Abstract
Glaucoma is among the leading causes of irreversible blindness worldwide. If diagnosed and treated early enough, the disease progression can be stopped or slowed down. Therefore, it would be very valuable to detect early stages of glaucoma, which are mostly asymptomatic, by broad screening. This study examines different computational features that can be automatically deduced from images and their performance on the classification task of differentiating glaucoma patients and healthy controls. Data used for this study are 3 x 3 mm en face optical coherence tomography angiography (OCTA) images of different retinal projections (of the whole retina, the superficial vascular plexus (SVP), the intermediate capillary plexus (ICP) and the deep capillary plexus (DCP)) centered around the fovea. Our results show quantitatively that the automatically extracted features from convolutional neural networks (CNNs) perform similarly well or better than handcrafted ones when used to distinguish glaucoma patients from healthy controls. On the whole retina projection and the SVP projection, CNNs outperform the handcrafted features presented in the literature. Area under receiver operating characteristics (AUROC) on the SVP projection is 0.967, which is comparable to the best reported values in the literature. This is achieved despite using the small 3 × 3 mm field of view, which has been reported as disadvantageous for handcrafted vessel density features in previous works. A detailed analysis of our CNN method, using attention maps, suggests that this performance increase can be partially explained by the CNN automatically relying more on areas of higher relevance for feature extraction.
1. Introduction
Glaucoma is a group of neurodegenerative eye diseases that are among the leading causes of irreversible blindness worldwide [1]. It is characterized by the degeneration of retinal ganglion cells (RGCs) and their axons in the peripapillary retinal nerve fiber layer (pRNFL). Additionally, it leads to specific alterations of the optic disc with loss of neuroretinal rim tissue, leading to slow progressive, irreversible loss of function. Although its pathogenesis is not completely understood, reduced ocular blood flow is suspected to contribute to its onset and progression [2–4]. If diagnosed and treated early, the progression can be slowed down or even stopped. Diagnosis of glaucoma is currently done by quantifying optic disc changes, visual field defects, and elevated intraocular pressure.
Fundus photography and scanning laser tomography have been one of the first image-based methods used to investigate glaucoma. The 2D en face images obtained from these devices allow us to examine the shape of the optic nerve head (ONH), for instance the cup-to-disk ratio or the rim area [5–9].
With the invention of optical coherence tomography (OCT) [10], three-dimensional in vivo imaging of the retina became feasible. This allowed for the visualization of structural information of the different retinal layers. Consequently, the thickness of the retinal nerve fiber layer (RNFL) [11,12] and the ganglion cell layer with inner plexiform layer (GCIPL) and ganglion cell complex (GCC = mRNFL + GCIPL) [13–15] were discovered as biomarkers for glaucoma. Moreover, in addition to the two-dimensional measurements of the ONH, three-dimensional measurements like its volume have been investigated [16,17].
With the development of OCT angiography, a relatively recent extension of OCT, a non-invasive, three-dimensional method for assessing retinal vasculature became available. This allows studying how glaucoma affects the microvasculature of the eyes. A widely used biomarker is vessel density (VD) which was studied in different regions (macula and ONH) and different sectors and plexus [18–24].
Since most forms of glaucoma are asymptomatic until the disease has progressed to an advanced stage, regular screening could detect the disease at an early stage. This would allow to intervene early and delay or even prevent the onset of symptoms. However, broad screening could result in an unmanageable workload for physicians. To prevent this, computer-aided diagnosis could be used to counteract this increase in measurements. Initial work on automatic screening used the previously identified biomarkers as computational features for machine learning to classify whether an eye exhibits signs of glaucoma. This was done by using RNFL and GCIPL thickness maps extracted from OCT volumes [25–27], microvascular features extracted from en face OCTA ONH scans [28], or a combination of VD and thickness parameters [29,30]. While for both OCT and fundus imaging fully data-driven approaches were presented [31–35], prior work using only OCTA still relies on handcrafted features.
To close this research gap we study the discriminative power of automatically detected features using convolutional neural networks (CNNs) for the task of distinguishing glaucoma patients from healthy controls and compare these features with handcrafted features from the literature. We evaluate the different approaches on 3x3 mm macular projections using the whole retina, the superficial vascular plexus (SVP), the intermediate capillary plexus (ICP), and the deep capillary plexus (DCP).
2. Methods
In this section, we will first explain our method of automatically extracting features using CNNs. After that, we introduce two sets of handcrafted features we used to benchmark our approach.
