Abstract
Background.
The staging of pancreatic neuroendocrine tumors (PNETs) is continuously evolving. Mitotic count, as measured by hematoxylin and eosin (H&E) or Ki67 labeling index (Ki67LI), is the best predictor of disease biology. However, both of these methods have several limitations. Phosphorylated histone H3 (PHH3), a novel mitotic marker, is potentially more accurate and easier to evaluate. This study aimed to evaluate the prognostic impact of PHH3 on patients with PNETs.
Methods.
Clinicopathologic data and paraffin-embedded tissue were evaluated for 100 of the 247 PNET patients whose tumors were resected between 1998 and 2010. Mitotic counts were analyzed on H&E-, Ki67-, and PHH3- stained slides by two independent pathologists. Kaplan–Meier curves, log-rank tests, Cox regression models, and time-dependent receiver operative characteristics (ROC) curves were used to evaluate the prognostic power of these markers. An internal data cross-validation was performed to select the best cutoff.
Results.
Of the 100 PNET patients resected, 53 were men. The median age of the patients was 59 years (range 19–96 years). The median follow-up period was 68 months (range 3–186 months). The median time for evaluation of an H&E- or PHH3-stained slide was 3 min, relative to 15 min for Ki67. The findings showed H&E, Ki67, and PHH3 all to be excellent predictors of disease-specific survival (DSS). However, PHH3 was superior to H&E and Ki67 in predicting both disease-free survival (DFS) (p = 0.006) and DSS (p = 0.001). Evaluation of the PHH3 mitotic count showed 7 mitoses per 10 high-power fields (HPFs) to be the optimal cutoff for differentiating between low- and high-risk PNET patients.
Conclusions.
PHH3 is a better predictor of both DFS and DSS than H&E or Ki67 in PNET. In addition, PHH3 appears to be both easier to interpret and more accurate when compared to current prognostic markers.
Pancreatic neuroendocrine tumors (PNETs) are uncommon neoplasms, which account for approximately 3 % of all pancreatic malignancies.1,2 The majority of PNETs are nonfunctional and incidentally discovered.3,4 The incidence of PNETs has been trending up during the last few decades, which can be attributed in part to technological advances as well as to a more extensive use of imaging in clinical practice.5
The staging and grading of PNETs is continuously evolving6. Recently, the 2010 World Health Organization (WHO) proposed a revised grading system,7 classifying PNETs into three categories based on mitotic count performed on hematoxylin-eosin (H&E)-stained slides and the Ki67 labeling index. The grade is defined by the highest proliferation index. Neuroendocrine tumors (NETs) are graded as G1 [<2 mitoses per 10 high-powered fields (HPFs) and/or a Ki67 labeling index of 3 % or less] or G2 (2–20 mitoses and/or a Ki67 labeling index of 3–20 %). Neuroendocrine carcinomas (NECs) are defined as 21 or more mitoses and/or a Ki67 labeling index greater than 20 %.
Different staging systems have been proposed by the American Joint Committee on Cancer/International Union Against Cancer (AJCC/UICC) and the European Neuroendocrine Tumour Society (ENETS).8,9 The AJCC/UICC system is based on the tumor-node-metastasis (TNM) criteria used for ductal adenocarcinoma of the pancreas. This classification defines localized pNETs as stage 1, locally advanced resectable pNETs as stage 2, locally advanced unresectable pNETs as stage 3, and pNETs with distant metastases as stage 4.9 The prognostic validity of these systems, however, has not been validated in a large series of patients.
Currently, the WHO classification is the most widely accepted staging system. The proliferative rate is generally representative of the biology of the disease and correlates well with the clinical behavior of PNETs.10 Nevertheless, both methods (H&E mitotic index and Ki67LI) adopted by the WHO to assess proliferative index have significant disadvantages. First, counting mitoses, as identified on H&E-stained slides, is generally regarded as a technique with reduced specificity because early-prophase mitoses cannot be recognized and apoptotic bodies often are mistaken for mitotic figures.10,11 The Ki67LI has different drawbacks. The WHO suggests counting between 500 and 2000 cells to determine the percentage of Ki67-positive cells. This is very labor intensive and typically takes more than 15 min per slide. Second, Ki67 is less reflective of the true proliferation index because it stains not only cells in the M phase but also those in the S, G1, and G2 phases.12
The lack of a specific, time-efficient, and operator-in-dependent method to determine the proliferative rate in PNETs prompted us to explore alternative prognostic techniques. Phosphorylation of the amino terminus of the histone H3 (Ser 10) correlates closely with the M phase of the cell cycle.13 For this reason, antiphosphorylated histone H3 (PHH3) is considered a mitosis-specific marker, and anti-PHH3 has been used to estimate proliferation in a variety of neoplasms including meningiomas, astrocytomas, melanomas, and pulmonary carcinoids.14-18
Recently, findings have shown the proliferation index as measured by PHH3 to be comparable with the current grading techniques in PNET.19 In this study, we hypothesized that PHH3 represents a superior prognostic marker in patients with PNETs. Therefore, the first aim of this study was to determine the accuracy of PHH3 mitotic counts as a more time-efficient and specific prognostic marker in PNET than Ki67Li or mitotic counts on H&E. The second aim was to propose a new clinically relevant PNET classification based on specific cutoffs for PHH3.
