Abstract
The Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) protein family members have cytidine deaminase activity and can induce cytosine to uracil transition in nucleic acid. The main function of APOBEC3 (A3) proteins is to trigger an innate immune response to viral infections. Recent reports have shown that several APOBEC family proteins such as A3B can induce somatic mutations into genomic DNA and thus promote cancer development. However, the role of A3D on somatic mutations is unclear. Here, we identified the alternative splicing of A3D, and investigated each splice variant’s subcellular localization and role in DNA mutagenesis. We identified four A3D variants, which all have one or two cytidine deaminase domains. The full-length form of A3D (variant 1) and truncated forms of A3D (variant 2, 6, 7) showed the ability to induce C/G to T/A transitions in foreign DNA, genomic DNA and retained antiretroviral activity. Furthermore, we demonstrated that A3D and A3B could induce deletions that are possibly repaired by microhomology-mediated end joining (MMEJ). Taken together, our experiments illustrated that alternative splicing generates functional diversity of A3D, and some variants can act as DNA mutators in genomic DNA.
Keywords: Apolipoprotein B mRNA Editing Enzyme, Catalytic Polypeptide-Like 3D (APOBEC3D), mutations, cytidine deaminase, resistance, leukemia
Introduction
The apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) protein family consists of 11 members: APOBEC1, APOBEC2, seven APOBEC3s (A3s), APOBEC4, and activation-induced cytidine deaminase (AID) [1]. APOBEC1, A3s, and AID possess cytidine deaminase activity and are known to induce a cytosine (C) to uracil (U) transition which causes C to thymine (T) or guanine (G) to adenine (A) transition after DNA replication [2]. APOBEC1 deaminates cytidine to create a stop codon in ApoB mRNA, while physiological roles of APOBEC2 and APOBEC4 are not well characterized [2]. AID is responsible for somatic hypermutation (SHM) and class switch recombination (CSR), both of which contribute to immune globulin diversification [3]. Among seven A3 proteins-isoforms A, B, C, D, F, G, and H, A3B/D/F/G exhibit two cytidine deaminase domains (CD1 and CD2), whereas A3A/C/H have only one cytidine deaminase domain (CD2) [4]. Several reports have shown that CD1 functions in nucleic acid binding and viral encapsidation while CD2 is catalytically active in A3B, 3F, and A3G [5-10]. In terms of deaminase activity, A3 proteins intrinsically prefer certain bases surrounding the target: A3G prefers 5’-CC while other A3s prefer 5’-TC [11, 12]. The main function of A3s is to trigger an innate immune response to viral infection [13]; however, A3G is also able to restrict human immunodeficiency virus 1 (HIV-1) infection by catalyzing the deamination of HIV-1 minus strand cDNA during reverse transcription.
Recently, it was reported that several AID/APOBEC proteins can introduce somatic mutations into genomic DNA, which can promote cancer development. For example, AID-mediated somatic mutations contribute to B-cell lymphoma and gastric cancer [14] while A3B expression is upregulated in some other cancers. Lung, cervical, breast, bladder, and head and neck cancers display A3 mutation signatures [15, 16]. Interestingly, A3B overexpression in vitro leads to increased mutations in the TP53 gene and subsequently to its inactivation [14]. It has been recognized that A3B may be involved in genomic instability, cancer development, and drug resistance; however, the roles of other A3s are still unknown.
The APOBEC3D gene is located on chromosome 22q13.1 and comprised of 7 exons. The two cytidine deaminase domains, CD1 and CD2, are encoded by exon 3 and exon 6, respectively [17]. A3D is normally expressed in B lymphocytes and has anti-HIV-1 activity; however, its catalytic activity is weaker than that of other A3 proteins [1]. Although the role of A3D on cancer development is largely unknown, recent reports show that the expression of full-length A3D induces defects in cell cycle progression [18], suggesting that A3D can trigger a DNA damage response leading to cell cycle arrest due to DNA deamination. In this study, we tested our hypothesis that A3D is expressed and can induce genomic mutations in human cancer cells such as leukemia.
Experimental procedures
Cells
Jurkat (TIB-152), Raji (CCL-86), HL-60 (CCL-240), K562 (CCL-243), HEL (TIB-180), H1975 (CRL-5908), HCC827 (CRL-2868), HEK293T (CRL-3216), Phoenix-Ampho (CRL-3213), and HeLa (CRL-2) were purchased from American Type Culture Collection (ATCC, Manassas, VA). NB4, MOLM14, CHRF-288-11 (CHRF), and PC9 cells were kindly provided by Dr. Daniel G. Tenen. SET-2 cells were kindly provided by Dr. Kimiharu Uozumi. Mononuclear cells (MNCs) and polymorphonuclear leukocytes (PMNs) were isolated from healthy volunteers as previously described [19, 20].
Expression data and survival analyses
The normalized expression dataset as RSEM (RNA-seq.V2) and its correlation with overall survival from 200 primary acute myelogenous leukemia (AML) samples [21] were obtained from cBioPortal (http://www.cbioportal.org/) [22, 23] and 173 samples which have mRNA expression data were analyzed in GraphPad Prism 6 (GraphPad Software, La Jolla, CA).
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from leukemia and lung cancer cell lines as well as MNCs and PMNs using RNeasy Mini Kit (Qiagen, Valencia, CA). Complementary DNA was synthesized using SuperScript II (Thermo Fisher Scientific, Waltham, MA). Primers used to amplify A3D splicing variants were 5’-GGAGACTGGGACAAGCGTAT-3’ and 5’- GGATGACCAGGCAAAAGCAC -3’.
DNA constructs
Plasmids containing human APOBEC3B or APOBEC3G coding sequence were previously described [24]. Expression vectors for HA-tagged A3Dv1, v2 and v6 were generated by sub-cloning of coding sequences into pCAG-GS vector from human cell lines. To generate expression vectors for A3Dv7, coding sequences of exon 1 and exon 5 to exon 7 were released from pCAG-GS-A3Dv1 expression vector by PCR and assembled by the Gibson assembly method [25]. Coding sequences of APOBEC3Dv1, v2, v6, v7 or APOBEC3B were released and cloned into MigR1-IRES-EGFP vector to make retroviral constructs. The expression vector for UGI, pEF-UGI was provided by Dr. Ruben S. Harris [26]. pCAG-GS-A3Dv1-E80/264Q and pCAG-GS-A3Dv6-E264Q were generated by PCR based mutagenesis using pCAG-GS-A3Dv1, pCAG-GS-A3Dv6 as templates.
Quantitative Real-time PCR (Q-RT-PCR)
The mRNA expression levels of genes were measured by Q-RT-PCR using the Rotor Gene 6000 sequence detection system (Qiagen, Valencia, CA). RNA was reverse transcribed, and the resulting cDNA was used in amplification reactions with iTaq Universal SYBR Green Supermix (BIO-RAD, Hercules, CA). The expression levels of the glyceraldehyde 3-phosohate dehydrogenase (GAPDH) were used for normalization. Reactions were performed in technical triplicates. The primers for qPCR are listed in Table S2.
