Abstract
Fish papillomaviruses form a newly discovered group broadly recognized as the Secondpapillomavirinae subfamily. This study expands the documented genomes of the fish papillomaviruses from six to 16, including one from the Antarctic emerald notothen, seven from commercial market fishes, one from data mining of sea bream sequence data, and one from a western gull cloacal swab that is likely diet derived. The genomes of secondpapillomaviruses are ~6 kilobasepairs (kb), which is substantially smaller than the ~8 kb of terrestrial vertebrate papillomaviruses. Each genome encodes a clear homolog of the four canonical papillomavirus genes, E1, E2, L1, and L2. In addition, we identified open reading frames (ORFs) with short linear peptide motifs reminiscent of E6/E7 oncoproteins. Fish papillomaviruses are extremely diverse and phylogenetically distant from other papillomaviruses suggesting a model in which terrestrial vertebrate-infecting papillomaviruses arose after an evolutionary bottleneck event, possibly during the water-to-land transition.
Keywords: Papillomavirus, Trematomus bernacchii, Melanogrammus aeglefinus, Sparus aurata, Centropristis striata, Larus occidentalis
Introduction
Papillomaviruses have circular double-stranded DNA genomes. The Papillomaviridae family is comprises of two subfamilies (Firstpapillomavirinae and Secondpapillomavirinae) (Van Doorslaer et al., 2018a). The Firstpapillomavirinae encompass >50 genera that infect mammals and other terrestrial vertebrates, including various birds (Canuti et al., 2019; Prosperi et al., 2016; Truchado et al., 2018; Van Doorslaer et al., 2017b; Varsani et al., 2014), lizards (Agius et al., 2019), and snakes (Gull et al., 2012; Kubacki et al., 2018). Some are oncogenic and can have disease outcomes that are lethal to their host.
The discovery of divergent papillomaviruses in fish prompted the establishment of a novel subfamily, Secondpapillomavirinae, composed of one genus, Alefpapillomavirus. Since the discovery of the first fish papillomavirus in a gilthead sea bream (Sparus aurata) (Lopez-Bueno et al., 2016), other fish papillomaviruses have been recovered from commercial market samples of haddock (Melanogrammus aeglefinus), red snapper (Lutjanus campechanus), and rainbow trout (Oncorhynchus mykiss) (Tisza et al., 2020), as well as farmed wels catfish (Silurus glanis) (Surjan et al., 2021). In the cases of the sea bream and of the wels catfish, the papillomaviruses were identified in individuals exhibiting papillomatous lesions (Lopez-Bueno et al., 2016; Surjan et al., 2021).
Unlike mammalian papillomavirus genomes, which are generally about 8 kilobase pairs (kb) in length (Frias-De-Diego et al., 2019), papillomaviruses associated with fish are significantly smaller, with sizes of 5.6–6 kb (Lopez-Bueno et al., 2016). Previously reported fish papillomavirus genomes contain the core four open reading frames (ORFs) with predicted protein products that include two early genes, E1 and E2, which encode protein products involved in replication), and two late genes, L1 and L2, that encode viral capsid proteins (Van Doorslaer et al., 2018a).
Here we report the identification of ten novel fish-associated papillomaviruses in an endeavor to gain further insight into the Secondpapillomavirinae. Genomic characterization, including ORFs with features that resemble the papillomavirus E6/E7 proteins (which we call Oncoid), and phylogenetic analyses shed light on this highly divergent group of papillomaviruses.
Material and methods
Sampling and processing for viral DNA
Antarctic fish samples
As part of a study to identify papillomaviruses circulating in Antarctic fishes, we sub-sampled four species of fish that were collected for various ongoing studies on the biology of these animals. All samples were stored at −20°C till processed.
Two of these species were collected in the East Antarctic and two in the West Antarctic. An emerald notothen (Trematomus bernacchii) and a sharp-spined notothen (Trematomus pennellii) were captured from the Ross Sea (East Antarctica) during the austral summer of 2012–2013, and under the 2011/08R animal ethics permit (University of Canterbury, New Zealand). From these two notothens we subsampled the stomach, including its contents, liver and gills. The fish did not show any obvious external pathology. Approximately 0.5 cm3 of each subsampled tissue was homogenized in 20 ml of SM buffer (0.1M NaCl, 50mM Tris/HCl – pH 7.4, 10mM MgSO4) using a mortar and pestle. The resulting homogenate for each tissue sample was then centrifuged at 6000 g for 10 min to pellet cell debris, and the supernatant was sequentially filtered through 0.45 and then 0.22 μm syringe filters. Viral particles in the filtrate were precipitated with 15% w/v PEG 8000 overnight at 4°C. The resulting solution was centrifuged at 6000 × g for 20 min, the supernatant was discarded and the pellet was resuspended in 1 ml of SM buffer. 200 μl of this was used for viral DNA extraction with a High Pure Viral Nucleic Acid Kit (Roche Diagnostics, USA).
Skin samples of 3 mm x 3 mm were necropsied from eight crowned notothen (Trematomus scotti) and two painted notothen (Nototheniops larseni) collected in the West Antarctic Peninsula during the austral fall of 2018 according to protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Oregon, USA (#13–27RRAA). Due to the small sample size of these samples, each was individually homogenized in 200 μl of SM buffer, which was then directly used to extract viral DNA using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, USA).