METHODS
Patients
Institutional Review Board approval at the Massachusetts General Hospital was obtained for this study. Between 1998 and 2010, 247 patients with PNETs were surgically resected at the Massachusetts General Hospital. Paraffin-embedded tumor blocks were available for 100 of these patients. Clinicopathologic data for each patient were obtained from the hospital’s database including age, gender, size of the tumor, type of operation, presence of metastases at the time of resection, disease-free survival, and overall survival.
Histology
In this study, 5-μm sections were cut from paraffin-embedded tumor blocks. Slides were stained using standard immunohistochemical techniques described elsewhere.20 Rabbit monoclonal antihistone H3 (1:5000, abcam #ab32107) and rabbit monoclonal anti-Ki67 (Thermo #RM-9106-R7) were used as primary antibodies. A horseradish peroxidase anti-rabbit secondary antibody was used (Dako, Carpinteria, CA, USA, #K4003), and the signal was visualized using a 3,3′-diaminobenzidine (DAB) detection kit (Dako), with procedures performed according to the manufacturer’s instructions. Human tonsil was used as a positive control in all immunohistochemical reactions.
Slide Scoring
Mitotic Count Analysis (H&E and PHH3)
Mitotic counts were independently analyzed by two pathologists (V.D. and K.K.M.) on H&E and PHH3 IHC slides. The slide with the largest tumor surface area was selected, and 50 HPFs (×400 magnification; field size 0; 196 mm2) per slide were analyzed. The ten fields with the highest number of mitotic figures were identified, and the mitotic rate was expressed as the number of mitotic figures per 10 HPFs. Only cells with metaphase or anaphase morphology were considered positive. The average time for counting mitoses on either H&E or PHH3 slides was 3 min.
Ki67LI Determination
To calculate the Ki67LI, the highest labeling region (hotspot) was identified under low magnification. Two fields within the hotspot then were captured using the SC100 Olympus digital camera system (Olympus America, Center Valley, PA) and printed on A4-size paper using a Hewlett-Packard color printer. At least 500 (range 515–830) nuclei were analyzed, and the Ki67LI was calculated as follows: number of positive Ki67 nuclei/total number of nuclei. Both Ki67-positive lymphocytes and other non-neoplastic cells were excluded from the counts. The average time for determining Ki67LI was 15 min.
Statistical Analysis
Kaplan–Meier curves and log-rank tests were used to analyze patient survival. A Student’s t test was used to correlate tumor grades and nodal status. Multivariate analysis was performed using Cox proportional hazards models. Optimal cutoffs for disease-specific death (DSD) were determined using time-dependent receiver operative characteristics (t-ROC) curves 21 by averaging the false positive plus false-negative rates for each mitotic count of 2–15 mitoses per 10 HPFs. We performed an internal cross-validation by randomly selecting 80 patients and testing which relative minimum false-positive plus false-negative value better divided the patients previously excluded into high- and low-risk classes. This operation was repeated 100 times, and a frequency distribution was calculated and used to propose the optimal cutoff. All statistical analyses were performed using Prism 6.0 for Mac (GraphPad Software, Inc., La Jolla, CA) and R software (R Foundation for Statistical Computing, Vienna, Austria). Results were considered significant at a p value of 0.05 or lower.
RESULTS
Patient Characteristics
The tissues from 100 patients (53 men and 47 women) with a median age of 59 years (range 19–96 years) who underwent surgical resection of a PNET were retrieved for evaluation. Table 1 summarizes the patients’ clinicopathologic characteristics. Of the 100 patients, 87 had nonfunctioning tumors and 13 had functional PNETs. Of the 13 functional PNETs, 11 were insulinomas and 2 were glucagonomas. The median tumor size was 3 cm (range 0.7–16.0 cm). The median follow-up period was 68 months (range 3–186 months). The median overall survival for this patient population has not been reached.
