: 10.5582/irdr.2018.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lowenberg B., Muus P., Ossenkoppele G., Rousselot P., Cahn J.Y., Ifrah N., et al. Phase 1/2 study to assess the safety, efficacy, and pharmacokinetics of barasertib (AZD1152) in patients with advanced acute myeloid leukemia. Blood. 2011;118(23):6030–6036. doi: 10.1182/blood-2011-07-366930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boss D.S., Witteveen P.O., van der Sar J., Lolkema M.P., Voest E.E., Stockman P.K., et al. Clinical evaluation of AZD1152, an i.v. inhibitor of Aurora B kinase, in patients with solid malignant tumors. Ann Oncol. 2011;22(2):431–437. doi: 10.1093/annonc/mdq344. [DOI] [PubMed] [Google Scholar]
- 113.Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3(4):392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 114.Brenner C., Deplus R., Didelot C., Loriot A., Vire E., De Smet C., et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24(2):336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 116.Peter S., Bultinck J., Myant K., Jaenicke L.A., Walz S., Muller J., et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6(12):1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang Y., Do H., Tian X., Zhang C., Liu X., Dada L.A., et al. E3 ubiquitin ligase Mule ubiquitinates Miz1 and is required for TNFalpha-induced JNK activation. Proc Natl Acad Sci U S A. 2010;107(30):13444–13449. doi: 10.1073/pnas.0913690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao X., Heng J.I., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10(6):643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bedard M., Maltais L., Montagne M., Lavigne P. Miz-1 and Max compete to engage c-Myc: implication for the mechanism of inhibition of c-Myc transcriptional activity by Miz-1. Proteins. 2017;85(2):199–206. doi: 10.1002/prot.25214. [DOI] [PubMed] [Google Scholar]
- 120.Crawford L.J., Campbell D.C., Morgan J.J., Lawson M.A., Down J.M., Chauhan D., et al. The E3 ligase HUWE1 inhibition as a therapeutic strategy to target MYC in multiple myeloma. Oncogene. 2020;39(27):5001–5014. doi: 10.1038/s41388-020-1345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y., Sun X.X., Sears R.C., Dai MS. Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis. 2019;6(4):359–371. doi: 10.1016/j.gendis.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rabellino A., Melegari M., Tompkins V.S., Chen W., Van Ness B.G., Teruya-Feldstein J., et al. PIAS1 Promotes Lymphomagenesis through MYC Upregulation. Cell Rep. 2016;15(10):2266–2278. doi: 10.1016/j.celrep.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Melo J., Kim S.S., Lourenco C., Penn LZ. Lysine-52 stabilizes the MYC oncoprotein through an SCF(Fbxw7)-independent mechanism. Oncogene. 2017;36(49):6815–6822. doi: 10.1038/onc.2017.268. [DOI] [PubMed] [Google Scholar]
- 124.Jaenicke L.A., von Eyss B., Carstensen A., Wolf E., Xu W., Greifenberg A.K., et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61(1):54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 125.Devaiah B.N., Case-Borden C., Gegonne A., Hsu C.H., Chen Q., Meerzaman D., et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23(6):540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Devaiah B.N., Mu J., Akman B., Uppal S., Weissman J.D., Cheng D., et al. MYC protein stability is negatively regulated by BRD4. Proc Natl Acad Sci U S A. 2020;117(24):13457–13467. doi: 10.1073/pnas.1919507117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kato F., Fiorentino F.P., Alibes A., Perucho M., Sanchez-Cespedes M., Kohno T., et al. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget. 2016;7(47):77378–77388. doi: 10.18632/oncotarget.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou S., Zhang S., Wang L., Huang S., Yuan Y., Yang J., et al. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9(3):33. doi: 10.1038/s41389-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alqahtani A., Choucair K., Ashraf M., Hammouda D.M., Alloghbi A., Khan T., et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA. 2019;5(3):FSO372. doi: 10.4155/fsoa-2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun Y., Han J., Wang Z., Li X., Sun Y., Hu Z. Safety and Efficacy of Bromodomain and Extra-Terminal Inhibitors for the Treatment of Hematological Malignancies and Solid Tumors: A Systematic Study of. Clinical Trials. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.621093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takayama M., Taira T., Iguchi-Ariga S.M., Ariga H. CDC6 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc/Max complex. FEBS Lett. 2000;477(1-2):43–48. doi: 10.1016/s0014-5793(00)01756-7. [DOI] [PubMed] [Google Scholar]
- 133.Takayama M.A., Taira T., Tamai K., Iguchi-Ariga S.M., Ariga H. ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells. 2000;5(6):481–490. doi: 10.1046/j.1365-2443.2000.00338.x. [DOI] [PubMed] [Google Scholar]
