Abstract
Phages infecting Campylobacter jejuni are considered a promising intervention strategy at broiler farms, yet phage sensitivity of naturally occurring poultry isolates is not well studied. Here, we investigated phage sensitivity and identified resistance mechanisms of C. jejuni strains originating from Danish broilers belonging to the most prevalent MLST (ST) types. Determining plaque formation of 51 phages belonging to Fletchervirus or Firehammervirus showed that 21 out of 31 C. jejuni strains were susceptible to at least one phage. While C. jejuni ST-21 strains encoded the common phase variable O-methyl phosphoramidate (MeOPN) receptor of the Fletchervirus and were only infected by these phages, ST-45 strains did not encode this receptor and were exclusively infected by Firehammervirus phages. To identify internal phage resistance mechanism in ST-21 strains, we performed comparative genomics of two strains, CAMSA2002 sensitive to almost all Fletchervirus phages and CAMSA2038, resistant to all 51 phages. The strains encoded diverse clustered regularly interspaced short palindromic repeats (CRISPR) spacers but none matched the tested phages. Sequence divergence was also observed in a predicted SspE homolog and putative restriction modification systems including a methyl-specific McrBC endonuclease. Furthermore, when mcrB was deleted, CAMSA2038 became sensitive to 17 out of 43 phages, three being Firehammervirus phages that otherwise did not infect any ST-21 strains. Yet, 16 phages demonstrated significantly lower efficiencies of plating on the mcrB mutant suggesting additional resistance mechanism still restricting phage propagation in CAMSA2038. Thus, our work demonstrates that C. jejuni isolates originating from broilers may have acquired several resistance mechanisms to successfully prevent phage infection in their natural habitat.
Keywords: bacteriophage, Campylobacter jejuni, comparative genomics, phage sensitivity, phage resistance, MLST (multilocus sequence typing), McrBC, restriction modification system
Introduction
Campylobacter jejuni is a zoonotic pathogen and the major cause of food borne associated gastroenteritis in the European Union (EU) (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2019). Despite the implementation of numerous intervention strategies over the years, a decline in human cases of campylobacteriosis in Europe has not been achieved (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2019). In addition, human campylobacteriosis is a rising problem in the developing countries and associated with an increased mortality rate in children (Liu et al., 2012; Platts-Mills and Kosek, 2014). C. jejuni resides as a commensal in poultry and the main transmission route leading to human infection is the consumption of improperly handled or undercooked contaminated poultry meat (Young et al., 2007; Cody et al., 2019; European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2019). Thus, novel interventions such as applying phages for biocontrol in poultry farms have been proposed and are currently being explored (Carvalho C. M. et al., 2010; Kittler et al., 2013; Chinivasagam et al., 2020). Also, the effect of adding phages directly on chicken meat to reduce the number of C. jejuni has been investigated (Goode et al., 2003; Zampara et al., 2017). Successful implementation of such strategies requires a better understanding of phage sensitivity of the highly diverse C. jejuni originating from the poultry habitat.
The genetic relationship of Campylobacter isolates is commonly studied using multilocus sequence typing (MLST) that is based on the genetic variation present in the seven housekeeping genes (glyA, uncA, pgm, gltA, glnA, aspA, and tkt) (Dingle et al., 2001). MLST and population studies have demonstrated that C. jejuni is genetically highly diverse, resulting in a large number of sequence types (STs) that can be further organized and grouped into clonal complexes (Dingle et al., 2001). Source attribution analyses have shown that MLST ST-21 and to a lesser extent ST-45 are a particular problem for human campylobacteriosis in Denmark (Boysen et al., 2014).
Also, the surface of C. jejuni is highly variable. Not only are membrane proteins N-linked glycosylated, the bacterium is also surrounded by a capsule comprised of capsular polysaccharides (CPS) and beneath this, a layer of lipo-oligosaccharides (LOS) is present (Guerry and Szymanski, 2008). Furthermore, two polar flagella ensuring motility of the bacterium are also covered by glycans as a result of the O-linked glycosylation locus (Guerry and Szymanski, 2008). Since both the CPS and O-linked glycosylation loci are genetically highly variable among C. jejuni strains, the carbohydrates composing CPS and LOS are also different (Poly et al., 2011; Karlyshev et al., 2013; Hameed et al., 2020). In addition, homopolymeric tracts (usually a stretch of G’s) are present in several genes within these loci, promoting phase variable gene expression (Parkhill et al., 2000; Bayliss et al., 2012; Aidley et al., 2018). For example, transferases that modify backbone carbohydrates of the CPS with O-methyl phosphoramidate (MeOPN) and O-methyl groups are commonly found to be phase variable expressed in C. jejuni (McNally et al., 2007; Aidley et al., 2017). Since phase variation is a stochastic process arising from slipped strand mispairing during DNA replication (Van der Woude, 2011), some cells express the transferases whereas others do not, despite carrying the same genetic loci. Thus, phase variable expression of genes within these loci further contribute to the surface diversity of C. jejuni. While the genetic content of the CPS and LOS loci show some correlation with the MLST type (Dingle et al., 2001; Hameed et al., 2020), this is not fully investigated.
Bacteriophages infecting C. jejuni primarily belong to the Firehammervirus (former Cp220likevirus) or Fletchervirus (former Cp8unalike virus) genera and are commonly isolated from chickens (cecum and fecal samples) and related environments (Atterbury et al., 2003; Hansen et al., 2007; Carvalho C. et al., 2010; Owens et al., 2013; Hammerl et al., 2014; Janež et al., 2014; Javed et al., 2014; Sørensen et al., 2015). The Campylobacter phages are highly conserved within each genus, but share very limited inter-genera homology (Javed et al., 2014). Moreover, the genomes of both Fletchervirus and Firehammervirus phages are refractory to most restriction enzymes and were recently shown to contain unusual modifications such as the replacement of guanosine with deoxyinosine and deazaguanosine (Javed et al., 2014; Crippen et al., 2019). In terms of receptor recognition, it is currently only known that Firehammervirus phages are dependent on motile flagella for infection, but an actual receptor has not yet been identified (Sørensen et al., 2015). In contrast, the Fletchervirus phages rely on CPS for infection and the phase variable MeOPN modification has been identified as a phage receptor recognized by many of these phages (Sørensen et al., 2011; Holst Sørensen et al., 2012; Gencay et al., 2018). We recently demonstrated that Fletchervirus phages encode up to four different receptor binding proteins (RBP1 to RBP4) (Sørensen et al., 2021). While RBP1 is responsible for binding to the common phase variable MeOPN receptor, phase variable expression of RBP2 ensures binding to another currently unknown receptor when MeOPN is not expressed (Sørensen et al., 2021). Thus, as a counter-resistance mechanism to phase variable expression of the common MeOPN receptor, Fletchervirus phages encode multiple receptor binding proteins including two that are phase variably expressed creating phenotypically diverse phage populations (Sørensen et al., 2021).
While receptor mutants and altered surface structures are the first line of bacterial defense, other phage resistance mechanisms target the incoming phage DNA such as restriction and modification (R/M) systems (Labrie et al., 2010). Usually nucleobase methylation provides sequence specific modification of the bacterial genome, thus allowing the cognate restriction endonuclease to discriminate and destroy unmodified invading phage DNA (Labrie et al., 2010). Only one such mechanism has been identified in C. jejuni, where a phase variable type IIG restriction-modification system provided partial resistance to phages belonging to both Fletchervirus and Firehammervirus phages (Anjum et al., 2016). Still, C. jejuni phage resistance mechanisms have only been investigated in a limited number of studies, mostly demonstrating phase variable expression of surface structures and loss of motility as the responsible factors (Coward et al., 2006; Sørensen et al., 2011; Holst Sørensen et al., 2012; Aidley et al., 2017; Gencay et al., 2018). Thus, mainly phage resistance mechanisms associated with the first step in phage infection, i.e., binding to the C. jejuni surface have been identified so far.
The study of phage sensitivity and development of phage resistance in laboratory C. jejuni strains is not sufficient to provide a complete picture of phage sensitivity and resistance mechanisms of more diverse poultry isolates of C. jejuni. Here, we aim to investigate phage susceptibility and elucidate underlying phage resistance mechanisms in C. jejuni strains isolated from Danish poultry belonging to the most prevalent MLST types observed in Denmark by performing comparative genomics.
Materials and Methods
Bacterial Strains, Phages and Growth Conditions
All bacterial strains and C. jejuni phages are listed in the Supplementary Tables 1, 2, respectively. Thirty-one C. jejuni strains were isolated from Danish broiler chickens in 2007–9 and were selected to represent the most frequent sequence types (ST) in this reservoir (Boysen et al., 2014). C. jejuni strains were standardly grown on base II agar plates containing 5% calf blood at 41.6°C under microaerobic conditions (6% O2, 6% CO2, 88% H2N2). When appropriate chloramphenicol was added to plates at a final concentration of 20 μg/ml. For phage-related work C. jejuni strains were routinely cultivated in brain heart infusion (BHI) broth (Oxoid) supplemented with 10 mM MgSO4 and 1 mM CaCl2 (CBHI).