TABLE 1.
Clinical-pathological variables
| H&E mitotic count |
p | Ki67LI |
p | ||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G1 | G2 | G3 | |||
| Age (mean ± SEM) | 59.32 ± 1.6 | 56.58 ± 2.6 | 0.55 | 58.48 ± 1.7 | 59.19 ± 2.3 | 59.67 ± 5.1 | 0.81 |
| Overall = 58.78 ± 1.3 | |||||||
| Tumor size | 34.7 ± 2.5 | 50.6 ± 4.1 | 39.0 ± 3.2 | 38.6 ± 4.5 | |||
| Gender | |||||||
| M (n = 53) | 39 | 14 | 0.39 | 31 | 17 | 5 | 0.3 |
| F (n = 47) | 30 | 17 | 31 | 15 | 1 | ||
| LNM | |||||||
| No (n = 82) | 60 | 22 | 0.08 | 52 | 26 | 4 | 0.78 |
| Yes (n = 12) | 9 | 9 | 10 | 6 | 2 | ||
| Non-secreting | |||||||
| No (n = 87) | 59 | 28 | 0.75 | 54 | 27 | 6 | 0.58 |
| Yes (n = 13) | 10 | 3 | 8 | 5 | 0 | ||
| AJCC | |||||||
| Ia | 18 | 2 | 0.06 | 16 | 4 | 0 | 0.28 |
| Ib | 20 | 8 | 20 | 6 | 2 | ||
| IIa | 19 | 12 | 15 | 14 | 2 | ||
| IIb | 7 | 8 | 7 | 6 | 2 | ||
| IV | 5 | 1 | 4 | 2 | 0 | ||
| Median DFS (years) | Not reached | 5.2 | Not reached | 5.9 | 3.0 | ||
| Median DSS (years) | Not reached | Not reached | Not reached | 12.0 | Not reached | ||
SEM standard error of the mean, LNM lymph node metastasis, AJCC American Joint Committee on Cancer, DFS disease free survival, DSS disease specific death
Grading of Proliferation Using H&E and Ki67LI
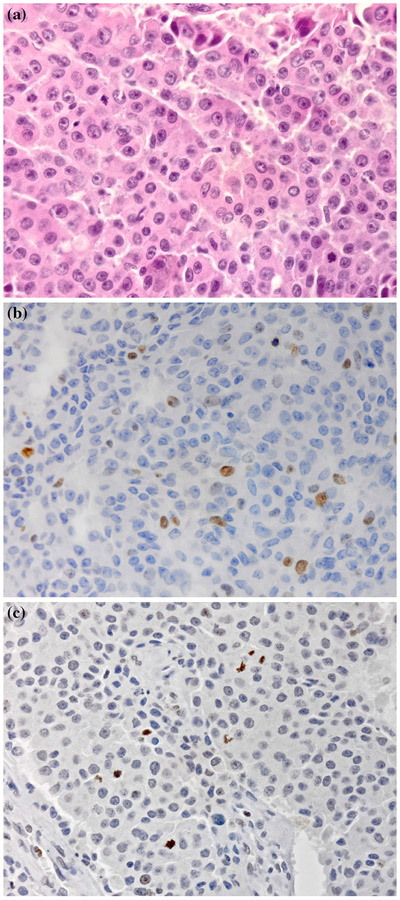
The representative patterns of H&E, Ki67, and PHH3 staining are shown in Fig. 1a-c. Grading using H&E mitotic counts and Ki67LI is summarized in Table 1. Neither technique had a significant correlation with age, gender, nodal status, presence of metastases, or AJCC status. We initially divided the patients into the three risk classes defined by WHO.
FIG. 1.