- 134.Cheng S.W., Davies K.P., Yung E., Beltran R.J., Yu J., Kalpana G.V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22(1):102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 135.Sammak S., Allen M.D., Hamdani N., Bycroft M., Zinzalla G. The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J. 2018;285(22):4165–4180. doi: 10.1111/febs.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Faiola F., Liu X., Lo S., Pan S., Zhang K., Lymar E., et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25(23):10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kurland J.F., Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68(10):3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 138.Dominguez-Sola D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448(7152):445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 139.Chen C.H., Lin D.S., Cheng C.W., Lin C.J., Lo Y.K., Yen C.C., et al. Cdc6 cooperates with c-Myc to promote genome instability and epithelial to mesenchymal transition EMT in zebrafish. Oncotarget. 2014;5(15):6300–6311. doi: 10.18632/oncotarget.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garcia-Sanz P., Quintanilla A., Lafita M.C., Moreno-Bueno G., Garcia-Gutierrez L., Tabor V., et al. Sin3b interacts with Myc and decreases Myc levels. J Biol Chem. 2014;289(32):22221–22236. doi: 10.1074/jbc.M113.538744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Conte M., Altucci L. Molecular pathways: the complexity of the epigenome in cancer and recent clinical advances. Clin Cancer Res. 2012;18(20):5526–5534. doi: 10.1158/1078-0432.CCR-12-2037. [DOI] [PubMed] [Google Scholar]
- 142.Sun K., Atoyan R., Borek M.A., Dellarocca S., Samson M.E., Ma A.W., et al. Dual HDAC and PI3K Inhibitor CUDC-907 Downregulates MYC and Suppresses Growth of MYC-dependent Cancers. Mol Cancer Ther. 2017;16(2):285–299. doi: 10.1158/1535-7163.MCT-16-0390. [DOI] [PubMed] [Google Scholar]
- 143.Hu C., Xia H., Bai S., Zhao J., Edwards H., Li X., et al. CUDC-907, a novel dual PI3K and HDAC inhibitor, in prostate cancer: Antitumour activity and molecular mechanism of action. J Cell Mol Med. 2020;24(13):7239–7253. doi: 10.1111/jcmm.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Oki Y., Kelly K.R., Flinn I., Patel M.R., Gharavi R., Ma A., et al. CUDC-907 in relapsed/refractory diffuse large B-cell lymphoma, including patients with MYC-alterations: results from an expanded phase I trial. Haematologica. 2017;102(11):1923–1930. doi: 10.3324/haematol.2017.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Younes A., Berdeja J.G., Patel M.R., Flinn I., Gerecitano J.F., Neelapu S.S., et al. Safety, tolerability, and preliminary activity of CUDC-907, a first-in-class, oral, dual inhibitor of HDAC and PI3K, in patients with relapsed or refractory lymphoma or multiple myeloma: an open-label, dose-escalation, phase 1 trial. Lancet Oncol. 2016;17(5):622–631. doi: 10.1016/S1470-2045(15)00584-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu X., Tesfai J., Evrard Y.A., Dent S.Y., Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278(22):20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Frank S.R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H.M., et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4(6):575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feris E.J., Hinds J.W., Cole MD. Formation of a structurally-stable conformation by the intrinsically disordered MYC:TRRAP complex. PLoS One. 2019;14(12) doi: 10.1371/journal.pone.0225784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei Y., Resetca D., Li Z., Johansson-Akhe I., Ahlner A., Helander S., et al. Multiple direct interactions of TBP with the MYC oncoprotein. Nat Struct Mol Biol. 2019;26(11):1035–1043. doi: 10.1038/s41594-019-0321-z. [DOI] [PubMed] [Google Scholar]
- 150.McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D., Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94(3):363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 151.Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.J., et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24(24):10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nikiforov M.A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., et al. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol. 2002;22(14):5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thomas L.R., Adams C.M., Wang J., Weissmiller A.M., Creighton J., Lorey S.L., et al. Interaction of the oncoprotein transcription factor MYC with its chromatin cofactor WDR5 is essential for tumor maintenance. Proc Natl Acad Sci U S A. 2019;116(50):25260–25268. doi: 10.1073/pnas.1910391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Macdonald J.D., Chacon Simon S., Han C., Wang F., Shaw J.G., Howes J.E., et al. Discovery and Optimization of Salicylic Acid-Derived Sulfonamide Inhibitors of the WD Repeat-Containing Protein 5-MYC Protein-Protein Interaction. J Med Chem. 2019;62(24):11232–11259. doi: 10.1021/acs.jmedchem.9b01411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aho E.R., Wang J., Gogliotti R.D., Howard G.C., Phan J., Acharya P., et al. Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity. Cell Rep. 2019;26(11):2916–2928. doi: 10.1016/j.celrep.2019.02.047. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 157.Zhang L., Riley-Gillis B., Vijay P., Shen Y. Acquired Resistance to BET-PROTACs (Proteolysis-Targeting Chimeras) Caused by Genomic Alterations in Core Components of E3 Ligase Complexes. Mol Cancer Ther. 2019;18(7):1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- 158.Soucek L., Whitfield J.R., Sodir N.M., Masso-Valles D., Serrano E., Karnezis A.N., et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27(5):504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu C.H., van Riggelen J., Yetil A., Fan A.C., Bachireddy P., Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. P Natl Acad Sci USA. 2007;104(32):13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]