Abstract
Impairment of the astrocytic glutamate transporter excitatory amino acid transporter 2 (EAAT2) is associated with neurological disorders such as Parkinson’s disease (PD), Alzheimer’s disease (AD), and manganism, a neurological disorder caused by overexposure to manganese (Mn) which shares the features of sporadic PD. Mechanisms of Mn-induced neurotoxicity include dysregulation of EAAT2 following activation of the transcription factor Yin Yang 1 (YY1) by transcriptional upregulation, but the posttranslational mechanisms by which YY1 is activated to repress EAAT2 remain to be elucidated. In the present study, we tested if Mn activates YY1 through posttranslational phosphorylation in cultured H4 human astrocytes, leading to EAAT2 repression. The results demonstrate that Mn exposure induced phosphorylation of YY1 at serine residues via kinases Aurora B kinase (AurkB) and Casein kinase II (CK2), leading to YY1 nuclear translocation, YY1/HDAC interactions, binding to the EAAT2 promoter, and consequent decreases in EAAT2 promoter activity and mRNA/protein levels. Although further studies are warranted to fully elucidate the mechanisms of Mn-induced YY1 phosphorylation and resultant EAAT2 impairment, our findings indicate that serine phosphorylation of YY1 via AurkB and CK2 is critical, at least in part, to its activation and transcriptional repression of EAAT2.
Keywords: Manganese, EAAT2, YY1, AurkB, CK2
1. Introduction
Impairment of excitatory amino acid transporter 2 (EAAT2), the astrocytic glutamate transporter which reuptakes over 90 % of synaptic glutamate, has been implicated in several neurological disorders including manganism, a neurological syndrome caused by overexposure to the heavy metal manganese (Mn) (Lee et al., 2009; Sidoryk-Wegrzynowicz et al., 2009). Mn is an essential enzymatic cofactor and trace metal, but chronic overexposure to Mn through environmental and occupational settings may lead to neurotoxicity (Sarkar et al., 2018). While the mechanisms of Mn neurotoxicity are not fully understood, our previous studies have shown that Mn targets astrocytes to dysregulate glutamate transporters at the transcriptional level, decreasing EAAT2 mRNA and protein levels and impairing astrocytic glutamate uptake with ensuing excitotoxicity (Johnson et al., 2018; Karki et al., 2015a). Notably, Mn-induced EAAT2 repression is mediated by transcriptional increase of the transcription factor (TF) Yin Yang 1 (YY1). The EAAT2 promoter contains a YY1 consensus site, and its mutation restores Mn-reduced EAAT2 expression and attenuates neurotoxicity (Karki et al., 2014; Pajarillo et al., 2020). However, whether posttranslational modulations (PTMs) of YY1 play a role in Mn-induced dysregulation of EAAT2 is currently unknown.
Mn-induced YY1 activation may occur via PTMs such as phosphorylation, which modulate YY1’s transcriptional activity and interaction with epigenetic modifiers such as histone deacetylases (HDACs) (Dong et al., 2017; Pflum et al., 2001; Wu and Li, 2017). Importantly, studies have demonstrated that YY1 is a substrate for the kinases Aurora B kinase (AurkB), Polo-like kinase 1 (PLK1) and Casein kinase 2 (CK2), and YY1 phosphorylation occurs at serine/threonine residues (Kassardjian et al., 2012; Riman et al., 2012; Rizkallah et al., 2011). As PTMs of TFs can impact their transcriptional activity, it is possible that Mn may induce posttranslational phosphorylation of YY1 via these kinases, increasing its nuclear localization, interaction with HDACs, and binding to the EAAT2 promoter, leading to EAAT2 repression.
In the present study, we investigated the role of posttranslational phosphorylation in Mn-induced YY1 activation. Our results demonstrate that Mn induces YY1 phosphorylation at serine residues, leading to YY1 nuclear translocation, YY1/HDAC interaction, YY1 binding to the EAAT2 promoter, and repression of EAAT2. Pharmacological inhibition along this pathway attenuated Mn-induced YY1 activation and restored EAAT2. Taken together, our novel findings demonstrate that posttranslational YY1 phosphorylation via kinases AurkB and CK2 is critically involved in Mn-induced YY1 activation and EAAT2 repression.
2. Materials and methods
2.1. Materials
Cell culture media and reagents were purchased from Gibco (Carlsbad, CA). Manganese (II) chloride (MnCl2), protease inhibitor cocktail, poly-L-lysine, and dimethyl sulfoxide (DMSO) were purchased from MilliporeSigma (St. Louis, MO). Antibodies for EAAT2 (ab41621), phosphoserine (ab9332) and goat anti-rabbit IgG (ab97051) were obtained from Abcam (Cambridge, MA). Antibodies for YY1 (sc-7341), HDAC1 (sc-81598), HDAC3 (sc-376957), β-actin (sc-47778), and histone H4 (sc-25260) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). ChIP-validated EAAT2 primers and biotinylated oligonucleotides were obtained from Invitrogen (Carlsbad, CA). The AurkB inhibitor Hesperadin (Hesp) and the CK2 inhibitor CX-4945 (CX) were purchased from Cayman Chemical (Ann Arbor, MI). All chemicals were prepared in phosphate-buffered saline (PBS), double-distilled H2O or DMSO and diluted to working concentrations in Opti-MEM prior to use.