2.1. Automatically extracted features: convolutional neural networks
Different convolutional neural network (CNN) architectures were examined for automatic feature extraction. All of the models consist of a block of convolutional layers that extract the features from a two-dimensional input image followed by the actual classification. The networks used in this study are DenseNet121, DenseNet161 [36], ResNet-152 [37], ResNext-50-32x4d [38] and WideResNet-101-2 [39]. All of the models were pretrained on ImageNet. Since these pretrained models require the input to be a three-channel (RGB) image, the one-dimensional en face OCTA images were replicated three times such that the resulting images also had three channels. Moreover, the last layer was changed to have a single output indicating whether the patient suffers from glaucoma (label 1) or not (label 0). The training of these networks then consisted of two steps. In the first step, all layers, except the last one, were frozen and the network was trained until the validation loss did not decrease for three epochs. This was done because the last layer had to be randomly initialized in contrast to the frozen layers. By keeping the pretrained feature extraction constant, a classification based on these pretrained features was learned. Next, all layers were unfrozen and additional training was performed until a total of 200 epochs was reached. With this second step, the network could adapt the features and the classification to the different content of the input data. The loss was calculated using binary cross-entropy and optimized using stochastic gradient descent (SGD) with a momentum of 0.9. The batch size was set to 10, which was the maximum number of images that fitted into memory, and three different learning rates ( , and ) were investigated. The mean and standard deviation of the pixel intensities of all images was computed before training and used to normalize the input images to have zero mean and a standard deviation equal to one. To deal with class imbalance, class weights were assigned to be inversely proportional to the total amount of images per class. This was necessary because our dataset had more en face OCTA images from glaucoma patients, which would bias the network towards classifying images as glaucomatous based on relative frequencies and thus lower the impact of actual pathological features in the image content. One drawback of CNNs is, that they can be considered a black-box approach. So it is be interesting to know which beneficial features were found on the 3 3 mm scans. To investigate this in greater detail, we applied the Gradient-weighted Class Activation Mapping (Grad-CAM) [40] algorithm to the predictions of the CNNs on the test set. The authors of Grad-CAM assume that the last convolutional layer of a CNN shows the best compromise between high-level semantics and detailed spatial information. They therefore use the gradient information flowing into this layer to assign importance values to each neuron for a particular classification target. These are then used to produce a class-discriminative localization map highlighting the important regions in the image for the prediction of a certain class.
2.2. Handcrafted features: Ong et al
To the best of our knowledge, there are few approaches known in literature for the distinction between healthy controls and glaucoma patients that use only OCTA data for this task. One example of handcrafted features that matched our dataset was presented by Ong et al. [28]. Although they presented their features for the region around the ONH, we adapted them and employed them on the macular region. In their work, they extracted the optic disc microvascular region for feature computation. Because the ONH is not included in our study, the features are directly applied to the macular region. They propose a set of local and global features for classification. The local features consist of Haralick’s information measures of correlation texture and features [41], inverse difference normalized and inverse difference moment normalized features [42], local structure mean, local structure standard deviation, and local structure deviation [28]. Global features included the mean, standard deviation, skewness, kurtosis, and entropy. They also proposed a thresholded cumulative count of microvascular pixels which was not used in this study because there was no microvascular region extracted.
After extracting these handcrafted features, we trained support vector machines (SVMs) to perform the classification task. Different kernels (linear, rbf and sigmoid) and regularization parameter (0.5, 1.0, 2.0 and 10.0) were tested. For the sigmoid kernel also different independent terms of the kernel function (0.0, 0.5, 1.0) were employed. As for the CNNs, class weights were used and the images were normalized in the same way.
2.3. Handcrafted features: vessel density
The vessel density (VD) is the most studied biomarker for glaucoma in OCTA images [18–21,24,43–47]. In order to compute the VD in this study, the histograms of all images were adjusted to match the histogram of a pre-selected scan in a first step. This procedure was done to neglect effects like different illumination across images such that the parameters of subsequent algorithms yield more reliable results once picked. After that, contrast limited adaptive histogram equalization (CLAHE) [48] is used in order to improve the contrast of the images, followed by the well-known Vesselness filter [49] to enhance the vessel structure in the images. As a last preprocessing step, hysteresis-thresholding was employed in order to create a binary image. This binary image is used to calculate the VD by dividing the number of pixels occupied by vessels by the total number of pixels in the image.
For the classification, we trained different SVMs on this feature with the same settings as described in the subsection above. This was done despite the fact that for a one-dimensional feature vector some of the parameters don’t matter and a SVM is unnecessary general. But since we also use multidimensional handcrafted features vectors for the evaluation, where a SVM is reasonable, we wanted to have the same learning strategy for the sake of an equal comparison.
3. Evaluation
In this section, we first describe the data used in this study and then the experiments performed on this data using the methods described in the section above.
