Abstract
In vivo propagation of [PSI+], an aggregation-prone prion isoform of the yeast release factor Sup35 (eRF3), has previously been shown to require intermediate levels of the chaperone protein Hsp104. Here we perform a detailed study on the mechanism of prion loss after Hsp104 inactivation. Complete or partial inactivation of Hsp104 was achieved by the following approaches: deleting the HSP104 gene; modifying the HSP104 promoter that results in low level of its expression; and overexpressing the dominant-negative ATPase-inactive mutant HSP104 allele. In contrast to guanidine-HCl, an agent blocking prion proliferation, Hsp104 inactivation induced relatively rapid loss of [PSI+] and another candidate yeast prion, [PIN+]. Thus, the previously hypothesized mechanism of prion dilution in cell divisions due to the blocking of prion proliferation is not sufficient to explain the effect of Hsp104 inactivation. The [PSI+] response to increased levels of another chaperone, Hsp70-Ssa, depends on whether the Hsp104 activity is increased or decreased. A decrease of Hsp104 levels or activity is accompanied by a decrease in the number of Sup35PSI+ aggregates and an increase in their size. This eventually leads to accumulation of huge agglomerates, apparently possessing reduced prion forming capability and representing dead ends of the prion replication cycle. Thus, our data confirm that the primary function of Hsp104 in prion propagation is to disassemble prion aggregates and generate the small prion seeds that initiate new rounds of prion propagation (possibly assisted by Hsp70-Ssa).
Prions (37) are protein isoforms that are capable of reproducing themselves by converting normal proteins of the same primary structure into a prion state. In mammals, including humans, the prion protein PrPSc is associated with infectious neurodegenerative diseases, such as mad cow disease (see reference 38 for a review). In yeast and fungi, prions serve as protein-based genetic elements, inherited via cytoplasm in a non-Mendelian fashion (see references 5, 46, and 52 for reviews). Prions form insoluble proteinase-resistant aggregates in vivo, in contrast to their normal (nonprion) counterparts, which are usually soluble. In vitro, prion proteins form amyloid-like polymers. It has been suggested that in vivo replication of prion conformation occurs by a nucleated polymerization mechanism (25, 26). This relates prion phenomena to other amyloidoses and neural inclusion disorders (see reference 21 for a review). An alternative model explains replication of prion conformation via a monomer-directed or template-assisted conformational switch in the heterodimer, suggesting that aggregate formation occurs as a consequence of the conformational switch (see reference 17 for a review). Recent data indicate that in vitro propagation of the yeast prion amyloids may combine features of both models and therefore could be termed a nucleated conformational conversion (45).
Since prion propagation apparently operates at the level of protein folding and assembly, it is likely that chaperone proteins, which assist in the protein folding and assembly and disassembly events, are involved in this process. Indeed, propagation of the yeast prion [PSI+], an aggregation-prone isoform of the translational termination factor Sup35 (eRF3), is modulated by the chaperones of Hsp100 (8) and Hsp70 (7, 18, 31) families. Among these, the effect of Hsp104 is the most striking. Propagation of [PSI+] appears to require an intermediate amount of Hsp104: both inactivation and overproduction of Hsp104 cure yeast cells of [PSI+] (8). Hsp104 is also involved in propagation of another yeast prion, [URE3] (29), as well as in propagation of the candidate prion [PIN+] (9). [URE3] is an aggregation-prone isoform of the Ure2 protein involved in regulation of nitrogen metabolism (53), while [PIN+] is a non-Mendelian element of an unidentified molecular nature that influences the efficiency of the de novo appearance of [PSI+] (9, 10). Thus, Hsp104 is likely to play a universal role in the replication of yeast prions of various origins.
Yeast Hsp104 protein is a homohexameric ATPase (33) of the evolutionarily conserved ClpB/Hsp100 family (43), which functions in solubilization and refolding of aggregated proteins damaged by heat stress (15, 32) as well as in protection of yeast cells from some other protein-damaging stresses (40). This provides an explanation for [PSI+] curing by excess Hsp104. Indeed, it has been confirmed that Hsp104 overproduction in [PSI+] (that is, prion-containing) strains results in the shift of Sup35 from the insoluble (aggregated) to the soluble fraction (34, 35). It is more difficult to explain why moderate levels of Hsp104 are required for [PSI+] propagation. The following models for this phenomenon have been discussed (see reference 52 for a review).
Model I states that Hsp104 is required for converting the cellular Sup35 protein into a partially unfolded intermediate, which serves as a substrate for prion conversion (8). Although this model in its original form did not necessitate Hsp104 involvement in the actual process of prion conversion, further modification of the model was proposed (34, 46), suggesting that Hsp104 might also facilitate prion conversion by combining the partially unfolded Sup35 molecules into oligomeric complexes. Indeed, Sup35 amyloid formation in vitro appears to proceed via oligomers of partially unfolded Sup35 molecules (45). In either version of the model, the absence of Hsp104 would block formation of the new prion molecules without affecting the preexisting prion aggregates. Therefore, this model predicts that prion loss induced by inactivation of Hsp104 should occur in a slow generation-dependent manner by dilution of unaltered preexisting aggregates in cell divisions.
Model II asserts that Hsp104 is responsible for production of prion seeds, that is, breaking the prion aggregates down into the smaller oligomers. Although this model in its original form stated that the absence of Hsp104 affects prion partitioning and segregation to the daughter cells (25, 35), it is also clear that seeds are required to initiate the new rounds of prion replication. Thus, this model predicts that the size of preexisting prion aggregates should increase due to continuous prion conversion in the absence of the Hsp104-mediated seeding. In addition to the loss of preexisting aggregates by dilution, their transmissibility could be impaired by increased size, resulting in rapid loss of active prion units.
In agreement with model II, in vitro formation of the Sup35 amyloid does not require Hsp104 (14, 45). However, it has been shown that the Sup35 protein, isolated from the heterologous system for in vitro experiments, is already partially unfolded, which could also explain a lack of Hsp104 requirement for in vitro amyloid formation (45). Thus, in vivo experiments are required to choose between the models.
A chemical agent, guanidine-HCl (GuHCl), has been identified (49) which causes [PSI+] loss in a strict generation-dependent manner, apparently by a dilution mechanism due to a blocking of [PSI+] proliferation (12). GuHCl also cures [URE3] (53) and [PIN+] (9). While GuHCl increases Hsp104 expression (8, 27), this increase is not sufficient to explain the prion-curing effect of GuHCl (9, 12). GuHCl is a protein-denaturing agent. However, the concentrations used in prion-curing experiments (1 to 5 mM) were not high enough to cause protein denaturing. On the other hand, these concentrations of GuHCl inhibited the ATPase activity of Hsp104 in vitro (15). This led to the suggestion that GuHCl cures yeast cells of prions by inactivating Hsp104 (12, 15).
Here we performed a detailed study of prion loss after Hsp104 inactivation. Kinetic parameters of prion loss and physical characteristics of the Sup35PSI+ aggregates confirm that Hsp104 inactivation causes rapid increase of aggregate size and loss of prion replicating ability, which are difficult to explain by a simple dilution mechanism. Thus, consequences of Hsp104 inactivation are different from those observed in the case of GuHCl treatment and are consistent with the model postulating the role of Hsp104 in the production of new prion seeds.
MATERIALS AND METHODS
Yeast strains.
The genotypes of Saccharomyces cerevisiae strains are shown in Table 1. The weak [PSI+] strain OT55 (also called [PSI+]1-1-74-D694), strong [PSI+] strain OT56 (also called [PSI+]7-74-D694), [psi− PIN+] strain OT60, and [psi− pin−] strain GT17 (1, 31) are previously described isogenic derivatives of 74-D694 (8). The strains GT81, which is a self diploid of GT56-35D, and GT82, which is a self diploid of GT56-11D, were described previously (6). GT81-1C (MATa) (7) and GT81-1D (MATα) (7) are haploid meiotic spore clones of GT81. GT234 is a [psi− pin−] meiotic segregant of GT81, cured of [PSI+] and [PIN+] by GuHCl. GT84 (7) and GT87 (this study) are derivatives of GT81 and GT82, respectively, in which a piece of DNA corresponding to the portion of the HSP104 promoter and open reading frame (ORF) (except for the C-terminal region) was removed from one homolog and replaced by the URA3 gene as described previously (8). The hsp104Δ::URA3 disruption allele was verified by Southern hybridization. GT241-2B and GT241-3B are hsp104Δ meiotic spore clones of GT84. GT238-1A and GT238-6C are hsp104Δ meiotic spore clones of GT84 bearing the plasmid pGHPM104 (see below). R. Wickner kindly provided strain YHE64 (called OT113 in our collection), which contains the yeast prion [URE3-2] (13). The hsp104Δ derivative of YHE64 was constructed by replacing the portion of the HSP104 promoter and ORF (except for the C-terminal region) with the LEU2 gene by using the technique described previously (8).
TABLE 1.
S. cerevisiae strains
| Names (synonym) | Genotype or description | Source or reference |
|---|---|---|
| OT60 ([psi−]-74-D694) | MATaade1–14 his3 leu2 trp1 ura3 [psi−PIN+] | 1 |
| OT55 ([PSI+]1-1-74-D694) | Weak [PSI+] derivative of OT60 | 31 |
| OT56 ([PSI+]7-74-D694) | Strong [PSI+] derivative of OT60 | 31 |
| GT17 | MATaade1-14 his3 leu2 trp1 ura3 [psi−pin−] | 1 |
| GT81 | MATa/MATα ade1-14/ade1-14 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 lys2/lys2 [PSI+PIN+] | 6 |
| GT82 | MATa/MATα ade1-14/ade1-14 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 [PSI+PIN+] | 6 |
| GT84 | Heterozygous hsp104Δ::URA3 derivative of GT81 | 7 |
| GT87 | Heterozygous hsp104Δ::URA3 derivative of GT82 | This study |
| GT81-1C | MATa haploid meiotic spore clone of GT81 | 7 |
| GT234 | MATα ade1-14 his3 leu2 trp1 ura3 lys2 [psi−pin−] | This study |
| GT238-1A, -6C | MATa and MATα hsp104Δ::URA3 [PMOD-HSP104] haploid meiotic spore clones of GT84 | This study |
| GT241-2B, -3B | MATa and MATα hsp104Δ::URA3 [psi−] haploid meiotic spore clones of GT84 | This study |
| YHE64 (OT113) | MATα ura1 leu2 trp1 [URE3-2] | 13 |
Plasmids.
