Significance
This work demonstrates microbial biosynthesis of bisbenzylisoquinoline (bisBIA) alkaloids. We show that several didomain epimerases can function in yeast to epimerize the nonnative substrate N-methylcoclaurine, an essential step in bisBIA biosynthesis. The N-methylcoclaurine epimerase activity was increased 10-fold by combining individual reductase and oxidase domains from different plant species. Strain engineering and optimization of media and growth conditions increased the bisBIA titer over 10,000-fold. We show that strains can be engineered to primarily produce one bisBIA product by selection of the cytochrome P450 variant that couples the monomer BIA subunits. We then leverage our bisBIA biosynthetic strain as a platform for the screening of other plant enzymes to produce two additional plant natural products de novo in a heterologous host.
Keywords: bisbenzylisoquinoline alkaloids, plant natural products, metabolic engineering, protein engineering, benzylisoquinoline alkaloids
Abstract
Benzylisoquinoline alkaloids (BIAs) are a diverse class of medicinal plant natural products. Nearly 500 dimeric bisbenzylisoquinoline alkaloids (bisBIAs), produced by the coupling of two BIA monomers, have been characterized and display a range of pharmacological properties, including anti-inflammatory, antitumor, and antiarrhythmic activities. In recent years, microbial platforms have been engineered to produce several classes of BIAs, which are rare or difficult to obtain from natural plant hosts, including protoberberines, morphinans, and phthalideisoquinolines. However, the heterologous biosyntheses of bisBIAs have thus far been largely unexplored. Here, we describe the engineering of yeast strains that produce the Type I bisBIAs guattegaumerine and berbamunine de novo. Through strain engineering, protein engineering, and optimization of growth conditions, a 10,000-fold improvement in the production of guattegaumerine, the major bisBIA pathway product, was observed. By replacing the cytochrome P450 used in the final coupling reaction with a chimeric variant, the product profile was inverted to instead produce solely berbamunine. Our highest titer engineered yeast strains produced 108 and 25 mg/L of guattegaumerine and berbamunine, respectively. Finally, the inclusion of two additional putative BIA biosynthesis enzymes, SiCNMT2 and NnOMT5, into our bisBIA biosynthetic strains enabled the production of two derivatives of bisBIA pathway intermediates de novo: magnocurarine and armepavine. The de novo heterologous biosyntheses of bisBIAs presented here provide the foundation for the production of additional medicinal bisBIAs in yeast.
Benzylisoquinoline alkaloids (BIAs) are a class of plant natural products (PNPs) that includes numerous medicinally important compounds, including the analgesic morphine, the histological stain berberine, and the antitussive noscapine. Included within the BIA family are the bisbenzylisoquinoline alkaloids (bisBIAs), which are produced through the coupling of two 1-benzylisoquinoline monomers, of which nearly 500 examples are known (1). Several natural and semisynthetic bisBIAs have been historically important, including tubocurarine, a natural BIA from Chondrodendron tomentosum used as an arrow poison (2), and cisatracurium, a semisynthetic derivative of the BIA tetrahydropapaverine from Papaver somniferum (opium poppy) used as a muscle relaxant during surgery and mechanical ventilation (3).
Due to the chemical complexity of BIAs, commercially viable chemical syntheses have yet to be developed, and this class of compounds is sourced primarily from plant biomass. However, this supply is hampered by low yield and purity, requiring laborious extraction and purification procedures, and faces variability caused by weather and climate change. Furthermore, harvesting of wild medicinal plants can damage their populations and the ecosystems of which they are a part, as has occurred with the bisBIA-producing plant Stephania tetrandra (4 –6). To overcome these limitations, engineered Saccharomyces cerevisiae strains have recently been reported that heterologously biosynthesize several BIAs de novo, including hydrocodone (7), berberine (8), and noscapine (9). These strains have served as valuable platforms for the production of additional pathway derivatives that would be challenging to produce synthetically (10, 11). However, to date, no microbial strains have been reported that produce any of the bisBIAs de novo.
One group of bisBIAs has drawn interest for the reported medicinal properties of both the parent plant species and the isolated compounds. This group of compounds is formed by the 3'–4′ coupling of two N-methylcoclaurine (NMC) monomers in either the (R) or (S) configuration, forming four structurally identical, diastereomeric linear bisBIAs. One of these diastereomers has only been prepared synthetically (12), while the other three have been identified in and isolated from plant sources: magnoline from Magnolia fuscata (13); berbamunine from a variety of Berberis species (14); and guattegaumerine from Mosannona depressa (15). Several Berberis species have been reported to have medicinal effects (16), most notably antidiabetic properties (17), and berbamunine has been shown to have dopaminergic properties (18). G. gaumeri has been used to treat hypercholesterolemia (19), and guattegaumerine has been shown to have antitumor (20) and neuroprotectant (21) properties. While synthetic strategies toward these compounds have been developed (22 –25), these procedures involve numerous steps, chiral resolutions, and often rely on BIAs isolated from plant biomass as starting materials.
To develop a more efficient platform for the synthesis of bisBIAs and their derivatives, we engineered S. cerevisiae strains that produce guattegaumerine and berbamunine de novo. To accomplish this, we first generated a strain that produces high titers of (S)-NMC de novo, which we used as a platform to determine the activities of downstream enzymes in vivo. Through strain engineering, optimization of fermentation conditions, and protein engineering, we were able to increase the total bisBIA titer over 10,000-fold to over 100 mg/L. By altering the cytochrome P450 that performs the final coupling reaction in the biosynthesis of our bisBIAs, we were able to effect a nearly complete switch in the product ratio from >98% guattegaumerine to >99% berbamunine. Finally, we leveraged our de novo bisBIA strains to produce two additional pathway derivatives, magnocurarine and armepavine, de novo.
Results
Construction of a Yeast Strain that Produces (S)-N-methylcoclaurine De Novo.
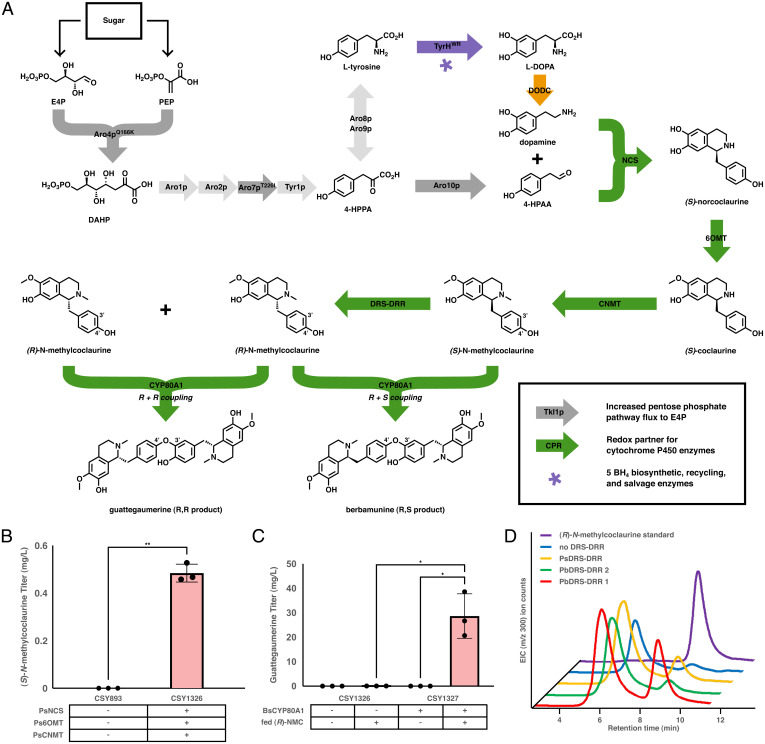
The biosynthesis of the bisBIAs guattegaumerine and berbamunine proceeds via a P450-mediated dimerization of two NMC monomers (Fig. 1A ). Guattergaumerine is the product of two (R)-NMC monomers, while berbamunine is the product of one (S)-NMC monomer joining with one (R)-NMC monomer. While the enzyme(s) responsible for the epimerization of (S)-NMC to (R)-NMC in planta have yet to be elucidated, the biosynthetic steps for the production of (S)-NMC are known. We therefore began our efforts toward the heterologous biosynthesis of bisBIAs by generating an S. cerevisiae strain that produces (S)-NMC de novo, intending to use this strain as a platform to explore biosynthesis routes for (R)-NMC.
Fig. 1.
Biosynthetic pathway for de novo bisBIA biosynthesis. (A) Biosynthetic pathway from fed sugar to the bisBIA products berbamunine and guattegaumerine. The color of the arrows represents the species of origin of the enzyme employed: light gray, unmodified native yeast enzyme; dark gray, modified native yeast enzyme; green, enzyme of plant origin; orange, enzyme of bacterial origin; purple, enzyme of mammalian origin. (B) De novo production of (S)-N-methylcoclaurine in engineered yeast strains. Strains were cultured in synthetic complete media with 2% dextrose at 30 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) Production of the bisBIA guattegaumerine in a yeast strain expressing BsCYP80A1 and fed exogenous (R)-N-methylcoclaurine. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at 30 °C for 96 h before being pelleted, resuspended in an equal volume of 200 µM (R)-NMC in 50 mM HEPES at pH 7.6, and incubated an additional 72 h at 30 °C before LC-MS/MS analysis of the media using the MRM transitions reported in SI Appendix, Table S5. (D) De novo production of (R)-N-methylcoclaurine in strain CSY1326 expressing PsCPR and different DRS-DRR enzymes off of low-copy plasmids. Strains were cultured in selective media (YNB-DO) with 2% dextrose at 30 °C for 96 h before extraction, purification, and analysis by normal-phase chiral chromatography. Data for B and C are presented as mean values ± the SD of three biologically independent samples. Asterisks represent Student’s two-tailed t test: *P < 0.05, **P < 0.01. Exact P values are given in SI Appendix, Table S6. Source data underlying B and C are provided in Dataset S1.
