Abstract
A method for the continuous detection of heart rate (HR) in signals acquired from patients using a sensor mat comprising a nine-element array of fiber Bragg gratings during routine magnetic resonance imaging (MRI) procedures is proposed. The method is based on a deep learning neural network model, which learned from signals acquired from 153 MRI patients. In addition, signals from 343 MRI patients were used for result verification. The proposed method provides automatic continuous extraction of HR with the root mean square error of 2.67 bpm, and the limits of agreement were -4.98–5.45 bpm relative to the reference HR.
1. Introduction
The usefulness of fiber-optic vital sign sensors has been confirmed several times in laboratory evaluations [1–5] and clinical assessments [6–8]. In particular, we have previously proved the efficacy of a fiber-optic sensor system in monitoring patients using a magnetic resonance imaging (MRI) scanner [9]. For patients with claustrophobic tendencies, an MRI examination is a stressful experience due to the limited space around them, isolation from the environment, and noise generated by the scanner [10]. A high level of anxiety usually manifests itself through elevated heart activity and breathing [11,12]. By monitoring these indicators, the MRI operator can control the current condition of the patient, decide whether to continue or stop the examination, and recommend therapy to the patient to complete his/her MRI [13]. In addition to claustrophobic patients, it is also recommended to monitor neonates, children, patients with disabilities, and others who are unable or may not be able to communicate or use the alarm button during MRI [14].
Fiber-optic technology is both immune to MRI electromagnetic field penetration and neutral to imaging quality [15,16]. The simplest sensor designs with a single fiber Bragg grating (FBG) exhibit the ability to acquire respiratory curves and ballistocardiographic (BCG) signals from the human body at full MRI compatibility [17,18]. FBGs are particularly attractive owing to their excellent multiplexing and self-referencing capabilities, which allow for addressing several sensing elements with a single optical fiber [19–21]. In the literature, details on sensors based on arrays of 12 [22] and 40 [23] FBGs, as well as concepts of monitoring many individuals with the use of FBGs connected in series [24] are provided. In general, the intention of the multi-FBG sensor constructors was to improve the reliability by acquiring signals from a number of localizations on the body and aggregating them into one noise-free signal used for the detection of physiological parameters.
In 2010, Hao et al. proposed a sensor system employing 12 FBGs arranged in a 3 × 4 array, connected in series, embedded in arc-shaped metal bridges, and deployed on a bed beneath the mattress supporting a patient [22]. The authors performed laboratory evaluations on 10 subjects and showed a maximum error of ±1 respirations per minute (rpm) compared with manual counting. They noted the sensor’s ability to measure HR; however, corroborating data have not been made available to date. As with most of the information presented in this paper, data processing is also briefly described. The initial stage of data processing included standard blocks such as normalization and filtration, while further processing was based on wavelet decomposition to remove signals other than those induced by respiration. Subsequently, an autocorrelation block was applied to observe how the signal changed over time, and the RR was determined from the pool of periodic components. The authors did not report whether the multi-point measurement was used to automatically select the best signal for detecting RR or only to present RR acquired from multiple locations in the patient.
In 2012, Allsop et al. presented an innovative system for respiratory function monitoring comprising an array of 40 FBGs connected in series and embedded into a specially designed textile vest [23]. The sensor system was distinguished by a high accuracy but was relatively complex, and the authors aimed at advanced measurements of three-dimensional volumetric changes of the human torso rather than common RR monitoring. The purpose of laboratory evaluations was to test the sensing vest on a small group of subjects (n = 5) with a large anatomical variation, and the vest was able to work with various sizes and changes in the torso. The study yielded the volumetric error of 6% when comparing the volume data obtained from a spirometer with the volume estimated using the synchronous data from the sensing vest. It is noteworthy that the extensive algorithm allowed the authors to convert the wavelengths to curvatures and generate the shape from the sensing array. By combining all of the curvature values, coupled with the known spatial dimensions, the authors were able to generate the corresponding x, y, and z coordinates of the sensor array and reconstruct the entire sensing array in real-time in three-dimensional space. This enabled them to calculate the volume of the various sections of the system and, consequently, observe the volumetric changes of the torso.
In 2017, Fajkus et al. developed a novel measuring probe based on two FBGs encapsulated inside a polydimethylsiloxane (PDMS) polymer [24]. The polymeric shell was used to increase the temperature sensitivity of the probe and ensure its nonreactivity to human skin. The use of two FBG elements in the probe enabled the authors to measure strain and temperature simultaneously if the FBGs differed in strain and thermal responsivities [25]. The authors achieved different strain and thermal responsivities of the FBGs in the encapsulation process and through a specific shape of the measuring probe. Laboratory evaluation included 10 subjects, and the results showed maximum errors of approximately ±1 rpm and ±4 bpm compared with reference respiration and ECG monitors, respectively. The authors also measured the body temperature of the subjects and obtained the maximum error of 0.36% relative to the measurement with a reference thermometer. Besides the possibility of measuring RR, HR, and body temperature simultaneously, another advantage of the probe was the provision of fiber-optic connectors on both sides of the FBGs connected in series. This allowed the authors to connect up to 32 probes in series, i.e., up to 64 sensors, with a measurement window with a spectral width of approximately 1.9 nm assigned to each probe. The authors presented the concept of a 4-channel system to interrogate 128 measuring probes (4 × 32), but its physical implementation has not been reported so far.
It is clear from the above discussion that techniques for efficient processing signals from multi-FBG vital sign sensors are still just beginning to develop. In this paper, we introduce a method for continuous detection of heart rate (HR), which allows for utilizing the advantages of fiber-optic sensors with multi-point measurement. The innovative character of the presented work is the combination of FBG sensing and deep learning temporal convolutional network (TCN) signal processing for HR detection.
2. Methods
2.1. Sensor mat
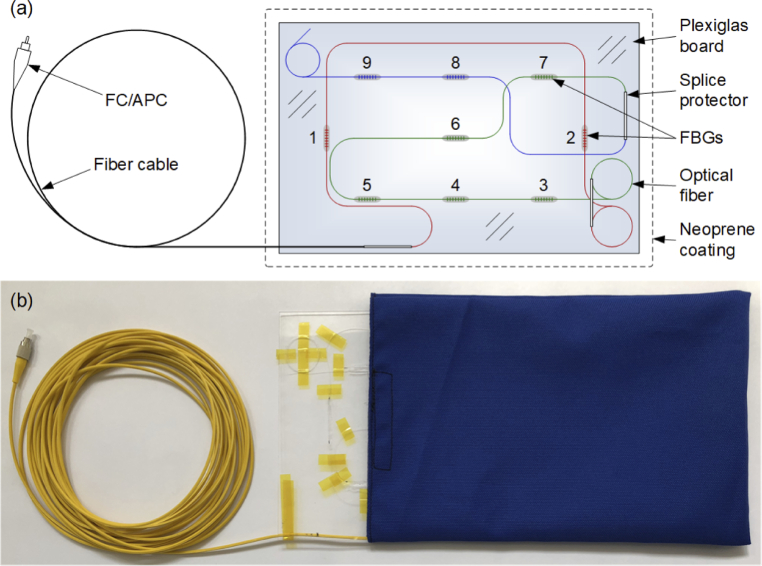
To develop the detection method, we constructed a sensor mat based on a nine-element array of FBGs (3 × 3), in which two of the FBGs were arranged orthogonally to the others, as shown in Fig. 1(a). The FBGs are marked with numbers 1–9 in order from the lowest to the highest value of the Bragg wavelength λB. The FBGs were attached to a springy Plexiglas board to perform a multi-point measurement of strains induced by body movements, including those exerted on the board by the work of the heart. Previously, we observed the best BCG signal acquisition when using FBGs placed along the longitudinal axis of the body [26]. However, FBGs placed along the transverse axis of the body may be crucial in the acquisition of the respiratory signal due to the reduced contribution of heart-induced artifacts to the entire mechanically disturbed signal. The variety in positioning of the FBGs relative to the body axes provides an opportunity to gain wider insight into heart- and lung-induced body motions and enables the selection of the most useful signals to detect HR and respiratory rate (RR). Nevertheless, the scope of this study is focused on HR detection issues.
