Abstract
The chondroitin sulfate proteoglycan versican is important for embryonic development and several human disorders. The versican V1 splice isoform is widely expressed and cleaved by ADAMTS proteases at a well-characterized site, Glu441-Ala442. Since ADAMTS proteases cleave the homologous proteoglycan aggrecan at multiple sites, we hypothesized that additional cleavage sites existed within versican. We report a quantitative label-free approach that ranks abundance of liquid chromatography-tandem mass spectrometry (LC-MS/MS)-identified semi-tryptic peptides after versican digestion by ADAMTS1, ADAMTS4 and ADAMTS5 to identify site-specific cleavages. Recombinant purified versican V1 constructs were digested with the recombinant full-length proteases, using catalytically inactive mutant proteases in control digests. Semi-tryptic peptide abundance ratios determined by LC-MS/MS in ADAMTS:control digests were compared to the mean of all identified peptides to obtain a z-score by which outlier peptides were ranked, using semi-tryptic peptides identifying Glu441 -Ala442 cleavage as the benchmark. Tryptic peptides with higher abundance in control:ADAMTS digests supported cleavage site identification. We identified several novel cleavage sites supporting the ADAMTS1/4/5 cleavage site preference for a P1-Glu residue in proteoglycan substrates. Digestion of proteins in vitro and application of this z-score approach is potentially widely applicable for mapping protease cleavage sites using label-free proteomics.
Keywords: Proteolysis, terminomics, Metalloprotease, ADAMTS, Proteoglycan, Versican
Graphical Abstract
Introduction
Versican is a large chondroitin sulfate (CS) proteoglycan in the pericellular and extracellular matrix (ECM) that plays an essential role during embryogenesis. Vcanhdf homozygous mutant embryos, which lack all versican isoforms, die by 10.5 days of gestation (E10.5) with severe cardiac anomalies [1, 2] and fail to form the primary vasculature and primitive blood cells [3], underscoring the necessity of versican for formation of the entire cardiovascular system. Conditional deletion of versican identified a role in skeletal development [4], and isoform-specific, cleavage-resistant or hypomorphic mutants have further defined the roles of versican in limb, neural and cardiovascular development [5–9]. The versican core protein bears several covalently-attached CS chains and forms high-molecular weight complexes with the glycosaminoglycan (GAG) hyaluronan (HA). The swelling capacity of versican-HA complexes provides compression resistance and viscoelasticity to numerous adult tissues. Prior work also suggests that versican regulates HA macromolecular assembly and stability [3, 9]. Versican contributes to regulation of cellular processes such as adhesion, proliferation, migration and apoptosis and to physiological processes such as ovulation and wound healing [10–14]. As an essential component of the pericellular matrix of several cell types, it is involved in phenotype regulation, such as the fibroblast to myofibroblast transition [15, 16], and an excess of pericellular versican leads to de-differentiation of uterine smooth muscle cells by interfering with focal adhesion formation [12]. Versican is implicated in cancer angiogenesis and metastasis [17–22], in the pathogenesis of atherosclerosis and aortic aneurysms [23–26], and in the immune and stromal responses to viral pneumonia [27, 28]. Thus, the levels of versican in tissues and the mechanisms of its turnover have high biological and disease significance.
Multiple isoforms of versican arise from alternative splicing of large exons encoding the GAG attachment domains, termed GAGα and GAGβ [29]. These isoforms are V0 (containing both GAGα and GAGβ), V1 (containing GAGβ only), V2 (containing GAGα only) and V3, lacking both GAG domains [8, 30]. Another isoform, V4, comprising the first 398 amino acids of the GAGβ region, was described in human breast cancer [31]. In addition to GAG attachment domains, versican has N- and C- terminal globular domains, named G1 and G3, respectively. The G1 domain mediates the interaction with HA, and thus brings versican in proximity to the cell surface via HA interactions with its cell-surface receptors. The G3 domain binds to a variety of extracellular matrix (ECM) proteins [32], linking the pericellular HA-versican matrix to interstitial ECM networks.
Proteolysis is a major mechanism of post-translational regulation of versican. Mouse mutants have unequivocally established that members of the A Disintegrin-like And Metalloprotease with Thrombospondin type 1 motifs (ADAMTS) family of metalloproteinases are essential for versican turnover in vivo [33, 34]. In particular, genetic deletion/haploinsufficiency of Adamts1, 5, 9, and 20 in mice results in developmental anomalies affecting many organ systems [8, 33]. These include the cardiovascular system, where ventricular, aortic and valvular defects occur in association with increased versican immunostaining and reduced immunoreactivity with a versican neoepitope antibody, anti-DPEAAE [35–39]. ADAMTS5 proteolysis of versican detected by anti-DPEAAE is also implicated in the formation of cerebral cavernous malformations [40] and in multiple myeloma [20]. Anti-DPEAAE reacts with the new C-terminus generated at an ADAMTS cleavage site in the versican GAGβ domain, which occurs at the Glu441-Ala442 bond (human V1 isoform sequence enumeration, the corresponding V0 isoform sequence is Glu1428-Ala1429) [16]. Indeed, recent work has shown that an ADAMTS deficiency-dependent anomaly, soft tissue syndactyly, occurs in homozygotes of two independently generated mouse strains, VcanR and VcanAA, that render the Glu441-Ala442 site uncleavable by ADAMTS proteases [7, 41]. An ADAMTS cleavage site occurring at Glu405-Gln406 was described in the GAGα domain (human V0, V2 isoform sequence enumeration) [42]. However, it is not known whether ADAMTS proteases cleave versican at other sites. In one report, short versican synthetic peptides were digested with ADAMTS1 to identify cleavages of the Glu950-Gly951 (V0/V2 versican enumeration) and Tyr1410-Ile1411/Tyr423-Ile424 bond (V0/V1 versican enumeration) [43]. However, whether these cleavages occur in the native, glycosylated versican core protein has not been determined.
Here, we have tested the hypotheses that, a. Versican is cleaved at additional sites in its core protein, and b. That ADAMTS1, ADAMTS4 and ADAMTS5, which are the best-studied ADAMTS proteases known to cleave versican [44], will show site-specific preferences. To address these hypotheses, we digested full-length recombinant V1 as well as a truncated recombinant V1 variant (V1–5GAG) [45], both purified from mammalian cells, with purified recombinant ADAMTS1, ADAMTS4 and ADAMTS5, using their active site mutants as the respective controls, and applied liquid-chromatography-tandem mass spectrometry (LC-MS/MS) in a novel label-free quantitative approach to identify new versican cleavage sites.
Experimental Methods:
Protein expression and purification.
cDNA constructs encoding human ADAMTS1, ADAMTS4 and ADAMTS5 with a C-terminal FLAG tag (DYKDDDDK) in pEGFP-N1 vector were described previously [44, 46]. The corresponding active site mutants ADAMTS1 E402→Q, ADAMTS4 E362→Q, and ADAMTS5 E411→Q, henceforth referred to as the respective “EQ mutants”, were generated using site-directed mutagenesis and the mutations were confirmed by Sanger sequencing. The constructs were transiently transfected in HEK293T cells using polyethylenimine (PEI) (Polysciences GmbH, Germany) and purified from the medium by anti-FLAG immunoaffinity as previously described [44]. Their activity was determined under kinetic equilibrium conditions by active-site titration with known concentrations of TIMP-3 (Bio-Techne, Cat. no.:973-TM-010, Bio-Techne) using quenched-fluorescent peptides as previously described [44, 46, 47]. The concentration of active site mutants was measured by optical absorbance at 280 nm using extinction coefficients (E1%, 1 cm) as predicted by the ProtParam Tool (ExPasy): 1.373 (ADAMTS1), 1.1190 (ADAMTS4), and 1.220 (ADAMTS5), respectively. DNA and protein concentrations were measured using a NanoDrop ND-2000 UV-visible spectrophotometer (Thermo Fisher Scientific, Nottingham, UK).