3.1. Data
259 eyes of 199 subjects of the Erlanger Glaucoma Registry (Erlangen Glaucoma Registry, ISSN 2191-5008, CS-2011; NTC00494923) were analyzed retrospectively: 75 eyes of 74 healthy subjects and 184 eyes of 125 glaucoma patients. The cohort consisted of 98 male and 101 female subjects. The average age was 50.6 years with a standard deviation of 21.3 years. All subjects received a standardized ophthalmological examination including measurement of intraocular pressure (IOP) by Goldmann tonometry, fundus photography, and automated visual field testing. Measurements of retinal nerve fiber layer thickness (RNFL), retinal ganglion cell layer (RGC), inner nuclear layer (INL), and Bruch’s Membrane Opening-Minimum Rim Width (BMO-MRW) were done by Heidelberg OCT II Spectralis (version 1.9.10.0, Heidelberg Engineering, Heidelberg, Germany, Glaucoma Premium Module). En face OCTA imaging was done using Heidelberg Spectralis II OCT (Heidelberg, Germany). Images were recorded with a angle and a lateral resolution of m/pixel, resulting in a retinal section of mm. The superficial vascular plexus (SVP), the intermediate capillary plexus (ICP), deep capillary plexus (DCP), and the whole retina (retina = SVP + ICP + DCP) were automatically segmented and projected using the manufacturer’s software. The projected enface OCTA images consist of 512 A-scans per B-scan and 512 consecutive B-scans.
The study has been approved by the ethics committee of the university of Erlangen-Nuremberg and performed in accordance to the tenets of the Declaration of Helsinki. Informed written consent was obtained from each participant.
3.2. Experiments
In this study, a five-fold cross-validation was performed. For one fold the data described above was split into 60% training set, 20% validation set, and 20% test set, with all eyes from one patient only belonging to a single set. The different architectures of the different methods as described in section Methods were then trained on the training set and evaluated on its validation set. The area under receiver operating characteristics (AUROC) was chosen as a metric for the validation. This procedure is then repeated five times such that each image appeared once in the validation set and once in the test set. After the five folds had been trained and evaluated, the mean AUROC per architecture over the validation results of the five folds was computed and the one, out of the three feature extracting methods, that yielded the highest mean AUROC was chosen for the final evaluation on the test set. This five-fold cross-validation was done for each of the three plexus and the whole retina.
4. Results and discussion
The best-performing model that was chosen for each method and plexus can be looked up in Table 1. The AUROC values on the test sets for the different methods and folds and the mean and standard deviation over all folds are displayed in Table 2.
Table 1. Architecture chosen per method and plexus (lr=learning rate, c=regularization parameter, coef0=independent kernel function term).
| CNN | SVM (VD) | SVM (Ong) | |
|---|---|---|---|
| Retina | DenseNet161 (lr=0.001) | Sigmoid kernel (c=10, coef0=0.5) | Linear kernel (c=10) |
| SVP | DenseNet161 (lr=0.001) | Linear kernel (c=1) | Linear kernel (c=1) |
| ICP | WideResNet-101-2 (lr=0.001) | Sigmoid kernel (c=0.5, coef0=0.0) | Linear kernel (c=10) |
| DCP | DenseNet161 (lr=0.001) | Linear kernel (c=1) | Linear kernel (c=2) |
Table 2. AUROC values on the test sets for the different methods and folds and the mean and standard deviation over all folds.
| CNN | SVM (Ong) | SVM (VD) | ||
|---|---|---|---|---|
| Fold 1 | Retina | 0.951 | 0.841 | 0.795 |
| SVP | 0.984 | 0.959 | 0.814 | |
| ICP | 0.937 | 0.883 | 0.814 | |
| DCP | 0.910 | 0.889 | 0.768 | |
|
| ||||
| Fold 2 | Retina | 0.928 | 0.827 | 0.805 |
| SVP | 0.980 | 0.950 | 0.850 | |
| ICP | 0.921 | 0.886 | 0.760 | |
| DCP | 0.877 | 0.886 | 0.760 | |
|
| ||||
| Fold 3 | Retina | 0.939 | 0.795 | 0.784 |
| SVP | 0.995 | 0.984 | 0.827 | |
| ICP | 0.957 | 0.971 | 0.892 | |
| DCP | 0.931 | 0.935 | 0.847 | |
|
| ||||
| Fold 4 | Retina | 0.886 | 0.699 | 0.645 |
| SVP | 0.921 | 0.879 | 0.676 | |
| ICP | 0.825 | 0.841 | 0.681 | |
| DCP | 0.879 | 0.926 | 0.571 | |
|
| ||||
| Fold 5 | Retina | 0.913 | 0.781 | 0.785 |
| SVP | 0.957 | 0.939 | 0.795 | |
| ICP | 0.935 | 0.957 | 0.948 | |
| DCP | 0.957 | 0.948 | 0.841 | |
|
| ||||
| Mean (Std) | Retina | 0.923 0.022 | 0.789 0.050 | 0.763 0.059 |
| SVP | 0.967 0.026 | 0.942 0.035 | 0.793 0.061 | |
| ICP | 0.915 0.046 | 0.912 0.048 | 0.826 0.077 | |
| DCP | 0.910 0.031 | 0.917 0.025 | 0.757 0.100 | |
Here we can see several things. First of all, the CNNs and the features proposed by Ong et al. performed better than the VD across all plexus. This might be because 3 3 mm scans were used and several studies showed that the most vulnerable areas for glaucoma are mostly outside the 3 3 mm area but inside the 6 6 mm area and thus the larger field sizes yield higher AUROC values than the small ones [46,47]. Moreover, on our data, the VD on the ICP performed best for the discrimination between glaucoma patients and healthy controls. Takusagawa et al. [18] reported on 6 6 mm scans an AUROC of 0.96 for the VD computed from the SVP. They also reported that on their data the VD was only slightly decreased for the ICP and not decreased on the DCP. Rabiolo et al. [50] evaluated different methods for the binarization of the vessel structure on 6 6 mm and reported an AUROC value of 0.94 on the SVP and of 0.99 on the DCP. But there were no exact values for the VD in the ICP found in the literature.