Representative images of a pancreatic neuroendocrine tumor (PNET) tissue section (×100 magnification) stained with a hematoxylin-eosin (H&E), b Ki67, and c phosphorylated histone H3 (PHH3)
H&E, PHH3, and Ki67LI as Predictors of Disease-Free Survival (DFS)
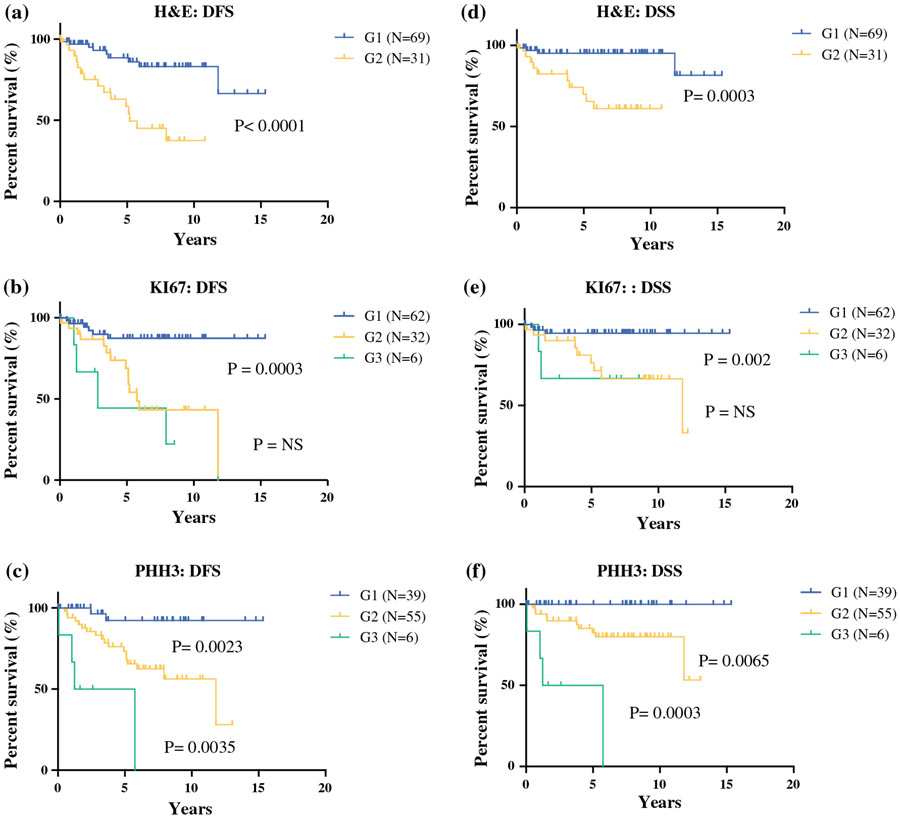
All three methods used in our study proved to be predictive of disease recurrence (G1 vs G2 H&E method, p < 0.0001; G1 vs G2 Ki67 method, p = 0.0003; G2 vs G3 Ki67 method, p = not significant; G1 vs G2 PHH3 method, p = 0.0023; G2 vs. G3 PHH3 method, p = 0.0035) (Fig. 2a-c).
FIG. 2.
Disease-free survival (DFS) of pancreatic neuroendocrine tumor (PNET) patients. The patients were divided into three risk classes (G1, G2, and G3) accordingly to the tumor grade, determined by counting the mitotic figures in a hematoxylin-eosin (H&E)-, b Ki67-, c or phosphorylated histone H3 (PHH3)-stained tumor sections. Survival differences were calculated using Kaplan–Meier curves (log-rank test: p = p value). Disease-specific survival (DSS) of PNET patients. The patients were divided into three risk classes (G1, G2, and G3) according to the tumor grade, determined by counting the number of mitotic figures in d H&E-, e Ki67-, or f PHH3-stained tissue sections. Survival differences were calculated using Kaplan–Meier curves (log-rank test, p = p value)
H&E, PHH3, and Ki67LI as Predictors of Disease-Specific Survival (DSS)
To evaluate the prognostic significance of the methods used, we analyzed the DSS using Kaplan–Meier curves and the log-rank test (Fig. 2). The proliferation indices evaluated by H&E, Ki67, and PHH3 all were excellent predictors of patient survival (G1 vs G2 H&E method, p = 0.0003; G1 vs G2 Ki67 method, p = 0.002; G2 vs G3 Ki67 method, p = not significant; G1 vs G2 PHH3 method, p = 0.0065; G2 vs G3 PHH3 method, p = 0.0003) (Fig. 2d-f).
Multivariate Analysis Evaluating the Prognostic Power of PHH3 Compared With H&E and Ki67
We tested whether PHH3 was a stronger prognostic tool for predicting DFS and DSS than H&E or Ki67. Applying a Cox regression model, PHH3 better predicted both DFS (p = 0.006) and DSS (p = 0.001). Furthermore, we were able to demonstrate similar results when the Akaike Information Criterion (AIC) was used (Table 2). In the multivariate analysis, only PHH3 and nodal status were statistically significant predictors of both DFS (Table 3) and DSS (Table 4).
TABLE 2.
Multivariate analysis to compare prognostic power of PHH3 versus H&E + Ki67
| HE + ki67 | PHH3 | |
|---|---|---|
| DSS | 89.82325 | 84.27353 |
| DFS | 85.50381 | 83.5987 |
Akaike Information Criterion was used to compare the prognostic power of PHH3 compared to both H&E and Ki67. In this statistical model, lower numbers represent a superior prognostic power
TABLE 3.