We generated an (S)-NMC–producing strain by starting from an S. cerevisiae strain that had previously been engineered to produce (S)-reticuline de novo (CSY1171) (10). Specifically, we generated the (S)-NMC–producing strain CSY1326 via the CRISPR-Cas9–mediated excision of a biosynthetic cassette encoding the genes for P. somniferum cytochrome P450 reductase (PsCPR), Eschscholzia californica N-methylcoclaurine-3′-hydroxylase (EcCYP80B1), and P. somniferum 3′-hydroxy-N-methylcoclaurine-4′-O-methyltransferase (Ps4’OMT). The enzymes encoded by these three genes are responsible for the conversion of (S)-NMC to (S)-reticuline. We expected that without these genes present, the strain would accumulate higher levels of (S)-NMC. Our engineered (S)-NMC–producing strain CSY1326, the parent (S)-reticuline–producing strain CSY1171, and CSY893—a control strain lacking the heterologous genes necessary for (S)-NMC biosynthesis (Coptis japonica norcoclaurine synthase [CjNCS], P. somniferum norcoclaurine 6-O-methyltransferase [Ps6OMT], and P. somniferum coclaurine N-methyltransferase [PsCNMT])—were cultured in synthetic complete media for 96 h at 30 °C and the supernatant was subsequently analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) for production of (S)-NMC. We observed that (S)-NMC accumulated to titers of 0.482 mg/L from strain CSY1326 and 0.208 mg/L from the parent strain CSY1171; no accumulation of (S)-NMC was observed from the control strain CSY893 (Fig. 1B ). In contrast, under these conditions we observed that reticuline accumulated to titers of 0.233 mg/L in the parent strain CSY1171 with no quantifiable reticuline detected from our engineered strain CSY1326, indicating that the pathway flux had been nearly entirely redirected to (S)-N-methylcoclaurine rather than reticuline ( SI Appendix, Fig. S1). The base (S)-NMC–producing strain CSY1326 thus demonstrated a 2.3-fold increase in (S)-NMC titer relative to the parent strain and afforded sufficient production of this key metabolite for further downstream pathway engineering for bisBIA production.
Expression of BsCYP80A1 in Yeast Results in Guattegaumerine Production from Fed (R)-NMC.
While the enzyme(s) responsible for the epimerization of (S)-NMC to (R)-NMC in planta are still unknown, the enzyme responsible for the subsequent and final step in the biosynthesis of the bisBIAs guattegaumerine and berbamunine in planta has been identified in Berberis stolonifera to be the cytochrome P450 berbamunine synthase (BsCYP80A1) (26). However, previous reports focused primarily on its characterization in vitro after expression from Berberis tissue, with successful heterologous activity reported only in vitro after expression from insect cells (27). BsCYP80A1 has previously been shown to be expressed successfully in S. cerevisiae but no activity was observed (28). In order to determine the feasibility of our proposed biosynthetic route for the bisBIAs guattegaumerine and berbamunine, we examined whether we could express active BsCYP80A1 in vivo in S. cerevisiae before attempting to identify an enzyme capable of catalyzing the epimerization of (S)-NMC to (R)-NMC in vivo.
To test the activity of BsCYP80A1 in vivo, we chromosomally integrated BsCYP80A1 into the (S)-NMC–producing strain CSY1326 to generate strain CSY1327 and expressed PsCPR from a low-copy plasmid in this strain. The P. somniferum cytochrome P450 reductase (CPR) was selected as the redox partner for BsCYP80A1 as the two enzymes had previously been shown to be functional together in vitro (29). This strain was cultured in synthetic complete media supplemented with 200 µM (R)-NMC for 120 h at 30 °C and the supernatant subsequently analyzed by LC-MS/MS for production of the bisBIAs guattegaumerine and berbamunine. However, we observed no production of guattegaumerine or berbamunine under these conditions. A similar apparent lack of activity in S. cerevisiae was reported for another plant P450 involved in the biosynthesis of a different BIA, the corytuberine synthase from C. japonica (CjCYP80G2), which converts (S)-reticuline to corytuberine in the biosynthesis of magnoflorine (30). While the authors originally saw no activity in vivo from CjCYP80G2 when (R,S)-reticuline was supplemented into the growth media, they were able to produce corytuberine by growing the cells to saturation and then transferring them to a buffered solution containing (R,S)-reticuline. The authors concluded that the initial apparent lack of activity of CjCYP80G2 was due to poor uptake of (R,S)-reticuline by the yeast cells at the low pH of the growth media, an issue that was alleviated by using a higher pH buffer solution containing (R,S)-reticuline for the resuspension. Based on the structural similarity between (R)-NMC and reticuline, the uptake of (R)-NMC into yeast cells may be similarly hindered at low pH.
We examined the activity of BsCYP80A1 when expressed in yeast cells under buffered conditions. Strain CSY1327 harboring PsCPR on a low-copy plasmid was cultured in synthetic complete media for 96 h at 30 °C. The cells were pelleted, resuspended in a solution of 200 µM (R)-NMC in 50 mM HEPES buffered to pH 7.6 as was previously performed with (R,S)-reticuline (30), and allowed to incubate for 72 h at 30 °C. The supernatant of the buffered solution was analyzed by LC-MS/MS for production of the bisBIAs guattegaumerine and berbamunine. Under these assay conditions, we observed production of the bisBIA guattegaumerine, but no berbamunine, by LC-MS/MS (Fig. 1C ), as verified by comparison with authentic standards of guattegaumerine and berbamunine ( SI Appendix, Fig. S2). We therefore concluded that BsCYP80A1 is functional in our engineered (S)-NMC–producing yeast strain. Given that protonated tertiary amines are known to have pKa values typically around 10 to 11, the resuspension of the tertiary amines (R,S)-reticuline and (R)-NMC in pH 7.6 HEPES buffer likely did not significantly alter the protonation state of these substrates. Curiously, no such resuspension in buffer was seen to be necessary for the internalization of the similar substrates norlaudanosoline and laudanosoline in a previous report (31). The mechanism for the increased internalization that we observed for (R)-NMC and that was previously reported for (R,S)-reticuline is therefore likely to be due to more substrate specific impacts on metabolite transporters from the altered conditions presented during the HEPES buffer resuspension.
Construction of a Yeast Strain That Produces (R)-NMC De Novo.
We next sought to identify an enzyme or enzymes capable of the epimerization of (S)-NMC to (R)-NMC to complete the de novo biosynthesis of the bisBIAs guattegaumerine and berbamunine. The epimerization of the structurally similar (S)-reticuline to (R)-reticuline is performed by the didomain enzyme dehydroreticuline synthase–dehydroreticuline reductase (DRS-DRR) (7), also referred to as reticuline epimerase (REPI) (32) or STORR [(S)- to (R)-reticuline] (33). DRS-DRR variants from P. somniferum and related plant species have been functionally expressed in S. cerevisiae strains harboring full biosynthetic pathways to catalyze the conversion of (S)-reticuline to (R)-reticuline and enable de novo biosynthesis of morphinan alkaloids. Furthermore, recent reports have demonstrated that some DRS-DRR variants are capable of converting (S)-NMC to (R)-NMC in vitro (34, 35). Given the reported substrate promiscuity of DRS-DRR variants in vitro on (S)-NMC, we sought to identify a variant with sufficient in vivo activity on (S)-NMC to enable downstream bisBIA biosynthesis.
In order to identify an enzyme that would enable (R)-NMC biosynthesis, we screened DRS-DRR variants in our (S)-NMC–producing yeast strain. We expressed three DRS-DRR variants, two from Papaver bracteatum (Pbr.89405 and Pbr.12180, hereafter referred to as PbDRS-DRR 1 and 2, respectively) and one from P. somniferum (Pso.2062398, hereafter referred to as PsDRS-DRR), from low-copy plasmids in strain CSY1326 (Fig. 1D ). These strains were grown, the alkaloids extracted, purified by preparative LC, the fractions containing NMC concentrated and resuspended, and the concentrate subsequently analyzed by chiral normal-phase LC-MS for production of (S)-NMC and (R)-NMC. We observed conversion of (S)-NMC to (R)-NMC by all three DRS-DRRs, with the DRS-DRR from PbDRS-DRR 1 yielding the highest ratio of (R)-NMC:(S)-NMC at 63:37. The higher apparent activity of PbDRS-DRR 1 compared with PbDRS-DRR 2 and PsDRS-DRR could be explained by PbDRS-DRR 1 having a higher catalytic efficiency than the other two enzymes screened, higher expression in S. cerevisiae, or a mix of both factors. These results indicate that PbDRS-DRR 1 can serve as the missing epimerase necessary to complete the de novo heterologous biosynthesis of the bisBIAs guattegaumerine and berbamunine.
Alternate CPR Variants and Growth Conditions Enable De Novo bisBIA Biosynthesis in Yeast.
Having demonstrated that PbDRS-DRR 1 can convert (S)-NMC to (R)-NMC in vivo, we examined whether this DRS-DRR variant in combination with BsCYP80A1 would achieve de novo bisBIA production in yeast. We chromosomally integrated PbDRS-DRR 1 and PsCPR into the (S)-NMC–producing yeast strain harboring BsCYP80A1 (CSY1327) to generate strain CSY1328. However, neither bisBIA product was observed from this strain. We suspected that the lack of bisBIA accumulation was a result of lower expression achieved for PbDRS-DRR 1 when chromosomally integrated rather than encoded on a low-copy plasmid. Thus, we created a bisBIA-producing strain (CSY1329) in which PsCPR was chromosomally integrated into strain CSY1327 and which harbored a low-copy plasmid encoding PbDRS-DRR 1. Analysis of the supernatant from this strain indicated production of the bisBIA guattegaumerine at titers of 6 µg/L (Fig. 2A ).
