Abstract
Background: Neuroinflammation is a key feature shared by most, if not all, neuropathologies. It involves complex biological processes that act as a protective mechanism to fight against the injurious stimuli, but it can lead to tissue damage if self-perpetuating. In this context, microglia, the main cellular actor of neuroinflammation in the brain, are seen as a double-edged sword. By phagocyting neuronal debris, these cells can not only provide tissue repair but can also contribute to neuronal damage by releasing harmful substances, including inflammatory cytokines. The mechanisms guiding these apparent opposing actions are poorly known. The endocannabinoid system modulates the release of inflammatory factors such as cytokines and could represent a functional link between microglia and neuroinflammatory processes. According to transcriptomic databases and in vitro studies, microglia, the main source of cytokines in pathological conditions, express the cannabinoid type 1 receptor (CB1R).
Methods: We thus developed a conditional mouse model of CB1R deletion specifically in microglia, which was subjected to an immune challenge (peripheral lipopolysaccharide injection).
Results: Our results reveal that microglial CB1R differentially controls sickness behavior in males and females.
Conclusion: These findings add to the comprehension of neuroinflammatory processes and might be of great interest for future studies aimed at developing therapeutic strategies for brain disorders with higher prevalence in men.
Keywords: microglia, CB1 receptor, inflammation, sickness behavior, cytokine
Introduction
Brain inflammation is a common process to all neuropathologies, which can ameliorate or worsen disease progression, depending on the pathophysiological context.1,2 Yet, molecular mechanisms underlying this dual action are still unclear, making any therapeutic strategy targeting neuroinflammatory processes elusive. Microglia are the resident macrophages of the central nervous system (CNS) and are key players in brain inflammation. Under basal conditions, microglia are in a dynamic state of surveillance of their environment.3 When faced with injury or disease, they reorient their phenotype and function to restore brain homeostasis.4 Among the various actions that microglia can initiate, they release proinflammatory cytokines, such as interleukin-1 beta (IL-1β), interleukin-6 (IL-6 and tumor necrosis factor alpha (TNF-α), to adapt neuronal activity and subsequently the behavior, to cope with the pathological situation.5–7 However, if chronically stimulated, microglial reactivity can become aberrant, and eventually leads to neuronal alterations and behavioral deficits.7 Understanding the mechanisms involved in the regulation of neuroinflammation, and in particular cytokine production by microglia, is therefore of high interest.
The endocannabinoid system (ECS) is a potent regulator of neuroinflammation.8–10 It can control microglial function and therefore represents a promising target for treating CNS dysfunction.10,11 The ECS is a complex system, including endogenous lipid ligands called endocannabinoids, such as anandamide (AEA) and 2-arachidonoylglycerol (2-AG).12 Endocannabinoids bind to cannabinoid type 1 and type 2 receptors (CB1R and CB2R, respectively), both of which are G protein coupled.13,14 The expression pattern of CBRs differs among cell types within the CNS. CB1R is abundantly found on neurons and is more moderately expressed on astrocytes and microglia.15,16 Conversely, CB2R expression is primarily restricted to microglia.10,17–19 Hence, the regulatory role of CB2R on neuroinflammation has been extensively studied.20–22 Yet, and while some evidence points for a role of CB1R in these mechanisms, only few studies have addressed the specific role of CB1R in regulating microglial function and neuroinflammation.23–27 CB1R activation can skew microglial phenotype toward anti-inflammatory/prophagocytic profile in vitro.25 Moreover, full CB1R knockout (KO) mice do not develop the whole range of symptoms of sickness behavior normally observed following challenge with the endotoxin lipopolysaccharide (LPS).24 CB1R KO mice also have significantly lower plasma levels of the proinflammatory cytokines TNF-α and IL-6 compared to control animals after LPS treatment.24 Furthermore, selective CB1R antagonists successfully reduce sickness symptoms after an LPS injection. When the CB1R antagonist, rimonabant, is administered directly into the brain, to avoid any systemic effect, the sickness response is still significantly suppressed, pointing toward the importance of brain CB1R.27 Overall, CB1R receptor influences neuroinflammation and related sickness behavior; however, whether and how microglial CB1R is involved in these processes remain to be understood.
In this study, we developed an inducible KO mouse line in which CB1R was specifically deleted in CX3CR1-positive cells (mainly microglia in the brain).28,29 After assessing the potential impact of this transgenic manipulation on basal behavior, we subjected mice to an LPS challenge and monitored sickness behavior as well as brain proinflammatory cytokine production. Since CB1R receptors are differentially expressed between males and females30 and because inflammatory processes are sex dependent,31,32 we studied the potential sexual dimorphism of microglial CB1R action. Our results show that brain response to inflammation is microglial CB1R dependent in males (both for behavioral alterations and cytokine production), while it is microglial CB1R independent in females.
Materials and Methods
Animals
All studies were approved by the local and national ethics committee for care and use of animals (No. A1644), and were performed according to the Quality Reference System of INRA. Male and female mice, aged 2–5 months, were used in all experiments. They were maintained under 12-h light/12-h dark cycle with standard diet and water ad libitum.
Cx3cr1-CB1R-KO mice were generated using the CRE/loxP system as previously described.33 Cx3cr1-CreERT2 mice (purchased from Jackson Lab) (Yona et al.)34 were crossed with mice carrying the “floxed” CB1R gene (CB1Rf/f),35 using a three-step backcrossing procedure to obtain CB1Rf/f;CX3CR1-CreERT2 mice. As CreERT2 protein is inactive in the absence of tamoxifen treatment,29 deletion of the CB1R gene was obtained by daily i.p. injection of tamoxifen (1 mg dissolved at 10 mg/mL in 90% corn oil and 10% ethanol; Sigma-Aldrich, Lyon, France) for 8 consecutive days. The vehicle was prepared similarly, but without the tamoxifen. The mice were injected i.p. with either 100 μL of vehicle or tamoxifen. Mice were used starting 3 weeks after the last tamoxifen injection to target microglia selectively due to their slower turnover rate compared with peripheral myeloid cells.28,29,33 Mice treated with tamoxifen and vehicle are called KO and wild-type (WT), respectively.
Genotyping was performed by polymerase chain reaction (PCR) as described for CB1Rf/f.35 PCR was performed with following primer set (forward primer: TTCCCGCAGAACCTGAAGATGTTCG and reverse primer: GGGTGTTATAAGCAATCCCCAGAAATGC) for genotyping of CX3CR1-Cre/ER mouse line. All lines were in a mixed genetic background, with a predominant C57BL/6NCrlBR contribution. The experimental and control groups for each genotype are littermates that were randomized to each group. Experimenters were always blind to genotype and treatment.
LPS preparation and administration
LPS (Escherichia coli 0127: B8; Sigma-Aldrich) was diluted in saline to a concentration of 0.5 mg/mL. The endotoxin was injected at 4 μL/g of body weight, resulting in a final dose of 2 mg/kg, based on pilot experiments and on the literature.36–40 At the beginning of the light phase, the mice were weighed and injected i.p. with either LPS (2 mg/kg) or saline (0.9%) as a control.41–43
Behavioral experiments
All behavioral experiments were performed in the morning, under conditions of dim light and low noise, as previously described.44,45 Behavior was videotaped to be scored later by a trained observer blind to drug treatments, using the Smart software (Panlab, Barcelona, Spain). All testing equipment was thoroughly cleaned between each session.
Open field
To assess locomotor activity and anxiety level, mice were placed in a rectangular cage (40×40 cm) with surrounding walls that prevent escape for 10 min. Total traveled distance, time, and distance in the center were calculated. A decrease in time spent in the center of the arena (at least 10 cm away from the wall) is considered an index of anxiety-like behavior, independent of locomotor activity.
Elevated plus maze
The maze was made of two opposing open arms (30×8 cm) and two opposing closed arms (30×8×15 cm) connected by a central platform (8×8 cm) and elevated 120 cm above the floor. Each mouse was placed in the center of the maze facing an open arm and the percentage of time spent in open arms was assessed during a 5-min period. An entry was scored as such only when the mouse placed all four limbs into any given arm. A decrease in time spent in open arms is considered an index of anxiety-like behavior, independent of locomotor activity.44,46,47
Tail suspension test
An adhesive tape was placed on the mouse tail (distance from the tip=2 cm) and hooked to a horizontal ring stand bar placed 30 cm above the floor. The test was conducted for a period of 6 min in a visually isolated area. Mice demonstrated several escape attempts interspersed with immobility periods during, which they hung passively and completely motionless. Data are represented as the time spent immobile.47
Y maze
The Y maze was used to assess spatial working memory as previously described.48–51 Each arm was 34 cm long, 8 cm wide, and 14 cm high. The floor of the maze was covered with corn cob litter, which was mixed between each trial to remove olfactory cues. Visual cues were placed in the testing room and kept constant during the whole test. In the first trial, one arm was closed with a guillotine door and mice were allowed to visit two arms of the maze for 5 min. After a 30-min intertrial interval, mice were placed back in the start arm and allowed free access to the three arms for 5 min. Start and closed arms were randomly assigned for each mouse. Data are presented as the percentage of time spent exploring the novel and the familiar arms during the second trial.
Food restriction
Basal food intake was measured over a 24-h period for all mice. Food was then removed and put back 24 h later. Food intake was measured 24 h after food retrieval.
Behavioral analyses listed above have been performed from the least stressful to the most stressful, to limit any bias: Y maze → open field (OF) → elevated plus maze (EPM) → tail suspension test (TST) → food restriction.
Sickness behavior
At 8 am (beginning of the light phase), the mice were weighed and injected with either 100 μL saline (0.9%) or LPS (2 mg/kg). Locomotor activity was measured in smart cages for the next 2 h (Smart V3.0; Panlab, USA). Mice body weight and food intake were measured hourly for 12 h. Twenty-four hours postinjection, the mice were weighed, their food intake was determined, and their body fat was measured using a body composition analyzer (Minispec LF90II BCA-analyzer; Burker).
