Abstract
The current COVID-19 pandemic caused by SARS-CoV-2 is raging, seriously threatening people's lives. The establishment of rapid and accurate pathogen detection technology is not only critical in this epidemic, but also a reminder that we must always be prepared for possible future outbreaks. Therefore, we developed a Palm Germ-Radar (PaGeR) device for rapid and simple detection of COVID-19 from extracted patient sample RNA by RT-LAMP. The whole procedure of rapid COVID-19 detection is based on 4 simple steps: inactivation, extraction, amplification, and detection. SARS-CoV-2 down to 1 copy/μL could be detected selectively with naked-eye. Three detection methods (colorimetric, fluorometric and lateral dipstick readout) could be performed in PaGeR instrument. By employing the PaGeR, we successfully detected SARS-CoV-2 in clinical RNA samples isolated from swab specimens. The results showed that 15 out of 17 COVID-19 patients were diagnosed as positive while all 55 normal samples were diagnosed as negative. Therefore, the developed PaGeR instrument can realize the detection of COVID-19 with easily visualized results, providing a promising instrument for rapid detection in the community as well as at home.
Keywords: SARS-CoV-2, COVID-19, RT-LAMP, Rapid detection, Palm germ-radar
Graphical abstract
1. Introduction
The Coronavirus Disease 2019 (COVID-19), caused by a new strain of coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is first identified in December 2019 in Wuhan, China (Wu et al., 2020a; Wu et al., 2020b). Since then, COVID-19 is tipping the world into a volatile and dangerous phase, and the coronavirus outbreak is officially labeled a pandemic by the World Health Organization (WHO) in March 2020 (Cucinotta and Vanelli, 2020). By the end of 2020, nearly 100 million people in over 220 countries had been infected around the world, causing more than 2 million deaths (WHO, COVID-19 situation dashboard). Nowadays, COVID-19 is the greatest threat facing the world since the Second Would War, rendering major impact on global health, economy and life quality (Chakraborty and Maity, 2020; Wang et al., 2020). Early diagnosis, treatment, and isolation of the disease and virus carrier can effectively reduce virus transmission. Therefore, a rapid, sensitive, responsive and reliable diagnostic strategy will be necessary and essential for this moment.
Currently, the diagnosis of COVID-19 mainly contains three categories, which can be combined for diagnosis in some scenarios: 1) Clinical manifestations such as fever and respiratory symptoms, combining with chest computed tomography (CT) scans (Huang et al., 2020a; Zhu et al., 2020a). Proper screening by the physicians and healthcare is of great importance in controlling virus spread, however, cross-infection is likely to occur in an outbreak area and the testing capacity is limited by the CT equipment. 2) Serology detections include colloidal gold immune-chromatography, ELISA, etc. (Zhang et al., 2020) Although antibody diagnosis is rapid, simplified and no equipment required, their potential utility in SARS-CoV-2 infection may be limited by sensitivity, because it is not easy to detect in the early stage or incubation period of infection for not enough antibody in blood samples. 3) Nucleic acid amplification tests (NAATs) (Carter et al., 2020; Esbin et al., 2020). Because of its high sensitivity combined with the high specificity of the fluorescent probe, the quantitative reverse transcriptase PCR (qRT-PCR) method is considered as the “gold standard” for COVID-19 diagnosis in clinical and large-scale screening (Udugama et al., 2020). However, qRT-PCR requires expensive equipment, well-trained technician as well as hours of work from collecting nasopharyngeal or oropharyngeal swab samples to reading out of the results, which limits its broad application in the communities and homes under COVID-19 global pandemic condition. Hence, there is an urgent need for responsive and simple method with the help of easy to use equipment for COVID-19 detection.
The development of isothermal amplification techniques has extended the application of NAATs to detect pathogens with high sensitivity, such as helicase-dependent amplification (HDA) (Barreda-Garcia et al., 2018), strand displacement amplification (SDA) (Toley et al., 2015), recombinase polymerase amplification (RPA) (Nassir et al., 2020) and loop-mediated isothermal amplification (LAMP) (Augustine et al., 2020), etc. In particular, LAMP is a promising method for NAATs which employs 3 pairs of primers to recognize target DNA with highly improved sensitivity and specificity compared to conventional PCR. Typically, more than 109-fold amplification of the target region can be achieved within 1 h by LAMP amplification and the optical reading of LAMP detection results is relatively simple. In addition, LAMP Bst (Bacillus stearothermophilus) DNA polymerase could tolerate the common inhibitory compounds presenting in clinical samples which typically inhibit PCR. Therefore, since its invention in 2000, a series of LAMP-based protocol had been successfully developed for detecting infectious diseases, such as avian influenza (Imai et al., 2007), Middle East respiratory syndrome (MERS) (Shirato et al., 2014), human immunodeficiency virus (HIV) (Curtis et al., 2008) and African swine fever (ASF) (Ye et al., 2019), etc. Some of them can achieve their applications in point-of-care testing (POCT) detection (Nguyen et al., 2019; Yao et al., 2020).
RT-LAMP has not escaped attention that it can be applied in SARS-CoV-2 specific detection, since it allows RNA reverse transcription (SARS-CoV-2 is a single-stranded RNA virus) and DNA amplification within the same temperature (El-Tholoth et al., 2020; Jiang et al., 2020; Lalli et al., 2020; Yan et al., 2020; Lai et al., 2021; Yu et al., 2020). For instance, Park et al. developed a RT-LAMP method for SARS-CoV-2 detection in the early stage of the pandemic outbreak (Park et al., 2020). Huang et al. employed RT-LAMP for SARS-CoV-2 detection with a limit of 80 copies per mL sample (Huang et al., 2020b). Thi et al. developed a colorimetric RT-LAMP and LAMP-sequencing for SARS-CoV-2 detection with clinical samples (Dao Thi et al., 2020). Zhu et al. have introduced RT-LAMP to nanoparticle-based lateral flow biosensor for diagnosis of COVID-19 (Zhu et al., 2020b). Recently, RT-LAMP coupled with clustered regularly interspaced palindromic repeats (CRISPR)/Cas technology has been applied to detect SARS-CoV-2(Broughton et al., 2020; Joung et al., 2020; Wang et al., 2021). For instance, Broughton et al. employed CRISPR-Cas12-based DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter) for SARS-CoV-2 detection (Broughton et al., 2020). Zhang et al. developed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) method for one-pot testing (Joung et al., 2020). However, most of the methods described above utilized pH indicator or SYBR nucleic acid dyes, resulting in unsatisfying distinguish capacity when the signal was weak. More importantly, the current experimental procedures associating with RT-LAMP cannot be performed using a small commercial instrument, the corresponding virus detection cannot be conveniently performed at home or in the community, which prominently affect the wide application of LAMP method.