2.2. Cell culture
H4 human astrocytes were obtained from American Type Culture Collection (ATCC, Manassas, VA) and grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin-streptomycin. Cultures were maintained at 37 °C in a 95 % air, 5 % CO2 incubator.
For treatment, H4 astrocytes were exposed to Mn (250 μM) for 3 h to assess YY1 phosphorylation. When applicable, kinase inhibitors, such as the AurkB inhibitor Hesp (250 nM) and CK2 inhibitor CX (10 μM), were added to the pre-treatment medium for 1 h prior to Mn exposure. Optimal concentrations and time were determined prior to Hesp and CX treatment of astrocytes.
2.3. Luciferase assay
H4 astrocytes were transfected with 0.5 μg of EAAT2 luciferase vectors (a gift from Dr. Baldwin, UNC Chapel Hill) using Lipofectamine 3000 (Invitrogen, Waltham, MA) in MEM supplemented with 5% FBS. Following transfection, cells were treated with the designated compounds and luciferase activity was measured using the Bright-Glo luciferase kit (Promega Co, Madison, WI) in accordance with the manufacturer’s protocol.
2.4. Western blot analysis
Following treatment with the designated compounds, cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing a protease inhibitor cocktail. Protein concentration was determined via bicinchoninic acid assay (ThermoFisher Scientific, Rockford, IL) and 30 μg of protein samples were prepared and run on a 10 % SDS-PAGE gel, then electrophoretically transferred to a nitrocellulose membrane. Western blot analysis was performed using primary antibodies at a 1:1000 dilution and horseradish peroxidase-conjugated secondary antibodies at a 1:5000 dilution. Proteins were detected using the SuperSignal™ West Pico PLUS chemiluminescent detection kit (ThermoFisher Scientific) and a Molecular Imager ChemiDoc XRS + System (BioRad, Hercules, CA).
2.5. Quantitative real-time PCR (qPCR)
Following treatment of H4 astrocytes with the indicated compounds, total RNA was extracted from cells using the RNeasy Mini RNA Isolation kit (Qiagen, Ann Arbor, MI). Two μg of purified RNA were reverse-transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) and qPCR was performed using the CFX96 real-time PCR detection system (BioRad). The primer pairs were as follows: EAAT2, 5′-CAA CAG AGC CCT CTC TGA ATA C-3′ (forward) and 5′- GTA GGG TGG ATG GGA TAC AAT G -3′ (reverse); GAPDH, 5′-CTC TGC TCC TCC TGT TCG AC -3′ (forward) and 5′-GCG CCC AAT ACG ACC AAA TC-3′ (reverse). The mRNA levels were analyzed with Bio-Rad CFX Manager Version 3.1 using GAPDH as an internal control.
2.6. Co-Immunoprecipitation
After cellular fractionation, 400 μg of nuclear protein was incubated with 2 μg of the designated antibodies, then 20 μL of A/G sepharose beads (Abcam) were added to each sample and incubated overnight. Lysates were centrifuged at 2,000 × g for 2 min at 4 °C, then beads were washed three times with RIPA buffer and centrifuged 2,000 × g for 2 min at 4 °C. After the addition of Laemmli buffer, samples were run on a 10 % SDS-PAGE gel for western blot analysis.
2.7. Immunocytochemistry
Cells were cultured on poly-l-lysine–coated glass coverslips, then fixed in 4% paraformaldehyde and washed with PBS. Cells were then permeabilized in 0.1 % Triton X-100 and incubated with blocking buffer (10 % normal goat serum in PBS) at room temperature for 1 h, followed by washing and overnight incubation with YY1 primary antibody (1:250). Fluorescent-conjugated Alexa Fluor® 488 anti-mouse secondary antibody was added, and samples were mounted onto slides with DAPI Fluoromount-G ® (Southern Biotech, Birmingham, AL) for imaging. Cellular localization was analyzed using an Stellaris confocal microscope (Leica Microsystems, Buffalo Grove, IL).
2.8. Chromatin immunoprecipitation (ChIP) assay
ChIP analysis was performed utilizing an EZ-ChIP chromatin immunoprecipitation kit (MilliporeSigma), according to the manufacturer’s instructions. One % of the supernatant was saved as input and the remainder of samples were incubated overnight at 4 °C with YY1 antibody (Santa Cruz) or rabbit IgG as a negative control. Following crosslink reversal and DNA purification, qPCR was performed with EAAT2 primers: 5′-CTG GGC GCA TCG CTC TCT C-3′ (forward); 5′-GTA AGC CCT TTA GCG CCT CAA CGG G-3′ (reverse). The end products of qPCR were resolved using agarose gel electrophoresis.