Constructs encoding full-length versican V1 and V1–5GAG (comprising amino acids 21–694 of V1) with a C-terminal tandem myc-His6 tag were described previously [44, 45]. V1 and V1–5GAG were purified from the medium of transiently transfected HEK293T cells using HiTrap DEAE Sepharose and Ni-Sepharose column chromatography (GE Healthcare) as previously described [44, 45].
Versican digestion:
All digests were done in TNC buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM CaCl2) at 37°C. ADAMTS1 (final concentration 140 nM), ADAMTS4 (5 nM) and ADAMTS5 (5 nM) were incubated either with V1 (150 nM) or V1–5GAG (310 nM) for 2 h at 37°C before addition of EDTA (50 mM).
Proteomics analysis by LC-MS/MS.
Approximately 6 μg ADAMTS-digested versican V1 or V1–5GAG was lyophilized in a SpeedVac evaporator, reconstituted in 50 μL of 6 M urea, 100 mM Tris, pH 7.0, reduced using 5 mM dithiothreitol at 60° C and alkylated with 20 mM iodoacetamide in the dark at room temperature (RT). The urea concentration was reduced to 1.2 M by diluting the sample with 100 mM ammonium bicarbonate and the pH was adjusted to below 8. The samples were subjected to overnight trypsin digestion (Trypsin Gold, mass spectrometry grade, Promega V5280) using a trypsin/protein ratio of 1:25 at 37°C. The resulting peptide mixture was desalted using Pierce C18 spin columns (Thermo Fisher Scientific), lyophilized in a SpeedVac evaporator and reconstituted in 30 μL 1% acetic acid. Peptides were analyzed on a Thermo Ultimate 3000 UHPLC in-line with a ThermoFisher Scientific Fusion Lumos tribrid mass spectrometer. The HPLC column was a Dionex 15 cm x 75 μm id Acclaim Pepmap C18, 2μm, 100 Å reversed phase capillary chromatography column. Five μL volumes of the samples were injected and the peptides eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μL/min were introduced into the source of the mass spectrometer. The nanospray ion source was operated at 1.9 kV. The digest was analyzed using a 90-minute data-dependent method with a 3 sec window of selection for the most abundant ions to undergo 35% collision-induced dissociation with an isolation window of 0.7 m/z for MS/MS. Dynamic exclusion was enabled with a repeat count of 1 and ions within 10 ppm of the fragmented mass were excluded for 60 seconds. MS/MS spectra were matched to the respective construct sequences using Proteome Discoverer 2.3. Dynamic modifications included oxidation (methionine), acetylation (peptide N-termini), Gln to pyro-Glu cyclization (Q N-termini) and the static modification used was carboxyamidomethylation of cysteine. The search included semi-tryptic peptides and allowed for up to 3 missed tryptic cleavages. Peptides sequences were validated using a false discovery rate (FDR) of 1% for high-confidence peptides and 5% for medium-confidence peptides against a decoy database. Chromatographic retention time alignment was used across samples for accurate label-free quantitation comparison and increased peptide identifications. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium vie the PRIDE [48] partner repository with the dataset identifier PXD025456 and 10.6019/PXD025456.
Protein and peptide data were analyzed using Microsoft Excel. High-confidence peptides were sorted as fully tryptic based on the presence of a lysine or arginine at expected sites, i.e., in the protein sequence preceding the peptide N-terminal residue or present as the last residue within the peptide, or as semi-tryptic, i.e., lacking one or both of these residues at the expected N- and /or C-terminal positions. Peptide abundance ratios were quantified in Proteome Discoverer 2.4 (ThermoFisher) using the label-free quantitation method and were then log base-2-transformed (log2) in Excel to reduce variations between peptides. Log2-transformed ratios were used to calculate the z-score of significance, an internal value for each experiment used in place of typical t-tests of significance which are applied to experiments containing replicates [49]. Log2 transformed peptide abundance values and Z-scores were tested for normal distribution by generation of histograms and by using the Anderson-Darling, D’Agostino and Pearson, Shapiro-Wilk, and Kolmogorov-Smirnov tests for normality in GraphPad Prism 9.0.0. Each measured semi-tryptic peptide abundance ratio in ADAMTS digest vs. EQ control was compared to the mean standard deviation of all identified peptides in each experiment to identify outlier semi-tryptic peptides. Specifically, the abundance ratio z-score for a peptide was calculated by subtracting the peptide ratio from the average ratio of all quantified peptides and dividing that by the standard deviation of all quantified peptide ratios. This score thus represents the number of standard deviations each peptide ratio is from the mean of the normal values. Z-scores were calculated for semi-tryptic peptides and separately for outlier tryptic peptides (using control:digest ratios) as absolute values. Z-scores and peptide ratios (log2 transformed) were plotted as scatter charts using GraphPad Prism 9.0.0 software. Peptides with a z-score > 2 (meaning the ratios were greater than 2 standard deviations from the mean) were considered significant [49]. Peptides which were found exclusively in the protease or control digests were arbitrarily assigned scaled fold-changes of 100; these peptides were excluded from the calculations made to obtain mean or standard deviation of all peptides as the basis for the z-score. A minimum peptide abundance cut-off was applied at an intensity of 1×104 to exclude comparison of peptides which were close to the limit of detection (intensity of 1×103).