Phage Propagation, Titration and Host Range Determination by Spot Assays
Phages were propagated and titrated as previously described (Gencay et al., 2017; Sørensen et al., 2017). Briefly, for the phage propagation, the plate lysis method was performed using corresponding C. jejuni propagation strains and original phage stocks. Propagated phage stocks were stored at 4°C. Phage titration (enumeration) and host range analyses were performed using spot assays (plaque assays) on bacterial lawns. Briefly, C. jejuni strains were standardly grown overnight, harvested into CBHI and adjusted to an optical density at 600 nm (OD600) of 0.35. Bacterial cultures were then incubated for 4 h at 41.6°C under microaerobic conditions before 500 μl was mixed with 5 ml NZCYM overlay agar (NZCYM broth [Sigma], 0.6% agar) tempered to 45°C and subsequently poured onto premade NZCYM basal agar plates (1.2% agar, 10 μg/ml vancomycin). Plates were then dried for 45 min in a flow hood. Phage stocks were tenfold serial diluted in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl, pH 7.5) up to 10–7, and three aliquots of 10 μl of the undiluted stock (100) and each serial dilution were spotted on the bacterial lawns made with relevant C. jejuni strains. Plates were incubated for 18–24 h at 41.6°C under microaerobic conditions. Following incubation plaques were counted and the mean plaque forming units per ml (pfu/ml) was calculated. For the host range analyses, all experiments were performed in duplicate and the data represent the mean of two or three independent experiments.
Genome Sequencing, Assembly and Comparative Genomics
Selected C. jejuni strains isolated from broiler chickens were genome sequenced using the Illumina platform (Illumina HiSeq) and assembled using CLC genomics workbench version 9.0.1. Average coverage of the genomes ranged from 80-fold to 269-fold. The genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). CPS loci were fully assembled using CLC genomics workbench version 9.0.1 by mapping reads to complete C. jejuni CPS reference sequences downloaded from GenBank. A CPS reference sequence was identified by blasting (BLASTN) annotated contigs containing CPS genes against all C. jejuni genomes available in the NCBI database and choosing the closest matching complete CPS sequence. Fully assembled CPS loci were subsequently confirmed by mapping sequencing reads and annotated using RASTtk (Brettin et al., 2015). The complete annotated CPS loci can be found as an appendix at Mendeley Data: https://data.mendeley.com/datasets/2n9r622g8d/1. Comparison of complete CPS loci was illustrated using Easyfig (Sullivan et al., 2011).
CAMSA2002 and CAMSA2038 were additionally sequenced on a PacBio RS II device (Pacific Biosciences, Menlo Park, CA, United States) using P6/C4 chemistry. One SMRT cell was used for each C. jejuni genome. Average coverage for CAMSA2002 was 535-fold while the average coverage for CAMSA2038 was 820-fold. Complete de novo genomes of these two strains were assembled using SMRT Analysis version 2.3 and the HGAP3 algorithm. Annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
Comparative genomics and McrBC protein sequence alignment was performed using the CLC genomics main workbench 20 and the Whole Genome Alignment plugin. Conserved domains in selected hypothetical protein sequences were identified using InterPro (Mitchell et al., 2019) and HHpred (Zimmermann et al., 2018). Pairwise comparisons between divergent genes were further conducted using BLASTN available at NCBI.
PCR and Sequencing of PolyG Regions in cj1421, cj1422, and cj1426 Homologs
PolyG regions in genes homologous to cj1421, cj1422, and cj1426 from C. jejuni NCTC11168 were amplified and sequenced (Sanger sequencing) using the primers cj1421F and cj1421R (cj1421), cj1422F and cj11422R (cj1422) and cj1426F and cj1426R (cj1426). Primer sequences are listed in Supplementary Table 3.
Phage Adsorption Assay
Phage adsorption assays were performed as previously described with minor modifications (Baldvinsson et al., 2014). Briefly, C. jejuni were grown overnight on BA plates and harvested in CBHI. Cells were pelleted by centrifugation at 6,000 × g for 5 min and re-suspended in CBHI. This washing procedure was repeated twice (total number of washes, three), and the final cell suspension was adjusted to an OD600 of 0.4. Bacteriophages were added to the bacterial suspension at a final concentration of 106 PFU/ml (multiplicity of infection [MOI] of approximately 0.0025) and incubated at 37°C with gentle shaking (50–80 rpm) for a total of 90 min. Samples containing free-phages were collected at 0, 15, 30, 60, and 90 min, filtered through an 0.22-μm sterile syringe filter (Millipore), and stored at 4°C until enumeration (plaque assay). Experiments were repeated twice and for each experiment, the calculated phage pfu/ml at time zero was designated as 100% free phages. The percentage of free phages was calculated for the remaining time points according to time zero. All experiments were performed in duplicate and the data represent the mean percentages of free phages and standard deviations thereof from the two independent experiments.
Motility Assay
Motility assays were performed as previously described (Sørensen et al., 2011). Briefly, C. jejuni strains were grown under standard conditions for 18–24 h, harvested into BHI and adjusted to an OD600 of 0.1. One microliter of the bacterial suspension was then placed in the center of five to six Heart infusion broth (HIB) (Difco) 0.25% agar plates that had been pre-dried for 45 min in a flow hood. Plates were then incubated under standard growth conditions and growth zones demonstrating movement in the soft agar were measured after 24 h. Measurements and standard deviations represent the mean counts from two independent experiments.
Construction of mcrB Deletion Mutant in C. jejuni CAMSA2038
A deletion mutant of mcrB in CAMSA2038 was constructed by replacing most of mcrB (203 bp to 2009 bp) with a cat cassette by homologous recombination. The vector construct used for homologous recombination was created by fusing three PCR amplicons as described below using In-fusion cloning (Takara). A PCR fragment (778 bp) of the flanking region upstream the mcrB deletion was amplified from C. jejuni CAMSA2038 using the primers FLOR06_OH2_F2 and FLOR06_OH2_R2. A PCR fragment (878 bp) of the flanking region downstream the mcrB deletion was amplified using the primers FLOR06_OH1_F4 and FLOR06_OH1_R3. The cat gene (723 bp) was amplified from pRY109 (Yao et al., 1993) using the primers FLOR06_Cat1_fwd and FLOR06_Cat2_rev. The cat gene was then inserted between the upstream and downstream mcrB deletion fragments and simultaneously inserted into the pET28a+ vector (Novagen) using the In-Fusion kit according to the manufacturer’s instructions. The resulting plasmid pFLOR06 was transformed into E. coli Stellar™ Competent Cells (Takara) according to manufacturer’s instructions and verified by sequencing. Purified pFLOR06 plasmid was transformed into C. jejuni CAMSA2038 by electroporation to allow homologous recombination of the flanking mcrB regions to create the CAMSA2038ΔmcrB. The mutant was verified by PCR and sequencing using the primers FLOR06_UP1_fwd, FLOR06_UP2_fwd, and FLOR06_DOWN1_rev. Plasmids and primers are listed in Supplementary Table 3.
Statistical Analyses
Data in the graphs show the mean values of two independent experiments with standard deviations depicted as error bars. When appropriate one-way ANOVA (GraphPad Prism 9.2.0) was used to assess if differences were statistical significant.
Results
Multilocus Sequence Typing ST-Type of Campylobacter jejuni Correlate With Sensitivity to Specific Phage Genera
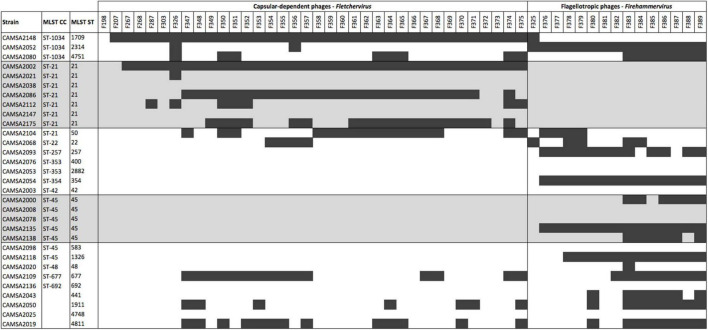
To identify putative novel phage resistance mechanisms in C. jejuni, we obtained a collection of 31 strains from Statens Serum Institute, which were isolated from broilers in Denmark between 2007 and 2009 and had previously been characterized by MLST typing. These strains represent the ST-types commonly found in broilers in Denmark and included several strains of the two dominant ST-types, ST-21 and ST-45. We then determined phage susceptibility by screening these strains for plaque formation using our collection of 51 C. jejuni phages belonging to either Fletchervirus or Firehammervirus. This host range analysis showed that 21 strains belonging a variety of STs were infected by at least one phage and most of the strains were sensitive to several phages. In contrast, ten strains belonging to different ST-types were completely resistant to all 51 phages (Figure 1). Most ST’s showed a mixed pattern of phage sensitivity, however interestingly, the phage sensitive ST-21 strains were only infected by Fletchervirus phages dependent on the CPS for infection. On the contrary, strains belonging to ST-45 were only infected by Firehammervirus phages. While our Fletchervirus phages are all dependent on the CPS for infection, the Firehammervirus are flagellotropic phages dependent on motile flagella (Sørensen et al., 2015). Thus, our results suggest a correlation between sensitivity toward a specific phage genus and the two major C. jejuni MLST types, ST-21 and ST-45, often associated with human disease.
FIGURE 1.
Phage host range analysis of C. jejuni broiler isolates characterized by MLST typing. MLST ST-21 and ST-45 isolates are highlighted in gray. Black bars indicate plaque formation detected. CC: clonal complex, ST: sequence type.