The CNNs and the SVM trained on the features presented by Ong et al. performed best on the SVP. Moreover, we can see that both yield approximately the same results for the ICP (CNN: 0.915; SVM(Ong): 0.912) and the DCP (CNN: 0.910; SVM(Ong): 0.917) but the CNNs outperform the SVM(Ong) approach on the SVP (CNN: 0.967; SVM(Ong): 0.942) and especially on the whole retina projection (CNN: 0.923; SVM(Ong): 0.789).
An interesting point here is that the SVM(Ong) approach yielded weak results on the whole retina projection in comparison to its mean AUROC values for the different plexus. A reason for this behavior might be because in the projections of the different plexus, individual vessels are better distinguishable from each other and are not superimposed over each other. Since the features proposed by Ong et al. are mainly textural features, this clearer visualization of the vessel network might be beneficial for automatic classification.
Table 2 shows that the CNN using the SVP achieves the highest AUROC values, which are comparable to the values reported in the literature for larger and/or other regions of the human retina.
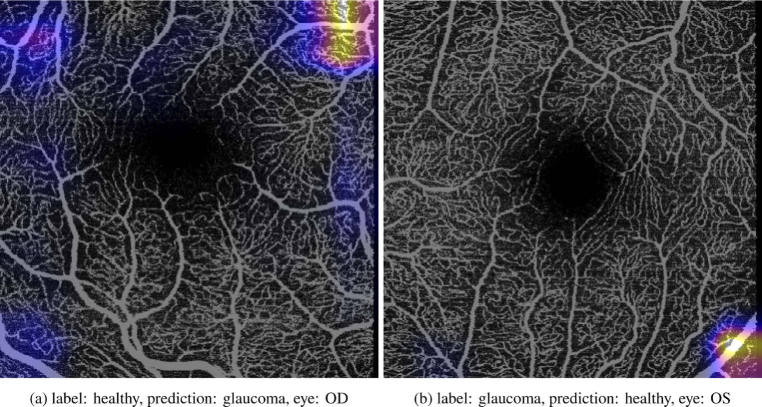
Representative visual examples for the Grad-CAM heatmaps for the classification of the glaucoma class for correctly classified samples and falsely classified samples can be found in Fig. 1 and Fig. 2 respectively. There has been some debate on how reliable the interpretation of these saliency maps are [51]. However, when looking at Fig. 1 and Fig. 2 it seems plausible that in our case they highlight the regions in the images that contribute to classifying an image as glaucomatous. First of all, in the images that were classified as not exhibiting signs of glaucoma (e.g. being healthy) there are no or only small regions that are highlighted. On the other side, if an image was classified as glaucomatous, the number of highlighted regions increased and are larger. Assuming that these highlights show the regions that influence the classification decision, it is striking that the highlights seem to be in regions where vessels are sparser. Moreover, the network seems to focus mostly on the corners and borders of the images, which is in accordance with the aforementioned studies that showed that the vessel density works better in regions farther from the macula. This leads us to the conclusion that vessel density as a feature does not perform worse in general on the smaller scan sizes, but rather the regions in which it was calculated on were too large and/or did not fit. Consequently, improving the region selection for the computation of the vessel density might improve its performance for classification. Further research is warranted to investigate whether these conclusions drawn from the saliency maps hold.
Fig. 1.
Grad-CAM heatmaps for correctly classified samples in the macular area of the test set. Darker/brighter colors indicate regions with lower/higher attention. No color indicates that there was no attention calculated by the Grad-CAM algorithm.
Fig. 2.
Grad-CAM heatmaps for falsely classified samples in the macular area of the test set. Darker/brighter colors indicate regions with lower/higher attention. No color indicates that there was no attention calculated by the Grad-CAM algorithm.