Immunoblotting
HEK293T cells were transfected with pCAG-GS-A3Dv1, v2, v6, v7 or A3B using TransIT-X2 (Mirus, Madison, WI). After 48 hours of incubation, cells were lysed in cell lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, protease inhibitor cocktail set III (EMD Millipore, Darmstadt, Germany) and 1 mM PMSF). Protein lysates (20 μg) were loaded on 10% SDS-polyacrylamide gels and transferred on PVDF membranes (Millipore, Bedford, MA). Anti-HA antibody (H3663) (Sigma-Aldrich, St. Louis, MO), anti- β-Actin antibody (sc-47778) (Santa-Cruz, Santa Cruz, CA) and Anti-p24 antibody (ab9071) (Abcam, Cambridge, UK) were used at 1:10000, 1:500 and 1:1000 dilutions, respectively.
Immunofluorescence staining and confocal microscopy
HeLa cells were transfected with pCAG-GS-A3Dv1, v2, v6, v7, A3B or empty control using TransIT-X2 (Mirus, Madison, WI). After 48 hours of incubation, cells were fixed in 4% paraformaldehyde and blocked in PBS with 5% normal horse serum (Life Technologies, Palo Alto, CA) and 0.25% Triton X-100 (Sigma-Aldrich, St. Louis, MO). Cells were incubated with anti-HA-tag antibody at 1:1000 dilution (ab18181) (Abcam, Cambridge, UK), followed by anti-mouse IgG antibody conjugated with Alexa488 at 1:400 dilution (A21202) (Life Technologies, Palo Alto, CA). Slides were mounted with VECTASHIELD with DAPI (Vector Labs, Burlingame, CA), and images were captured using Zeiss LSM 510 Meta microscope (Zeiss, Oberkochen, GER).
3D-PCR and clonal sequencing
All primers used for 3D-PCR are listed in Table S2. For the foreign DNA editing assay, HEK293T cells were co-transfected with pcDNA3.1(−)-EGFP, pEF-UGI, and a pCAG expression vector for A3Dv1, v2, v6, v7, A3B or empty control using TransIT-X2 (Mirus, Madison, WI). 4 days after transfection, DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) was used for the first round of 3D-PCR with following reaction profile: 30 sec at 98 °C; 30 cycles of 10 sec at 98 °C; 30 sec at 65.7 °C; 30 sec at 72 °C; and followed by 2 min at 72 °C. The first round PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and 25 ng of purified PCR products were used as templates for the second round of 3D-PCR, which was performed with HotStar HiFidelity Polymerase Kit (Qiagen, Valencia, CA). The reaction profile was: 5 min at 95 °C; 35 cycles of 15 sec at 86.3-87.9 °C; 1 min at 62 °C; 80 sec at 72 °C; and followed by 10 min at 72 °C. The second PCR products derived from a denaturation temperature (Td) of 86.8 °C in cells expressing A3D or A3B, and Td of 87.9 °C in mock cells were gel extracted using Gel Extraction Kit (Qiagen, Valencia, CA). The extracted second round PCR products were cloned using pGEM-T Easy Vector System (Promega, Madison, WI) and sequenced. For the genomic TP53 DNA editing assay, HEK293T cells were retrovirally infected with MigR1-IRES-EGFP expressing A3Dv1, v2, v6, v7, A3B or empty control, and EGFP positive cells were sorted using FACSAria (BD Biosciences, Franklin Lakes, NJ) 72 hours after infection. DNA was isolated 4 weeks after retroviral infections using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). The first round of 3D-PCR with the following reaction profile was performed: 30 sec at 98 °C; 30 cycles of 10 sec at 98 °C; 30 sec at 60 °C; 30 sec at 72 °C; and followed by 2 min at 72 °C. 25 ng of purified first round PCR products were used as templates for the second round of 3D-PCR with the following reaction profile: 5 min at 95 °C; 35 cycles of 15 sec at 83.8-85.3 °C; 1 min at 62 °C; 80 sec at 72 °C; and followed by 10 min at 72 °C. The amplifications of second round PCR products from the lowest Td in each group were cloned and sequenced. The primers for 3D-PCR are listed in Table S2.
Infectivity Assay using ΔVif HIV-1 with luciferase reporter
We prepared luciferase reporter viruses without Vif by co-transfection of pNL43/ΔEnv-Luc plus pVSV-G with expression vectors for A3G or A3D variants using X-tremeGene DNA transfection kit (Roche, Basel, Switzerland). We collected viruses from the supernatant 48 hours after transfection and measured virus titer with an enzyme- linked immunosorbent assay kit for the p24 antigen (RETRO-TEK, ZeptoMetrix, Buffalo, NY). We then challenged an adjusted amount of viruses to HEK293T cells. 24 hours after infection, cells were lysed in passive lysis buffer (Promega, Madison, WI), and their luciferase activities were measured by ARVO microplate reader (Perkin-Elmer, Waltham, MA). We presented values as percent infectivity normalized to the value of empty vector sample.
HIV-1 gag sequencing
Genomic DNA was extracted from HEK293T cells infected with VSV-G pseudotyped Vif-deficient HIV-1 that contained lentivirus for A3G or A3D variants after 2 days of infection using the QuickGene DNA whole blood kit S (KURABO, Osaka, Japan). We amplified HIV-1 gag sequences using regular PCR with the KOD FX Neo (ToYoBo, Osaka, Japan). The primer set is available in Table S2. We cloned amplicons in T-vector pMD20 (Takara Bio, Kyoto, Japan) and picked 5 colonies to sequence using the 3130xl Genetic Analyzer (Applied Biosystems, Waltham, MA).
Flow cytometry
HEK293T cells transiently transfected with expression constructs containing A3Dv1, v2, v6, v7, A3B or an empty control were analyzed by LSRII flow (BD Biosciences, Franklin Lakes, NJ) at 4 days after transfection. We used FlowJo (FLOWJO, LLC, Ashland, OR) to analyze data.
Statistical analysis
Differences between the experimental groups were tested with Student’s t-test. Differences between the survival curves were tested with log-rank test (Mantel-Cox). P-values of less than 0.05 were considered statistically significant.