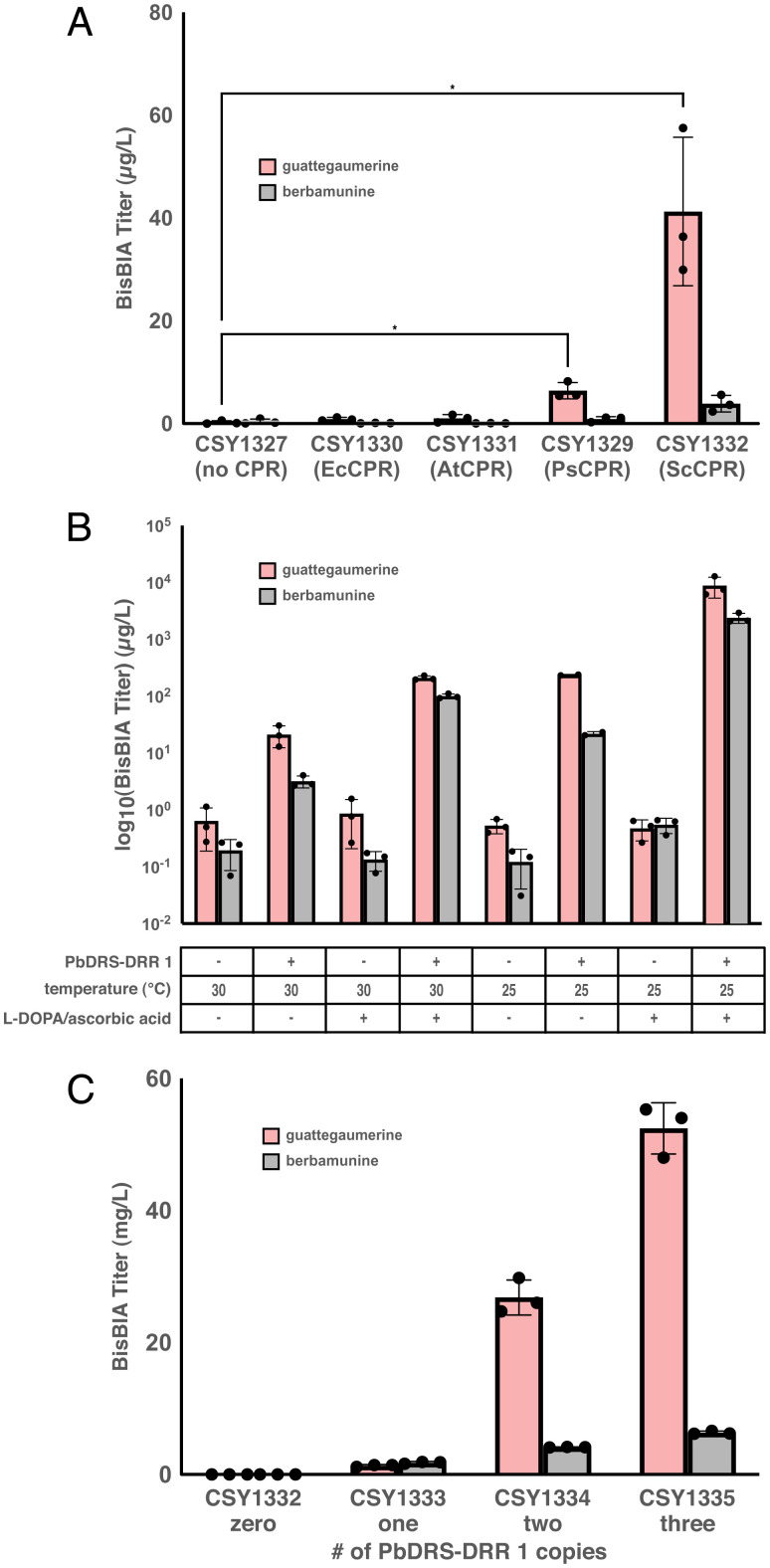
Fig. 2.
Strain and media optimization to increase bisBIA titers. (A) De novo production of bisBIAs from yeast strains expressing PbDRS-DRR 1 from a low-copy plasmid and different integrated CPRs. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at 30 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (B) De novo production of bisBIAs from engineered yeast strains at different temperatures and media supplementation (2 mM L-DOPA, 10 mM ascorbic acid) and with or without PbDRS-DRR 1 expressed from a low-copy plasmid. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at the indicated temperature and media supplementation conditions for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) De novo production of bisBIAs from engineered strains expressing varying copy numbers of PbDRS-DRR 1. Strains were cultured in synthetic complete media with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. Data are presented as mean values ± the SD of three biologically independent samples. Asterisks represent Student’s two-tailed t test: *P < 0.05. Exact P values are given in SI Appendix, Table S6. Source data underlying A–C are provided in Dataset S1.
We next sought to increase the titer of guattegaumerine produced, as well as to achieve the de novo biosynthesis of the alternate bisBIA, berbamunine. When BsCYP80A1 is expressed homologously from Berberis cell culture, ∼90% of the total bisBIA titer is observed to be berbamunine (27). Thus, we were surprised to observe only guattegaumerine produced in our base de novo bisBIA-producing yeast strain and examined whether strain engineering and alteration of culture conditions could result in production of berbamunine and how these factors might alter the ratio of the two bisBIA products. We first explored the impact of alternative CPRs on bisBIA titer and ratio. We chromosomally integrated genes for three alternate CPRs from E. californica (EcCPR; CSY1330), Arabidopsis thaliana (AtCPR; CSY1331), and S. cerevisiae (ScCPR; CSY1332) into the (S)-NMC–producing yeast strain harboring BsCYP80A1 (CSY1327). No bisBIA accumulation was observed from the strains expressing either EcCPR or AtCPR; however, the strain expressing ScCPR (CSY1332) produced over sixfold higher guattegaumerine than strain CSY1329 (41 µg/L versus 6 µg/L) and produced low levels of berbamunine (4 µg/L).
We next explored the impact of media and growth conditions on the titer and ratio of the bisBIA products. The first dedicated intermediate in the biosynthesis of BIAs, including the bisBIAs, is norcoclaurine, which is formed by the enzymatic Pictet–Spengler reaction between L-DOPA and 4-hydroxyphenylacetic acid. We therefore examined whether supplementation of the synthetic complete media with L-DOPA would result in increased bisBIA titers. For these experiments, strains were cultured in synthetic complete media with the indicated supplementation for 96 h at either 25 or 30 °C and the supernatant subsequently analyzed by LC-MS/MS for production of guattegaumerine or berbamunine. The addition of L-DOPA and ascorbic acid to the synthetic complete media increased the titer of guattegaumerine roughly 10-fold to over 200 µg/L and berbamunine over 30-fold to over 100 µg/L (Fig. 2B ). We thus observed the ratio of guattegaumerine to berbamunine was altered from roughly 7:1 without the addition of L-DOPA and ascorbic acid to 2:1 upon their addition. Reducing the temperature during growth from 30 to 25 °C resulted in an over 10-fold increase in the guattegaumerine titer to 235 µg/L and a sevenfold increase in the berbamunine titer to 22 µg/L. Finally, we examined the impact of combining L-DOPA and ascorbic acid supplementation and reduction of the growth temperature to 25 °C on bisBIA titers. The results indicate an over 400-fold increase of guattegaumerine titer to nearly 9 mg/L and a nearly 750-fold increase of berbamunine titer to ∼2.4 mg/L, indicating a synergistic effect on bisBIA production from these two alterations in growth conditions. We also examined the effect of these modifications in media conditions and growth temperature on the production of the upstream metabolite (S)-N-methylcoclaurine. While the control strain, CSY893, produced no detectable (S)-NMC and the parent strain, CSY1171, produced 0.37 mg/L (S)-NMC, a value comparable to the 0.208 mg/L observed in this strain without the decreased growth temperature and L-DOPA/ascorbic acid supplementation to the media, our initial (S)-NMC biosynthesizing strain, CSY1326, furnished 63 mg/L (S)-NMC ( SI Appendix, Fig. S3), a 131-fold improvement over the concentration originally measured (Fig. 1B ). This indicates that these modifications improved the supply of the precursor (S)-NMC to the extent necessary to see the dramatic increases in bisBIA titers that we observed.
Optimization of DRS-DRR Copy Number Increases bisBIA Production in Yeast.
We examined the impact of varying DRS-DRR copy number on the accumulation and ratio of bisBIA products in our engineered yeast strains. We chromosomally integrated PbDRS-DRR 1 into the (S)-NMC–producing yeast strain harboring BsCYP80A1 and ScCPR (CSY1332) to generate strain CSY1333. Strain CSY1333 achieved a guattegaumerine titer of 1.35 mg/L (an 85% reduction compared to CSY1332 expressing PbDRS-DRR 1 from a low-copy plasmid) and a berbamunine titer of 1.8 mg/L (a 25% reduction compared to CSY1332 expressing PbDRS-DRR 1 from a low-copy plasmid) (Fig. 2C ). Thus, strain CSY1333 was the first strain resulting in a higher titer of berbamunine than guattegaumerine. The data indicate that the expression of the DRS-DRR enzyme may be a limiting step in the production of bisBIA products in this pathway, given the reduction in overall bisBIA titer observed upon integrating PbDRS-DRR 1 into the genome versus expressing it from a low-copy plasmid.