Iba-1 immunostaining
Twenty-four hours post-LPS treatment, mice were deeply anesthetized with isoflurane and transcardially perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Each brain was removed, postfixed in PFA overnight at 4°C, and cryoprotected in 30% sucrose at 4°C. Immunohistochemistry experiments were performed on free-floating coronal 30 μm cryostat slices using the streptavidin-biotin-immunoperoxidase technique.52 After washing off the cryoprotectant solution, slices were incubated 30 min in PBS containing 0.3% of H2O2 to quench endogenous peroxidase activity. Samples were incubated in a blocking solution containing 3% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS for 45 min at room temperature (RT), and immunostained with primary anti-Iba-1 antibody (1:1000; overnight at 4°C; Wako, Neuss, Germany) followed by biotinylated goat anti-rabbit antibody (1:2000; Invitrogen, Saint Aubin, France), 2 h at RT. Avidin-biotin peroxidase (Vectastain ABC kit Biovalley) diluted in PBS according to manufacturer's instructions (Vector Laboratories) was added for 1 h at RT. The peroxidase reaction product was developed using diaminobenzidine and nickel-enhanced glucose oxidase method, giving a black precipitate.
Stereological counting of Iba-1 immunoreactive cells in the hippocampus and hypothalamus
We assessed the density of Iba-1-positive cells 24 h post-LPS injection, a time point at which microglial proliferation starts to be measurable.53 We used the unbiased stereological sampling method based on optical dissector stereological probe to quantify Iba-1-immunoreactive cells in the hippocampus and hypothalamus, as previously described.52 Hippocampus slices were analyzed between the stereotaxic coordinates −0.80 to −2.07 mm from Bregma. In the hypothalamus, we focused on the arcuate nucleus of the hypothalamus (ARH; −1.07 to −2.07 from Bregma) and paraventricular nucleus (PVN; −0.37 to −1.07 mm from Bregma). Stereological analysis was performed using an Olympus BX51 microscope with a motorized Z and X-Y stage encoders linked to a computer-assisted stereological system (Mercator digital imaging system; Explora Nova, La Rochelle, France).
The volume of each structure was calculated using the formula V(structure)=ΣS td, where ΣS is the sum of surface areas (μm2) and t the average section thickness (and d the distance between the sections). The average section thickness (t) was estimated to 10 μm after immunohistochemistry processing and guard zones of 2 μm were used to ensure that top and bottom of sections were never included in the analysis. From a random start position, a computer-generated sampling grid placed the counting frames that were used to count the cells per mm2 for each brain region per animal in images at 40×magnification.
Where Iba-1 immunoreactivity was present only in cellular ramifications, cells were not considered positive microglia. Microglia were counted in the whole slice thickness, and individual cells were taken into account only if more than half of the cell was inside the two consecutive considered boundaries. Split cell counting error was corrected by using the Abercrombie formula. To estimate the number of Iba-1-immunoreactive cells, we used the following formula: N=V(structure) [ΣQ−/ΣV(dis)], where N is the estimation of the number of Iba-1-immunoreactive cells, V the volume of the structure, ΣQ− the number of cells counted in the frames, and ΣV(dis) the total volume of the frames. Mean cell number per plane and standard errors of the mean (SEMs) were then calculated for each group of mice.
CB1R immunostaining and electron microscopy imaging
Sections of the hypothalamus containing the arcuate nucleus region (−1.07 to −2.07 from Bregma; female mice treated with LPS) were washed in Tris-buffered saline (TBS) and incubated with 0.1% solution of NaBH4 for 15 min. After washes in TBS, sections were incubated in a blocking buffer of TBS containing 10% BSA and 0.01% Triton X-100 for 1 h at RT. Sections were incubated with the primary goat polyclonal anti-CB1 receptor antibody (CB1-Go-Af450; 1:100; Frontier Institute) in blocking buffer overnight at 4°C for 2 days. After 15 min at RT, the sections were rinsed with TBS and incubated in a 1.4 nm gold-labeled donkey anti-goat immunoglobulin-G secondary antibody (1:100; Jackson Lab) in blocking buffer at RT for 3 h.
After washes in TBS, sections were postfixed with 0.3% glutaraldehyde for 1 h at RT. Section were washed in double-distilled water twice in a 3% sodium acetate solution, and then incubated with a silver enhancement of gold particle kit (HQ Silver; Nanoprobes, Inc., Yaphank, NY) for about 12 min in the dark. Several washes in a 3% sodium acetate solution were performed (until the solution was no longer viscous), and then tissues were washed in phosphate buffer (PB). After washes in PBS, tissues were incubated for 1 h in a solution containing 3% potassium ferrocyanide in 0.1 M PB buffer combined with an equal volume of 4% aqueous osmium tetroxide. After washes with double-distilled water, the sections were incubated with a filtered thiocarbohydrazide solution for 20 min at RT (1% thiocarbohydrazide in ddH2O placed in a 60°C oven for 1 h).
Then washes in double-distilled water were performed and sections were placed in 2% osmium tetroxide in double-distilled water for 30 min, at RT. Sections were washed in double-distilled water, dehydrated in increasing concentrations of ethanol and then immersed in propylene oxide. Tissues were impregnated in Durcupan resin overnight at RT and polymerized between ACLAR sheets at 55°C for 72 h. Ultrathin sections were cut at 70 nm using a Leica ARTOS 3D ultramicrotome. Pictures finally were acquired at a magnification of×10,000 using a JEOL JEM-1400 transmission electron microscope (80 kV) equipped with a 10.7 MP Gatan Orius SC1000A1 camera. Microglia have a dark, electron-dense cytoplasm. Their cell bodies are recognized by their small size and triangular shape, distinctive heterochromatin pattern, long stretches of endoplasmic reticulum, association with extracellular space pockets, frequent lysosomes and endosomes, and interactions with synapses and other elements of the neuropil.54
Quantitative real-time PCR
Expression of cytokine transcripts was measured 2 h post-LPS injection, at the peak of production.41,55–57 Hypothalamus and hippocampus samples were homogenized in Tri-reagent (Euromedex); RNA was isolated using a standard chloroform/isopropanol protocol58 and purified by incubation with Turbo DNA-free (Fisher Scientific). RNA was processed and analyzed following an adaptation of published methods.59 Complementary DNA (cDNA) was synthesized from 2 μg of total RNA using Maxima Reverse Transcriptase (Fisher Scientific) and primed with oligo-dT primers (Fisher Scientific) and random primers (Fisher Scientific). Quantitative polymerase chain reaction (qPCR) was performed using a LightCycler® 480 Real-Time PCR System (Roche, Meylan, France).
qPCR reactions were done in duplicate for each sample, using transcript-specific primers, cDNA (4 ng) and LightCycler 480 SYBR Green I Master (Roche), in a final volume of 10 μL. For the determination of the reference gene, the refFinder method was used.60 Relative expression analysis was normalized against one or two reference genes, depending on the tissue. The beta2-microglobulin (B2M) or tubulin alpha 4 a (Tuba4a) and actin beta (Actb) genes were used as reference genes for hypothalamus. The B2M or glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and succinate dehydrogenase complex subunit (Sdha) genes were used as reference genes for hippocampus. The relative level of expression was calculated using the comparative 2−ΔΔCT method (Xie et al.60).
The following primers were used: B2M (Mm00437762); Tuba4a (NM_007393); Actb (NM_007393); Gapdh (NM_0080); Sdha (NM_023281); IL-6 (Mm00446190_M1); TNF-α (Mm00443258_M1); IL-1β (Mm00434228_M1); interleukin-1 receptor antagonist (IL-1RA, il1rn gene; NM_031167); interleukin-10 (il10; NM_010548); transforming growth factor beta 1 (tgfb1; NM_011577); toll-like receptor 2 (tlr2; NM_011905); and toll-like Receptor 4 (tlr4; NM_021297).
Microglial isolation and sorting
Brains were homogenized in Hanks' balanced salt solution (HBSS), pH 7.4, passing through a 70 μm nylon cell strainer. Homogenates were centrifuged at 600 g for 6 min. Supernatants were removed and cell pellets were resuspended in 70% isotonic Percoll (GE Healthcare, Aulnay sous Bois, France). Single-cell suspensions were prepared and centrifuged over a 37%/70% discontinuous Percoll gradient (GE Healthcare) at 2000 g for 20 min and mononuclear cells were isolated from the interface.
For each brain extraction, ∼3×105 cells were isolated. Cells were washed and incubated with anti-CD16/CD32 antibody (eBiosciences, Paris, France) to block Fc receptors for 10 min on ice. Cells were washed and then incubated for 45 min with the appropriate conjugated antibodies: anti-CD11b-APC and anti-CD45-PerCP Cy5.5 (eBiosciences). Cells were washed and then suspended in PBS/BSA 0.1% for analysis. Nonspecific binding was assessed by using nonspecific, isotype-matched antibodies. Cells were sorted using a FACS Aria 5-Blue 2-Violet 2-Red laser configuration (BD Biosciences) (Supplementary Fig. S1).
Plasma cytokine assay
Cytokine assays were performed as previously described.48 The limit of detection was 1.1 pg/mL for IL-6, 2.3 pg/mL for TNF-α, and 5.4 pg/mL for IL-1β. Briefly, samples diluted 1/2 were added to a 96-well microtiter plate (25 μL/well) coated with beads (Millipore), covered with aluminum foil, and incubated overnight on a shaker at 4°C in the dark. After removal of sample using a magnet, beads were incubated with detection antibodies for1 h at RT, while shaking, followed by streptavidin-PE for 30 min. The beads were then resuspended in 150 μL sheath fluid and analyzed using the BioPlex 200 system (Bio-Rad). The reader was set to read a minimum of 50 beads with an identical fluorescence expressed as the median fluorescence intensity. Median fluorescence intensity readings were converted to pg/mL using calibration curves prepared with cytokine standards included in the kit.
Data analysis
All data were visualized and analyzed using GraphPad 9. Normality and homoscedasticity of data were assessed by Shapiro-Wilk and Brown-Forsythe tests, respectively. The results presented in Figure 1 were analyzed using two-way analysis of variance (ANOVA) followed by Tukey's multiple comparison post-hoc test when interaction was statistically positive (genotype and treatment or genotype and sex as factors). All other data were analyzed using three-way ANOVA followed by Tukey's multiple comparison post-hoc test when interaction was statistically positive (genotype, sex, and treatment as factors). All data were expressed as mean±SEM. A p<0.05 was considered statistically significant.
FIG. 1.