The centromeric plasmids pYS104 (URA3), containing the HSP104 gene under its own promoter, pH28 (HIS3) and pYSGal-104 (URA3), bearing the HSP104 gene under the galactose-inducible GAL1,10 (GAL) promoter, plasmid pGAL-SSA1 (URA3), bearing the SSA1 gene under the GAL promoter, and matching control plasmids with the GAL promoter, pLA1 (HIS3) and pRS316GAL (URA3), were described earlier (see reference 7 for sources). All these plasmids except pRS316GAL were provided by S. Lindquist. The plasmid pKT218,620, provided by S. Lindquist, is a pYS104 derivative with two Lys-to-Thr substitutions generated at codons 218 and 620 of the HSP104 gene. These substitutions inactivate both ATP binding sites of Hsp104 (33). The plasmid pUK21-KT218,620 was constructed by inserting the XhoI-EcoRI fragment of HSP104 obtained from the plasmid pKT218,620 into XhoI-EcoRI-digested plasmid pUK21 (50). The HIS3-based plasmid pLA1-HSP104-KT and URA3-based plasmid pRS316GAL-HSP104-KT, which express the HSP104-KT218,620 double mutant allele under the GAL promoter, were constructed by inserting the 3.2-kb BamHI-SpeI fragment of mutant HSP104 from pUK21-KT218,620 into BamHI-SpeI-digested pLA1 and pRS316GAL, respectively. The TRP1-based centromeric plasmids pFL39-GAL-HSP104 and pFL39-GAL-HSP104-KT were constructed by inserting the XhoI-SacI GAL-HSP104 fragment from plasmids pH28 and pLA1-KT218,620, respectively, into SacI-SalI-digested pFL39 (3). The pHGPm104 plasmid is a derivative of pH28, containing a portion of the endogenous HSP104 promoter inserted between the GAL promoter and the HSP104 ORF in the same orientation. To construct this plasmid, the 291-bp piece of HSP104 immediately adjacent to the 5′ end of the HSP104 ORF was PCR amplified from pYS104 by using the primers HSP104-300 (5′-GAGGATCCATGCCAGAATTTTCTAGAAGGG) and HSP104(-10)-REV (5′-TCGGATCCATATTTTATGGTACGTGTAGTTG), purchased from GIBCO-BRL (Gaithersburg, Md.) and containing BamHI extensions (restriction sites are underlined). The PCR fragment was digested with BamHI and inserted into the BamHI site of pH28. The resulting chimeric GAL-HSP104 promoter (PMOD) assures constitutive HSP104 expression at 4 to 5% (see below) of the normal level and is affected by neither galactose nor heat shock (data not shown). The construction of centromeric plasmid pLSpSUP35NM-GFP, containing the SUP35NM region fused in frame to superglow green fluorescent protein (GFP) and expressed under the SUP35 endogenous promoter, has been described previously (2). The multicopy 2μm DNA-based LEU2 plasmid pSTR7, bearing the SUP35 gene under its own promoter, and centromeric HIS3-based plasmid pLA1-SUP35, bearing the SUP35 gene without the C terminus under the GAL promoter, were used for the induction of [PSI+] appearance in the [psi− PIN+] strain as described earlier (7).
Media and growth conditions.
Yeast cultures were grown at 30°C unless otherwise noted. Standard yeast media and standard procedures for yeast cultivation, phenotypic and genetic analysis, transformation, sporulation, and dissection were used (19). Synthetic media lacking adenine, histidine, leucine, lysine, tryptophan, and uracil are designated −Ade, −His, −Leu, −Lys, −Trp, and −Ura, respectively. In all cases where the carbon source is not specifically indicated, 2% glucose (Glu) was used. The synthetic medium containing 2% galactose (Gal) or Gal and 2% raffinose (Gal+Raf) instead of glucose was used to induce the GAL promoter. Liquid cultures were grown with at least a 1/5 liquid/flask volumetric ratio in a shaking incubator (200 to 250 rpm).
Assays for [PSI+], [PIN+], and [URE3].
The presence of [PSI+] was detected by its ability to suppress the ade1-14 (UGA) mutant allele, as described previously (8). The [psi−] ade1-14 strains are unable to grow on −Ade medium and exhibit dark-red color on organic complete (YPD) medium, while [PSI+] ade1-14 strains are able to grow on −Ade after 2 to 3 days (strong [PSI+]) or 7 to 10 days (weak [PSI+]) and exhibit a light-pink color on YPD. The mosaic colonies containing both [PSI+] and [psi−] cells were detected as sectored pink-red colonies on YPD medium. The presence of [PIN+], which controls the ability of overproduced Sup35 to induce de novo formation of [PSI+] (9), was monitored by one of the following assays depending on the strains used. (i) The [psi−] strains bearing the galactose-inducible plasmid pLA1-SUP35 were incubated on galactose medium lacking His (−His/Gal medium) and velveteen replica plated onto −Ade/Glu medium. [PIN+] was detected by growth on −Ade medium after 10 to 14 days of incubation due to induction of [PSI+]. (ii) The [psi−] strains lacking pLA1-SUP35 were mated to the strain [psi− pin−] GT234 bearing the multicopy SUP35 plasmid, pSTR7. The [PIN+] diploids, in contrast to [pin−] diploids, grew on −Ade medium after 10 to 14 days of incubation due to induction of [PSI+]. (iii) To check the [PSI+] strains for the presence of [PIN+], they were first cured of [PSI+] by galactose-inducible wild-type Hsp104 (8), which does not cure yeast cells of [PIN+] (reference 9 and confirmed below). At least two independently cured [psi−] derivatives were usually analyzed for each [PSI+] clone, always with the same result. These [psi−] derivatives were either transformed individually with the multicopy SUP35 plasmid pSTR7 (in the case of diploid strains) or were mated to the [psi− pin−] GT234 strain bearing pSTR7 (in the case of haploid strains). The presence of [PIN+] in the resulting transformants or diploids was detected as described above.
The presence of [URE3] was detected by the ability of the ura2 strain to grow on −Ura medium supplied with ureidosuccinic acid (USA), as described previously (53).
Prion rescue in hsp104Δ progeny.
The [PSI+ PIN+] lys2/lys2 diploid strain GT84, heterozygous for hsp104Δ::URA3 disruption, was sporulated and dissected by using a micromanipulator Ergaval Series 10 from Carl Zeiss, Jena, Germany. To rescue [PSI+] in the spores, they were cell-to-cell mated directly to the MATa HSP104+ LYS2+ [psi− pin−] strain GT17. Only MATα hsp104Δ::URA3 spores were able to form Ura+ Lys+ diploids with GT17. These diploids were tested for the presence of [PSI+] as described above. As only one fourth of all spores contained both MATα and hsp104Δ::URA3, analysis of prion maintenance in the mitotic progeny of individual spores, especially for periods longer than two generations, would become too laborious without prior identification of the spores of the desired genotype. To identify MATα cells, the newly isolated spores were placed immediately next to a dense patch of MATα cells on YPD medium that had been prepared 2 to 3 h earlier to allow α-factor to be secreted and diffused through the medium. The spores were monitored for 2 to 5 h. The MATα spores were distinguished by their ability to initiate bud formation, in contrast to G1-arrested (shmoo) MATa spores (47). To identify the hsp104Δ::URA3 (Ura+) cells, the MATα spores were transferred onto −Ura medium with triple amounts of all necessary amino acids and quadruple amounts of adenine and methionine. Only Ura+ cells were able to continue budding on this medium. These cells were allowed to divide for the desired number of generations. The number of generations was calculated as log2N, where N is the number of cells in a microcolony counted under microscopic examination. The individual cells from each microcolony were then picked up and cell-to-cell mated to GT17 on YPD medium. After 2 to 3 days, colonies were picked, checked on −Ura-Lys medium, lacking both uracil and lysine, to confirm diploid status, and tested for the presence of [PSI+] and [PIN+] as described above.
Plate assays for prion curing.
Qualitative analysis of prion curing was performed by transforming the corresponding prion-containing yeast strains with plasmids bearing the wild-type or mutant HSP104 gene under either the endogenous or the GAL promoter. The matching plasmids without HSP104 were used as negative controls. For plasmids bearing HSP104 or HSP104-KT under the endogenous promoter, resulting transformants were cured of the plasmid and then tested for the presence of the corresponding prion ([PSI+] or [PIN+]) as described above. For the plasmids bearing HSP104 or HSP104-KT under the GAL promoter, transformants were velveteen replica plated onto Gal medium selective for the plasmid in order to induce the GAL promoter. After 3 to 4 days of incubation, transformants were velveteen replica plated onto Glu medium selective for the plasmid and allowing scoring for the corresponding prion, [PSI+] or [URE3], as described above. In this way, only cells which retained the plasmid throughout the whole period of induction on Gal medium were scored. At least eight independent transformants were tested for each strain-plasmid combination.
Quantitation of prion curing.
For quantitative analysis of prion curing by overproduced wild-type or mutant Hsp104, transformants containing the plasmids with GAL-HSP104 or GAL-HSP104-KT constructs were grown in synthetic liquid Glu medium selective for the plasmid, washed, and inoculated into synthetic plasmid-selective Gal+Raf medium at the starting concentration of 105 cells/ml. Growth was monitored by counting the cells, and cultures were maintained in the exponential phase of growth by periodically diluting them with fresh medium. Aliquots were taken after certain periods of time and were plated onto synthetic plasmid-selective Glu medium. Colonies were counted and analyzed for the presence of [PSI+] and [PIN+] as described above. In addition to complete [PSI+] and [psi−] colonies, a small fraction of the mosaic colonies was uncovered that produced both light-pink ([PSI+]) and red ([psi−]) sectors. Each mosaic colony was counted as half [PSI+] and half [psi−]. The number of generations (G) for the time period t was calculated according to the following formula: G = log2(Ct/C0) where Ct is the concentration of the viable plasmid-containing cells at time point t and C0 is the concentration of the viable plasmid-containing cells at the starting point. Concentrations of viable plasmid-containing cells were determined from the numbers of colonies grown on selective medium. Curing by 5 mM GuHCl was quantified in exactly the same way except that the strains did not contain the galactose-inducible plasmids, both cultures growing in synthetic Gal+Raf medium and cultures growing in YPD medium were analyzed for the comparison, aliquots were plated onto YPD medium, and all viable cells rather than only plasmid-containing cells were counted to calculate the numbers of generations.
DNA and protein analysis.