Fig. 1.
Multi-point measurement mat: (a) design and (b) physical realization.
Using the phase mask technique [27], we inscribed nine FBGs into the cores of three sections of single-mode fibers with the following configurations of the Bragg wavelengths and distances between the FBGs:
-
•
Section I (marked red in Fig. 1(a)): FBG #1: λB1 = 1526 nm, distance of 36 cm, FBG #2: λB2 = 1530 nm.
-
•
Section II (marked green): FBG #3: λB3 = 1538 nm, distance of 7 cm, FBG #4: λB4 = 1541 nm, distance of 7 cm, FBG #5: λB5 = 1546 nm, distance of 17 cm, FBG #6: λB6 = 1551 nm, distance of 10 cm, FBG #7: λB7 = 1556 nm.
-
•
Section III (marked blue): FBG #8: λB8 = 1560 nm, distance of 7 cm; FBG #9: λB9 = 1564 nm.
The spectral distances of Bragg wavelengths were arranged to avoid the problem of spectral overlapping during measurements. Prior to the inscription of the gratings, the optical fibers were hydrogen-loaded at 110 bar at room temperature for two weeks to enhance their photosensitivity. Then, the three series of FBGs were inscribed using a UV pulsed KrF excimer laser operating at 248 nm with the pulse energy of 3 mJ and pulse repetition of 500 Hz. The growth of the gratings was monitored during the fabrication process to obtain similar reflectivity for each of the FBGs. Finally, the optical fibers were annealed at 100 °C for 48 h to enhance the stability of the FBG parameters [28].
The optical fiber ends were spliced together and protected with shrink tubes; hence, all the FBGs were connected in series. This FBG assembly was arranged on a Plexiglas board with dimensions of A4 paper and thickness of 2 mm. The FBGs were attached to the board using epoxy adhesive. The fiber was looped in the case of excess. The loops and splice protectors were taped onto the board. The FBG series was spliced one-sidedly with a 10-m fiber cable pigtail terminated with a fiber-optic connector for angled physical contact (FC/APC). The Plexiglas board was inserted into a neoprene coating from which the fiber cable emerged, as shown in Fig. 1(b). For a better view, the Plexiglas board is shown partially slid out of the neoprene coating.
2.2. Experimental setup
A broadband superluminescent light-emitting diode (SLED) source by DenseLight Semiconductors, with the 3-dB wavelength range of 1515–1575 nm, was used to illuminate the FBGs through an embedded optical circulator. Spectral signals reflected from the FBGs were analyzed using the I-MON 256 monitor by Ibsen Photonics based on fused silica transmission gratings to split the spectra onto an array of 255 diodes. The monitor provides real-time spectral measurement in the range of 1525–1570 nm at a frequency of up to 6 kHz and wavelength fit resolution below 0.5 pm. Data from the diode array were transmitted via a USB connection to a laptop personal computer (PC) and processed to recover the instantaneous positions of the FBG reflection peaks. Deviations from the wavelength linearity of the diode array were corrected using polynomial functions in the analysis software. Because the average wavelength between successive diodes in the array is approximately 170 pm, and the full width at half maximum is approximately 300 pm for each of the FBGs used in the sensor mat, a single FBG reflection peak is sampled using 6–8 wavelength values. 2-3 of these values correspond to the points laying on the peak within the FWHM, and the remaining values correspond to the points laying on the slopes on either side of the peak, i.e., beyond the FWHM. The analysis software determines the center frequency of an FBG by fitting a Gaussian curve using the least-squares method. The time-dependent changes in the Bragg wavelength are the input signals for further processing steps.
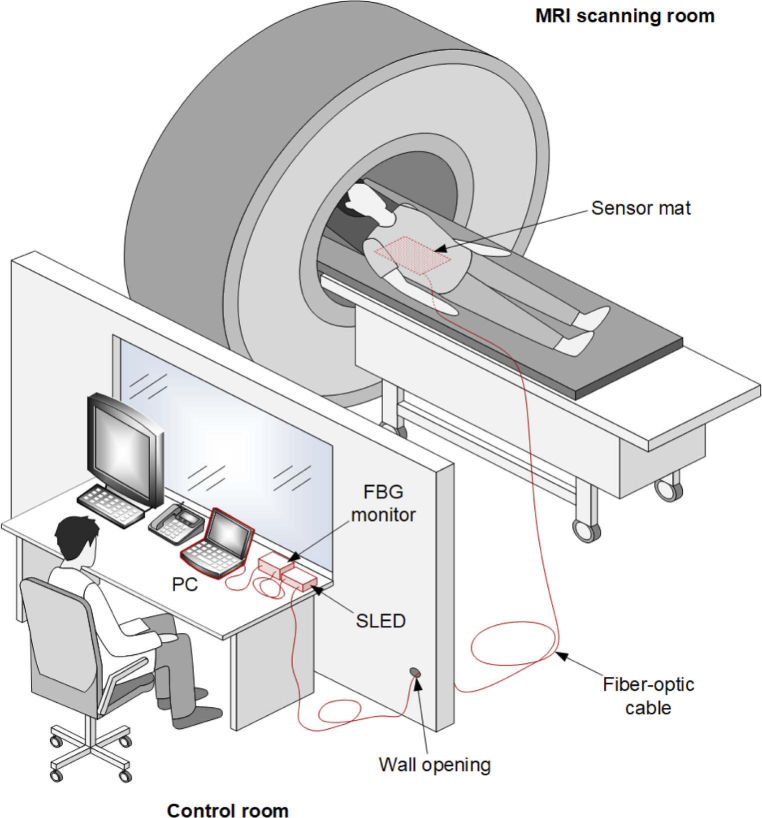
The experimental setup was evaluated during routine MRI procedures in the configuration shown in Fig. 2. In this study, 1.5-T and 3-T MRI units, specifically, model Vantage Elan 1.5T and model Vantage Titan 3.0T, by Canon Medical Systems, were employed. Measurement data acquired from MRI patients were used to develop a deep learning neural network model in the range of network learning, validation, and testing. Deep learning is a widely used technique for signal or image processing and is readily used in the field of biomedical photonics [29–31]. The basic assumption of the study was to limit the interference in MRI procedures to the minimum required. Therefore, the only nonstandard action was to place the sensor mat under the patient’s back. We did not decide to record physiological signals or parameters using a reference measurement system, as this would require patient preparation and extend the MRI procedures. The use of additional equipment, for example, MRI-compatible carbon electrodes, could also adversely affect study-associated stress levels in patients. Instead, we tested the proposed online HR detection method indirectly, using an additional developed auxiliary offline method. First, we verified the offline method using HR detected from both the optical signal and electrocardiographic (ECG) signal acquired from volunteers outside the MRI environment. In this case, the HR detected from the ECG signal was the reference for the HR detected from the optical signal coming from the sensor mat. Then, the HR obtained using the online method was compared with that obtained using the previously verified offline method. In this case, the offline method was used as a reference for the online method.