Deletion of microglial CB1R does not affect basal behavior in males and females. (A) EM images showing immunostained CB1R in WT (left image) and in CX3CR1-CB1R KO (right image) female mice. Blue arrows indicate CB1R staining in microglial cells. Green arrows indicate CB1R staining in other cell types. (B) Total traveled distance (in cm) in the OF arena [genotype effect, F(1,47)=0.02, p=0.87; sex effect, F(1,47)=1.93, p=0.17; interaction, F(1,47)=0.4, p=0.53]. N=12–14 per group. (C, D) Time (in seconds) (C) and distance (in cm) traveled (D) in the center of the OF arena. (C) Genotype effect, F(1,48)=0.56, p=0.46; sex effect, F(1,48)=0.33, p=0.57; interaction, F(1,48)=2.61, p=0.11. N=12–14 per group; (D) genotype effect, F(1,47)=0.41, p=0.53; sex effect, F(1,47)=0.19, p=0.66; interaction, F(1,47)=4.25, *p=0.045. Tukey's multiple comparisons post-hoc analysis did not reveal any significant difference between groups. N=12–14 per group. (E) Percentage of time spent in the open arm of the EPM [genotype effect, F(1,48)=1.13, p=0.29; sex effect, F(1,48)=1.74, p=0.19; interaction, F(1,48)=0.17, p=0.68]. N=12–14 per group. (F) Time (in seconds) spent immobile in the TST [genotype effect, F(1,46)=0.016, p=0.89; sex effect, F(1,46)=0.28, p=0.6; interaction, F(1,46)=0.085, p=0.77]. N=11–14 per group. (G, H) Percentage of time spent in the novel or familiar arm of the Y maze for males (G) and females (H). (G) Genotype effect, F(1,47)=0.02, p=0.89; arm effect, F(1,47)=27.11, ***p<0.0001; interaction, F(1,47)=1.39, p=0.24; (H) genotype effect, F(1,50)=0, p>0.99; arm effect, F(1,50)=13.94, ***p=0.0005; interaction, F(1,50)=0.44, p=0.51. N=12–14 per group. (I) Total traveled distance (in cm) in the Y maze [genotype effect, F(1,48)=1.84, p=0.18; sex effect, F(1,48)=0.66, p=0.43; interaction, F(1,48)=0.82, p=0.37]. N=12–14 per group. (J, K) FI (in g) after a 24-h period of food restriction in males (J) and in females (K). (J) Genotype effect, F(1,44)=5.08, *p=0.03; food restriction effect, F(1,44)=131.9, ***p<0.0001; interaction, F(1,44)=1.13, p=0.26; (K) genotype effect, F(1,48)=0.12, p=0.73; food restriction effect, F(1,48)=277.4, ***p<0.0001; interaction, F(1,48)=0.3, p=0.59. *p<0.05, ***p<0.001. CB1R, cannabinoid type 1 receptor; EM, electron microscopy; EPM, elevated plus maze; KO, knockout; m, microglia; ma, myelinated axons; OF, open field; TST, tail suspension test. Color images are available online.
Results
Deletion of CB1R in CX3CR1-positive cells does not alter basal behavioral responses
Depending on their cellular location, CB1R receptors regulate a plethora of behaviors ranging from memory processes to food intake and anxiety.46,61–65 To study the physiological role of CX3CR1-CB1R on behavior, we submitted conditional mutant mice, expected to lack CB1R receptors in CX3CR1-positive cells (“KO”), to a series of tests to assess locomotion, affective behavior and cognitive abilities, as well as food intake.
According to public RNAseq databases, CB1R RNA is expressed in microglia,30 yet at a very low level. In certain cells, the expression levels of CB1 receptors can be very low (below threshold of detection and/or quantification), but still maintain functional relevance. For instance, CB1 receptors expressed in astrocytes are extremely difficult to detect by conventional approaches, such as light microscopy immunohistochemistry or in situ hybridization.62 However, immunogold electron microscopy (EM) allows the detection of such low levels of astroglial CB1 receptor protein.33,66,67 Thus, we adopted this approach to detect CB1 receptor in microglial cells. CB1R immunogold particles were present in several cell types, which notably included microglia in WT mice (Fig. 1A, left image), whereas CX3CR1-CB1R KO mice were deficient in CB1R protein, specifically within microglial cells (Fig. 1A, right).
We then studied the behavioral phenotype of the animals. Neither CX3CR1-KO males nor females displayed locomotor deficits as they traveled the same distance in the OF arena compared to their respective wild-type littermate controls (Fig. 1B). Likewise, anxiety levels were not altered by the mutation as both mutant and control mice spent the same amount of time in the center area of the OF (Fig. 1C, D) and their behavior did not differ in the EPM (Fig. 1E). We could not observe any sex or genotype effect in the TST (Fig. 1F), or in learning and memory abilities in the Y maze task. In the latter, males and females of both genotypes spent significantly more time in the novel arm, as a marker of proper spatial working memory abilities (Fig. 1G, H), with no difference in the distance traveled (Fig. 1I). We finally measured food intake under free-feeding and after 24 h of fasting. Both males and females significantly increased their food intake after fasting. We also found a global genotype effect on food intake in males, but not in females. Yet, the deletion of microglial CB1R did not affect the food intake response following fasting, neither in males nor in females (Fig. 1J, K).
CB1R in CX3CR1-positive cells differentially controls sickness behavior in males and females
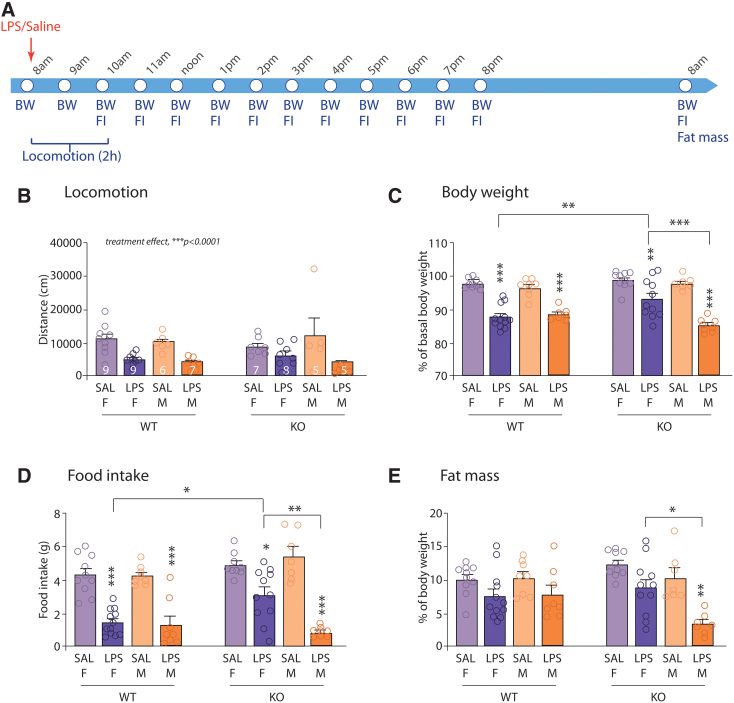
Both microglia and the ECS are involved in the brain response to systemic inflammatory challenges.9,68–71 We thus assessed the ability of the endotoxin LPS to induce a sickness response in CX3CR1-CB1R-KO and WT mice (Fig. 2A).5,6,41,72 As expected, both male and female mice reduced their locomotor activity over the first 2 h post-LPS injection. This behavioral response was not altered in CX3CR1-CB1R-KO mice (Fig. 2B). The endotoxin also significantly reduced body weight and food intake in WT mice, when measured 24 h post-treatment, independent of the sex (Fig. 2C, D). However, the effects of LPS were significantly dampened when knocking out microglial CB1R in female mice, specifically for body weight and food intake (Fig. 2C–E), while they were exacerbated in CX3CR1-CB1R-KO male mice with a further decrease in fat mass (Fig. 2E).
FIG. 2.
Microglial CB1R is implicated in the behavioral and physiological response to a peripheral immune challenge. (A) Experimental design. (B) Total distance (in cm) traveled by male and female mice over the 2-h period following LPS injection [genotype effect, F(1,48)=0.1640, p=0.6873; sex effect, F(1,48)=0.47, p=0.49; treatment effect, F(1,48)=27.91, ***p<0.0001; genotype×sex, F(1,48)=0.034, p=0.85; genotype×treatment, F(1,48)=0.04, p=0.84; sex×treatment, F(1,48)=2.84, p=0.098; genotype×sex×treatment, F(1,48)=2.7, p=0.11]. Males: n=5–7; females: n=7–9. (C) Body weight loss following LPS injection in males and females, expressed as the percentage of basal body weight [genotype effect, F(1,65)=2.13, p=0.15; sex effect, F(1,65)=9.10, **p=0.0036; treatment effect, F(1,65)=144.5, ***p<0.0001; genotype×sex, F(1,65)=8.13, **p=0.006; genotype×treatment, F(1,65)=0.02, p=0.89; sex×treatment, F(1,65)=2.75, p=0.1; genotype×sex×treatment, F(1,65)=8.98, **p=0.0039. Tukey's multiple comparisons post-hoc analysis: F-WT-SAL vs. F-WT-LPS: ***p<0.0001; M-WT-SAL vs. M-WT-LPS: ***p=0.0002; F-KO-SAL vs. F-KO-LPS: **p=0.0029; M-KO-SAL vs. M-KO-LPS: ***p<0.0001; F-WT-LPS vs. F-KO-LPS: **p=0.0028; F-KO-LPS vs. M-KO-LPS: ***p<0.0001]. Males: n=7–8; females: n=10–12. (D) Cumulative FI (in g) over the 24 h following LPS injection in males and females [genotype effect, F(1,65)=8.19, **p=0.0056; sex effect, F(1,65)=3.16, p=0.08; treatment effect, F(1,65)=134.9, ***p<0.0001; genotype×sex, F(1,65)=2.025, p=0.16; genotype×treatment, F(1,65)=0.26, p=0.61; sex×treatment, F(1,65)=7.32, **p=0.0087; genotype×sex×treatment, F(1,65)=6.42, *p=0.014. Tukey's multiple comparisons post-hoc analysis: F-WT-SAL vs. F-WT-LPS: ***p<0.0001; M-WT-SAL vs. M-WT-LPS: ***p<0.0001; F-KO-SAL vs. F-KO-LPS: *p=0.012; M-KO-SAL vs. M-KO-LPS: ***p<0.0001; F-WT-LPS vs. F-KO-LPS: *p=0.014; F-KO-LPS vs. M-KO-LPS: **p=0.0014]. Males: n=7–8; females: n=9–12. (E) Fat mass measured 24 h post-LPS injection and expressed as the percentage of mice body weight [genotype effect, F(1,65)=0.07, p=0.79; sex effect, F(1,65)=5.31, *p=0.02; treatment effect, F(1,65)=26.06, ***p<0.0001; genotype×sex, F(1,65)=6.83, *p=0.011; genotype×treatment, F(1,65)=3.33, p=0.072; sex×treatment, F(1,65)=1.23, p=0.27; genotype×sex×treatment, F(1,65)=1.33, p=0.25. Tukey's multiple comparisons post-hoc analysis: M-KO-SAL vs. M-KO-LPS: **p=0.0028; F-KO-LPS vs. M-KO-LPS: **p=0.0016]. Males: n=7–8; females: n=10–12. *p<0.05, **p<0.01; ***p<0.001. LPS, lipopolysaccharide. Color images are available online.