Commercial market fishes
Whole uncleaned black sea bass (Centropristis striata) (n=3) and haddock (Melanogrammus aeglefinus) (n=4) specimens were purchased from Maryland (USA) fish vendors between 2018–2019. The fishing location of these fishes is unknown. Approximately 0.5 g of scales, muscle, liver, and small samples of other tissues were combined, macerated and resuspended in 15 ml Dulbecco’s PBS with Triton™ X-100 (MilliporeSigma, USA) detergent (1% w/v). To this solution and 0.02% Benzonase® Nuclease (MilliporeSigma, USA). The mixture was vortexed followed by incubation in a 37°C water bath for 30 min. NaCl was added to a final concentration of 0.85M and the lysate was clarified by centrifugation for 5 min at 5000 × g. The supernatant was then transferred to a fresh siliconized tube and the centrifugation step was repeated. The resulting supernatant was used for iodixanol/OptiPrep™ (MilliporeSigma, USA) ultracentrifugal step gradients using the protocol outline in Tisza et al. (Tisza et al., 2020). From the gradient fractions, viral nucleic acids were extracted following a standard DNA extraction protocol (Tisza et al., 2020). Sequencing reads for these samples were deposited in NCBI under the BioProject PRJNA393166.
Western gull cloacal swab samples
Western gulls (Larus occidentalis) are generalist predators that have diverse diets including fish, crustaceans and other birds (Pierotti and Annett, 1995). Cloacal swabs were collected in 2011 from 42 Western gull chicks on the South Farallon Islands (part of the Farallon Islands National Wildlife Refuge) located 42km west of San Francisco. The samples were collected as part of an avian gut microbial study. The swabs were stored in RNAlater and guanidinium isothiocyanate buffer. Viral DNA was extracted from 200μl of the lysate using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, USA).
Enrichment of circular nucleic acids and high throughput sequencing
For each sample, 1–5μl of the viral DNA extract were used to preferentially amplify circular DNA by rolling circle amplification (RCA) using a TempliPhi 100 kit (GE Healthcare, USA).
The RCA product was then used to generate Illumina sequencing libraries (2×150bp) and sequenced on either Illumina 4000 or NextSeq500 sequencers.
De novo assemblies from high-throughput sequencing and identification of fish papillomaviruses
Sequence reads were filtered for quality, trimmed using Trimmomatic v0.39 (Bolger et al., 2014) or FastP (Chen et al., 2018), and then de novo assembled using metaSPades v3.12 (Bankevich et al., 2012) or Megahit (Li et al., 2015). Contigs >1000 nts were analyzed and processed through the Cenote-Taker, virus discovery and annotation pipeline (Tisza et al., 2020). An un-annotated papillomavirus sequence fragment (GenBank accession number FLSL01000248) was detected in a sea bream metagenomic survey. A complete circular map for the virus was curated back to parent read sets (BioProject PRJEB7439) using CLC Genomics Workbench v.21 (http://www.clcbio.com/products/clc-genomics-workbench/).
Genome amplification and verification
Antarctic fish samples
A 5,752 nt contig assembled from the viral DNA of the emerald notothen stomach sample displayed similarities to papillomavirus sequences. This contig was determined to be circular based on terminal redundancy resulting in a putative 5675 nt circular molecule sequence. Based on the sequence of this contig, back-to-back primers in the L1 gene region were designed (BiS_F: 5-’AAC GAC ATG CTA CTG GTA TCA GAC ATC TGG-3’; BiS_R: 5’-CAA TGA TCA TGA AGT TGG AGT CTC CAG CAT C-3’) and used in a polymerase chain reaction (PCR) to recover and verify the papillomavirus genome sequence. The PCR reaction was performed using Kapa HiFi polymerase (Kapa Biosystems, USA) as per manufacturer’s recommendations. The amplicon was run on a 0.7% agarose gel, excised, purified and cloned using pJET1.2 plasmid (Thermo Fisher, USA). The recombinant plasmid was Sanger sequenced at Macrogen Inc (South Korea) by primer walking.
No papillomavirus-like sequences were identified in sharp-spined notothen, crowned notothen or painted notothen tissue samples.
Western gull cloacal swab samples
A 6072 nt contig with similarities to fish papillomaviruses was identified in a Western gull cloacal swab. This contig was determined to be circular based on terminal redundancy resulting in a putative 5994 nt circular molecule sequence. Based on the sequence of this contig, back-to-back primers in the L1 gene region were designed (W11C2_F: 5-’GTC TTC CCG TAA GAC GTG TGG CTG C-3’; W11C2_R: 5-’GCA GGC TGA CTG TGG TGA TTC TTA GGT G-3’) and used to screen the 42 western gull chick swab DNA extracts. Of the 42, only one (sample ID 3_WEGU 2011_C) was found to be positive by PCR using Kapa HiFi polymerase (Kapa Biosystems, USA) as per manufacturer’s recommendations. The amplicon was run on a 0.7% agarose gel, excised, purified and cloned using pJET1.2 plasmid (Thermo Fisher, USA). The recombinant plasmid was Sanger sequenced at Macrogen Inc (South Korea) by primer walking.
