Abstract
Steric-blocking oligonucleotides (SBOs) are short, single-stranded nucleic acids designed to modulate gene expression by binding to RNA transcripts and blocking access from cellular machinery such as splicing factors. SBOs have the potential to bind to near-complementary sites in the transcriptome, causing off-target effects. In this study, we used RNA-seq to evaluate the off-target differential splicing events of 81 SBOs and differential expression events of 46 SBOs. Our results suggest that differential splicing events are predominantly hybridization driven, whereas differential expression events are more common and driven by other mechanisms (including spurious experimental variation). We further evaluated the performance of in silico screens for off-target splicing events, and found an edit distance cutoff of three to result in a sensitivity of 14% and false discovery rate (FDR) of 99%. A machine learning model incorporating splicing predictions substantially improved the ability to prioritize low edit distance hits, increasing sensitivity from 4% to 26% at a fixed FDR of 90%. Despite these large improvements in performance, this approach does not detect the majority of events at an FDR <99%. Our results suggest that in silico methods are currently of limited use for predicting the off-target effects of SBOs, and experimental screening by RNA-seq should be the preferred approach.
Keywords: off-target effects, steric-blocking oligonucleotides, splice-switching
Introduction
Antisense oligonucleotides (ASOs) are short, single-stranded nucleic acids designed to bind to a target transcript using Watson–Crick base pairing [1–3]. They have been successfully used as therapeutics by downregulating gene expression [4] or modulating other processes such as splicing [5].
As with other therapeutic compounds, ASOs can cause off-target effects. These can be grouped into effects caused by unintended hybridization to RNA regions that are similar to the ASO target sequence (known as hybridization-dependent off-target events), sequence-dependent effects resulting from ASO-protein interactions [6], and sequence-independent effects resulting from the chemical properties of the ASO or delivery system. Hybridization-driven effects depend on the sequence, and so it has been suggested that they can be identified through in silico screens for near-complementary sites in the transcriptome [7].
Previous studies have assessed the hybridization-dependent off-target effects of ASOs designed to degrade the target transcripts through RNase H cleavage. Oligonucleotides acting through this mechanism are known as gapmers, owing to the presence of DNA bases flanked by modified bases [1,8]. One study used microarrays to assess the off-target gene expression changes of two 13-mer locked nucleic acid gapmers [9]. They calculated the edit distance, a count of the number of mismatches or gaps between the ASO and RNA sequence, to all genes, and found that 139/256 (54%) of all downregulated genes were at an edit distance of zero or one. Similar results were observed in a study of two 17-mer locked nucleic acid gapmers [10]. Others have used quantitative PCR dose–response experiments to investigate off-targets nominated from in silico screens of 96 high-viability gapmers [11]. In this study, 97/832 (11.7%) predicted off-target sites had a reduction in gene expression with potency within 10-fold of the intended target transcript.
Steric-blocking oligonucleotides (SBOs) are fully modified ASOs that do not contain any DNA bases. Without DNA bases, RNAse H does not recognize the binding between the oligonucleotide and mRNA, and so the target mRNA is not degraded. SBOs instead act by blocking access to regulatory proteins and modifying RNA secondary structure [12,13]. These properties have been exploited therapeutically to modulate splicing [5,14,15] or to increase expression [16]. SBO-mediated splicing modulation has also been successfully used for ultra-rapid development of a tailor-made treatment for a rare disorder [17] and similar “N-of-1” therapeutic applications may become more commonplace in the near future. These valuable therapeutic applications strongly motivate understanding SBO off-target effects.
Because not all potential binding sites overlap a regulatory element, hybridization-dependent off-target effects are expected to be less frequent than for gapmers. One group used reverse transcription PCR to quantify splicing changes at off-target binding sites with low predicted minimum free energy, a measure of the stability of the ASO/RNA complex, and observed a splicing change for 22/108 (20.4%) exons [18].
In this study, we use RNA-seq to comprehensively characterize the off-target splicing effects of 81 SBOs. Differential expression off-target effects are characterized for a subset of 46 SBOs with sufficient biological replicates (see Materials and Methods section). By assessing off-target effects transcriptome-wide, not only at near-complementary sites, we are able to evaluate the sensitivity and specificity of in silico methods. To our knowledge, this is the most extensive characterization of off-target transcriptome changes induced by SBOs.
Materials and Methods
SBO design and synthesis
Eighty-one PS-MOE steric-blocking oligos of lengths 16 to 20 were designed using a variety of approaches. Forty SBOs were designed to hybridize to specific parts of the transcriptome (5′ UTR N = 2, 3′ UTR N = 6, coding exonic N = 23, intronic N = 9). Of the remaining 41, 31 were designed as “promiscuous” SBOs with more than one exact match in the transcriptome, 3 were designed as nontargeting controls with no binding sites at edit distance two or lower, and 2 as sense oligonucleotides. The remaining five SBOs were designed to contain motifs of selected RNA-binding proteins (HuR, HNRNPK, HNRNPA1, and RBFOX2), with the sequences selected from eCLIP peaks or to maximize the DeepBind score [19] for the selected protein. All SBOs were tested as part of R&D experiments, and did not go through the rigorous testing for on-target activity adopted for therapeutic programs (ie, systematic replication by independent experiments and measurement of transcript/protein levels by orthogonal experimental methods).
SBOs were synthesized at Integrated DNA Technologies using solid-supported methods in an oligonucleotide synthesizer. 2′-O-methoxyethyl (MOE) nucleotide phosphoramidites of adenosine (A), guanosine (G), 5-methyl cytidine (5-methyl C), and thymidine (T) were used in iterative detritylation, coupling, capping, and sulfurization steps. Protecting groups were removed from the oligonucleotide and the support was cleaved before desalting. The amount of salt-free oligonucleotide in the aqueous solution was measured using UV absorbance of the solution. The tube was then dried by vacuum and the resulting oligonucleotide pellet was resuspended in 1 × TE buffer, pH 8.0, to a final concentration of 100 μM, using the known amount of salt-free oligonucleotide in the tube.