CB1R in CX3CR1-positive cells promote LPS-induced brain cytokine production in both males and females
LPS-induced sickness behavior partly relies on the production of proinflammatory cytokines by microglia.73 We thus assessed the expression levels of the three main proinflammatory factors known to drive the brain inflammatory response following intraperitoneal LPS administration, that is, IL-1β, IL-6, and TNF-α. Cytokine expression was measured in the hypothalamus and hippocampus, two brain regions that regulate sickness behavior, including decrease in food intake and body weight.41,73,74
In the hypothalamus, LPS significantly increased the production of IL-1β and TNF-α messenger RNA (mRNA) 2 h post-injection, in both WT males and females (Fig. 3A), while IL-6 transcripts were significantly upregulated in WT males only (Fig. 3A). Deletion of microglial CB1R dampened IL-1β mRNA synthesis in both males and females, as well as IL-6 mRNA synthesis in males. There was no effect of the genotype on TNF-α production (Fig. 3A). IL-1β in females and both IL-1β and TNF-α in males were still overexpressed at 24 h post-LPS injection, yet we could not observe a genotype effect (Fig. 3B). Conversely, IL-6 mRNA levels were significantly different between CX3CR1-CB1R-KO and WT mice 24 h post-LPS (Fig. 3B).
FIG. 3.
Microglial CB1R promotes the synthesis of proinflammatory cytokines in the hypothalamus of male mice following LPS administration. (A) qPCR quantification of IL-1β, IL- and TNF-α mRNA in the hypothalamus of male and female mice, 2 h post-LPS [il-1b: genotype effect, F(1,58)=3.61, p=0.06; sex effect, F(1,58)=0.0058, p=0.81; treatment effect, F(1,58)=37.97, ***p<0.0001; genotype×sex, F(1,58)=0.76, p=0.39; genotype×treatment, F(1,58)=4.34, *p=0.04; sex×treatment, F(1,58)=0.011, p=0.92; genotype×sex×treatment, F(1,58)=0.18, p=0.67; Tukey's multiple comparisons post-hoc analysis: F-WT-SAL vs. F-WT-LPS: *p=0.022; M-WT-SAL vs. M-WT-LPS: ***p=0.0003; il-6: genotype effect, F(1,57)=17.46, ***p=0.0001; sex effect, F(1,57)=4.87, *p=0.03; treatment effect, F(1,57)=29.49, ***p<0.0001; genotype×sex, F(1,57)=2.55, p=0.12; genotype×treatment, F(1,57)=14.27, ***p=0.0004; sex×treatment, F(1,57)=3.92, p=0.053; genotype×sex×treatment, F(1,57)=3.23, p=0.077; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001; F-WT-LPS vs. M-WT-LPS, **p=0.005; M-WT-LPS vs. M-KO-LPS: ***p<0.0001; tnf-a: genotype effect, F(1,58)=3.29, p=0.07; sex effect, F(1,58)=0.029, p=0.86; treatment effect, F(1,58)=38.50, ***p<0.0001; genotype×sex, F(1,58)=0.39, p=0.53; genotype×treatment, F(1,58)=3.82, p=0.055; sex×treatment, F(1,58)=0.0001, p=0.99; genotype×sex×treatment, F(1,58)=0.11, p=0.73]. Males: n=8–11; females: n=6–9. (B) qPCR quantification of IL-1β, IL-6, and TNF-α mRNA in the hypothalamus of male and female mice, 24 h post-LPS [il-1b: genotype effect, F(1,66)=0.42, p=0.52; sex effect, F(1,66)=0.07, p=0.79; treatment effect, F(1,66)=22.62, ***p<0.0001; genotype×sex, F(1,66)=0.84, p=0.36; genotype×treatment, F(1,66)=3.8, p=0.055; sex×treatment, F(1,66)=2.15, p=0.15; genotype×sex×treatment, F(1,66)=4.8, *p=0.03; Tukey's multiple comparisons post-hoc analysis: F-WT-SAL vs. F-WT-LPS: *p=0.025, F-KO-SAL vs. F-KO-LPS: **p=0.009; il-6: genotype effect, F(1,73)=6.33, *p=0.014; sex effect, F(1,73)=0.039, p=0.84; treatment effect, F(1,73)=3.015, p=0.087; genotype×sex, F(1,73)=3.6, p=0.06; genotype×treatment, F(1,73)=1.11, p=0.29; sex×treatment, F(1,73)=0.03, p=0.86; genotype×sex×treatment, F(1,73)=0.11, p=0.75; tnf-a: genotype effect, F(1,68)=0.48, p=0.49; sex effect, F(1,68)=21.64, ***p<0.0001; treatment effect, F(1,68)=10.14, **p=0.0022; genotype×sex, F(1,68)=0.54, p=0.46; genotype×treatment, F(1,68)=4.16, *p=0.045; sex×treatment, F(1,68)=10.88, **p=0.0015; genotype×sex×treatment, F(1,68)=4.28, *p=0.04; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS, ***p=0.0005, F-WT-LPS vs. M-WT-LPS: ***p<0.0001]. Males: n=5–11; females: n=10–13. *p<0.05; **p<0.01; ***p<0.001. IL-1β, interleukin-1 beta; IL-6, interleukin-6; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction; TNF-α, tumor necrosis factor alpha. Color images are available online.
In the hippocampus, LPS significantly induced the production of IL-1β, IL-6, and TNF-α mRNA in males only, 2 h post-injection (Fig. 4A). Knocking out CB1R in CX3CR1-positive cells reduced the synthesis of IL-1β and TNF-α mRNA in males (Fig. 4A). Besides a significant increase in the expression of IL-6 transcripts in females, deletion of microglial CB1R did not dramatically affect the production of cytokines in the hippocampus of females (Fig. 4A). Proinflammatory cytokines were still overexpressed 24 h post-LPS injection in males (Fig. 4B), with no overt effect of the genotype.
FIG. 4.
Microglial CB1R promotes the synthesis of proinflammatory cytokines in the hippocampus of male mice following LPS administration. (A) qPCR quantification of IL-1β, IL-6, and TNF-α mRNA in the hippocampus of male and female mice, 2 h post-LPS [il-1b: genotype effect, F(1,46)=0.88, p=0.35; sex effect, F(1,46)=5.1, *p=0.029; treatment effect, F(1,46)=28.20, ***p<0.0001; genotype×sex, F(1,46)=4.82, *p=0.03; genotype×treatment, F(1,46)=0.78, p=0.38; sex×treatment, F(1,46)=6.52, *p=0.014; genotype×sex×treatment, F(1,46)=4.15, *p=0.047; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001, F-WT-LPS vs. M-WT-LPS: **p=0.0014; il-6: genotype effect, F(1,46)=1.74, p=0.19; sex effect, F(1,46)=4.36, *p=0.04; treatment effect, F(1,46)=83.6, ***p<0.0001; genotype×sex, F(1,46)=0.033, p=0.86; genotype×treatment, F(1,46)=0.90, p=0.35; sex×treatment, F(1,46)=7.23, **p=0.0099; genotype×sex×treatment, F(1,46)=0.020, p=0.88; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001; F-KO-SAL vs. F-KO-LPS: *p=0.012; M-KO-SAL vs. M-KO-LPS: ***p<0.0001; tnf-a: genotype effect, F(1,47)=8.54, **p=0.0053; sex effect, F(1,47)=15.35, ***p=0.0003; treatment effect, F(1,47)=54.42, ***p<0.0001; genotype×sex, F(1,47)=6.5, *p=0.014; genotype×treatment, F(1,47)=6.92, *p=0.011; sex×treatment, F(1,47)=14.43, ***p=0.0004; genotype×sex×treatment, F(1,47)=8.97, **p=0.004; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001; F-WT-LPS vs. M-WT-LPS: ***p<0.0001; M-WT-LPS vs. M-KO-LPS: ***p<0.0001]. Males and females: n=6–7. (B) qPCR quantification of IL-1β, IL-6, and TNF-α mRNA in the hippocampus of male and female mice, 24 h post-LPS [il-1b: genotype effect, F(1,59)=0.65, p=0.42; sex effect, F(1,59)=6.8, *p=0.011; treatment effect, F(1,59)=18.31, ***p<0.0001; genotype×sex, F(1,59)=0.17, p=0.68; genotype×treatment, F(1,59)=6.01, *p=0.017; sex×treatment, F(1,59)=0.039, p=0.84; genotype×sex×treatment, F(1,59)=4.06, *p=0.049; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: **p=0.0057; il-6: genotype effect, F(1,61)=0.006, p=0.93; sex effect, F(1,61)=7.76, **p=0.007; treatment effect, F(1,61)=6.69, *p=0.012; genotype×sex, F(1,61)=1.48, p=0.23; genotype×treatment, F(1,61)=2.045, p=0.16; sex×treatment, F(1,61)=1.87, p=0.18; genotype×sex×treatment, F(1,61)=0.64, p=0.43; tnf-a: genotype effect, F(1,58)=0.06, p=0.81; sex effect, F(1,58)=5.93, *p=0.018; treatment effect, F(1,58)=12.71, ***p=0.0007; genotype×sex, F(1,58)=0.06, p=0.8; genotype×treatment, F(1,58)=4.89, *p=0.03; sex×treatment, F(1,58)=0.015, p=0.9; genotype×sex×treatment, F(1,58)=5.05, *p=0.028; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: *p=0.022]. Males: n=5–9; females: n=7–12. *p<0.05, **p<0.01; ***p<0.001. Color images are available online.