2.9. DNA affinity purification assay (DAPA)
The DAPA assay was performed using the μMACS FactorFinder Kit (Miltenyi Biotec, Inc., Auburn, CA). After cellular fractionation, 50 μg of nuclear extract were incubated with 1.5 μg of biotinylated oligonucleotides in binding buffer for 20 min. After the addition of 100 μL streptavidin microbeads, the samples were incubated for an additional 10 min. and the reaction mixture was applied onto a pre-equilibrated micro-column. After four washes of 100 μL low- and high-salt buffers, proteins were eluted and analyzed by western blotting.
2.10. Statistical analysis
All data are expressed as mean ± standard deviation (SD). Statistical analyses were carried out via student t-test or one-way analysis of variance (ANOVA), followed by a Tukey’s post hoc test using GraphPad Prism Software version 6.0 (San Diego, CA). A p-value of less than 0.05 (p < 0.05) was considered statistically significant.
3. Results
3.1. Mn phosphorylates YY1 at serine residues via AurkB and CK2 in H4 astrocytes
It has been reported that the kinases AurkB and CK2 phosphorylate YY1 in other cellular contexts, such as mitosis or cell cycle progression (Kassardjian et al., 2012; Riman et al., 2012). In the present study, we utilized selective kinase inhibitors to test if a pathologically relevant concentration of Mn (250 μM) induced YY1 phosphorylation at serine residues via the kinases AurkB and/or CK2 (Karki et al., 2015a; Ke et al., 2019; Kumar et al., 2014; Song et al., 2017). As antibodies for phospho-YY1 are not commercially available, we pulled down with YY1 antibody, then utilized the precipitates to western blot for phosphoserine. Our results showed that pre-treatment of H4 astrocytes with the AurkB inhibitor Hesp (250 nM) partially, but significantly attenuated YY1 phosphorylation at serine residues (Fig. 1A). Pre-treatment of astrocytes with the CK2 inhibitor CX (10 μM, 1 h) also attenuated Mn-induced YY1 phosphorylation at serine residues. (Fig. 1B) (Hauf et al., 2003; Pierre et al., 2011). Interestingly, inhibition of PLK1, a kinase which phosphorylates YY1 at threonine 39 (Rizkallah et al., 2011), did not attenuate Mn-induced YY1 phosphorylation, indicating that PLK1 does not mediate YY1 phosphorylation in Mn neurotoxicity (data not shown).
Fig. 1.
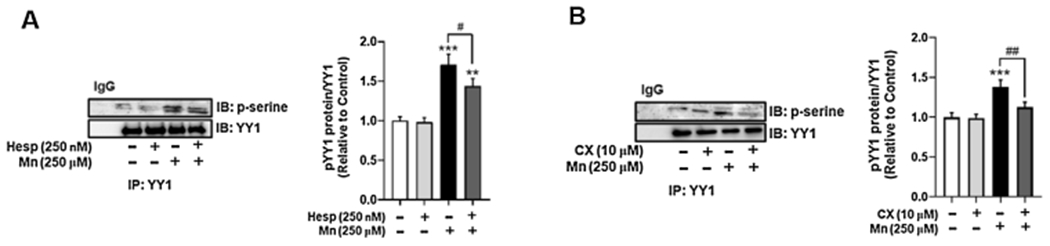
Mn phosphorylates YY1 at serine residues via AurkB and CK2 in H4 astrocytes. (A) Astrocytes were pre-treated with the AurkB inhibitor Hesperadin (Hesp, 250 nM, 1 h), then exposed to Mn(250 μM, 3 h) and immunoprecipitated with YY1 antibody, followed by western blotting for phosphoserine. (B) Astrocytes were pre-treated with the CK2 inhibitor CX-4945 (CX, 10 μM, 1 h), then exposed to Mn (250 μM, 3 h) and immunoprecipitated for YY1, followed by western blotting for phosphoserine. (**p < 0.01, ***p < 0.001, compared to the control; #p < 0.05, ##p < 0.01, ANOVA followed by Tukey’s post hoc test; N = 3).
3.2. Mn-induced YY1 phosphorylation via AurkB and CK2 leads to YY1 nuclear translocation
To determine if Mn-induced YY1 phosphorylation mediated its cellular localization, we tested if treatment with AurkB and CK2 inhibitors could attenuate Mn-induced YY1 nuclear translocation. Pre-treatment of H4 astrocytes with the AurkB inhibitor Hesp attenuated Mn-induced YY1 nuclear translocation (Fig. 2A), and the CK2 inhibitor CX also exerted similar effects (Fig. 2B). Inhibiting both AurkB and CK2 mitigated Mn-induced YY1 nuclear translocation (Fig. 2C), indicating that Mn-induced YY1 phosphorylation induced YY1’s translocation to the nucleus, where it functions as a TF for its downstream target genes.
Fig. 2.