RNA-seq experiments
A total of 105 RNA-seq experiments were performed in HepG2 cells (N = 48), HEK293T cells (N = 53), or PXB cells (human hepatocytes isolated from a repopulated mouse liver, N = 4) [20]. Each experiment compared samples transfected with an SBO to mock-transfected samples. Two to six (median 3) biological replicates were used per condition. SBO concentrations were chosen to maximize the on-target effect for several therapeutic programs (data not shown).
Oligonucleotide transfections were performed by reverse transfection in HepG2 and HEK293T cells using Lipofectamine RNAiMAX reagent (ThermoFisher Scientific). In brief, 300 pmol of oligonucleotide was mixed with 200 μL of OptiMEM reduced serum medium (ThermoFisher Scientific) in a 12-well plate. Three microliters of RNAiMAX was then added to each well, mixed by rocking, and allowed to incubate for 20 min before addition of cells. After the incubation, 3 × 105 cells in 800 μL of media without antibiotics (DMEM +10% FBS for HepG2, IMDM + 10% cosmic calf serum for HEK293T) was added to each well and mixed by rocking, giving a final oligonucleotide concentration of 300 nM. Cells were then incubated for 48 h, after which RNA extractions were performed either manually using the Qiagen RNeasy Mini Kit (Qiagen) or using the QIAcube automated extraction system according to the manufacturer's recommended protocols (inclusive of DNase digestion steps).
For PXB cells, oligonucleotide transfections were performed by forward transfection using Lipofectamine RNAiMAX reagent. In brief, 6.5 × 104 PXB cells were first seeded in 96-well plates in a volume of 100 μL antibiotic-free PXB culture medium plus 10% FBS. After incubation for 24 h, 50 pmol of oligonucleotide was mixed with 20 μL of OptiMEM reduced serum media combined with 0.3 μL of Lipofectamine RNAiMAX and allowed to incubate for 20 min. After Lipofectamine incubation, OptiMEM/Lipofectamine/oligonucleotide mixtures were added to the wells (417 nM final concentration of oligonucleotide), mixed by rocking, and returned to the cell culture incubator for 48 h. For RNA extractions, the Qiagen RNeasy Mini Kit was used according to the manufacturer's recommended protocol with the following exceptions. Initially, 100 μL of buffer RLT plus beta-mercaptoethanol was added to each well containing cells in the plate and pipetted up and down to lyse cells. Six replicate wells with 100 μL of lysate were then combined together and an equal volume of 70% ethanol was added to the lysate. Two separate centrifugations were performed to process the entire volume of lysate/ethanol mixture. The remainder of the protocol was followed according to the manufacturer's recommended conditions (inclusive of DNase digestion steps).
RNA quality was assessed using a Bioanalyzer 2100, and samples with RNA integrity score <7.5 were dropped. RNA-seq libraries were prepared with the NEBNext Ultra II Directional RNA preparation kit, with polyA selection performed using the NEBNext Poly(A) mRNA Magnetic Isolation module. Sequencing was carried out on an Illumina HiSeq 2500 or NovaSeq 6000 sequencer.
Cell viability assay
HepG2 cells were reverse transfected with SBOs using RNAiMAX in 96-well plates. About 20,000 HepG2 cells were seeded per well. SBOs were transfected at 400 nM and cells were incubated for 48 h at 37°C and 0.5% CO2. Viability was assessed using the CellTiter-Fluor Cell Viability Assay (Promega) and performed in quadruplicate. To calculate SBO viability scores, background fluorescence was first subtracted from the fluorescence in all wells with SBOs. Negative and positive controls on each plate were then used to create a linear mapping to a reference dataset, ensuring that the reported values are comparable across experiments.
Validation by reverse transcription PCR
Two SBOs that target the same exon and overlap by 11 bp, DG768 and DG783, were chosen for validation by reverse transcription PCR (RT-PCR). Four exons where at least one of the SBOs had an edit distance of four or five and a large splicing change was observed by RNA-seq were considered as off-target sites. For each exon–SBO pair, the original SBO was tested alongside an SBO designed to be an exact match to the off-target binding site. In parallel, the SBOs designed to hybridize to the off-target binding intervals were tested against the on-target exon of DG768 and DG783.
Oligonucleotide transfections were performed as described previously in triplicate reactions on separate days. Forty-eight hours after transfection, cells were harvested and RNA extracted using the Qiagen RNeasy Mini Kit (Qiagen) according to the manufacturer's recommended protocol. A total of 2 μg of total RNA was then used for reverse transcription with the SuperScript IV VILO master mix with EZ DNase enzyme kit (ThermoFisher). The cDNA was then diluted to a final volume of 200 μL in nuclease free water.
PCRs for CEP290, NPAS2, and TMPO were performed using KOD Hot Start DNA polymerase (Millipore Sigma) according to the manufacturer's recommended protocol using 5 μL of diluted cDNA. Cycling conditions were as follows: 95°C for 2 min, followed by 35 cycles of 95°C for 20 s, 60°C for 10 s, and 70°C for 15 s. PCRs for WDR35 and DYNC1I2 were performed using Phusion High-Fidelity PCR Master Mix (NEB) according to the manufacturer's recommended protocol using 5 μL of diluted cDNA. Cycling conditions were as follows: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 62°C for 20 s, and 72°C for 15 s. A final extension step of 5 min was also performed.
After amplification, PCR products were diluted 1:5 and subsequently analyzed using the LabChip GX Touch Nucleic Acid Analyzer (Perkin Elmer) according to the manufacturer's recommended protocol.