Cytokine-induced sickness behavior is a fully reversible phenomenon, and involves the action of anti-inflammatory cytokines that target IL-1 specifically, such as IL-1RA, or that have more generalized antagonist effects, such as interleukin-10 (IL-10) and transforming growth factor beta 1 (TGFβ1) (Dantzer, 2008). We thus tested whether our observations could rely on the exacerbation of the anti-inflammatory response in CX3CR1-CB1R-KO mice.
To this aim, we analyzed the mRNA expression of IL-1RA (il-1rn), IL-10, and TGFβ1 in the hypothalamus and hippocampus of both males and females, 2 and 24 h post-LPS injection (Supplementary Figs. S2 and S3). In the hypothalamus, LPS induced the expression of TGFβ1 and IL-1RA in females, and of TGFβ1 in males when measured 2 h post-treatment (Supplementary Fig. S2A). Twenty-four hours post-LPS injection, we did not observe any modification of cytokine production (Supplementary Fig. S2B). In the hippocampus, the endotoxin did not affect anti-inflammatory cytokine production, neither at 2 h nor at 24 h post-injection (Supplementary Fig. S3). Finally, we could not observe any effect of CB1R deletion on the expression levels of these cytokines, except a slight reduction of IL-RA transcripts in females.
This finding suggests that microglial CB1R-mediated proinflammatory response was not due to an exacerbated anti-inflammatory response.
Finally, we tested whether knocking out CB1R could differentially affect the expression of LPS receptors, namely TLR4 and TLR2,75 by quantifying their transcripts in all experimental conditions. LPS increased the expression of both TLR2 and TLR4 in the hypothalamus of males and females, and of TLR2 only in the hippocampus, 2 h post-treatment (Supplementary Fig. S4A, C). At 24 h, only TLR4 in the hippocampus was significantly increased in both sexes (Supplementary Fig. S4B, D).
Moreover, TLRs were expressed to the same extent in the hypothalamus of males and females, while they were overexpressed in the hippocampus of males compared to females, 2 h post-LPS treatment (Supplementary Fig. S4). Finally, we could not observe any effect of microglial CB1R deletion on the expression of the receptors neither in males nor in females (Supplementary Fig. S4), suggesting that a differential expression of LPS receptors does not account for the differential effect of the endotoxin on cytokine production and sickness behavior.
Deletion of CB1R in CX3CR1-positive cells does not affect microglial density in the hippocampus and hypothalamus of mice
Since male CX3CR1-CB1R-KO mice displayed altered LPS-induced production of cytokines and knowing that microglia are the main source of cytokines in response to LPS,5,76 we then carried out stereological counting of Iba-1-positive cells to evaluate microglial density in the hippocampus and hypothalamus of male and female mice (Fig. 5). We focused on two nuclei of the hypothalamus: the ARH and PVN, which are key structures in the regulation of sickness behavior.77,78 Our results show that LPS significantly increased microglial density only in the hippocampus of male mice at 24 h post-treatment, independent of the genotype. Moreover, deletion of microglial CB1R had no effect on the number of microglia in the hippocampus or PVN, neither in control condition nor following LPS administration (Fig. 5A, C). An effect of the genotype was observed in the ARH, yet post-hoc analyses did not reveal any significant difference between groups (Fig. 5B).
FIG. 5.
Deletion of microglial CB1R receptors does not affect microglial density. (A) Stereological counting of Iba-1-positive cells in the HPC, ARH, and PVN nuclei of the hypothalamus in male and female mice [HPC: genotype effect, F(1,49)=0.49, p=0.49; sex effect, F(1,49)=0.68, p=0.41; treatment effect, F(1,49)=22.54, ***p<0.0001; genotype×sex, F(1,49)=0.0012, p=0.97; genotype×treatment, F(1,49)=0.21, p=0.64; sex×treatment, F(1,49)=7.86, **p=0.0072; genotype×sex×treatment, F(1,49)=1.28, p=0.26; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: **p=0.008; ARH: genotype effect, F(1,27)=4.46, p=0.04; sex effect, F(1,27)=2.81, p=0.11; treatment effect, F(1,27)=0.055, p=0.82; genotype×sex, F(1,27)=0.07, p=0.79; genotype×treatment, F(1,27)=0.17, p=0.68; sex×treatment, F(1,27)=0.35, p=0.56; genotype×sex×treatment, F(1,27)=0.1, p=0.75; PVN: genotype effect, F(1,38)=0.005, p=0.94; sex effect, F(1,38)=0.76, p=0.39; treatment effect, F(1,38)=0.094, p=0.76; genotype×sex, F(1,38)=0.88, p=0.35; genotype×treatment, F(1,38)=0.46, p=0.50; sex×treatment, F(1,38)=0.35, p=0.56; genotype×sex×treatment, F(1,38)=0.0007, p=0.98]. Males: n=4–7; females: n=4–11. (B) Representative images of Iba1 immunostaining in the hippocampus, ARH and PVN nuclei. Scale bar=150 μm. ARH, arcuate nucleus of the hypothalamus; PVN, paraventricular nucleus. Color images are available online.
Deletion of CB1R in CX3CR1-positive cells does not potently affect LPS-induced peripheral cytokine production
To rule out any peripheral effect of CB1R deletion, we finally measured the circulating levels of blood proinflammatory cytokines in male and female mice treated with the endotoxin. We withdrew blood samples at 2 h post-LPS injection (Fig. 6). IL-1β, IL-6, and TNF-α proteins were overall significantly more abundant in LPS-treated male and female mice than in controls. CB1R deletion in CX3CR1-positive cells did not hamper IL-6 and TNF-α protein levels, different from what was observed in the brain (Fig. 6B, C). IL-1β production was modulated in an opposite way in both male and female CX3CR1-CB1R-KO mice, but expression levels of the cytokine stayed overall very low compared to IL-6 and TNF-α (Fig. 6A–C).
FIG. 6.

Microglial CB1R does not affect the synthesis of blood proinflammatory cytokines following LPS administration. (A–C) Bioplex quantification of IL-1β (A), IL-6 (B), and TNF-α (C) proteins in the blood of male and female mice, 2 h post-LPS [il-1b: genotype effect, F(1,47)=0.05, p=0.83; sex effect, F(1,47)=1.39, p=0.24; treatment effect, F(1,47)=38.95, ***p<0.0001; genotype×sex, F(1,47)=8.17, **p=0.0063; genotype×treatment, F(1,47)=0.38, p=0.54; sex×treatment, F(1,47)=0.61, p=0.44; genotype×sex×treatment, F(1,47)=10.62, **p=0.0021; Tukey's multiple comparisons post-hoc analysis: F-WT-SAL vs. F-WT-LPS: ***p<0.0001; M-KO-SAL vs. M-KO-LPS: **p=0.01; F-WT-LPS vs. M-WT-LPS: *p=0.0104; F-WT-LPS vs. F-KO-KPS: **p=0.0041; il-6: genotype effect, F(1,49)=1.94, p=0.17; sex effect, F(1,49)=50.53, ***p<0.0001; treatment effect, F(1,49)=137.7, ***p<0.0001; genotype×sex, F(1,49)=0.38, p=0.54; genotype×treatment, F(1,49)=1.93, p=0.17; sex×treatment, F(1,49)=50.53, ***p<0.0001; genotype×sex×treatment, F(1,49)=0.39, p=0.53; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001; M-KO-SAL vs. M-KO-LPS: ***p<0.0001; F-WT-LPS vs. M-WT-LPS: ***p<0.0001; F-KO-LPS vs. M-KO-LPS: ***p<0.0001; tnf-a: genotype effect, F(1,47)=3.48, p=0.07; sex effect, F(1,47)=26.33, ***p<0.0001; treatment effect, F(1,47)=85.71, ***p<0.0001; genotype×sex, F(1,47)=2.28, p=0.14; genotype×treatment, F(1,47)=3.98, p=0.052; sex×treatment, F(1,47)=27.91, ***p<0.0001; genotype×sex×treatment, F(1,47)=2.097, p=0.15; Tukey's multiple comparisons post-hoc analysis: M-WT-SAL vs. M-WT-LPS: ***p<0.0001; M-KO-SAL vs. M-KO-LPS: ***p<0.0001; F-WT-LPS vs. M-WT-LPS: ***p<0.0001; F-KO-LPS vs. M-KO-LPS: *p=0.0103]. Males: n=4–9; females: n=5–9. *p<0.05, **p<0.01; ***p<0.001. Color images are available online.
Discussion
The aim of this study was to assess the role of microglial CB1R on basal behavior and in the regulation of brain inflammation, in male and female mice. Although CB1R expression is extremely low in microglia, our data showed that CX3CR1-CB1R-KO mice are deficient in CB1R proteins, specifically within microglial cells. These mice display behavioral alterations indicating a role of microglial CB1R signaling in sickness behavior. Microglial CB1R does not seem to be involved in the regulation of locomotion, cognition, and anxiety-like behavior under normal physiological conditions. In response to an immune challenge, however, both male and female CX3CR1-CB1R-KO mice displayed a decrease in the production of central proinflammatory cytokines in relation to their WT counterparts. In addition, this decrease was correlated to enhanced sickness behavior (further decrease in fat mass) in males, while females were protected (dampening of body weight and food intake changes). We thus revealed (1) a potential role for microglial CB1R in the brain response to an immune challenge and (2) a microglial CB1R-dependent sexual dimorphism in this process.
The role of microglia in the regulation of basal behavior is still a matter of debate. Several studies have shown that altering microglial function in adult male mice leads to deficits in multiple learning tasks, such as motor learning, auditory-cued fear conditioning, and novel object recognition.29 Proper microglial function is also required for the adaptation to a stressful or enriched environment.79–81 Conversely, Elmore et al. showed that depleting the CNS in microglia, using a CSF1R inhibitor (CSF1R being essential for microglial maintenance and long-term survival (Erblich et al.82; Ginhoux et al.83), does not affect cognition and motor functions.84 Hence, understanding the role of microglia on basal behavior requires more investigation. With these data, we add novel information that deleting CB1R specifically in microglia has no impact on locomotion, anxiety levels, spatial working memory, and fasting-induced food intake behavior at steady state.