Results
Alternative splicing of A3D in human primary cells and cancer cell lines
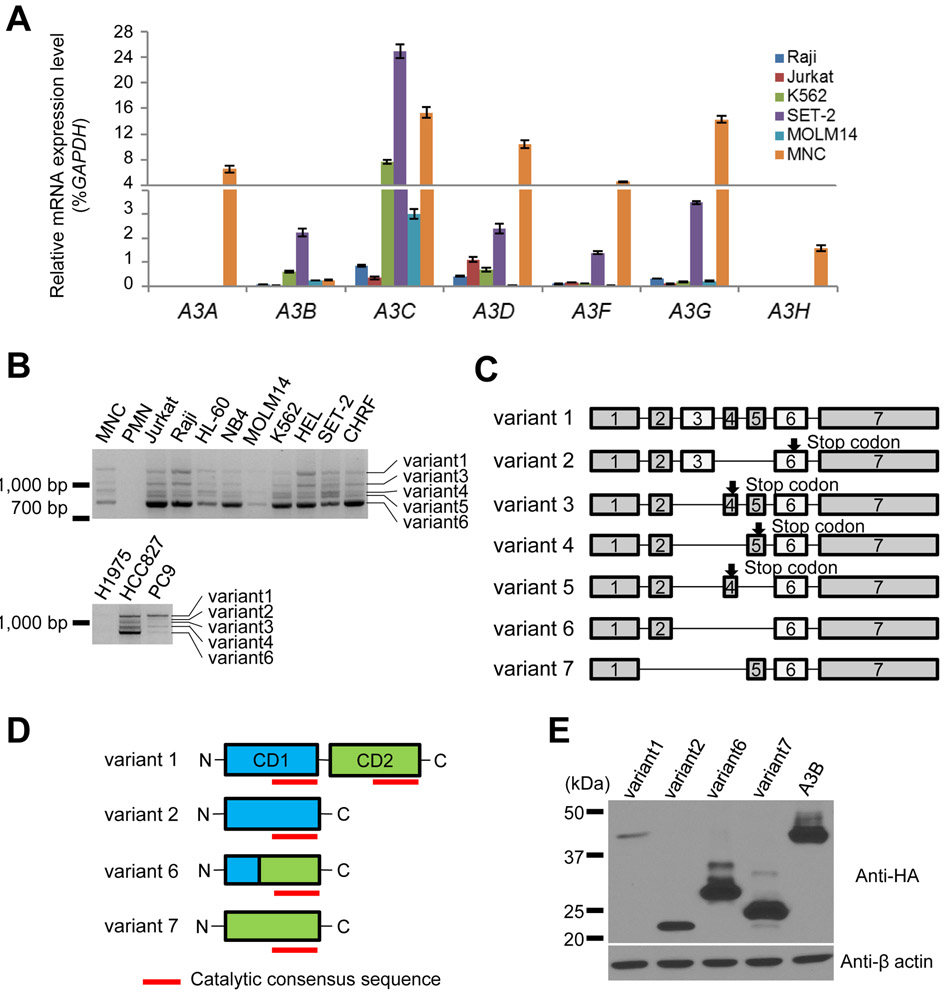
As an initial step to understand the roles of A3 on leukemia development, we used Q-RT-PCR to survey the mRNA levels of A3 members in a small panel of leukemia cell lines, together with normal MNC isolated from healthy volunteers. While A3A and A3H were not detected, other A3 genes were expressed in all leukemia cells (Fig. 1A). Next, we analyzed previously-published The Cancer Genome Atlas (TCGA) data from patients with AML [21] to find somatic alterations in APOBEC3 genes using cBioPortal [22, 23]. We found that each APOBEC3 was upregulated by 4-7%, and 19% of patients had an upregulation of at least one APOBEC3 (Supplementary Fig. S1A). Overall survival of patients with APOBEC3 upregulation was lower than those without upregulation (p=0.0256, Hazard ratio 1.614, 95%CI; 1.072 to 2.829) (log-rank test) (Supplementary Fig. S1B), suggesting an important role of A3s on prognosis in patients with AML. As it has been shown that A3A, A3B, and A3D show capacity to inflict DNA damage [18], we decided to focus on A3D because its role in cancer development and drug resistance are unknown. Our attempt to generate A3D expression constructs revealed that there are multiple bands in PCR products (Fig. 1B) from normal human cells (peripheral blood mononuclear cells: MNCs, polymorph nuclear leukocytes: PMNs), leukemia and lung cancer cell lines. Following cloning of all transcripts, we identified 5 transcript variants (A3Dv1, v3, v4, v5, and v6) from leukemia cell lines. According to Ensembl (http://ensemblgenomes.org/), 3 transcripts have been reported so far (A3D-201, 202, and 203). A3Dv1 corresponds to either A3D -201 and A3Dv6 to A3D-202. Although we were unable to detect A3D-203 in our analysis, we included this transcript for further study (A3Dv7). We also included another variant (A3Dv2), which lacks exon 4 and exon 5, that was detected in lung cancer cell lines (Fig. 1B and manuscript in preparation). These variants were numbered in descending order of PCR products length. We found that A3Dv1 was a full-length mRNA containing all 7 exons and harbored two cytidine deaminase domains (CD1 and CD2) (Fig. 1C and 1D). Protein products from A3Dv2 and A3Dv7 harbored a single CD1 and CD2 catalytic domain, respectively (Fig. 1D). A product from A3Dv6 was predicted to harbor a single hybrid catalytic domain (Fig. 1D). Products from A3Dv3, A3Dv4, and A3Dv5 had neither domain, suggesting that these variants are not functional. Therefore, we tested 4 A3D variants (A3Dv1, v2, v6, v7) in further studies (Fig. 1D).
Figure 1. Expression of APOBEC3 mRNA and alternative splicing variants of A3D in leukemia cell lines and normal MNC.
(A) Quantitative RT-PCR for APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H in leukemia cell lines and normal MNCs. The endogenous glyceraldehyde 3-phosohate dehydrogenase (GAPDH) was used for normalization. Error bars indicate S.D. (B) RT-PCR analysis of APOBEC3D gene in leukemia and lung cancer cell lines, normal mononuclear cells (MNCs) and normal polymorph nuclear leukocyte (PMNs). (C) Alternative splicing variants of APOBEC3D. The exons encode cytidine deaminase (CD) enzymatic site are shown as white boxes. (D) Structure of four A3D variants which have CD domains and expected to have enzymatic activity. Red lines indicate the catalytic consensus sequence. (E) Immunoblotting analysis of HA-tagged A3D variants and A3B (top panel) and β-actin (bottom panel). Protein samples were collected from transfected HEK293T cells.
To assess the protein stability of these 4 variants, we generated expression constructs encoding each variant with an HA-tag and transfected them into HEK293T cells. We also generated constructs expressing A3B as control. We detected A3Dv1 (46 kDa), A3Dv2 (21 kDa), A3Dv6 (30 kDa), and A3Dv7 (23 kDa) as expected (Fig. 1E).