Campylobacter jejuni ST-21 Strains Encode the Common O-Methyl Phosphoramidate Phage Receptor Recognized by Fletchervirus Phages Whereas the ST-45 Strains Do Not
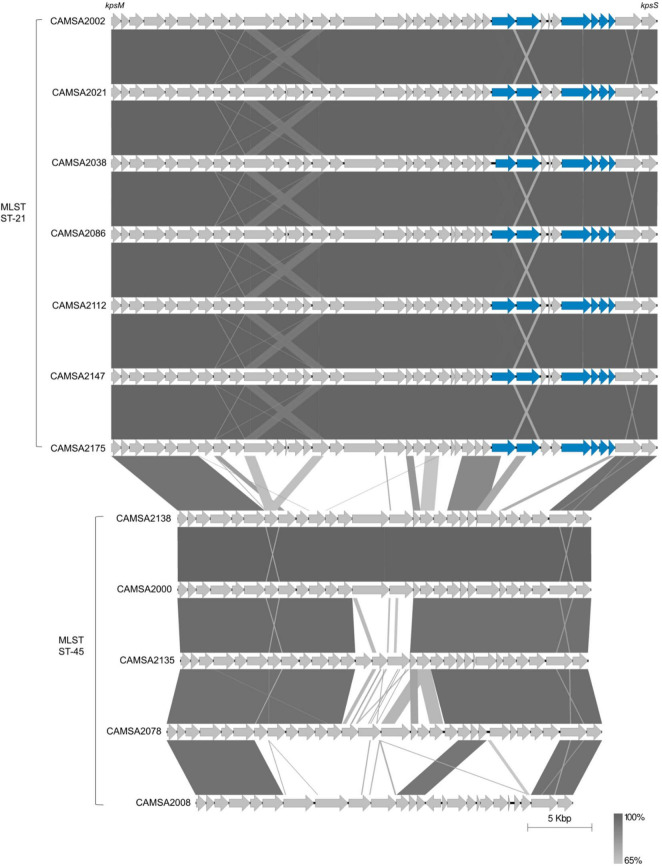
To further investigate the genetic relationship between ST-21 and ST-45, we genome sequenced all strains belonging to these MLST types. As Fletchervirus phages are dependent on CPS for infection, we fully assembled and performed comparative genomics of the CPS loci encoded by the ST-21 and ST-45 strains (Figure 2). Our analysis demonstrated that all ST-21 strains encoded identical CPS loci typical of the Penner serotype HS2 group, similar to the CPS locus of the well-characterized C. jejuni NCTC11168 strain. The HS2 CPS locus encodes all biosynthesis genes needed to produce the MeOPN modification and two MeOPN transferases cj1421 and cj1422 responsible for attaching MeOPN to GalfNAc and heptose residues in the CPS, respectively. We previously demonstrated that MeOPN forms a receptor recognized by several Fletchervirus phages both in C. jejuni strains NCTC11168 and NCTC12662 (Sørensen et al., 2011; Holst Sørensen et al., 2012; Gencay et al., 2018). Thus, all ST-21 strains encode the common MeOPN phage receptor of the Fletchervirus phages. On the contrary, the ST-45 strains encode diverse CPS loci, but interestingly no MeOPN biosynthesis or transferase genes were found. These results show that the common MeOPN phage receptor is not present in any of the ST-45 strains, explaining their resistance toward the CPS-dependent Fletchervirus phages.
FIGURE 2.
Comparison of CPS loci of ST-21 and ST-45 C. jejuni strains. CPS loci are compared using Easyfig (Sullivan et al., 2011). Genes are indicated by gray arrows except for MeOPN biosynthesis and transferase genes that are shown as blue arrows. Complete CPS loci sequences can be found at Mendeley Data: https://data.mendeley.com/datasets/2n9r622g8d/1 or are derived from the following: CAMSA2002: GenBank accession no. CP071460.1, CAMSA2038: GenBank accession nos. CP071459.1 and CAMSA2138: GenBank accession no. NZ_QELH01000003.1.
Phase Variable Expression of Capsular Polysaccharide Modifications Is Not Responsible for Phage Resistance in ST-21 Strains CAMSA2038 and CAMSA2147 Toward Fletchervirus Phages
Although all ST-21 strains encode the common MeOPN phage receptor, three of these strains (CAMSA2021, CAMSA2038, and CAMSA2147) are resistant to phage infection (Figure 1). We previously showed that phase variable expression of the MeOPN transferases cj1421 and cj1422 and the 6-O-Me transferase cj1426 lead to rapid phage resistance development in strain NCTC11168 (Sørensen et al., 2011; Holst Sørensen et al., 2012; Aidley et al., 2017). This was caused by variations in the N-terminal polyG tracts turning the expression of these genes either off (cj1421) or on (cj1422 and cj1426) promoting the loss or masking of the MeOPN receptor, respectively. To investigate if phase variation could be involved in generating phage resistance in these three ST-21 strains, we compared the polyG tract length in the cj1421, cj1422, and cj1426 homologs with sequencing data obtained following PCR amplification of polyG tract regions in these genes (Table 1). As a control, we also included the highly phage sensitive CAMSA2002 strain in our analysis. For all strains, varying polyG tracts length were detected, suggesting mixed populations with some cells expressing the transferases while others did not. Overall, expression of cj1421 was mostly turned on in all four strains leading to expression of the MeOPN receptor attached to GalfNAc. In contrast, variable tract lengths were detected in cj1422 and cj1426 turning expression of the genes both on and off, hence the presence of MeOPN attached to Hep and the 6-O-Me group varied. Thus, these results indicate that phase variable expression of surface structures leading to loss or masking of the MeOPN receptor could not be solely responsible for the observed phage resistant phenotypes. To further investigate if phage resistance was associated with lack of phage binding to the surface of the phage resistant ST-21 strains, we used phage F367 as a representative Fletchervirus and determined adsorption to CAMSA2002 (phage sensitive), CAMSA2038 (phage resistant) and CAMSA2147 (phage resistant). Our data showed that phage F367 indeed bind to both resistant strains (CAMSA2038 and CAMSA2147) at levels comparable to the phage sensitive CAMSA2002 (Figure 3). Thus, our data demonstrate that phage resistance toward Fletchervirus in CAMSA2038 and CAMSA2147 is not associated with lack of phage binding due to phase variable expression of surface structures and involves potentially novel internal phage resistance mechanisms.
TABLE 1.
PolyG tract lengths in phase variable CPS genes previously associated with phage resistance in C. jejuni as detected during genome sequencing or after complete CPS locus assembly and following PCR amplification.
| C. jejuni strain | Phage sensitivity | Origin of polyG tract | PolyG tract length and gene expression state (on/off) |
||
| MeOPN-GalfNAc transferase cj1421 homolog (gene) | MeOPN-Hep transferase cj1422 homolog (gene) | 6-O-Me transferase cj1426 homolog (gene) | |||
| CAMSA2002 | Sensitive | Genome sequence | (DDR89_07185) 9 G’s - on | (DDR89_07190) 9 G’s - on | (DDR89_07210) 9 G’s – on |
| PCR | 9 G’s - on | 10 G’s - off | aND | ||
| CAMSA2021 | Resistant | Complete CPS locus assembly | (CAMSA2021_28) 9 G’s - on | (CAMSA2021_27) 9 G’s - on | (CAMSA2021_23) 10 G’s - on |
| PCR | 9 G’s - on | 10 G’s - off | 10 G’s - on | ||
| CAMSA2038 | Resistant | Genome sequence | (DDV75_07200) 9 G’s - on | (DDV75_07205) 8 G’s - off | (DDV75_07225) 10 G’s - on |
| PCR | 10 G’s - off | 9 G’s - on | 10 G’s - on | ||
| CAMSA2147 | Resistant | Complete CPS locus assembly | (CAMSA2147_28) 9 G’s - on | (CAMSA2147_27) 9 G’s - on | (CAMSA2147_22, CAMSA2147_23) 8 G’s - off |
| PCR | 9 G’s - on | 9 G’s - on | 11 G’s - off | ||
aCould not be determined.
FIGURE 3.
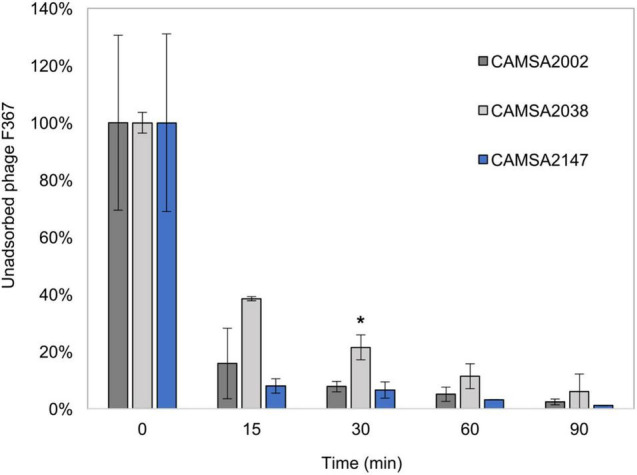
Phage F367 adsorption assay with CAMSA2002, CAMSA2038, and CAMSA2147. Unabsorbed phage F367 is illustrated in percentage over time when incubated with C. jejuni strains CAMSA2002 (phage sensitive), CAMSA2038 (phage resistant), and CAMSA2147 (phage resistant). The data represent the mean values and standard deviations from two independent experiments. *Significant different F367 absorption levels (one-way ANOVA, P-value 0.0274) was observed only after 30 min when comparing CAMSA2038 (mean: 21%) to CAMSA2002 (mean: 8%).