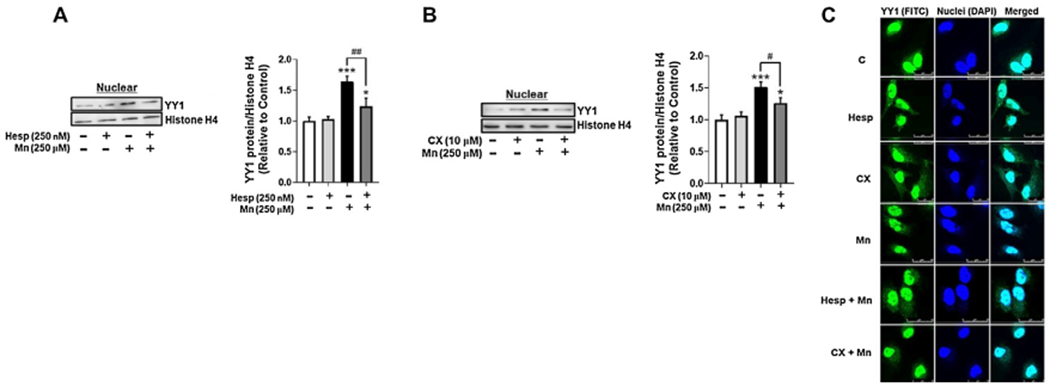
Mn-induced YY1 phosphorylation via AurkB and CK2 leads to YY1 nuclear translocation. (A) Astrocytes were pre-treated with Hesp (250 nM, 1 h) and exposed to Mn (250 μM, 3 h), then cells were fractionated and analyzed for YY1 protein levels in the nuclear fraction by western blotting. Nuclear protein histone H4 was used as a loading control. (B) Astrocytes were pre-treated with CX (10 μM, 1 h) and exposed to Mn (250 μM, 3 h), then cells were fractionated and analyzed for YY1 protein expression in the nuclear fraction by western blotting. (C) Astrocytes were pre-treated with Hesp or CX and exposed to Mn for the indicated time periods, then analyzed via immunocytochemistry (ICC) for YY1 nuclear expression. Scale: 0-25 μM. (*p < 0.05, ***p < 0.001, compared to the control; #p < 0.05, ##p < 0.01, ANOVA followed by Tukey’s post hoc test; N = 3).
3.3. Mn-induced YY1 phosphorylation via AurkB and CK2 increases interaction between YY1 and HDACs in H4 astrocytes
Our previous studies show that YY1 and HDAC 1 and 3 form a complex which binds to the EAAT2 consensus sequence (Karki et al., 2015b). Given that PTMs of TFs can modulate their function and interaction with other proteins such as epigenetic modifiers HDACs, we tested if inhibition of AurkB and CK2 could block the Mn-induced interaction of YY1 and HDACs. Results demonstrated that pre-treatment of astrocytes with the AurkB inhibitor Hesp attenuated Mn-induced YY1/HDAC1 complex formation (Fig. 3A), and the CK2 inhibitor CX inhibited YY1/HDAC3 complex formation (Fig. 3B).
Fig. 3.
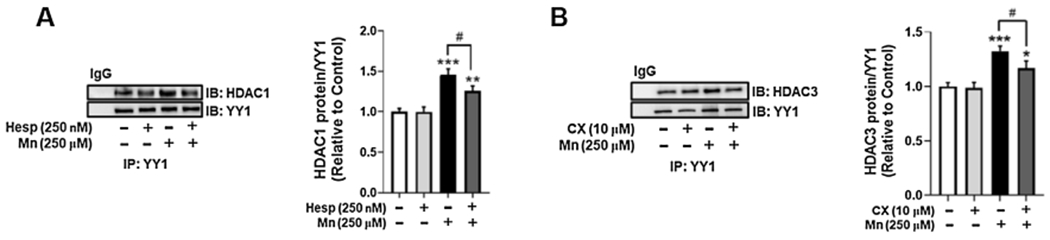
Mn-induced YY1 phosphorylation increases interaction between YY1 and HDACs in H4 astrocytes. (A) Astrocytes were pre-treated with Hesp (250 nM, 1 h), then exposed to Mn (250 μM, 3 h) and immunoprecipitated for YY1, followed by western blotting for HDAC1. (B) Astrocytes were pre-treated with CX (10 μM, 1 h), then exposed to Mn (250 μM, 3 h) and co-immunoprecipitated for YY1/HDAC3 interaction. (*p < 0.05, **p < 0.01, ***p < 0.001, compared to the control; #p < 0.05, ANOVA followed by Tukey’s post hoc test; N = 3).
3.4. Mn-induced YY1 phosphorylation via AurkB and CK2 increases YY1 binding to the EAAT2 promoter in H4 astrocytes
We have previously reported that Mn increases YY1 binding to the EAAT2 promoter, but the mechanisms by which this binding occurs are not completely understood (Karki et al., 2014). Thus, we tested if Mn-induced YY1 phosphorylation via AurkB and CK2 modulates YY1 binding to its consensus site on the EAAT2 promoter. Pre-treatment of astrocytes with the AurkB inhibitor Hesp attenuated Mn-induced YY1 binding to its consensus sequence in the EAAT2 promoter by DAPA (Fig. 4A) and ChIP assays (Fig. 4B), respectively. Pre-treatment of astrocytes with the CK2 inhibitor CX (Fig. 4C and D) showed similar effects, indicating that Mn-induced YY1 phosphorylation is critical for YY1 binding to the EAAT2 promoter.