Subcellular localization of full-length and truncated isoforms of A3D
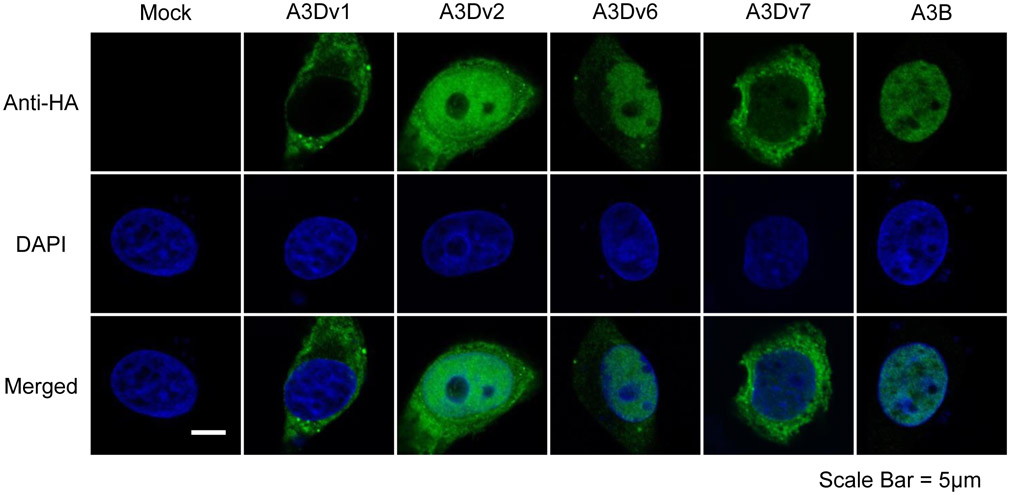
It has been shown that A3D, A3F, and A3G are located in the cytoplasm, A3B in the nucleus, and A3A, A3C, and A3H, each with a single deaminase domain, are present in both compartments [18]. Thus, we examined where the cellular localization will be for the full length A3D (A3Dv1) protein and truncated A3D (A3Dv2, v6, v7) proteins. HeLa cells were transfected with pCAG-GS-A3Dv1, v2, v6, v7, A3B or empty control and analyzed 2 days after transfection. Confocal microscopy analysis revealed that full-length A3Dv1 was located in the cytoplasm while A3B was located in the nucleus as previously shown [18] (Fig. 2). Although all truncated isoforms (A3Dv2, v6, 7v) showed cell-wide distribution. A3Dv2 and A3Dv6 were predominantly located in the nucleus, and A3Dv7 was primarily located in the cytoplasm (Fig. 2).
Figure 2. Subcellular localization of A3D variants and A3B in HeLa cells.
Confocal immunofluorescence images of HA-tagged A3 variants in transfected HeLa cells. HA-tagged A3s (top panels; green), nuclear stained with DAPI (middle panels; blue), and merged images (bottom panels) are shown. Scale bar = 5 μm.
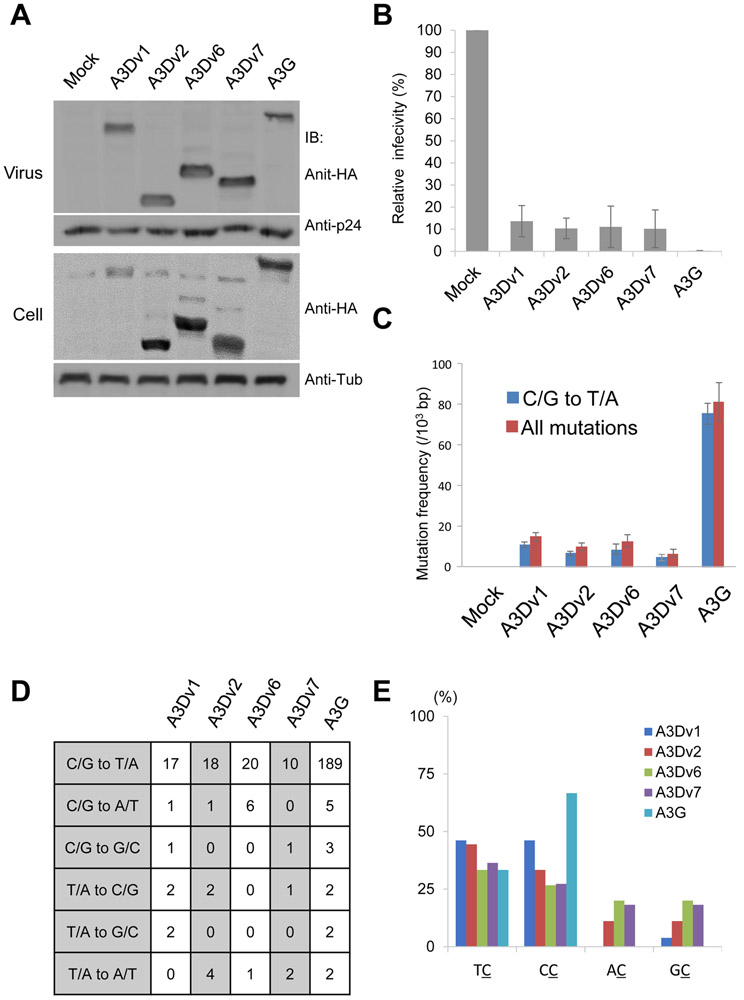
Anti HIV-1 activity of A3D variants
APOBEC3 proteins including A3D are originally shown to inhibit Vif-deficient HIV-1 infectivity [27]. We tested the anti-viral activity of A3D variants against VSV-g pseudotyped HIV-1 ΔVif viruses using luciferase reporter assays. All A3D variants were detected in the viral supernatant (Fig. 3A). A3Dv1 inhibited HIV-1 infectivity less effectively than A3G as previously reported [27], whereas the other A3D variants (A3Dv2, v6, v7) showed similar antiviral activity against HIV-1 (Fig 3B). These data suggest that A3D variants retain binding ability to HIV-1 gag and RNA for incorporation into the budding HIV-1 virion and show active anti-HIV-1 activity. To analyze the pattern of mutation, we cloned and sequenced HIV-1 gag which was amplified by regular PCR method using the genomic DNA of infected cells. We detected 81.9 and 11.4 mutations per 103 base pairs (bps) in HIV-1 gag derived from A3G or A3Dv1 containing viruses, respectively. A3Dv2, v6 and v7 induced 7.2, 9.2, and 5.1 mutations per 103 bps in HIV-1 gag, respectively (Fig. 3C). Most of these mutations were C/G to T/A transitions (Fig. 3C, 3D). A3G preferred CC dinucleotide context, whereas A3D variants targeted TC and CC dinucleotides (Fig. 3E). No mutations were detected in the PCR products of mock cells.
Figure 3. Antiviral activity of A3D and its variants on HIV-1.
(A) We transfected pNL4-3/Δenv/Δvif-Luc and pVSV-g with expression vectors for APOBEC3D variants and concentrated viral supernatant by ultracentrifugation. Viral concentrates and cell lysates were subjected to immunoblotting using indicated antibodies. Tub: Tubulin (B) Luciferase activity of HIV in the presence of APOBEC3D variants or APOBEC3G was measured with a luminometer and presented as relative values normalized to the value without APOBEC3 expression in the producer cells. Values represent the average of three independent experiments and error bars indicate S.D. (C) Mutation frequencies in HIV-1 gag PCR products derived from infected cells. 5 clones (2500 bps) were sequenced in each group. C/G to T/A mutation frequencies (blue bar) and mutation frequencies including all patterns (red bar) are shown. Error bars indicate S.E.M. (D) Mutation patterns of edited HIV-1 gag sequences. 5 clones (2500 bps) were sequenced for each group. (E) Dinucleotide patterns in edited HIV-1 gag. The rates of indicated dinucleotide sequence at C/G to T/A mutations.