Motility of ST-21 and ST-45 Strains Does Not Correlate With Sensitivity Toward Firehammervirus Phages
While the receptor recognized by Firehammervirus phages has not been identified, it was previously demonstrated that lack or reduced motility can lead to phage resistance in C. jejuni toward this group of phages (Coward et al., 2006; Sørensen et al., 2015). Since all ST-21 strains are resistant to all Firehammervirus phages, we compared the motility of the ST-21 strains with motility patterns observed for the ST-45 that are only infected by this group of phages. However, even though motility varied between strains, the motility of ST-21 strains was comparable to that observed for strains belonging to ST-45 (Supplementary Figure 1). Thus, absence of or reduced motility is not responsible for Firehammervirus phage resistance observed in the ST-21 strains. To further investigate if other differences related to flagella biosynthesis could play a role in Firehammervirus phage resistance, we compared the flagellar O-linked glycosylation loci of the ST-21 and ST-45 strains. Similar to our results obtained from the comparison of the CPS loci, the flagellar O-linked glycosylation loci of all the ST-21 strains was similar to the O-linked glycosylation encoded by C. jejuni NCTC11168, whereas this locus differs among the ST-45 strains (data not shown). Since not much is known about the receptor of Firehammervirus, we could not conclude if specific components related to flagella synthesis and glycosylation was responsible for the differences in Firehammervirus sensitivity of the ST-21 and ST-45 strains. However, our results show that motility per se was not impaired in any of the strains.
The Genomes of Phage Sensitive CAMSA2002 and Phage Resistant CAMSA2038 Only Differ in Few Genes
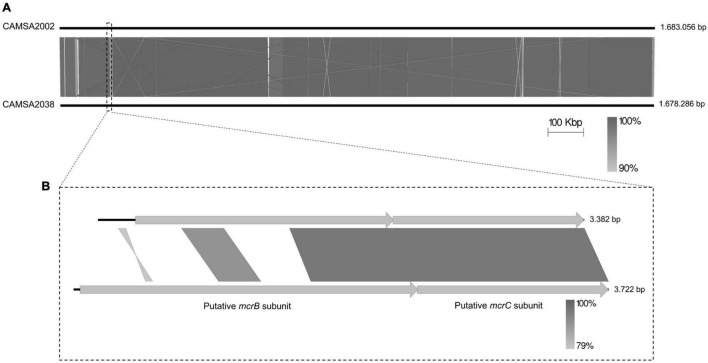
To identify possible genetic changes responsible for phage resistance in the ST-21 strain CAMSA2038, we performed comparative genomics of CAMSA2038 with the phage sensitive ST-21 CAMSA2002. Overall, we found that the genomes were highly conserved over the entire length on the chromosome including the CPS and O-linked glycosylation locus (99.57 average nucleotide identity, 99.11 alignment percentage) (Figure 4A). In addition, the phage sensitive CAMSA2002 contains a plasmid with high sequence similarity to the pTet plasmid of C. jejuni strain 81–176 (Supplementary Figure 2), a plasmid encoding tetracycline resistance as it contains a tetO gene (Bacon et al., 2000). The pTet plasmid found in CAMSA2002 (38.0 kb) is, however, substantially smaller compared to pTet in 81–176 (45.2 kb) and it does not encode the tetO gene. Interestingly, only few genes differed between the CAMSA2002 and CAMSA2038 when not considering SNP mutations (Table 2). Also, sequence divergence was detected in the clustered regularly interspaced short palindromic repeats (CRISPR) region of these two strains. In the following, we will present the major genetic differences detected in the phage sensitive CAMSA2002 and phage resistant CAMSA2038.
FIGURE 4.
Whole genome comparison of CAMSA2002 and CAMSA2038. (A) The genome sequences are depicted as solid lines and the genome sizes are listed in bp. (B) Alignment of mcrBC genes encoded by CAMSA2002 and CAMSA2038 demonstrating divergence in the N-terminal region responsible for DNA binding. Comparisons are performed using Easyfig (Sullivan et al., 2011).
TABLE 2.
Divergent genes found at corresponding genomic positions in CAMSA2002 and CAMSA2038.
| Phage sensitive CAMSA2002 |
Phage resistant CAMSA2038 |
Similarity |
||||||
| Gene | Position | Product | Gene | Position | Product | Query cover | Similarity | E-value |
| DDR89_00040 | 12.642–14.393 | DUF262 domain-containing protein | DDV75_00040 | 12.653–14.344 | DUF262 domain-containing protein | No significant similarity | ||
| DDR89_00180 | 46.404–50.144 | Eco57I restriction-modification methylase domain-containing protein | DDV75_00180 | 46.521–50.294 | Eco57I restriction-modification methylase domain-containing protein | 88% | 94.5% | 0.0 |
| DDR89_00185 | 50.147-52.291 | AAA family ATPase | DDV75_00185 | 50.693–52.141 | Pentapeptide repeat-containing protein | No significant similarity | ||
| DDR89_00190 | 52.276–52.788 | Hypothetical protein | Not found in CAMSA2038 | |||||
| DDR89_00700 | 141.547–143.345 | AAA family ATPase | DDV75_00695 | 140.472–142.823 | McrB family protein | 54% | 95.6% | 0.0 |
| DDR89_03100 | 588.738–592.166 | Hypothetical protein | DDV75_03095 | 587.945–588.988 | Type I restriction enzyme HsdR N-terminal domain-containing protein | No significant similarity | ||
| DDR89_05110 | 983.669–987.490 | N-6 DNA methylase | DDV75_05120 | 980.543–984.562 | N-6 DNA methylase | 84% | 99.7% | 0.0 |
| DDV75_06135 | 1.187.171–1.187.380 | 4-oxalocrotonate tautomerase family protein | Not found in CAMSA2002 | |||||
| DDR89_06630 | 1.275.681–1.276.898 | DUF2920 family protein | DDV75_06645 | 1.272.961–1.273.551 | DUF2920 family protein | 37% | 97.0% | 0.0 |
| – | 1.496.383–1.497.142 | CRISPR region | – | 1.492.448–1.493.011 | CRISPR region | 100% | 88.1% | 3e-143 |
| DDR89_08675 | 1.674.475–1.675.119 | Outer membrane beta-barrel protein | DDV75_08685 | 1.670.356–1.671.000 | Outer membrane beta-barrel protein | No significant similarity | ||
Clustered Regularly Interspaced Short Palindromic Repeats Is Not Associated With Phage Resistance in CAMSA2038
Sequence analysis of the CRISPR region identified more spacers in the phage sensitive CAMSA2002 strain (six spacers) compared to the phage resistant CAMSA2038 (three spacers) (Table 3). Only one spacer sequence was present in both CAMSA2002 and CAMSA2038 whereas the sequences of the remaining spacers differed between the strains (Table 3). The identical spacer shows 100% similarity to phage DA10, an unclassified phage belonging to the Myoviridae family unrelated to the Fletchervirus and Firehammervirus (Hooton et al., 2020). Furthermore, this spacer sequence also showed a match (28 out of 30 bp) to gene PC5_00034 (hypothetical protein) encoded by Campylobacter phage PC5 belonging to the Fletchervirus genus (Janež et al., 2014). However, the PC5_00034 gene is only found in the PC5 and DA10 genomes, and not present in any other currently available Fletchervirus phage genomes. Another spacer sequence found in the phage sensitive CAMSA2002 strain showed some sequence similarity to a conserved putative phosphatidyl-serine decarboxylase encoded by all Fletchervirus phages (Table 3). But the similarity was rather weak (23–25 bp out of 30 bp) explaining why this spacer does not protect against Fletchervirus phage infection, as CAMSA2002 is sensitive to 34 phages belonging to this genus. The remaining spacers found in CAMSA2038 and two spacers in CAMSA2002 show 100% sequence similarity to the genome of phage DA10. These findings show that CAMSA2002 and CAMSA2038 mainly contain sequences originating from phage DA10 in their CRISPR array. In conclusion, in silico analysis and host range data support that CRISPR is not responsible for the phage resistant nature of CAMSA2038.
TABLE 3.
CRISPR region comparison of CAMSA2002 and CAMSA2038 and spacer sequence matches to publicly available Campylobacter phage genomes.
| C. jejuni strain | CRISPR spacer sequence | Campylobacter phage genome match (GenBank accession no.) | Position in phage genome | Match identities | E-value |
| CAMSA2002 | CTACAAGAATGAGGATGATGATATTTTACAa) | DA10 (MN530981.1) | 30585-30556 | 30/30 bp (100%) | 3e-6 |
| PC5 (KX229736.1) | 24059-24088 | 28/30 bp (93%) | 0-16 | ||
| TCCATTCTCATGAAATATTTAGCCATTATT | PC5 (KX229736.1) PC14 (KX236333) NCTC12673 (FN667788) | 69233.69261 112354.112382 77362.77390 | 25/30 (73%) | 2e-06 | |
| vB_CjeM_Los1 (KX879627) F355 (MT863718) F356 (MT863719) F370 (MT863727) F371 (MT863728) F372 (MT863729) | 29450.29478 5957.5984 5956.5983 5953.5981 5953.5981 5953.5981 | 24/30 (70%) | 2e-06 | ||
| CP30A (NC_018861) CP81 (FR823450) CPX (NC_016562) CP8 (KF148616) F207 (MT863714) F336 (MT863715) F348 (MT863716) F352 (MT863717) F357 (MT863720) F358 (MT863721) F360 (MT863722) F361 (MT863723) F365 (MT863724) F367 (MT863725) F368 (MT863726) F374 (MT863730) F375 (MT863731) | 59057.79085 98709.98737 29738.29766 29972.30000 5952.5980 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 5953.5981 | 23/30 bp (66%) | 8e-06 | ||
| TTAAATTTCAAAGATGAGAGTATAGCTAA | DA10 (MN530981.1) | 24586-24558 | 28/29 bp (97%) | 0-002 | |
| GTTTCCTTCCTATTTGCTCTATACTCTAAA | None | – | – | ||
| TTAGCAACTTATAATAACTCTAATGTTATT | DA10 (MN530981.1) | 3577-3548 | 30/30 bp (100%) | 3e-6 | |
| GCCCCTGCTTTTGATTGAACAAAGCAGCCA | None | – | – | ||
| CAMSA2038 | TTTCCAAAGTTTCATTAGTTGAATTTAACT | DA10 (MN530981.1) | 33283-33312 | 30/30 bp (100%) | 3e-6 |
| CTACAAGAATGAGGATGATGATATTTTACAa) | DA10 (MN530981.1) | 30585-30556 | 30/30 bp (100%) | 3e-6 | |
| PC5 (KX229736.1) | 24059-24088 | 28/30 bp (93%) | 0-16 | ||
| CGCAACTGGTAGCACTTTAACAACTACAGAA | DA10 (MN530981.1) | 9061-9031 | 31/31 bp (100%) | 3e-6 |
CRISPR repeat sequence: GTTTTAGTCCCTTTTTAAATTTCTTTATGGTAAAAT.
a)Identical spacer sequence in CAMSA2002 and CAMSA2038.