Standard procedures were used for isolation of DNA, restriction digestion, ligation, and bacterial transformation (39). Enzymes were purchased from New England Biolabs and GIBCO-BRL. The thermal cycler was from Ericomp, Inc. Plasmid copy number was determined by Southern hybridization with the labeled 0.5-kb PvuI-PvuII fragment of pH28 containing the HSP104 C-terminal region present in both the disrupted chromosomal copy of HSP104 and the plasmid copy of HSP104. Yeast DNA cut with PvuII contained two fragments homologous to this probe: a 4.5-kb chromosomal fragment, used as a loading control, and a 1.3-kb plasmid fragment. Chemiluminescent labeling and hybridization were performed according to Amersham protocols. Protein isolations and analyses were performed according to previously described techniques (see reference 31).
Antibodies.
The monoclonal mouse antibody to Hsp104 was a gift of S. Lindquist. The rabbit polyclonal antibodies to tagged Sup35NM were produced by Cocalico, Inc. Albumin and proteases were removed from the serum by using Affi-Gel (Blue) from Bio-Rad according to Bio-Rad protocols. Bacterial expression vector used to produce the His6-tagged Sup35NM fragment was constructed by P. A. Bailleul-Winslett via insertion of the 0.75-kb EcoRI-HpaI fragment from the plasmid pCEN-GAL-SUP35 (11) into the plasmid pET-32b(+), purchased from Novagen and cut with EcoRI and XhoI (XhoI-generated sticky ends were blunted with DNA polymerase I). The resulting construct contains essentially all of the Sup35NM region, with an N-terminal extension, including the His6, Trx, and S tags, under the control of the T7 promoter. T7 expression was induced by isopropyl-β-d-thiogalactopyranoside in Escherichia coli strain AD494 (DE3), purchased from Novagen and bearing the T7 RNA polymerase gene under control of the Plac promoter. The tagged Sup35NM protein was isolated and purified by chromatography on Ni2+ columns according to the Novagen protocols, with slight modifications. Western blotting and reaction to antibodies were performed according to Amersham protocols. The ECL detection kit from Amersham was used. Densitometry measurements were performed by using the program Alphaimager 2000 from the Alpha Innotech Corporation.
Fluorescence microscopy.
Live images of GFP distribution in the exponentially growing yeast cells containing the plasmid pLSpSup35NM-GFP were obtained as described previously (2). The fixed cells for GFP imaging and immunostaining were prepared by adding formaldehyde directly to the culture, up to a final concentration of 4%, and incubating cultures for 15 min at 25°C. To destroy the cell wall, cells were then gently spun down, washed twice in solution B (100 mM potassium phosphate buffer, pH 7.5, with 1.2 M sorbitol), resuspended in 1 ml of the same solution, treated by adding 2 μl of 2-mercaptoethanol and 20 μl of a lyticase (1 mg/ml), incubated for 30 min at 37°C, precipitated again, and washed twice with solution B. For immunostaining, fixed cells with destroyed cell walls were resuspended in 100 μl of solution F (100 mM potassium phosphate buffer, pH 7.4, 1 mg of bovine serum albumin/ml, 15 mM sodium azide, 15 mM sodium chloride) containing the Sup35 antibody, incubated in the dark for 1 h, washed 10 times with solution F, and resuspended in solution F containing rhodamine-conjugated anti-mouse secondary antibodies purchased from Sigma. After 1 h of incubation in the dark, cells were washed 10 times with solution F and resuspended in phenylenediamine mounting solution (1 mg of p-phenylenediamine/ml [Sigma] in 1× phosphate-buffered saline and 90% glycerol) (36). Preparation for imaging was accomplished by placing an aliquot of cells onto a glass slide and sealing the coverslip to the slide with clear nail polish. The samples were scanned using a Zeiss LSM510 UV confocal laser scanning microscope (Carl Zeiss Inc., New York, N.Y.), and image analysis was conducted using the Zeiss LSM Image browser (Carl Zeiss, Jena, Germany) as described previously (2). The excitation wavelength was 543 nm for the helium-neon laser (rhodamine fluorescence) and 488 nm for the argon laser used to visualize GFP.
RESULTS
Hsp104Δ causes rapid loss of [PSI+] and [PIN+].
It has been shown previously that hsp104Δ cures yeast cells of [PSI+] (8) and [PIN+] (9). To determine whether prion loss occurs by simple blocking of prion proliferation (which would result in slow generation-dependent kinetics) or by alteration of the preexisting prion aggregates (which would cause rapid loss of a prion), we performed an analysis of hsp104Δ-induced prion loss in cell generations. The diploid strain GT84 ([PSI+ PIN+] HSP104+/hsp104Δ) was sporulated and dissected, and the mitotic progeny of hsp104Δ spores was analyzed for the ability to rescue [PSI+] and [PIN+] in genetic crosses to the [psi− pin−] HSP104+ strain, GT17, as described in Materials and Methods. Our data (Table 2) confirm that [PSI+] can be rescued in all the hsp104Δ spores and that essentially all the mitotic progeny of these spores maintain [PSI+] for the first two mitotic divisions after meiosis (with the exception of one mosaic diploid colony originating from rescuing the second-division progeny). Significant loss of [PSI+] begins in the third division, which produces about 16% of the [psi−] cells. By the seventh generation, only 14% of the cells remained [PSI+]. The loss of [PIN+] was even more rapid: only 11% of the [PSI+] cells and none of the [psi−] cells retained [PIN+] after three divisions in the absence of the HSP104 gene (Table 2).
TABLE 2.
Loss of [PSI+] and [PIN+] in the progeny of hsp104Δ spores
| No. of generations after meiosis | No. of spores (microcolonies) analyzed | No. (%) of [PSI+] cells | No. of cells analyzed | Distribution of [PIN+] amonga:
|
|||
|---|---|---|---|---|---|---|---|
| [PSI+]
|
[psi−]
|
||||||
| No. (%) [PIN+] | Total no. of [PSI+] analyzed | No. (%) [PIN+] | Total no. of [psi−] analyzed | ||||
| 0 | 27 | 27 (100) | 27 | NT | NA | ||
| 1 | 3 | 6 (100) | 6 | NT | NA | ||
| 2.0–2.3 | 13 | 47.5 (99) | 48 | NT | NA | ||
| 3.0–3.3 | 7 | 50.5 (84) | 60 | 2 (11) | 19 | 0 | 3 |
| 4.0–4.2 | 4 | 35 (56) | 62 | 1 (25) | 4 | 0 | 5 |
| 7.1–7.6 | 4 | 13 (14) | 95 | 0 | 5 | 4 (9) | 44 |
NT, not tested; NA, not applicable.
To interpret these data, one should compare the kinetics of [PSI+] loss to the behavior of Hsp104 protein during sporulation and germination. It is known (40) and confirmed by us (Fig. 1A) that Hsp104 levels are increased during sporulation. Protein analysis data show that transition from sporulating to the germinating cultures is accompanied by a significant decrease in Hsp104 levels (Fig. 1A), possibly due to a partial loss of cytoplasm in the process of spore formation. However, average levels of the Hsp104 protein in the population of germinating ascospores originating from the heterozygous HSP104+/hsp104Δ diploid are about the same as those in the population of germinating ascospores originating from the isogenic homozygous HSP104+/HSP104+ diploid (Fig. 1A), despite the fact that about half of the spores in the former population lack the HSP104 gene. In contrast, the approximately twofold difference in Hsp104 protein levels between these populations becomes evident after the first two mitotic divisions following germination (Fig. 1A). This confirms that the HSP104+ and hsp104Δ ascospores originating from the Hsp104+ diploid initially contain approximately equal amounts of the Hsp104 protein, and loss of Hsp104 occurs in mitotic cell divisions following germination. This observation easily explains retention of [PSI+] by the hsp104Δ ascospores.
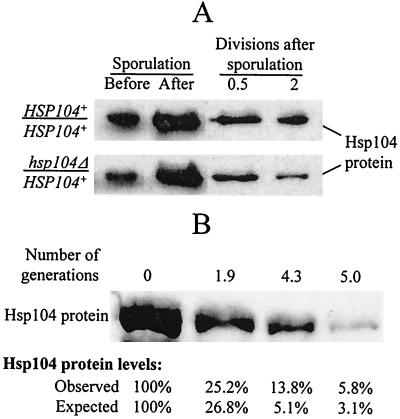
FIG. 1.
Loss of Hsp104 protein after inactivation of the HSP104 gene. (A) Fluctuations of Hsp104 levels during sporulation and germination. The diploid cultures of GT81 (HSP104+/HSP104+) and GT84 (HSP104+/hsp104Δ) were sporulated in liquid sodium acetate medium. The ascii were destroyed by lyticase treatment followed by intense vortexing, and the ascospores were shifted to the synthetic glucose medium to germinate and resume growth. Aliquots of cultures were taken at the following time points: (i) before sporulation; (ii) after 3 days of incubation in the sporulation medium, when more than 90% of cells produced ascii; and (iii) at various time points after the shift to glucose medium. Proteins were isolated, run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and reacted to Hsp104 antibody. Coomassie staining was used to verify that equal amounts of total protein were loaded in each case. Both strains contain similar amounts of Hsp104 protein at the earlier stages of germination. However, the difference between the GT81-originated culture (containing only HSP104+ cells) and the GT84-originated culture (containing a mixture of HSP104+ and hsp104Δ cells) becomes evident after two mitotic divisions following germination. (B) Loss of Hsp104 protein after inactivation of the HSP104 gene in the mitotically dividing cells occurs by dilution rather than by degradation. The hsp104Δ strains GT241-2B and GT241-3B were transformed separately with the plasmid pH28 (HIS3-GAL-HSP104). Cultures were grown in −His/Gal+Raf for 20 to 22 h to induce GAL-HSP104 and then were washed and shifted to −His/Glu, where GAL-HSP104 is repressed. The control experiment (not shown) confirmed that no detectable Hsp104 is produced by GAL-HSP104 on glucose medium. Total protein lysates were collected at various time points. Proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were reacted to Hsp104-specific antibodies. Relative Hsp104 protein amounts were quantified by using densitometry as described in Materials and Methods. Observed levels are reported as the percentage of protein remaining relative to point 0 (time of shift to −His/Glu). Expected values in percentages (E) was determined from the equation E = 100/(2X) where X is the number of generations, under the assumption that no protein degradation occurs. Data confirm that a decrease in Hsp104 levels occurs due to dilution rather than degradation.