Fish papillomavirus annotation, and sequence analysis
A dataset of all previously reported fish-associated papillomaviruses (n=6) was compiled from GenBank (downloaded on 28 June 2021). Open reading frames in the ten papillomaviruses identified in this study were first determined using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) with manual input for putative introns and splice acceptor / donor sites coupled with similarity searches using HHpred (Gabler et al., 2020; Zimmermann et al., 2018). MacVector v. 18.1.5 was used to scan candidate “Oncoid” proteins for the following short linear motifs: Rb interaction: (LI)XCX(ED) or (DEN)(LIMV)XX(LM)(FY)D; casein kinase 2 phosphorylation: (ST)XX(DE); cell division sequence motif (CDSM): DXXCX(TES)X1–8(DE)(DETS)(DE); zinc coordination: CXXC(X4–40C)XXC or CXCXXC; leucine interaction motif: LXXLLX (where X ≠ proline); PDZ interaction motifs: class 1 (STC)X(ACVILF)*, (VLIFY)X(ACVILF)*, class 3 (DE)X(ACVILF) (Kumar et al., 2020). RNA hairpins were predicted using MXfold2 http://www.dna.bio.keio.ac.jp/mxfold2/(Sato et al., 2021).
The pairwise identities of the full papillomavirus genomes and the E1, E2, L1 and L2 amino acid sequences were determined using SDT v1.2 (Muhire et al., 2014). A neighbor-joining phylogenetic tree of the complete papillomavirus genomes was generated. Genomes were aligned with MUSCLE (Edgar, 2004) inferred with Jukes-Cantor substitution model in MEGA 5.2 (Tamura et al., 2013) and midpoint rooted.
Since the E1 protein is the most conserved protein across all papillomaviruses with high similarity in the helicase domain, representative papillomavirus E1 amino acid sequences were downloaded from the Papillomavirus Episteme (PaVE) (Van Doorslaer et al., 2017a) and aligned together with all available fish papillomavirus E1 sequences using MAFFT (Katoh and Standley, 2013). The resulting alignment was trimmed using trimAL (Capella-Gutierrez et al., 2009) with gappyout function and this was then used to infer a maximum-likelihood phylogenetic tree using best substitution model LG+G4 with IQ-Tree v2.1.3 (Minh et al., 2020).
To gain better insights into the evolutionary relationships among fish papillomaviruses, the amino acid sequences of the most conserved proteins, i.e., E1, E2 and L1 of the fish and avian papillomaviruses were concatenated and “block” aligned using MUSCLE (Edgar, 2004). This alignment was then used to infer a maximum-likelihood phylogenetic tree using partition models (Chernomor et al., 2016) Q.yeast+F+I+G4 for the E1 protein, Q.pfam+F+I+G4 for the E2 protein and rtREV+F+G4 for the L1 protein in IQ-Tree v2.1.3 (Minh et al., 2020).
All phylogenetic trees were visualized in iTOL v6 (Letunic and Bork, 2019).
Results and discussion
Discovery of novel fish papillomaviruses
Here we present the genomes of ten new papillomaviruses from emerald notothen (n=1), black sea bass (n=3), haddock (n=4), sea bream (n=1), and from a cloacal swab sample of a western gull (n=1).
The emerald notothen papillomavirus genome (5.6 kb; GenBank accession MZ447865) (Figure 1A and B), was identified and recovered from the stomach tissue of an individual in whose liver we had previously identified a novel polyomavirus (Van Doorslaer et al., 2018b). Simultaneous co-infection with both a papillomavirus and a polyomavirus has been noted previously in a gilthead sea bream (Lopez-Bueno et al., 2016). No papillomaviruses were identified in the other Antarctic fish (sharp-spined, crowned and painted notothens).
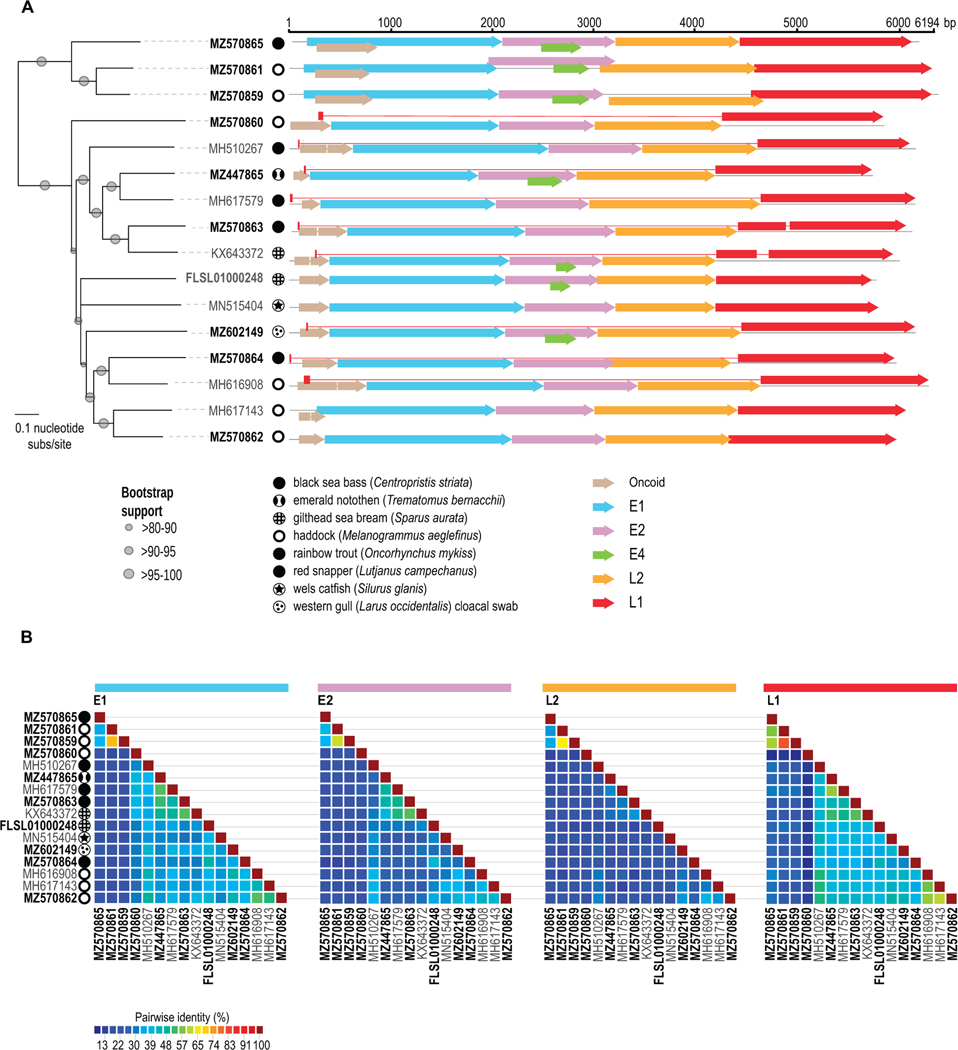
Figure 1:
Phylogenetic and genome analyses of all known fish papillomaviruses. Those recovered in this study are shown in bold. A. Neighbor-joining tree of all fish papillomavirus genomes andtheirgenomic organization, with colored arrows representing inferred protein coding sequences. B. Pairwise comparisons of the E1, E2, L2 and L1 amino acid sequences.