Results:
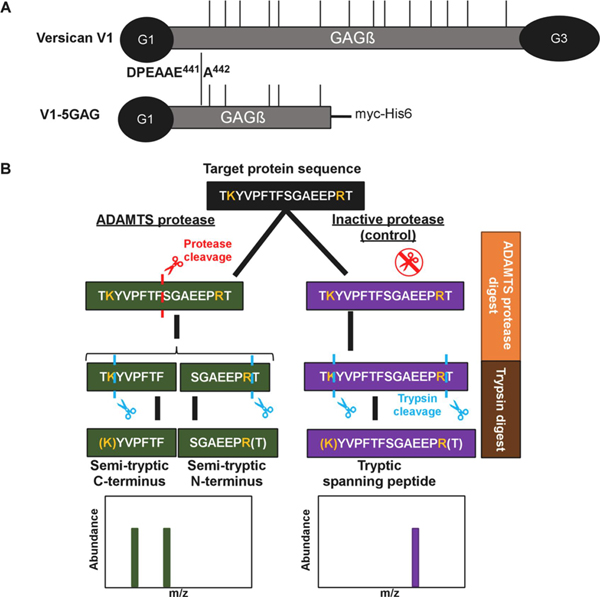
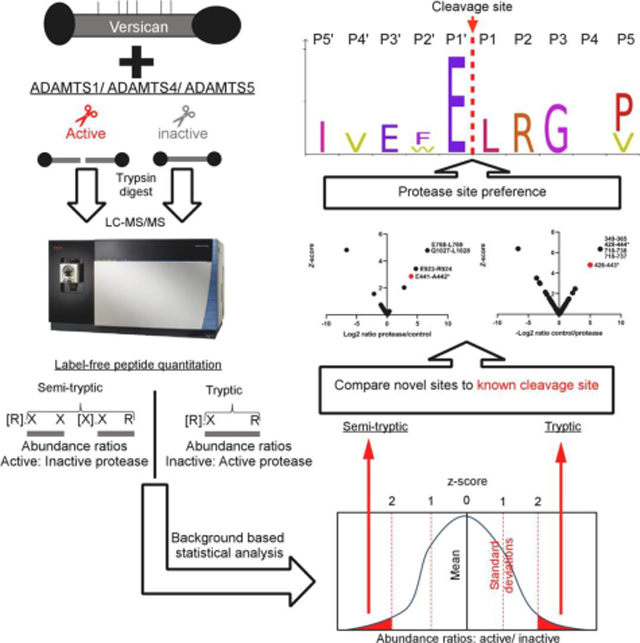
Full-length recombinant versican (V1) and a recombinant C-terminally truncated versican V1 isoform lacking the G3 domain and bearing only the most N-terminal 5 CS-chains (V1–5GAG, amino acids 21–694 of V1) [45] were digested with ADAMTS1, ADAMTS4, and ADAMTS5 using the respective active site (EQ) mutants as controls (Figure 1A). Both versican constructs contain the cognate ADAMTS cleavage site at Glu441-Ala442. These digests were subsequently digested with trypsin, used as the working protease for LC-MS/MS in the experimental workflow (Figure 1B). A non-tryptic terminus in a peptide with higher abundance in ADAMTS digests was considered as indicating a putative cleavage site for ADAMTS1, ADAMTS4 or ADAMTS5, specifically, peptides with z-score >2. Conversely, tryptic peptides with a z-score >2 and higher abundance in the control digests were designated as potentially spanning ADAMTS cleavage sites and were compared to the semi-tryptic peptides for sequence overlap (Figure 1B).
Figure 1. Experimental strategy.
A. Versican constructs used in the present analysis. GAG chains are indicated with thin vertical lines. B. Schematic of the experimental design, illustrated with a hypothetical example comparing digestion with an active ADAMTS protease vs a catalytically inactive control, followed by cleavage site identification using label-free proteomics
Novel ADAMTS cleavage sites in versican V1–5GAG.
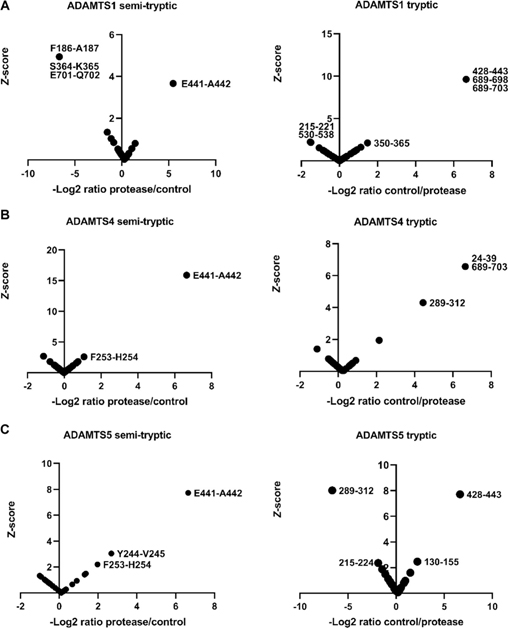
Since the z-score method relies on normal distribution of peptide abundance, distribution histograms were generated using each peptide z-score (from non-singleton peptides, i.e. using only peptides found in both the ADAMTS and EQ digests) and all but three (representing V1–5GAG digests) contained a sufficient number of peptides (≥30) to demonstrate normal distribution (Supplemental Figure 1,3). 4 significance tests were performed on the three datasets which contained fewer than 30 peptides showing that they also followed a normal distribution (Supplemental Figure 2). As expected, and essentially validating the approach, a semi-tryptic peptide indicating cleavage at Glu441-Ala442 in the GAGβ domain was found exclusively in the digests with ADAMTS1, ADAMTS4 and ADAMTS5 but not in the controls (Supplemental Table 1, Figure 2A–C, left-hand panels). Moreover, we identified a tryptic peptide spanning this cleavage site exclusively in each of the corresponding control digests with catalytically inactive ADAMTS1, 4 and 5 (Supplemental Table 1, Figure 2A–C, right-hand panels). Digestion of V1–5GAG with ADAMTS1, ADAMTS4 and ADAMTS5 identified 4 semi-tryptic peptides with a z-score indicating higher abundance in the ADAMTS digest than in the control, suggesting 2 putative novel cleavage sites (Supplemental Table 1, Figure 2A–C). Additional semi-tryptic peptides identified two novel cleavage sites, namely Phe253-His254 (ADAMTS4 and ADAMTS5) and Tyr244-Val245 (ADAMTS5 only) (Fig. 2B, C). The peptides identifying the Phe253-His254 site were not however identical, since the relevant peptide in the ADAMTS5 digest had two missed cleavages. A peptide with high abundance in the protease digests indicated cleavage at Glu701-Gln702, which is located in the myc tag of V1–5GAG, and hence, was not considered further. A few tryptic peptides were identified with higher abundance in the ADAMTS1 and ADAMTS5 digests that did not have sequence overlap with the semi-tryptic peptides, and possibly represent trypsin-like cleavages of the ADAMTS proteases (Fig. 2A, C, right-hand panels). Specifically, the ADAMTS1 digest had two highly abundant tryptic peptides spanning residues 215–221 and 530–538 (Fig. 2A, left-hand panel). Interestingly, the tryptic peptide 215–221 overlapped with the tryptic peptide showing the same z-score significance in the ADAMTS5 digest (residues 215–224) but differed in a single missed cleavage (Fig. 2C, left-hand panel). A tryptic peptide with higher abundance in the ADAMTS5 digest spanned residues 289–312, which matched a peptide found significantly higher in the control digest of ADAMTS4 (Figure 2, Supplemental Table 1).
Figure 2. Digestion of versican V1–5GAG with ADAMTS1, ADAMTS4 and ADAMTS5.
V1–5GAG was digested with A. ADAMTS1, B. ADAMTS4, and C. ADAMTS5 for 2 h at 37°C. Semi-tryptic peptides present at higher levels in digests with the active proteases identified the cleavage sites that are shown in the left-hand panels in A-C, whereas the panels on the right show tryptic peptides having higher abundance in the control digests.
Tryptic peptides with higher abundance spanning residues 350–365, 689–698, 689–703 in the ADAMTS1 control digest, 24–39, 289–312, and 689–703 in the ADAMTS4 control digest and 130–155 in the ADAMTS5 control digest which do not contain any known cleavages or experimentally determined semi-tryptic peptides with high z-score in this study, were also identified (Figure 2A–C, right-hand panels, Supplemental Table 1).
Novel ADAMTS cleavage sites in versican V1.