There are several limitations to our study. We did not differentiate between different sub-types and severity stages of glaucoma. So it would be interesting to apply the automatic feature extraction to these more challenging tasks. Another point to notice is that we only discriminated glaucoma patients from a healthy control group and did not include other diseases like age-related macula degeneration (AMD) or diabetic retinopathy (DR) in this second group. It would be an interesting study to see how the different features perform in this more realistic setup. However, the Erlanger glaucoma registry that we worked with only provided data for glaucoma patients and the healthy control group but no other diseases. Another limitation is that for the vessel density, several studies have shown that the 6 6 mm region is more susceptible to changes. Our study was performed on the smaller 3 3 mm images because a larger amount of scans was available. Therefore, it would be interesting whether the features proposed by Ong et al. and the automated features extracted from the CNN could yield even better results if trained on the larger 6 6 mm regions, or if this is only beneficial for classification based on vessel density. However, we have seen that the AUROC values using the CNNs on the smaller scan size are already comparable to the ones achieved by the vessel density on the larger scan size. Consequently, it is feasible to use 3 3 mm scan sizes for the glaucoma detection on OCTA images when choosing appropriate features. Since CNNs are a black-box approach we can conclude that the automatically derived features in the 3 3 mm area allow a good distinction between glaucoma patients and healthy controls, but those features are difficult to interpret. So it would be interesting as a future work to investigate, especially in the SVP, what changes appear and if those could be used as an interpretable biomarker. We did a first step in this direction by computing saliency maps on the test set, but further research has to be conducted in order to demonstrate that the conclusions drawn from them are correct.
Overall this study shows that the automatically extracted features perform no less and in certain plexus even much better than the handcrafted features available for OCTA images for the distinction between glaucoma patients and healthy subjects on 3 3 mm macular scans. With these automatically extracted features we achieved AUROC values that are comparable to the best values reported in the literature, although using the smaller 3 3 mm scan sizes that have performed considerably worse in previously published studies.
5. Summary and conclusion
In this study, we examined handcrafted and automatically trained features for the classification of glaucoma patients. We show that the best performance was achieved on the SVP using the automatically extracted features. Moreover, AUROC values can be achieved that are comparable to the best-reported values in the literature, even on 3 3 mm scan sizes, which were shown to perform worse than larger scans for the vessel density, because the area of the macula that is most vulnerable to glaucomatous vascular damage is outside the central 3 3 mm region. Further analysis of the CNN using attention maps showed that the network focuses on areas where there are fewer vessels, but that these regions are mostly at the border and the corners of the scans and therefore can not be picked up so well by the mean of the vessel density over a larger area.
Acknowledgments
We are grateful to the NVIDIA corporation for supporting our research with the donation of a Quadro P6000 GPU.
Funding
Deutsche Forschungsgemeinschaft10.13039/501100001659 (MA 4898/12-1); European Research Council10.13039/501100000781 (810316).
Disclosures
The authors declare no conflicts of interest.
Data availability
Data underlying the results presented in this paper are not publicly available at this time.
References
- 1.Tham Y.-C., Li X., Wong T. Y., Quigley H. A., Aung T., Cheng C.-Y., “Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis,” Ophthalmology 121(11), 2081–2090 (2014). 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 2.Galassi F., Giambene B., Varriale R., “Systemic vascular dysregulation and retrobulbar hemodynamics in normal-tension glaucoma,” Invest. Ophthalmol. Visual Sci. 52(7), 4467–4471 (2011). 10.1167/iovs.10-6710 [DOI] [PubMed] [Google Scholar]
- 3.Bonomi L., Marchini G., Marraffa M., Bernardi P., Morbio R., Varotto A., “Vascular risk factors for primary open angle glaucoma: the Egna-Neumarkt study,” Ophthalmology 107(7), 1287–1293 (2000). 10.1016/S0161-6420(00)00138-X [DOI] [PubMed] [Google Scholar]
- 4.Tobe L. A., Harris A., Hussain R. M., Eckert G., Huck A., Park J., Egan P., Kim N. J., Siesky B., “The role of retrobulbar and retinal circulation on optic nerve head and retinal nerve fibre layer structure in patients with open-angle glaucoma over an 18-month period,” Br. J. Ophthalmol. 99(5), 609–612 (2015). 10.1136/bjophthalmol-2014-305780 [DOI] [PubMed] [Google Scholar]