The question remains whether Hsp104 protein is quickly degraded once ascospores resume vegetative growth or whether its levels decrease slowly due to dilution of preexisting Hsp104 in cell divisions. Direct measurements of the Hsp104 protein levels in the mixed population originated from the sporulating HSP104+/hsp104Δ culture are not sensitive enough to answer this question due to the presence of a large number of HSP104+ cells continuously producing Hsp104. In order to study the kinetics of Hsp104 loss in mitotically dividing yeast cells, we induced expression of the GAL-HSP104 construct in the hsp104Δ strain (isogenic to GT84, used in the rescue experiment) on Gal+Raf medium and measured Hsp104 levels at various periods of time after GAL-HSP104 expression was turned off by shifting to Glu medium. Our data (Fig. 1B) are consistent with the model suggesting that degradation of Hsp104 essentially does not occur during at least the first several generations after loss of the expressed HSP104 gene, so that cellular levels of Hsp104 decrease primarily due to dilution of preexisting Hsp104 protein in cell divisions. This suggests that the two-division lag in [PSI+] loss observed in the rescue experiment is most likely explained by the necessity to drop Hsp104 levels below 25% of the normal level to initiate [PSI+] loss. After this, [PSI+] is lost in a relatively rapid mode, as demonstrated by the data in Table 2.
Prion curing by ATPase-inactive Hsp104 (Hsp104-KT) resembles prion curing by hsp104Δ.
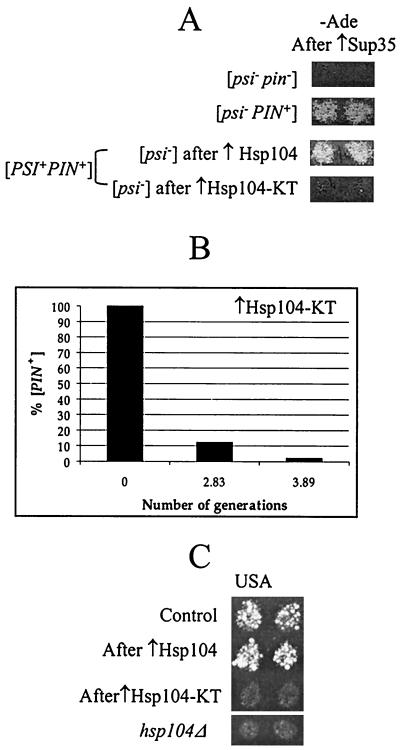
A double Lys-to-Thr substitution at residues 218 and 620 of Hsp104 inactivates both nucleotide binding domains (NBD) and results in the loss of ATP binding activity and the ability to hydrolyze ATP (33, 42). When the plasmid containing the double-mutant allele of HSP104 (designated HSP104-KT hereafter) is introduced into a wild-type HSP104+ [PSI+] cell, loss of [PSI+] results (8). It has been proposed that [PSI+] curing is due to a dominant-negative phenotype, that is, inactivation of wild-type Hsp104 in the presence of mutant Hsp104 (8). Alternatively, one could suggest that the ability of excess Hsp104 to cure [PSI+] is not related to its ATPase activity. To distinguish between these explanations, we have checked the ability of Hsp104-KT to cure yeast cells of [PIN+] and [URE3]. While all three elements are cured by hsp104Δ, only [PSI+] is cured by Hsp104 overproduction (8, 9, 29). Our results demonstrate that in contrast to overproduced wild-type Hsp104, mutant Hsp104-KT cures yeast cells of both [PIN+] (Fig. 2A) and [URE3] (Fig. 2C). [PIN+] was curable by Hsp104-KT in both [PSI+ PIN+] (Fig. 2A) and [psi− PIN+] (Fig. 2B) strains. Quantitation of [PIN+] curing in the [psi− PIN+] strain OT60 by using the galactose-inducible GAL-HSP104-KT construct demonstrated that [PIN+] loss after Hsp104-KT induction is very rapid and is nearly complete by the third generation (Fig. 2B), similar to the rapid loss of [PIN+] in hsp104Δ strains discussed previously (Table 2). These data confirm that excess Hsp104-KT cures [PSI+] by a mechanism similar to that of Hsp104 inactivation rather than to that of wild-type Hsp104 overproduction.
FIG. 2.
Curing of [PSI+], [PIN+], and [URE3] by ATPase-inactive mutant Hsp104 (Hsp104-KT). (A) Hsp104-KT cures both [PSI+] and [PIN+]. OT55 ([PSI+ PIN+]) containing the pLA1-SUP35 plasmid was transformed with either pYS104 (↑Hsp104) or pKT218,620 (↑Hsp104-KT), which led to a loss of [PSI+]. pYS104 or pKT218,620 was subsequently lost and the presence of [PIN+] was determined by Sup35 induction as described in Materials and Methods. Control strains GT17 ([psi− pin−]) and OT60 ([psi− PIN+]) are shown for comparison. (B) Quantitation of [PIN+] curing in the [psi− PIN+] strain OT60 transformed with the plasmid pRS316GAL-HSP104-KT. Expression of the GAL-HSP104-KT construct was induced on −Ura/Gal medium. Cells were plated onto −Ura/Glu medium after various periods of time. The presence of [PIN+] in resulting colonies was determined in the cross to the tester strain GT234/pSTR7, as described in Materials and Methods. [PIN+] curing is very rapid and nearly complete by the third generation. (C) Hsp104-KT cures [URE3]. The [URE3] strain YHE64 was transformed with the TRP1 plasmids pFL39-GAL-HSP104 (↑Hsp104), pFL39-GAL-HSP104-KT (↑Hsp104-KT), and a matching control plasmid. Cultures were induced on −Trp/Gal medium and were velveteen replica plated onto −Trp −Ura + USA/Glu medium selective for [URE3] (see Materials and Methods). In contrast to transient overproduction of wild-type Hsp104, transient overproduction of Hsp104-KT cures yeast cells of [URE3]. For comparison, the hsp104Δ derivative of YHE64 on −Ura + USA/Glu medium is shown.
Mutant Hsp104-KT interferes with the activity of wild-type Hsp104.
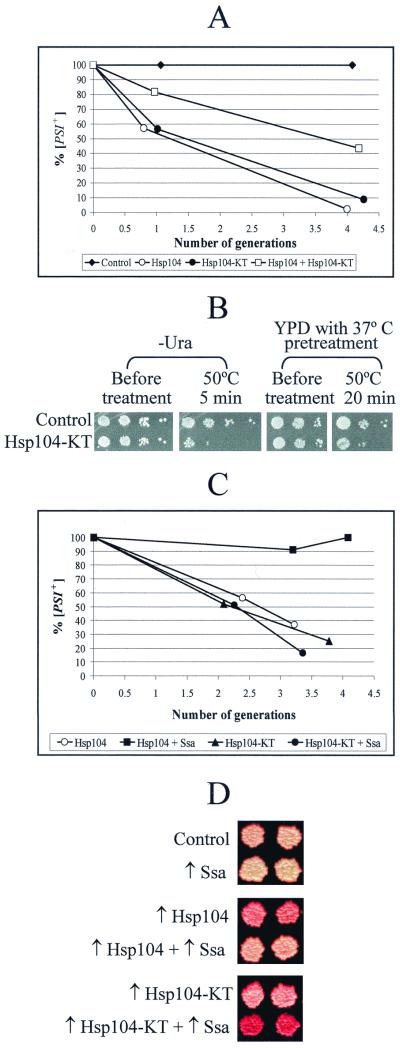
One possible mechanism for the mutant Hsp104-KT to cure [PSI+] is to interfere directly with the activity of wild-type Hsp104. This could be achieved by inhibiting assembly of the functional Hsp104 hexameric units. Indeed, amino acid substitutions in the second NBD of Hsp104 (including K620T, used in this work) were previously shown to be impaired in homohexamer formation in vitro (42). It is likely that in vivo Hsp104-KT interacts with wild-type Hsp104 but prevents the joining of new molecules to heteromultimeric complexes so that formation of functional Hsp104 hexamers becomes inefficient. If so, this effect should be partly overcome by increased concentrations of wild-type Hsp104. Likewise, Hsp104-KT should partly ameliorate the [PSI+] curing effect of excess wild-type Hsp104. To check this, we overexpressed galactose-inducible GAL-HSP104 and GAL-HSP104-KT constructs separately and together in the [PSI+] strain OT56. [PSI+] curing by wild-type or mutant Hsp104 alone proceeded with similar rapid kinetics, so that less than 60% of cells in each culture remained [PSI+] one generation after galactose induction and less than 10% remained [PSI+] after four generations (Fig. 3A). In contrast, more than 80% of cells remained [PSI+] after one generation, and more than 40% remained [PSI+] after four generations in the culture which overproduced both wild-type and mutant Hsp104 together (Fig. 3A). Therefore, wild-type and mutant Hsp104 interfere with each other's [PSI+] curing effects, as would be expected if mutant Hsp104-KT acts by inactivating wild-type Hsp104.
FIG. 3.
Interactions of wild-type Hsp104, mutant Hsp104-KT, and Hsp70-Ssa. (A) Antagonistic interaction between mutant and wild-type Hsp104 in [PSI+] curing. The [PSI+ PIN+] strain OT56 was transformed with pH28 (Hsp104), pLA1-HSP104-KT (Hsp104-KT), a combination of both plasmids, or a control matching plasmid. Cultures were induced in liquid −Ura-His/Gal+Raf medium (see Materials and Methods) and plated onto YPD medium at various time points to detect [PSI+] curing. Both the wild-type and mutant Hsp104 proteins cure [PSI+] almost completely by the fourth generation of growth. Coexpression of wild-type Hsp104 and mutant Hsp104 reduces [PSI+] curing efficiency. (B) Antagonistic interactions between mutant and wild-type Hsp104 in thermotolerance. Strain OT60 was transformed with the URA3 plasmid pKT218,620 bearing HSP104-KT under the endogenous HSP104 promoter (Hsp104-KT) or matching control plasmid (Control). Transformants were grown either in the synthetic −Ura medium (which is selective for the plasmid and causes slight induction of Hsp104) or in YPD medium at 25°C. In YPD cultures, HSP104 expression was induced by a 30-min pretreatment at 37°C prior to heat shock (40). Cultures were heat shocked at 50°C, and serial dilutions were spotted onto −Ura medium. Plates were photographed after 3 days of incubation. The plasmid pKT218,620 greatly decreases the thermotolerance of the culture, confirming the dominant-negative effect of Hsp104-KT. The same result was reproduced with strain GT17 (not shown). (C) Effects of Hsp104-Ssa on [PSI+] curing: the quantitative assay. Yeast strain, OT56; plasmids, pH28 + pRS316GAL (Hsp104), pH28 + pGAL-SSA1 (Hsp104 + Ssa), pLA1-HSP104-KT + pRS316GAL (Hsp104-KT), and pLA1-HSP104-KT + pGAL-SSA1 (Hsp104-KT + Ssa). Production of the Hsp104, Hsp104-KT, and Hsp70-Ssa proteins was induced in the liquid −Ura-His/Gal+Raf medium. The number of generations after induction is shown. Excess Hsp70-Ssa protects [PSI+] from curing by excess Hsp104 but not from curing by excess Hsp104-KT. (D) Effects of Hsp70-Ssa on [PSI+] curing: the color assay. Yeast strain, OT56; plasmids, pLA1 (Control), pGAL-SSA1 (↑Ssa), pH28 (↑Hsp104), pH28 + pGAL-SSA1 (↑Hsp104 + ↑Ssa), pLA1-HSP104-KT (↑Hsp104-KT), and pLA1-HSP104-KT + pGAL-SSA1 (↑Hsp104-KT + ↑Ssa). Transformants were grown on solid −Ura-His/Gal medium and were then velveteen replica plated onto YPD medium and analyzed by color phenotype. A lighter color is indicative of more intense suppression, apparently due to a larger proportion of [PSI+] cells in the culture. Results confirm that excess Hsp70-Ssa decreases the [PSI+] curing effect of ↑Hsp104 but increases the [PSI+] curing effect of ↑Hsp104-KT.