Here, we developed a Palm Germ-Radar (PaGeR) device for rapid and simple detection of SARS-CoV-2 from extracted patient sample RNA by RT-LAMP. The detections of SARS-CoV-2 have been demonstrated with high selectivity and sensitivity, and SARS-CoV-2 down to 1 copy/μL could be detected after reaction optimization. Through this method, we detected SARS-CoV-2 RNA in clinical RNA samples isolated from swab specimens. The result demonstrated that PaGeR is consistent with the normalization instrument. Among 17 patient samples, 15 samples tested positive in both PaGeR and the real-time quantitative fluorescence amplification instrument. In addition, the result of the pharyngeal swabs taken from the volunteers showed that none of the 55 negative samples were tested positive. The PaGeR device provides a rapid, affordable, user-friendly and reliable diagnostic strategy for COVID-19 detection. Therefore, the developed PaGeR instrument could realize the diagnosis of COVID-19, providing a promising instrument for rapid detection in the community as well as at home.
2. Materials and methods
Chemicals and reagents. Diethyl pyrocarbonate (DEPC)-treated water and DNAaway were obtained from Thermo Fisher (MA, USA). Betaine was obtained from Sigma-Aldrich (MO, USA). 10x isothermal amplification buffer, MgSO4, WarmStart Bst 2.0 DNA polymerase, and WarmStart RTx reverse transcriptase were obtained from New England BioLabs (MA, USA). 20x EvaGreen® dye was obtained from Biotium (CA, USA). The viral transport medium (VTM) was obtained from Qingdao Hope Bio-Technology (Qingdao, China). Primers were synthesized by Sangon Biotech (Table S1) (Shanghai, China). All chemicals used were of analytical grade unless specified otherwise. Viral genomic RNA was extracted by MiniBSET viral RNA extraction Kit, which was obtained from Takara Bio (Beijing, China). Disposable nucleic acid detection strip and the matching buffer were obtained from Yousida Bio (Hangzhou, China).
Nucleic acid preparation. The pUC57 plasmid containing SARS-CoV-2 (Genbank accession: NC_045512.2) open reading frame (ORF1ab; 13201-13766) or nucleoprotein gene (N gene; 28267-29527) sequence was synthesized by Sangon Biotech. The standard plasmid was diluted from 106 copies/μL to 0.1 copy/μL with 10-fold serial dilution for sensitivity assays. The COVID-19 in vitro transcribed RNA reference material was obtained from the National Institute of Metrology (Beijing, China). The COVID-19 RNA was diluted in DEPC-treated water to working concentrations. Influenza A virus (H1N1) RNA, influenza B virus RNA, human coronavirus OC43 (hCoV-OC43) cDNA, and MERS-CoV cDNA, which were donated by the Qingdao International Travel Healthcare Center, were employed for specificity assays.
Real-time LAMP and RT-LAMP assay for SARS-CoV-2. In order to setup the mixture in room temperature, WarmStart enzyme (RTx and Bst 2.0) was employed in the reaction. The WarmStart enzyme contained a reversibly-bound aptamer, which inhibits polymerase activity at room temperature. The LAMP reaction was carried out in a total 20 μL reaction mixture containing 2 μL 10x isothermal amplification buffer, 6 mM MgSO4, 1.4 mM dNTP, 1 M betaine, 2 μL 10x Primer mixture (Table S2), and 8 U of WarmStart Bst 2.0 DNA polymerase. For RT-LAMP, 6 U of WarmStart RTx reverse transcriptase was added into the mixture. When using quantitative fluorescence amplification, 0.5 μL 20x EvaGreen® dye was added. Fluorescence signal measurement was performed by Real-Time PCR System (Roche LightCycler480 II for proof-of-concept and ABI QuantStudio 5 for human clinical samples) at 30 s intervals under 65°C. Afterward, additional heat inactivation at 80°C for 5 min if needed. The LAMP products were subsequently identified by 2% gel electrophoresis to confirm the amplification result.
RNA extraction from human clinical samples. Samples of nasopharyngeal swab were collected and transferred into a tube containing VTM, then transported in sterile containers to the diagnostic laboratory within a few hours. Sample RNA was inactivated and then extracted by the Takara MiniBSET viral RNA extraction Kit in a biosafety level 2 (BSL-2) cabinet and stored at −80°C for future use (above steps were completed with the assistance of staffs from the Customs). Negative nasopharyngeal swabs were obtained from healthy donors in the Single-Cell Center laboratory with the approval of Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (QIBEBT, CAS) Institutional Review Board. Healthy donors have signed written informed consents. Clinical swab samples from COVID-19 patients were collected in Qingdao International Travel Healthcare Center after virus inactivation.