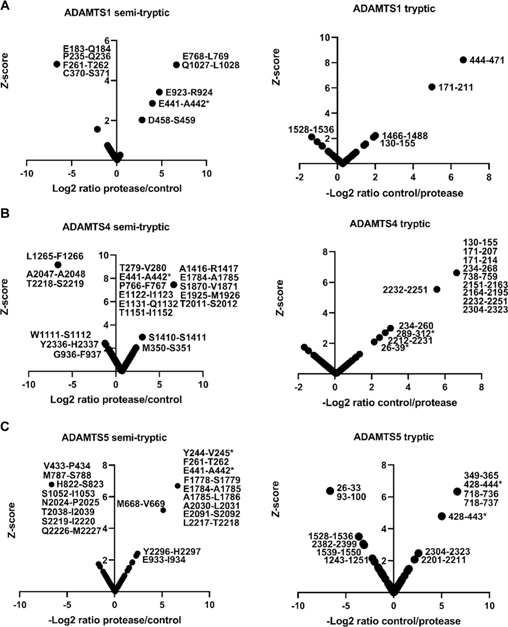
As above, normal distribution was ascertained from histograms generated from peptide z-scores (using non-singleton peptides). Each dataset contained a sufficient number of peptides (≥30) for this analysis (Supplemental Figure 3) therefore, QQ plots and significance tests were not necessary to prove normal distribution. While V1–5GAG contains the previously described Glu441-Ala442 cleavage site in the truncated βGAG region, cleavage sites further downstream of the full-length sequence would not be detected. Digestions of full-length versican V1 with the ADAMTS proteases were therefore analyzed using identical methods as for V1–5GAG. These digests identified 42 semi-tryptic peptides, of which 30 had higher abundance in ADAMTS digests than the EQ controls and corresponded to 27 unique predicted cleavage sites (Figure 3A–C and Supplemental Table 2). In addition to peptides identifying the Glu441-Ala442 bond, other significant peptides with equal or higher z-scores were identified (Table 1). Many of these peptides indicated cleavage sites with Glu at the P1 position and a small hydrophobic residue at the P1′ position, e.g., Glu768-Leu769 (ADAMTS1), Glu1784-Ala1785 (ADAMTS4 and 5), Glu1122-Ile1123 and Glu1925-Met1926 (ADAMTS4), and Glu933-Ile934 (ADAMTS5), all of which were located in the GAGβ domain. Furthermore, each of the three proteases generated other semi-tryptic peptides with high z-scores whose sequences indicated distinct cleavage site preferences, such as Glu923-Arg924 (ADAMTS1), Glu1131-Gln1132 (ADAMTS4) and Glu2091-Ser2092 (ADAMTS5) (Figures 3A–C, left-hand panels, Supplementary Figure 4). The cleavages deduced from semi-tryptic peptides with high z-scores are summarized in Figure 4.
Figure 3. Digestion of versican V1 with ADAMTS1, ADAMTS4 and ADAMTS5.
Versican V1 was digested with A. ADAMTS1, B. ADAMTS4, and C. ADAMTS5 for 2 h at 37°C. Semi-tryptic peptides present at higher levels in the digest with the active proteases identified the cleavage sites that are shown in the left-hand panels in A-C, whereas the panels on the right show tryptic peptides having higher abundance in the control digests. * indicates peptides that were also found in the V1–5GAG digest in Figure 2.
Table 1.
Semi-tryptic peptides indicating novel cleavage sites in versican V1
| Protease | Semi-tryptic peptide sequence | Cleavage site position | Protease:control | z- score |
|---|---|---|---|---|
| ADAMTS1 | [E].LESPNVATSSDSGTR.[K] | E768-L769 | 100 | 4.78 |
| ADAMTS1 | [Q].LVTVSSSVVPVLPSAVQK.[F] | Q1027-L1028 | 100 | 4.78 |
| ADAMTS1 | [E].RLGEPNYGAEIR.[G] | E923-R924 | 27.09 | 3.42 |
| ADAMTS1 | [K].HLVTTVPKDPEAAE.[A] | E441-A442 | 15.74 | 2.86 |
| ADAMTS4 | [T].VGELQAAWR.[N] | T279-V280 | 100 | 7.45 |
| ADAMTS4 | [K].HLVTTVPKDPEAAE.[A] | E441-A442 | 100 | 7.45 |
| ADAMTS4 | [P].FELESPNVATSSDSGTR.[K] | P766-F767 | 100 | 7.45 |
| ADAMTS4 | [E].IESETTSEEQIQEEK.[S] | E1122-I1123 | 100 | 7.45 |
| ADAMTS4 | [R].QEVNPVRQEIESETTSEE.[Q] | E1131-Q1132 | 100 | 7.45 |
| ADAMTS4 | [T].IFDSQTFTETELK.[T] | T1151-I1152 | 100 | 7.45 |
| ADAMTS4 | [A].RAYGFEMAK.[E] | A1416-R1417 | 100 | 7.45 |
| ADAMTS4 | [K].HAGPSFQPEFSSGAEE.[A] | E1784-A1785 | 100 | 7.45 |
| ADAMTS4 | [S].VMSPQDSFK.[E] | S1870-V1871 | 100 | 7.45 |
| ADAMTS4 | [K].LEPSEDDGKPELLEE.[M] | E1925-M1926 | 100 | 7.45 |
| ADAMTS4 | [T].SERPTLSSSPEINPETQAALIR.[G] | T2011-S2012 | 100 | 7.45 |
| ADAMTS5 | [Y].VDHLDGDVFHLTVPSK.[F] | Y244-V245 | 100 | 6.69 |
| ADAMTS5 | [F].TFEEAAK.[E] | F261-T262 | 100 | 6.69 |
| ADAMTS5 | [K].HLVTTVPKDPEAAE.[A] | E441-A442 | 100 | 6.69 |
| ADAMTS5 | [R].TQEEYEDKKHAGPSFQPEF.[S] | F1778-S1779 | 100 | 6.69 |
| ADAMTS5 | [K].HAGPSFQPEFSSGAEE.[A] | E1784-A1785 | 100 | 6.69 |
| ADAMTS5 | [R].TQEEYEDKKHAGPSFQPEFSSGAEEA.[L] | A1785-L1786 | 100 | 6.69 |
| ADAMTS5 | [A].LIRGQDSTIAASEQQVAAR.[I] | A2030-L2031 | 100 | 6.69 |
| ADAMTS5 | [E].SVEGTAIYLPGPDR.[C] | E2091-S2092 | 100 | 6.69 |
| ADAMTS5 | [L].TSILSHEEQMFVNR.[V] | L2217-T2218 | 100 | 6.69 |
Residues written in bold text indicate the cleaved peptide bond. Z-scores for peptides identifying the Glu441–A442 site are shown in bold text.
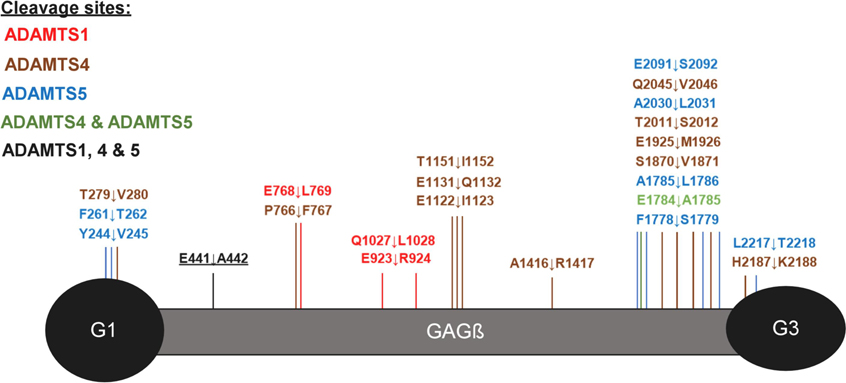
Figure 4. Overview of ADAMTS cleavage sites in versican.