Multivariate analysis to predict DFS
| Coef | exp (coef) | se (coef) | z | p value | |
|---|---|---|---|---|---|
| PHH3 | 0.23479 | 1.26465 | 0.06404 | 3.666 | 0.000246 |
| N | 3.4546 | 31.64569 | 1.00423 | 3.44 | 0.000582 |
| Secreting | 1.44525 | 4.24293 | 0.98396 | 1.469 | 0.141884 |
| Age | −0.03284 | 0.96769 | 0.03055 | −1.075 | 0.282335 |
| Size | −0.00237 | 0.99763 | 0.01174 | −0.202 | 0.839963 |
Cox-regression analysis to compare the prognostic power of PHH3 versus clinical variables
TABLE 4.
Multivariate analysis to predict DSS
| Coef | exp (coef) | se (coef) | z | p value | |
|---|---|---|---|---|---|
| PHH3 | 0.235584 | 1.265647 | 0.062085 | 3.795 | 0.000148 |
| N | 3.366723 | 28.983406 | 1.00343 | 3.355 | 0.000793 |
| Secreting | 1.419066 | 4.13326 | 0.950097 | 1.494 | 0.13528 |
| Age | −0.028037 | 0.972352 | 0.030892 | −0.908 | 0.364084 |
| Size | −0.004137 | 0.995871 | 0.012639 | −0.327 | 0.743421 |
Cox-regression analysis to compare the prognostic power of PHH3 versus clinical variables
Determination of an Optimal Cutoff Using PHH3
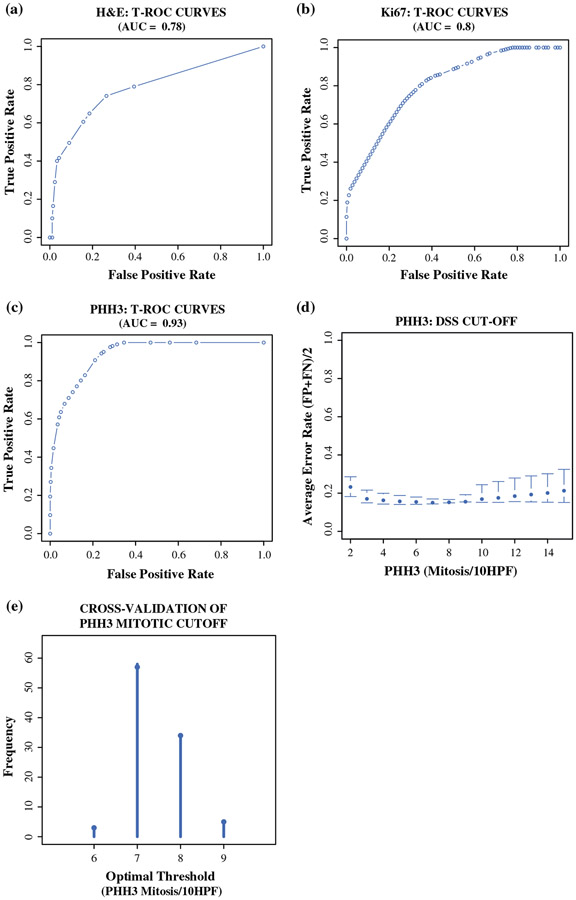
Using time-dependent receiver operative characteristics (t-ROC) curves, we evaluated all three markers of proliferation. The findings showed that PHH3 had the highest sensitivity and specificity with a greatest area under the curve (AUC) value (PHH3 AUC, 0.93; H&E AUC, 0.78; Ki67 AUC, 0.80).
Because PHH3 was found to be more efficient and more specific, we aimed to determine prognostic PHH3 cutoffs for predicting DFS and DSS. Using t-ROC curves, we were able to determine an optimal mitotic cutoff of 0–15 mitoses per 10 HPFs (Fig. 2d) and to maximize the sensitivity and specificity in predicting the DSS 10 years after surgical resection. By minimizing the sum of the false-positive and false-negative rates obtained by the PHH3 t-ROC curve, we obtained a distribution of candidate cutoffs for distinguishing between low- and high-risk patients (Fig. 3d). A relative minimum was observed within a range of 6–9 mitoses per 10 HPFs, whereas below and above this range, the average of false-positive plus false-negative rates increased, as shown in Fig. 3d.