CB1R is a potent regulator of behavior, ranging from learning and memory to mood control (for review, Busquets-Garcia et al.). Depending on the targeted cell type (neuronal subtypes or astrocytes) and the activity status of the animal, knocking out CB1R leads to complex outcomes on food intake, anxiety, or memory abilities.62,65,85–88 We here demonstrate that the specific deletion of CB1R in microglial cells determines behavioral alterations in animals undergoing an inflammatory reaction.
According to our results, the brain response to inflammation is microglial CB1R dependent in both males and females, revealing a proinflammatory role for the receptor. Our data are in line with many previous reports showing that the use of pharmacological agents that block CB1R activity or of full CB1R-KO protects from inflammation, both in vitro and in vivo.24,25,27,89,90 Surprisingly, we observed, however, that deletion of CB1R CX3CR1-positive cells uncoupled the behavioral and cytokine responses to an endotoxin challenge in males. Indeed, while male CX3CR1-CB1R-KO mice were sicker than WT littermates (further decrease in body weight, fat mass, and locomotion), the brain production of proinflammatory cytokines was dampened in the absence of microglial CB1R. We expected opposite results as decades of research on the molecular substrate of sickness have convincingly demonstrated that early increase in IL-1β, IL-6, or TNF-α production is required for inflammatory anorexia, metabolic alterations, or decrease in locomotion in response to LPS.5,6,41,72,91,92 It is known that peripheral cytokines induce the de novo synthesis and release of cytokines in the brain.92 Hence, the effect of microglial CB1R deletion on brain inflammation might rely on a differential effect of LPS on blood cytokine production. However, the inducible genetic approach we used allowed to stably delete the cnr1 gene in microglia, while macrophages are not durably affected,29 which was confirmed by the demonstration that CB1R deletion did not dramatically affect the peripheral inflammatory response.
However, when analyzing the plasma levels of IL-1β, we observed a significant increase of this cytokine in LPS-treated CX3CR1-CB1R KO males and a significant decrease in CX3CR1-CB1R KO female mice. We cannot exclude that this might contribute to the behavioral effects observed in CX3CR1-CB1R KO mice. One explanation could be that not all peripheral macrophages have recovered a WT phenotype at the time of LPS injection. However, in this case, we would also observe a significant effect on the expression levels of IL-6 and TNF-α, which also depend on macrophage reactivity.
The differential expression of IL-1β observed in male and female KO mice could also be the result of a differential control from the CNS. Indeed, it has been well established that the CNS can signal to the periphery to modulate inflammation through efferent hormonal and neuronal pathways, including the autonomic nervous system.93 Hence, the modulation of brain inflammation could affect the peripheral inflammatory response in return. That being said, when compared to IL-6 or TNF-α protein levels, IL-1β concentrations were very low and overall, both WT and KO mice display a clear inflammatory response at the periphery.
Another plausible explanation is that CB1R temporally controls microglial cytokine production, delaying or accelerating the synthesis of inflammatory factors in response to LPS. Hence, while 2 h postadministration is usually the peak of cytokine production in response to this dose and strain of LPS,42 it might be too early or too late in the case of microglial CB1R KO mice. A more precise time course analysis of brain cytokine production and behavioral deficit progression would be required to address this question.
Alternatively, the lack of CB1R might favor the production of “anti-inflammatory” cytokines on one hand, yet worsen sickness behavior through alternative unknown mechanisms on the other hand. Notwithstanding, we could not find any effect of microglial CB1R deletion on the production anti-inflammatory cytokines.
Another remarkable result is that LPS-induced behavioral deficits and cytokine mRNA production were, in most cases, always in comparable range between males and females. Sexual dimorphism of the brain and blood response to an immune challenge is still a matter of debate. Some groups have previously shown a stronger vulnerability of rodent males to immune reactivity, with greater sickness symptoms and brain cytokine release,94–96 while others could not find any sex effect.97 Unfortunately, mice data are still very scarce, and the outcome might depend on the strain of mice, the strain and dose of LPS, and the route of injection, among other factors.
In this study, we could not observe major sexual dimorphism in the behavioral response to LPS injection. Only the synthesis of IL-1β and TNF-α in the hippocampus, and of IL-6 in the hypothalamus, was reduced in females compared to males. The latter suggests that the production of brain cytokines is likely to be region specific following LPS administration. Microglia represent a rather homogeneous population in the adult male brain; however, no data are available for female mice.98–101 Hence, regional differences of microglial cells could account for the differences we observed.
Similarly, deleting microglial CB1R differentially affects brain inflammation between males and females. Numerous studies reported sex differences in the biological activity of the ECS.102 Regarding CB1R, sex differences in receptor availability have been observed in humans using positron emission tomography imaging and revealed 41% more CB1R receptors in the brain of men versus women.103
In mice, CB1R receptor density is significantly higher in the prefrontal cortex and amygdala of males than in females, while no difference could be observed in any of the other brain regions analyzed.104 The opposite results were observed in microglial cells of young mice, which express more CB1R mRNAs in females compared to males.30 Microglia also display hormone-mediated sexual dimorphism,105 ranging from regional cell density106–109 to functional responses in neuropathic pain,110 in chronic stress,111 and in their interaction with the gut microbiota.30 Our data suggest that CB1R activity is a sexually dimorphic mechanism in the context of brain inflammatory response to an immune challenge.
In conclusion, our findings pinpoint a role for microglial CB1R in brain inflammation in a sex-dependent manner. While the underlying mechanisms are still unknown, this evidence adds to the comprehension of neuroinflammatory processes and might be of great interest for future studies aiming at developing therapeutic strategies for brain disorders with higher prevalence in men, in which CB1R receptors play a role.
Supplementary Material
Acknowledgments
We also thank Atika Zouine and Vincent Pitard for their technical assistance at the flow cytometry facility, CNRS UMS 3427, Institut national de la santé et de la recherche médicale (INSERM) US 005, University of Bordeaux (Bordeaux, France). We are finally grateful to Patrick C. Nahirney at the Division of Medical Sciences, University of Victoria, for the use of his electron microscope.
Abbreviations Used
- Actb
actin beta
- ANOVA
analysis of variance
- ARH
arcuate nucleus of the hypothalamus
- B2M
beta2-microglobulin
- BSA
bovine serum albumin
- CB1R
cannabinoid type 1 receptor
- CB2R
cannabinoid type 2 receptor
- cDNA
complementary DNA
- CNS
central nervous system
- ECS
endocannabinoid system
- EM
electron microscopy
- EPM
elevated plus maze
- FDF
Fondation de France
- FI
food intake
- FRM
Fondation pour la Recherche Médicale
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- IL-1β
interleukin-1 beta
- IL-10
interleukin-10
- IL-1RA
interleukin-1 receptor antagonist
- IL-6
interleukin-6
- INRAE
Institut National de Recherche pour l'Agriculture, l'alimentation et l'environnement
- INSERM
Institut national de la santé et de la recherche médicale
- KO
knockout
- LPS
lipopolysaccharide
- m
microglia
- ma
myelinated axons
- mRNA
messenger RNA
- OF
open field
- PB
phosphate buffer
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PFA
paraformaldehyde
- PVN
paraventricular nucleus
- qPCR
quantitative polymerase chain reaction
- RT
room temperature
- Sdha
succinate dehydrogenase complex subunit
- SEM
standard error of the mean
- TBS
Tris-buffered saline
- TGFβ1
transforming growth factor beta 1
- TLR2
toll-like receptor 2
- TLR4
toll-like receptor 4
- TNF-α
tumor necrosis factor alpha
- TST
tail suspension test
- Tuba4a
tubulin alpha 4 a
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Funding for this research was provided by the Institut National de Recherche pour l'Agriculture, l'alimentation et l'environnement (INRAE), the Bordeaux University, the Fondation pour la Recherche Médicale (FRM DEQ20170336724, to S.L.), the Fondation de France (FDF No. 00070700, to S.L.), and the Nouvelle Région Aquitaine (2017-1R30237-00013179, to S.L.). J.D.M. and Z.A. were supported by the European Commission Erasmus Mundus programme (Neurasmus 2016–2059 EMJMD). L.M. was supported by the FDF. Q.L. was supported by the Region Ile de France (PICRI, the Cerebral Palsy Foundation No. 13020605, to A.N.) and FRM (FDT20170437075, to Q.L.), D.C. and G.M. are supported by INSERM. F.D. is funded by Idex/University of Bordeaux and benefits from the Bordeaux Neurocampus Graduate Program (EUR 17-EURE-0028 project). K.P. and M.C. are supported by doctoral scholarships from Fonds de recherche du Québec—Santé (FRQS). M.-E.T. is a Tier II Canada Research Chair in Neurobiology of Aging and Cognition. This work benefited from the NutriNeuro's animal house and behavioral facilities supported by INRAE, University Bordeaux, Bordeaux INP, Nouvelle Région Aquitaine and Labex Brain (ANR-10-LABX-43). This work also benefited from the support of the Transcriptomic facility funded by Inserm and LabEX BRAIN ANR-10-LABX-43, thanks to T. Leste-Lasserre and the personnel of the platform of the Neurocentre Magendie Inserm U1215, and from the mouse genotyping platform, thanks to Delphine Gonzales and the personnel of the platform, funded by INSERM and Labex Brain (ANR-10-LABX-43).
Supplementary Material
Cite this article as: De Meij J, Alfanek Z, Morel L, Decoeur F, Leyrolle Q, Picard K, Carrier M, Aubert A, Séré A, Lucas C, Laforest G, Helbling J-C, Tremblay M-E, Cota D, Moisan M-P, Marsicano G, Layé S, Nadjar A (2021) Microglial cannabinoid type 1 receptor regulates brain inflammation in a sex-specific manner, Cannabis and Cannabinoid Research 6:6, 488–507, DOI: 10.1089/can.2020.0170.