A Putative SspE Homolog Shows Sequence Divergence Between CAMSA2002 and CAMSA2038
Phage sensitive CAMSA2002 and phage resistant CAMSA2038 both encode a DUF262-containing protein (DDR89_00040 in CAMSA2002, DDV75_00040 in CAMSA2038) in the same location of the genomes, but the protein sequences show no similarity (Table 2). We therefore conducted a detailed in silico analysis to predict putative functions of these two proteins (Supplementary Table 4). In CAMSA2038, the DUF262 is present in the N-terminal of DDV75_00040, but the protein also contains a DUF1524 located in the C-terminal and is predicted by HHpred as a SspE homolog across the entire protein length. In contrast, the corresponding protein DDR89_00040 in CAMSA2002 contains a smaller N-terminal DUF262, but no domains in the C-terminal and thus only the N-terminal region was predicted as a SspE homolog (Supplementary Table 4). Recently, SspE coupled with SspABCD was identified as a phage defense system, where SspE senses and nicks phage DNA not containing phosphorothioate (PT) modifications of the sugar-phosphate DNA backbone produced by SspABCD (Xiong et al., 2020). While the DUF262 encode NTPase and PT sensing activity, the DUF1524 domain is responsible for the nicking endonuclease activity. Thus, both domains are needed to ensure the overall structure, enzymatic and phage resistance activities of SspE (Xiong et al., 2020). It is therefore likely that only CAMSA2038 encodes a true SspE homolog (Supplementary Table 4). Furthermore, SspE only exerts its phage resistance properties when SspABCD is present. It is currently not known if C. jejuni DNA contains PT modifications needed for SspE to distinguish and recognize foreign DNA and thus to function. We therefore searched the CAMSA2038 genome for hypothetical proteins containing DUF499 (SspD) and DUF4007 (SspB) domains, however, none such proteins were found. Thus, future experimental verification is needed to show if SspE indeed is associated with phage resistance in CAMSA2038.
Several Potential Restriction-Modification Systems Differ Between CAMSA2002 and CAMSA2038
Several divergent genes of CAMSA2002 and CAMSA2038 encode putative components of restriction-modification (RM) systems (Table 2). Both CAMSA2002 and CAMSA2038 encode homologs of the RM type IIG system encoded by cj0031 in C. jejuni NCTC11168 (DDR89_00180 in CAMSA2002, DDV75_00180 in CAMSA2038). However, significant sequence divergence is observed in the C-terminal region of these genes. cj0031 encodes both the endonuclease and methylase in a single gene and variations in the C-terminal region has been associated with differences in site-specific methylation (Anjum et al., 2016). Thus, DDR89_00180 and DDV75_00180 may methylate different and potential novel sites in CAMSA2002 and CAMSA2038. In addition, the phage resistant CAMSA2038 contains a polyG tract in DDV75_00180 promoting phase variable expression of the gene.
Other differences include a Type I restriction enzyme HsdR N-terminal domain-containing protein (DDV75_03095) encoded by CAMSA2038 that is not found in CAMSA2002 (Table 2). Type I restriction-modification systems are multi-functional complexes encode by three hsd genes, hsdR (restriction of unmethylated DNA), hsdM (modification) and hsdS (recognize target sequence) where the gene products form a pentameric R2M2S complex (Cooper et al., 2017). Both CAMSA2002 and CAMSA2038 encode other hdsR, hsdM and hsdS genes, thus the role of this hypothetical HdsR protein remains unclear.
Finally, CAMSA2038 encodes a putative McrB homolog (DDV75_00695) of the McrBC type IV restriction endonuclease system, while CAMSA2002 in the corresponding position on the chromosome encodes a pseudogene (DDR89_00700) annotated as a putative AAA family ATPase (Table 2). Comparison of the two genes demonstrated a query coverage of 54% with 96% similarity, suggesting that CAMSA2002 also encodes a putative McrB homolog, yet with a significant different sequence (Figure 4B). Further In silico analysis demonstrated that the putative mcrB gene in CAMSA2002 contains frameshift mutations thereby resulting in the annotation as a pseudogene. Thus, while both CAMSA2002 and CAMSA2038 encode mcrC homologs forming the restriction endonuclease, only CAMSA2038 seems to encode a complete mcrB homolog responsible for the DNA binding (Figure 4B). Aligning the mcrB genes from CAMSA2002 and CAMSA2038 showed that the far C-terminals were highly conserved, but the N-terminals showed significant sequence divergence (Figure 4B). The N-terminal of the McrB subunit comprises the actual DNA binding region while the C-terminal region contains an AAA + GTPase domain responsible for GTP hydrolysis (Gast et al., 1997; Pieper and Pingoud, 2002; Loenen et al., 2014). Thus, the McrB encoded by CAMSA2038 may recognize a different methylation site or different type of DNA modification as compared to McrB in CAMSA2002. An additional small conserved region was observed near the middle of the two mcrB genes (position 965–1235 bp in DDV75_00695), suggesting that conservation of this region could be necessary for McrB to establish its function. Further analysis demonstrated that this region covers the beginning of the GTPase domain (data not shown).
In summary, in silico analysis indicate that different types of RM systems could be associated with phage resistance in CAMSA2038.
The McrBC Type IV Restriction Endonuclease Promotes Phage Resistance in Campylobacter jejuni Strain CAMSA2038
In E. coli, McrBC forms a methyl-specific endonuclease specifically cleaving modified DNA (Zagorskaitė et al., 2018). While McrB homologous are found in several C. jejuni strains, not much is known about their function and role in this species (Gardner and Olson, 2012). To investigate the role of McrBC in relation to phage resistance in C. jejuni we created a mcrB (DDV75_00695) deletion mutant in CAMSA2038 thereby rendering the McrBC complex non-functional. We then tested the mutant for phage sensitivity by determining plaque formation using 43 Fletchervirus and Firehammervirus phages from our collection. Our data showed that CAMSA2038ΔmcrB was sensitive to 17 out of the 43 phages tested (Table 4). Fourteen Fletchervirus phages were able to infect CAMSA2038 in the absence of mcrB. However, in all cases we observed reduced efficiencies of plating as compared to the number of plaques formed on the phage propagation hosts. In addition, plaque formation varied between experiments. Also, three Firehammervirus phages (F377, F378, and F379) were able to infect and produce plaques on the mcrB mutant. None of the ST-21 strains including CAMSA2038 were otherwise infected by Firehammervirus phages. While plaque formation of phage F377 and F379 also demonstrated reduced and variable efficiencies, F378 was able to consistently form plaques at level almost equal to what is observed on its propagation host. Thus, our data demonstrate that McrBC cause phage resistance towards both Fletchervirus and Firehammervirus phages in C. jejuni strain CAMSA2038.
TABLE 4.
Phage sensitivity of CAMSA2038ΔmcrB.
| Genus and receptor type | Phage | Propagation host | CAMSA2038 | CAMSA2038 Δ mcrB |
| Fletchervirus | F347 | 7,0 | – | – |
| CPS-dependent | F348 | 8,0 | – | – |
| phages | F349 | 5,8 | – | 4,3* |
| F350 | 6,1 | – | 4,8* | |
| F351 | 6,4 | – | 4,9* | |
| F352 | 8,5 | – | 5,5* | |
| F353 | 9,0 | – | NC plaques | |
| F354 | 8,8 | – | NC plaques | |
| F355 | 9,0 | – | – | |
| F356 | 6,9 | – | – | |
| F357 | 9,7 | – | 8,2* | |
| F358 | 9,5 | – | – | |
| F359 | 8,1 | – | 6,2* | |
| F360 | 9,2 | – | – | |
| F361 | 9,4 | – | – | |
| F362 | 8,4 | – | 6,5* | |
| F363 | 6,9 | – | – | |
| F364 | 6,9 | – | 4,7* | |
| F365 | 9,0 | – | – | |
| F366 | 9,1 | – | 6,8* | |
| F367 | 9,4 | – | 6,7* | |
| F368 | 9,2 | – | 7,5* | |
| F369 | 7,8 | – | 5,3 | |
| F370 | 9,0 | – | – | |
| F371 | 9,1 | – | – | |
| F372 | 9,4 | – | – | |
| F373 | 8,2 | – | – | |
| F374 | 6,6 | – | – | |
| F375 | 9,3 | – | – | |
| Firehammervirus | F376 | 9,4 | – | – |
| Flagellotropic | F377 | 9,3 | – | 4,8* |
| phages | F378 | 9,5 | – | 8,8 |
| F379 | 9,8 | – | 4,2* | |
| F380 | 9,4 | – | – | |
| F381 | 8,8 | – | – | |
| F382 | 8,7 | – | – | |
| F383 | 9,1 | – | – | |
| F384 | 9,6 | – | – | |
| F385 | 9,6 | – | – | |
| F386 | 9,4 | – | – | |
| F387 | 9,2 | – | – | |
| F388 | 9,8 | – | – | |
| F389 | 9,5 | – | – |
Values represent log10 (pfu/ml). NC, non-countable plaques. –, no lysis or plaques detected. *Plaques detected in 1 or 2 out of three experiments.