We examined bisBIA production levels from strains with multiple copies of PbDRS-DRR 1 and BsCYP80A1 under similar assay conditions. We generated a strain with two chromosomally integrated copies of PbDRS-DRR 1 (CSY1334) and observed a 10-fold improvement in bisBIA levels relative to a strain with a single copy of PbDRS-DRR 1 (CSY1332), with guattegaumerine titer increased 20-fold to 27 mg/L and berbamunine titer increased 2.3-fold to 4 mg/L (Fig. 2C ). A strain with three chromosomally integrated copies of PbDRS-DRR 1 (CSY1335) resulted in an additional twofold increase in total bisBIA titer relative to the strain with two copies of PbDRS-DRR 1 (CSY1334) and a guattegaumerine titer of 52 mg/L and a berbamunine titer of over 6 mg/L. We measured the titers of residual (S)-NMC and (R)-NMC in these strains to determine the effect of increased DRS-DRR copy number on the (S)-NMC:(R)-NMC ratio and observed that the (S)-NMC:(R)-NMC ratio decreased from 48:1 in strain CSY1333 (containing one integrated copy of PbDRS-DRR 1) to 21:1 in strain CSY1334 (containing two integrated copies of PbDRS-DRR 1) to 1.7:1 in strain CSY1335 (containing three integrated copies of PbDRS-DRR 1) ( SI Appendix, Fig. S4). We also generated a strain with two chromosomally integrated copies of BsCYP80A1 (CSY1336) and observed a smaller, approximately twofold increase in total bisBIA titer relative to a strain with a single copy of BsCYP80A1 (CSY1332), from 6.04 mg/L (4.63 mg/L guattegaumerine and 1.41 mg/L berbamunine) to 11.7 mg/L (9.46 mg/L guattegaumerine and 2.24 mg/L berbamunine) ( SI Appendix, Fig. S5). We thus achieved a nearly 10,000-fold increase in the total bisBIA titer over our original de novo strain, increasing the titer from 6 µg/L to nearly 60 mg/L.
Expression of Engineered Split DRS and DRR Enzymes Further Increases bisBIA Production Levels.
Finally, we examined the impact of varying the individual DRS and DRR domains on the accumulation and ratio of bisBIA products from the engineered yeast strain. DRS-DRR enzymes are didomain enzymes, with the DRS domain performing the two-electron oxidation of (S)-NMC to produce the achiral intermediate 1,2-dehydro-N-methylcoclaurine, which is then stereospecifically reduced by the DRR domain to furnish (R)-NMC (Fig. 3A ). DRS-DRR variants have been identified from multiple plant species, each possessing their own DRS and DRR domains. As (S)-NMC is not known to be the native substrate of any of the DRS-DRR variants identified to date, it is not likely that any of these DRS-DRR variants are optimized for activity on (S)-NMC. Furthermore, the DRS and DRR domains of a given enzyme may each be suboptimal for activity on (S)-NMC and 1,2-dehydro-N-methylcoclaurine, respectively, to varying degrees. We thus examined whether higher activity might be achieved by combining DRS and DRR domains that originated from different DRS-DRR variants.
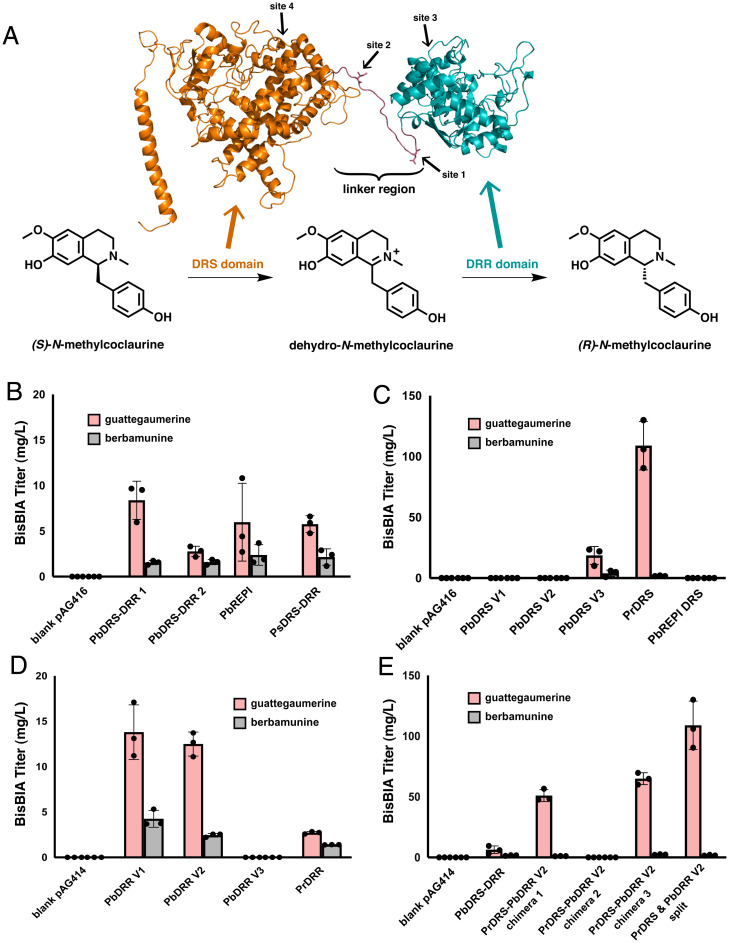
Fig. 3.
Engineering an optimized NMC epimerase for bisBIA production. (A) Homology model of PbDRS-DRR 1 and reaction schematic of the epimerization of (S)-N-methylcoclaurine to (R)-N-methylcoclaurine. The homology model was constructed with RaptorX (51) using CYP76AH1 from S. miltiorrhiza (44) as the basis of the DRS domain (orange) and 4,2’,4’,6’-tetrahydroxychalcone reductase from Medicago sativa (52) as the basis of the DRR domain (teal). By analogy to the reactions performed on the native substrate of PbDRS-DRR 1, (S)-reticuline, the DRS domain (orange) oxidizes (S)-N-methylcoclaurine to the intermediate dehydro-N-methylcoclaurine, which is then reduced by the DRR domain (teal) to the product (R)-N-methylcoclaurine. The four sites explored in C–E for cleaving the DRS and DRR domains of PbDRS-DRR 1 apart are indicated on the homology model with arrows. (B) De novo production of bisBIAs in yeast strains expressing natural DRS-DRR fusion proteins. Strains were cultured in selective media (YNB-Ura) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) De novo production of bisBIAs in yeast strains expressing PbDRR V2 with different DRS domains as individual polypeptides. Strains were cultured in selective media (YNB-Ura-Trp) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (D) De novo production of bisBIAs in yeast strains expressing PbDRS V3 with different DRR domains as individual polypeptides. Strains were cultured in selective media (YNB-Ura-Trp) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (E) De novo production of bisBIAs in yeast strains expressing the two best performing DRS and DRR domains. The activity of the PrDRS and PbDRR V2 domains were tested as chimeric fusion proteins (sequences provided in SI Appendix, Table S7) and individual polypeptides. The native fused PbDRS-DRR 1 construct that was employed in previous figures is also shown for comparison. Strains were cultured in selective media (YNB-Ura for fusion proteins, YNB-Ura-Trp for split constructs) with 2% dextrose with 2 mM L-DOPA and 10 mM ascorbic acid at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. Data in B–E are presented as mean values ± the SD of three biologically independent samples. Source data underlying B–E are provided in Dataset S1.
We first compared the bisBIA levels produced by natural DRS-DRR fusions. DRS-DRR variants were expressed from low-copy plasmids in strain CSY1336, as it is the strain which produced the highest total bisBIA titer without any integrated copies of a DRS-DRR (see SI Appendix, Fig. S5). We expressed the three DRS-DRR variants previously examined—PbDRS-DRR 1, PbDRS-DRR 2, and PsDRS-DRR—and an additional previously reported variant (32) with less than 60% sequence similarity to the three aforementioned variants from P. bracteatum (hereafter referred to as PbREPI)—from low-copy plasmids in strain CSY1336. We observed the highest total bisBIA titer (9.92 mg/L) from PbDRS-DRR 1 and lower titers from PbDRS-DRR 2 (4.37 mg/L), PsDRS-DRR (7.91 mg/L), and PbREPI (8.36 mg/L) (Fig. 3B ). Given that our prior optimization efforts had been performed with PbDRS-DRR 1 and it afforded the highest total bisBIA titer of the DRS-DRR variants screened, we selected this variant as the basis for our screening of the independent DRS and DRR domains.