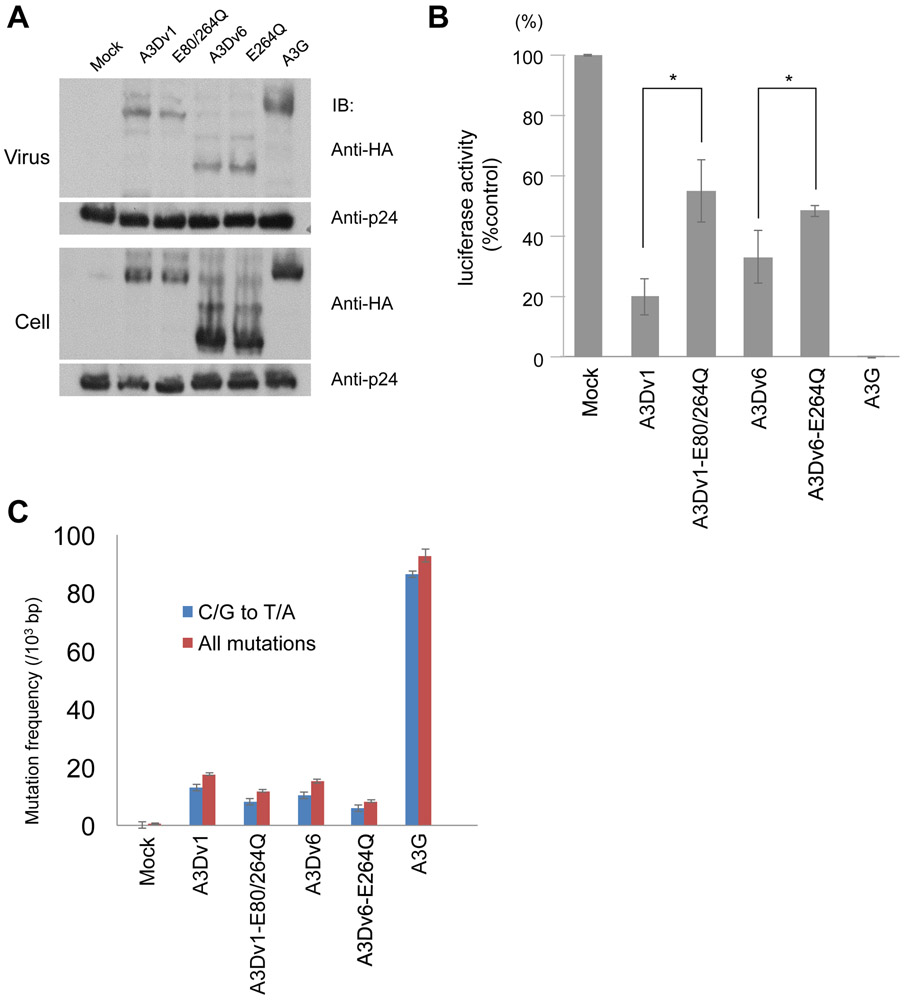
To confirm emergence of mutations are mediated by catalytic activity we generated constructs expressing A3Dv1-E80/264Q and A3Dv6-E264Q, which harbor missense mutations at catalytic domains (E80Q for CD1 and E264Q for CD2) and impair catalytic activity. Protein expression was similar in all extracts. (Fig. 4A). Antiviral activity was partially reversed by A3Dv1-E80/264Q or A3Dv6-E264Q compared to wild type A3Dv1 or A3Dv6 (Fig. 4B). In terms of frequency of mutations, we detected 11.0 and 1.5 mutations per 103 bps in HIV-1 gag derived from A3Dv1 or A3Dv1-E80/264Q containing viruses, respectively. A3Dv6 and A3Dv1-E264Q induced 6.5 and 2.0 mutations per 103 bps in HIV-1 gag, respectively (Fig. 4C). These results indicate that mutations induced by A3Ds depend on their catalytic activity.
Figure 4. Antiviral activity of A3Ds harboring mutation at catalytic domains on HIV-1.
(A) We transfected pNL4-3/Δenv/Δvif-Luc and pVSV-g with expression vectors for APOBEC3D variants and concentrated viral supernatant by ultracentrifugation. Viral concentrates and cell lysates were subjected to immunoblotting using indicated antibodies. (B) Luciferase activity of HIV in the presence of APOBEC3D variants or APOBEC3G was measured with a luminometer and presented as relative values normalized to the value without APOBEC3 expression in the producer cells. Values represent the average of three independent experiments and error bars indicate S.D. Asterisks* on the error bar indicate P<0.05 (Student’s t-test). (C) Mutation frequencies in HIV-1 gag PCR products derived from infected cells. 5 clones (2,500 bps) were sequenced in each group. C/G to T/A mutation frequencies (blue bar) and mutation frequencies including all patterns (red bar) are shown. Error bars indicate S.E.M.