- 5.Miglior S., Guareschi M., Gomarasca S., Vavassori M., Orzalesi N., “Detection of glaucomatous visual field changes using the moorfields regression analysis of the heidelberg retina tomograph,” Am. J. Ophthalmol. 136(1), 26–33 (2003). 10.1016/S0002-9394(03)00084-9 [DOI] [PubMed] [Google Scholar]
- 6.Kruse F. E., Burk R. O., Völcker H.-E., Zinser G., Harbarth U., “Reproducibility of topographic measurements of the optic nerve head with laser tomographic scanning,” Ophthalmology 96(9), 1320–1324 (1989). 10.1016/S0161-6420(89)32719-9 [DOI] [PubMed] [Google Scholar]
- 7.Rohrschneider K., Burk R. O., Völcker H. E., “Reproducibility of topometric data acquisition in normal and glaucomatous optic nerve heads with the laser tomographic scanner,” Graefe’s Arch. Clin. Exp. Ophthalmol. 231(8), 457–464 (1993). 10.1007/BF02044232 [DOI] [PubMed] [Google Scholar]
- 8.Mikelberg F. S., Parfitt C. M., Swindale N. V., Graham S. L., Drance S. M., Gosine R., “Ability of the Heidelberg retina tomograph to detect early glaucomatous visual field loss,” J. glaucoma 4(4), 242–247 (1995). 10.1097/00061198-199508000-00005 [DOI] [PubMed] [Google Scholar]
- 9.Hatch W. V., Flanagan J. G., Etchells E. E., Williams-Lyn D. E., Trope G. E., “Laser scanning tomography of the optic nerve head in ocular hypertension and glaucoma,” Br. J. Ophthalmol. 81(10), 871–876 (1997). 10.1136/bjo.81.10.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee M. R., Flotte T., Gregory K., Puliafito C. A., Fujimoto J. G., “Optical coherence tomography,” Science 254(5035), 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengtsson B., Andersson S., Heijl A., “Performance of time-domain and spectral-domain optical coherence tomography for glaucoma screening,” Acta Ophthalmol. 90(4), 310–315 (2012). 10.1111/j.1755-3768.2010.01977.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grewal D. S., Tanna A. P., “Diagnosis of glaucoma and detection of glaucoma progression using spectral domain optical coherence tomography,” Curr. opinion ophthalmology 24(2), 150–161 (2013). 10.1097/ICU.0b013e32835d9e27 [DOI] [PubMed] [Google Scholar]
- 13.Mwanza J.-C., Durbin M. K., Budenz D. L., Sayyad F. E., Chang R. T., Neelakantan A., Godfrey D. G., Carter R., Crandall A. S., “Glaucoma diagnostic accuracy of ganglion cell–inner plexiform layer thickness: comparison with nerve fiber layer and optic nerve head,” Ophthalmology 119(6), 1151–1158 (2012). 10.1016/j.ophtha.2011.12.014 [DOI] [PubMed] [Google Scholar]
- 14.Kotowski J., Folio L. S., Wollstein G., Ishikawa H., Ling Y., Bilonick R. A., Kagemann L., Schuman J. S., “Glaucoma discrimination of segmented cirrus spectral domain optical coherence tomography (SD-OCT) macular scans,” Br. J. Ophthalmol. 96(11), 1420–1425 (2012). 10.1136/bjophthalmol-2011-301021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeoung J. W., Choi Y. J., Park K. H., Kim D. M., “Macular ganglion cell imaging study: glaucoma diagnostic accuracy of spectral-domain optical coherence tomography,” Invest. Ophthalmol. Visual Sci. 54(7), 4422–4429 (2013). 10.1167/iovs.12-11273 [DOI] [PubMed] [Google Scholar]
- 16.Mwanza J.-C., Oakley J. D., Budenz D. L., Anderson D. R., T. N. D. S. Group C. O. C., “Ability of cirrus HD-OCT optic nerve head parameters to discriminate normal from glaucomatous eyes,” Ophthalmology 118(2), 241–248.e1 (2011). 10.1016/j.ophtha.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medeiros F. A., Zangwill L. M., Bowd C., Vessani R. M., Susanna Jr R., Weinreb R. N., “Evaluation of retinal nerve fiber layer, optic nerve head, and macular thickness measurements for glaucoma detection using optical coherence tomography,” Am. J. Ophthalmol. 139(1), 44–55 (2005). 10.1016/j.ajo.2004.08.069 [DOI] [PubMed] [Google Scholar]
- 18.Takusagawa H. L., Liu L., Ma K. N., Jia Y., Gao S. S., Zhang M., Edmunds B., Parikh M., Tehrani S., Morrison J. C., Huang D., “Projection-resolved optical coherence tomography angiography of macular retinal circulation in glaucoma,” Ophthalmology 124(11), 1589–1599 (2017). 10.1016/j.ophtha.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lommatzsch C., Rothaus K., Koch J., Heinz C., Grisanti S., “Octa vessel density changes in the macular zone in glaucomatous eyes,” Graefe’s Arch. Clin. Exp. Ophthalmol. 256(8), 1499–1508 (2018). 10.1007/s00417-018-3965-1 [DOI] [PubMed] [Google Scholar]
- 20.Rao H. L., Pradhan Z. S., Weinreb R. N., Reddy H. B., Riyazuddin M., Dasari S., Palakurthy M., Puttaiah N. K., Rao D. A., Webers C. A., “Regional comparisons of optical coherence tomography angiography vessel density in primary open-angle glaucoma,” Am. J. Ophthalmol. 171, 75–83 (2016). 10.1016/j.ajo.2016.08.030 [DOI] [PubMed] [Google Scholar]
- 21.Rao H. L., Pradhan Z. S., Weinreb R. N., Riyazuddin M., Dasari S., Venugopal J. P., Puttaiah N. K., Rao D. A., Devi S., Mansouri K., Webers C. A., “A comparison of the diagnostic ability of vessel density and structural measurements of optical coherence tomography in primary open angle glaucoma,” PLoS One 12(3), e0173930 (2017). 10.1371/journal.pone.0173930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao H. L., Pradhan Z. S., Weinreb R. N., Riyazuddin M., Dasari S., Venugopal J. P., Puttaiah N. K., Rao D. A., Devi S., Mansouri K., Webers C. A., “Vessel density and structural measurements of optical coherence tomography in primary angle closure and primary angle closure glaucoma,” Am. J. Ophthalmol. 177, 106–115 (2017). 10.1016/j.ajo.2017.02.020 [DOI] [PubMed] [Google Scholar]