We have also asked whether mutant Hsp104-KT would interfere with the activity of wild-type Hsp104 in the temperature tolerance assays. Our results (Fig. 3B) demonstrate that the presence of Hsp104-KT decreases induced thermotolerance of yeast cultures previously shown to result from activity of wild-type Hsp104 (40). This agrees with observations of another group (44) that detected an inhibitory effect of the single NBD-defective Hsp104 mutants on thermotolerance in vivo. These data confirm that mutant Hsp104-KT acts by inactivating wild-type Hsp104 rather than by specifically interfering with prion propagation. This enabled us to use Hsp104-KT as a tool for reversible inactivation of Hsp104 in the yeast cell, in parallel with the other approaches that eliminate or decrease Hsp104 function.
Interactions of Hsp70-Ssa with wild-type and mutant Hsp104.
Another chaperone, Hsp70-Ssa, has been shown to protect [PSI+] from curing by excess wild-type Hsp104 but not from curing by hsp104Δ (31). In order to examine whether Hsp70-Ssa had the ability to protect cells from [PSI+] curing by mutant Hsp104, the GAL-SSA1 construct was induced simultaneously with either wild-type GAL-HSP104 or mutant GAL-HSP104-KT in the [PSI+] strain OT56 (Fig. 3D). As expected, excess Hsp70-Ssa protected [PSI+] from excess wild-type Hsp104 but not from excess Hsp104-KT. Moreover, [PSI+] curing by Hsp104-KT appeared to be slightly more efficient in the presence of excess Hsp70-Ssa. Yeast cultures that underwent simultaneous overexpression of Hsp70-Ssa and wild-type Hsp104 exhibited lighter color on YPD medium compared to that of cultures that overexpressed Hsp104 alone. This corresponds to a higher retention of [PSI+]-mediated nonsense suppression in the presence of excess Hsp70-Ssa. However, efficiency of [PSI+]-mediated nonsense suppression was reduced in the cultures that underwent simultaneous overexpression of Hsp70-Ssa and Hsp104-KT compared to that of the cultures that overexpressed Hsp104-KT alone. This reduction resulted in a more intense red color on YPD medium (Fig. 3D). Thus, Hsp70-Ssa exhibits opposite effects on [PSI+] curing by overproduced wild-type and mutant Hsp104.
Comparison of the kinetic parameters of prion curing by excess Hsp104, inactivation of Hsp104, and GuHCl.
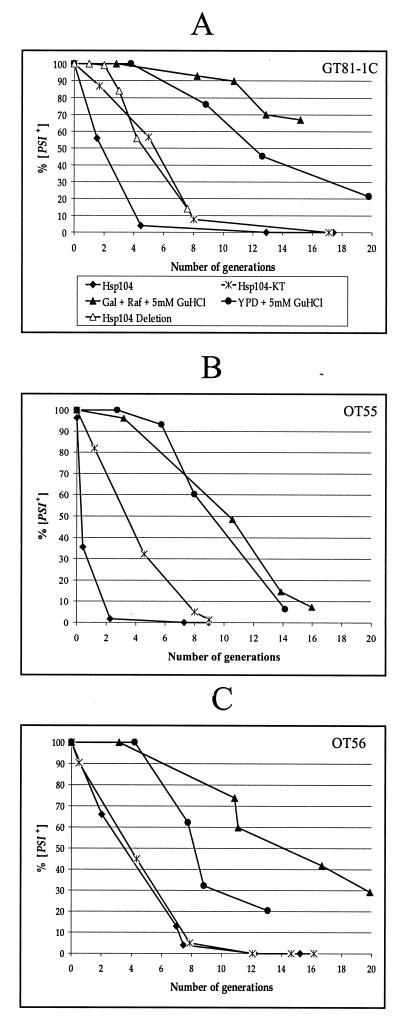
GuHCl, an agent that blocks [PSI+] proliferation (12), has previously been hypothesized to act by inactivating Hsp104 (12, 15). To check this, we compared kinetic parameters of [PSI+] curing by wild-type Hsp104, mutant Hsp104-KT, and GuHCl in the [PSI+ PIN+] strains GT81-1C (Fig. 4A), OT55 (Fig. 4B), and OT56 (Fig. 4C). As wild-type and mutant Hsp104 were induced in the synthetic Gal+Raf medium, the GuHCl-induced curing of [PSI+] was also studied in both YPD (as described previously) and Gal+Raf media. Although GT81-1C and OT56 were more resistant to the [PSI+] curing effect of GuHCl in Gal+Raf medium than in YPD medium, it did not change the general picture. In all cases, no statistically significant [PSI+] loss was observed for the first 4 to 5 (strong [PSI+] strains GT81-1C, Fig. 4A, and OT56, Fig. 4C) or 3 to 4 (weak [PSI+] strain OT55, Fig. 4B) cell divisions in the presence of GuHCl, confirming the existence of a lag period apparently required for dilution of preexisting [PSI+] replicating units (12). In contrast, cultures overexpressing either wild-type or mutant Hsp104 did not exhibit this lag. Usually the [psi−] cells were detected in the very first cell divisions after induction (Fig. 4). The rates of [PSI+] curing by mutant Hsp104-KT in the strain GT81-1C and by hsp104Δ in the progeny of the isogenic diploid GT84 were similar (the data for hsp104Δ, listed in Table 2, are also shown in Fig. 4A for the comparison). The only difference was a short two-division lag phase observed in the case of hsp104Δ, which should be attributed to the necessity to dilute the preexisting Hsp104, as argued above (Fig. 1). It is worth noting that the weak [PSI+] strain OT55 was more sensitive to the curing effect of wild-type Hsp104 but not to the curing effect of Hsp104-KT than was the isogenic strong [PSI+] strain OT56 (Fig. 4B and C). This further confirms a difference between the mechanisms of [PSI+] curing effects of wild-type Hsp104 and mutant Hsp104-KT.
FIG. 4.
Comparison of kinetic parameters of [PSI+] curing by Hsp104 overproduction, Hsp104 inactivation, and GuHCl treatment. Yeast strains were GT81-1C (A), OT55 (B), and OT56 (C). Curing conditions included induction of GAL-HSP104 (Hsp104), induction of GAL-HSP104-KT (Hsp104-KT), growth in the presence of 5 mM GuHCl in rich YPD medium (YPD + 5 mM GuHCl) or in synthetic Gal+Raf medium (Gal+Raf + 5 mM GuHCl), and deletion of the HSP104 gene (Hsp104 Deletion). Designations are shown in the box in panel A. See Materials and Methods for procedures. Data for hsp104Δ are from Table 2. These results were obtained by dissecting the diploid GT84, which is isogenic to GT81-1C.
Effects of wild-type and mutant Hsp104 on [PIN+] were also monitored in the strains OT55 and OT56 (Table 3). In strong agreement with previous data (9), overproduction of wild-type Hsp104 did not cure yeast cells of [PIN+] even after seven generations. In contrast, [PIN+] curing by Hsp104-KT was as rapid in [PSI+] strains (Table 3) as in [psi−] strains (Fig. 2B). Therefore, [PIN+] was lost even faster than [PSI+], in agreement with the results observed for hsp104Δ (see above and Table 2). The [pin−] cells were detected as early as in the first generation after Hsp104-KT induction (Table 3). Quite remarkably, [PIN+] appeared to be more frequently retained in the cells remaining [PSI+] than in the cells becoming [psi−]: in strain OT56, [PIN+] was still retained by some [PSI+] cells after eight generations, yet it was entirely lost from [psi−] cells after four generations (Table 3). This is in agreement with preferential coretention and coloss of [PSI+] and [PIN+] after GuHCl treatment that was reported previously (10).
TABLE 3.
Distribution of [PIN+] among [PSI+] and [psi−] colonies obtained after excess Hsp104 or excess Hsp104-KT treatment
| Strain | Induced construct | No. of generations | Distribution ofa:
|
|||
|---|---|---|---|---|---|---|
| [PSI+]
|
[psi−]
|
|||||
| No. (%) [PIN+] | Total no of [PSI+] analyzed | No. (%) [PIN+] | Total no. of [psi−] analyzed | |||
| OT55 | GAL-HSP104 | 0.4 | NT | 8 (100) | 8 | |
| Weak | 2.3 | 8 (100) | 8 | 8 (100) | 8 | |
| [PSI+] | 7.3 | NT | 8 (100) | 8 | ||
| OT55 | GAL-HSP104-KT | 1.2 | 16 (80) | 20 | 1 (33) | 3 |
| 4.0 | 2 (8) | 24 | 1 (6) | 16 | ||
| 8.0 | NT | 0 | 3 | |||
| 9.0 | NT | 0 | 5 | |||
| OT56 | GAL-HSP104 | 2.0 | 8 (100) | 8 | 8 (100) | 8 |
| Strong | 7.0 | NT | 8 (100) | 8 | ||
| [PSI+] | 7.5 | NT | 8 (100) | 8 | ||
| OT56 | GAL-HSP104-KT | 0.5 | NT | 2 (29) | 7 | |
| 4.3 | 5 (21) | 24 | 0 | 26 | ||
| 7.9 | 5 (21) | 24 | 0 | 21 | ||
NT, not tested.
[PSI+] maintenance at low Hsp104 levels.