References
- 1. Sochocka M, Diniz BS, Leszek J. Inflammatory response in the CNS: friend or foe? Mol Neurobiol. 2017;54:8071–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ransohoff RM, Brown MA. Innate immunity in the central nervous system. J Clin Invest. 2012;122:1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. [DOI] [PubMed] [Google Scholar]
- 4. Färber K, Kettenmann H. Physiology of microglial cells. Brain Res Brain Res Rev. 2005;48:133–143. [DOI] [PubMed] [Google Scholar]
- 5. Dantzer R, O'Connor JC, Freund GG, et al. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nadjar A, Bluthé R-M, May MJ, et al. Inactivation of the cerebral NFkappaB pathway inhibits interleukin-1beta-induced sickness behavior and c-Fos expression in various brain nuclei. Neuropsychopharmacology. 2005;30:1492–1499. [DOI] [PubMed] [Google Scholar]
- 7. Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav Immun. 2011;25:181–213. [DOI] [PubMed] [Google Scholar]
- 8. Correa F, Mestre L, Molina-Holgado E, et al. The role of cannabinoid system on immune modulation: therapeutic implications on CNS inflammation. Mini Rev Med Chem. 2005;5:671–675. [DOI] [PubMed] [Google Scholar]
- 9. Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat Rev Drug Discov. 2008;7:438–455. [DOI] [PubMed] [Google Scholar]
- 10. Stella N. Endocannabinoid signaling in microglial cells. Neuropharmacology. 2009;56(Suppl 1):244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lisboa SF, Gomes FV, Terzian ALB, et al. The endocannabinoid system and anxiety. Vitam Horm. 2017;103:193–279. [DOI] [PubMed] [Google Scholar]
- 12. Di Marzo V. Endocannabinoid signaling in the brain: biosynthetic mechanisms in the limelight. Nat Neurosci. 2011;14:9–15. [DOI] [PubMed] [Google Scholar]
- 13. Lutz B, Marsicano G, Maldonado R, et al. The endocannabinoid system in guarding against fear, anxiety and stress. Nat Rev Neurosci. 2015;16:705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parsons LH, Hurd YL. Endocannabinoid signalling in reward and addiction. Nat Rev Neurosci. 2015;16:579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Núñez E, Benito C, Pazos MR, et al. Cannabinoid CB2 receptors are expressed by perivascular microglial cells in the human brain: an immunohistochemical study. Synapse. 2004;53:208–213. [DOI] [PubMed] [Google Scholar]
- 16. Walter L, Franklin A, Witting A, et al. Nonpsychotropic cannabinoid receptors regulate microglial cell migration. J Neurosci. 2003;23:1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mecha M, Carrillo-Salinas FJ, Feliú A, et al. Microglia activation states and cannabinoid system: therapeutic implications. Pharmacol Ther. 2016;166:40–55. [DOI] [PubMed] [Google Scholar]
- 18. Guida F, Luongo L, Boccella S, et al. Palmitoylethanolamide induces microglia changes associated with increased migration and phagocytic activity: involvement of the CB2 receptor. Sci Rep. 2017;7:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galiègue S, Mary S, Marchand J, et al. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. [DOI] [PubMed] [Google Scholar]
- 20. Estrada JA, Contreras I. Endocannabinoid receptors in the CNS: potential drug targets for the prevention and treatment of neurologic and psychiatric disorders. Curr Neuropharmacol. 2020;18:769–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galán-Ganga M, Del Río R, Jiménez-Moreno N, et al. Cannabinoid CB2 receptor modulation by the transcription factor NRF2 is specific in microglial cells. Cell Mol Neurobiol. 2020;40:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanaka M, Sackett S, Zhang Y. Endocannabinoid modulation of microglial phenotypes in neuropathology. Front Neurol. 2020;11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cabral GA, Marciano-Cabral F. Cannabinoid receptors in microglia of the central nervous system: immune functional relevance. J Leukoc Biol. 2005;78:1192–1197. [DOI] [PubMed] [Google Scholar]
- 24. Duncan M, Galic MA, Wang A, et al. Cannabinoid 1 receptors are critical for the innate immune response to TLR4 stimulation. Am J Physiol Regul Integr Comp Physiol. 2013;305:R224–R231. [DOI] [PubMed] [Google Scholar]
- 25. Mecha M, Feliú A, Carrillo-Salinas FJ, et al. Endocannabinoids drive the acquisition of an alternative phenotype in microglia. Brain Behav Immun. 2015;49:233–245. [DOI] [PubMed] [Google Scholar]
- 26. Navarro G, Borroto-Escuela D, Angelats E, et al. Receptor-heteromer mediated regulation of endocannabinoid signaling in activated microglia. Role of CB1 and CB2 receptors and relevance for Alzheimer's disease and levodopa-induced dyskinesia. Brain Behav Immun. 2018;67:139–151. [DOI] [PubMed] [Google Scholar]
- 27. Steiner AA, Molchanova AY, Dogan MD, et al. The hypothermic response to bacterial lipopolysaccharide critically depends on brain CB1, but not CB2 or TRPV1, receptors. J Physiol (Lond). 2011;589:2415–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldmann T, Wieghofer P, Müller PF, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. [DOI] [PubMed] [Google Scholar]
- 29. Parkhurst CN, Yang G, Ninan I, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thion MS, Low D, Silvin A, et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 2018;172:500..e16–516.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. [DOI] [PubMed] [Google Scholar]
- 32. Lasselin J, Lekander M, Axelsson J, et al. Sex differences in how inflammation affects behavior: what we can learn from experimental inflammatory models in humans. Front Neuroendocrinol. 2018;50:91–106. [DOI] [PubMed] [Google Scholar]
- 33. Han J, Kesner P, Metna-Laurent M, et al. Acute cannabinoids impair working memory through astroglial CB1 receptor modulation of hippocampal LTD. Cell. 2012;148:1039–1050. [DOI] [PubMed] [Google Scholar]
- 34. Yona S, Kim K-W, Wolf Y, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marsicano G, Goodenough S, Monory K, et al. CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science. 2003;302:84–88. [DOI] [PubMed] [Google Scholar]
- 36. Cazareth J, Guyon A, Heurteaux C, et al. Molecular and cellular neuroinflammatory status of mouse brain after systemic lipopolysaccharide challenge: importance of CCR2/CCL2 signaling. J Neuroinflammation. 2014;11:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fu HQ, Yang T, Xiao W, et al. Prolonged neuroinflammation after lipopolysaccharide exposure in aged rats. PLoS One. 2014;9:e106331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ip JPK, Noçon AL, Hofer MJ, et al. Lipocalin 2 in the central nervous system host response to systemic lipopolysaccharide administration. J Neuroinflammation. 2011;8:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saraiva C, Barata-Antunes S, Santos T, et al. Histamine modulates hippocampal inflammation and neurogenesis in adult mice. Sci Rep. 2019;9:8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao X, Yan M, Wang L, et al. Homeostasis imbalance of microglia and astrocytes leads to alteration in the metabolites of the kynurenine pathway in LPS-induced depressive-like mice. Int J Mol Sci. 2020;21:1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Layé S, Gheusi G, Cremona S, et al. Endogenous brain IL-1 mediates LPS-induced anorexia and hypothalamic cytokine expression. Am J Physiol Regul Integr Comp Physiol. 2000;279:R93–R98. [DOI] [PubMed] [Google Scholar]
- 42. Madore C, Joffre C, Delpech JC, et al. Early morphofunctional plasticity of microglia in response to acute lipopolysaccharide. Brain Behav Immun. 2013;34:151–158. [DOI] [PubMed] [Google Scholar]
- 43. Mingam R, De Smedt V, Amédée T, et al. In vitro and in vivo evidence for a role of the P2X7 receptor in the release of IL-1 beta in the murine brain. Brain Behav Immun. 2008;22:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lafourcade M, Larrieu T, Mato S, et al. Nutritional omega-3 deficiency abolishes endocannabinoid-mediated neuronal functions. Nat Neurosci. 2011;14:345–350. [DOI] [PubMed] [Google Scholar]
- 45. Madore C, Leyrolle Q, Morel L, et al. Essential omega-3 fatty acids tune microglial phagocytosis of synaptic elements in the mouse developing brain. Nat Commun. 2020;11:6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosch-Bouju C, Larrieu T, Linders L, et al. Endocannabinoid-mediated plasticity in nucleus accumbens controls vulnerability to anxiety after social defeat stress. Cell Rep. 2016;16:1237–1242. [DOI] [PubMed] [Google Scholar]
- 47. Dinel A-L, André C, Aubert A, et al. Cognitive and emotional alterations are related to hippocampal inflammation in a mouse model of metabolic syndrome. PLoS One. 2011;6:e24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moranis A, Delpech J-C, De Smedt-Peyrusse V, et al. Long term adequate N-3 polyunsaturated fatty acid diet protects from depressive-like behavior but not from working memory disruption and brain cytokine expression in aged mice. Brain Behav Immun. 2012;26:721–731. [DOI] [PubMed] [Google Scholar]
- 49. Labrousse VF, Nadjar A, Joffre C, et al. Short-term long chain omega3 diet protects from neuroinflammatory processes and memory impairment in aged mice. PLoS One. 2012;7:e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delpech J-C, Thomazeau A, Madore C, et al. Dietary N-3 PUFAs deficiency increases vulnerability to inflammation-induced spatial memory impairment. Neuropsychopharmacology. 2015;40:2774–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Labrousse VF, Leyrolle Q, Amadieu C, et al. Dietary omega-3 deficiency exacerbates inflammation and reveals spatial memory deficits in mice exposed to lipopolysaccharide during gestation. Brain Behav Immun. 2018;73:427–440. [DOI] [PubMed] [Google Scholar]
- 52. Madore C, Nadjar A, Delpech J-C, et al. Nutritional N-3 PUFAs deficiency during perinatal periods alters brain innate immune system and neuronal plasticity-associated genes. Brain Behav Immun. 2014;41:22–31. [DOI] [PubMed] [Google Scholar]
- 53. Furube E, Kawai S, Inagaki H, et al. Brain region-dependent heterogeneity and dose-dependent difference in transient microglia population increase during lipopolysaccharide-induced inflammation. Sci Rep. 2018;8:2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nahirney PC, Tremblay M-E. Brain ultrastructure: putting the pieces together. Front Cell Dev Biol. 2021;9:629503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alperina E, Idova G, Zhukova E, et al. Cytokine variations within brain structures in rats selected for differences in aggression. Neurosci Lett. 2019;692:193–198. [DOI] [PubMed] [Google Scholar]