All A3D variants induce mutations in foreign DNA
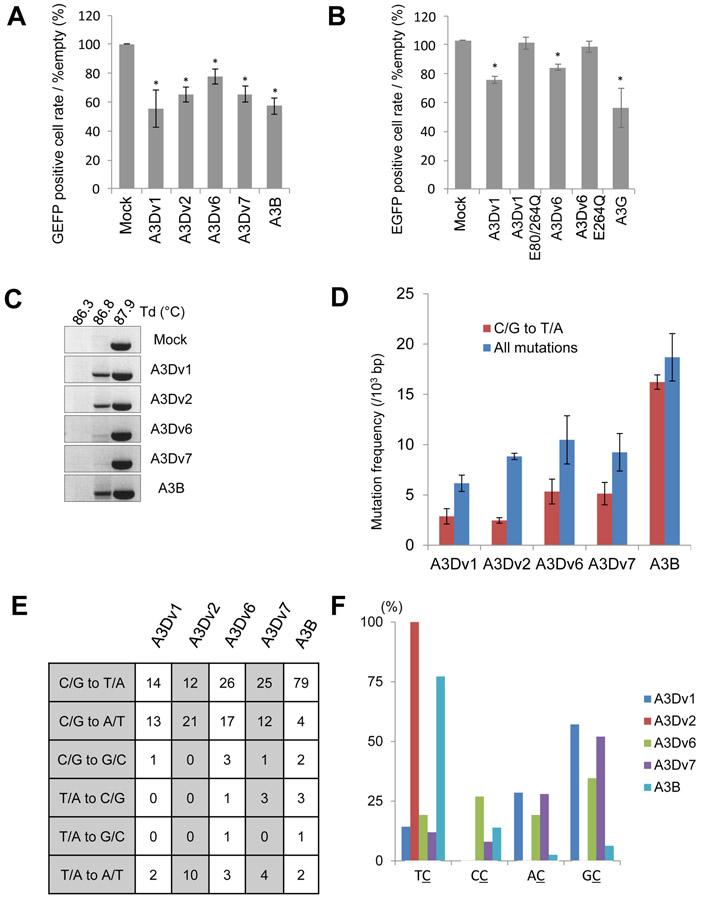
We then asked whether full-length and truncated forms of A3D have ability to induce mutations in DNA. To examine whether A3D proteins induce mutations in foreign DNA in human cells, we co-transfected HEK293T cells with an expression vector for EGFP, an expression vector for Uracil-DNA glycosylase inhibitor (UGI), and pCAG-GS containing A3Dv1, v2, v6, v7, A3B, or empty control as previously described [24]. Interestingly, flow cytometry analysis revealed that the cells transfected with A3D variants and A3B had lower EGFP positive cell rate than cells transfected with empty pCAG-GS vector as a control after 4 days (Fig. 5A). To confirm catalytic activity is crucial for mutagenesis, we used A3Dv1-E80Q/E264Q and A3Dv6-E264Q as negative controls. The EGFP positive cell rate remained similar in cells transfected with A3Dv1-E80Q/E264Q or A3Dv6-E264Q, suggesting that catalytic activity is crucial for attenuate EGFP activity, possibly induced by mutagenesis. To test this possibility, DNA was isolated from these cells for 3D-PCR analysis as previously described [24]. PCR products were detected at a denaturation temperature (Td) of 87.9 °C in all samples. Amplification at Td of 86.8 °C was lost in control cells, whereas PCR products were still amplified in the cells transfected with all A3D variants and A3B expression vectors (Fig. 5C). To confirm that mutations were introduced in the EGFP gene, we cloned and sequenced PCR products at Td of 86.8 °C in cells expressing either A3D variants or A3B and PCR product of mock cells at Td of 87.9 °C as a control. Mutation frequencies were 6.1-10.5 per 103 bps in cells transfected with each A3D variant, whereas there were 18.7 mutations per 103 bps in cells transfected with A3B (Fig. 5D and 5E). No mutations were detected in the PCR products of mock cells at Td of 87.9 °C (data not shown). Only approximately 30-60% of mutations were C/G to T/A transitions in cells transfected with A3D variants compared to 90% of mutations in cells transfected with A3B (Fig. 5D). In terms of target dinucleotide sequences, A3B and A3Dv2 preferred 5’-TC, while other A3D variants did not show preference (Fig. 5F). We also observed clones with 47- to 334- bps deletions in all A3D variants or A3B expressing cells, but not in mock cells (Table 1). Interestingly, 1- to 9- bases micro homologous sequences in the broken ends before joining were detected in some clones. Existence of these micro homologous sequences suggests that DNA double strand breaks were repaired by microhomology-mediated end joining (MMEJ).
Figure 5. Foreign DNA editing assay by A3Ds and A3B.
(A) EGFP positive cell rates and (B) EGFP mean fluorescence intensity (MFI) compared to mock cells analyzed by flow cytometry 4 days after transfection in co-transfected HEK293T cells. Asterisks* on the error bar indicate P<0.05 (Student’s t-test). (C) Analysis of 3D-PCR products in HEK293T cells expressing A3D variants, A3B or empty as control. 3D-PCR for EGFP gene at indicated denaturation temperatures (Td) was performed. (D) Mutation frequencies in 3D-PCR products in HEK293T cells expressing A3D variants and A3B. 10 clones (4870 bps) were sequenced in each group. C/G to T/A mutation frequencies (blue bar) and mutation frequencies including all patterns (red bar) are shown. Error bars indicate S.E.M. (E) Mutation patterns of edited EGFP gene sequences derived from 3D-PCR products. (F) Dinucleotide patterns in edited EGFP gene. The rates of indicated dinucleotide sequence at C/G to T/A mutations.
Table 1: Deletions in amplifications from 3D-PCR foreign DNA editing assay.
Deletions were detected in amplifications from 3D-PCR foreign DNA editing assay. Numbers in deleted location in EGFP column show numbers from protein coding region. Four of these clones harboring microhomology longer than 5 nucleotides.
| Clone | Deleted location in EGFP |
Deleted length (bp) |
Microhomology |
|---|---|---|---|
| A3Dv1-8 | 58 -> 309 | 252 | GACGGC |
| A3Dv1-17 | 147 -> 193 | 47 | none |
| A3Dv1-29 | 149 -> 421 | 273 | CA |
| A3Dv2-13 | 83 -> 416 | 334 | none |
| A3Dv6-2 | 161 -> 431 | 271 | none |
| A3Dv6-3 | 118 -> 319 | 202 | none |
| A3Dv6-6 | 107 -> 190 | 84 | none |
| A3Dv6-7 | 88 -> 229 | 142 | none |
| A3Dv7-14 | 55 -> 182 | 128 | C |
| A3Dv7-17 | 96 -> 344 | 249 | CGAGGGCGA |
| A3Dv7-22 | 56 -> 470 | 415 | none |
| A3Dv7-86 | 115 -> 195 | 81 | ACCTACGGC |
| A3B-8 | 158 -> 473 | 316 | GCA |
| A3B-26 | 102 -> 344 | 243 | CGAGGGCGA |
| A3B-28 | 91 -> 238 | 148 | none |
A3D induced mutations in TP53 gene in human cells
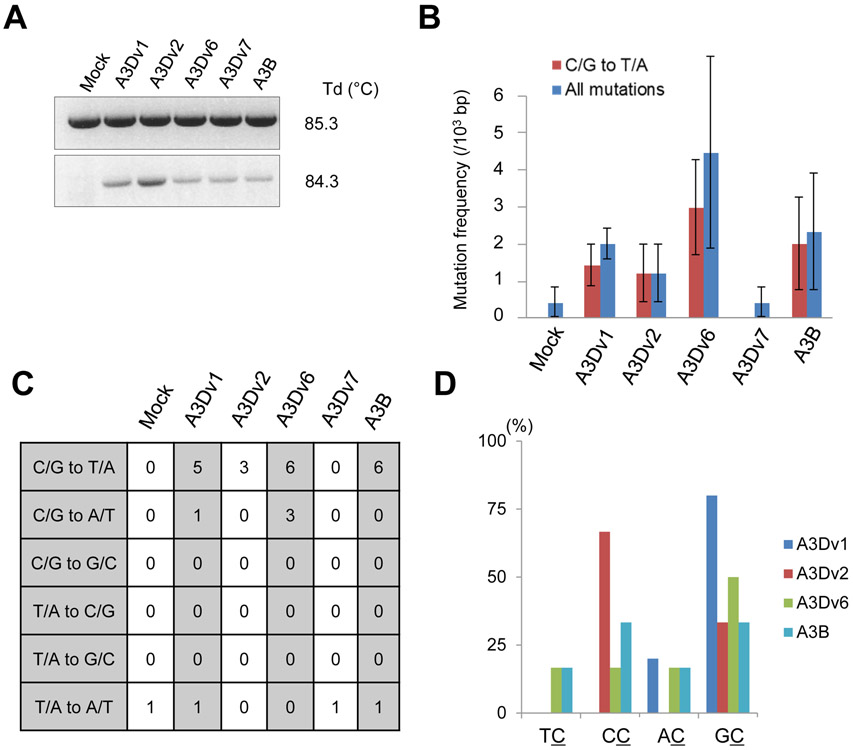