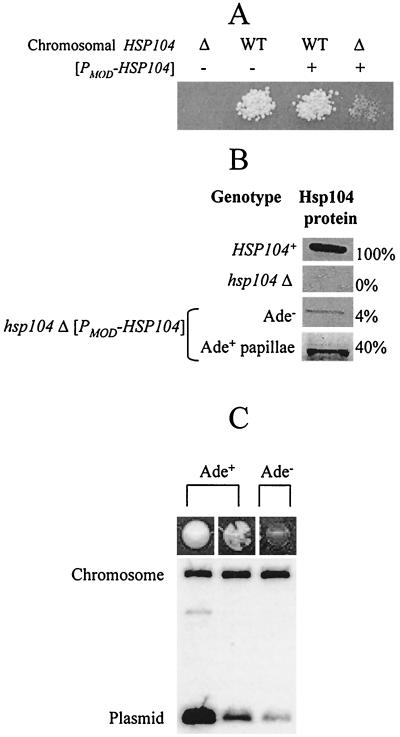
To directly monitor [PSI+] aggregates in the conditions of Hsp104 depletion, we developed a system that enables us to generate yeast cells expressing Hsp104 at levels which are below normal wild-type levels. For this purpose, the [PSI+ PIN+] HSP104+/hsp104Δ diploids GT84 and GT87 were transformed with the centromeric HIS3 plasmid bearing HSP104 under a modified promoter, [PMOD-HSP104] (see Materials and Methods). Resulting transformants were sporulated and dissected. The clones, originated from hsp104Δ spores not containing [PMOD-HSP104], were completely [psi−] (that is, nonleaky Ade−). In contrast, all the clones, originated from the spores bearing both hsp104Δ and [PMOD-HSP104], were Ade− but produced Ade+ papillae on −Ade medium (Fig. 3A). If spore clones were mated to the HSP104+ [psi−] strain, most of the progeny was Ade−, but a small fraction of the progeny turned out to be Ade+, and these Ade+ cells contained GuHCl-curable [PSI+] (data not shown). Individual colonies obtained by subcloning the hsp104Δ [PMOD-HSP104] spore clones in conditions selective for the plasmid have lost the ability to produce papillae and to rescue [PSI+] in the cross to the HSP104+ [psi−] strain (data not shown). This indicates that the progeny of [PMOD-HSP104] spores is able to maintain [PSI+] for a certain period of time but eventually loses it.
Cultures, originated from Ade+ papillae of the hsp104Δ [PMOD-HSP104] spore clones, were heterogenous and produced three different types of colonies on YPD medium: light-pink Ade+, sectored red-pink (also called mosaic) Ade+, and red Ade− colonies (Fig. 3D). The light-pink and sectored red-pink colonies always retained the [HIS3 PMOD-HSP104] plasmid, while red colonies could be both His+ (plasmid-containing) and His− (containing no plasmid). The light-pink colonies and lighter sectors of red-pink colonies remained unstable and produced all three phenotypes in the further subcloning steps (not shown). Frequency of spontaneous mitotic loss of the plasmid was significantly lower in the cells originating from light-pink colonies than that of sectored red-pink His+ colonies and red His+ colonies (data not shown). Southern hybridization analysis demonstrated that light-pink isolates contain more copies of the [HIS3 PMOD-HSP104] plasmid per cell than sectored red-pink isolates, while the latter ones contain more plasmid copies per cell than did red isolates (Fig. 5C). This confirms that the suppressor (Ade+) phenotype is apparently due to retention of [PSI+] in the cells bearing larger numbers of [PMOD-HSP104] copies. Direct measurements of Hsp104 levels show that the original hsp104Δ [PMOD-HSP104] spore clones and their red (Ade−) derivatives, still containing the plasmid, express 4 to 5% of the normal levels of Hsp104, while the Ade+ papillae and their light-pink derivatives express 35 to 40% of the normal levels of Hsp104 (see examples in Fig. 5B). This correlates well with the data presented above (Fig. 1B and Table 2) indicating that [PSI+] loss begins once Hsp104 levels drop below 25%.
FIG. 5.
[PSI+] maintenance at low Hsp104 levels. (A) A representative tetratype tetrad obtained from dissection of GT84 (HSP104+/hsp104Δ) transformed with the [PMOD-HSP104] plasmid. WT, wild type. (B) Levels of the Hsp104 protein in the spore clones shown in panel A as well as those in the Ade+ papillae obtained from the hsp104Δ [PMOD-HSP104] spore clone. Protein amounts, determined by densitometry (as described in Materials and Methods), are relative to wild-type Hsp104 levels (100%). (C) Plasmid copy number analysis. DNA was isolated from the following three types of colonies, originated from the Ade+ papillae of the hsp104Δ [PMOD-HSP104] spore clone: (i) colonies retaining the persistent Ade+ phenotype (light pink); (ii) red-pink sectored Ade+ colonies; and (iii) Ade− colonies (dark red), originated from the same hsp104Δ [PMOD-HSP104] (Ade+ papillae) spore clone. Southern blot hybridization was performed on the HSP104 probe that identifies both the vestige sequence of the disrupted chromosomal copy of HSP104 (4.5-kb band, used as a loading control) and the plasmid-borne [PMOD-HSP104] (1.3-kb band). Higher intensity of suppression (lighter color) correlates with higher copy number.
Decreased Hsp104 levels or activity results in decreased number and increased size of prion aggregates.
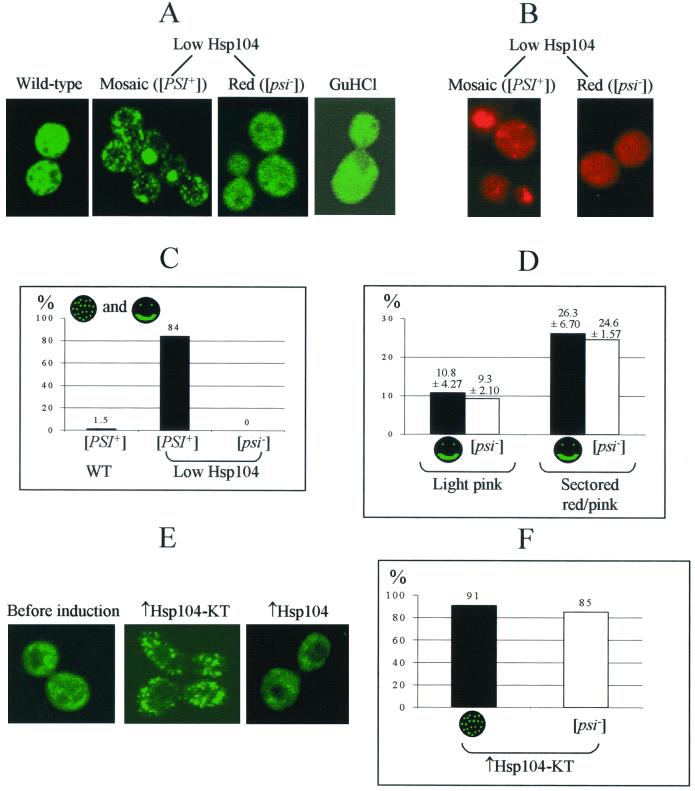
Cultures with decreased levels of Hsp104 and cultures overexpressing the dominant-negative allele of HSP104 provide a unique opportunity to visualize prion aggregates in the process of their loss due to decreased Hsp104 levels or activity. For this purpose we used a SUP35NM-GFP fusion construct under the control of the endogenous SUP35 promoter (PSUP35). The previously described constructs, overexpressing Sup35NM-GFP from the strong constitutive or inducible promoter, produced large fluorescent aggregates in the [PSI+] but not the [psi−] cells (34). In contrast, the moderate levels of Sup35NM-GFP production from PSUP35 normally gave rise to a large number of very small Sup35NM-GFP aggregates, which were almost uniformly distributed throughout the HSP104+ [PSI+] control cells (Fig. 6A and E; see also reference 2 for a comparison). This makes it difficult to distinguish between [psi−] and [PSI+] cells by using the PSUP35-SUP35NM-GFP construct in normal growth conditions, although such a distinguishment could be improved by freezing the Sup35NM-GFP aggregates and making them more visible after treatment with protein synthesis inhibitors (2). However, this feature makes the PSUP35-SUP35NM-GFP construct a sensitive tool for identifying the conditions which reduce numbers and increase sizes of the Sup35NM-GFP aggregates in the yeast cells.
FIG. 6.
Visual detection of the Sup35PSI+ aggregates. (A and B) Effects of Hsp104 depletion. Yeast strains were GT81-1C (wild type); [PSI+] and [psi−] derivatives of GT238–1A (hsp104Δ [PMOD-HSP104]) (low Hsp104); and GT81-1C grown in the presence of 5 mM GuHCl (GuHCl). [PSI+] aggregates tagged by GFP were visualized by fluorescence microscopy using the LEU2 plasmid pLSpSUP35NM-GFP (A) or by immunostaining with Sup35-specific primary rabbit antibodies and rhodamine-labeled secondary antibodies (B), as described in Materials and Methods. Exponential cultures were grown in the synthetic complete glucose medium with 5 mM GuHCl (GuHCl) or in the −Leu-His/Glu medium (all other strains). Representative examples are shown. The Sup35NM-GFP aggregates become larger and less numerous in the [PSI+] low-Hsp104 strain. (C) Comparison of the frequencies of the cells containing easily detectable medium-size Sup35NM-GFP aggregates or huge Sup35NM-GFP agglomerates in the [PSI+] and [psi−] deivatives of the low-Hsp104 strain, GT238-1A, and in the isogenic control wild-type [PSI+] strain, GT81-1C. From 70 to 325 cells were screened in each case. (D) Comparison of the frequencies of the cells containing huge Sup35NM-GFP agglomerates and frequencies of the [psi−] (red) colonies produced by two [PSI+] isolates of the low-Hsp104 strain, GT238-1A. Correlation between these numbers suggests that huge Sup35 agglomerates possess reduced ability to transmit the prion state. Limits of variation (95%) are shown. (E and F) Effects of the overexpressed dominant-negative Hsp104-KT protein on Sup35NM-GFP aggregate size. The yeast strain used was GT81-1C; the inducible constructs were URA3-GAL-HSP104-KT (↑Hsp104-KT) and URA3-GAL-HSP104 (↑Hsp104). Media were −Ura-Leu/Glu (before induction) and −Ura-Leu/Gal+Raf after 18 h of incubation (↑Hsp104-KT and ↑Hsp104). After induction, about 91% of 50 cells screened in the ↑Hsp104-KT culture exhibited detectable medium-size Sup35NM-GFP aggregates. No aggregates of this type were observed in the culture overexpressing wild-type Hsp104. In the same conditions, about 85% of cells of the ↑Hsp104-KT culture formed [psi−] colonies.