Fig. 2.
Experimental setup within MRI procedures.
2.3. Detection method
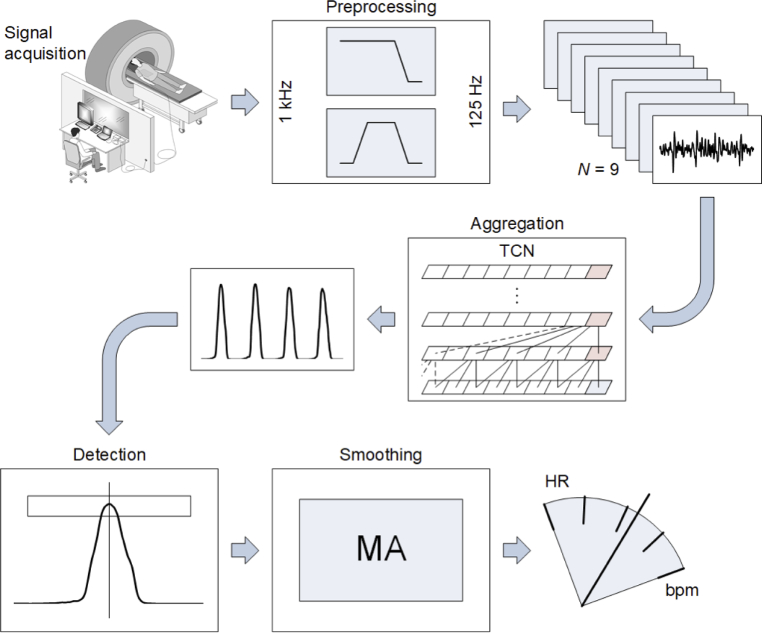
The optical signal from the sensor mat was processed according to the scheme shown in Fig. 3 through the following main functional blocks:
-
•
Preprocessing;
-
•
Aggregation;
-
•
Detection of characteristic points;
-
•
Smoothing.
Fig. 3.
Block diagram of signal processing
The original signals were recorded at the sampling rate of 1 kHz. Preprocessing included downsampling by the factor of 8 with a decimation filter, prefiltering with a bandpass filter at 6 and 20 Hz cut-off frequencies, and normalization. The filters removed unwanted frequency components such as slowly varying components originating from temperature changes as well as high-frequency noise. The normalization procedure utilized moving-window normalization so that the signal could be analyzed continuously. The parameters used for normalization, i.e., the median and interquartile range (IQR), were determined in a moving window time range. The median was subtracted from the input signal, whereas IQR was used to scale the values of the signal. The normalization window length was a parameter of the method, which was set to 3 s based on a series of trials.
The next processing block formed an aggregated detection function. A deep learning TCN was used to generate a single trace from N = 9 input signals coming from the FBGs while removing any noise that might appear on one of them. The actual HR detection was performed using an aggregated detection function. Descriptions of the structure of the network and the learning process are provided in sections 2.4–2.6.
The characteristic points are the notches occurring periodically in the analyzed signal, caused by the mechanical activity of the heart. The greatest effect in the BCG signal recorded by the FBGs was caused by the contraction of the ventricles [17]. The detection of the characteristic points of the BCG waves allowed us to determine the duration of the cardiac cycle. A given value of the analyzed signal was classified as a potential characteristic point if it was the maximum value within an interval of a specified width with the center at the time of occurrence of this value. The width of this interval was derived from the dominant frequency component of the signal, specifically, the pitch, according to the procedure described in [32].
In the smoothing block, the sequence of the characteristic points was converted into a sequence of intervals, producing a tachogram. Then, the intervals were averaged with 3-element moving average (MA).
2.4. Network building
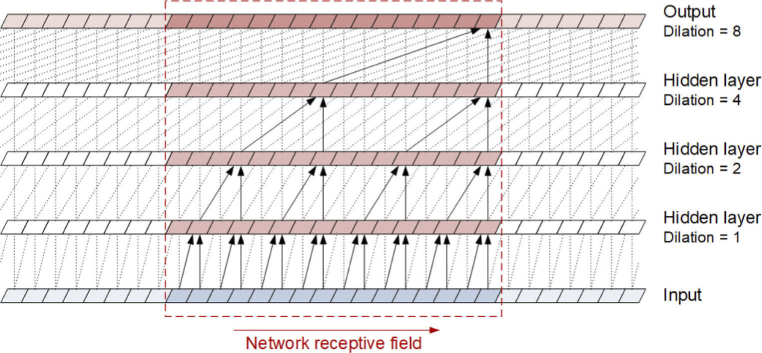
As previously mentioned, we used TCN in the proposed detection method. This architecture was distinguished by causal convolutions, so there was no information leakage from the future to the past values of the input sequences [33]. Moreover, the TCN might accept a sequence of any length and map it to an output sequence of the same length. An additional advantage of the TCN architecture is the ability to create networks with a large history size, that is, the ability of the network to look very distant from current sequence values. The data flow in a TCN is schematically shown in Fig. 4. To facilitate understanding of the operation principle, the network is simplified to 4 layers and the kernel size 2, although the TCN model used in the study and described in section 2.6 was of greater size and complexity. The figure shows the structure of the information flow from the input sequence, through the successive layers to the output sequence, including the time dependence of elements of the input sequence that form the receptive field of the network. The scheme does not reflect the full complexity of the network, such as the number of input sequences and filters in individual layers as well as the construction of residual blocks that make up the network layers; all of these details are described in [33]. The main component of the residual blocks are dilated convolutions. Their use is to obtain dilations between sequence elements selected as the inputs to higher layers, and the dilations enable the network receptive filed to grow proportionally to the square of the number of layers. The TCN can accept an input sequence of any length, although the implementation of the network requires the provision of the input sequence in portions to optimize computations.
Fig. 4.
Scheme of the data flow in a TCN
During the study, the possibility of using the long short-term memory (LSTM) network as well as the combination of TCN and LSTM architectures were also recognized. However, no better results were obtained compared with those using only the TCNs.
In the proposed approach, the TCN architecture generated a detection function, i.e., a signal in which characteristic points related to HR were detected. The input of the network was the signals coming from the sensor mat, which had been preprocessed as described above. The TCN architecture converted N = 9 input sequences (N is equal to the number of FBG elements) into one output sequence, which had the same length as the input sequence.