We next examined the impact of expressing the DRS and DRR domains as separate polypeptides on bisBIA titers. We expressed the individual PbDRS (PbDRS V3) and PbDRR (PbDRR V2) domains from PbDRS-DRR 1 from low-copy plasmids and used these two domains as the basis for our screening of nonnatural partner domains. In order to determine if there was an effect on the activity of either domain by the linker region connecting the two domains in the wild-type fused PbDRS-DRR (Fig. 3A ), we also expressed additional PbDRS and PbDRR domains which each incorporated differing amounts of the linker region (see Fig. 3A for the sites of cleavage of these two domains; the exact sequences of each domain are given in SI Appendix, Table S7). We first screened one DRR variant with multiple DRS variants in strain CSY1336. PbDRR V2 was expressed from a low-copy plasmid and five DRS domains—PbDRS V1, PbDRS V2, PbDRS V3, Papver rhoeas DRS (PrDRS), and PbREPI DRS—were expressed individually from an orthogonal low-copy plasmid. In contrast to the other reticuline epimerases examined, which consist of P450 oxidase and NADPH reductase didomain fusions, in P. rhoeas the DRS and the DRR domains are produced as separate monomers (32). The individual PbDRS and PbDRR domains from PbDRS-DRR 1 when expressed together resulted in double the titer of total bisBIAs to the native, fused PbDRS-DRR 1 at 14.9 mg/L total bisBIAs for the split domain versus 7.9 mg/L total bisBIAs for the fused enzyme despite producing similar ratios of bisBIAs (5:1 guattegaumerine:berbamunine for the split domains versus 4:1 guattegaumerine:berbamunine for the fused enzyme). However, the combination of the PbDRR V2 and PrDRS domains resulted in nearly 14-fold higher total bisBIAs levels at 110 mg/L with a guattegaumerine:berbamunine ratio of greater than 60:1 (Fig. 3C ). We next screened one DRS variant with multiple DRR variants under similar assay conditions. PbDRS V3 was expressed from a low-copy plasmid, and four DRR domains—PbDRR V1, PbDRR V2, PbDRR V3, and P. rhoeas DRR (PrDRR)—were expressed individually from an orthogonal low-copy plasmid. We observed that PbDRR V1 and PbDRR V2 furnished similar titers of total bisBIAs, at 18.0 mg/L for PbDRR V1 and 14.9 mg/L for PbDRR V2, with a guattegaumerine:berbamunine ratio of 3:1 seen with PbDRR V1 and 5:1 for PbDRR V2 (Fig. 3D ). We also examined the combination of PrDRS, the best identified individual DRS domain, with each of the DRR domains to confirm that none performed better in combination with PrDRS than with PbDRS V3 ( SI Appendix, Fig. S6). Given that there was no statistically significant difference observed in the titers observed with PbDRR V1 or PbDRR V2 in combination with PbDRS V3 or PrDRS, we selected PbDRR V2 for subsequent studies, as it was the domain used in the test with which we obtained our highest total bisBIA titer observed, 110 mg/L, when paired with PrDRS.
We then explored whether the PrDRS and PbDRR V2 domains exhibited higher activity as individual enzymes or fused into a single polypeptide in a form resembling most natural DRS-DRR enzymes. We designed three different fusions of PrDRS and PbDRR V2: chimera 1 contains a 36 base pair (bp) linker, derived from the region joining the DRS and DRR domains in PbDRS-DRR 1, between the two domains; chimera 2 contains no linker between the two domains; and chimera 3 contains an 84 bp linker between the two domains. Each fused variant was encoded on a low-copy plasmid and expressed in strain CSY1336. While PrDRS-PbDRR V2 chimera 2 was inactive, chimera 1 produced 52 mg/L of total bisBIAs (50.9 mg/L guattegaumerine and 1.1 mg/L berbamunine) and chimera 3 produced 67.2 mg/L of total bisBIAs (64.8 mg/L guattegaumerine and 2.4 mg/L berbamunine), both of which were substantially less than the 110 mg/L total bisBIAs observed from the strain in which the two domains were expressed independently. We created a genetic module encoding independent genes for PrDRS, PbDRR V2, and a second copy of BsCYP80A1 (as a second copy of this gene was present in CSY1336, which served as the base strain for screening DRS and DRR variants) and integrated this module into strain CSY1332 to generate strain CSY1337. Strain CSY1337 produced a bisBIA titer of over 94 mg/L, with over 92 mg/L guattegaumerine and 2 mg/L berbamunine, a ratio of guattegaumerine to berbamunine of 46:1. These data indicate that there is a lower reduction in bisBIA titers when integrating the independent PrDRS and PbDRR V2 domains compared to expression off of low-copy plasmids (15% total bisBIA titer decrease) than that observed with the fused PbDRS-DRR 1 enzyme (70% total bisBIA titer decrease).
An Alternate CYP80A1 Variant Preferentially Produces Berbamunine Over Guattegaumerine.
Having developed a strain (CSY1337) that produces a high titer of guattegaumerine de novo, we sought to modify this strain to instead produce primarily berbamunine. While strain CSY1333 produced a higher titer of berbamunine than guattegaumerine, the titers of both were relatively modest (1.8 mg/L berbamunine and 1.35 mg/L guattegaumerine) compared with our highest yielding guattegaumerine strains. The highest berbamunine titer obtained with any of our strains was 6.3 mg/L with strain CSY1335, in contrast to the 108.8 mg/L guattegaumerine titer obtained with strain CSY1336 expressing PrDRS and PbDRR V2 from low-copy plasmids. Since berbamunine is produced by the CYP80A1-mediated coupling of one equivalent each of (S)-N-methylcoclaurine and (R)-N-methylcoclaurine, both of which are biosynthetic intermediates for the production of guattegaumerine, we hypothesized that both are produced in sufficient quantities in our engineered strains to furnish a higher titer of berbamunine than we had thus far observed.
We screened other CYP80A1 variants in a strain producing high titers of (S)-NMC and (R)-NMC to determine if a different variant of this enzyme would result in a higher titer of berbamunine. We removed both copies of BsCYP80A1 from strain CSY1337 to generate strain CSY1339. We investigated four alternate CYP80A1 variants for their ability to produce berbamunine: NnCYP80A1, from Nelumbo nucifera; VvCYP80A1, from Vitis vinifera; MaCYP80A1 from Mahonia aquifolium; and XsCYP80A1, from Xanthorhiza simplicissima. The transcripts for MaCYP80A1 and XsCYP80A1 were obtained from a transcriptome search and were partially incomplete, lacking reads corresponding to a signal peptide or N-terminal integral membrane anchor common to most plant P450s. An amino acid sequence alignment of the translations of the partial MaCYP80A1 and XsCYP80A1 transcripts to the full length BsCYP80A1 was generated (36). The findings from the sequence alignment demonstrated that both MaCYP80A1 and XsCYP80A1 share close sequence homology to the core P450 domain of BsCYP80A1. Our group has previously generated functional plant P450 chimeras with improved activity by swapping the N-terminal integral membrane portion (residues 1 to 86) of one P450 variant with that of another known to express and function well in yeast in vivo (7, 37). We attempted this strategy with MaCYP80A1 and XsCYP80A1 using BsCYP80A1 as the donor for our N terminus sequence. As such, the first 86 residues from BsCYP80A1 were appended to each to generate Bs-MaCYP80A1 and Bs-XsCYP80A1. NnCYP80A1, VvCYP80A1, Bs-MaCYP80A1, and Bs-XsCYP80A1 were expressed from high-copy plasmids in strain CSY1339 for 96 h at 25 °C in synthetic complete media without tryptophan and supplemented with L-DOPA and ascorbic acid, and the supernatant was then screened by LC-MS/MS for de novo production of berbamunine. While NnCYP80A1, VvCYP80A1, and Bs-XsCYP80A1 produced no observable berbamunine, Bs-MaCYP80A1 produced 25.4 mg/L berbamunine with no quantifiable guattegaumerine observed to accumulate (Fig. 4B ). We also measured the titers of residual (S)-NMC and (R)-NMC in this top de novo berbamunine strain (CSY1339 expressing Bs-MaCYP80A1 off of a high-copy plasmid) and our top de novo guattegaumerine strain (CSY1337; see Expression of Engineered Split DRS and DRR Enzymes Further Increases bisBIA Production Levels section). Our top de novo guattegaumerine strain produced 0.831 mg/L (S)-NMC and 0.687 mg/L (R)-NMC, while our top de novo berbamunine strain produced 1.868 mg/L (S)-NMC and 0.112 mg/L (R)-NMC ( SI Appendix, Fig. S7).
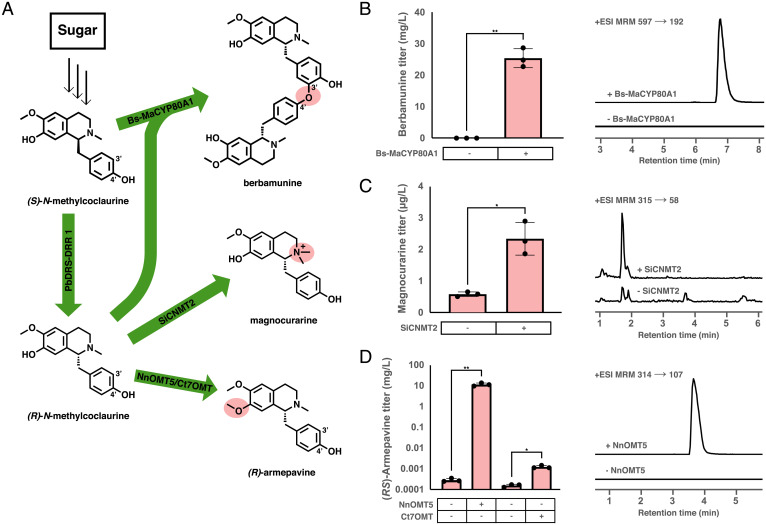
Fig. 4.
De novo biosynthesis of derivatives of the guattegaumerine biosynthetic pathway. (A) Biosynthetic scheme showing the enzymatic biosyntheses of three derivatives accessible from intermediates of the guattegaumerine biosynthetic pathway: berbamunine, formed via the coupling of one (S)-N-methylcoclaurine monomer with one (R)-N-methylcoclaurine; magnocurarine, formed via the N-methylation of (R)-N-methylcoclaurine to afford the quaternary amine product; (R)-armepavine, formed from the 7-O-methylation of (R)-N-methylcoclaurine. (B) De novo production of berbamunine in a yeast strain expressing the engineered Bs-MaCYP80A1 chimeric protein and representative LC-MS/MS traces. Strains were cultured in selective media (YNB-Trp) with 2% dextrose at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (C) De novo production of magnocurarine in an engineered yeast strain expressing SiCNMT2 and representative LC-MS/MS traces. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. (D) De novo production of armepavine in engineered yeast strains expressing NnOMT5 and Ct7OMT and representative LC-MS/MS traces. The titers reported here are the total armepavine concentration [the sum of (S)-armepavine and (R)-armepavine]. Strains were cultured in selective media (YNB-Ura) with 2% dextrose at 25 °C for 96 h before LC-MS/MS analysis of the growth media using the MRM transitions reported in SI Appendix, Table S5. Data in B–D are presented as mean values ± the SD of three biologically independent samples. Asterisks represent Student’s two-tailed t test: *P < 0.05, **P < 0.01. Exact P values are given in SI Appendix, Table S6. Source data underlying B–D are provided in Dataset S1.