FIG. 3.
a-c Time-dependent receiver operative characteristics (t-ROC) curves were used to determine the sensitivity and specificity of hematoxylin-eosin (H&E)-, Ki67-, and phosphorylated histone H3 (PHH3) in predicting long-term survival (10 years). The area under the curve (AUC) in each graph estimates the prognostic power of each marker. d The optimal cutoff for the novel grading system proposed for pancreatic neuroendocrine tumor (PNET) patients was determined by plotting the mean of false-positive (FP) and false-negative (FN) rates for each mitotic count from a range of 2–15 mitoses per 10 high-power fields (HPFs) counted in PHH3-stained slides. The range of 6–9 mitoses per 10 high-power fields (HPFs) is the range that minimizes the average of FP and FN rates. e To find the optimal cutoff inside, an internal cross-validation was applied, which showed 7 mitoses per 10 HPFs to be the most frequent cutoff
To assess the best cutoff in our cohort of patients, we randomly selected 80 of the 100 cases and then tested its prognostic power on those patients previously excluded. We repeated this process 100 times to achieve a robust internal cross-validation simulating an external patient cohort. Figure 3e represents the frequency distribution of all the potential cutoffs observed in our simulation. We found 7 mitoses per 10 HPFs to be the most frequent cutoff observed, followed by 8 mitoses per 10 HPFs. Based on these data, we propose 7 mitoses per 10 HPFs as the optimal prognostic cutoff for PNET patients.
DISCUSSION
Controversy surrounds the management of pancreatic neuroendocrine tumors, especially incidentally detected, relatively small, nonfunctioning PNETs. It is not always clear which of these neoplasms should be resected. The timing and frequency of post-resection surveillance also are debated.
Currently, follow-up management and surveillance is guided by the WHO classification of PNETs. The WHO classification requires accurate mitotic counts, typically based on examination of H&E-stained slides. However, this platform has the disadvantage that reliable identification of mitotic figures on H&E is challenging even for an experienced pathologist because apoptotic bodies can be mistaken for mitoses.
The other method advocated by the WHO for grading PNETs is the Ki67LI, which is considered complementary to counting mitoses on H&E. Although the WHO does not endorse the one technique over the other, it strongly recommends that the higher grade should be used, and thus the use of both techniques is encouraged. However, Ki67LI is significantly more time-consuming than H&E or PHH3. For Ki67LI to be accurate, at least 500 nuclei must be counted. Whether this is performed manually or with an automated system, a minimum of 10–15 min is needed per clinical case. This makes the method impractical for busy pathologists. Although automated platforms may considerably shrink the time required for evaluation, the current algorithms do not distinguish tumor cells from Ki67-positive lymphocytes and other non-neoplastic cells. Thus, an experienced operator is critical to the analysis. Furthermore, although automated systems have been available for Almost a decade, we know of no institution in the United States that has adopted this technique (personal communications).
The anti-PHH3 antibody is able to overcome many of these shortcomings. By specifically staining phosphorylated H3, a protein detected only during the M phase, it eliminates the ambiguity of H&E scoring. Furthermore, PHH3 brightly stains cells in mitosis but not apoptotic cells. Due to its higher specificity and the excellent visual contrast between positive and negative cells, reading mitotic counts with PHH3 is both easier and less time consuming than standard H&E staining. The PHH3 and H&E evaluations were both performed in approximately 3 min. However, the predictive value of PHH3 was superior to that of H&E staining. In comparison, the Ki67 counts required 15 min on average, again with a predictive value inferior to that of PHH3.
In addition to its improved specificity for staining mitotic cells, the PHH3 mitotic rate count has a low interobserver variation. This has been observed in a number of studies, including a recent investigation of PNETs that reported almost perfect (k > 0.98) interobserver agreement.19 More importantly, as a consequence of its high specificity for mitoses, PHH3 is a more accurate representation of the tumor’s biology. This is likely to be the basis for its superior performance in predicting patient survival.
The importance of a grading system resides in its clinical utility. This study aimed to identify a platform that is easy to use and reproducible, but also one that can reliably guide therapeutic management. The more extensive use of imaging techniques has increased the number of PNETs diagnosed incidentally3. The management of small, asymptomatic nonfunctional PNETs is debatable. On the one hand, a curative resection, albeit curative, comes with potentially significant risks. On the other hand, it remains difficult to identify tumors with indolent biology on the basis of serial imaging studies and fine-needle aspiration (FNA).