To develop a model that converts a set of signals coming from the FBGs into a single detection function, the open-source machine learning library PyTorch was used. This environment is extremely flexible because it is essentially an automatic differentiation system [34]. PyTorch enables optimization of models with a user-defined structure described in the Python programming language. This, in turn, allowed us to customize the target functions used in the learning process. In cases where the task of the model was to generate a sequence that was similar to a given target sequence, the first-choice function was the root mean square error (RMSE). In the presented modification of the target function, the RMSE calculation step was preceded by the synchronization of the signals using cross-correlation. This introduced an additional degree of freedom during the optimization, as the model did not have to generate a signal that was perfectly synchronized with the target signal. The resulting signal might be shifted in time by some amount with respect to the target signal. The parameter of the learning process was the time range in which synchronization was sought, i.e., the synchronization time. Additionally, the initial portion of the waveforms was not included in the RMSE calculation. This allowed the initial startup of the network. The length of this initial portion was defined as a divider of the length of the compared sequences and was a parameter of the learning process, that is, the idle divider. In the process of weight optimization of the neural network, the objective function is differentiated. Therefore, in addition to the development of the signal synchronizing code itself, it was necessary to provide the possibility of differentiating the code using the tape-based automatic differentiation system of PyTorch.
Because the detection method was implemented in C# in the .Net platform environment, the TCN model was used from the level of this platform via open neural network exchange (ONNX) technology. ONNX is an open-source software library that allows for running neural network models on various hardware and operating system environments, freely available on the ONNX website and via GitHub.
2.5. Training set
To train the neural network, a method for creating target signals was developed. The software used to create the training set had the form of the R computing environment script. Ultimately, the network was to operate in the continuous (i.e., online) regime; however, when generating the training set, this assumption was not necessary.
In the first step of the training waveform preparation, each signal obtained from the sensor mat was converted into a detection function. The block producing the detection function consisted of a bandpass filter, derivative operation, square operation, and final low-pass filter. The cut-off frequencies of the input bandpass filter were 3 and 10 Hz, whereas the final low-pass filter had the cut-off frequency of 4 Hz. The next stage of processing was the detection of the local maxima of the detection function, which was identical to the local maxima detection described in section 2.3.
The purpose of the next step in creating target signals was to determine the HR for each of the detection functions that originated from the FBG signals. This procedure was implemented in C# language in the form of a software library. The library was used as a tool from the level of the R language script used to create the training set. The variability measure for each of the obtained HR signals was calculated in the form of standard deviation and its quantile at the level of 0.3. This constituted a threshold for the criterion of selection of signals used for further processing steps; specifically, signals with greater variability than the limit value were rejected. The justification for this procedure was that possible errors in signal detection usually significantly increased the variability of the signal in relation to the real physiological signal. Subsequently, the detection functions of the signals that had passed the selection were added together to form an aggregated detection function, and then the local maxima of this aggregated function were detected. The generated sequence of maxima, that is, a set of potential characteristic points, was an input for the pulse generator. The generator created an impulse train, constructed from Dirac delta functions, which were then filtered with a low-pass filter to create the target signal for the learning process. This signal was similar to the detection function; however, it did not contain fluctuations in the amplitude of the pulses, and the signal values between the pulses were very close to 0.
Each element of the training set consisted of 16-s portions of the input signals and the target signal that resulted from the operation of the procedure described above. There were 153 sets of inputs and the same number of target outputs. The inputs for the developed model were portions of recordings acquired from 153 patients (98 females and 55 males) aged 48.80 ± 15.43 (M ± SD) years using the FBG-based sensor mat placed under the patients’ backs during routine MRI procedures.
2.6. Training
The Adam optimizer was employed during the training process (beta1 = 0.9, beta2 = 0.999) [35]. The network-based models were tested with many combinations of network sizes and other metaparameters. The final model was developed using the values listed in Table 1. The learning rate is a standard tuning parameter of the optimization algorithms, and determines how much the model should be changed at each iteration of the learning process when the gradient of the loss function is estimated while moving toward the minimum of this function. The validation fraction defined the fraction of the validation set count in the total number of learning cases. The batch size was the number of cases taken to determine the value of the objective function in each iteration of the learning process. The dropout is the probability of random zeroing of a network parameter. The clip is a gradient clipping-by-norm method threshold that prevents large change in the weight values during training when the loss function landscape forms a cliff. The kernel size, layers, and filters per layer parameters define the shape of the TCN model [33]. The synchronization time is the width of the signal synchronization interval in the objective function computation in seconds. The idle divider was the length of the initial fragment ignored when calculating the RMS in the objective function computation, defined as a divider of the length of the compared sequences.
Table 1. Metaparameters of the learning process.
| Parameter | Value |
|---|---|
| Learning rate | 0.0005 |
| Validation fraction | 0.2 |
| Batch size | 32 |
| Dropout | 0.25 |
| Clip | 0.1 |
| Kernel size | 6 |
| Layers | 6 |
| Filters per layer | 16 |
| Synchronization time | 0.23 |
| Idle divider | 4 |
3. Results
Verification of the HR detection method proposed for the multi-FBG-based sensor was carried out in two stages. In the first stage, we compared the HR traces obtained from the optical signal using the auxiliary offline method with those obtained from the ECG signal. In the second stage, we used the auxiliary offline HR detection method to verify the method for continuous HR detection, i.e., the online method. Thus, in both stages, an essential element of the analysis was the auxiliary offline method, which was first verified and then used as a reference for the proposed online method. The successive steps of the auxiliary offline HR detection method were similar to the procedure of creating target signals for the training set for the machine learning process described in section 2.5, except that the obtained characteristic point sequence was not used as a trigger for the pulse generator, but was directly converted into an HR signal.
In the auxiliary offline method, the signals obtained from the sensor mat were converted into the detection function, followed by the detection of local maxima of the detection function and determination of the HR signal expressed in beats per minute (bpm). For all of the obtained HR signals, measures of their variability were calculated in the form of standard deviations and their quantiles at the level of 0.3. Traces with a variability lower than the determined threshold were added together to create an aggregated detection function. Next, the characteristic points were detected in this function, then, the correction of the characteristic points sequence was applied, and, consequently, the corrected points were used for the final HR detection. The correction was based on the analysis of the intervals between the points, particularly their variability. The variability, expressed as the standard deviation, was checked in the queue of the k last characteristic points. For each of these points, the variability of intervals, expressed as a standard deviation, including the considered point and its nearest neighboring points (k points in total), was computed and compared with the variability excluding a given point. If the ratio of these values indicates a significant decrease in variability after removing the midpoint, the point is removed. It was assumed that a characteristic point was removed if the variability decreased to 2/3 of the initial value and below. The number of points used for the variability computation k was a metaparameter of the method and was set to five.