RNA-seq analysis
Reads were aligned with HISAT2 v2.1.0 [21] to obtain full alignments for differential splicing analyses. The alignment index was generated by combining Gencode v25 annotations with Intropolis [22] splice junctions (filtered to junctions supported by at least two samples and five reads, with one end annotated in Gencode v25, and spliced in at least 0.01% of the time). The first 20 million reads for each sample were first aligned to detect novel splice sites, and these splice sites were used as input for a final HISAT2 run to align all reads. The samples had a median of 60.2 million mapped paired-end reads. Quality control was performed by assessing read coverage, percentage duplicated reads, and 5′ to 3′ read bias with FastQC v0.11.8 and RSeQC v2.6.4.
Quantifying exon usage
We developed a novel method for quantifying exon usage based on spliced reads. For each exon, a set of possible upstream donors and downstream acceptors was compiled from all overlapping annotated Gencode v27 transcripts. Splice junctions mapping from any of these splice sites to the exon boundaries were counted as inclusion reads, I, whereas splice junctions mapping directly from upstream splice sites to downstream ones without including the exon were counted as exclusion reads, E. The splice junctions themselves do not need to be consistent with an annotated transcript, but both the 5′ and the 3′ end of the junction must be annotated as a splice site.
Because every mRNA molecule can result in at most two inclusion reads and one exclusion read, the inclusion reads were divided by two. Percent spliced in (PSI) of the exon can then be calculated as the ratio of inclusion reads to total reads.
In the case where the exon of interest shares exactly one splice site with another exon, reads mapping to the end with the shared splice junction cannot be used to distinguish between the two exons and are not counted. Only the end with the alternative splice site end is used, and spliced reads mapping to the alternative boundary of the other annotated exon are counted as exclusion reads. Because only one end of the exon is considered, each mRNA molecule can give rise to at most one inclusion read, and the inclusion read count is not divided by two.
If the exon shares its acceptor site with another exon, and its donor site with a different one, PSI cannot be quantified and such exons are dropped from the analysis. This was the case for 19,072/241,061 (7.9%) unique exons in Gencode v27.
Detecting splicing changes
To test for significant differences in PSI between treated and control samples, we used a bootstrap test. If the observed difference in PSI was owing to chance, it was comparable with the differences seen if the treated and control sample labels were shuffled. To simulate this scenario, we used a two-step bootstrap procedure that starts by randomly selecting samples with replacement, and then samples reads with replacement.
At each bootstrap iteration, treated and control samples were first sampled with replacement. The reads for these samples were pooled to a common set of reads. For each “treated” and “control” sample, reads were bootstrapped from this pooled set. The observed difference in PSI (dPSI) between the actual treated and control samples was compared with the simulated differences to obtain an empirical P value. Each exon was initially run for 1,000 bootstrap iterations. If the P value was <0.05, 49,000 additional bootstrap iterations were performed to obtain a more accurate P value estimate.
Detecting expression changes
To estimate transcript abundances for differential expression analysis, reads were pseudo-aligned to Gencode v27lift37 with Kallisto v0.46.0 [23], using 1,000 bootstrap iterations and sequence-based bias correction. Transcript-level counts were aggregated to gene-level counts with tximport v1.10.1. DESeq2 v1.22.0 [24] was used to test for differential expression between treated and control samples. Multiple testing correction was performed using the Benjamini–Hochberg method [25].
Experiment reproducibility
Average transcripts per million and PSI estimates were compared between repeated experiments using Spearman correlation. To assess reproducibility of differential expression and splicing results, log fold changes and dPSI were compared. Spearman correlation was computed between experiments for genes/exons with a significant difference between treated and control samples in at least one of the experiments. We further used the observed absolute effect size (log fold change or dPSI) as a predictor of significant events in the replication experiment. Sensitivity and false discovery rate (FDR) was evaluated at different thresholds.
Quantifying effects by edit distance
We computed the minimal edit distance between each SBO and different regions in the transcriptome. The edit distance between two strings is a count of the minimum number of insertions, deletions, or substitutions needed to transform one string into the other. For each SBO and site (eg, exon), we searched for the interval where the SBO could hybridize with the lowest edit distance, up to a maximum of edit distance four for SBOs of length 16 or 17 and a maximum of five for SBOs of length 18 or longer. Sites without any binding site at the maximum edit distance or lower were labeled as background.
To quantify splicing events by edit distance, we calculated the edit distance to all nonterminal in-frame exons, as skipping an out-of-frame exon is likely to trigger nonsense-mediated decay. We further removed all exons with a total read coverage <15 for either cases or controls owing to low power (34,004–47,427/78,541 filtered in-frame exons, median of 36,889). Multiple testing corrections were performed on the remaining exons using the Benjamini–Hochberg method. For each edit distance bin, we counted the proportion of exons where a significant (q < 0.05) change in splicing was observed.
To quantify differential expression events, comparisons with fewer than three case or control samples were removed because of low power, resulting in 64 comparisons of 46 unique SBOs. Thirty-three of the SBOs were designed to hybridize to a specific region of the transcriptome (3′ UTR N = 6, coding exonic N = 20, intronic N = 7), two were designed as sense oligonucleotides, three as nontargeting controls, three as “promiscuous” SBOs with exact complementarity to many regions, and five to contain RNA-binding protein motifs. We calculated the minimal edit distance between the SBO and the full gene (pre-mRNA or mature principal APPRIS v27 transcript), 5′’ UTR, 3′ UTR, and out-of-frame exons. For each category, the proportion of significant changes in gene expression was calculated within each edit distance bin.
Sequence dependence of expression events
RNA-seq experiments were carried out with five SBOs targeting the same exon such that four SBOs overlap each other in two sets of two. Treated samples were compared with mock-transfected samples, and the Spearman correlation of log-fold changes was computed for all pairs of SBOs. To compare the overlap of differential expression events with absolute log-fold change >1, the Jaccard index was computed for all pairs of SBOs.