Fig. 4.
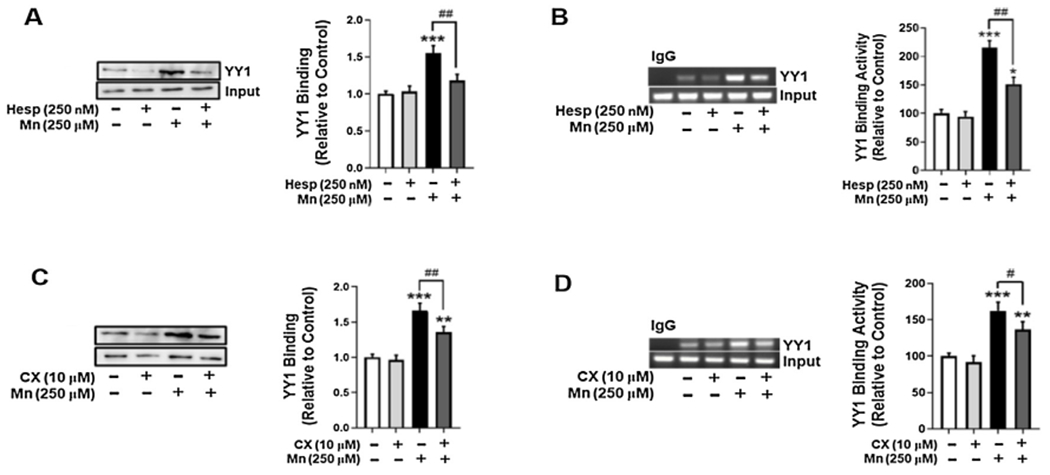
Mn-induced YY1 phosphorylation via AurkB and CK2 increases YY1 binding to the EAAT2 promoter in H4 astrocytes. (A) Astrocytes were pre-treated with Hesp (250 nM, 1 h) and exposed to Mn (250 μM, 6 h), then cells were fractionated and the DAPA assay was performed to assess YY1 binding to the EAAT2 promoter. (B) Astrocytes were treated with Hesp and Mn for the indicated time periods, followed by the ChIP assay to assess YY1 binding to EAAT2 promoter. (C) Astrocytes were pre-treated with CX (10 mM, 1 h) and exposed to Mn (250 μM, 6h), then cells were fractionated and the DAPA assay was performed. (D) Astrocytes were treated with CX and Mn for the indicated time periods and the ChIP assay was performed. (*p < 0.05, **p < 0.01, ***p < 0.001, compared to the control; #p < 0.05, ##p < 0.01, ANOVA followed by Tukey’s post hoc test; N = 3).
3.5. Inhibition of AurkB and CK2 attenuates Mn-induced EAAT2 repression in H4 astrocytes
Currently, the role of YY1 phosphorylation in Mn-induced EAAT2 repression is not well understood (Alexander and Rizkallah, 2017). In the present study, we tested if inhibition of YY1 phosphorylation could attenuate Mn-induced decreases in EAAT2 promoter activity and expression. Pre-treatment of astrocytes with the AurkB inhibitor Hesp attenuated Mn-decreased EAAT2 promoter activity (Fig. 5A) and mRNA (Fig. 5B)/protein (Fig. 5C) levels. Pre-treatment of astrocytes with the CK2 inhibitor CX also attenuated Mn-induced decreases in EAAT2 promoter activity (Fig. 5D), mRNA (Fig. 5E), and protein levels (Fig. 5F).
Fig. 5.