Seven other papillomavirus genomes were identified in sequence data generated from commercial market fish. Three genomes were identified in black sea bass (5.7–6 kb; GenBank accession MZ570863–MZ570865) and four genomes from haddock(5.7–6.1 kb; GenBank accession MZ570859 and MZ570862) (Figure 1A and B). An additional genome was detected in a dataset for a red snapper, but it was nearly identical to a sequence we previously reported from a haddock (MH616908). The red snapper and haddock specimens were purchased from different fish markets and were subjected to sequencing in different runs six months apart. The observation illustrates the problem that surface cross-contamination between different fish species is highly likely in the context of a fish market. It thus not possible to confidently assign host tropism for the market-derived fish papillomaviruses.
One papillomavirus more similar to those from fish compared to those from birds and other terrestrial vertebrates was identified in a cloacal swab from a chick of a seabird, the western gull (6 kb; GenBank accession MZ602149) sampled on the Farallon Islands, USA in 2011. This virus likely originated from a fish that was ingested as part of the bird’s diet (Figure 1).
A TBLASTN survey of the GenBank nr database revealed an un-annotated papillomavirus-like sequence sea bream (Sparus aurata) dataset (5.6 kb; GenBank accession FLSL01000248; BioProject PRJEB7439; BioSample: SAMEA2826833) (Figure 1).
To gain a better understanding of the genome organization of all the fish papillomaviruses we re-analyzed previously published fish papillomavirus sequences together with the ones we report in this study. Several papillomaviruses identified in commercial market rainbow trout, red snapper, and haddock had previously been annotated using an automated computational method, Cenote-Taker v1 (Tisza et al., 2020). Manual re-annotation confirmed the presence of the major ORFs encoding the canonical E1, E2, L2, and L1 proteins in these samples (Figure 1).
In well-studied mammalian papillomaviruses, L1 mRNAs typically initiate within the E7 ORF and a large intron encompassing the entire early region and L2 is spliced out. This positions a highly conserved ATG codon found near the 5’ end of the L1 ORF near the 5’ end of the L1 mRNA.
The L1 ORFs of many fish papillomaviruses lack a suitable ATG initiator codon at the 5’ end of the L1 ORF. It has been proposed that some bird papillomaviruses, such as puffin papillomavirus 1, initiate translation of their L1 protein from a non-canonical GTG initiation codon (Canuti et al., 2019). This is a surprising proposition, in the sense that the major capsid protein must be expressed at high levels in the late phase of the papillomavirus life cycle and GTG initiation codons are not used efficiently (Kearse and Wilusz, 2017). Inspection of the puffin papillomavirus 1 map suggests the alternative explanation that a traditional ATG initiation codon might be encoded on the first exon of a hypothetical L1 mRNA, creating a novel L1 leader peptide at map positions 7600–7662. We invoke a similar solution to the L1 initiation codon puzzle for several fish papillomaviruses (Figure 1). In some cases, the hypothetical L1 initiator ATG codon is generated by splicing.
The L2 minor capsid proteins of mammalian papillomaviruses have an N-terminal polybasic motif, which is cleaved by host furin proteases during the infectious entry process, and a conserved Cys-X5-Cys motif (Richards et al., 2006). In all available fish papillomavirus L2 protein sequences, the familiar di-cysteine motif is found near the C-terminus and a potential polybasic furin cleavage site is located between the two cysteines (Duckert et al., 2004; Gabler et al., 2020; Zimmermann et al., 2018).