Predicting splicing changes
We evaluated the sensitivity and FDR of different edit distance-based and energy-based methods for predicting off-target events. In-frame exons with a statistically significant change in PSI and an absolute dPSI of at least 0.5 were labeled as positive off-target hits. The longest exact match between the SBO and exon was computed by taking the reverse complement of the SBO and finding the longest common substring. The delta G of SBO/exon hybridization was computed using the RNA-cofold and RNA-plex methods included in ViennaRNA v2.4.14 [26], with a padding of 50 bp around the exon sequence. Exons longer than 1000 bp were excluded.
Combined splicing effect and binding affinity model
To investigate the predictive ability of a model that factors in both splicing as well as binding affinity we trained a Gradient-Boosted Decision Tree (GBDT) using the LightGBM algorithm [27]. We first created high-quality splice junction annotations using Gencode v27 and Intropolis [22]. For all genomic locations within protein-coding genes, we labeled whether it lied within an intron or exon, or if it corresponded to an acceptor or donor splice junction. We trained a Deep Convolutional Neural Network (CNN) that took the raw genomic sequence in a large window (16 kbp) and predicted the splicing annotations for every position in the input. We then used this CNN to predict the change in splicing caused by the SBO, assuming perfect hybridization to the exon. These predicted splicing scores combined with the predicted delta G, PSI of control samples, and features describing the relative position of the SBO with respect to the exon were then fed into the GBDT to predict the likelihood of an exon–SBO pair being an off-target hit. The model was trained and evaluated on exon–SBO pairs at an edit distance of five or lower (exon body or 200 bp into flanking intron). Exon–SBO pairs with a significant change in dPSI >0.2 were labeled as positives, and all other exons with coverage of at least 15 reads for both treated and control samples were used as negatives. We split the dataset with a 70:15:15 split for training, validation, and testing ensuring that no SBO or exon appears in multiple splits. Having data splits fully disjointed by both SBO and exon sequence, we can get an unbiased estimate of the model's generalization performance on new unseen examples.
Results
We systematically assessed the off-target effects of splice-switching SBOs through 105 RNA-seq experiments with 81 different SBOs (Supplementary Table S1). Forty of the SBOs were designed to hybridize to specific places in the transcriptome (5′ UTR N = 2, 3′ UTR N = 6, coding exonic N = 23, intronic N = 9), for a total of 16 different genes. Of the remaining 41, 3 were designed as nontargeting controls, 2 as sense oligonucleotides, 31 as “promiscuous” SBOs with complementarity to many parts of the transcriptome (2–30,670 exact matches in the transcriptome, median of 4), and 5 to contain RNA-binding protein motifs (Supplementary Fig. S1, Supplementary Tables S2, and S3).
For each RNA-seq experiment, treated samples were compared with mock-transfected samples and putative off-target events were identified through differential splicing analyses (Fig. 1, Materials and Methods section). Differential expression events were assessed for a subset of 46 SBOs with sufficient number of replicates. The experiments had a median of 322.5 (range 3–5,403) differentially expressed genes (q < 0.05) with a reduction in expression of at least 50% or increase of at least 2 × . Differential splicing events were less frequent, with a median of 5 (range 0–140) differentially used exons with a large change in PSI (absolute dPSI >0.5). As expected based on a hybridization and sequence-specific mechanism, nontargeting control oligos had the lowest numbers of differential splicing (median = 0) and differential expression (median = 9.5) events (Table 1).
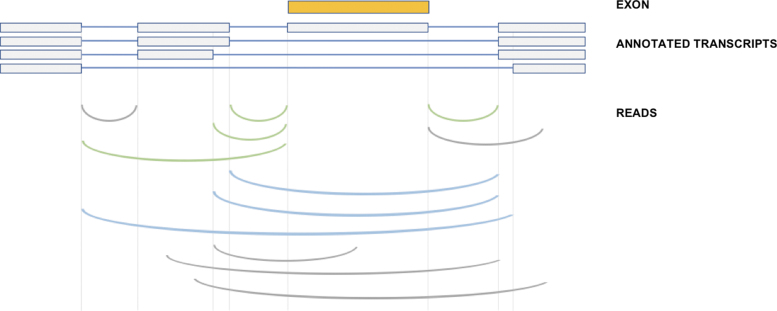
FIG. 1.
Quantification of percent spliced in for each exon. Overlapping transcripts are used to identify a set of upstream and downstream splice sites, and spliced reads mapping between these and the exon boundaries are counted as inclusion reads (green). Reads mapping directly between the upstream and downstream splice sites are counted as exclusion reads (blue). Reads where one or both ends does not match an annotated splice junction are not counted (gray).
Table 1.
Number of Significant Differential Expression Events (Absolute Log2 Fold Change >1) and Differential Splicing Events (Absolute Difference Percent Spliced in >0.5) by Category of Steric-Blocking Oligonucleotide
| Analysis | Category | Q1 | Median | Q3 | No. of SBOs | No. of experiments | Median replicates |
|---|---|---|---|---|---|---|---|
| Expression | 3′ UTR | 565.5 | 823 | 1330.75 | 6 | 8 | 3 |
| Expression | Coding exonic | 63.75 | 397.5 | 1190.25 | 20 | 30 | 3 |
| Expression | Intronic | 29 | 137 | 750 | 7 | 9 | 3 |
| Expression | Nontargeting control | 6 | 9.5 | 53.75 | 3 | 4 | 3 |
| Expression | Promiscuous | 127 | 838 | 2263 | 3 | 6 | 3 |
| Expression | RBP motif | 8 | 239 | 1580 | 5 | 5 | 3 |
| Expression | Sense | 106.5 | 171 | 235.5 | 2 | 2 | 3 |
| Splicing | 3′ UTR | 5 | 10 | 52.75 | 6 | 8 | 3 |
| Splicing | 5′ UTR | 10.25 | 12.5 | 14.75 | 2 | 2 | 2 |
| Splicing | Coding exonic | 1 | 7 | 17 | 23 | 33 | 3 |
| Splicing | Intronic | 0 | 1 | 2 | 9 | 13 | 3 |
| Splicing | Nontargeting control | 0 | 0 | 0 | 3 | 5 | 3 |
| Splicing | Promiscuous | 2 | 6 | 21 | 31 | 37 | 2 |
| Splicing | RBP motif | 0 | 3 | 41 | 5 | 5 | 3 |
| Splicing | Sense | 0 | 0 | 0 | 2 | 2 | 3 |
SBOs, steric-blocking oligonucleotides.