To assess whether A3D can induce mutations in genomic DNA, we generated HEK293T cells stably expressing EGFP and either A3Dv1, v2, v6, v7, or A3B. EGFP positive cells were sorted 72 hours after retroviral infection and cultured. 4 weeks after retroviral infection, genomic DNA was extracted and subjected to 3D-PCR to detect whether mutations were introduced in the TP53 gene locus. The amplifications existed at a Td of 84.3 °C in cells expressing either an A3D variant or A3B; however, no bands were detected in DNA from mock cell cultures (Fig. 6A). Sequencing of the PCR products at the lowest Td revealed that the mutation frequency in the TP53 gene was 0.4-4.5 per 103 bps in cells expressing A3D variants and A3B (Fig. 6B). The rates of C/G to T/A transition in these mutations were 70%, 100%, 67% and 85% in the cells expressing A3Dv1, v2, v6 and A3B, respectively (Fig. 6B and 6C). However, mutations in the cell expressing A3Dv7 had no difference compared to mock cells (Fig. 6B and 6C). Unexpectedly, a preference for 5’-TC was not observed in cells expressing A3D variants and A3B (Fig. 6D). The preferred target of A3Dv1 and A3Dv6 was 5’-GC dinucleotide while A3Dv2 preferred 5-CC (Fig. 5D). Clones that harbored deletions in the TP53 gene were also observed (Table 2). Only 1- to 3- bps homologous sequences internal to the broken ends before joining were present in the TP53 gene.
Figure 6. 3D-PCR for TP53 gene.
(A) Agarose gel analysis of 3D-PCR products in HEK293T cells stably expressing A3D variants, A3B or empty as control. 3D-PCR for TP53 gene at indicated Td was performed. (B) Mutation frequencies in 3D-PCR products in HEK293T cells stably expressing EGFP and A3D variants or A3B. Error bars indicate S.E.M. (C) Mutation patterns of edited TP53 gene sequences derived from 3D-PCR products. 5-7 clones (2500-3500 bps) were sequenced in each group. (D) Dinucleotide patterns in edited TP53 gene. The rates of indicated dinucleotide sequence at C/G to T/A mutations. A3Dv7 is not shown because no mutations were detected in cells expressing A3Dv7 in 3D-PCR for TP53 gene.
Table 2: Deletions in amplifications from 3D-PCR genomic DNA editing assay.
Numbers in deleted location in TP53 column show position in TP53 DNA sequence (NG_017013.2).
| Clone | Deleted location in TP53 |
Deleted length (bp) |
Microhomology |
|---|---|---|---|
| A3Dv1-4 | 18635 -> 18869 | 235 | none |
| A3Dv1-20 | 18730 -> 18798 | 69 | CG |
| A3Dv2-17 | 18799 -> 18917 | 119 | none |
| A3Dv2-19 | 18754 -> 19045 | 292 | T |
| A3Dv2-21 | 18747 -> 19086 | 340 | T |
| A3Dv6-5 | 18855 -> 18855 | 1 | none |
| A3Dv6-6 | 18776 -> 18875 | 100 | GG |
| A3Dv6-7 | 18780 -> 19029 | 250 | G |
| A3Dv6-8 | 18684 -> 18870 | 187 | none |
| A3Dv7-15 | 18741 -> 19028 | 288 | CTT |
| A3Dv7-16 | 18830 -> 19037 | 208 | none |
| A3B-5 | 18731 -> 19049 | 319 | none |
| A3B-21 | 18738 -> 18885 | 148 | none |
Discussion
Among AID/APOBEC family members, AID is best described in its oncogenic role in hematopoietic malignancies besides A3B in solid tumors such as breast cancers [15]. AID can induce c-myc/IgH which is characteristic of Burkitt’s lymphoma [28]. AID is upregulated by the BCR-ABL1 kinase and contributes to the aggressive phenotypes observed in B-ALL [29]. Furthermore, AID promotes B lymphoid blast crisis and drug resistance to ABL inhibitors in chronic myeloid leukemia (CML) [30]. Recent reports indicate that A3 signatures are observed in ALL and CLL [31, 32]; however, the role of A3 proteins in leukemogenesis, disease progression, and drug resistance mechanisms are largely unknown.
Compared to its definitive role of A3B as a DNA mutator, the roles of other A3s are not completely elucidated. A recent pan-cancer transcriptomic analysis demonstrated that only A3B expression correlates with cell cycle and DNA repair genes, while other A3 genes are involved in immune function [33]. However, as the authors themselves pointed out, the co-expression data on cancer cell line cohorts do not reflect the TCGA datasets. In addition, other APOBEC3 members such as A3A, A3G, and A3H are reportedly involved in tumorigenesis [17, 34, 35]. Indeed, our analysis using TCGA data from patients with AML revealed that APOBEC3D was upregulated by 5% of patients (Supplementary Figure. S1A). Furthermore, we cannot exclude the possibility that A3 family members may serve as both genetic mutators and immune modulators. Further investigations will be required to answer this question.
Previous reports and our results demonstrated that there are at least 4 variants (A3Dv1, 2, 6, and 7) that harbor functional deaminase domains (Fig. 1C). A3Dv1 has both CD1 and CD2, A3Dv2 has CD1, and A3Dv6 and 7 have CD2 (Fig. 1C). Consistent with the previous report [18], A3Dv1 is located in the cytoplasm and A3Dv2 in the nucleus (Fig. 2). Interestingly, A3Dv6 and A3Dv7 showed different distribution even though both variants possess CD2. One explanation is that the nuclear export signal exists in exon 5, which A3Dv1 and A3Dv7 have; however, no such sequences were detected in exon 5, implying that this change in localization is independent of exportin. Functionally, all truncated A3D proteins (A3Dv2, v6, v7) have ability to induce mutations in foreign DNA and HIV DNA (Fig. 3C, 4D), suggesting that both CD1 and CD2 have cytidine deaminase activity. Consistent with this, our experiments using deaminase activity-dead mutants (A3Dv1-E80/264Q and A3Dv6-E264Q) support that A3Ds’ ability to induce mutations relies on catalytic activity (Fig. 4 and Fig. 5B). It should be noted that a preference for 5’-TC was not observed in all A3D variants except for A3Dv2 against foreign DNA (Fig. 5F) and genomic DNA (Fig. 6D). These results suggest that mutations mediated by A3D may not fall into the APOBEC mutation signatures.