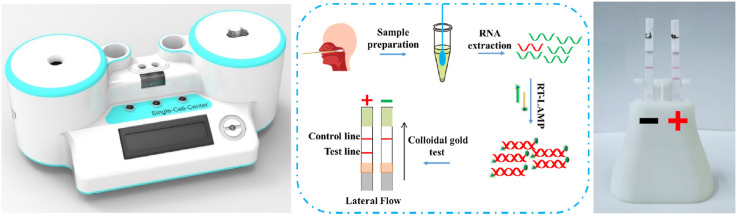
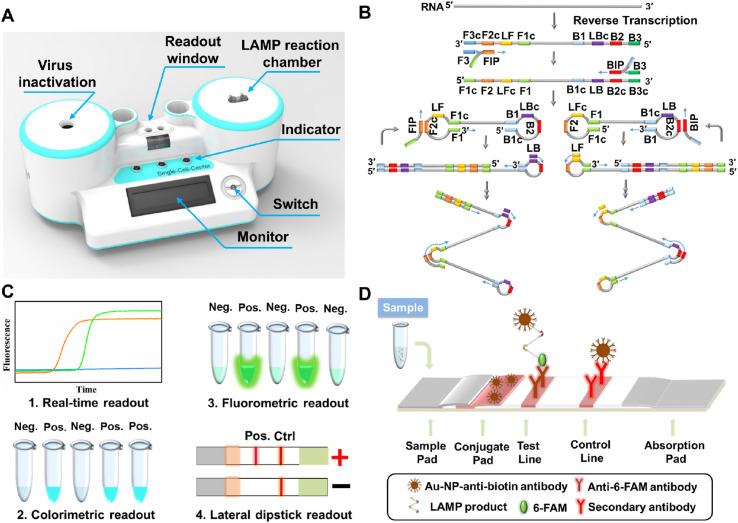
Assembly of PaGeR platform. As shown in Fig. 1 A, the PaGeR platform for COVID-19 detection is composed of three parts: the heating part, programed control part, and detection part. (1) The heating part contained two similar modules, one for virus inactivation and the other for LAMP reaction. It contained a metal heat-conductive base plate with an integrated heating rod to form a heating cavity and a DS18B20 digital thermal sensor for feedback of cavity temperature. (2) The programed control part was programed in STC 89C52RC (HongJing Tech, Jiangsu, China) Micro-Controller-Unit using C++, which contained control of power management, heating elements, light source switch, as well as monitor for temperature and steps. (3) The detection part supported direct results readout with naked eyes. For fluorescence detection, a 375 nm LED array was lay under the readout window, the result could be directly visualized after LAMP reaction. For lateral flow dipstick detection, the RT-LAMP products in matching buffer were added to the strip. A single control band indicated a negative result, whereas two bands contained test line band and control line band indicated a positive result.
Fig. 1.
A PaGeR device for COVID-19 detection by RT-LAMP. (A) Schematic illustration of the PaGeR device, which was composed of two similar heating parts for virus inactivation and RT-LAMP reaction, one programmed control part (microcontroller unit), and one result readout part. (B) Principle of the RT-LAMP reaction with loop primers. Specially designed nested primers combined with target regions formed hairpins to permit subsequent amplification. Forward Inner Primer (FIP) consists of a F2 region at the 3'end and a F1c region at the 5'end. Backward Inner Primer (BIP) consists of a B2 region at the 3'end and a B1c region at the 5'end. Forward Outer Primer (F3) region is complementary to the F3c region of the template sequence. Backward Outer Primer (B3) is complementary to the B3c region of the template sequence. (C) Four types of result readout methods: real-time fluorescence readout, fluorometric readout, colorimetric readout, and lateral dipstick readout. (D) The principle of visual lateral flow nucleic acid detection after RT-LAMP. Neg., negative; Pos., positive; Ctrl, control.
The whole system work in concert to realize isothermal amplification and result readout. The 3D-printed and metal structures were designed in AutoCAD 2019 (Autodesk, CA, USA). A 3D-printed outer shell was printed with photosensitive resin in Stereo Lithography. The detail of interior structure, such as virus inactivation, LAMP reaction chamber, monitor and other structure, was shown in the supplementary Component 1. Besides, we had also uploaded the files to GitHub (https://github.com/qibebt-microfluidics/PaGeR) so that users could download. The 12V DC power supply (Jing Sai Electronics, Guangzhou, China) was mounted and distributed to the individual components with an AC power jack locating at the rear of the platform, allowing the instrument to be plugged into a standard 220V AC socket. The maximum power consumption during operation did not exceed 30 W, thus battery power or small solar system could be easily compatible with the generator.
Visual RT-LAMP detection by PaGeR. For PaGeR assays, the laboratory safety should be at BSL-2 and personal protection at BSL-3. After specimens were transported to the lab, the swab sample was inactivated in the left heating cavity then transferred to the biosafety cabinet for RNA extraction and reaction mixture preparation. A total of 10 μL reaction mixture contained modified primers was incubated at the right heating cavity at 65°C for 40 min. After completion of the RT-LAMP step, the amplicon was added to the sample pad of lateral flow strip, then the strip was added to the matching buffer tube which contained 100 μL matching buffer for lateral flow. The visualized result would be displayed within 1–2 min a single band close to the absorption pad (control line) indicated a negative result, whereas two bands containing both control line and test line indicated a positive result.
Real-time qRT-PCR assay. The qRT-PCR assay was performed with 2x Luna® Universal One-Step RT-qPCR Kit (NEB) on an ABI QuantStudio 5 (Applied Biosystems) with standard protocol. Primers of N gene sequence were provided by China CDC (Centers for Disease Control). The program was set for 10 min at 55°C for the RT step, then thermal cycling of PCR with 40 cycles was carried out for sample verification.
3. Results
Principle of PaGeR Platform for RT-LAMP detection. As shown in Fig. 1A, the proposed COVID-19 detection platform utilized RT-LAMP reaction to detect RNA of the SARS-CoV-2. The structure of the device had two heating parts: one for virus inactivation, another for RT-LAMP reaction. After the nasopharyngeal or oropharyngeal swabs were transferred to the laboratory by VTM, a series of operations such as virus inactivation, RT-LAMP reaction as well as result readout could be completed on the PaGeR platform.
In the RT-LAMP reaction (Fig. 1B), 4 different primers (FIP, BIP, F3, and B3) were employed to amplify 6 distinct regions on the target gene, while an additional pair of loop primers (LF and LB) could further accelerate the reaction. Afterward, the RT-LAMP result could be read out by a variety of methods (Fig. 1C). i) Real-time readout. Amplification can be recorded precisely by real-time quantitative analysis with the presence of fluorescence nucleic acid dye such as EvaGreen. ii) Colorimetric readout. Colorimetric pH indicators such as malachite green (MG) are utilized for visual end-point assessment of LAMP products. Nucleic acid amplification process required the attachment of dNTP to the template, during which the protons (H+) were released. As the LAMP assay proceeds, the pH was dropped displaying a visible color change from colorless to blue, which could be easily distinguished by the naked eye (Nzelu et al., 2016). iii) Fluorometric readout. If the colorimetric regent is replaced by fluorescence metal indicator (calcein) and the Mn2+ was added into the reaction mixture, the reaction end-point could be observed by UV light pen (365 nm). iv) Lateral dipstick readout. More intuitively, the reaction result could be shown on lateral flow coupled with nucleic acid biosensor.