Discussion
Despite implementation of numerous intervention strategies during the last decades, C. jejuni remains the major cause of foodborne gastroenteritis in EU as well as in developing countries (Platts-Mills and Kosek, 2014; European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2019). While phage therapy is a promising strategy to reduce C. jejuni in primary production and on food, not much is known about phage sensitivity and phage resistance mechanisms in naturally occurring isolates originating from poultry and broilers. Previous studies using laboratory C. jejuni strains have shown that phase variable expression of surface structures such as a common phage receptor, and loss of motility cause resistance toward Fletchervirus and Firehammervirus phages infecting C. jejuni (Coward et al., 2006; Sørensen et al., 2011; Holst Sørensen et al., 2012; Aidley et al., 2017; Gencay et al., 2018). In contrast, only a few internal phage resistance mechanisms have been described in C. jejuni involving induction of a Mu-like prophage and phase variable expression of a type IIG RM system able to partially restrict invading phage DNA (Scott et al., 2007; Anjum et al., 2016). Here, we determine the phage susceptibility of 31 C. jejuni isolates originating from Danish broilers and use comparative genomics to identify several putative phage resistance mechanisms. Furthermore, we provide experimental evidence that the type IV McrBC restriction endonuclease acts as a phage resistance mechanism in C. jejuni.
Population studies of C. jejuni strains using MLST have demonstrated a large genetic diversity within this species (Dingle et al., 2002). However, the MLST types ST-21 and ST-45 are the two most frequent STs recorded in the pubMLST database (4% each; February 6, 2021)1. Despite a large geographical variation in MLST distributions, most studies find that ST-21 and ST-45 are often associated with foodborne gastroenteritis and represent the most prevalent STs both in broilers and human cases of campylobacteriosis (Cody et al., 2012; McCarthy et al., 2012; Griekspoor et al., 2015; Mossong et al., 2016). It is therefore important to investigate the phage susceptibility of particularly C. jejuni ST-21 and ST-45 strains originating from the broiler habitat to support successful implementation of phage therapy against C. jejuni at farm level. Using 51 phages we found a correlation between sensitivity to a phage genus and the ST-type as our ST-21 strains were only infected by Fletchervirus phages, whereas ST-45 strains were exclusively infected by phages belonging to the Firehammervirus genus. ST-21 and ST-45 represent two genetically distinct generalist linages that have emerged independently, but both having a wide range of host animals (Sheppard et al., 2014; Yahara et al., 2017). It is thus intriguing to speculate if the two distinct Campylobacter phage genera have evolved to specifically target one of these major lineages. Yet, the number of ST-21 (seven) and ST-45 (five) strains included in our analysis was rather limited, thus it will be important to test the phage susceptibility of more such isolates to verify if a correlation indeed exist.
A number of other studies have investigated the phage susceptibility of several diverse C. jejuni isolates originating from the chicken/broiler habitat (Loc Carrillo et al., 2005; Hansen et al., 2007; Hammerl et al., 2014; Janež et al., 2014; Nowaczek et al., 2019). In all these studies, several isolates demonstrated complete resistance toward the phages applied, however, often only a limited number of phages were tested, which could explain the high level of resistance. Interestingly, in one study nine phages were screened against 49 C. jejuni strains of both human (24 strains) and broiler (25 strains) origin, and while 80% of the strains of human origin were susceptible to phage infection only 44% of the C. jejuni strains of broiler origin could be infected by the phages (Janež et al., 2014). We also observed that 10 out of 31 strains were completely resistant to phage infection despite including 51 phages in our analysis. These phage resistant strains belonged to several diverse ST types and were also found within the ST-21 and ST-45 groups such as CAMSA2038 (ST-21). Such observations suggest that the C. jejuni strains present in broilers may have acquired several phage resistance mechanisms in order to survive the phage predation in their natural habitat. Thus, this could represent a challenge for the future phage applications targeting C. jejuni in broilers and supports the need for investigating underlying resistance mechanisms in these isolates.
Our comparative genomics identified several RM systems that were divergent between the phage sensitive CAMSA2002 and phage resistant CAMSA2038 strains, including the type IV restriction endonuclease McrBC cleaving only modified DNA. While McrBC is widespread in C. jejuni, little is known about its function, but it was previously noted that expression of mcrC was 1.75-fold upregulated in C. jejuni NCTC11168 during infection with the Fletchervirus phage NCTC12673 (Sacher et al., 2018). We here experimentally confirm that McrBC of CAMSA2038 acts as a novel phage resistance mechanism in C. jejuni targeting phages belonging to both Fletchervirus and Firehammervirus. The genomes of both Fletchervirus and Firehammervirus phages are highly modified containing unusual bases, but also other types of DNA modifications may exist due to their refractory nature toward several restriction nucleases (Javed et al., 2014; Crippen et al., 2019). In E. coli, McrBC was shown to specifically cleave phage genomes containing 5-hydroxymethycytosine, 5-methylcytosine and N4-methylcystosine (Zagorskaitė et al., 2018). McrB recognizes these modified bases by a base flipping mechanism where cytosine is flipped out of the DNA helix and positioned within the binding pocket of McrB for further discrimination (Sukackaite et al., 2012). In E. coli, the pocket size and pocket residues distinguish pyrimidines from purines to pre-select cytosine and is large enough to accommodate cytosine, 5mC, 5hmC, or 4mC, but cannot fit glycosylated derivatives (Sukackaite et al., 2012; Zagorskaitė et al., 2018). Interestingly, the McrB subunit in C. jejuni CAMSA2038 (784 aa) is almost double the size of the McrB subunit in E. coli K12 (459 aa), mainly due to a larger N-terminal region responsible for DNA binding. It is tempting to speculate that the larger N-terminal in CAMSA2038 creates a different pocket size, pocket residues or structural organization, allowing another type of base discrimination, and consequently different or larger modified bases to be recognized (Zagorskaitė et al., 2018) such as modifications encoded by the Fletchervirus and Firehammervirus phages. Indeed, it is interesting that McrBC is able to target the DNA of both Fletchervirus and Firehammervirus phages, as these two genera are highly distinct genetically and the base substitutions already identified are diverse, i.e., deoxyinosine versus deazaguanine (Crippen et al., 2019). This supports that McrBC also in C. jejuni may recognize a more common DNA modification found in some phages of both genera. Most type IV RM systems described so far indeed recognize and cleave the common m6A, 5mC, or 5hmC modifications despite demonstrating huge diversity (Loenen et al., 2014), and it is currently unknown if our C. jejuni phages contain such DNA modifications. However, the discovery of other type IV RM systems such as the PvuRts1 family and GmrSD that recognize and cleave larger glycosylated bases (Janosi et al., 1994; Bair and Black, 2007; Weigele and Raleigh, 2016) demonstrate that other types of modified DNA certainly can be targeted by this group of enzymes. Especially the observation that our Firehammervirus phage F378 showed consistent plaque formation on the CAMSA2038ΔmcrB mutant at frequencies comparable to its propagation host, clearly demonstrate McrBC acting as the major defense mechanism in CAMSA2038 against this phage. We are currently investigating DNA modifications and modification motifs present in phage F378 to further elucidate the type of DNA modification recognized by McrBC in C. jejuni.