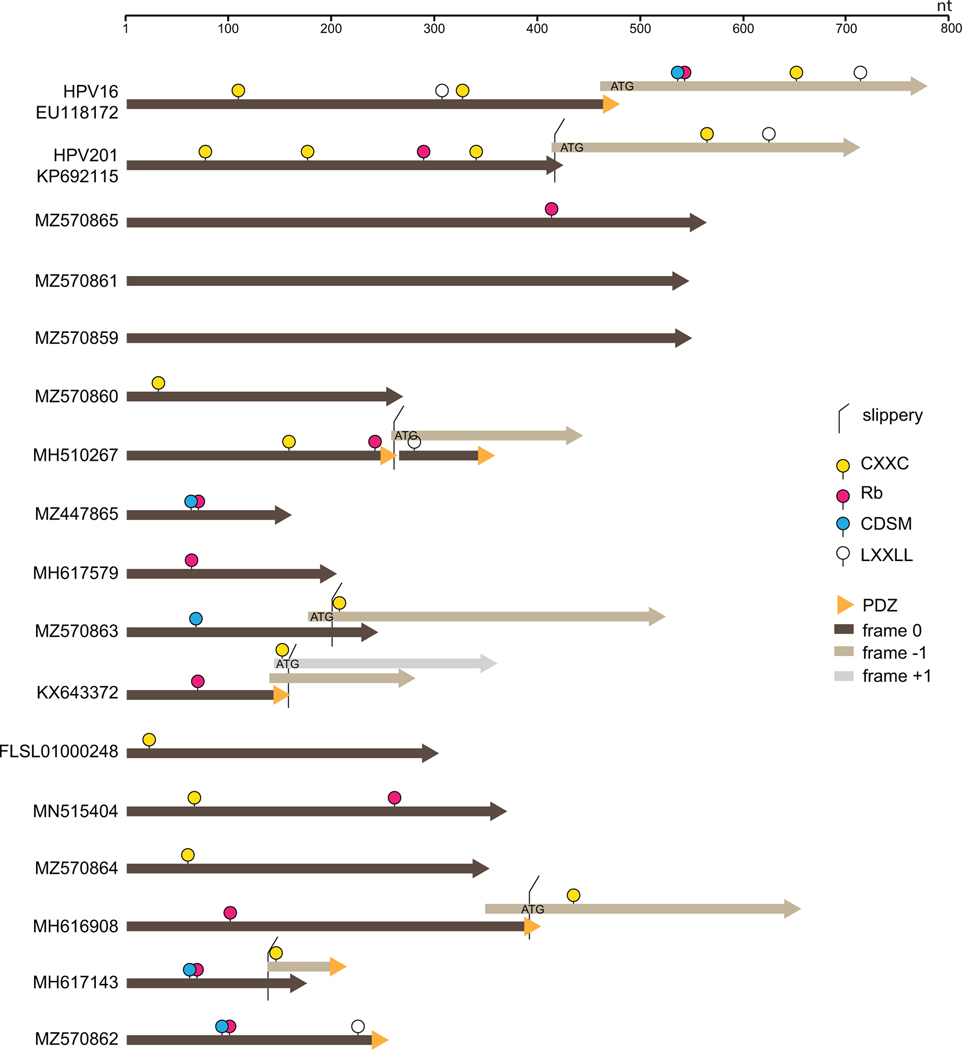
Cancer-causing HPVs encode two oncogenes, E6 and E7. A hallmark feature of E6 is a C-terminal PDZ-interaction motif and a hallmark feature of E7 oncogenes is an LXCXE motif followed by a potential casein kinase 2 (CK2) phosphorylation site that drives interactions with Rb and related tumor suppressor proteins (Suarez and Trave, 2018). The Rb-interaction motif often overlaps a cell-division sequence motif (Figge and Smith, 1988). The core fold of both E6 and E7 is anchored by sets of paired CXXC motifs that coordinate a zinc ion, supporting speculation that the two genes might have arisen through duplication of a single ancestral zinc-binding protein (Van Doorslaer, 2013). This hypothesis has been difficult to explore because E6 and E7 share no discernible primary sequence similarity with one another beyond their shared CXXC motifs. There is also staggering sequence diversity within each gene class. In an arbitrary example, the E6 proteins of Alphapapillomavirus HPV16 (EU118173) and Gammapapillomavirus HPV201 (KP692115) share only 26% amino acid identity. Moreover, the HPV201 E6 protein lacks the hallmark PDZ-interaction motif and instead encodes a hallmark pRb interaction motif that is missing from HPV201 E7 (Figure 2).
Figure 2:
Short linear motifs observed in candidate “Oncoid” proteins. Potential protein sequences encoded by open reading frames found near the 5’ end of the E1 ORF were scanned for the presence of various short linear peptide motifs of interest (see Materials and Methods for search syntax). The E6 and E7 oncoproteins of HPVs 16 and 201 are shown for reference.
All observed fish papillomaviruses encode at least one short ATG-initiated ORF near the 5’ end of the E1 ORF (Figure 1). In light of the poor conservation of HPV E6/E7 proteins, it was unsurprising that attempts to align protein sequences encoded by the E6/E7-syntenic fish papillomavirus ORFs against known terrestrial papillomavirus oncogenes did not reveal discernible similarities. We therefore resorted to scanning the genes for short linear peptide motifs. About half of the candidate fish papillomavirus “Oncoid” genes encode either paired CXXC motifs or a Rb-interaction motif or both (Figure 2). In several instances (e.g., MH617143 MH617579, and MZ447865) the LXCXE-CK2-CXXC segment gives high-probability hits for the solved structure of HPV16 E7 in HHPred searches (Gabler et al., 2020; Zimmermann et al., 2018).