Indeed, we have observed that the unstable [PSI+] culture originating from the low-Hsp104 (hsp104Δ [PMOD-HSP104]) strain exhibits a mode of Sup35NM-GFP aggregation that is markedly different from that of the isogenic [PSI+] culture with normal levels of Hsp104. While about 98.5% of cells in the normal [PSI+] culture contained almost indistinguishable small aggregates diffused all over the cell cytoplasm, about 84% of cells in the low-Hsp104 [PSI+] cultures contained either medium-size aggregates (present in a smaller number per cell but clearly visible due to larger average aggregate size) or very large agglomerates (usually only one or very few per cell) (Fig. 6A and C). Some cells contain these agglomerates in the form of bars or rings (not shown). Neither medium-size Sup35NM-GFP aggregates nor huge Sup35NM-GFP agglomerates were ever detected in the red (that is, completely cured [psi−]) derivatives of the same low-Hsp104 strain. This confirms that appearance of the visible aggregates is [PSI+] dependent and is not caused simply by the Sup35NM-GFP folding defect in the absence of Hsp104 function.
Quite remarkably, the percentage of cells containing huge agglomerates was proportional to the stability of [PSI+] in the low-Hsp104 cultures. The light-pink low-Hsp104 isolate that produced about 9% of [psi−] (red) colonies contained about 11% of cells with one or very few huge Sup35NM-GFP agglomerates, while the sectored red-pink low-Hsp104 isolate that produced about 25% of [psi−] colonies contained about 26% of cells with one or very few huge Sup35NM-GFP agglomerates (Fig. 6D). While such an exact correspondence of these numbers could be a coincidence, the tendency observed clearly indicates that increased aggregate size correlates with decreased ability to transmit the prion state, suggesting that huge agglomerates represent the dead ends of the prion replication cycle.
As one could suggest that behavior of the endogenous Sup35 protein is different from that of the Sup35NM-GFP fusion construct, we have also attempted to identify the endogenous Sup35 aggregates by immunostaining. Compared to GFP tagging, this procedure produced less clear images and more intense backgrounds that could be due in part to lower specificity of the polyclonal Sup35 antibody. However, we were able to identify the large Sup35 aggregates in the [PSI+] low-Hsp104 culture (Fig. 6B), thus confirming that data obtained by the GFP-based approach reflect actual behavior of the Sup35 protein in the [PSI+] low-Hsp104 cells.
To confirm that increased aggregate size is indeed associated with the loss of Hsp104 activity, we used an alternative approach to inactivate Hsp104: overproduction of the dominant-negative mutant Hsp104-KT. Indeed, induction of the GAL-HSP104-KT construct in the [PSI+] strain also resulted in decreased numbers and increased sizes of the Sup35NM-GFP aggregates: after 18 h of incubation on Gal medium, each cell of the predominant (91%) class contained relatively low numbers of medium-size aggregates similar to those detected in the majority of the cells in the low-Hsp104 culture and clearly distinguishable from the numerous small aggregates detected in the isogenic [PSI+] control (Fig. 6E and F). As only 15% of cells in the culture that had undergone Hsp104-KT induction for that period of time were still able to form the [PSI+] colonies, it is clear that at least some of the aggregates observed lost the ability to transmit the prion state. No huge agglomerates were observed in these experimental conditions. This could be due to the fact that the relatively short time period that passed after Hsp104-KT induction is insufficient for the prion aggregates to grow into huge agglomerates.
In contrast, the overproduction of wild-type Hsp104 (also curing yeast cells of [PSI+] with about the same efficiency as that of Hsp104-KT; see Fig. 4) did not increase aggregate size (Fig. 6E), which agrees with the previously reported ability of Hsp104 to solubilize prion aggregates (34, 35). In the same way, no medium size aggregates or huge agglomerates were detected in the isogenic (GT81-1C) yeast culture growing in the presence of GuHCl (Fig. 6A). This confirms that loss of [PSI+] in conditions other than Hsp104 depletion does not necessarily proceed via the large aggregate stage.
DISCUSSION
Comparison between prion-curing effects of Hsp104 inactivation and GuHCl.
If the function of Hsp104 is required for formation of new prion molecules (8, 34), one would expect that prion loss following Hsp104 inactivation would occur by dilution of preexisting prion units. Such a mechanism has previously been reported for the prion-curing agent GuHCl (12). [PSI+] loss in the presence of GuHCl follows slow generation-dependent kinetics with a long lag period that is required to decrease the average copy number of the prion aggregates. Calculations, based on kinetics of GuHCl-induced loss, suggested that the [PSI+] strain used in that work contains about 60 proliferating Sup35PSI+ units per cell (12). Kinetics of [PSI+] curing in our strains (see Fig. 4) was generally similar to those reported previously (12), although weak [PSI+] was lost faster than the strong one, as expected (see Fig. 4). It appears that our strong [PSI+] strains contain about the same number of proliferating [PSI+] units as the strains used by Eaglestone et al. (12). However, [PSI+] loss induced by Hsp104-KT did not show a significant lag period and followed rapid kinetics, strikingly different from that of GuHCl-induced loss in the same conditions (Fig. 4). [PSI+] loss after elimination of the HSP104 gene has exhibited a short lag period for two cell divisions (Table 2 and Fig. 4A), most likely due to the fact that the Hsp104 protein is very stable and remains in the cell for a long time after elimination of the HSP104 gene, so that Hsp104 concentration is decreased only due to protein dilution in the cell divisions (Fig. 1B). After Hsp104 protein had been diluted below 25% of the normal wild-type level, the prion loss in hsp104Δ cells followed rapid kinetics, indistinguishable from prion loss induced by Hsp104-KT (Fig. 4A and Table 2). This agrees with the results of another experiment indicating that cultures expressing Hsp104 at 35 to 40% of the normal levels (on average) are capable of maintaining the [PSI+] state in the majority of the cells (Fig. 5B).
Striking differences in kinetic parameters of [PSI+] loss after GuHCl and Hsp104 inactivation contradict the previous model, suggesting that GuHCl cures yeast cells of [PSI+] (and possibly of the other prions) by inactivating Hsp104 (12, 15). Moreover, [PIN+] loss after Hsp104 inactivation was even more rapid than [PSI+] loss (Fig. 2B and Tables 2 and 3). There could be two possible explanations for such a discrepancy. One suggests that the long lag in GuHCl-mediated curing is not necessarily due to dilution of multiple prion units. Rather, it is due to the necessity to inactivate most of the Hsp104 molecules. Then GuHCl could still act by Hsp104 inactivation and Hsp104 inactivation would result primarily in blocking [PSI+] proliferation. In this case, the average number of prion aggregates per cell should be small (no more than 4 to 8 units per cell for [PSI+] and even less for [PIN+]), explaining the absence of lag in prion loss after Hsp104 inactivation. However, this model would predict that GuHCl, like inactivated Hsp104, would cure low-copy-number prions ([PIN+]) faster than the high-copy-number ones ([PSI+]). In contrast, low concentrations (1 mM) of GuHCl usually cured [PSI+] but not [PIN+] (9), while at higher concentrations of GuHCl (5 mM), [PSI+] and [PIN+] appeared to be preferentially lost together (10). These experiments were performed in a 74-D694 genotypic background and employed the set of [PSI+] and [PIN+] isolates which overlapped one used in this work. It should also be noted that a large average number of [PSI+] aggregates per cell, deduced from GuHCl experiments, also agrees with indirect calculations performed by other means (28) as well as with the visual observations of the Sup35-GFP clumps. We favor an alternative explanation which suggests that [PSI+] loss after Hsp104 inactivation is not explained by simple dilution in cell divisions, as in the case of GuHCl. In addition to the proliferation defect, the state of preexisting prion units is somehow affected by the lack of Hsp104. This would also mean that either the GuHCl curing effect on [PSI+] is primarily not due to Hsp104 inactivation or that GuHCl-mediated inactivation of Hsp104 is incomplete and is sufficient only for blocking the proliferation of [PSI+] but not for altering the parameters of preexisting Sup35PSI+ aggregates.
Reverse correlation between Hsp104 activity and aggregate size.
Alteration of the Sup35PSI+ aggregates in Hsp104-defective strains has also been confirmed by visual detection. In both [PSI+] strains with low levels of Hsp104 protein (Fig. 6A) and [PSI+] strains overexpressing Hsp104-KT (Fig. 6E), the GFP-tagged Sup35PSI+ aggregates were fewer and larger than those of the isogenic strains with normal levels of Hsp104. No such effect was observed for excess Hsp104 (Fig. 6E) or GuHCl (Fig. 6A) in the same strain. Reverse correlation between Hsp104 activity and aggregate size agrees with the previously proposed Hsp104 role in aggregate disassembly (25, 35). Our data also suggest a reverse correlation between size of the aggregate and its ability to transmit the [PSI+] state. Several additional pieces of evidence support this notion. Overproduction of the Sup35NM-GFP protein in [PSI+] cells increases aggregate size (see reference 2 for a comparison) and causes partial loss of [PSI+] (R. D. Wegrzyn and Y. O. Chernoff, unpublished data). Treatment with the anticytoskeletal drug latrunculin A results in aggregate dissipation, sometimes leading to the appearance of large amorphous agglomerates, and induces loss of [PSI+] (2). The Sup35 protein with deletion of amino acids 22 to 69 can form unstable [PSI+] derivatives ([PSI+]Δ22–69) which are not maintained in nonselective conditions. These [PSI+]Δ22–69-bearing cells contain larger Sup35 aggregates than the isogenic cells with full-size [PSI+] prions (A. Borchsenius, R. D. Wegrzyn, G. Newnam, S. Inge-Vechtomov, and Y. O. Chernoff, unpublished data). Moreover, some of these cells exhibit huge agglomerates similar to those observed in the low-Hsp104 [PSI+] cells. Taken together, these data strongly suggest that huge aggregates represent dead ends of [PSI+] propagation, while in vivo prion proliferation probably occurs via small oligomers. This also agrees with recent observations indicating that in vitro formation of the Sup35NM amyloid proceeds via oligomeric intermediates (45).
Model for Hsp104 role in prion propagation.
Our results confirm that a decrease in Hsp104 levels or activity results in a decreased number and increased size of prion aggregates, leading to the loss of prion reproductive activity. In general, this is consistent with the previously hypothesized role of Hsp104 in the production of the new prion seeds (25, 35). However, one important clarification should be made. The original model proposed that the Hsp104-mediated seeding is required primarily for prion partitioning and segregation. However, our results show that prion loss caused by Hsp104 inactivation is rapid and cannot be explained simply by a segregation defect (Fig. 4 and Table 2). Most likely, prion aggregates constantly undergo cycles of assembly and disassembly in the yeast cell. The chaperone helpers regulate these processes, with Hsp104 being a primary catalyst of the disassembly step. Lack or shortage of Hsp104 function results in formation of the large aggregates with reduced ability to transmit the prion state.