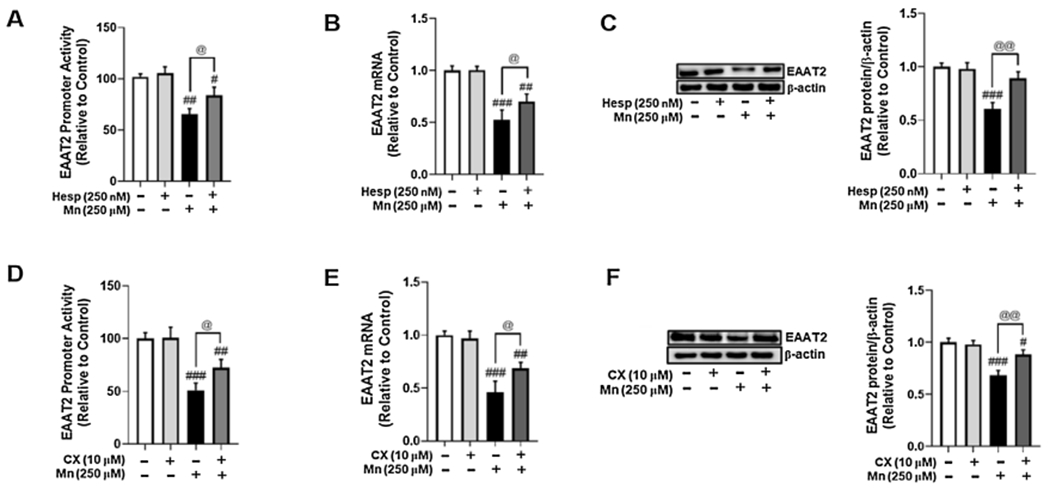
Inhibition of AurkB and CK2 attenuates Mn-induced EAAT2 repression in H4 astrocytes. (A) Astrocytes were transfected overnight with an EAAT2 promoter vector, then treated with Hesp (250 nM, 1 h) prior to Mn exposure (250 μM, 6 h), followed by the luciferase assay. (B) Astrocytes were pre-treated with Hesp (250 nM, 1 h) and exposed to Mn (250 μM, 12 h), followed by measurement of EAAT2 mRNA levels by real-time qPCR (normalized to GAPDH). (C) Astrocytes were treated with Hesp and Mn as previously indicated, followed by measurement of EAAT2 protein levels by western blotting. (D) Astrocytes were transfected overnight with an EAAT2 promoter vector, then treated with CX (10 μM, 1 h) prior to Mn exposure (250 μM, 6 h), followed by the luciferase assay. (E) Astrocytes were pre-treated with CX (10 μM, 1 h) prior to Mn exposure (250 μM, 12 h), followed by measurement of EAAT2 mRNA levels by real-time qPCR (normalized to GAPDH). (F) Astrocytes were treated with CX and Mn as previously indicated, followed by measurement of EAAT2 protein levels by western blotting. (# p < 0.05, ##p < 0.01, ###p < 0.001; @p < 0.05, @@p < 0.01, ANOVA followed by Tukey’s post hoc test; N = 3).
4. Discussion
Our novel findings demonstrate that Mn phosphorylates YY1 at serine residues via AurkB and CK2 in human H4 astrocytes. Mn-induced phosphorylation of YY1 led to YY1 nuclear translocation and increased interaction with epigenetic modifiers HDACs 1 and 3. Further, Mn-induced YY1 phosphorylation led to increased YY1 binding to the EAAT2 promoter and consequent decreases in EAAT2 promoter activity and expression. Accordingly, inhibition of AurkB and CK2 activity attenuated YY1 activation and restored EAAT2, suggesting that these kinases could be potential molecular targets in the development of neurotherapeutics to treat manganism and other YY1-related disorders.
As dysregulation of the astrocytic glutamate transporter EAAT2 has been implicated in several neurological disorders, it is critical to delineate the mechanisms by which Mn activates YY1 to impair EAAT2 (Kim et al., 2011; Lin et al., 2012). PTMs have emerged as key regulators of TF function and activity, and several studies have demonstrated that the kinases AurkB and CK2 phosphorylate YY1 in other cellular contexts (Wu and Li, 2017; Kassardjian et al., 2012; McNeil et al., 1998). Mn-induced YY1 phosphorylation appears to be serine-residue specific since only AurkB and CK2 mediated Mn-induced YY1 phosphorylation, but inhibition of the kinase PLK1, which is known to phosphorylate YY1 at threonine 39 (Rizkallah et al., 2011) failed to attenuate Mn’s phosphorylation of YY1. These results provide novel information that Mn phosphorylates YY1 at serine residues to repress EAAT2 and lead to excitoxicity.
Studies have demonstrated that posttranslational modifications can impact YY1’s cellular localization, transcriptional activity, and interaction with other proteins (Wu and Li, 2017; Kassardjian et al., 2012; Takasaki et al., 2007). The current study demonstrates that Mn induces YY1 phosphorylation at serine residues within 3 h, leading to YY1 nuclear translocation. The selective kinase inhibitors Hesp (AurkB) and CX (CK2) attenuated these effects, suggesting that serine phosphorylation of YY1 via AurkB and CK2 is a critical mediator of Mn-induced YY1 cellular localization and EAAT2 transcriptional activity. It is possible that Mn activates other signaling proteins that phosphorylate YY1 to decrease EAAT2 expression in astrocytes. We have reported previously that Mn phosphorylated MAPK/ERK and p38 (Kim et al., 2019). It is also possible that CK2 can phosphorylate MAPK/ERK which in turn, decreases EAAT2 expression. Further investigation is warranted to better understand the precise mechanisms of signaling pathways involved in regulating YY1 phosphorylation and EAAT2 expression.