A haddock-associated papillomavirus, MH617143, encodes a downstream Oncoid ORF with a potential zinc-coordinating motif and a C-terminal PDZ interaction motif. There are no suitable splice acceptor signals that would potentiate expression of the downstream ORF as a second exon. In the specific cases of sea bass and sea bream papillomaviruses (MZ570863 and KX643372, respectively) the overlap between the upstream ATG-initiated Oncoid ORF and an overlapping downstream ORF have a −1 frame relationship with a TTTAAAC “slippery” motif in the overlap as well as a predicted RNA pseudoknot just downstream of the first ORF stop codon (Figure 2). These features closely resemble the organization of the programmed −1 ribosomal frameshifting machinery that promotes expression of the fused ORF1ab polyprotein in coronaviruses (Bhatt et al., 2021; Brierley et al., 1992). We hypothesize that fish papillomavirus species with apparently split Oncoid ORFs might express a single fused Oncoid protein via translational frameshifting. Interestingly, nearly all HPVs exhibit a −1 overlap between the E6 and E7 ORFs and, in some cases, the overlap includes a canonical TTTAAAC slippery motif and a predicted RNA pseudoknot just downstream of the E6 stop codon. It would be interesting to experimentally test the hypothesis that some HPVs express a fused E6:E7 protein via programmed −1 ribosomal frameshift mechanism (Harger et al., 2002).
Phylogenetic and genetic similarity analyses
The genome-wide phylogeny showed two major fish papillomavirus clades, one containing emerald notothenpapillomavirus and another comprised of two haddock-associated papillomaviruses (MZ570861 and MZ570859) and one sea bass papillomavirus (MZ570865) (Figure 1A). A distinctive feature of the second clade is that the candidate Oncoid ORF is overprinted in the +1 frame of the E1 ORF (Figure 1). The smaller clade comprising three viruses, with overprinted Oncoid genes, share 59–75% genome-wide similarity (Supplementary data 1). Members of the larger clade, which houses all other known fish papillomavirus genomes, share 58–66% genome-wide similarity within the group and 57–60% between members of the two groups. The E1 and L1 genes are the most conserved genes among all the terrestrial vertebrate papillomaviruses and this is also the case for the fish group (Figure 1). The four core proteins share the following pairwise identities among the fish papillomavirus group: E1: 23–73%, E2:21–65%, L2: 17–83% and L1: 21–69% (Figure 1B).
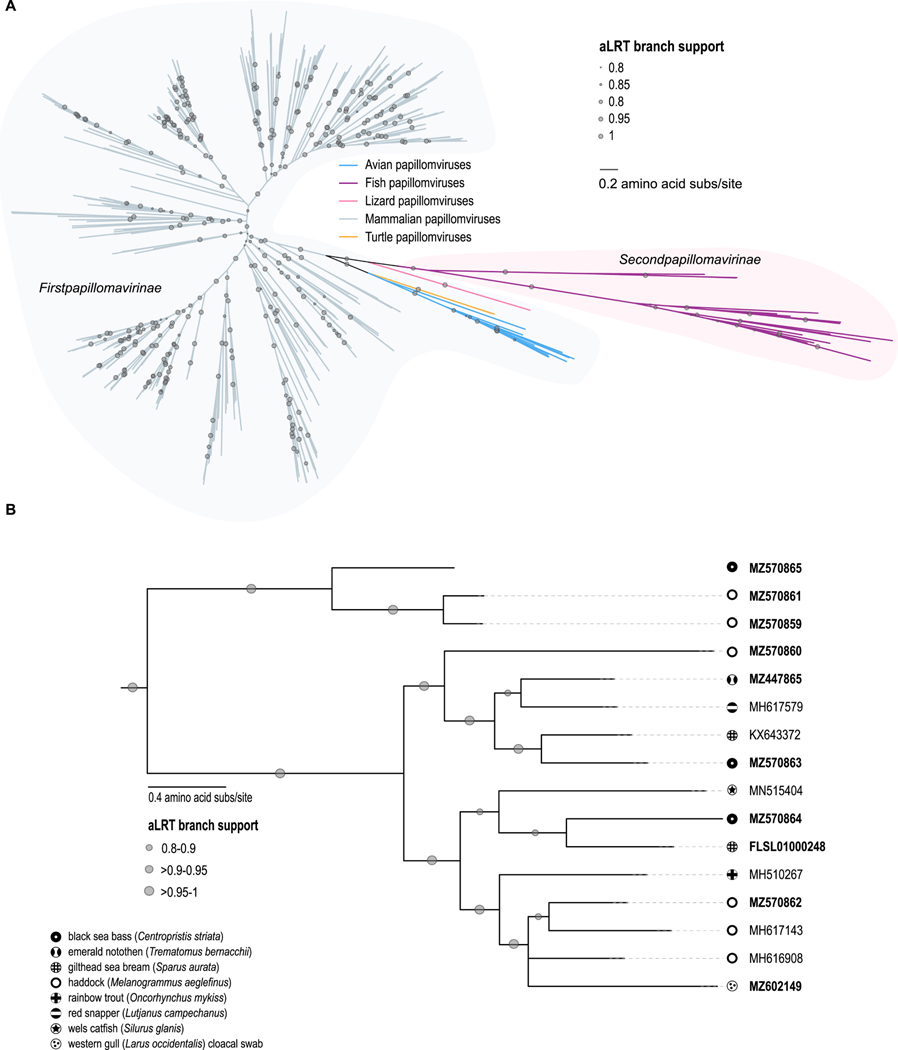
Analysis of E1 proteins from papillomaviruses across all vertebrate host groups revealed that papillomaviruses from fish form a distantly related group of viruses sister to avian and reptile papillomaviruses (Figure 3A). A maximum likelihood phylogeny of the E1, E2 and L1 proteins from all fish papillomaviruses (Figure 3B), rooted with the avian papillomaviruses, confirmed the two major fish papillomavirus clades observed in the genome-wide phylogeny (Figure 1A). It should be noted that the Western gull cloacal swab-derived papillomavirus is most closely related to papillomaviruses from haddock, gilthead sea bream and black seabass (Figure 1A and 3B), neither of which are found in the Farallon Island foraging area. Fish that are part of the primary diet of Western gulls are northern anchovy (Engraulis mordax), juvenile rockfish (Sebastes spp.), and other observed fish prey include Pacific whiting (Merluccius productus), Jack mackerel (Trachurus symmetricus), Pacific saury (Cololabis saira), midshipman (Poricthys spp.), white croaker (Genyonemus lineatus), spotted cusk eel (Chilara taylori), and jacksmelt (Atherinopsis californiensis) (Ainley, 1990). Papillomaviruses from the same host species do not always cluster together, for example the three isolates recovered from sea bass resolved in separate clades. A similar situation was also observed for the haddock papillomaviruses. However, an important caveat is that the sea bass and haddock samples were collected from a commercial market and the fish samples might have been cross contaminated by serial handling of different fish species. This caveat is less likely to apply to the two highly divergent papillomaviruses identified in single-species surveys of sea bream (KX643372 and FLSL01000248). Based on current data the fish papillomavirus phylogeny appears to not follow the host phylogeny given the multiple placements of viruses from a single host across the papillomavirus phylogeny, and therefore infers that host switching, recombination and/or multiple within host divergence events have occurred. A phenomenon that has been observed in other papillomavirus groups such as those from Weddell seals (Smeele et al., 2018).
Figure 3:
Phylogenetic analyses of core fish papillomavirus proteins. A. Unrooted Maximum likelihood phylogeny of E1 proteins from all known papillomaviruses highlighting the highly divergent fish papillomavirus sub-family (Secondpapillomavirinae) compared to the terrestrial vertebrate papillomavirus sub-family (Firstpapillomavirinae). B. Maximum-likelihood phylogeny of concatenated E1, E2 and L1 amino acid sequences of the fish papillomaviruses and rooted with the avian papillomavirus sequences.