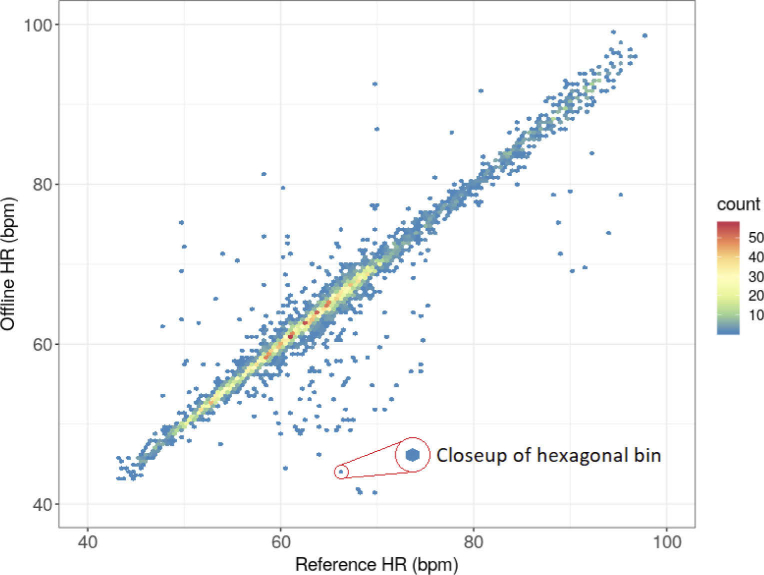
The first stage of the validation was conducted using an ECG recorder as a reference to verify the auxiliary offline method. ECG signals were acquired from 16 subjects (2 females and 14 males) aged 25.56 ± 10.03 (M ± SD) years using an ECG module of the medical monitoring subsystem described in [36]. Figure 5. shows the scatter plot, or 2D histogram, of the HR values obtained using the auxiliary offline method against the reference ECG-based HR values. The count of results is assigned to hexagonal bins; the number of bins in both the vertical and horizontal directions is set to 120. The bins contain HR counts corresponding to the detected heartbeats. Since the presented method was developed to measure HR continuously, we treated HR as a signal. There may be a different time interval between each heartbeat and the previous one. Thus, each successive reading of the HR value may differ from the previous one. An HR recording during the MRI procedure for one subject contains many HR values corresponding to the detected heartbeats. The longer the registration, the more HR values. The number of results in each bin was coded according to the color of the points. The bins shown in Fig. 5 contain HR counts, the number of which corresponds to the number of detected heartbeats.
Fig. 5.
Scatter plot (2D histogram) of the HR values obtained by the auxiliary offline method against the reference HR values determined from the ECG signal.
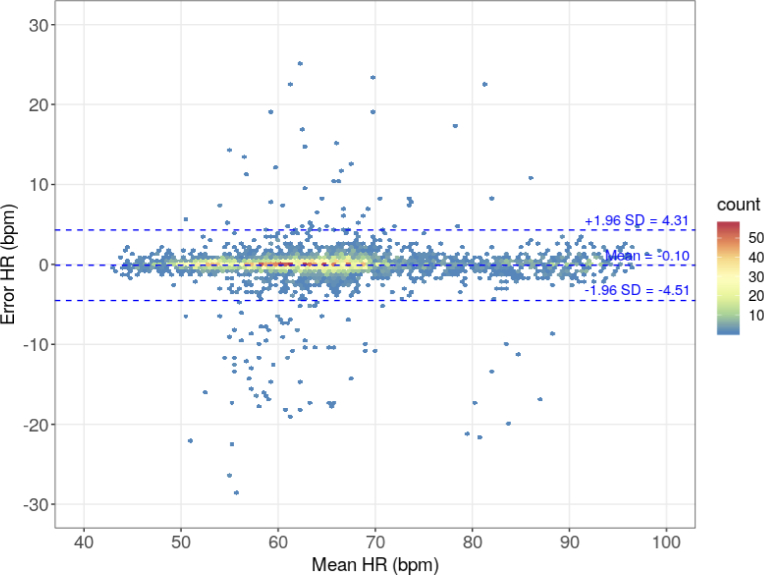
Figure 6 shows the modified Bland–Altman plot. As in the classic Bland–Altman plot, the X-axis is the average of the compared values, and the Y-axis is their difference. The individual points of the graph correspond to the number of results in hexagonal bins; the number of hexagonal bins in both the vertical and horizontal directions is set to 120. The number of results in each bin was coded according to the color of the points. The Bland–Altman plot shows the limits of agreement at -4.51 and 4.31 bpm with the mean error of -0.10 bpm. The RMSE obtained from comparing the auxiliary offline method results and HR computed from the ECG acquisitions is 2.25 bpm.
Fig. 6.
Bland-Altman plot of the HR values obtained from the auxiliary offline method against the reference HR values determined from the ECG signal.
The auxiliary offline HR detection method was used in the second stage of testing to verify the online method. A total of 343 MRI patients (220 females and 123 males) aged 49.29 ± 14.50 (M ± SD) years were included in this stage. It should be noted that these were different patients than those whose recordings were used to learn the network. The experimental protocol complied with the Declaration of Helsinki and was approved by the Ethics Committee of the Military Institute of Aviation Medicine (Decision 10/2015). The study was performed in the Laboratory of Computed Tomography and Magnetic Resonance in Bialystok, a part of TMS Diagnostyka Sp. z o.o., in accordance with the relevant guidelines and regulations. The subjects were informed of the purpose and nature of the study and signed their informed consent for study participation and the use of physiological parameters in the data analysis. The interference with MRI procedures was minimal; it only included the presence of the sensor mat under the patients’ backs.
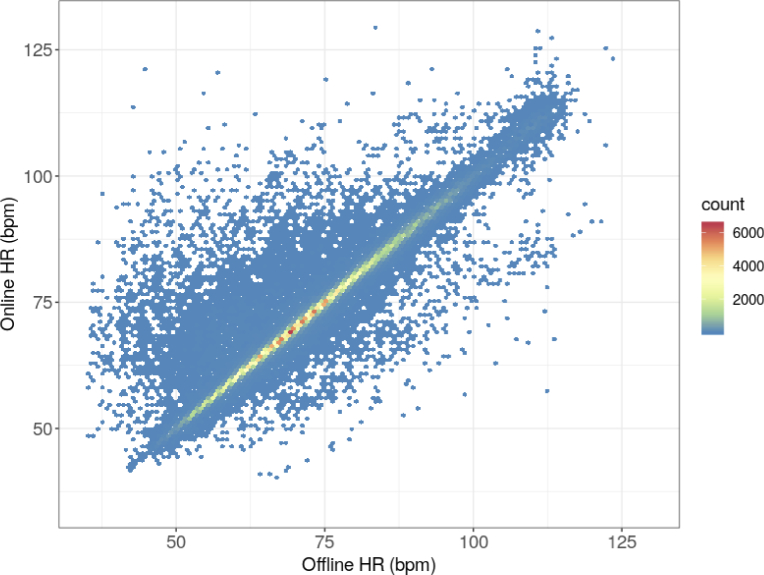
The analysis performed to verify the method included 368,954 HR values. Figure 7. shows the scatter plot of the HR values obtained using the online method against the reference HR values obtained with the previously verified auxiliary method. The count of results is assigned to hexagonal bins; the number of bins in both the vertical and horizontal directions is set to 120. As previously, the bins contain HR counts, the number of which corresponds to the number of detected heartbeats.
Fig. 7.
Scatter plot (2D histogram) of the HR values obtained by the online method against the reference HR values obtained by the previously verified auxiliary method.