Our previous studies have shown that YY1 interacts with epigenetic modifiers HDACs 1 and 3 to repress EAAT2 promoter activity in primary astrocytes, and treatment with HDAC inhibitors restored expression of astrocytic glutamate transporter GLT-1, the rodent homolog of human EAAT2, in mice (Johnson et al., 2018; Karki et al., 2014). As inhibition of AurkB and CK2 attenuated Mn-induced YY1/HDAC interaction, it is likely that posttranslational YY1 phosphorylation is critical for YY1’s interaction with epigenetic modifier HDACs. Notably, inhibition of AurkB- and CK2-mediated YY1 phosphorylation attenuated Mn-induced YY1 binding to its consensus site in the EAAT2 promoter and consequent decreases in EAAT2 promoter activity and mRNA/protein levels. The present findings do not provide direct evidence for the role of YY1 phosphorylation in Mn-induced decreases in EAAT2 expression via AurkB and CK2, as it is possible that these kinases may modulate EAAT2 via other unknown pathways, but support that YY1 phosphorylation is important for Mn-induced YY1 binding to and consequent impairment of EAAT2.
Although several studies have reported that phosphorylation of YY1 decreased its DNA binding activity during mitosis (Alexander and Rizkallah, 2017; Rizkallah and Hurt, 2009), other studies have shown that phosphorylation of YY1 increased its stability and activity as a transcriptional repressor (Wu and Li, 2017), corroborating with our findings. As YY1 is a multifunctional TF which can act as a transcriptional activator or repressor, depending on cellular context, the outcomes of YY1 phosphorylation may reflect the complex and dynamic processes involved in transcriptional regulation. Further investigation is warranted to understand the mechanisms by which Mn induces YY1 phosphorylation and which residues regulate YY1’s cellular localization and activity as a transcriptional repressor of EAAT2.
Taken together, our novel findings demonstrate that Mn activates YY1 at least in part via phosphorylation at serine residues via AurkB and CK2, leading to decreases in EAAT2 promoter activity and mRNA/protein levels as shown in the following model (Fig. 6). The results from the present study provide novel insight into the mechanisms of Mn-induced YY1 activation and provide new molecular targets in the development of therapeutics to treat neurological disorders associated with glutamate transporter impairment.
Fig. 6.
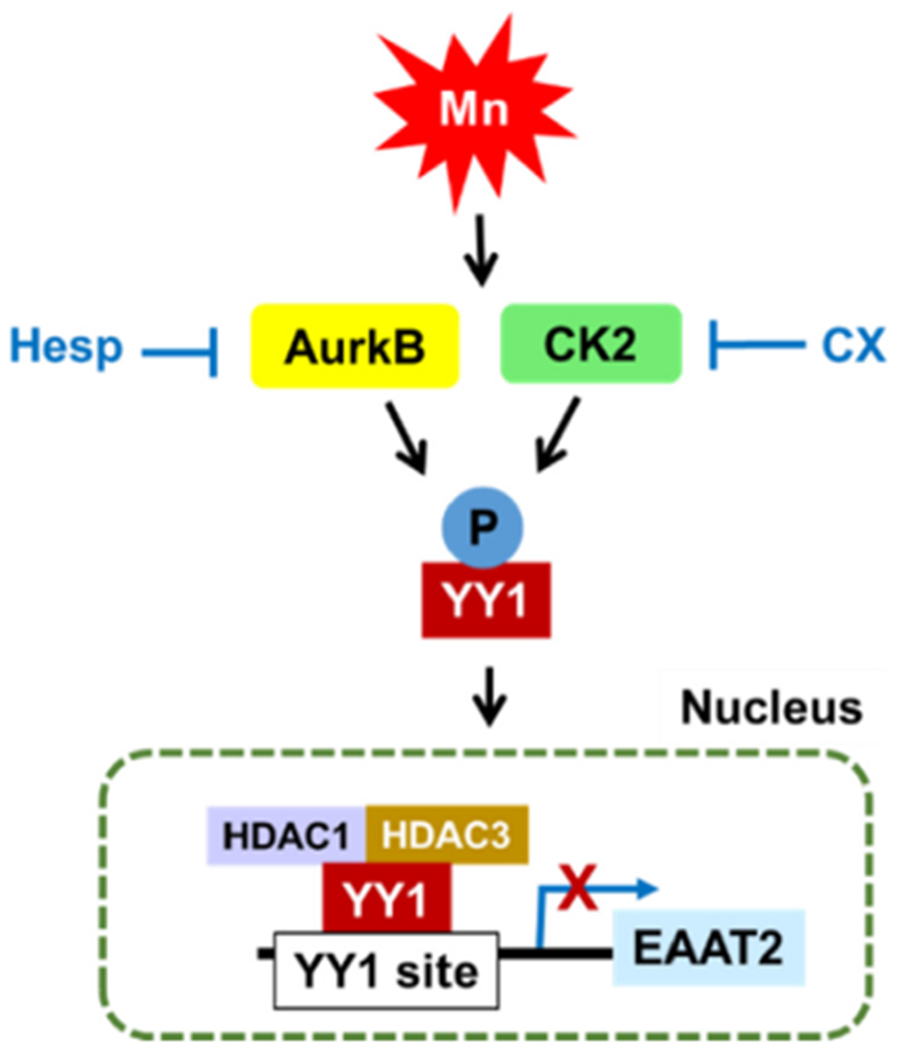
Proposed mechanism of Mn-induced EAAT2 repression via YY1 phosphorylation in astrocytes. Mn induces phosphorylation of YY1 at serine residues via the kinases AurkB and CK2, leading to YY1 nuclear translocation, YY1/HDAC interaction, YY1 binding to the EAAT2 promoter, followed by consequent repression of EAAT2.