Resistance caused by type I-III RM systems against phage infection is rarely as efficient as preventing the phage from binding such as by altering or removing the phage receptor (Labrie et al., 2010). While these RM systems decrease the chance of productive phage infection, they rarely stop infection completely, as the phage DNA can become modified by host methylases, thus rendering host endonucleases ineffective against the phage progeny (Sneppen et al., 2015). This was observed in C. jejuni NCTC11168, where the type IIG RM system only lead to partial resistance against Fletchervirus and Firehammervirus phages (Anjum et al., 2016). Here, we identified putative type I and II RM systems in the phage resistant C. jejuni strain (CAMSA2038) that could play a role in generating phage resistance, including a homolog of the type IIG RM system. Thus, the presence of several RM systems targeting invading phage DNA may have complementary effects to consequently prevent phage infection more effectively as observed for CAMSA2038. This is supported by our observation that most Fletcherviruses and Firehammervirus showed a much lower efficiency of plating on the mcrB mutant in CAMSA2038 compared to their propagation hosts, suggesting that additional factors are still restricting efficient phage replication in CAMSA2038. For many of these phages, we also observed variable results, i.e., plaque formation occurring in some experiments while not in others, in addition to smaller sized plaques being formed on CAMSA2038ΔmcrB (data not shown). We have previously observed variation in plaque formation of Fletchervirus phages due to phase variable expression of CPS surface structure including the common MeOPN phage receptor recognized by this group of phages (Sørensen et al., 2011; Holst Sørensen et al., 2012; Gencay et al., 2018). But, also in CAMSA2038 the type IIG RM system is phase variably expressed due to the presence of a polyG tract as observed in C. jejuni strain NCTC11168 (Anjum et al., 2016). Indeed, smaller plaque sizes have been associated with low adsorption rates (Gallet et al., 2011), but could also reflect progeny phages circumventing type I and II RM systems due to methylation by the corresponding host methylases. We previously demonstrated that phase variable expression of CPS surface structures acts as a major defense mechanism against the Fletchervirus phages also in the chicken gut (Sørensen et al., 2011; Holst Sørensen et al., 2012; Aidley et al., 2017; Gencay et al., 2018). Yet, we recently discovered that phages belonging to this genus can encode up to four different receptor binding protein which allows these phages to infect C. jejuni in the absence of the common phase variable MeOPN receptor (Sørensen et al., 2021). This supports the need for complementary phage defense mechanisms in C. jejuni to efficiently prevent infection by this group of phages. Resistance toward flagellotropic C. jejuni phages has mostly been associated with loss of motility in laboratory settings (Coward et al., 2006; Scott et al., 2007; Baldvinsson et al., 2014). Motility is however, a key feature for establishing colonization of C. jejuni in the chicken gut (Young et al., 2007; Epps et al., 2013), thus additional defense mechanism such as McrBC targeting such phages, i.e., Firehammervirus are expected to be present in poultry isolates.
In conclusion, our work demonstrates that C. jejuni isolates originating from broilers may have acquired several resistance mechanisms to efficiently prevent phage infection in their natural habitat and that the study of such isolates may reveal novel phage resistance mechanisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the Supplementary Material and below: https://data.mendeley.com/datasets/2n9r622g8d/1, Mendeley Data.
Author Contributions
MS, YG, EN, and LB contributed to the conceptualization and funding acquisition. MS, YG, JK, EN, and LB contributed to the methodology and resources. MS, YG, FF, MC, MM, and JK contributed to the investigation. MS and YG contributed to the formal analysis and supervision. MS and LB contributed to the writing – original draft. All authors contributed to the article and approved the submitted version.
Conflict of Interest
YG is currently employed by SNIPR Biome. The salary of MS was partially funded by Intralytix, Inc., but the company had no influence on the design of the study nor any impact on the conclusions of the presented work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank laboratory technician Vi Phuong Thi Nguyen for excellent technical assistance. We also thank Jonas Torgny Björkman for technical assistance on whole genome sequencing and initial assembly of some of the CAMSA strains and Cedric Woudstra for assistance with drafting figures in Easyfig. We are thankful to Anna Bratus-Neuenschwander and additional staff of the Functional Genomics Center Zurich for their excellent technical assistance with SMRT genome sequencing of CAMSA2002 and CAMSA2038.
Footnotes
Funding
This work was funded by the Danish AgriFish Agency of Ministry of Environment and Food (34009-14-0873), the European Union’s Horizon 2020 Marie Skladowska Curie Individual Fellowship (705817) and by Intralytix, Inc.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.780559/full#supplementary-material
References
- Aidley J., Sørensen M. C. H., Bayliss C. D., Brøndsted L. (2017). Phage exposure causes dynamic shifts in the expression states of specific phase-variable genes of Campylobacter jejuni. Microbiology 163 911–919. 10.1099/mic.0.000470 [DOI] [PubMed] [Google Scholar]
- Aidley J., Wanford J. J., Green L. R., Sheppard S. K., Bayliss C. D. (2018). PhasomeIt: an ‘omics’ approach to cataloguing the potential breadth of phase variation in the genus Campylobacter. Microb. Genom. 4:e000228. 10.1099/mgen.0.000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum A., Brathwaite K. J., Aidley J., Connerton P. L., Cummings N. J., Parkhill J., et al. (2016). Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168. Nucleic Acids Res. 44 4581–4594. 10.1093/nar/gkw019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atterbury R. J., Connerton P. L., Dodd C. E., Rees C. E., Connerton I. F. (2003). Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69 4511–4518. 10.1128/aem.69.8.4511-4518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon D. J., Alm R. A., Burr D. H., Hu L., Kopecko D. J., Ewing C. P., et al. (2000). Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68 4384–4390. 10.1128/iai.68.8.4384-4390.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair C. L., Black L. W. (2007). A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 366 768–778. 10.1016/j.jmb.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldvinsson S. B., Sørensen M. C. H., Vegge C. S., Clokie M. R. J., Brøndsted L. (2014). Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Appl. Environ. Microbiol. 80 7096–7106. 10.1128/AEM.02057-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss C. D., Bidmos F. A., Anjum A., Manchev V. T., Richards R. L., Grossier J. P., et al. (2012). Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 40 5876–5889. 10.1093/nar/gks246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen L., Rosenquist H., Larsson J. T., Nielsen E. M., Sørensen G., Nordentoft S., et al. (2014). Source attribution of human campylobacteriosis in Denmark. Epidemiol. Infect. 142 1599–1608. 10.1017/S0950268813002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C., Susano M., Fernandes E., Santos S., Gannon B., Nicolau A., et al. (2010). Method for bacteriophage isolation against target Campylobacter strains. Lett. Appl. Microbiol. 50 192–197. 10.1111/j.1472-765X.2009.02774.x [DOI] [PubMed] [Google Scholar]
- Carvalho C. M., Gannon B. W., Halfhide D. E., Santos S. B., Hayes C. M., Roe J. M., et al. (2010). The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 10:232. 10.1186/1471-2180-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinivasagam H. N., Estella W., Maddock L., Mayer D. G., Weyand C., Connerton P. L., et al. (2020). Bacteriophages to control Campylobacter in commercially farmed broiler chickens, in Australia. Front. Microbiol. 11:632. 10.3389/fmicb.2020.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody A. J., Maiden M. C., Strachan N. J., McCarthy N. D. (2019). A systematic review of source attribution of human campylobacteriosis using multilocus sequence typing. Euro. Surveill. 24:1800696. 10.2807/1560-7917.ES.2019.24.43.1800696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody A. J., McCarthy N. M., Wimalarathna H. L., Colles F. M., Clark L., Bowler I. C., et al. (2012). A longitudinal 6-year study of the molecular epidemiology of clinical campylobacter isolates in Oxfordshire, United Kingdom. J. Clin. Microbiol. 50 3193–3201. 10.1128/JCM.01086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L. P., Roberts G. A., White J. H., Luyten Y. A., Bower E. K., Morgan R. D., et al. (2017). DNA target recognition domains in the Type I restriction and modification systems of Staphylococcus aureus. Nucleic Acids Res. 45 3395–3406. 10.1093/nar/gkx067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward C., Grant A. J., Swift C., Philp J., Towler R., Heydarian M., et al. (2006). Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl. Environ. Microbiol. 72 4638–4647. 10.1128/AEM.00184-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippen C. S., Lee Y. J., Hutinet G., Shajahan A., Sacher J. C., Azadi P., et al. (2019). Deoxyinosine and 7-Deaza-2-Deoxyguanosine as carriers of genetic information in the DNA of Campylobacter viruses. J. Virol. 93 e1111–e1119. 10.1128/JVI.01111-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Ure R., Wagenaar J. A., Duim B., Bolton F. J., et al. (2002). Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8 949–955. 10.3201/eid0809.020122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Wareing D. R., Ure R., Fox A. J., Bolton F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39 14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epps S. V., Harvey R. B., Hume M. E., Phillips T. D., Anderson R. C., Nisbet D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 10 6292–6304. 10.3390/ijerph10126292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC] (2019). The European union one health 2018 Zoonoses report. EFSA J. 17:e05926. 10.2903/j.efsa.2019.5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet R., Kannoly S., Wang I. N. (2011). Effects of bacteriophage traits on plaque formation. BMC Microbiol. 11:181. 10.1186/1471-2180-11-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. P., Olson J. W. (2012). Barriers to horizontal gene transfer in Campylobacter jejuni. Adv. Appl. Microbiol. 79 19–42. 10.1016/b978-0-12-394318-7.00002-4 [DOI] [PubMed] [Google Scholar]
- Gast F. U., Brinkmann T., Pieper U., Kruger T., Noyer-Weidner M., Pingoud A. (1997). The recognition of methylated DNA by the GTP-dependent restriction endonuclease McrBC resides in the N-terminal domain of McrB. Biol. Chem. 378 975–982. 10.1515/bchm.1997.378.9.975 [DOI] [PubMed] [Google Scholar]