Putative ADAMTS-mediated cleavages identified by the z-score method are shown on a graphic representation of versican V1. The canonical cleavage site (E441↓A442) is underlined and labeled in black lettering.
Thirty-three significant tryptic peptides were identified in the versican V1 digest of which 26 were higher in abundance in the controls than in the ADAMTS protease digests and corresponded to 24 unique sequences (Figure 3A–C, right-hand panels) (Supplemental Table 2). Of these, only one tryptic peptide (residues 444–471) spanned the region of a putative ADAMTS1 cleavage site, specifically spanning the novel Asp458-Ser459 site (Supplemental Table 2). The ADAMTS1 digestion identified 4 tryptic peptides significantly more abundant in the EQ control, two of which matched tryptic peptides that had higher scores in the ADAMTS4 digests (Figure 3), i.e., residues 171–211 and 130–155. These three peptides are located within the V1–5GAG sequence, but only the peptide spanning residues 130–155 was detected in V1–5GAG digests. Two significant ADAMTS1-generated tryptic peptides (spanning residues 26–39 and 289–312) matched those found in the V1–5GAG digestion. Two tryptic peptides spanned the location of a putative ADAMTS4 and ADAMTS5 cleavage site (Phe253-His254) identified in the V1–5GAG digests (Figure 2). There were 7 tryptic peptides with high z-scores in the ADAMTS digests of versican-V1 that could represent trypsin-like activities of the ADAMTS proteases and 18 semi-tryptic peptides with high z-scores in the control digests whose origins are unclear.
Reviewing the data from the V1–5GAG and V1 digests together suggests that novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavages occur throughout the full-length proteoglycan sequence but are predominantly located in the GAGβ region in two distinct clusters, one located centrally, and the other toward the C-terminus. In the case of ADAMTS1, three cleavage sites were identified that were exclusive to this protease, and the corresponding semi-tryptic peptides had a higher z-score than the semi-tryptic peptide identifying the Glu441-Ala442 site (Figure 4, Table 1). The deduced cleavages at Glu768-Leu769 and Gln1027-Leu1028 share Leu in the P1′ position and were absent in the control digest. One ADAMTS4 cleavage site is two amino acids away from the Glu768-Leu769 site. Many of the ADAMTS4 and ADAMTS5 cleavage sites cluster around the C-terminal region of the GAGβ domain between residues 1778–2091 (Figure 4). Here these proteases share a cleavage site at Glu1784-Ala1785 which resembles the Glu441-Ala442 site at positions P1, P1′, and at P3 (both sites contain an Ala at P3). Among the three proteases tested, ADAMTS4 exclusively preferred the residue region 1122–1151 in the GAGβ domain where 3 cleavage sites were identified within 30 amino acids of each other, Glu1122-Ile1123, Glu1131-Gln1132, and Thr1151-Ile1152.
Although all the semi-tryptic peptides with significant z-scores identify potential ADAMTS cleavage sites in versican, we considered further refinement of our approach in light of the availability of a validated cleavage at the Glu441-Ala442 site, which furthermore, has demonstrated biological relevance. Specifically, we ranked the highest priority novel cleavage sites based on their z-scores relative to the Glu441-Ala442 site and whether or not additional evidence supporting the cleavage was provided by a tryptic peptide having a significantly higher abundance in the control digests (Table 1). ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites previously found in the literature and the MEROPS database (https://www.ebi.ac.uk/merops/) were compiled into iceLogo plots to determine the sequence preferences at positions P10′-P10 (Supplemental Figure 4). Only 10 other cleavage sites have been described for ADAMTS1 to date, whereas ADAMTS4 and ADAMTS5 have 76 and 53 previously identified cleavage sites, respectively, in a variety of substrates (Supplemental File 1). Therefore, the iceLOGO plots for ADAMTS4 and ADAMTS5 have >95% confidence while ADAMTS1 does not reach the 30-cleavage site threshold previously determined to provide the necessary statistical confidence [50]. Addition of the putative cleavage sites found in this study to the iceLogo analysis was done to refine the positional preferences compared to that which was previously known, although the refined consensus we obtained essentially supports that previously established (Supplemental Figure 4). Many of the putative cleavage events identified in the present study were marked by a glutamic acid residue in the P1 position (3 out of 5 for ADAMTS1, 5 out of 15 for ADAMTS4, and 4 out of 12 for ADAMTS5), consistent with an earlier description of these enzymes as glutamyl endopeptidases [51]. In comparison, all the 4 major ADAMTS4/5 cleavage sites in aggrecan have glutamic acid at P1, as well as a small hydrophobic amino acid at P1′ [44]. Small hydrophobic amino acids were frequently found at P1′ in our analysis, consistent with previously identified cleavage sites (Supplemental Figure 4). ADAMTS4 seems to prefer bulkier amino acids such as phenylalanine at P1′, as previously observed from screening a library of 13-mer peptides [52]. In our analysis, a phenylalanine in the P1′ position was observed in the Pro766-Phe767 V1 cleavage site. Compared with ADAMTS5, ADAMTS4 is indeed characterized by a deeper and larger S1′ pocket which can accommodate a bulkier side chain [53].
Importantly, some of the peptides detected in this study as arising from ADAMTS activity were incidentally reported in different proteomic analyses of human tissues, although not in the course of seeking ADAMTS or other protease substrates (Supplemental Table 3), strongly suggesting that the experimentally-defined cleavages we observed occur naturally (Figure 4, Supplemental Table 2).