If LB primer is labeled with biotin at the 5′ end, while LF primer is modified with 6-FAM (6- Carboxyfluorescein) at the 5′ end, the LAMP product could form a double-labeled detectable product with biotin and 6-FAM. As the product running to the detection region, a product labeled with 6-FAM could be captured by the anti-6-FAM antibody located on the test line, while excess anti-biotin-Au-NP conjugates will running continuously and be captured by the secondary antibody located on the control line. In the no template control (NTC), there was no conjugate that was captured on the test line, only a red band on the control line (Fig. 1D). In this work, three ways of result read out were employed in the PaGeR instrument, only the real-time fluorescence detection was not included.
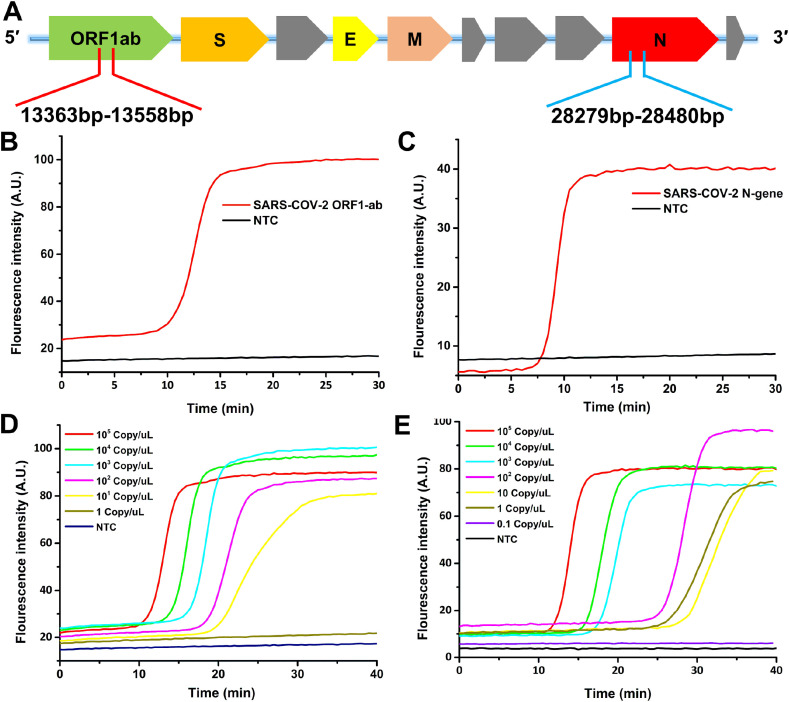
Primer design and sensitivity of LAMP detection. The LAMP primers were designed with PrimerExplorer V5 (http://primerexplorer.jp/lampv5e/index.html) on the basis of the SARS-CoV-2 ORF1ab region from 13363 to 13558 and N gene region from 28279 to 28480 (Fig. 2 A). Loop primers were designed after four basic primers (B3, F3, BIP, and FIP) were selected. The primer sequences were shown in Table S1. We first investigated the feasibility of designed primers using recombinant plasmid which contained this two region sequence. In the verification stage, the reaction was conducted for 60 min at 65°C followed by enzyme inactivation at 80°C for 2 min. As shown in Fig. 2B and C, the fluorescence intensity of SARS-CoV-2 (ORF1ab and N gene) was increased rapidly within 15 min and remained at this intensity over time, while no significant signal increased in the NTC tube. The result illustrated that primers designed in this study were suitable for SARS-CoV-2 detection.
Fig. 2.
The sensitivity of LAMP detection. (A) The detection region for primers design in SARS-CoV-2 RT-LAMP reaction. (B) The feasibility of LAMP primer for SARS-CoV-2 ORF1ab fragment. (C) The feasibility of LAMP primer for SARS-CoV-2 N gene fragment. (D) Real-time fluorescence intensity of ORF1ab sensitivity test with 10-fold serial diluted plasmid. (E) Real-time fluorescence intensity of N gene sensitivity test with 10-fold serial diluted plasmid.
Afterward, the sensitivity of LAMP assay was explored by testing a serial 10-fold dilution DNA plasmid (from 105 copies/μL to 0.1 copy/μL). With the optimization of the experimental condition such as reaction temperature, Mg2+ and betaine concentration, we could detect ORF1ab sequence down to 5 copies/μL while N gene sequence down to 1 copy/μL. As shown in Fig. 2D and E, fluorescence intensity of ORF1ab had been significantly increased with a concentration range from 105 copies/μL to 5 copies/μL in less than 40 min. However, the concentration of 1 copy/μL had no significant signal increase compared with the NTC tube. Similarly, fluorescence intensity of N gene showed an apparent increase from 105 copies/μL to 1 copy/μL, while the concentration of 0.1 copy/μL had no significant signal increase compared with the NTC tube. After testing these two primer sets, we found that the sensitivity of N gene was better than ORF1ab, as N gene could be detected down to 1 copy/μL. In addition, the calibration curve could be generated by correlating the threshold time (the time required to reach saturation) with the logarithm of the DNA copy number. As shown in Fig. S1, the linear regression plot was conducted. As reaction time was set as 40 min, the detection threshold could reach 0.11 copies/μL for N gene and 2.0 copies/μL for ORF1ab from the calibration curve. The detection ability of target sequence was also affected by target GC percentage and melting temperature of primers. Target N gene sequence with GC percentage around 50–60% (normal sequence) was more suitable for LAMP reaction than the target sequence with GC percentage around 35–45% (AT rich sequence) in the ORF1ab region. Therefore, we choose the N gene as the subsequent LAMP reaction target.