- Gencay Y. E., Birk T., Sørensen M. C., Brøndsted L. (2017). Methods for isolation, purification, and propagation of bacteriophages of campylobacter jejuni. Methods Mol Biol. 1512 19–28. 10.1007/978-1-4939-6536-6_3 [DOI] [PubMed] [Google Scholar]
- Gencay Y. E., Sørensen M. C. H., Wenzel C. Q., Szymanski C. M., Brøndsted L. (2018). Phase variable expression of a single phage receptor in Campylobacter jejuni NCTC12662 influences sensitivity toward several diverse CPS-dependent phages. Front Microbiol. 9:82. 10.3389/fmicb.2018.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode D., Allen V. M., Barrow P. A. (2003). Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69 5032–5036. 10.1128/AEM.69.8.5032-5036.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor P., Engvall E. O., Åkerlind B., Olsen B., Waldenström J. (2015). Genetic diversity and host associations in Campylobacter jejuni from human cases and broilers in 2000 and 2008. Vet. Microbiol. 178 94–98. 10.1016/j.vetmic.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Guerry P., Szymanski C. M. (2008). Campylobacter sugars sticking out. Trends Microbiol. 16 428–435. 10.1016/j.tim.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Hameed A., Woodacre A., Machado L. R., Marsden G. L. (2020). An updated classification system and review of the lipooligosaccharide biosynthesis gene locus in Campylobacter jejuni. Front. Microbiol. 11:677. 10.3389/fmicb.2020.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerl J. A., Jäckel C., Alter T., Janzcyk P., Stingl K., Knüver M. T., et al. (2014). Reduction of Campylobacter jejuni in broiler chicken by successive application of group II and group III phages. PLoS One 9:e114785. 10.1371/journal.pone.0114785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen V. M., Rosenquist H., Baggesen D. L., Brown S., Christensen B. B. (2007). Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 7:90. 10.1186/1471-2180-7-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Sørensen M. C., van Alphen L. B., Fodor C., Crowley S. M., Christensen B. B., Szymanski C. M., et al. (2012). Phase variable expression of capsular polysaccharide modifications allows Campylobacter jejuni to avoid bacteriophage infection in chickens. Front. Cell Infect. Microbiol. 2:11. 10.3389/fcimb.2012.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton S., D’Angelantonio D., Hu Y., Connerton P. L., Aprea G., Connerton I. F. (2020). Campylobacter bacteriophage DA10: an excised temperate bacteriophage targeted by CRISPR-cas. BMC Genomics 21:400. 10.1186/s12864-020-06808-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janež N., Kokošin A., Zaletel E., Vranac T., Kovač J., Vučković D., et al. (2014). Identification and characterisation of new Campylobacter group III phages of animal origin. FEMS Microbiol. Lett. 359 64–71. 10.1111/1574-6968.12556 [DOI] [PubMed] [Google Scholar]
- Janosi L., Yonemitsu H., Hong H., Kaji A. (1994). Molecular cloning and expression of a novel hydroxymethylcytosine-specific restriction enzyme (PvuRts1I) modulated by glucosylation of DNA. J. Mol. Biol. 242 45–61. 10.1006/jmbi.1994.1556 [DOI] [PubMed] [Google Scholar]
- Javed M. A., Ackermann H. W., Azeredo J., Carvalho C. M., Connerton I., Evoy S., et al. (2014). A suggested classification for two groups of Campylobacter myoviruses. Arch. Virol. 159 181–190. 10.1007/s00705-013-1788-2 [DOI] [PubMed] [Google Scholar]
- Karlyshev A. V., Quail M. A., Parkhill J., Wren B. W. (2013). Unusual features in organisation of capsular polysaccharide-related genes of C. jejuni strain X. Gene 522 37–45. 10.1016/j.gene.2013.03.087 [DOI] [PubMed] [Google Scholar]
- Kittler S., Fischer S., Abdulmawjood A. (2013). Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Appl. Environ. Microbiol. 79 7525–7533. 10.1128/AEM.02703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie S., Samson J., Moineau S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8 317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- Liu L., Johnson H. L., Cousens S., Perin J., Scott S., Lawn J. E., et al. (2012). Child health epidemiology reference group of WHO and UNICEF. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379 2151–2161. 10.1016/S0140-6736(12)60560-1 [DOI] [PubMed] [Google Scholar]
- Loc Carrillo C., Atterbury R. J., El-Shibiny A., Connerton P. L., Dillon E., Scott A., et al. (2005). Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71 6554–6563. 10.1128/aem.71.11.6554-6563.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. A. M., Dryden D. T. F., Raleigh E. A., Wilson G. G., Murray N. E. (2014). Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 42 3–19. 10.1093/nar/gkt990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N. D., Gillespie I. A., Lawson A. J., Richardson J., Neal K. R., Hawtin P. R., et al. (2012). Molecular epidemiology of human Campylobacter jejuni shows association between seasonal and international patterns of disease. Epidemiol. Infect. 140 2247–2255. 10.1017/S0950268812000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally D. J., Lamoureux M. P., Karlyshev A. V., Fiori L. M., Li J., Thacker G., et al. (2007). Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282 28566–28576. 10.1074/jbc.M704413200 [DOI] [PubMed] [Google Scholar]
- Mitchell A. L., Attwood T. K., Babbitt P. C., Blum M., Bork P., Bridge A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47 D351–D360. 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossong J., Mughini-Gras L., Penny C., Devaux A., Olinger C., Losch S., et al. (2016). Human Campylobacteriosis in Luxembourg, 2010-2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci. Rep. 6:20939. 10.1038/srep20939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczek A., Urban-Chmiel R., Dec M., Puchalski A., Stępień-Pyśniak D., Marek A., et al. (2019). Campylobacter spp. and bacteriophages from broiler chickens: characterization of antibiotic susceptibility profiles and lytic bacteriophages. Microbiologyopen 8:e784. 10.1002/mbo3.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens J., Barton M. D., Heuzenroeder M. W. (2013). The isolation and characterization of Campylobacter jejuni bacteriophages from free range and indoor poultry. Vet. Microbiol. 162 144–150. 10.1016/j.vetmic.2012.08.017 [DOI] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403 665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- Pieper U., Pingoud A. (2002). A mutational analysis of the PD.D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry 41 5236–5244. 10.1021/bi0156862 [DOI] [PubMed] [Google Scholar]
- Platts-Mills J. A., Kosek M. (2014). Update on the burden of Campylobacter in developing countries. Curr. Opin. Infect. Dis. 27 444–450. 10.1097/QCO.0000000000000091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F., Serichatalergs O., Schulman M., Ju J., Cates C. N., Kanipes M., et al. (2011). Discrimination of major capsular types of Campylobacter jejuni by multiplex PCR. J. Clin. Microbiol. 49 1750–1757. 10.1128/JCM.02348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher J. C., Flint A., Butcher J., Blasdel B., Reynolds H. M., Lavigne R., et al. (2018). Transcriptomic analysis of the Campylobacter jejuni response to T4-Like phage NCTC 12673 infection. Viruses 10:332. 10.3390/v10060332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. E., Timms A. R., Connerton P. L., Loc Carrillo C., Adzfa Radzum K., Connerton I. F. (2007). Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:e119. 10.1371/journal.ppat.0030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Cheng L., Méric G., de Haan C. P., Llarena A. K., Marttinen P., et al. (2014). Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol. Ecol. 23 2442–2451. 10.1111/mec.12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneppen K., Semsey S., Seshasayee A., Krishna S. (2015). Restriction modification systems as engines of diversity. Front. Microbiol. 6:528. 10.3389/fmicb.2015.00528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. C., Gencay Y. E., Brøndsted L. (2017). Methods for initial characterization of Campylobacter jejuni bacteriophages. Methods Mol. Biol. 1512 91–105. 10.1007/978-1-4939-6536-6_9 [DOI] [PubMed] [Google Scholar]
- Sørensen M. C., van Alphen L. B., Harboe A., Li J., Christensen B. B., Szymanski C. M., et al. (2011). Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 193 6742–6749. 10.1128/JB.05276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. C. H., Gencay Y. E., Birk T., Baldvinsson S. B., Jäckel C., Hammerl J. A., et al. (2015). Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophages. PLoS One 10:e0116287. 10.1371/journal.pone.0116287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. C. H., Vitt A., Neve H., Soverini M., Ahern S. J., Klumpp J., et al. (2021). Campylobacter phages use hypermutable polyG tracts to create phenotypic diversity and evade bacterial resistance. Cell Rep. 35:109214. 10.1016/j.celrep.2021.109214 [DOI] [PubMed] [Google Scholar]
- Sukackaite R., Grazulis S., Tamulaitis G., Siksnys V. (2012). The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 40 7552–7562. 10.1093/nar/gks332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Woude M. W. (2011). Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14 205–211. 10.1016/j.mib.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Weigele P., Raleigh E. A. (2016). Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 116 12655–12687. 10.1021/acs.chemrev.6b00114 [DOI] [PubMed] [Google Scholar]
- Xiong X., Wu G., Wei Y., Liu L., Zhang Y., Su R., et al. (2020). SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat. Microbiol. 5 917–928. 10.1038/s41564-020-0700-6 [DOI] [PubMed] [Google Scholar]
- Yahara K., Méric G., Taylor A. J., de Vries S. P., Murray S., Pascoe B., et al. (2017). Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ. Microbiol. 19 361–380. 10.1111/1462-2920.13628 [DOI] [PubMed] [Google Scholar]
- Yao R., Alm R. A., Trust T. J., Guerry P. (1993). Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130 127–130. 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- Young K., Davis L., DiRita V. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5 665–679. 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
- Zagorskaitė E., Manakova E., Sasnauskas G. (2018). Recognition of modified cytosine variants by the DNA-binding domain of methyl-directed endonuclease McrBC. FEBS Lett. 592 3335–3345. 10.1002/1873-3468.13244 [DOI] [PubMed] [Google Scholar]
- Zampara A., Sørensen M. C. H., Elsser-Gravesen A., Brøndsted L. (2017). Significance of phage-host interactions for biocontrol of Campylobacter jejuni in food. Food Control 73 1169–1175. 10.1016/j.foodcont.2016.10.033 [DOI] [Google Scholar]
- Zimmermann L., Stephens A., Nam S. Z., Rau D., Kübler J., Lozajic M., et al. (2018). A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430 2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the Supplementary Material and below: https://data.mendeley.com/datasets/2n9r622g8d/1, Mendeley Data.