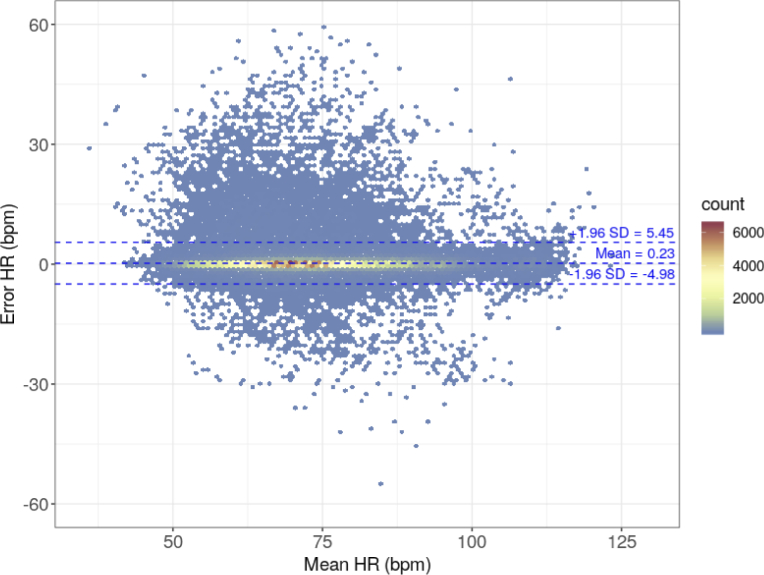
Figure 8 shows the modified Bland–Altman plot. As in the classic Bland–Altman plot, the X-axis is the average of the compared values, and the Y-axis is their difference. The individual points of the graph correspond to the number of results in hexagonal bins; the number of hexagonal bins in both the vertical and horizontal directions is set to 120. The number of results in each bin was coded according to the color of the points. The chart also shows the limits of agreement at -4.98 and 5.45 bpm, and the mean error of 0.23 bpm. The RMSE obtained by comparing the HR results of the online method and the auxiliary offline method is 2.67 bpm.
Fig. 8.
Bland-Altman plot of the HR values obtained by the online method against the reference HR values obtained by the previously verified auxiliary method.
When analyzing Figs. 7 and 8, particular attention should be paid to the color of the bins. Although most of the points (i.e., measurements) are near the x = y line, there are few points with significant error, which are unfortunately well highlighted. This is the result of the color coding of the number of measurements as even a bin of one measurement is displayed in the plot. The color of these outliers corresponds to the lowest number of cases. The bins corresponding to low errors most often have a color that reflects a greater number of measurements. The same effect is seen in Figs. 5 and 6; however, it is less pronounced owing to the overall lower number of measurements. The excess of points above the zero-error line, i.e., prevalence of positive errors, noticeable in Fig. 8, does not have any deeper physical sense, as there are very few points compared to those with low error within the limits of agreement range.
4. Discussion
The multiplication of FBG elements has long been known and used in distributed monitoring of the condition of mechanical structures of bridges, tunnels, or chimneys [37,38] as well as the electrical parameters of submersible pumps, wind turbines, or power networks [39]. Although FBG-based vital sign sensors have been developed in recent years, only a few research groups have proposed multi-FBG sensor solutions. As shown in the “Introduction” section, it is difficult to compare the HR detection method proposed in this paper with the methods presented by other authors [22–24]. This is mainly due to the fact that those authors had different goals and did not try to solve the problems associated with the automatic selection of the best signal for detecting the parameter needed. Our experience shows that the whole process of HR detection in FBG signals is complex, mainly due to small levels of the amplitude of the notches related to the mechanical activity of the heart in relation to the noise and resolution of the measurement system. The use of multiple sources of signals, or multi-point measurement by an array of FBGs, is one of the ways to mitigate this disadvantage. However, construction of multi-FBG sensors contributes to the need for development of an effective method for aggregating information from multiple sources, which allows the removal of noise and enhances the useful characteristics of the signals.
In the process of designing the sensor mat, the most important aspect was to match the FBG parameters to the specification of the FBG monitor and to minimize the risk of errors in the HR detection method. Thus, the Bragg wavelengths of all the FBGs had to be within the operating range of the FBG monitor of 45 nm, and could not exceed this range due to heart- or lung-induced strains experienced by the FBGs. On the other hand, the spectral distance between the neighboring FBGs as well as their distribution along the fiber were designed to avoid spectral overlapping when the subject was lying on the sensor mat, and the heart-induced signals were acquired from the subject. A series of preliminary tests allowed for determining the spectral parameters of the FBGs. The grating reflectivity and bandwidth were selected according to the technical specification of the FBG monitor and based on the authors’ experience to properly apply the Gaussian fit, and finally to obtain the best possible accuracy of wavelength measurement. It is worth to mention that the presented method does not need very accurate absolute Bragg wavelength measurement because the HR signal is derived from relative Bragg wavelength changes. Hence, the designed parameters are not crucial until the spectra do not overlap each other and the reflectivity spectra of FBGs are not saturated and thus are no longer Gaussian.
The amplitude of the signal strongly depends on the axis along which an FBG is attached. Better results are achieved when the BCG signal is recorded by an FBG located along the longitudinal axis of the body as the heart significantly expands and contracts along longitudinal and sagittal axes. The expansion and contraction of the heart along the transverse axis of the body is less significant. This confirms both the well-known results from the 50s [40] and more recent research on three-dimensional ballistocardiography in microgravity [41–43]. Therefore, 7 of 9 FBGs of the sensor mat are oriented longitudinally on the Plexiglas board. 2 FBGs are oriented transversely in order to reduce contribution of heart-induced artifacts to the respiratory signal that is planned to be studied in the future.
In the study, a particular effort was made to ensure the repeatability of measurement and reproducibility of the sensor fabrication process. Therefore, the phase mask technique was used to inscribe the Bragg gratings into the fiber core, which currently provides the best reproducibility. In this technique, the Bragg wavelength of a grating is determined by the phase mask period, and its repeatability does not exceed ± 0.2 nm. During the grating growth process in the phase mask technique, the bandwidth and reflectivity are precisely controlled by monitoring the reflection spectrum of the FBG. These parameters can be tailored with a deviation of no more than a few percent. The measurement repeatability of the Bragg wavelength using the I-MON 256 monitor is on average 3 pm and does not exceed the value of 5 pm.
The Plexiglas board was machine cut, while the sensor mat was assembled by hand and hence every sensor manufactured in this way may be slightly different. However, in the process of optimizing the FBG layout, we manufactured a number of sensors, including several pieces with almost identical layout and parameters of the FBGs, and we did not notice any differences in their operation significant for the quality of measurements. The satisfactory repeatability of measurement was confirmed by the number of subjects involved in the study.
The method and evaluation had limitations; The method is limited by the possibility of obtaining details of the neural network operation. The neural network will always remain a “black box” that can respond in an unpredictable manner to some input data. An example of such a reaction is the autopilot system in Tesla’s car, in which a deep neural network identifying the lane marking in the camera images directed the car into the wrong lane when three small stickers were placed in an intersection [44,45]. Thus, neural networks can be tricked through “adversarial” images. For the presented measurement system, intentionally generating artifacts in the network input signals seems unlikely, but it is possible to imagine a situation in which the waveform generating unstable network operation arises naturally. However, the fact that the network performance was verified in a large series of trials, in which the network received data unknown during the learning process, suggests that the risk of unexpected operations is negligible.
Another limitation is related to the verification of the proposed method using an auxiliary method. As mentioned in the “Methods” section, this was due to the lack of a reference signal measurement during routine MRI procedures, in which 496 recordings were obtained. However, a high agreement of the auxiliary method, i.e., RMSE of 2.25 bpm, was achieved compared with the measurements obtained with ECG being the standard for precise HR determination. Therefore, the approach correctly verified the proposed method in the presented MRI application.