Besides, we also examined the sensitivity of RT-LAMP (N gene) by using the COVID-19 in vitro transcribed RNA reference material (Fig. S2), showing a similar result consistent with the above LAMP reaction. Thus, the above results validated that primers used in this study were able to detect SARS-CoV-2 with high sensitivity.
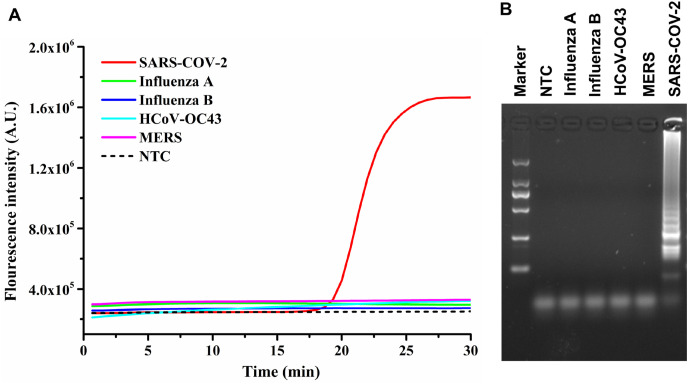
The specificity of COVID-19 RT-LAMP assay. Herein, the specificity of the assay was evaluated with SARS-CoV-2 and other four different viruses (Influenza A virus, influenza B virus RNA, hCoV-OC43, and MERS-CoV). The concentration of DNA/RNA for the specificity tests was 104 copies/μL for each virus. As shown in Fig. 3 A, only the red line with SARS-CoV-2 sample showed obvious change in fluorescence intensity. However, the other four different virus samples were as smoothing as the NTC. Afterward, LAMP products were subjected to 2% agarose gel electrophoresis for further proof of specificity (Fig. 3B). A typical ladder pattern was observed on the gel electrophoresis demonstrated that only SARS-CoV-2 could induce the RT-LAMP reaction. The high specificity of the developed method fully benefited the inherent LAMP reaction mechanism, since amplification could only occur when all primers were simultaneously bound to the target sequence.
Fig. 3.
The specificity of SARS-CoV-2 RT-LAMP assay. (A) The real-time fluorescence curves for SARS-CoV-2 and four other viruses. Only the target virus showed obvious change in fluorescence intensity. (B) Agarose gel electrophoresis image of LAMP products. The marker lane was DL2000 Takara ladder marker.
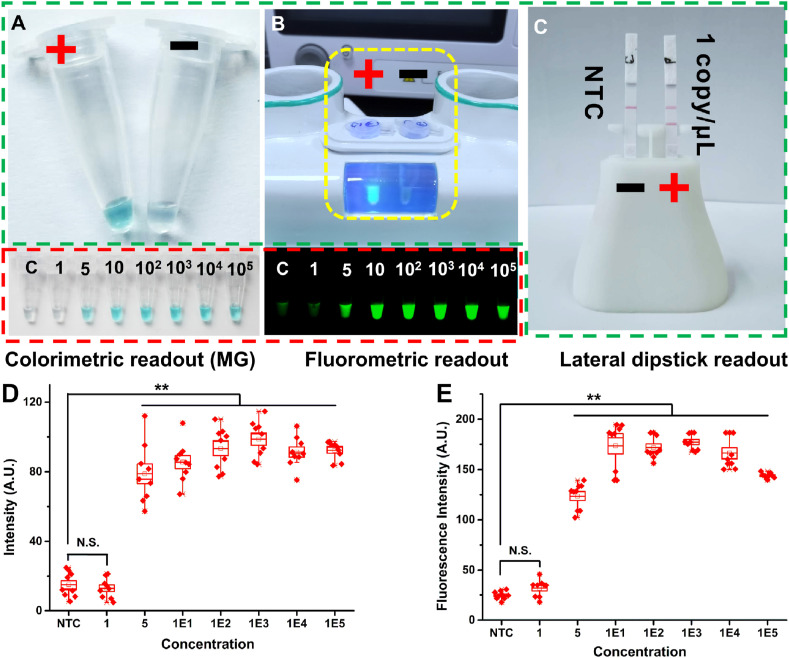
Three methods for RT-LAMP result readout by PaGeR platform. As described before, the detection of LAMP reaction could be performed by four main methods. Three detection methods (colorimetric readout, fluorometric readout and lateral dipstick readout) could be performed in PaGeR instrument (Fig. 4 , highlight with green dashed line), except for the real-time quantitative detection. As a pH-sensitive indicator, MG had been successfully employed in visual end-point detection of LAMP product. As shown in Fig. 4A, the color of MG-LAMP could be changed to light blue by naked eye when LAMP reaction was proceeded while the color could not be changed showing a colorless result when there was no reaction (Fig. 4A). Another detection method was calcein-based fluorometric detection, which could be achieved by UV light (365 nm) embedded in PaGeR instrument. Calcein was quenched by Mn2+ at the beginning of the reaction. However, the Mn2+ were gradually consumed by generating Mn2P2O7, making the fluorescence of calcein increase when the reaction proceeds. As shown in Fig. 4B, the left tube was a positive result which showed a strong fluorescence signal while the right tube was a negative result which showed a colorless signal (within the yellow dashed line).
Fig. 4.