Currently, the size of the PNET is the most widely adopted criterion for determining which patients should undergo pancreatic resection or enucleation. The National Comprehensive Cancer Network (NCCN, Fort Washington, PA) guidelines suggest that some patients with a nonfunctional tumor smaller than 2 cm in diameter could be followed clinically (depending on surgical risk, site of the tumor, and presence of comorbidities), whereas patients with a tumor having a diameter greater than 2 cm should be considered surgical candidates.21
Several studies have reported a significant correlation between tumor size and patient survival.22,23 These results, however, have been questioned by other studies, including one study that evaluated more than 3800 PNET patients in a multivariate analysis.24
In our series, only mitotic count, and not size, was a predictor of patients’ survival. Analyzing the t-ROC curves for PHH3 mitotic counts, we identified an optimal G2 cutoff at 7 mitoses per 10 HPFs. None of the patients with nonfunctional tumors under this cutoff died of the disease during a follow-up period of longer than 10 years. Because the proposed cutoff of fewer than 7 mitoses per 10 HPFs is able to identify tumors with an indolent biology, this classification could be used to guide the intensity of surveillance after resection. The NCCN guidelines suggest follow-up evaluation by history and physical examination, appropriate biochemical markers, and multiphasic computed tomography (CT) or magnetic resonance imaging (MRI) 3–12 months after tumor resection (or earlier if the patient presents with symptoms). Between 1 and 10 years after resection, follow-up evaluation is recommended every 6–12 months. The NCCN recommendations, however, are supported by a category 2A level of evidence (non-uniform consensus based on low-level evidence).21 Patients with a PHH3 grade 1 tumor (number of mitoses <7) might need less frequent surveillance over time.
We are aware that the data from this study have several limitations. Our cutoff of 7 mitoses per 10 HPFs has only been internally validated and not yet validated in an independent set of PNET patients. Validation in a large external cohort of PNETs would be essential before consideration of this methodology and cutoff in the clinical setting.
The clinical application of a scoring system based on PHH3 mitotic counts has several appealing features. First, it provides the pathologist with a faster and more efficient tool for the classification of PNETs. The high interobserver reproducibility could make this prognostic technique available to hospitals that do not have a team of dedicated gastrointestinal pathologists. Second, it more accurately defines the biology of the disease, which could guide pre- and postoperative therapy and follow-up evaluation.
In conclusion, this study demonstrated that mitotic counts determined by PHH3 staining are an excellent prognostic marker of both DFS and DSS in PNET patients. We identified 7 mitoses per 10 HPFs as the cutoff for the selection of patients who will require a more aggressive therapeutic intervention and follow-up evaluation. We demonstrated that PHH3 is a more robust means of predicting disease-specific death than H&E mitotic index or Ki67LI. Although further prospective clinical studies will be necessary to validate our findings, we hope the findings will stimulate research in this field.
ACKNOWLEDGMENT
The Loeffler Family and other generous patients provided philanthropic support for this study. Matteo Ligorio is the recipient of American-Italian Cancer Foundation Post-Doctoral Fellowship. Vincenzo Villani is the recipient of a Research Fellowship from the Centro per la Comunicazione e la Ricerca of the Collegio Ghislieri of Pavia.
Footnotes
CONFLICTS OF INTEREST There are no conflicts of interest.
REFERENCES
- 1.Franko J, Feng W, Yip L, Genovese E, Moser AJ. Nonfunctional neuroendocrine carcinoma of the pancreas: incidence, tumor biology, and outcomes in 2158 patients. J Gastrointest Surg. 2010;14:541–8. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence B, Gustafsson BI, Chan A, Svejda B, Kidd M, Modlin IM. The epidemiology of gastroenteropancreatic neuroendocrine tumors. Endocrinol Metabol Clin N Am. 2011;40:1–18, vii. [DOI] [PubMed] [Google Scholar]