- 23.Yip V. C., Wong H. T., Yong V. K., Lim B. A., Hee O. K., Cheng J., Fu H., Lim C., Tay E. L., Loo-Valdez R. G., Teo H. Y., Lim A., Yip L. W., “Optical coherence tomography angiography of optic disc and macula vessel density in glaucoma and healthy eyes,” J. glaucoma 28(1), 80–87 (2019). 10.1097/IJG.0000000000001125 [DOI] [PubMed] [Google Scholar]
- 24.Akil H., Huang A. S., Francis B. A., Sadda S. R., Chopra V., “Retinal vessel density from optical coherence tomography angiography to differentiate early glaucoma, pre-perimetric glaucoma and normal eyes,” PLoS One 12(2), e0170476 (2017). 10.1371/journal.pone.0170476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asaoka R., Murata H., Hirasawa K., Fujino Y., Matsuura M., Miki A., Kanamoto T., Ikeda Y., Mori K., Iwase A., Shoji N., Inoue K., Yamagami J., Araie M., “Using deep learning and transfer learning to accurately diagnose early-onset glaucoma from macular optical coherence tomography images,” Am. J. Ophthalmol. 198, 136–145 (2019). 10.1016/j.ajo.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 26.Muhammad H., Fuchs T. J., De Cuir N., De Moraes C. G., Blumberg D. M., Liebmann J. M., Ritch R., Hood D. C., “Hybrid deep learning on single wide-field optical coherence tomography scans accurately classifies glaucoma suspects,” J. glaucoma 26(12), 1086–1094 (2017). 10.1097/IJG.0000000000000765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J., Kim Y. K., Park K. H., Jeoung J. W., “Diagnosing glaucoma with spectral-domain optical coherence tomography using deep learning classifier,” J. glaucoma 29(4), 287–294 (2020). 10.1097/IJG.0000000000001458 [DOI] [PubMed] [Google Scholar]
- 28.Ong E. P., Cheng J., Wong D. W., Liu J., Tay E. L., Yip L. W., “Glaucoma classification from retina optical coherence tomography angiogram,” in 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), (IEEE, 2017), pp. 596–599. [DOI] [PubMed] [Google Scholar]
- 29.Bowd C., Belghith A., Proudfoot J. A., Zangwill L. M., Christopher M., Goldbaum M. H., Hou H., Penteado R. C., Moghimi S., Weinreb R. N., “Gradient-boosting classifiers combining vessel density and tissue thickness measurements for classifying early to moderate glaucoma,” Am. J. Ophthalmol. 217, 131–139 (2020). 10.1016/j.ajo.2020.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopinath K., Sivaswamy J., Mansoori T., “Automatic glaucoma assessment from angio-OCT images,” in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), (IEEE, 2016), pp. 193–196. [Google Scholar]
- 31.Christopher M., Belghith A., Bowd C., Proudfoot J. A., Goldbaum M. H., Weinreb R. N., Girkin C. A., Liebmann J. M., Zangwill L. M., “Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs,” Sci. Rep. 8(1), 16685 (2018). 10.1038/s41598-018-35044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H., Li L., Wormstone I. M., Qiao C., Zhang C., Liu P., Li S., Wang H., Mou D., Pang R., Yang D., Zangwill L. M., Moghimi S., Hou H., Bowd C., Jiang L., Chen Y., Hu M., Xu Y., Kang H., Ji X., Chang R., Tham C., Cheung C., Shu Wei Ting D., Yin Wong T., Wang Z., Weinreb R. N., Xu M., Wang N., “Development and validation of a deep learning system to detect glaucomatous optic neuropathy using fundus photographs,” JAMA Ophthalmol. 137(12), 1353–1360 (2019). 10.1001/jamaophthalmol.2019.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson A. C., Jammal A. A., Berchuck S. I., Mariottoni E. B., Medeiros F. A., “Assessment of a segmentation-free deep learning algorithm for diagnosing glaucoma from optical coherence tomography scans,” JAMA Ophthalmol. 138(4), 333–339 (2020). 10.1001/jamaophthalmol.2019.5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maetschke S., Antony B., Ishikawa H., Wollstein G., Schuman J., Garnavi R., “A feature agnostic approach for glaucoma detection in oct volumes,” PLoS One 14(7), e0219126 (2019). 10.1371/journal.pone.0219126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russakoff D. B., Mannil S. S., Oakley J. D., Ran A. R., Cheung C. Y., Dasari S., Riyazzuddin M., Nagaraj S., Rao H. L., Chang D., Chang R. T., “A 3d deep learning system for detecting referable glaucoma using full oct macular cube scans,” Trans. Vis. Sci. Tech. 9(2), 12 (2020). 10.1167/tvst.9.2.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G., Liu Z., Van Der Maaten L., Weinberger K. Q., “Densely connected convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017), pp. 4700–4708. [Google Scholar]
- 37.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016), pp. 770–778. [Google Scholar]
- 38.Xie S., Girshick R., Dollár P., Tu Z., He K., “Aggregated residual transformations for deep neural networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017), pp. 1492–1500. [Google Scholar]
- 39.Zagoruyko S., Komodakis N., “Wide residual networks,” arXiv preprint arXiv:1605.07146 (2016).