Three methods for RT-LAMP result readout by PaGeR platform. (A) The MG colorimetric detection method for RT-LAMP. (B) The calcein-based fluorometric detection method for RT-LAMP (within the yellow dashed line). (C) Visual lateral dipstick biosensor detection. All three ways could be detected by the PaGeR (highlighted with green dashed line). The sensitivity tests of colorimetric and fluorometric detection were highlighted with red dashed line. (D) The intensity for 10-fold serial diluted plasmid by colorimetric detection. (E) The fluorescence intensity with 10-fold serial diluted plasmid by calcein-based fluorometric detection. Data were averages of three independent experiments, each consisting of three technical replicates. Data were shown as the mean ± SD. **p < 0.01 (Student's t-test). C, control; P, positive. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The dispersion graphs of MG colorimetric detection and calcein-based fluorometric detection were shown in Fig. 4D and E. For the MG detection, we obtain the individual value of R (red), G (green), and B (blue) of the color. Since the G and B values for MG were almost the same between tubes, only R value was counted to reflect the intensity of MG results. Also, the fluorescence signal intensity of fluorometric readout was obtained by grayscale graph. When the concentration of virus exceeded 5 copies/μL, the intensity was significantly increased compared to the NTC group (p < 0.01). However, there was no obvious change of the signal between the 1 copy/μL group and the NTC group by both detection methods. The colorimetric method detection objects are based on H+-induced pH change. When the sample concentration was low, the LAMP reaction does not reach the threshold for pH change, resulting in a failure to distinguish from a negative sample within a reaction time. In addition, in clinical application, many factors might interfere pH change. The colorimetric and fluorometric ways were judged depending on naked eyes, which were potentially subjective. For example, the sensitivity test of these two methods were highlighted with red dotted line in Fig. 4. With naked-eye we could detect virus sequence down to 5 copies/μL. Nevertheless, some of the 1 copy/μL sample showed a significant color change, while some of them could only show a relatively light color change within the predetermined time (40 min), which meant that there was little difference in color between samples and NTC. This feature makes the subjective judgment inconspicuous at low nucleic acid concentrations, which means the lateral dipstick detection method could provide a more objective result. As shown in Fig. 4C, the lateral dipstick could detect virus sequence down to 1 copy/μL. Visualization detection was achieved by employing an anti-6-FAM antibody to capture 6-FAM labeled nucleic acid product (test line), whereas excess anti-biotin-Au-NP conjugates would generate a signal when captured by the secondary antibody located on the control line (control line). Therefore, with the most intuitive result presentation, the lateral flow strip was finally employed in the following PaGeR platform test.
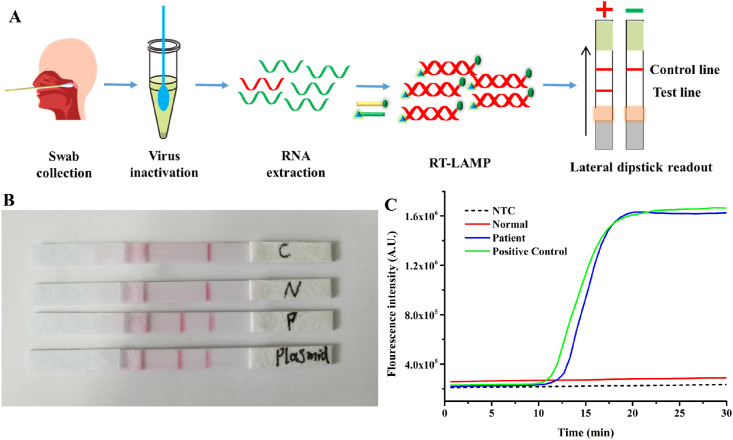
RT-LAMP assay for clinical samples by PaGeR platform. Based on the established method, collected samples could be transported to the PaGeR instrument for the amplification reaction. The schematic diagram of the detection process was shown in Fig. 5 A. And the whole process of detection by PaGeR from swab collection to results readout was shown in Video S1. Firstly, the collected samples from nasopharyngeal and oropharyngeal flocked swabs were inactivated by heating the sample at 95°C for 5 min in PaGeR built-in heating system. After treatment, swab RNA extraction was done by RNA extraction kit. Once the virus RNA was extracted, it was added to the reaction mixture contained modified primers. By heating at 65°C for 40 min, one-step RT and LAMP reaction could be completed. Finally, visualized results of lateral flow were read simply by the naked eye. A single band of control line indicated a negative result, whereas two bands indicated a positive result. Totally, the whole process including swab sample collection (3–5 min), RNA extraction (10–15 min), one-step RT-LAMP reaction (40 min) and lateral dipstick readout (less than 5 min) could be completed around 1 h by employing the PaGeR platform.
Fig. 5.
RT-LAMP assay for clinical sample by PaGeR platform. (A) Schematic diagram of COVID-19 detection process from sample collection to result readout. Among them, virus inactivation as well as RT-LAMP amplification and result reading could be achieved by PaGeR. The detection result of PaGeR system (B) compared with Real-Time LAMP system(C). C, NTC; N, negative sample; P, positive sample.
To further investigate the diagnostic accuracy and reliability of PaGeR for clinical samples, we first tested one patient sample and one normal sample (healthy volunteer) by employing both PaGeR system and Real-Time LAMP system (Real-Time PCR system with isothermal program). As shown in Fig. 5B, detection strips reacting with patient sample and positive plasmid captured the corresponding labeled probe and appeared two bands containing test line and control line. While the strips reacting with control and normal samples appeared only one band on the Test line. Furthermore, by employing Real-Time PCR (LAMP) system, the fluorescence intensity of the reaction mixture from patient and positive plasmid was increased, while no significant signal increased in reaction mixture from neither control nor normal sample (Fig. 5C).