There could be at least two nonexclusive explanations for the loss of prion-converting activity by these large aggregates. First, the total surface of all aggregates in the cell decreases dramatically with increase of the average aggregate size. This would decrease the fraction of prion protein that is able to interact with the soluble nonprion molecules. Second, the intracellular location of the large aggregates could also be altered so that they become inaccessible to the soluble Sup35 molecules. As a result, prion reproduction is inhibited and nonprion protein molecules are rapidly accumulated. Such a model agrees with the recent observations suggesting that huge inclusion bodies in aggregation-related disorders are not toxic, while proliferation of amyloid is probably associated with the smaller intermediates (see reference 21 for a review).
The fate of the large aggregates remains unclear. As they do not possess prion activity anymore, they cannot be monitored by genetic means. It is possible that the unrestricted increase of the aggregate size results in eventual elimination of these aggregates from the population due to death of the cells containing these large aggregates. Indeed, it has been observed that a significant fraction of cells containing huge ring-like aggregates of the overproduced Sup35-GFP are unable to form viable colonies (54). It is also possible that large aggregates eventually lose their high-order organization and proteinase resistance, which leads to their destruction by the proteolytic systems.
Interaction of Hsp104 and Hsp70-Ssa in prion maintenance.
The Hsp70-Ssa protein has previously been shown to protect [PSI+] from curing by excess Hsp104 (31). Excess Ssa1 protein also increased nonsense suppression by [PSI+] (31), while mutation in the SSA1 gene decreased [PSI+] stability and, in combination with the deletion of another member of Hsp70 family, SSA2, led to [PSI+] elimination (18). These data suggest that Hsp70-Ssa protein promotes [PSI+] propagation. However, excess Ssa1 protein failed to protect [PSI+] from curing by hsp104 deletion (31) or by Hsp104-KT (Fig. 3C). Moreover, excess Ssa1 protein enhanced an anti-[PSI+] effect of Hsp104-KT (Fig. 3D). This indicates that a role of Hsp70-Ssa protein in [PSI+] maintenance is complex and depends on its functional interaction with Hsp104.
We suggest that the primary Hsp70-Ssa effect on prion aggregates is opposite to that of Hsp104. While Hsp104 catalyzes aggregate disassembly, Hsp70-Ssa promotes new rounds of aggregate assembly. In this way concerted action of Hsp104 and Hsp70-Ssa results in prion proliferation. Increased levels of Hsp104 result in intensification of aggregate disassembly that might lead to eventual monomerization and loss of prion- forming activity. However, increased levels of Hsp70-Ssa, resulting in intensified assembly of new aggregates, could counteract such a process. In contrast, increased Hsp70-Ssa assembly-promoting function in a low-Hsp104 background would result in facilitated accumulation of the large aggregates, losing prion activity. This explains the opposite effects of excess Ssa1 on [PSI+] curing by Hsp104 overproduction and Hsp104 inactivation, which have been observed in our experiments.
At first glance, such a model would contradict the previously reported role of Hsp70-Ssa in solubilization of heat-damaged aggregated proteins. In that case, Hsp104 initiates the aggregate breakdown, while Hsp70-Ssa catalyzes the protein refolding back to the normal (soluble) state (15). However, this difference could be due to unusual features of prions differing from the components of amorphous aggregates of heat-damaged misfolded proteins. As we have discussed previously (31), Hsp70-Ssa apparently does not recognize a prion as an abnormal or misfolded protein. Rather, it might recognize it as a damaged intracellular structural complex which needs protection and recovery.
It has been reported that [PSI+] response to excess Hsp70-Ssa is strain specific: the [PSI+]-mediated nonsense suppression was enhanced in some strains (31) and antagonized in other strains (8, 27) by overproduced Ssa1. Moreover, excess Hsp70-Ssa affected some, but not all, [PSI+] isolates in the same genetic background (24). These discrepancies could potentially be explained by the differences in average aggregate size and the effective number of the prion replicating units that could be controlled by both genetic composition of the yeast strain (e.g., via the Hsp104/Hsp70 ratio) and by specific patterns of the prion strain. In this case, prions with the larger number of smaller seeds would be curable by excess Hsp104, which causes further dissociation of the seeds to monomers, but strengthened by excess Hsp70-Ssa, which counteracts this effect. On the other hand, prions with the smaller number of larger seeds would be insensitive to excess Hsp104 but curable by excess Hsp70-Ssa (that makes seeds even larger and inactive). At least in one case, such a correlation was indeed observed. The unstable [PSI+] prions formed by the heterologous Sup35NM (or Sup35N) domain of Pichia methanolica in S. cerevisiae (6) are insensitive to excess Hsp104 (23), are curable by excess Hsp70-Ssa (24), and exhibit smaller numbers and larger average sizes of the GFP-tagged aggregates than the endogenous stable S. cerevisiae [PSI+] prions in the same genotypic background (E. Lewitin and Y. O. Chernoff, unpublished data). The experiments aimed at further testing this model are presently under way.
Our data confirm that the chaperone machinery of the yeast cell is adjusted to levels that are optimal for prion reproduction. It remains a mystery how such an adjustment could have evolved. Recent evidence, pointing to the involvement of chaperones of the Hsp100 and Hsp70 groups in control of aggregation of poly-Gln proteins of various origins (4, 20, 22, 30, 41, 51), supports evolutionary conservation of this regulatory mechanism. Further understanding of the chaperone-based regulation of aggregate propagation can therefore help in the fight against amyloidoses and other aggregation-related disorders. Moreover, prion modulation by the stress-regulated chaperones provides a mechanism for environmental changes to cause inherited protein alterations. If protein-based genetic variations play a beneficial role by increasing the inherited variability of population in the changing environment as hypothesized recently (5, 48), such a mechanism could significantly influence our understanding of interactions between organism and environment in the process of evolution.
ACKNOWLEDGMENTS
We thank S. Lindquist for plasmids and antibodies, R. Wickner for the yeast strain YHE64, P. Bailleul-Winslett and E. Lewitin for help with plasmid constructions, K. Allen for critical reading of the manuscript, and S. Woodard for technical advice with fluorescence microscopy experiments.
This work was supported in part by NIH grant R01GM58763 to Y.O.C.
REFERENCES
- 1.Bailleul P A, Newnam G P, Steenbergen J N, Chernoff Y O. Genetic study of interactions between the cytoskeletal assembly protein sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81–94. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailleul-Winslett P A, Newnam G P, Wegrzyn R D, Chernoff Y O. An anti-prion effect of the anticytoskeletal drug latrunculin A in yeast. Gene Expr. 2000;9:145–156. doi: 10.3727/000000001783992650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 4.Chai Y, Koppenhafer S L, Bonini N M, Paulson H L. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernoff Y O. Mutation processes at the protein level: is Lamarck back? Mutat Res. 2001;488:39–64. doi: 10.1016/s1383-5742(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 6.Chernoff Y O, Galkin A P, Lewitin E, Chernova T A, Newnam G P, Belenkiy S M. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 7.Chernoff Y O, Newnam G P, Kumar J, Allen K, Zink A D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch I L, Bradley M, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derkatch I L, Bradley M E, Masse S V, Zadorsky S P, Polozkov G V, Inge-Vechtomov S G, Liebman S W. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaglestone S S, Ruddock L W, Cox B S, Tuite M F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edskes H K, Gray V T, Wickner R B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;97:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of Saccharomyces cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 15.Glover J R, Lindquist S. Hsp104, Hsp70 and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:1–20. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 16.Goloubinoff P, Mogk A, Zvi A P, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison P M, Bamborough P, Daggett V, Prusiner S B, Cohen F E. The prion folding problem. Curr Opin Struct Biol. 1997;7:53–59. doi: 10.1016/s0959-440x(97)80007-3. [DOI] [PubMed] [Google Scholar]
- 18.Jung G, Jones G, Wegrzyn R D, Masison D C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 20.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 21.Koo E H, Lansbury P T, Kelly J W. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci USA. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushnirov V V, Kochneva-Pervukhova N V, Chechenova M B, Frolova N S, Ter-Avanesyan M D. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushnirov V V, Kryndushkin D S, Boguta M, Smirnov V N, Ter-Avanesyan M D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 25.Kushnirov V V, Ter-Avanesyan M D. Structure and replication of yeast prions. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 26.Lansbury P T, Caughey B. The chemistry of scrapie reaction: the “ice 9” metaphor. Chem Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 27.Lindquist S, Patino M M, Chernoff Y O, Kowal A S, Singer M A, Lee K-H, Blake T, Liebman S W. The role of Hsp104 in stress tolerance and [PSI+] propagation in Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1995;60:451–460. doi: 10.1101/sqb.1995.060.01.050. [DOI] [PubMed] [Google Scholar]
- 28.McCready S J, Cox B S, McLaughlin C S. The extrachromosomal control of nonsense suppression in yeast: an analysis of the elimination of [psi+] in the presence of a nuclear gene PNM. Mol Gen Genet. 1977;150:265–270. doi: 10.1007/BF00268125. [DOI] [PubMed] [Google Scholar]
- 29.Moriyama H, Edskes H K, Wickner R B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchowski P J, Schaffar G, Sittler A, Wanker E E, Hayer-Hartl M K, Hartl F U. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 33.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 34.Patino M M, Liu J J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 35.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 36.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 37.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner S B, Scott M R, De Armond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satyal S H, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer J M, Morimoto R I. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;23:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schirmer E C, Queitsch C, Kowal A S, Parsell D A, Lindquist S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem. 1998;273:15546–15552. doi: 10.1074/jbc.273.25.15546. [DOI] [PubMed] [Google Scholar]
- 43.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 44.Schirmer E C, Ware D M, Queitsch C, Kowal A S, Lindquist S L. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci USA. 2001;98:914–919. doi: 10.1073/pnas.031568098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 46.Serio T R, Lindquist S L. Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- 47.Sprague G F., Jr Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- 48.True H L, Lindquist S L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 49.Tuite M F, Mundy C R, Cox B S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 51.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 52.Wickner R B, Chernoff Y O. Prions of fungi: [URE3], [PSI] and [Het-s] discovered as heritable traits. In: Prusiner S B, editor. Prion biology and diseases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 229–272. [Google Scholar]
- 53.Wickner R B. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 54.Zhou P, Derkatch I L, Liebman S W. The relationship between visible intracellular aggregates that appear following overexpression of Sup35, and the yeast prion-like elements [PSI+] and [PIN+] Mol Microbiol. 2000;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]