HIGHLIGHTS.
Mn activates YY1 through posttranslational phosphorylation in astrocytes.
Mn phosphorylates YY1 at serine residues via kinases Aurora B kinase (AurkB) and Casein kinase II (CK2).
Mn reduces EAAT2 expression via YY1 phosphorylation and subsequent YY1 nuclear translocation.
Acknowledgements
The study was supported in part by National Institutes of Health Research Grants, NIEHS R01 ES024756, R01 ES031282, R01 ES010563, R01 ES020852, NIMHD U54 MD007582, and NCI SC1 CA200519.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alexander KE, Rizkallah R, 2017. Aurora a phosphorylation of YY1 during mitosis inactivates its DNA binding activity. Sci. Rep 7 (1), 10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, et al. 2017. YY1 promotes HDAC1 expression and decreases sensitivity of hepatocellular carcinoma cells to HDAC inhibitor. Oncotarget 8 (25), 40583–40593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, et al. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol 161 (2), 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J Jr, et al. 2018. Valproate and sodium butyrate attenuate manganese-decreased locomotor activity and astrocytic glutamate transporters expression in mice. Neurotoxicology 64, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki P, et al. 2014. Yin Yang 1 is a repressor of glutamate transporter EAAT2, and it mediates manganese-induced decrease of EAAT2 expression in astrocytes. Mol. Cell. Biol 34 (7), 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki P, et al. 2015a. Genetic dys-regulation of astrocytic glutamate transporter EAAT2 and its implications in neurological disorders and manganese toxicity. Neurochem. Res 40 (2), 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki P, et al. 2015b. Transcriptional regulation of the astrocytic excitatory amino acid transporter 1 (EAAT1) via NF-kappaB and Yin Yang 1 (YY1). J. Biol. Chem 290 (39), 23725–23737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassardjian A, et al. 2012. The transcription factor YY1 is a novel substrate for Aurora B kinase at G2/M transition of the cell cycle. PLoS One 7(11) p. e50645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke T, et al. 2019. Role of astrocytes in manganese neurotoxicity revisited. Neurochem. Res 44 (11), 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, et al. 2011. Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: opportunities for developing novel therapeutics. J. Cell. Physiol 226 (10), 2484–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Pajarillo E, Rizor A, Son DS, Lee J, Aschner M, Lee E, 2019. LRRK2 kinase plays a critical role in manganese-induced inflammation and apoptosis in microglia. PLoS One 14 (1) e0210248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KK, et al. 2014. Cellular manganese content is developmentally regulated in human dopaminergic neurons. Sci. Rep 4, 6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, et al. 2009. Estrogen and tamoxifen reverse manganese-induced glutamate transporter impairment in astrocytes. J. Neurochem 110 (2), 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, et al. 2012. Glutamate transporter EAAT2: a new target for the treatment of neurodegenerative diseases. Future Med. Chem 4 (13), 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil S, et al. 1998. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem 68 (4), 500–510. [PubMed] [Google Scholar]
- Pajarillo E, et al. 2020. Astrocyte-specific deletion of the transcription factor Yin Yang 1 in murine substantia nigra mitigates manganese-induced dopaminergic neurotoxicity. J. Biol. Chem 295 (46), 15662–15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum MK, et al. 2001. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem 276(50), 47733–47741. [DOI] [PubMed] [Google Scholar]
- Pierre F, et al. 2011. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem 356 (1–2), 37–43. [DOI] [PubMed] [Google Scholar]
- Riman S, et al. 2012. Phosphorylation of the transcription factor YY1 by CK2alpha prevents cleavage by caspase 7 during apoptosis. Mol. Cell. Biol 32 (4), 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah R, Hurt MM, 2009. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol. Biol. Cell 20 (22), 4766–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah R, et al. 2011. The transcription factor YY1 is a substrate for Polo-like kinase 1 at the G2/M transition of the cell cycle. PLoS One 6 (1) p. e15928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, et al. 2018. Manganese exposure induces neuroinflammation by impairing mitochondrial dynamics in astrocytes. Neurotoxicology 64, 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoryk-Wegrzynowicz M, et al. 2009. Manganese disrupts astrocyte glutamine transporter expression and function. J. Neurochem 110 (3), 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, et al. 2017. FOXO3 promoted mitophagy via nuclear retention induced by manganese chloride in SH-SY5Y cells. Metallomics 9 (9), 1251–1259. [DOI] [PubMed] [Google Scholar]
- Takasaki N, et al. 2007. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 26 (6), 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XY, Li T, 2017. A casein kinase II phosphorylation site in AtYY1 affects its activity, stability, and function in the ABA response. Front. Plant Sci 8, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]