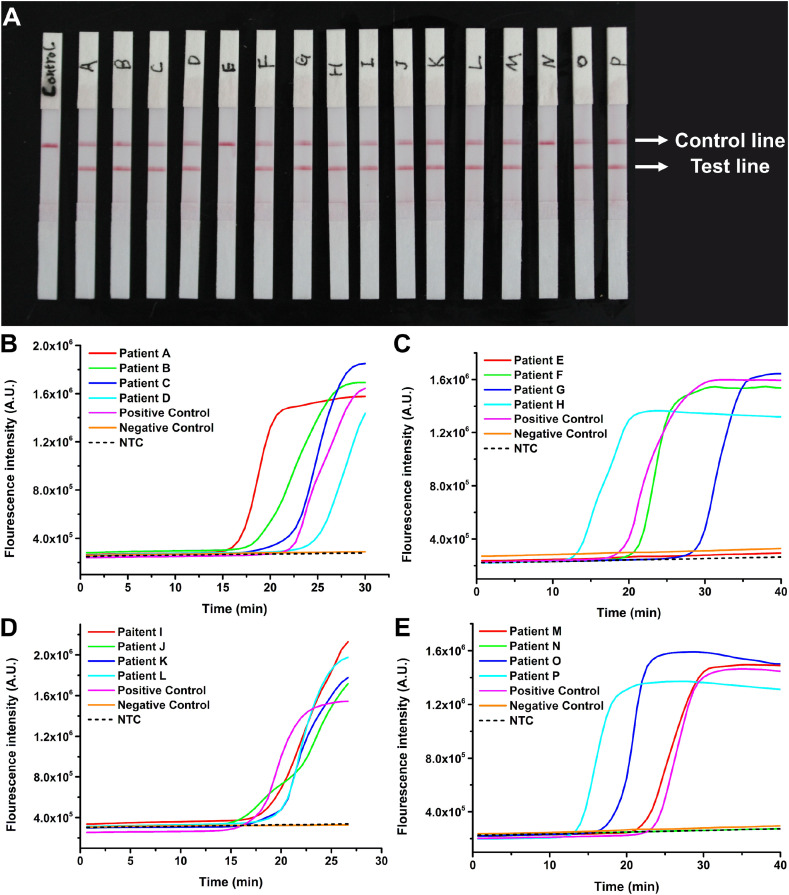
Afterward, 70 respiratory swab samples from 16 patients and 54 healthy donors were then investigated by PaGeR. The negative samples from healthy donors and clinical samples from COVID-19 patients were diagnosed by RT-qPCR with the standard protocol prior to detection using PaGeR. Among the 16 samples from patients, 14 of them directly showed two bands, which contained a test line and a control line, indicating that the test result was positive (Fig. 6 A). Only patient E and patient N did not show positive results, probably because the viral concentration was too low to be detected with this method. While all the 54 uninfected samples were showed one band on the control line with a negative result (data not shown). In addition, the fluorescence intensity of 16 patients and 54 healthy donors (divide into 6 groups) were shown in Fig. 6B–E and Fig. S3, which indicated that detection using PaGeR achieved consistent results relative to that with Real-Time LAMP system. Together with the two samples in the validation phase, the accuracy of PaGeR could reach 97.2% with a total of 72 subjects enrolled in the study (Fig. S4). The sensitivity of PaGeR platform was 88.2% (15/17) and the specificity was 100% (55/55). The positive predictive value (PPV) could reach 100% (15/15) while the negative predictive value (PPV) was 96.5% (55/57), which verified the possibility of PaGeR platform for COVID-19 detection in clinical samples.
Fig. 6.
The results of 16 COVID-19 infected patient samples by PaGeR. (A) 14 of 16 samples directly showed two bands. Only patient E and patient N did not show two bands. (B–E) The results of 16 COVID-19 infected patient samples by Real-Time PCR System. (B) The real-time fluorescence curves for SARS-CoV-2 positive control, four patient samples, negative control (healthy sample) and NTC. (C–E) Other 12 patient samples real-time fluorescence curves. Only samples from patient E and patient N show no significant increase of fluorescence intensity.
4. Discussion
The COVID-19 pandemic is still ongoing. Early diagnosis, treatment, and vaccine can effectively reduce virus transmission. RT-PCR is the gold standard for SARS-CoV-2 determination with high sensitivity. However, the qRT–PCR-based assays need expensive laboratory instrumentation, and the detection needs to be performed in public health laboratories currently. The PaGeR device provides a rapid, affordable, user-friendly and reliable diagnostic strategy for COVID-19 detection from extracted patient sample RNA by RT-LAMP. Firstly, the PaGeR device can be integrated into a 23 × 11 × 9 cm3 suitcase, thus acting as a potential mini-instrument for economical and operation simple POCT detection outside the clinical laboratory. Secondly, the cost of PaGeR device is less than $400, and a single RT-LAMP reaction only cost less than $3. The cost of lateral dipstick employed in our assays is estimated to be $2 per test. Thirdly, the developed PaGeR device allows detection of SARS-CoV-2 with approximately 1 h. To sum up, our PaGeR is a promising choice for rapid detection in the community as well as at home.
Since there are some commercial colorimetric LAMP Kits (e.g., NEB WarmStart™ Colorimetric LAMP 2 × Master Mix) applicable to SARS-CoV-2 detection, colorimetric methods for RT-LAMP amplification have been mostly reported in COVID-19 detection (El-Tholoth et al., 2020; Lalli et al., 2020; Huang et al., 2020b; Park et al., 2020; Dao Thi et al., 2020). The sensitivity of colorimetric detection varies from 0.08 copy/μL to 5 copies/μL (calculated into copies per μL) (Table S3). The extreme sensitivity of detection also acts as a double-edged sword, which can cause false positive results (e.g., 0.08 copies/μL) (Huang et al., 2020b). In addition, the detection using colorimetric method are based on H+-induced pH change, so the elution buffers for various RNA extraction kits will affect the result. The fluorometric method detection objects are not affected by pH, making the reaction conditions more tolerant for acids and bases. The sensitivity of detections can reach 0.5 copies/μL to 8 copies/μL (Table S3) (Jiang et al., 2020; Yan et al., 2020). However, it requires a supplementary equipment to detect the fluorescence. RT-LAMP coupled with CRISPR/Cas technology can detect SARS-CoV-2 with high specificity and the detection result can also be performed by fluorometric and lateral dipstick methods (Broughton et al., 2020; Joung et al., 2020; Wang et al., 2021). The sensitivity is similar to the LAMP method, and in some cases better than the RT-LAMP method (Table S3). However, the assays require a two-step thermostatic amplification process, a 65°C LAMP reaction and a 37°C CRISPR reaction.
The self-developed PaGeR device supports three methods (colorimetric readout, fluorometric readout and lateral dipstick readout) for rapid and simple detection of COVID-19 by RT-LAMP. Among them, the lateral dipstick method has a higher sensitivity than other two methods (Fig. 4). So the lateral dipstick method was employed in further detection using clinical samples. The accuracy of PaGeR could reach 97.2% with 72 clinical samples (Fig. S4). Our PaGeR device can be reconfigured within days to use. We have used a 4-step standard process method: inactivation, extraction, amplification and detection. We fabricate the device with two temperature units, one for virus inactivation and the other for LAMP amplification. The virus inactivation step is set up to protect the operator from infection by the virus within the sample, if any. In the case of self-testing, no inactivation step is required. Further work will focus on rapid one-step sample processing (e.g., direct addition to the RT-LAMP system after 5 min at 95°C), which will shorten the steps of the assay. Also, the future development of microfluidic technology and lyophilized reagents can enable POCT detection outside of the laboratory in a more friendly way to users.