We assessed the reproducibility of RNA-seq by conducting repeated experiments in HepG2 cells with four SBOs, each with four to six replicates. Transfections were performed on different days by different people, and sequencing carried out in separate batches. Average expression and PSI estimates were highly reproducible (Spearman correlations of 0.88–0.91; Supplementary Fig. S2), and the effect sizes of genes and exons with a significant change in at least one of the experiments displayed a good degree of correlation (Spearman correlations of 0.34–0.77; Supplementary Fig. S3). A total of 11,084 of 32,172 (34.5%) significant differential expression events and 1,588 of 8,440 (18.8%) differential splicing events were shared between the two experiments. Differential splicing events with a large decrease in PSI were generally consistent between experiments. Several of these events occur at high edit distances, and selected ones were experimentally validated (Supplementary Fig. S4; Fig. S7).
Next, we sought to quantify off-target events at near-complementary binding sites. To quantify the similarity between the SBO and off-target binding site, we counted the number of mismatches or gaps (edit distance) between the SBO and the reverse complement of the binding site. We computed the edit distance between the 81 SBOs and all in-frame nonterminal exons, and evaluated the probability of observing an off-target effect for edit distance zero to five. Exons without a low edit distance binding site were included in the analysis to provide an estimate of the background rate of events.
Splicing changes are strongly enriched at low edit distances, with decreasing probability of observing a difference and smaller effect sizes as the edit distance increases (Fig. 2A). At exons that contain a binding site perfectly complementary to the SBO, the probability of seeing a change in PSI of at least 0.2 is ∼35%, although this could be partially driven by the selection of SBOs with an on-target effect. When restricting the analysis to SBOs that were not designed to hybridize to one location in the transcriptome, the probability drops to 16% (Supplementary Fig. S5). This result is consistent with previous screens of steric-blocking oligos [28,29], where not all exonic binding SBOs alter splicing.
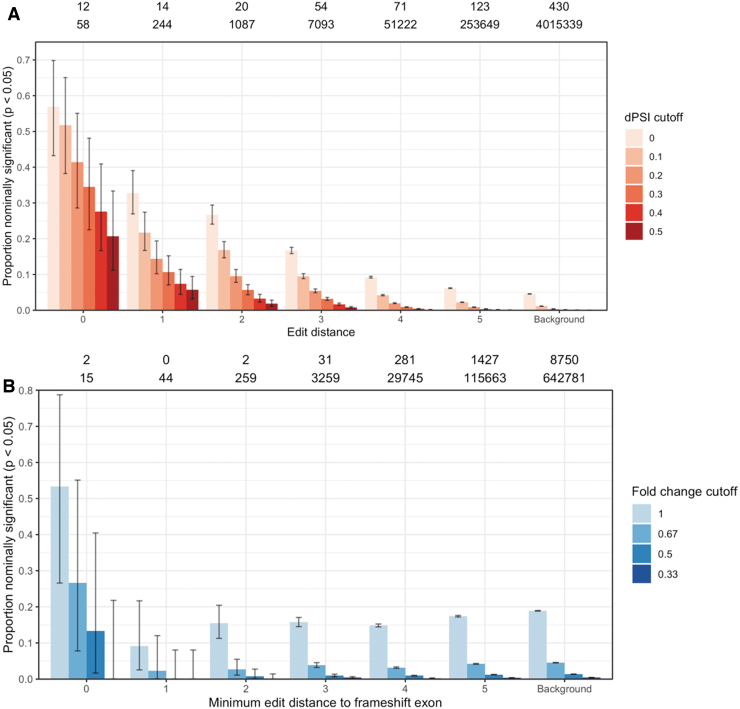
FIG. 2.
Proportion of potential binding sites resulting in a splicing (A) and expression (B) change, broken down by edit distance and effect size. The numbers above the bars give the number of significant (q < 0.05) events with absolute dPSI >0.5 (A) or fold change <0.5 (B) and the total number of events by edit distance. Error bars show 95% binomial proportion confidence intervals. dPSI, difference in percent spliced in.
Although the probability of observing a change in splicing drops off with more gaps and mismatches between the SBO and off-target binding site, even high edit distances have an enrichment of events compared with exons without an off-target binding site. Exons at edit distance five have a fivefold enrichment of large splicing changes compared with background exons. Because of the larger number of candidate exons, most differential splicing events occur at high edit distances (Supplementary Table S4). A total of 678 of 724 (93.6%) large splicing changes occur at exons with an edit distance of three or higher. Similar results were observed when restricting the analysis to the subset of 46 SBOs used for differential expression (Supplementary Fig. S6).
To confirm that the off-target splicing events at high edit distances can be caused by direct hybridization of the SBO, we picked four exon–SBO pairs at edit distances of four and five and validated the effect by RT-PCR. For each exon, the off-target SBO was tested along with an SBO designed to be fully complementary to the off-target binding site. In all cases, the fully complementary SBO caused a larger change in splicing than the partially complementary SBO. When the fully complementary SBOs were tested against the on-target site of the original SBOs, four of five caused partial skipping (Supplementary Fig. S7).