Discussion
Cleavage of versican at the Glu441-Ala442 site by ADAMTS1 and ADAMTS4 was first described two decades ago [16] and since then, abundant evidence has indicated that this proteolytic event is important in many morphogenetic and disease contexts [7, 11, 12, 23, 24, 28, 35–39, 41, 54–59] via a dual effect, i.e., on versican clearance, and by generation of a bioactive-terminal fragment, versikine, that is proapoptotic during web regression [11, 20] and presents a damage-associated molecular pattern in multiple myeloma [20]. Recently, the biological significance of cleavage at this site was supported by recapitulation of soft-tissue syndactyly (an anomaly seen in several ADAMTS mutants that is associated with reduced anti-DPEAAE staining [11, 20, 60]), in VcanR and VcanAA mutants [7, 41]. These findings present a stringent proof for biological impact of cleavage at Glu441-Ala442. Since ADAMTS1,4,5,9, 15 and 20 each cleaves versican at the Glu441-Ala442 bond [11, 16, 20, 61–64], the neoepitope anti-DPEAAE antibody is widely used as an indicator of versican proteolysis by these ADAMTS proteases. The possibility that ADAMTS proteases may cleave versican at multiple sites was initially suggested by the observation that ADAMTS4 and 5, the best characterized proteoglycanases to date, cleaved aggrecan at multiple sites [65–67] and supported by the observation that VcanR/R and VcanAA/AA mice [7, 41] have a relatively mild phenotype. Although these mutants have undergone limited analysis to date, it is known that they lack cleft palate, a phenotype which is invariably lethal in mice at birth and seen in combined Adamts9 and Adamts20 mutants, where it is associated with abnormal versican accumulation in the developing palate [55, 58, 60, 68]. Moreover, female VcanR/R and VcanAA/AA mice are fertile, whereas impaired ovulation is associated with reduced versican proteolysis in Adamts1 mutants [69, 70]. Female VcanR/R and VcanAA/AA mice deliver pups apparently without impairment, whereas female mice with conditional deletion of Adamts9 in smooth-muscle cells have dystocia and cannot give birth as a result of versican accumulation in the myometrium [12]. The lack of absolute correlation between the phenotypes resulting from ADAMTS-deficiency and Glu441-Ala442 cleavage-resistant versican in mice was not unexpected, since versican-degrading ADAMTS proteases cleave other substrates including, but not limited to other aggregating proteoglycans (e.g., aggrecan, brevican), small leucine-rich proteoglycans such as biglycan and decorin, matrilin-3, thrombospondin-1 and syndecan-4 [52, 71–75]. Another potential explanation, which the present analysis addresses, is that individual ADAMTS proteases may recognize additional specific sites in the versican core protein such that abrogation of cleavage at the canonical Glu441-Ala442 site has limited impact.
To address this possibility, we developed a simple, quantitative label-free LC/MS-MS method to compare versican peptides generated by wild-type ADAMTS proteases with those generated in the presence of their inactive counterparts. The validity of the approach was confirmed by detection of the Glu441-Ala442 site as a preferred cleavage site of ADAMTS1, ADAMTS4 and ADAMTS5. This is evident from the high z-score of the signature semi-tryptic peptide (2.86, 7.45 and 6.69 for ADAMTS1, ADAMTS4 and ADAMTS5 respectively), as well as the fact that it was detected only in the presence of these proteases and not the inactive mutants. Further validation was provided by detection of a tryptic peptide spanning the Glu441-Ala442 site exclusively in the presence of inactive ADAMTS proteases.
One strength of this study, therefore, is the repeated internal validation of the method by identification of Glu441-Ala442 as a cleavage site in the experiments with each ADAMTS proteases digesting two distinct versican constructs. Moreover, the analysis utilized full-length ADAMTS proteases as well as full-length versican purified from mammalian cells rather than from prokaryotic systems or synthetic peptides, which was important since ADAMTSs require long-range interactions with versican [44] as well as the presence of GAGs for maximal activity [45]. In contrast to these strengths, the present study is limited by intrinsic shortcomings of bottom-up proteomics, which is that peptides that are too large, too small, have an inappropriate m/z ratio, or carry unspecified post-translational modifications, would not be identified or detected. These limitations are particularly relevant for peptides in the GAGβ domain where we observed an abundance of negatively charged residues, but a lower frequency of Lys/Arg than is the norm (relevant to the use of trypsin as the working protease). Specifically, G1 and G3 domains contain an average of 1 Lys/Arg every 8–10 residues whereas the GAGβ domain averages 1 Lys/Arg every 17–18 residues with gaps as large as 75 amino acid residues between Lys/Arg residues, and is extensively modified by O- and N-glycosylation. These limitations can be addressed in the future by using alternative working proteases, and by inclusion of specialized approaches to identify glycopeptides [76]. The use of trypsin as the working protease for LC/MS/MS introduces a potential caveat in interpretation, i.e., we assume that all trypsin-like cleavages arose from trypsin used in the workflow, whereas one or more of the tested ADAMTS proteases could have an Arg or Lys preference at the P1 position, which would not be detected with our semi-tryptic peptide approach. This caveat can be mitigated by labeling protein N-termini after digestion with the ADAMTS proteases but prior to trypsin digestion, as utilized in N-terminomics/degradomics approaches such as Terminal Amine Isotopic Labeling of Substrates (TAILS) [77–79] as applied recently [80], or by using a different working protease. Despite these limitations, some of which are inherent to any LC-MS/MS approach, the z-score strategy as described here is broadly applicable to identifying cleavages of any substrate or an ensemble of a small number of purified substrates by any purified protease other than one with a trypsin-like preference, although as noted above, N-terminal labeling would mitigate this shortcoming. Indeed, N-terminal labeling is the basis for the well-established methods TAILS [78, 79] and ATOMS (Amino-Terminal Oriented Mass Spectrometry of Substrates) [81, 82] in which isotopically-labeled individual samples (such as one sample digested with a protease along with a control) can be duplexed or multiplexed for LC-MS-MS. These methods have been previously used to classify protease-generated termini as well as cleaved spanning peptides [83, 84]. Whereas multiplexing is a significant strength of these labeling methods, our simple label-free approach can be readily applied by most labs, since neither proteome labeling nor selective enrichment of N- or C-termini is needed.
Conclusion
Here, we have described both a label-free approach for identifying cleavage sites in protease substrates, and application of the method to uncover potentially novel ADAMTS cleavage sites in the proteoglycan versican. Whereas, the z-score approach can be used in a fully agnostic manner, we were fortunate in the present study to have a known cleavage site as a benchmark for experimental validation which could allow further refinement of semi-tryptic peptides in essentially a 2-step approach. The present analysis has identified several novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican, suggesting that these proteases, despite sharing activity at the Glu441-Ala442 site, target discrete additional sites in the versican core protein. In the future, occurrence and tissue distribution of the highest ranked putative cleavages can be determined by generating neo-epitope antibodies to the new C- and/or N-termini. It remains to be determined if V1 fragments generated by these newly identified proteolytic events are endowed with biological activity.
Supplementary Material
Highlights:
Versican proteolysis is biomedically significant, but few cleavage sites are known.
A novel z-score-based label-free proteomics method was used to define new cleavages.
Cleavage of two versican constructs by three ADAMTS proteases was tested.
A well-characterized versican cleavage site at Glu441-Ala442 was the benchmark.
Novel cleavage sites for ADAMTS1, ADAMTS4 and ADAMTS5 were identified.
Significance:
Versican abundance and turnover are relevant to the pathogenesis of several human disorders. Versican is cleaved by A Disintegrin-like And Metalloprotease with Thrombospondin type 1 motifs (ADAMTS) family members at Glu441-Ala442, generating a bioactive proteoform called versikine, but additional cleavage sites and the site-specificity of individual ADAMTS proteases is unexplored. Here, we used label-free proteomics approach to identify versican cleavage sites for 3 ADAMTS proteases, applying a novel z-score-based statistical approach to compare the protease digests of versican to controls (digests with inactivate protease) using the known protease cleavage site as a benchmark. We identified 21 novel cleavage sites that had a comparable z-score to the benchmark. Given the functional significance of versikine, they represent potentially significant cleavages and helped to refine a substrate site preference for each protease.The z-score approach is potentially widely applicable for discovery of site-specific cleavages within an purified protein or small ensemble of proteins using any protease.
Acknowledgments
Funding was provided by the NIH-NHLBI (HL107147 and HL141130), the American Heart Association-Paul G. Allen Frontiers Group Distinguished Investigator Award and the British Heart Foundation (PG/18/15/33566, and FS/IBSRF/20/25032). The Orbitrap Fusion Lumos mass spectrometer was purchased via an NIH shared instrument grant, 1S10OD023436-0.