- 56. Skelly DT, Hennessy E, Dansereau M-A, et al. A systematic analysis of the peripheral and CNS effects of systemic LPS, IL-1B, TNF-α and IL-6 challenges in C57BL/6 mice. PLoS One. 2013;8:e69123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Teeling JL, Cunningham C, Newman TA, et al. The effect of non-steroidal anti-inflammatory agents on behavioural changes and cytokine production following systemic inflammation: implications for a role of COX-1. Brain Behav Immun. 2010;24:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. [DOI] [PubMed] [Google Scholar]
- 59. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. [DOI] [PubMed] [Google Scholar]
- 60. Xie F, Xiao P, Chen D, et al. MiRDeepFinder: a MiRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84. [DOI] [PubMed] [Google Scholar]
- 61. Bellocchio L, Soria-Gómez E, Quarta C, et al. Activation of the sympathetic nervous system mediates hypophagic and anxiety-like effects of CB1₁ receptor blockade. Proc Natl Acad Sci U S A. 2013;110:4786–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Busquets-Garcia A, Bains J, Marsicano G. CB1 receptor signaling in the brain: extracting specificity from ubiquity. Neuropsychopharmacology. 2018;43:4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dubreucq S, Durand A, Matias I, et al. Ventral tegmental area cannabinoid type-1 receptors control voluntary exercise performance. Biol Psychiatry. 2013;73:895–903. [DOI] [PubMed] [Google Scholar]
- 64. Hao S, Avraham Y, Mechoulam R, et al. Low dose anandamide affects food intake, cognitive function, neurotransmitter and corticosterone levels in diet-restricted mice. Eur J Pharmacol. 2000;392:147–156. [DOI] [PubMed] [Google Scholar]
- 65. Rey AA, Purrio M, Viveros M-P, et al. Biphasic effects of cannabinoids in anxiety responses: CB1 and GABA(B) receptors in the balance of GABAergic and glutamatergic neurotransmission. Neuropsychopharmacology. 2012;37:2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gutiérrez-Rodríguez A, Bonilla-Del Río I, Puente N, et al. Localization of the cannabinoid type-1 receptor in subcellular astrocyte compartments of mutant mouse hippocampus. Glia. 2018;66:1417–1431. [DOI] [PubMed] [Google Scholar]
- 67. Jimenez-Blasco D, Busquets-Garcia A, Hebert-Chatelain E, et al. Glucose metabolism links astroglial mitochondria to cannabinoid effects. Nature. 2020;583:603–608. [DOI] [PubMed] [Google Scholar]
- 68. DiPatrizio NV, Piomelli D. The thrifty lipids: endocannabinoids and the neural control of energy conservation. Trends Neurosci. 2012;35:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Layé S, Nadjar A, Joffre C, et al. Anti-inflammatory effects of omega-3 fatty acids in the brain: physiological mechanisms and relevance to pharmacology. Pharmacol Rev. 2018;70:12–38. [DOI] [PubMed] [Google Scholar]
- 70. Mazier W, Saucisse N, Gatta-Cherifi B, et al. The endocannabinoid system: pivotal orchestrator of obesity and metabolic disease. Trends Endocrinol Metab. 2015;26:524–537. [DOI] [PubMed] [Google Scholar]
- 71. Prinz M, Jung S, Priller J. Microglia biology: one century of evolving concepts. Cell. 2019;179:292–311. [DOI] [PubMed] [Google Scholar]
- 72. Konsman JP, Parnet P, Dantzer R. Cytokine-induced sickness behaviour: mechanisms and implications. Trends Neurosci. 2002;25:154–159. [DOI] [PubMed] [Google Scholar]
- 73. Dantzer R. Cytokine-induced sickness behavior: where do we stand? Brain Behav Immun. 2001;15:7–24. [DOI] [PubMed] [Google Scholar]
- 74. Schwartz MW, Woods SC, Porte D, et al. Central nervous system control of food intake. Nature. 2000;404:661–671. [DOI] [PubMed] [Google Scholar]
- 75. Good DW, George T, Watts BA. Toll-like receptor 2 is required for LPS-induced toll-like receptor 4 signaling and inhibition of ion transport in renal thick ascending limb. J Biol Chem. 2012;287:20208–20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Griton M, Konsman JP. Neural pathways involved in infection-induced inflammation: recent insights and clinical implications. Clin Auton Res. 2018;28:289–299. [DOI] [PubMed] [Google Scholar]
- 78. Konsman JP, Dantzer R. How the immune and nervous systems interact during disease-associated anorexia. Nutrition. 2001;17:664–668. [DOI] [PubMed] [Google Scholar]
- 79. Maggi L, Scianni M, Branchi I, et al. CX(3)CR1 deficiency alters hippocampal-dependent plasticity phenomena blunting the effects of enriched environment. Front Cell Neurosci. 2011;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Milior G, Lecours C, Samson L, et al. Fractalkine receptor deficiency impairs microglial and neuronal responsiveness to chronic stress. Brain Behav Immun. 2016;55:114–125. [DOI] [PubMed] [Google Scholar]
- 81. Rogers JT, Morganti JM, Bachstetter AD, et al. CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J Neurosci. 2011;31:16241–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Erblich B, Zhu L, Etgen AM, et al. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 2011;6:e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ginhoux F, Greter M, Leboeuf M, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Elmore MRP, Najafi AR, Koike MA, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hao S, Dey A, Yu X, et al. Dietary obesity reversibly induces synaptic stripping by microglia and impairs hippocampal plasticity. Brain Behav Immun. 2016;51:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bellocchio L, Lafenêtre P, Cannich A, et al. Bimodal control of stimulated food intake by the endocannabinoid system. Nat Neurosci. 2010;13:281–283. [DOI] [PubMed] [Google Scholar]
- 87. Lafenêtre P, Chaouloff F, Marsicano, G. Bidirectional regulation of novelty-induced behavioral inhibition by the endocannabinoid system. Neuropharmacology. 2009;57:715–721. [DOI] [PubMed] [Google Scholar]
- 88. Metna-Laurent M, Soria-Gómez E, Verrier D, et al. Bimodal control of fear-coping strategies by CB1₁ cannabinoid receptors. J Neurosci. 2012;32:7109–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lopez-Rodriguez AB, Siopi E, Finn DP, et al. CB1 and CB2 cannabinoid receptor antagonists prevent minocycline-induced neuroprotection following traumatic brain injury in mice. Cereb Cortex. 2015;25:35–45. [DOI] [PubMed] [Google Scholar]
- 90. Suárez J, Rivera P, Aparisi Rey A, et al. Adipocyte cannabinoid CB1 receptor deficiency alleviates high fat diet-induced memory deficit, depressive-like behavior, neuroinflammation and impairment in adult neurogenesis. Psychoneuroendocrinology. 2019;110:104418. [DOI] [PubMed] [Google Scholar]
- 91. Kent S, Bret-Dibat JL, Kelley KW, et al. Mechanisms of sickness-induced decreases in food-motivated behavior. Neurosci Biobehav Rev. 1996;20:171–175. [DOI] [PubMed] [Google Scholar]
- 92. Dantzer R, Kelley KW. Twenty years of research on cytokine-induced sickness behavior. Brain Behav Immun. 2007;21:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Waldburger J-M, Firestein GS. Regulation of peripheral inflammation by the central nervous system. Curr Rheumatol Rep. 2010;12:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cai KC, van Mil S, Murray E, et al. Age and sex differences in immune response following LPS treatment in mice. Brain Behav Immun. 2016;58:327–337. [DOI] [PubMed] [Google Scholar]
- 95. Rossetti AC, Paladini MS, Trepci A, et al. Differential neuroinflammatory response in male and female mice: a role for BDNF. Front Mol Neurosci. 2019;12:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mouihate A, Chen X, Pittman QJ. Interleukin-1beta fever in rats: gender difference and estrous cycle influence. Am J Physiol. 1998;275:R1450–R1454. [DOI] [PubMed] [Google Scholar]
- 97. Schneiders J, Fuchs F, Damm J, et al. The transcription factor nuclear factor interleukin 6 mediates pro- and anti-inflammatory responses during LPS-induced systemic inflammation in mice. Brain Behav Immun. 2015;48:147–164. [DOI] [PubMed] [Google Scholar]
- 98. Grabert K, Michoel T, Karavolos MH, et al. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hammond TR, Dufort C, Dissing-Olesen L, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253..e6–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Keren-Shaul H, Spinrad A, Weiner A, et al. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169:1276..e17–1290.e17. [DOI] [PubMed] [Google Scholar]
- 101. Masuda T, Sankowski R, Staszewski O, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. [DOI] [PubMed] [Google Scholar]
- 102. Farhang B, Diaz S, Tang SL, et al. Sex differences in the cannabinoid regulation of energy homeostasis. Psychoneuroendocrinology. 2009;34(Suppl 1):S237–S246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Laurikainen H, Tuominen L, Tikka M, et al. ; Sex difference in brain CB1 receptor availability in man. Neuroimage. 2019;184:834–842. [DOI] [PubMed] [Google Scholar]
- 104. Castelli MP, Fadda P, Casu A, et al. Male and female rats differ in brain cannabinoid CB1 receptor density and function and in behavioural traits predisposing to drug addiction: effect of ovarian hormones. Curr Pharm Des. 2014;20:2100–2113. [DOI] [PubMed] [Google Scholar]
- 105. Tay TL, Carrier M, Tremblay M-È. Physiology of microglia. Adv Exp Med Biol. 2019;1175:129–148. [DOI] [PubMed] [Google Scholar]
- 106. Lenz KM, Nugent BM, Haliyur R, et al. Microglia are essential to masculinization of brain and behavior. J Neurosci. 2013;33:2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mouton PR, Long JM, Lei D-L, et al. Age and gender effects on microglia and astrocyte numbers in brains of mice. Brain Res. 2002;956:30–35. [DOI] [PubMed] [Google Scholar]
- 108. Nelson LH, Warden S, Lenz KM. Sex differences in microglial phagocytosis in the neonatal hippocampus. Brain Behav Immun. 2017;64:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem. 2012;120:948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sorge RE, Mapplebeck JCS, Rosen S, et al. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci. 2015;18:1081–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bollinger JL, Bergeon Burns CM, Wellman CL. Differential effects of stress on microglial cell activation in male and female medial prefrontal cortex. Brain Behav Immun. 2016;52:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.