The proposed method for detecting HR developed using machine learning allowed us to achieve satisfactory results for the automatic continuous extracting of HR using the all-dielectric array of FBG sensing elements with RMSE of 2.67 bpm and the limits of agreement of -4.98 and 5.45 bpm comparing with the auxiliary method. The proposed method does not require significant computing power; hence, a commercial off-the-shelf personal computer is sufficient for its application. Thus, alongside the inherent electromagnetic immunity of fiber-optic sensors, the method can be easily integrated into an unobtrusive continuous monitoring system of MRI patients.
Funding
Narodowe Centrum Badań i Rozwoju10.13039/501100005632 (PBS3/B9/41/2015).
Disclosures
The authors declare no conflicts of interest.
Data availability
The source code for data preparation, neural network learning, and HR detection method software is available at GitHub [46–49]. Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (873.8KB, pdf) for supporting content.
References
- 1.Tan F., Chen S., Lyu W., Liu Z., Yu C., Lu C., Tam H.-Y., “Non-invasive human vital signs monitoring based on twin-core optical fiber sensors,” Biomed. Opt. Express 10(11), 5940–5952 (2019). 10.1364/BOE.10.005940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R., Zhao J., Sun Y., Yu H., Zhou N., Zhang H., Jia D., “Wearable respiration monitoring using an in-line few-mode fiber Mach-Zehnder interferometric sensor,” Biomed. Opt. Express 11(1), 316–329 (2020). 10.1364/BOE.376782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett A., Beiderman Y., Agdarov S., Beiderman Y., Hendel R., Straussman B., Zalevsky Z., “Monitoring of vital bio-signs by analysis of speckle patterns in a fabric-integrated multimode optical fiber sensor,” Opt. Express 28(14), 20830–20844 (2020). 10.1364/OE.384423 [DOI] [PubMed] [Google Scholar]
- 4.Chen S., Tan F., Lyu W., Yu C., “Ballistocardiography monitoring system based on optical fiber interferometer aided with heartbeat segmentation algorithm,” Biomed. Opt. Express 11(10), 5458–5469 (2020). 10.1364/BOE.403086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao T., Fu X., Zhan J., Chen K., Li Z., “Vital signs monitoring using the macrobending small-core fiber sensor,” Opt. Lett. 46(17), 4228–4231 (2021). 10.1364/OL.428664 [DOI] [PubMed] [Google Scholar]
- 6.Chen Z., Lau D., Teo J. T., Ng S. H., Yang X., Kei P. L., “Simultaneous measurement of breathing rate and heart rate using a microbend multimode fiber optic sensor,” J. Biomed. Opt. 19(5), 057001 (2014). 10.1117/1.JBO.19.5.057001 [DOI] [PubMed] [Google Scholar]
- 7.Leitão C., Ribau V., Afreixo V., Antunes P., André P., Pinto J. L., Boutouyrie P., Laurent S., Bastos J. M., “Clinical evaluation of an optical fiber-based probe for the assessment of central arterial pulse waves,” Hypertens. Res. 41(11), 904–912 (2018). 10.1038/s41440-018-0089-2 [DOI] [PubMed] [Google Scholar]
- 8.Nedoma J., Martinek R., Fajkus M., Brablik J., Kahankova R., Fridrich M., Kostelansky M., Hanzlikova P., Vojtisek L., Behbehani K., “A novel FBG-based triggering system for cardiac MR imaging at 3 Tesla: a pilot pre-clinical study,” IEEE Access 8, 181205–181223 (2020). 10.1109/ACCESS.2020.3028224 [DOI] [Google Scholar]
- 9.Dziuda Ł., Zieliński P., Baran P., Krej M., Kopka L., “A study of the relationship between the level of anxiety declared by MRI patients in the STAI questionnaire and their respiratory rate acquired by a fibre-optic sensor system,” Sci. Rep. 9(1), 4341 (2019). 10.1038/s41598-019-40737-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlander B. M., Engvall J., Ericsson E., “Anxiety during magnetic resonance imaging of the spine in relation to scanner design and size,” Radiography 26(2), 110–116 (2020). 10.1016/j.radi.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 11.van Minde D., Klaming L., Weda H., “Pinpointing moments of high anxiety during an MRI examination,” Int. J. Behav. Med. 21(3), 487–495 (2014). 10.1007/s12529-013-9339-5 [DOI] [PubMed] [Google Scholar]
- 12.Sardinha A., Freire R. C., Zin W. A., Nardi A. E., “Respiratory manifestations of panic disorder: causes, consequences and therapeutic implications,” J. Bras. Pneumol. 35(7), 698–708 (2009). 10.1590/S1806-37132009000700012 [DOI] [PubMed] [Google Scholar]
- 13.Tugwell J. R., Goulden N., Mullins P., “Alleviating anxiety in patients prior to MRI: a pilot single-centre single-blinded randomised controlled trial to compare video demonstration or telephone conversation with a radiographer versus routine intervention,” Radiography 24(2), 122–129 (2018). 10.1016/j.radi.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 14.Shellock F. G., Magnetic resonance procedures: health effects and safety , 1st ed. (CRC, 2001). [Google Scholar]
- 15.Dziuda Ł., “Fiber-optic sensors for monitoring patient physiological parameters: a review of applicable technologies and relevance to use during magnetic resonance imaging procedures,” J. Biomed. Opt. 20(01), 1 (2015). 10.1117/1.JBO.20.1.010901 [DOI] [PubMed] [Google Scholar]
- 16.Ladrova M., Martinek R., Nedoma J., Hanzlikova P., Nelson M. D., Kahankova R., Brablik J., Kolarik J. “Monitoring and synchronization of cardiac and respiratory traces in magnetic resonance imaging: a review,” IEEE Rev. Biomed. Eng. (to be published). [DOI] [PubMed]
- 17.Dziuda Ł., Skibniewski F. W., Krej M., Baran P. M., “Fiber Bragg grating-based sensor for monitoring respiration and heart activity during magnetic resonance imaging examinations,” J. Biomed. Opt. 18(5), 057006 (2013). 10.1117/1.JBO.18.5.057006 [DOI] [PubMed] [Google Scholar]
- 18.Nedoma J., Fajkus M., Martinek R., Nazeran H., “Vital sign monitoring and cardiac triggering at 1.5 Tesla: a practical solution by an MR-ballistocardiography fiber-optic sensor,” Sensors 19(3), 470 (2019). 10.3390/s19030470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abad S., López-Amo M., Araújo F. M., Ferreira L. A., Santos J. L., “Fiber Bragg grating–based self-referencing technique for wavelength-multiplexed intensity sensors,” Opt. Lett. 27(4), 222–224 (2002). 10.1364/OL.27.000222 [DOI] [PubMed] [Google Scholar]
- 20.Kashyap R., Fiber Bragg Gratings , 2nd ed. (Academic, 2009). [Google Scholar]
- 21.Sahota J. K., Gupta N., Dhawan D., “Fiber Bragg grating sensors for monitoring of physical parameters: a comprehensive review,” Opt. Eng. 59(06), 1 (2020). 10.1117/1.OE.59.6.060901 [DOI] [Google Scholar]