Footnotes
conflicts of interest
None of the authors have any conflicts of interest in relation to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mjaatvedt CH, et al. , The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev Biol, 1998. 202(1): p. 56–66. [DOI] [PubMed] [Google Scholar]
- 2.Yamamura H, et al. , A heart segmental defect in the anterior-posterior axis of a transgenic mutant mouse. Dev Biol, 1997. 186(1): p. 58–72. [DOI] [PubMed] [Google Scholar]
- 3.Nandadasa S, et al. , The versican-hyaluronan complex provides an essential extracellular matrix niche for Flk1(+) hematoendothelial progenitors. Matrix Biol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choocheep K, et al. , Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J Biol Chem, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dours-Zimmermann MT, et al. , Versican V2 assembles the extracellular matrix surrounding the nodes of ranvier in the CNS. J Neurosci, 2009. 29(24): p. 7731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatano S, et al. , Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology, 2012. 22(9): p. 1268–77. [DOI] [PubMed] [Google Scholar]
- 7.Islam S, et al. , Accumulation of versican facilitates wound healing: implication of its initial ADAMTS-cleavage site. Matrix Biol, 2019. [DOI] [PubMed] [Google Scholar]
- 8.Nandadasa S, Foulcer S, and Apte SS, The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol, 2014. 35: p. 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suwan K, et al. , Versican/PG-M Assembles Hyaluronan into Extracellular Matrix and Inhibits CD44-mediated Signaling toward Premature Senescence in Embryonic Fibroblasts. J Biol Chem, 2009. 284(13): p. 8596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunning KR, et al. , Activation of Mouse Cumulus-Oocyte Complex Maturation In Vitro Through EGF-Like Activity of Versican. Biol Reprod, 2015. 92(5): p. 116. [DOI] [PubMed] [Google Scholar]
- 11.McCulloch DR, et al. , ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell, 2009. 17(5): p. 687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mead TJ, et al. , ADAMTS9-Regulated Pericellular Matrix Dynamics Governs Focal Adhesion-Dependent Smooth Muscle Differentiation. Cell Rep, 2018. 23(2): p. 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang BL, et al. , Versican G3 domain enhances cellular adhesion and proliferation of bovine intervertebral disc cells cultured in vitro. Life Sci, 2003. 73(26): p. 3399–413. [DOI] [PubMed] [Google Scholar]
- 14.Yang BL, et al. , Cell adhesion and proliferation mediated through the G1 domain of versican. J Cell Biochem, 1999. 72(2): p. 210–20. [DOI] [PubMed] [Google Scholar]
- 15.Carthy JM, et al. , Versican V1 Overexpression Induces a Myofibroblast-Like Phenotype in Cultured Fibroblasts. PLoS One, 2015. 10(7): p. e0133056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandy JD, et al. , Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS- 4. J Biol Chem, 2001. 276(16): p. 13372–8. [DOI] [PubMed] [Google Scholar]
- 17.Asano K, et al. , Stromal Versican Regulates Tumor Growth by Promoting Angiogenesis. Sci Rep, 2017. 7(1): p. 17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanhchaksai K, et al. , Host stromal versican is essential for cancer-associated fibroblast function to inhibit cancer growth. Int J Cancer, 2016. 138(3): p. 630–41. [DOI] [PubMed] [Google Scholar]
- 19.Gao D, et al. , Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res, 2012. 72(6): p. 1384–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope C, et al. , Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood, 2016. 128(5): p. 680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricciardelli C, et al. , The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev, 2009. 28(1–2): p. 233–45. [DOI] [PubMed] [Google Scholar]
- 22.Zheng PS, et al. , Versican/PG-M G3 domain promotes tumor growth and angiogenesis. FASEB J, 2004. 18(6): p. 754–6. [DOI] [PubMed] [Google Scholar]
- 23.Cikach FS, et al. , Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight, 2018. 3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didangelos A, et al. , Novel role of ADAMTS-5 protein in proteoglycan turnover and lipoprotein retention in atherosclerosis. J Biol Chem, 2012. 287(23): p. 19341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemire JM, et al. , Versican/PG-M isoforms in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol, 1999. 19(7): p. 1630–9. [DOI] [PubMed] [Google Scholar]
- 26.Wight TN and Merrilees MJ, Proteoglycans in atherosclerosis and restenosis: key roles for versican. Circ Res, 2004. 94(9): p. 1158–67. [DOI] [PubMed] [Google Scholar]
- 27.Boyd DF, et al. , Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature, 2020. 587(7834): p. 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon M, et al. , ADAMTS5 Is a Critical Regulator of Virus-Specific T Cell Immunity. PLoS Biol, 2016. 14(11): p. e1002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dours-Zimmermann MT and Zimmermann DR, A novel glycosaminoglycan attachment domain identified in two alternative splice variants of human versican. J Biol Chem, 1994. 269(52): p. 32992–8. [PubMed] [Google Scholar]
- 30.Zimmermann DR and Ruoslahti E, Multiple domains of the large fibroblast proteoglycan, versican. Embo J, 1989. 8(10): p. 2975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kischel P, et al. , Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int J Cancer, 2010. 126(3): p. 640–50. [DOI] [PubMed] [Google Scholar]
- 32.Wu YJ, et al. , The interaction of versican with its binding partners. Cell Res, 2005. 15(7): p. 483–94. [DOI] [PubMed] [Google Scholar]
- 33.Dubail J and Apte SS, Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol, 2015. 44–46: p. 24–37. [DOI] [PubMed] [Google Scholar]
- 34.Santamaria S and de Groot R, ADAMTS proteases in cardiovascular physiology and disease. Open Biol, 2020. 10(12): p. 200333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuis LE, et al. , Altered versican cleavage in ADAMTS5 deficient mice; a novel etiology of myxomatous valve disease. Dev Biol, 2011. 357(1): p. 152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis LE, et al. , Insufficient versican cleavage and Smad2 phosphorylation results in bicuspid aortic and pulmonary valves. J Mol Cell Cardiol, 2013. 60: p. 50–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kern CB, et al. , Versican proteolysis mediates myocardial regression during outflow tract development. Dev Dyn, 2007. 236(3): p. 671–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kern CB, et al. , Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev Dyn, 2006. 235(8): p. 2238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kern CB, et al. , Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol, 2010. 29(4): p. 304–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong CC, et al. , Cerebral cavernous malformations are driven by ADAMTS5 proteolysis of versican. J Exp Med, 2020. 217(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nandadasa S, B.d.R.C., Koch C, Tran-Lundmark K, Dours-Zimmermann MT, Zimemrmann DR, Valleix S, Apte SS, A new mouse mutant with cleavage-resistant versican and isoform-specific versican mutants demonstrate that proteolysis at the Glu441-Ala442 peptide bond in the V1 isoform is essential for interdigital web regression. Matrix Biology Plus, 2021. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westling J, et al. , ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem J, 2004. 377(Pt 3): p. 787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonsson-Rylander AC, et al. , Role of ADAMTS-1 in atherosclerosis: remodeling of carotid artery, immunohistochemistry, and proteolysis of versican. Arterioscler Thromb Vasc Biol, 2005. 25(1): p. 180–5. [DOI] [PubMed] [Google Scholar]