The De Novo bisBIA Strain Provides a Platform for the Biosynthesis of Related PNPs.
We next sought to leverage our de novo bisBIA biosynthetic S. cerevisiae strain to produce additional pathway derivatives of interest. The intermediate (R)-NMC is an important branch point in the biosynthesis of numerous bioactive natural products. Magnocurarine is a quaternary ammonium BIA from Magnolia obovata and other species that is formed by N-methylation of (R)-NMC and has been reported to have activity similar to that of curare, a nicotinic acetylcholine receptor inhibitor that has historically been used as an arrow poison (38). Armepavine is found in Daurian moonseed, Menispermum dauricium, among other species, that is made by the selective O-methylation of the 7-hydroxyl of NMC and has been reported to have anticancer activity (39).
To attempt the production of magnocurarine and armepavine de novo in yeast, the methyltransferases reported to produce each product were expressed in strain CSY1339. CNMT2 from Stephania intermedia (SiCNMT2) has been reported to N-methylate (R)-NMC in vitro to produce magnocurarine. SiCNMT2 was cloned onto a high-copy plasmid and expressed in strain CSY1339 for 96 h at 25 °C. While strain CSY1339 without SiCNMT2 accumulated only 0.57 µg/L magnocurarine in the supernatant, when SiCNMT2 was expressed magnocurarine accumulated to a titer of 2.3 µg/L, a fourfold improvement (Fig. 4C ). NnOMT5 from N. nucifera was reported to methylate the 7-hydroxyl group of (S)-N-methylcoclaurine in vitro, but its activity for (R)-N-methylcoclaurine was not evaluated (40). Another O-methyltransferase, Ct7OMT from Coptis teeta, has been reported to either 6-O-methylate or 7-O-methylate (S)-norcoclaurine to produce (S)-coclaurine or (S)-isococlaurine, respectively (41). We cloned NnOMT5 and Ct7OMT onto high-copy plasmids and expressed each plasmid independently in strain CSY1339 for 96 h at 25 °C. We observed accumulation of 12 µg/L and 11.1 mg/L armepavine in the supernatant upon expression of Ct7OMT and NnOMT5, respectively (Fig. 4D ). While NnOMT5 has previously only been reported to accept (S)-N-methylcoclaurine as a substrate, we purified NnOMT5 from Escherichia coli and confirmed its activity in vitro on (R)-N-methylcoclaurine to produce (R)-armepavine ( SI Appendix, Fig. S8). The yeast strains described here are therefore likely biosynthesizing a mixture of (S)-armepavine and (R)-armepavine de novo. We expressed our five CYP80A1 variants (BsCYP80A1, NnCYP80A1, VvCYP80A1, Bs-MaCYP80A1, and Bs-XsCYP80A1) off of high-copy plasmids in the armepavine strain expressing NnOMT5 and screened these strains for the production of the bisBIA dauricine, a chemotherapeutic produced in M. dauricium through the 3′–4′ coupling of two (R)-armepavine monomers (42), but did not observe any accumulation of dauricine.
Discussion
In this study, we demonstrate de novo microbial biosyntheses of bisBIA alkaloids. While the biosynthetic steps from the first dedicated intermediate, (S)-norcoclaurine, to (S)-N-methylcoclaurine were previously demonstrated in yeast, the ultimate step, the cytochrome P450 mediated coupling of two NMC monomers to form the bisBIA, had only been established in insect cell culture (27). Furthermore, the penultimate step, the epimerization of (S)-N-methylcoclaurine to (R)-N-methylcoclaurine, was not known to be catalyzed in vivo by any reported enzyme. We found that PbDRS-DRR and BsCYP80A1 function in yeast to catalyze the final two reaction steps, furnishing both guattegaumerine and berbamunine, albeit at an initially low titer of 6 µg/L total bisBIAs. Through strain engineering, protein engineering, and optimization of the fermentation conditions, we increased the titer over 18,000-fold in our highest producing strain, which produced 110 mg/L total bisBIAs.
Two of the modifications that together afforded the largest increase in titer were the addition of L-DOPA and ascorbic acid to the growth medium and the reduction of the growth temperature from 30 to 25 °C. L-DOPA is a precursor for dopamine, which is then coupled with 4-HPAA by norcoclaurine synthase to produce (S)-norcoclaurine, the first BIA intermediate in the bisBIA biosynthetic pathway (Fig. 1A ). Addition of L-DOPA to the media thus likely increases bisBIA titer by increasing the supply of precursors for bisBIA biosynthesis. While the titers of berbamunine and guattegaumerine were both increased upon supplementation of L-DOPA and ascorbic acid, the titer of berbamunine tended to increase more dramatically than that of guattegaumerine, thus serving overall to decrease the ratio of guattegaumerine to berbamunine and bring the two products closer to parity in production. As L-DOPA feeds into (S)-norcoclaurine and further downstream into (S)-N-methylcoclaurine, the substrate pool of (S)-NMC is likely increased more so than the titer of (R)-NMC, since epimerization of (S)-NMC to (R)-NMC appears to be the rate-limiting step in this pathway (see Fig. 2C ). The greater increase in the titer of berbamunine, being the product of the coupling of (S)-NMC with (R)-NMC, over guattegaumerine, the product of the coupling of two (R)-NMC monomers, is therefore in line with a putative greater increase in the (S)-NMC substrate pool over that of (R)-NMC. Because many of the enzymes in the bisBIA biosynthetic pathway are of plant origin, their optimal temperatures can be lower than the 30 °C temperature commonly used for growing S. cerevisiae, and the increase in bisBIA titer upon lowering the temperature to 25 °C is likely due to this temperature being closer to the optimal temperature for one or more of these enzymes. The degree to which each enzyme prefers this lower temperature likely differs, though, and therefore the overall impact on the product ratio of a temperature reduction is harder to predict ab initio; we observed an increase in the ratio of guattegaumerine to berbamunine produced upon reducing the growth temperature in each set of conditions we examined.
Berbamunine was observed to be the main bisBIA produced in B. stolonifera cell culture, with no guattegaumerine observed in the initial report (43). A later in vitro investigation of BsCYP80A1, the cytochrome P450 responsible for the C-O phenol-coupling reaction to form bisBIAs from NMC monomers, showed that BsCYP80A1 purified from B. stolonifera favored the synthesis of berbamunine over guattegaumerine by a factor of roughly 9:1 (27). However, the same study investigated BsCYP80A1 expressed from insect cell culture and assayed in vitro and found that it produced 62% guattegaumerine and 38% berbamunine. The authors found that the ratio of guattegaumerine to berbamunine varied with the ratio of (S)-NMC and (R)-NMC fed to the enzyme, as well as when alternate CPRs were used (27) and as the ratio of the CPR to the CYP80A1 varied (29). Taken together, these studies suggest that the entry or binding of the substrates to CYP80A1 is influenced by the CPR partner, and structural studies of CYP80A1 variants may indicate a possible mechanistic role. Our results further showed that BsCYP80A1 expressed from S. cerevisiae also favored the synthesis of guattegaumerine over berbamunine under almost all conditions examined, often in high ratios approaching 50:1 guattegaumerine:berbamunine in our highest titer strains tested. This observed difference in product ratios between BsCYP80A1 expressed heterologously and in planta might be caused by a difference in posttranslational modifications occurring in B. stolonifera versus the heterologous hosts. Given the effect that has been observed on the product ratio by the CPR partner used (29), it is also possible that quaternary interactions between the P450, CPR, and possible unknown additional partners that differ in planta and in heterologous hosts might influence the product ratio formed. Quaternary interactions between the CPR and another P450 in our pathway, the DRS domain of DRS-DRR, could explain the higher bisBIA titer we observed when expressing the DRS domain independently rather than fused to the DRR domain. Future work may focus on characterizing differences in structure, localization, and posttranslational modification that occur in planta versus in other cell systems to elucidate the mechanistic basis for this difference in product specificity.
Our engineering of the DRS-DRR enzyme to increase its efficiency also suggests that the substrate pool directly affects the product ratio observed. The improvements we made to DRS-DRR, both by splitting this enzyme into its two separate domains and examining combinations of individual domains from different plant sources, altered the total bisBIA titer observed. Given that no other alterations to the strain were made for these tests, differences in the total bisBIA titer were presumably due only to differences in the relative abundance of the (S)-NMC and (R)-NMC substrate pools available; identical cytochrome P450s, CPRs, and fermentation conditions were employed for all of these tests. Comparisons of the guattegaumerine production from the fused PbDRS-DRR construct (6.26 mg/L), the split PbDRS V3 and PbDRR V2 constructs (12.5 mg/L), the fused PrDRS-PbDRR V2 construct (64.8 mg/L for the best fusion examined), and the split PrDRS and PbDRR V2 constructs (109 mg/L) showed that the guattegaumerine titer increased significantly with optimization of the DRS-DRR activity. However, the berbamunine titer stayed relatively constant, varying from 1.64, to 2.44, to 2.40, to 1.77 mg/L for the four aforementioned constructs, respectively. We therefore suspect that increases in availability of (R)-NMC led primarily to increases in guattegaumerine, the product of the coupling of two (R)-NMC monomers, rather than berbamunine, the product of the coupling of one (S)-NMC monomer with one (R)-NMC monomer.