By contrast, expression changes do not have a clear association with edit distance (Fig. 2B and Supplementary Table S5). The background set of genes without a low edit distance hit have ∼19% probability of seeing a downregulation in expression, far higher than the expected type I error rate of 5%. Similar results were observed when looking at off-target binding sites in other regions of the gene and restricting to high-viability SBOs (Supplementary Figs. S8 and S9).
To assess whether differential expression events that do not depend on direct hybridization still depend on sequence, we used five SBOs designed to skip the same exon. The two pairs of overlapping SBOs had more similar expression changes than SBOs that do not overlap, although the concordance in differentially expressed genes is low for all pairs (Supplementary Fig. S10). Based on these results, we chose to focus on differential splicing events as a more direct readout of hybridization-dependent effects.
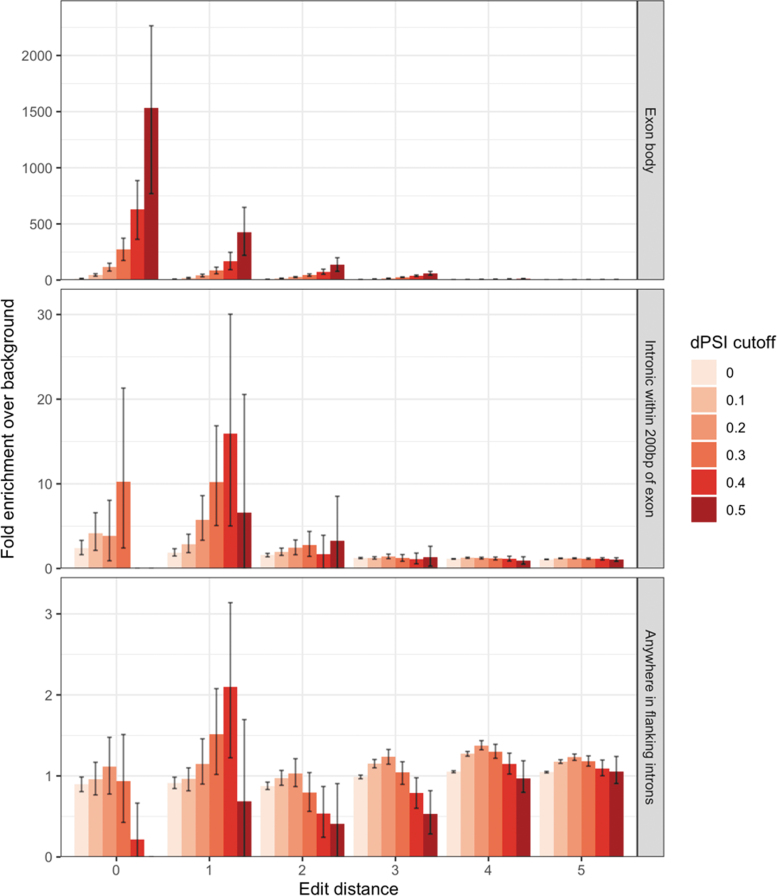
For gapmer oligonucleotides, it has previously been reported that intronic binding sites are more susceptible to off-target effects than exonic regions [30]. To investigate the effects of intronic off-target binding for SBOs, we repeated the differential splicing analysis when including the flanking intron (Fig. 3). The enrichment of significant events at low edit distances drops off as intronic sequences are included, consistent with there being fewer splicing enhancer and silencer elements in deep intronic regions [31].
FIG. 3.
Enrichment of hits compared with a background of hits at edit distance 6+ at different dPSI cutoffs, broken down by region. Error bars show 95% bootstrap confidence intervals.
The U.S. Food and Drug Administration (FDA) draft guidance for hepatitis B virus drugs suggested using in silico screens to identify potential off-target binding sites with three or fewer mismatches to an ASO [32]. Although splicing changes are more likely to occur if the edit distance between the SBO and exon is low, the majority of differential splicing events in our dataset still occur at higher edit distance sites. We therefore set out to evaluate the performance of different edit distance cutoffs for predicting off-target effects (Fig. 4 and Supplementary Table S6).
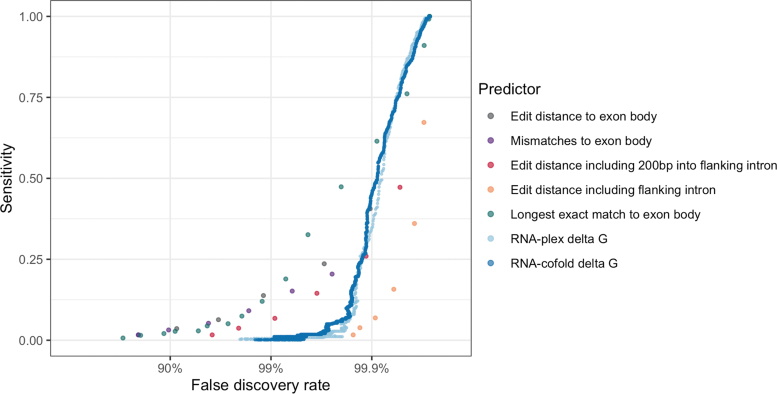
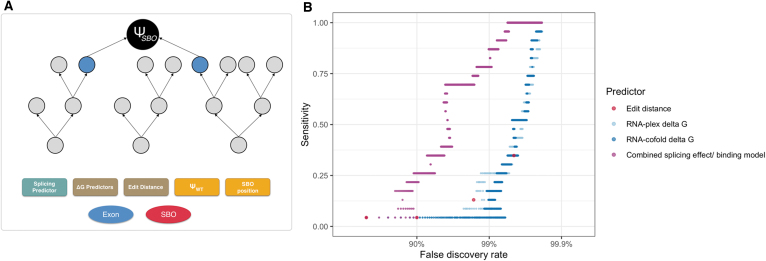
FIG. 4.