- 22.Hao J., Jayachandran M., Kng P. L., Foo S. F., Aung P. W., Cai Z., “FBG-based smart bed system for healthcare applications,” Front. Optoelectron. China 3(1), 78–83 (2010). 10.1007/s12200-009-0066-0 [DOI] [Google Scholar]
- 23.Allsop T., Bhamber R., Lloyd G., Miller M. R., Dixon A., Webb D., Ania Castanon J. D., Bennion I., “Respiratory function monitoring using a real-time three-dimensional fiber-optic shaping sensing scheme based upon fiber Bragg gratings,” J. Biomed. Opt. 17(11), 117001 (2012). 10.1117/1.JBO.17.11.117001 [DOI] [PubMed] [Google Scholar]
- 24.Fajkus M., Nedoma J., Martinek R., Vasinek V., Nazeran H., Siska P., “A non-invasive multichannel hybrid fiber-optic sensor system for vital sign monitoring,” Sensors 17(12), 111 (2017). 10.3390/s17010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokhtar M. R., Sun T., Grattan K. T. V., “Bragg grating packages with nonuniform dimensions for strain and temperature sensing,” IEEE Sens. J. 12(1), 139–144 (2012). 10.1109/JSEN.2011.2134845 [DOI] [Google Scholar]
- 26.Dziuda Ł., Skibniewski F. W., “A new approach to ballistocardiographic measurements using fibre Bragg grating-based sensors,” Biocybern. Biomed. Eng. 34(2), 101–116 (2014). 10.1016/j.bbe.2014.02.001 [DOI] [Google Scholar]
- 27.Markowski K., Perka A., Jędrzejewski K., Osuch T., “Custom FBGs inscription using modified phase mask method with precise micro- and nano-positioning,” Proc. SPIE 10031, 100311H (2016). 10.1117/12.2249381 [DOI] [Google Scholar]
- 28.Osuch T., Kossek T., Jedrzejewski K., Lewandowski L., “Thermal and aging tests of fiber Bragg gratings as wavelength standards,” Proc. SPIE 5951, 59510H (2005). 10.1117/12.622811 [DOI] [Google Scholar]
- 29.Wang H., Wu N., Zhao Z., Han G., Zhang J., Wang J., “Continuous monitoring method of cerebral subdural hematoma based on MRI guided DOT,” Biomed. Opt. Express 11(6), 2964–2975 (2020). 10.1364/BOE.388059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Moatti A., Zhang X., Ghashghaei H. T., Greenabum A., “Deep learning-based autofocus method enhances image quality in light-sheet fluorescence microscopy,” Biomed. Opt. Express 12(8), 5214–5226 (2021). 10.1364/BOE.427099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Silva T., Chew E. Y., Hotaling N., Cukras C. A., “Deep-learning based multi-modal retinal image registration for the longitudinal analysis of patients with age-related macular degeneration,” Biomed. Opt. Express 12(1), 619–636 (2021). 10.1364/BOE.408573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krej M., Baran P., Dziuda Ł., “Detection of respiratory rate using a classifier of waves in the signal from a FBG-based vital signs sensor,” Comput. Methods Programs Biomed. 177, 31–38 (2019). 10.1016/j.cmpb.2019.05.014 [DOI] [PubMed] [Google Scholar]
- 33.Bai S., Kolter J. Z., Koltun V., “An empirical evaluation of generic convolutional and recurrent networks for sequence modeling,” arXiv: 1803.01271v2 (2018).
- 34.Baydin A. G., Pearlmutter B. A., Radul A. A., Siskind J. M., “Automatic differentiation in machine learning: a survey,” arXiv: 1502.05767v4 (2015).
- 35.Kingma D. P., Ba J., “Adam: A Method for Stochastic Optimization,” arXiv:1412.6980v9 (2014).
- 36.Dziuda Ł., Krej M., Śmietanowski M., Sobotnicki A., Sobiech M., Kwaśny P., Brzozowska A., Baran P., Kowalczuk K., Skibniewski F. W., “Development and evaluation of a novel system for inducing orthostatic challenge by tilt tests and lower body negative pressure,” Sci. Rep. 8(1), 7793 (2018). 10.1038/s41598-018-26173-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Higuera J. M., Cobo L. R., Incera A. Q., Cobo A., “Fiber optic sensors in structural health monitoring,” J. Lightwave Technol. 29(4), 587–608 (2011). 10.1109/JLT.2011.2106479 [DOI] [Google Scholar]
- 38.Wu T., Liu G., Fu S., Xing F., “Recent progress of fiber-optic sensors for the structural health monitoring of civil infrastructure,” Sensors 20(16), 4517 (2020). 10.3390/s20164517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fusiek G., Niewczas P., “Photonic voltage transducer with lightning impulse protection for distributed monitoring of MV networks,” Sensors 20(17), 4830 (2020). 10.3390/s20174830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.March H. W., “Three-plane ballistocardiography: the effect of age on the longitudinal, lateral, and dorsoventral ballistocardiograms,” Circulation 12(5), 869–882 (1955). 10.1161/01.CIR.12.5.869 [DOI] [PubMed] [Google Scholar]
- 41.Prisk G. K., Verhaeghe S., Padeken D., Hamacher H., Paiva M., “Three-dimensional ballistocardiography and respiratory motion in sustained microgravity,” Aviat. Space Environ. Med. 72(12), 1067–1074 (2001). [PubMed] [Google Scholar]
- 42.Migeotte P.-F., Tank J., Pattyn N., Funtova I., Baevsky R., Neyt X., Prisk G. K., “Three dimensional ballistocardiography: methodology and results from microgravity and dry immersion,” Proc. 33rd Annual International Conference of the IEEE Eng. Med. Biol. Soc. (2011), pp. 4271–4274. [DOI] [PubMed] [Google Scholar]
- 43.Migeotte P.-F., Deliere Q., Tank J., Funtova I., Baevsky R., Neyt X., Pattyn N., “3D ballistocardiography in microgravity: comparison with ground based recordings,” Proc. 35th Annual International Conference of the IEEE Eng. Med. Biol. Soc. (2013), pp. 7012–7016. [DOI] [PubMed] [Google Scholar]
- 44.“Experimental security research of Tesla autopilot,” Research Rep. (Tencent Keen Security Lab, 2019). [Google Scholar]
- 45.Ackerman E., “Three small stickers in intersection can cause Tesla autopilot to swerve into wrong lane,” IEEE Spectrum (2019).
- 46.Krej M., “TCN code forked from github.com/locuslab/TCN adapted to interoperate with ONNX,” GitHub (2021) [accessed 20 Oct 2021], https://github.com/mkkmod/TCN/
- 47.Krej M., “Python code for learning TCN as the signal aggregator for the method of continuous HR detection in signals from the multi-FBG sensor,” GitHub (2021) [accessed 20 Oct 2021], https://github.com/mkkmod/OptoSigNN/
- 48.Krej M., “R language scripts used to create the learning set to learn the signal aggregator,” GitHub (2021) [accessed 20 Oct 2021], https://github.com/mkkmod/OptoHrSrcSigQuality/
- 49.Krej M., “C# language code of the method of continuous HR detection in signals from the multi-FBG sensor,” GitHub (2021) [accessed 20 Oct 2021], https://github.com/mkkmod/SasDspExts/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code for data preparation, neural network learning, and HR detection method software is available at GitHub [46–49]. Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.