- 40.Selvaraju R. R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D., “Grad-cam: Visual explanations from deep networks via gradient-based localization,” in Proceedings of the IEEE international conference on computer vision, (2017), pp. 618–626. [Google Scholar]
- 41.Haralick R. M., Shanmugam K., Dinstein I. H., “Textural features for image classification,” IEEE Transactions on Systems, Man, and Cybernetics, pp. 610–621 (1973).
- 42.Clausi D. A., “An analysis of co-occurrence texture statistics as a function of grey level quantization,” Can. J. Remote Sens. 28(1), 45–62 (2002). 10.5589/m02-004 [DOI] [Google Scholar]
- 43.Rao H. L., Kadambi S. V., Weinreb R. N., Puttaiah N. K., Pradhan Z. S., Rao D. A., Kumar R. S., Webers C. A., Shetty R., “Diagnostic ability of peripapillary vessel density measurements of optical coherence tomography angiography in primary open-angle and angle-closure glaucoma,” Br. J. Ophthalmol. 101(8), 1066–1070 (2017). 10.1136/bjophthalmol-2016-309377 [DOI] [PubMed] [Google Scholar]
- 44.Lee K., Maeng K. J., Kim J. Y., Yang H., Choi W., Lee S. Y., Seong G. J., Kim C. Y., Bae H. W., “Diagnostic ability of vessel density measured by spectral-domain optical coherence tomography angiography for glaucoma in patients with high myopia,” Sci. Rep. 10(1), 1–10 (2020). 10.1038/s41598-019-56847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J., Sebastian R. T., Chu C. J., McGregor F., Dick A. D., Liu L., “Reduced macular vessel density and capillary perfusion in glaucoma detected using OCT angiography,” Curr. Eye Res. 44(5), 533–540 (2019). 10.1080/02713683.2018.1563195 [DOI] [PubMed] [Google Scholar]
- 46.Hood D. C., Raza A. S., de Moraes C. G. V., Liebmann J. M., Ritch R., “Glaucomatous damage of the macula,” Prog. Retinal Eye Res. 32, 1–21 (2013). 10.1016/j.preteyeres.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moghimi S., Hou H., Rao H. L., Weinreb R. N., “Optical coherence tomography angiography and glaucoma: a brief review,” Asia-Pac. J. Ophthalmol. 8, 115–125 (2019). 10.22608/APO.201914 [DOI] [PubMed] [Google Scholar]
- 48.Pizer S. M., Amburn E. P., Austin J. D., Cromartie R., Geselowitz A., Greer T., ter Haar Romeny B., Zimmerman J. B., Zuiderveld K., “Adaptive histogram equalization and its variations,” Comput. Vision, Graphics, Image Processing 39(3), 355–368 (1987). 10.1016/S0734-189X(87)80186-X [DOI] [Google Scholar]
- 49.Frangi A. F., Niessen W. J., Vincken K. L., Viergever M. A., “Multiscale vessel enhancement filtering,” in International Conference on Medical Image Computing and Computer-assisted Intervention (Springer, 1998), pp. 130–137. [Google Scholar]
- 50.Rabiolo A., Gelormini F., Sacconi R., Cicinelli M. V., Triolo G., Bettin P., Nouri-Mahdavi K., Bandello F., Querques G., “Comparison of methods to quantify macular and peripapillary vessel density in optical coherence tomography angiography,” PLoS One 13(10), e0205773 (2018). 10.1371/journal.pone.0205773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viviano J. D., Simpson B., Dutil F., Bengio Y., Cohen J. P., “Saliency is a possible red herring when diagnosing poor generalization,” arXiv preprint arXiv:1910.00199 (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time.