Performance of different predictors at identifying off-target splicing events with a change in PSI of at least 0.5.
If only exonic sites with exact complementarity are considered as potential off-target binding sites, only 1.7% of all large off-target splicing (absolute dPSI >0.5) events are recovered at an FDR of 79%. Increasing the edit distance cutoff improves sensitivity, but results in an overwhelming FDR. Even at an edit distance of 5, only 40.6% of all differential splicing events are recovered, and 99.91% of predicted off-target sites do not have a change in splicing. We compared this performance with gapless edit distance, longest exact match, edit distance with flanking intronic sequence, and predicted minimum free energy (delta G) from RNA-cofold and RNA-plex. The free energy predictions capture potential G:U wobble base pairs [33].
None of the predictors are able to identify the majority of off-target events at a FDR <99%. Following the FDA guidance of considering off-target binding sites at three or fewer mismatches would only identify 9.1% of the true differential splicing events, and 98.3% of the identified sites would be false positives. Similar results are observed when removing the SBOs designed to hybridize to more than one region in the transcriptome (Supplementary Fig. S11).
Edit distance and minimum free energy predictions measure the likelihood of an SBO binding to a particular sequence, but do not capture any information on the expected splicing effect.
To explore whether in silico hits (edit distance five or lower) can be further prioritized based on the expected splicing effect, we trained a gradient-boosted tree (Materials and Methods section) to predict splicing changes with a change in PSI of 0.2 or more, using both splicing predictions and binding affinity predictions as input. This model shows a clear improvement over binding affinity alone when evaluated on a test set of unseen SBOs and exons, increasing the sensitivity at 90% FDR from 4.4% to 26.1% (Fig. 5). Because of the lack of a clear association with edit distance, we did not attempt to train a similar model for differential expression results.
FIG. 5.
(A) Illustration of the gradient-boosted tree model trained to prioritize in silico hits at an edit distance of five or lower. (B) Performance when evaluated on a test set of unseen SBOs and exons. Significant splicing changes with dPSI >0.2 were labeled positives. SBOs, steric-blocking oligonucleotides.
Discussion
Steric-blocking oligos designed to hybridize to one site in the transcriptome can cause off-target effects by binding to near-complementary sites. Our results suggest that expression changes are more common than splicing changes, and also that only off-target splicing effects are predominantly hybridization dependent. Changes in transcript levels are less reproducible, and may be driven by other off-target mechanisms, or indirect transcriptional effects of on-target effects, or confounders such as batch effects. Technical factors such as transfection efficiency could also affect the reproducibility of differential expression events. Given the enrichment of splicing effects at low edit distances to in-frame exons, we would expect that there is a set of genes downregulated owing to off-target binding to an out-of-frame exon, but this effect is not clear from the data because of a high background rate of differential expression events.
Limited reproducibility and the lack of a clear, hybridization-driven mechanism suggest that differential expression events need to be replicated before they can be considered true off-target effects. Differential splicing results, especially when of large effect, are more reproducible, although replication experiments could still be necessary to accurately detect smaller splicing changes.
Unlike for gapmers, hybridization to an off-target region is not enough for an SBO to cause an effect [34]. Intronic binding sites rarely lead to a change in splicing, and even the majority of exonic binding sites do not cause a large effect, making it difficult to predict off-target events based on binding affinity alone. A previous study of two gapmers found 54% of gene expression changes to occur at an edit distance of zero or one, with an FDR of 71% [9]. By contrast, we found an edit distance cutoff of one to the exon body to result in a sensitivity of 3.6% at an FDR of 91% for differential splicing events. Differential expression events are even more difficult to predict from in silico screens, as there is no clear association with edit distance.
These results suggest that in silico methods that consider all binding sites below a certain edit distance as potential off-target hits are of limited use for SBOs, as they are likely to only capture a small fraction of all off-target events and result in a high FDR. Empirically searching for these events using RNA-seq may be a superior approach to detecting these events, although follow-up experiments may be needed to replicate the findings. Better predictors that incorporate both binding affinity and splicing effects could also improve the utility of in silico methods.
Our study is retrospective, meaning that various experimental factors could not be carefully designed a priori but rather reflect what R&D experiments have been historically performed. It is also based on experiments at a relatively high SBO dose. Dose–response experiments would help characterize off-target effects at lower doses. Another limitation is that our study is limited to PS-MOE SBOs in three different cell types. We did not investigate the effect of different chemistries or delivery methods on off-target hybridization. Larger studies that include in vivo experiments will be needed to understand the extent of hybridization-dependent off-target effects for therapeutic compounds.
Supplementary Material
Acknowledgment
The authors thank the staff of The Centre for Applied Genomics for library preparation and sequencing.
Data Availability
The raw RNA-seq data is proprietary and will only be made available to not-for-profit groups that are not pursuing any therapeutic programs in competition with Deep Genomics. Processed data are provided in Supplementary Tables.
Author Disclosure Statement
All authors are current or former employees and shareholders in Deep Genomics Inc., Deep Genomics is developing steric-blocking oligonucleotide therapies.