Based on our findings that A3Dv6 is expressed in all leukemia cell lines we tested (Fig. 1A) and is located in the nucleus (Fig. 2), our initial hypothesis was that A3Dv6 would be the most powerful DNA mutator among A3D variants. A3Dv6 caused mutations in foreign DNA (Fig. 4D) and the genomic TP53 gene as well (Fig. 5B). These results support our hypothesis. We also confirmed previous reports [18, 26] demonstrating that full-length A3D (A3Dv1) is cytoplasmic (Fig. 2); however, it effectively induced genomic TP53 mutations (Fig. 5B, 5C). To explain these paradoxical results, we propose that at least two events may be required to induce genomic DNA mutations: 1) A3s should be shuttled into the nucleus to gain access to the genomic DNA; and 2) proper conditions should be provided for A3s to maximize their catalytic activity. One possibility is that A3D may need co-factors to recognize targeted sequences or augment catalytic activity. Further study will be required to answer these important questions.
We also demonstrated that A3D variants and A3B induced deletions in both foreign and genomic DNA (TP53) in addition to causing base-pair changes. Compared to base substitutions, which can be identified by A3’s target motifs, deletion caused by A3s may be overlooked particularly when they occur outside mutation clusters, “kataegis” [36]. In our study, 47- to 415- bps deletions and 1- to 9- bps microhomologus sequences were detected in the foreign DNA, whereas 1- to 340- bps deletions and 1- to 3- bps microhomologus sequences were detected in the genomic TP53 (Table 1, S1). Most likely these deletions were generated by MMEJ, a DNA repair mechanism for double strand breaks (DSBs) in genomic DNA, and our results suggest that A3s may be involved in this error-prone DSB repair [37]. Deletions induced by MMEJ can be genotoxic when they are introduced in tumor suppressor genes by suppressing their functions or activating oncogenic genes.
In summary, we demonstrated that A3D expressed in human normal MNCs and leukemia cells as A3B. A3D has at least 7 splicing variants, 5 of which are expressed in MNCs and leukemia cells. A3D variants (A3Dv1, v2, v6, v7) have ability to induce base substitutions in both foreign and genomic TP53 DNA. Moreover, it is probable that A3Dv6 is the strongest DNA mutator among A3D variants. Of note, we demonstrated that A3D as well as A3B can induce deletions possibly induced by MMEJ. Further studies will be required to elucidate the role of A3D on cancer development and progression by inducing somatic mutations.
Supplementary Material
Supplementary Table 1. Primers
Primers used to amplify A3D variants, mutagenesis, QT-PCR, and 3D-PCR.
Acknowledgements:
We thank Drs. Kimiharu Uozumi (Kagoshima University, Japan) and Daniel Tenen (Beth Israel Deaconess Medical Center, USA) for providing cell lines. Mihoko Yamamoto, Alistair J. Tan, Ayako Hirata, and Yuta Inoue for technical assistance. This work was supported by National Institution of Health (R21CA178301 and R01CA169259), American Cancer Society (RSG-13-047), and Harvard Stem Cell Institute Blood Program (DP-0110-12-00).
Footnotes
Conflict of interest
Dr. Kobayashi reports grants and personal fees from Boehringer Ingelheim, grants from Taiho Pharmaceutical, grants from MiNA therapeutics, personal fees from Pfizer, personal fees from Ono, personal fees from Chugai, personal fees from Astra Zeneca, personal fees from Roche, outside the submitted work. Dr. Yamashita reports personal fees from Takeda Pharm., outside the submitted work. The other authors declare no relevant conflict of interest.
References
- 1.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–77. [DOI] [PubMed] [Google Scholar]
- 2.Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med. 2015;21:274–84. [DOI] [PubMed] [Google Scholar]
- 3.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knisbacher BA, Gerber D, Levanon EY. DNA Editing by APOBECs: A Genomic Preserver and Transformer. Trends Genet. 2016;32:16–28. [DOI] [PubMed] [Google Scholar]
- 5.Xiao X, Yang H, Arutiunian V, Fang Y, Besse G, Morimoto C, et al. Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation. Nucleic Acids Res. 2017;45:7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Xiao X, Wolfe A, Chen XS. The in vitro Biochemical Characterization of an HIV-1 Restriction Factor APOBEC3F: Importance of Loop 7 on Both CD1 and CD2 for DNA Binding and Deamination. J Mol Biol. 2016;428:2661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–31. [DOI] [PubMed] [Google Scholar]
- 8.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, et al. Structural Insights into HIV-1 Vif-APOBEC3F Interaction. J Virol. 2015;90:1034–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–70. [DOI] [PubMed] [Google Scholar]
- 11.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77. [DOI] [PubMed] [Google Scholar]
- 12.Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience. 2015;2:320–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avesson L, Barry G. The emerging role of RNA and DNA editing in cancer. Biochim Biophys Acta. 2014;1845:308–16. [DOI] [PubMed] [Google Scholar]
- 15.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi S, Yamashita K, Takeoka T, Ohtsuki T, Suzuki Y, Takahashi R, et al. Calpain-mediated X-linked inhibitor of apoptosis degradation in neutrophil apoptosis and its impairment in chronic neutrophilic leukemia. J Biol Chem. 2002;277:33968–77. [DOI] [PubMed] [Google Scholar]
- 20.Will B, Siddiqi T, Jorda MA, Shimamura T, Luptakova K, Staber PB, et al. Apoptosis induced by JAK2 inhibition is mediated by Bim and enhanced by the BH3 mimetic ABT-737 in JAK2 mutant human erythroid cells. Blood. 2010;115:2901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinohara M, Io K, Shindo K, Matsui M, Sakamoto T, Tada K, et al. APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells. Sci Rep. 2012;2:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, , Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. [DOI] [PubMed] [Google Scholar]
- 26.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. Journal of virology. 2011;85:11220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldhahn N, Henke N, Melchior K, Duy C, Soh BN, Klein F, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med. 2007;204:1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebhandl S, Huemer M, Gassner FJ, Zaborsky N, Hebenstreit D, Catakovic K, et al. APOBEC3 signature mutations in chronic lymphocytic leukemia. Leukemia. 2014;28:1929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng JCF, Quist J, Grigoriadis A, Malim MH, Fraternali F. Pan-cancer transcriptomic analysis dissects immune and proliferative functions of APOBEC3 cytidine deaminases. Nucleic Acids Res. 2019;47:1178–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nature communications. 2016;7:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowarski R, Kotler M. APOBEC3 cytidine deaminases in double-strand DNA break repair and cancer promotion. Cancer Res. 2013;73:3494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Primers
Primers used to amplify A3D variants, mutagenesis, QT-PCR, and 3D-PCR.