- 3.Halfdanarson TR, Rabe KG, Rubin J, Petersen GM. Pancreatic neuroendocrine tumors (PNETs): incidence, prognosis, and recent trend toward improved survival. Ann Oncol. 2008;19:1727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wilde RF, Edil BH, Hruban RH, Maitra A. Well-differentiated pancreatic neuroendocrine tumors: from genetics to therapy. Nat Rev. 2012;9:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuo JH, Lee JA, Chabot JA. Nonfunctional pancreatic neuroendocrine tumors. Surg Clin N Am. 2014;94:689–708. [DOI] [PubMed] [Google Scholar]
- 6.Liu TC, Hamilton N, Hawkins W, Gao F, Cao D. Comparison of WHO Classifications (2004, 2010), the Hochwald grading system, and AJCC and ENETS staging systems in predicting prognosis in locoregional well-differentiated pancreatic neuroendocrine tumors. Am J Surg Pathol. 2013;37:853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosman FT, Carneiro F, Hruban RH, Theise N. WHO classification of tumours of the digestive system. 4th ed. Lyon: IARC Press; 2010. [Google Scholar]
- 8.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A (eds). AJCC cancer staging manual. New York: Springer; 2010. pp. 241–9. [Google Scholar]
- 9.Rindi G, Falconi M, Klersy C, Albarello L, Boninsegna L, Buchler MW, et al. TNM staging of neoplasms of the endocrine pancreas: results from a large international cohort study. J Natl Cancer Inst. 2012;104:764–77. [DOI] [PubMed] [Google Scholar]
- 10.McCall CM, Shi C, Cornish TC, Klimstra DS, Tang LH, Basturk O, et al. Grading of well-differentiated pancreatic neuroendocrine tumors is improved by the inclusion of both Ki67 proliferative index and mitotic rate. Am J Surg Pathol. 2013;37:1671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Tang LH, Klimstra DS. Gastroenteropancreatic neuroendocrine neoplasms: historical context and current issues. Semin Diagn Pathol. 2013;30:186–96. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- 13.Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci USA. 1998;95:7480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribalta T, McCutcheon IE, Aldape KD, Bruner JM, Fuller GN. The mitosis-specific antibody anti-phosphohistone-H3 (PHH3) facilitates rapid reliable grading of meningiomas according to WHO 2000 criteria. Am J Surg Pathol. 2004;28:1532–6. [DOI] [PubMed] [Google Scholar]
- 15.Colman H, Giannini C, Huang L, Gonzalez J, Hess K, Bruner J, et al. Assessment and prognostic significance of mitotic index using the mitosis marker phospho-histone H3 in low- and intermediate-grade infiltrating astrocytomas. Am J Surg Pathol. 2006;30:657–64. [DOI] [PubMed] [Google Scholar]
- 16.Casper DJ, Ross KI, Messina JL, Sondak VK, Bodden CN, McCardle TW, et al. Use of anti-phosphohistone H3 immunohistochemistry to determine mitotic rate in thin melanoma. Am J Dermatopathol. 2010;32:650–4. [DOI] [PubMed] [Google Scholar]
- 17.Nasr MR, El-Zammar O. Comparison of pHH3, Ki-67, and survivin immunoreactivity in benign and malignant melanocytic lesions. Am J Dermatopathol. 2008;30:117–22. [DOI] [PubMed] [Google Scholar]
- 18.Tsuta K, Raso MG, Kalhor N, Liu DD, Wistuba II, Moran CA. Histologic features of low- and intermediate-grade neuroendocrine carcinoma (typical and atypical carcinoid tumors) of the lung. Lung Cancer Amst Neth. 2011;71:34–41. [DOI] [PubMed] [Google Scholar]
- 19.Voss SM, Riley MP, Lokhandwala PM, Wang M, Yang Z. Mitotic count by phosphohistone H3 immunohistochemical staining predicts survival and improves interobserver reproducibility in well-differentiated neuroendocrine tumors of the pancreas. Am J Surg Pathol. 2015;39:13–24. [DOI] [PubMed] [Google Scholar]
- 20.Ogino T, Bandoh N, Hayashi T, Miyokawa N, Harabuchi Y, Ferrone S. Association of tapasin and HLA class I antigen downregulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res. 2003;9:4043–51. [PubMed] [Google Scholar]
- 21.Neuroendocrine tumors. NCCN Clinical Practical Guidelines in Oncology (NCCN Guidelines) version 12015. National Comprehensive Cancer Network, 2014. [Google Scholar]
- 22.Scarpa A, Mantovani W, Capelli P, Beghelli S, Boninsegna L, Bettini R, et al. Pancreatic endocrine tumors: improved TNM staging and histopathologic grading permit a clinically efficient prognostic stratification of patients. Mod Pathol. 2010;23:824–33. [DOI] [PubMed] [Google Scholar]
- 23.Gullo L, Migliori M, Falconi M, Pederzoli P, Bettini R, Casadei R, et al. Nonfunctioning pancreatic endocrine tumors: a multicenter clinical study. Am J Gastroenterol. 2003;98:2435–9. [DOI] [PubMed] [Google Scholar]
- 24.Bilimoria KY, Talamonti MS, Tomlinson JS, Stewart AK, Winchester DP, Ko CY, et al. Prognostic score predicting survival after resection of pancreatic neuroendocrine tumors: analysis of 3851 patients. Ann Surg. 2008;247(3):490–500. [DOI] [PubMed] [Google Scholar]