Funding Information
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Supplementary Material
References
- 1. Shen X and Corey DR. (2018). Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res 46:1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khvorova A and Watts JK. (2017). The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roberts TC, Langer R and Wood. MJA (2020). Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. [Epub ahead of print]; DOI: 10.1038/s41573-020-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, Planté-Bordeneuve V, Barroso FA, Merlini G, et al. (2018). Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med 379:22–31. [DOI] [PubMed] [Google Scholar]
- 5. Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, et al. (2017). Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med 377:1723–1732. [DOI] [PubMed] [Google Scholar]
- 6. Shen W, De Hoyos CL, Sun H, Vickers TA, Liang X-H and Crooke ST. (2018). Acute hepatotoxicity of 2′ fluoro-modified 5–10–5 gapmer phosphorothioate oligonucleotides in mice correlates with intracellular protein binding and the loss of DBHS proteins. Nucleic Acids Res 46:2204–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lindow M, Vornlocher H-P, Riley D, Kornbrust DJ, Burchard J, Whiteley LO, Kamens J, Thompson JD, Nochur S, et al. (2012). Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat Biotechnol 30:920–923. [DOI] [PubMed] [Google Scholar]
- 8. Crooke ST. (2004). Progress in antisense technology. Drug Deliv Syst Cancer Ther 55:61–95. [DOI] [PubMed] [Google Scholar]
- 9. Yoshida T, Naito Y, Yasuhara H, Sasaki K, Kawaji H, Kawai J, Naito M, Okuda H, Obika S and Inoue T. (2019). Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cells 24:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michel S, Schirduan K, Shen Y, Klar R, Tost J and Jaschinski F. (2021). Using RNA-seq to assess Off-target effects of antisense oligonucleotides in human cell lines. Mol Diagn Ther 25:77–85. [DOI] [PubMed] [Google Scholar]
- 11. Watt AT, Swayze G, Swayze EE and Freier. SM (2020). Likelihood of nonspecific activity of gapmer antisense oligonucleotides is associated with relative hybridization free energy. Nucleic Acid Ther. [Epub ahead of print]; DOI: 10.1089/nat.2020.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rigo F, Hua Y, Chun SJ, Prakash TP, Krainer AR and Bennett CF. (2012). Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol 8:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Havens MA and Hastings ML. (2016). Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res 44:6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dulla K, Aguila M, Lane A, Jovanovic K, Parfitt DA, Schulkens I, Chan HL, Schmidt I, Beumer W, et al. (2018). Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991 1655A>G LCA10 models. Mol Ther Nucleic Acids 12:730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komaki H, Nagata T, Saito T, Masuda S, Takeshita E, Sasaki M, Tachimori H, Nakamura H, Aoki and S'ichi Takeda Y. (2018). Systemic administration of the antisense oligonucleotide NS-065/NCNP-01 for skipping of exon 53 in patients with Duchenne muscular dystrophy. Sci Transl Med 10. [Epub ahead of print]; DOI: 10.1126/scitranslmed.aan0713. [DOI] [PubMed] [Google Scholar]
- 16. Lim KH, Han Z, Jeon HY, Kach J, Jing E, Weyn-Vanhentenryck S, Downs M, Corrionero A, Oh R, et al. (2020). Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat Commun 11:3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, Pendergast MK, Goldkind SF, Lee EA, et al. (2019). Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med 381:1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scharner J, Ma WK, Zhang Q, Lin K-T, Rigo F, Bennett CF and Krainer AR. (2020). Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res 48:802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alipanahi B, Delong A, Weirauch MT and Frey BJ. (2015). Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33:831–838. [DOI] [PubMed] [Google Scholar]
- 20. Hata K, Sayaka T, Takahashi M, Sasaki A, Umekawa Y, Miyashita K, Ogura K, Toshima G, Maeda M, Takahashi J and Kakuni M. (2020). Lipoprotein profile and lipid metabolism of PXB-cells, human primary hepatocytes from liver-humanized mice: proposal of novel in vitro system for screening anti-lipidemic drugs. Biomed Res 41:33–42. [DOI] [PubMed] [Google Scholar]
- 21. Kim D, Paggi JM, Park C, Bennett C and Salzberg SL. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nellore A, Jaffe AE, Fortin J-P, Alquicira-Hernández J, Collado-Torres L, Wang S, Phillips III RA, Karbhari N, Hansen KD, Langmead B and Leek. JT (2016). Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol 17:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bray NL, Pimentel H, Melsted P and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. [DOI] [PubMed] [Google Scholar]
- 24. Love MI, Huber W and Anders. S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benjamini Y and Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Society Series B (Methodological) 57:289–300. [Google Scholar]
- 26. Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF and Hofacker. IL (2011). ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q and Liu T-Y. (2017). LightGBM: A highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst 3146–3154. [Google Scholar]
- 28. Sinha R, Kim YJ, Nomakuchi T, Sahashi K, Hua Y, Rigo F, Bennett CF and Krainer AR. (2018). Antisense oligonucleotides correct the familial dysautonomia splicing defect in IKBKAP transgenic mice. Nucleic Acids Res 46:4833–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hua Y, Vickers TA, Baker BF, Bennett CF and Krainer. AR (2007). Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamola PJ, Kitson JDA, Turner G, Maratou K, Eriksson S, Panjwani A, Warnock LC, Douillard Guilloux GA, Moores K, et al. (2015). In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res 43:8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, et al. (2015). The human splicing code reveals new insights into the genetic determinants of disease. Science 347. [Epub ahead of print]; DOI: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Food and Drug Administration Center for Drug Evaluation and Research. (2018). Chronic Hepatitis B Virus Infection: Developing Drugs for Treatment: Guidance for Industry (Draft Guidance), FDA Maryland. accessed https://www.fda.gov/regulatory-information/search-fda-guidance-documents/chronic-hepatitis-b-virus-infection-developing-drugs-treatment.
- 33. SantaLucia J Jr and Hicks D. (2004). The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 33:415–440. [DOI] [PubMed] [Google Scholar]
- 34. Obad S, dos Santos C O, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data is proprietary and will only be made available to not-for-profit groups that are not pursuing any therapeutic programs in competition with Deep Genomics. Processed data are provided in Supplementary Tables.