Concluding remarks
Here we report nine novel fish papillomaviruses derived directly from fish tissues, one of which is the first identification of a fish papillomavirus in an Antarctic fish, and an additional papillomavirus from a western gull cloacal sample that groups with the fish papillomaviruses and was likely diet derived. These papillomaviruses belong to the Secondpapillomavirinae subfamily. Within an Antarctic context, this is the third animal species in which papillomaviruses have so far been identified; the others were firstpapillomaviruses found in Adélie penguin (Pygoscelis adeliae) (Van Doorslaer et al., 2017b; Varsani et al., 2014) and Weddell seal (Leptonychotes weddellii) (Smeele et al., 2018). It is apparent from the sample set described here that fishpapillomaviruses are highly diverse and likely present in diverse forms in many more, yet unsampled, fish species. Current genome sequence data suggest that all fish papillomaviruses derive from a common ancestor and are highly divergent from other papillomaviruses, however, more investigation on fish, and early diverging vertebrates such as sharks and rays as well as early branching terrestrial vertebrates such as amphibians, reptiles, and birds, is needed to gain insight into the origins, evolutionary relationships, and disease outcomes of the fish papillomaviruses in the context of the entire papillomavirus family.
Supplementary Material
Identification and genomic characterization of ten novel fish papillomaviruses
Identification of open reading frames with features reminiscent of E6/E7 oncoproteins
Fish papillomaviruses are highly diverse
Acknowledgements
The authors are grateful to Alison McBride, and Karl Munger for their advice on papillomavirus oncogenes. The field work in the Ross Sea was supported by a grant (K057) awarded to WD from Antarctica New Zealand. The field work on the West Antarctic Peninsula was supported by the National Science Foundation (NSF) polar program grant National Science Foundation grant OPP-1543383 (JHP, TD). The molecular work described in this study for the fish from Ross Sea was supported by the Center of Evolution and Medicine Venture Fund (Center of Evolution and Medicine, Arizona State University, USA) grant awarded to AV. The molecular data acquisition on fish from the West Antarctic Peninsula was supported by the NSF polar grant (OPP-1947040) awarded to TD, AV and JHP. In addition, the NSF polar grant (OPP-1947040) also partially supported SK, TD, JHP, RSF and AV. The commercial market fish work was funded in part by the NIH Intramural Research Program, with support from the NCI Center for Cancer Research awarded to CB. The collection of cloacal swabs from Western gulls was conducted by Point Blue Conservation Science with the support of the California Academy of Sciences and the U.S. Fish and Wildlife Service under cooperative agreement number 81640–5-J046.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agius JE, Phalen DN, Rose K, Eden JS, 2019. New insights into Sauropsid Papillomaviridae evolution and epizootiology: discovery of two novel papillomaviruses in native and invasive Island geckos. Virus Evol 5, vez051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainley DG, 1990. The feeding ecology of Farallon seabirds. Seabirds of the Farallon Islands, Ecology, Dynamics and Structure of an Upwelling-system Community. [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA, 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt PR, Scaiola A, Loughran G, Leibundgut M, Kratzel A, Meurs R, Dreos R, O’Connor KM, McMillan A, Bode JW, Thiel V, Gatfield D, Atkins JF, Ban N, 2021. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Jenner AJ, Inglis SC, 1992. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol 227, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuti M, Munro HJ, Robertson GJ, Kroyer ANK, Roul S, Ojkic D, Whitney HG, Lang AS, 2019. New Insight Into Avian Papillomavirus Ecology and Evolution From Characterization of Novel Wild Bird Papillomaviruses. Front Microbiol 10, 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T, 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J, 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ, 2016. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst Biol 65, 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P, Brunak S, Blom N, 2004. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 17, 107–112. [DOI] [PubMed] [Google Scholar]
- Edgar RC, 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge J, Smith TF, 1988. Cell-division sequence motif. Nature 334, 109. [DOI] [PubMed] [Google Scholar]
- Frias-De-Diego A, Jara M, Escobar LE, 2019. Papillomavirus in Wildlife. Frontiers in Ecology and Evolution 7, 406. [Google Scholar]
- Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Soding J, Lupas AN, Alva V, 2020. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 72, e108. [DOI] [PubMed] [Google Scholar]
- Gull JM, Lange CE, Favrot C, Dorrestein GM, Hatt JM, 2012. Multiple papillomas in a diamond python, Morelia spilota spilota. J Zoo Wildl Med 43, 946–949. [DOI] [PubMed] [Google Scholar]
- Harger JW, Meskauskas A, Dinman JD, 2002. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem Sci 27, 448–454. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE, 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubacki J, Ramsauer AS, Bachofen C, Favrot C, Nicolier A, Fraefel C, Tobler K, 2018. Complete Genome Sequence of a Boa (Boa constrictor)-Specific Papillomavirus Type 1 Isolate. Microbiol Resour Announc 7, e01159–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Gouw M, Michael S, Samano-Sanchez H, Pancsa R, Glavina J, Diakogianni A, Valverde JA, Bukirova D, Calyseva J, Palopoli N, Davey NE, Chemes LB, Gibson TJ, 2020. ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res 48, D296–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P, 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, Lam TW, 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. [DOI] [PubMed] [Google Scholar]
- Lopez-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, Alejo A, 2016. Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream. J Virol 90, 8768–8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, 2020. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire BM, Varsani A, Martin DP, 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9, e108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierotti RJ, Annett CA, 1995. Western Gull: Larus Occidentalis. American Ornithologists’ Union. [Google Scholar]
- Prosperi A, Chiari M, Zanoni M, Gallina L, Casa G, Scagliarini A, Lavazza A, 2016. Identification and Characterization of Fringilla coelebs Papillomavirus 1 (FcPV1) in Free-living and Captive Birds in Italy. J Wildl Dis 52, 756–758. [DOI] [PubMed] [Google Scholar]
- Richards RM, Lowy DR, Schiller JT, Day PM, 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103, 1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Akiyama M, Sakakibara Y, 2021. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat Commun 12, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeele ZE, Burns JM, Van Doorsaler K, Fontenele RS, Waits K, Stainton D, Shero MR, Beltran RS, Kirkham AL, Berngartt R, Kraberger S, Varsani A, 2018. Diverse papillomaviruses identified in Weddell seals. J Gen Virol 99, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez I, Trave G, 2018. Structural Insights in Multifunctional Papillomavirus Oncoproteins. Viruses 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjan A, Fonagy E, Eszterbauer E, Harrach B, Doszpoly A, 2021. Complete genome sequence of a novel fish papillomavirus detected in farmed wels catfish (Silurus glanis). Arch Virol 166, 2603–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang YS, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta’ala BA, Pletnikova O, Troncoso JC, Resnick SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB, 2020. Discovery of several thousand highly diverse circular DNA viruses. Elife 9, e51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchado DA, Moens MAJ, Callejas S, Perez-Tris J, Benitez L, 2018. Genomic characterization of the first oral avian papillomavirus in a colony of breeding canaries (Serinus canaria). Vet Res Commun 42, 111–120. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, 2013. Evolution of the papillomaviridae. Virology 445, 11–20. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, Chen Z, Bernard HU, Chan PKS, DeSalle R, Dillner J, Forslund O, Haga T, McBride AA, Villa LL, Burk RD, Ictv Report C, 2018a. ICTV Virus Taxonomy Profile: Papillomaviridae. J Gen Virol 99, 989–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Kraberger S, Austin C, Farkas K, Bergeman M, Paunil E, Davison W, Varsani A, 2018b. Fish polyomaviruses belong to two distinct evolutionary lineages. J Gen Virol 99, 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, Sun Q, Kaur R, Huyen Y, McBride AA, 2017a. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 45, D499–D506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Ruoppolo V, Schmidt A, Lescroel A, Jongsomjit D, Elrod M, Kraberger S, Stainton D, Dugger KM, Ballard G, Ainley DG, Varsani A, 2017b. Unique genome organization of non-mammalian papillomaviruses provides insights into the evolution of viral early proteins. Virus Evol 3, vex027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsani A, Kraberger S, Jennings S, Porzig EL, Julian L, Massaro M, Pollard A, Ballard G, Ainley DG, 2014. A novel papillomavirus in Adelie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J Gen Virol 95, 1352–1365. [DOI] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V, 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430, 2237–2243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.