Given that we initially observed a higher titer of berbamunine than guattegaumerine only in strain CSY1333, which contained a single integrated copy of unoptimized PbDRS-DRR 1, it is likely that this strain produced a lower titer of (R)-NMC than our subsequent strains, which contained either multiple copies of PbDRS-DRR 1 or our optimized split PrDRS and PbDRR V2 constructs, and our heterologous in vivo results are thus consistent with the earlier reported heterologous in vitro results. Ultimately, we were able to achieve a near complete inversion of the product ratio by substituting an alternate P450, Bs-MaCYP80A1, for BsCYP80A1, without making any other modification to the parent strain or substrate pool available. Bs-MaCYP80A1 has 94% amino acid identity with BsCYP80A1, differing in only 27 residues out of 487 ( SI Appendix, Fig. S9). While no protein structures are currently available for any CYP80A1, we performed an alignment of these CYP80A1s to the plant P450 with a crystal structure reported that is most similar to BsCYP80A1, CYP76AH1 from Salvia miltiorrhiza (SmCYP76AH1) ( SI Appendix, Fig. S9) (44). An examination of all residues within 5 Å of the active site of SmCYP76AH1 revealed no differences between BsCYP80A1 and Bs-MaCYP80A1 in these residues. It was also reported that SmCYP76AH1 has two largely hydrophobic access channels to the active site that might allow and regulate substrate entry and product exit. However, BsCYP80A1 and Bs-MaCYP80A1 did not differ in any of the residues reported to form these two access channels in SmCYP76AH1. Given the dissimilarity of BsCYP80A1 and SmCYP76AH1 (<40% identity), the structures of BsCYP80A1 and Bs-MaCYP80A1 might differ significantly from our homology model. Crystal structures could reveal key differences in the architecture of the active site and/or analogous access channels in BsCYP80A1 and Bs-MaCYP80A1, which might explain the striking difference in guattegaumerine:berbamunine ratio produced by these enzymes.
The availability of an engineered S. cerevisiae strain that produces bisBIAs de novo that resulted from this work also offers a platform for the production of additional bioactive molecules of interest in this broader family of alkaloids. We demonstrated this through the de novo biosynthesis of two additional alkaloids from intermediates in our bisBIA biosynthesis pathway: magnocurarine from (R)-NMC and armepavine from either (S)-NMC or (R)-NMC. Additional linear bisBIAs could be produced from NMC via alternative coupling chemistries, such as the sedative nelumboferine (45), which is formed by the 7–3′ coupling of two equivalents of (R)-NMC, or from the 3′–4′ coupling we demonstrated with guattegaumerine and berbamunine but employed on alternative substrates, such as the chemotherapeutic dauricine (42), which is formed from two equivalents of (R)-armepavine. While none of the CYP80A1 variants we screened afforded dauricine at this time (see The De Novo bisBIA Strain Provides a Platform for the Biosynthesis of Related PNPs section), we envision that our armepavine-producing strain will facilitate the de novo heterologous biosyntheses of this and other bisBIAs beyond the two we produced in this work. Furthermore, alternative coupling enzymes may be found or engineered which could enable the biosynthesis of cyclic bisBIAs, for example the arrow poison tubocurarine (2) or the antimalarial cycleanine (46). Finally, the application of recently described computational tools that enable the expansion of core heterologous pathways to new derivatives via incorporation of existing enzymatic activities (11, 47) can facilitate the production of additional new-to-nature pathway derivatives.
Materials and Methods
Additional materials and methods are provided in SI Appendix .
Growth Conditions for Metabolite Assays.
Metabolite production tests were performed in yeast nitrogen base-synthetic complete (YNB-SC) or yeast nitrogen base-dropout (YNB-DO) media with at least three replicates. Yeast colonies were inoculated into 300 μL media and grown in 2-mL deep-well 96-well plates covered with AeraSeal gas-permeable film (Excel Scientific). Cultures were then grown for 96 h at 25 or 30 °C (exact temperature is specified in each figure), 460 rpm, and 80% relative humidity in a Lab-Therm LX-T shaker (Adolf Kuhner).
Analysis of Metabolite Production.
Cultures were pelleted by centrifugation at 3,500 × g for 10 min at 4 °C, and 100 μL aliquots of the supernatant were removed for direct analysis. Metabolite production was analyzed by LC-MS/MS using an Agilent 1260 Infinity Binary high-performance liquid chromatography (HPLC) and an Agilent 6420 Triple Quadrupole mass spectrometer. Nonchiral chromatography was performed using a Zorbax EclipsePlus C18 column (2.1 × 50 mm, 1.8 μm; Agilent Technologies) with water with 0.1% vol/vol formic acid as solvent A and acetonitrile with 0.1% vol/vol formic acid as solvent B. The column was operated with a constant flow rate of 0.4 mL/min at 40 °C and a sample injection volume of 10 μL. Compound separation for the detection of norcoclaurine, N-methylcoclaurine, berbamunine/guattegaumerine, armepavine, or magnocurarine was performed using the following gradient: 0.00 to 0.20 min, 10% B; 0.20 to 10.00 min, 10 to 14% B; 10.00 to 11.00 min, 14 to 90% B; 11.00 to 12.00 min, 90% B; 12.00 to 12.20 min, 90 to 10% B; 12.20 to 12.70 min, equilibration with 10% B. The LC eluent was directed to the MS from 0.5 to 12.0 min operating with electrospray ionization (ESI) in positive mode, source gas temperature 350 °C, gas flow rate 11 L/min, and nebulizer pressure 40 psi. Metabolites were quantified by integrated peak area in MassHunter Workstation software (Agilent) based on the multiple reaction monitoring (MRM) parameters in SI Appendix, Table S5. Integrated peak areas were converted to titers by comparison to standard curves prepared using a chemical standard. Primary MRM transitions for berbamunine/guattegaumerine and magnocurarine were identified by analysis of a 0.1 mM standard in methanol using the MassHunter Optimizer software package (Agilent); all other MRM transitions used were previously reported and are described in SI Appendix, Table S5.
Chiral Analysis of (S)- and (R)-N-methylcoclaurine Production.
Yeast colonies were used to inoculate 5 mL YNB-Ura/Trp and grown overnight at 30 °C and 460 rpm. All 5 mL of the primary cultures was used to inoculate 500 mL YNB-Ura/Trp media in a 2-L Erlenmeyer flask and grown for 96 h at 30 °C and 220 rpm. The cultures were pelleted by centrifugation at 3,500 × g for 10 min at 4 °C. To the supernatant was added 30 mg Amberlite XAD-4 resin per mL, and the mixture was then incubated overnight at room temperature at 200 rpm. The supernatant was then decanted off and to the resin was added 50 mL methanol, and the mixture was then rotated overnight at room temperature. The methanol was then evaporated to dryness by rotary evaporation, and the residue was resuspended in 5 mL water with 0.1% formic acid. The concentrate was fractionated by reverse-phase HPLC on an Agilent 1200 Series LC using a Pursuit XRs-C18 column (50 × 10 mm, 5 μm; Agilent Technologies) with isocratic 15% methanol with 0.1% formic acid over 6.5 min with a flow rate of 5 mL/min and injection volume of 40 to 60 μL. Fractions were pooled, lyophilized, and resuspended in 300 μL isopropanol. Depending on concentration, 1 to 5 μL was injected onto a Lux Cellulose-1 column (150 × 2 mm, 3 µm; Phenomenex Technologies) and separated with isocratic 72% N-hexane, 28% isopropanol, 0.1% diethylamine with a flow rate of 0.3 mL/min and detection by MS and ultraviolet (UV) detection at 250 nm. MS detection was performed with an Agilent 6320 Ion Trap mass spectrometer with ESI source gas temperature 350 °C, gas flow of 10 L/min, nebulizer pressure 40 PSI, and isolation of m/z 330.1 with width 1.0. The retention time of (R)-N-methylcoclaurine was confirmed by comparison with an authentic (R)-N-methylcoclaurine standard. Individual chromatograms from chiral analysis were smoothed using the Gauss smoothing function in the Bruker DataAnalysis software (50 cycles).
For the strains described in SI Appendix, Figs. S4 and S7, a modified version of the above procedure was used. Cultures were grown as described in the Analysis of Metabolite Production section. Chiral resolution of (S)-NMC and (R)-NMC was performed using the LC method described for the Lux Cellulose-1 column but with metabolite detection performed on an Agilent 6420 Triple Quadrupole mass spectrometer in place of the Agilent 6320 Ion Trap mass spectrometer, using the MRM transitions described in SI Appendix, Table S5.
Protein Purification.
Plasmids containing the gene of interest in a pET28 expression vector (reference SI Appendix, Table S4 for a full list of plasmids used in this study) were used to transform E. coli BL21(DE3) (Invitrogen) competent cells via heat shock. Briefly, 1 ng plasmid DNA was added to a 50-µL aliquot of competent cells, the tube was chilled on ice for 15 min, placed in a 42 °C water bath for 35 s, then returned to ice for 2 min. Then, 250 µL super optimal broth with catabolite repression (SOC) media were added, and the tube was rotated at 37 °C for 45 min before being plated on an LB agar plate containing 50 µg/mL kanamycin. A single colony was then picked and used to inoculate a primary culture of 5 mL of LB media containing 50 µg/mL kanamycin which was then grown for 24 h. All 5 mL of this primary culture were then used to inoculate a secondary or expression culture of 500 mL of LB medium containing 50 µg/mL kanamycin. This expression culture was grown to an OD600 of 0.6 to 1.0 and then induced with IPTG at a final concentration of 0.1 mM. The expression culture was then grown at 16 °C for 20 h at 250 rpm, after which the culture was harvested by centrifugation (10 min at 3,500 × g in a 50 mL Falcon tube) and stored at −20 °C until lysis and purification.