5. Conclusion
In this work, we developed a PaGeR device for rapid and simple detection of COVID-19 from extracted patient sample RNA by RT-LAMP. The virus inactivation, the RT-LAMP reaction and the lateral dipstick result readout were successfully performed on this device. We successfully detected SARS-CoV-2 down to 1 copy/μL after reaction optimization. We further investigated the PaGeR platform to detect SARS-CoV-2 in clinical RNA samples isolated from swab specimens. The results showed that 15 out of 17 COVID-19 patients were diagnosed as positive while all 55 normal samples were diagnosed as negative.
Thus, this developed PaGeR device allowed detection of SARS-CoV-2 in a diagnostic test within approximately 1 h. It had several unique advantages including one-step RT-LAMP reaction, user-friendly procedure and without any expensive instruments. These advantages of the device provide great potential as a mini-instrument for economical and operation simple POCT detection, especially in resource-limited environments. Future work will be focused on increasing sensitivity and simultaneous detection of multiplex target genes such as Membrane protein (M gene), small envelope gene (E gene) or spike glycoprotein gene (S gene). In addition, convenient operation process and simplified PaGeR device would be considered to facilitate the use by non-professional individuals. Therefore, our developed PaGeR device can be employed to investigate the testing of nasal swabs for detection of nucleic acid from SARS-CoV-2, which could be further used for in vitro diagnosis of COVID-19 under clinical, environmental samples community as well as at home.
CRediT authorship contribution statement
Anle Ge: Methodology, Formal analysis, Validation, Writing - original draft. Fengyi Liu: Data curation, Formal analysis. Xindong Teng: Virus sample treatment, Software. Chaojie Cui: Data curation, Formal analysis. Fei Wu: PaGeR design. Wenjing Liu: Methodology. Yang Liu: Video capture, Writing - original draft. Xiaoguang Chen: Resources, Formal analysis, Jian Xu Conceptualization, Funding acquisition, Data curation. Bo Ma: Funding acquisition, Methodology, Writing - original draft.
Availability of data and materials
The detail of interior structure component and the control program in this work are available at GitHub (https://github.com/qibebt-microfluidics/PaGeR) to ensure reproducibility.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge the financial supports from National Key Research & Development Program (2017YFF0204601), National Natural Science Foundation of China (31800084, 21775155), Scientific Research Project of General Administration of Customs, CHINA (No. 2020HK134) and the Natural Science Foundation of Shandong Province (ZR2018ZC0128, ZR2019BC017).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113925.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B.D., Thakor A.S. Biology. 2020;9(8):182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreda-Garcia S., Miranda-Castro R., de-Los-Santos-Alvarez N., Miranda-Ordieres A.J., Lobo-Castanon M.J. Anal. Bioanal. Chem. 2018;410(3):679–693. doi: 10.1007/s00216-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty I., Maity P. Sci. Total Environ. 2020;728:138882. doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K.A., Rudolph D.L., Owen S.M. J. Virol. Methods. 2008;151(2):264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Krausslich H.G., Knop M., Anders S. Sci. Transl. Med. 2020;12(556) doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tholoth M., Bau H.H., Song J. ChemRxiv. 2020 [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. RNA. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Van Tu P., Tien N.T.K., Tashiro M., Odagiri T. J. Virol. Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jiang M., Pan W., Arasthfer A., Fang W., Ling L., Fang H., Daneshnia F., Yu J., Liao W., Pei H., Li X., Lass-Flörl C. Front. Cell. Infect. Microbiol. 2020;10(331) doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.-L.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.Y., Tang S.N., Lau Y.L. Am. J. Trop. Med. Hyg. 2021;105(2):375–377. doi: 10.4269/ajtmh.21-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli M.A., Langmade S.J., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. 2020. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir A.A., Baptiste M.J., Mwikarago I., Habimana M.R., Ndinkabandi J., Murangwa A., Nyatanyi T., Muvunyi C.M., Nsanzimana S., Leon M., Musanabaganwa C. 2020. medRxiv. [Google Scholar]
- Nguyen H.V., Nguyen V.D., Nguyen H.Q., Chau T.H.T., Lee E.Y., Seo T.S. Biosens. Bioelectron. 2019;141:111466. doi: 10.1016/j.bios.2019.111466. [DOI] [PubMed] [Google Scholar]
- Nzelu C.O., Caceres A.G., Guerrero-Quincho S., Tineo-Villafuerte E., Rodriquez-Delfin L., Mimori T., Uezato H., Katakura K., Gomez E.A., Guevara A.G., Hashiguchi Y., Kato H. Acta Trop. 2016;153:116–119. doi: 10.1016/j.actatropica.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., Maeng J.-S. J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Virol. J. 2014;11(1):139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toley B.J., Covelli I., Belousov Y., Ramachandran S., Kline E., Scarr N., Vermeulen N., Mahoney W., Lutz B.R., Yager P. Analyst. 2015;140(22):7540–7549. doi: 10.1039/c5an01632k. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. Biosens. Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.H., Chen X., Zhang X.L., Liu Q., Zhu J.H., Zhao W., Liu S.X., Sui G.D. ACS Sens. 2020;5(5):1354–1362. doi: 10.1021/acssensors.9b02595. [DOI] [PubMed] [Google Scholar]
- Ye X., Li L., Li J., Wu X., Fang X., Kong J. ACS Sens. 2019;4(11):3066–3071. doi: 10.1021/acssensors.9b01731. [DOI] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F., Du L., Jiang S. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The detail of interior structure component and the control program in this work are available at GitHub (https://github.com/qibebt-microfluidics/PaGeR) to ensure reproducibility.