- 44.Santamaria S, et al. , Exosites in Hypervariable Loops of ADAMTS Spacer Domains control Substrate Recognition and Proteolysis. Sci Rep, 2019. 9(1): p. 10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foulcer SJ, et al. , Determinants of versican-V1 proteoglycan processing by the metalloproteinase ADAMTS5. J Biol Chem, 2014. 289(40): p. 27859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santamaria S, et al. , Exosite inhibition of ADAMTS-5 by a glycoconjugated arylsulfonamide. Sci Rep, 2021. 11(1): p. 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knight CG, Willenbrock F, and Murphy G, A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett, 1992. 296(3): p. 263–6. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Riverol Y, et al. , The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res, 2019. 47(D1): p. D442–d450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramus C, et al. , Benchmarking quantitative label-free LC-MS data processing workflows using a complex spiked proteomic standard dataset. J Proteomics, 2016. 132: p. 51–62. [DOI] [PubMed] [Google Scholar]
- 50.Schauperl M, et al. , Characterizing Protease Specificity: How Many Substrates Do We Need? PLoS One, 2015. 10(11): p. e0142658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patwari P, et al. , Mannosamine inhibits aggrecanase-mediated changes in the physical properties and biochemical composition of articular cartilage. Arch Biochem Biophys, 2000. 374(1): p. 79–85. [DOI] [PubMed] [Google Scholar]
- 52.Hills R, et al. , Identification of an ADAMTS-4 cleavage motif using phage display leads to the development of fluorogenic peptide substrates and reveals matrilin-3 as a novel substrate. J Biol Chem, 2007. 282(15): p. 11101–9. [DOI] [PubMed] [Google Scholar]
- 53.Mosyak L, et al. , Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci, 2008. 17(1): p. 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown HM, et al. , ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod, 2010. 83(4): p. 549–57. [DOI] [PubMed] [Google Scholar]
- 55.Enomoto H, Nelson C, Somerville RPT, Mielke K, Dixon L, Powell K, Apte SS, Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development, 2010. 137: p. 4029–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fava M, et al. , Role of ADAMTS (A Disintegrin and Metalloproteinase With Thrombospondin Motifs)-5 in Aortic Dilatation and Extracellular Matrix Remodeling. Arterioscler Thromb Vasc Biol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gueye NA, et al. , Versican Proteolysis by ADAMTS Proteases and Its Influence on Sex Steroid Receptor Expression in Uterine Leiomyoma. J Clin Endocrinol Metab, 2017. 102(5): p. 1631–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nandadasa S, et al. , Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat Commun, 2019. 10(1): p. 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell DL, et al. , Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem, 2003. 278(43): p. 42330–9. [DOI] [PubMed] [Google Scholar]
- 60.Dubail J, et al. , A new Adamts9 conditional mouse allele identifies its non-redundant role in interdigital web regression. Genesis, 2014. 52(7): p. 702–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dancevic CM, et al. , Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. J Biol Chem, 2013. 288(52): p. 37267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Longpre JM, et al. , Characterization of proADAMTS5 processing by proprotein convertases. Int J Biochem Cell Biol, 2009. 41(5): p. 1116–26. [DOI] [PubMed] [Google Scholar]
- 63.Silver DL, Hou L, Somerville R, Young ME, Apte SS, Pavan WJ, The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet, 2008. 4(2): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Somerville RP, et al. , Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J Biol Chem, 2003. 278(11): p. 9503–13. [DOI] [PubMed] [Google Scholar]
- 65.Carlos Rodriguez-Manzaneque J, et al. , ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun, 2002. 293(1): p. 501–8. [DOI] [PubMed] [Google Scholar]
- 66.Gendron C, et al. , Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J Biol Chem, 2007. 282(25): p. 18294–306. [DOI] [PubMed] [Google Scholar]
- 67.Tortorella MD, et al. , Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS- 4). J Biol Chem, 2000. 275(24): p. 18566–73. [DOI] [PubMed] [Google Scholar]
- 68.Nandadasa S, Nelson CM, and Apte SS, ADAMTS9-Mediated Extracellular Matrix Dynamics Regulates Umbilical Cord Vascular Smooth Muscle Differentiation and Rotation. Cell Rep, 2015. 11(10): p. 1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mittaz L, et al. , Adamts-1 is essential for the development and function of the urogenital system. Biol Reprod, 2004. 70(4): p. 1096–105. [DOI] [PubMed] [Google Scholar]
- 70.Shindo T, et al. , ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest, 2000. 105(10): p. 1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee NV, et al. , ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. Embo J, 2006. 25(22): p. 5270–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melching LI, et al. , The cleavage of biglycan by aggrecanases. Osteoarthritis Cartilage, 2006. 14(11): p. 1147–54. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez-Manzaneque JC, et al. , Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. Int J Biochem Cell Biol, 2009. 41(4): p. 800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torres-Collado AX, et al. , ADAMTS1 interacts with, cleaves, and modifies the extracellular location of the matrix inhibitor tissue factor pathway inhibitor-2. J Biol Chem, 2006. 281(26): p. 17827–37. [DOI] [PubMed] [Google Scholar]
- 75.Matthews RT, et al. , Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem, 2000. 275(30): p. 22695–703. [DOI] [PubMed] [Google Scholar]
- 76.Noborn F, et al. , Expanding the chondroitin glycoproteome of Caenorhabditis elegans. J Biol Chem, 2018. 293(1): p. 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colige A, et al. , Proteomic discovery of substrates of the cardiovascular protease ADAMTS7. J Biol Chem, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kleifeld O, et al. , Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol, 2010. 28(3): p. 281–8. [DOI] [PubMed] [Google Scholar]
- 79.Kockmann T, Carte N, Melkko S & Keller U.a.d., Identification of Protease Substrates in Complex Proteomes by iTRAQ-TAILs on a Thermo Q Exactive Instrument in Neuromethods Grant JE, Li H, Editor. 2016, Springer Science+Business Media: New York. p. 187–207. [Google Scholar]
- 80.Martin DR, et al. , Proteomics identifies a convergent innate response to infective endocarditis and extensive proteolysis in vegetation components. JCI Insight, 2020. 5(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doucet A and Overall CM, Amino-Terminal Oriented Mass Spectrometry of Substrates (ATOMS) N-terminal sequencing of proteins and proteolytic cleavage sites by quantitative mass spectrometry. Methods Enzymol, 2011. 501: p. 275–93. [DOI] [PubMed] [Google Scholar]
- 82.Doucet A and Overall CM, Broad coverage identification of multiple proteolytic cleavage site sequences in complex high molecular weight proteins using quantitative proteomics as a complement to edman sequencing. Mol Cell Proteomics, 2011. 10(5): p. M110 003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prudova A, et al. , TAILS N-Terminomics and Proteomics Show Protein Degradation Dominates over Proteolytic Processing by Cathepsins in Pancreatic Tumors. Cell Rep, 2016. 16(6): p. 1762–73. [DOI] [PubMed] [Google Scholar]
- 84.Prudova A, et al. , TAILS N-terminomics of human platelets reveals pervasive metalloproteinase-dependent proteolytic processing in storage. Blood, 2014. 124(26): p. e49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.