Frozen pellets were then thawed and resuspended in 25 mL of Ni-nitrilotriacetic (Ni-NTA) equilibration buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 7.4) and lysed by sonication while kept on ice (Branson Sonifier 450, 0.5” horn, 50% duty cycle, 4 × 1 min with 2 min rests). Lysed cultures were then clarified by centrifugation (45 min at 35,000 × g at 4 °C), and the clarified lysate was purified by Ni-NTA affinity chromatography. Briefly, 1 mL Ni-NTA resin (Fisher Scientific) was equilibrated with at least 5 volumes of Ni-NTA equilibration buffer and then loaded with the clarified lysate. The loaded resin was then washed with at least 5 volumes of Ni-NTA wash buffer (50 mM sodium phosphate, 300 mM NaCl, 50 mM imidazole, pH 7.4) and then the bound protein was eluted with 5 volumes of Ni-NTA elution buffer (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, pH 7.4). The eluted fractions were then combined and concentrated using an Amicon 30 kDa cutoff spin filter (EMD Millipore) at 5,000 × g at 4 °C. Concentrated protein fractions were then exchanged into storage buffer (50 mM potassium phosphate, 100 mM NaCl, 10% glycerol, pH 7.5), split into separate aliquots, and stored at −20 °C until use.
In Vitro Bioconversions.
Analytical reactions were carried out at the 50 µL scale in triplicate. To a 1.5-mL Eppendorf tube was added 5 nmol substrate (final concentration of 100 µM), 1 µmol sodium ascorbate (final concentration of 25 mM), 5 nmol S-adenosylmethionine (final concentration of 100 µM), and 150 pmol of each purified methyltransferase enzyme (3 µM final concentration) in 50 mM potassium phosphate, pH 8.0. The reactions were shaken at 600 rpm at 37 °C for 2 h before being quenched with an equal volume of methanol, spun down at 20,000 × g for 10 min, and filtered prior to LC-MS analysis (see Analysis of Metabolite Production section above for details on LC-MS analysis conditions).
Software.
ChemDraw 18.0 was used for illustrating chemical structures. Pymol was used for visualizing protein structures. Clustal Omega was used to generate protein sequence alignments (48). ESPript was used for visualization of protein sequence alignments (49, 50). RaptorX was used to construct protein homology models (51). MassHunter Workstation (Agilent) was used to collect and analyze LC-MS/MS data. ChemStation (Agilent) and DataAnalysis (Bruker) were used to collect and analyze, respectively, chiral LC-MS data. GraphPad Prism 9 was used to generate bar graphs. Microsoft Excel 2016 was used to perform statistical analysis. Inkscape was used to create all figures shown in this work.
Supplementary Material
Acknowledgments
We thank Toni Kutchan (Donald Danforth Plant Science Center) for providing the standards of guattegaumerine and berbamunine. We thank A. Cravens for the yeast CRISPRm Cas9/single guide RNA (sgRNA) plasmids (pCS3410, 3411, 3414, and 3702) and the pCS4717 plasmid. We thank P. Srinivasan, O. Jamil, D. Kong, and M. Kaplan for their invaluable assistance in keeping research running smoothly despite interruptions from COVID-19. We thank P. Srinivasan for the helpful discussions and feedback in the preparation of this manuscript. Funding for this work was provided by NIH grants (grant to C.D.S., AT007886; fellowship to J.T.P., F32 AT009509 − 03) and the Joint Initiative for Metrology in Biology grant (for T.R.V.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of General Medical Sciences (NIGMS) or NIH.
Footnotes
Competing interest statement: C.D.S. is an inventor on a pending patent application and a founder and the CEO of Antheia, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112520118/-/DCSupplemental.
Data Availability
Data supporting the findings of this work are available within the paper and its SI Appendix . Source data for the figures in this paper and the SI Appendix are provided in Dataset S1.
Change History
June 17, 2022: The authors unintentionally referenced the former name Guatteria gaumeri as the source organism of guattegaumerine instead of the currently accepted name Mosannona depressa. The introduction has been updated.
References
- 1. Weber C., Opatz T., Bisbenzylisoquinoline Alkaloids (Elsevier Inc., ed. 1, 2019). [DOI] [PubMed] [Google Scholar]
- 2. Bisset N. G., Arrow and dart poisons. J. Ethnopharmacol. 25, 1–41 (1989). [DOI] [PubMed] [Google Scholar]
- 3. Wastila W. B., Maehr R. B., Turner G. L., Hill D. A., Savarese J. J., Comparative pharmacology of cisatracurium (51W89), atracurium, and five isomers in cats. Anesthesiology 85, 169–177 (1996). [DOI] [PubMed] [Google Scholar]
- 4. Wei G., Zhan-yun S., Chun-sheng L., Qiao-xian X., Resource investigation of wild Stephania tetrandra in Anhui and Jiangxi Province. Zhonghua Zhongyiyao Zazhi 25, 909–911 (2010). [Google Scholar]
- 5. Huang L. P., Liu X. Y., Li L. H., Investigation of wild Stephania tetrandra resources in the south Anhui mountainous region. Res. Pract. Chin. Med. 22, 22–24 (2008). [Google Scholar]
- 6. Qing Z., Wang D. Q., Resources of medicinal plants menispermaceae in Anhui Province. J. Anhui Tradit. Chin. Med. Coll. 26, 52–54 (2007). [Google Scholar]
- 7. Galanie S., Thodey K., Trenchard I. J., Interrante M. F., Smolke C. D., Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galanie S., Smolke C. D., Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb. Cell Fact. 14, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., et al. , Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E3922–E3931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valentic T. R., Payne J. T., Smolke C. D., Structure-guided engineering of a scoulerine 9-O-methyltransferase enables the biosynthesis of tetrahydropalmatrubine and tetrahydropalmatine in yeast. ACS Catal. 10, 4497–4509 (2020). [Google Scholar]
- 11. Hafner J., Payne J., MohammadiPeyhani H., Hatzimanikatis V., Smolke C., A computational workflow for the expansion of heterologous biosynthetic pathways to natural product derivatives. Nat. Commun. 12, 1760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kametani T., H. Iida, K. Sakurai, A total synthesis of magnoline. J. Chem. Soc. C, 500–501 (1969). [Google Scholar]
- 13. Proskurnina N. F., Orekhov A. P., Alkaloids of Magnolia fuscata . Bull. Soc. Chim. Fr. Mem. 5, 1357–1360 (1938). [Google Scholar]
- 14. Sarraf M., Beig Babaei A., Naji-Tabasi S., Investigating functional properties of barberry species: An overview. J. Sci. Food Agric. 99, 5255–5269 (2019). [DOI] [PubMed] [Google Scholar]
- 15. Dehaussy H., Tits M., Angenot L., Guattegaumerine, new bisbenzylisoquinoline alkaloid from Guatteria gaumeri . Planta Med. 49, 25–27 (1983). [DOI] [PubMed] [Google Scholar]
- 16. Mokhber-Dezfuli N., Saeidnia S., Gohari A. R., Kurepaz-Mahmoodabadi M., Phytochemistry and pharmacology of berberis species. Pharmacogn. Rev. 8, 8–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belwal T., et al. , Phytopharmacology and clinical updates of Berberis species against diabetes and other metabolic diseases. Front. Pharmacol. 11, 41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cortes D., Figadere B., Saez J., Protais P., Displacement activity of bisbenzylisoquinoline alkaloids at striatal 3H-SCH 23390 and 3H-raclopride binding sites. J. Nat. Prod. 55, 1281–1286 (1992). [DOI] [PubMed] [Google Scholar]
- 19. Resendiz J. S., Lerdo de Tejada A., Cholesterol-lowering effect of Guatteria gaumeri (preliminary report). J. Ethnopharmacol. 6, 239–242 (1982). [DOI] [PubMed] [Google Scholar]
- 20. Leclercq J., Quetin J., De Pauw-Gillet M. C., Bassleer R., Angenot L., Antimitotic and cytotoxic activities of guattegaumerine, a bisbenzylisoquinoline alkaloid. Planta Med. 53, 116–117 (1987). [DOI] [PubMed] [Google Scholar]
- 21. Lü Q., et al. , Guattegaumerine protects primary cultured cortical neurons against oxidative stress injury induced by hydrogen peroxide concomitant with serum deprivation. Cell. Mol. Neurobiol. 29, 355–364 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kametani T., Sakurai K., Iida H., [Total synthesis of berbamunine and its diastereoisomer (studies on the synthesis of heterocyclic compounds. CCLIV)]. Yakugaku Zasshi 88, 1163–1167 (1968). [DOI] [PubMed] [Google Scholar]
- 23. Kametani T., Iida H., Sakurai K., A total synthesis of magnoline. J. Chem. Soc. C Org. 3, 500 (1969). [Google Scholar]
- 24. Kametani T., Iida H., Sakurai K., Kano S., Ihara M., The nuclear magnetic resonance spectra and optical rotatory dispersion of berbamunine, magnoline, and two diastereomers. Chem. Pharm. Bull. (Tokyo) 17, 2120 (1969). [Google Scholar]
- 25. Gawley R. E., Smith G. A., Formal synthesis of the bisbenzylisoquinoline alkaloid berbamunine by asymmetric substitution of chiral organolithium compounds. ARKIVOC 2011, 167–179 (2011). [PMC free article] [PubMed] [Google Scholar]
- 26. Stadler R., Zenk M. H., The purification and characterization of a unique cytochrome P-450 enzyme from Berberis stolonifera plant cell cultures. J. Biol. Chem. 268, 823–831 (1993). [PubMed] [Google Scholar]
- 27. Kraus P. F. X., Kutchan T. M., Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C–O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera . Proc. Natl. Acad. Sci. U.S.A. 92, 2071–2075 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trenchard I., Engineering Saccharomyces cerevisiae for the Production of Plant-Derived Pharmaceuticals (Stanford University, 2014). [Google Scholar]
- 29. Rosco A., Pauli H. H., Priesner W., Kutchan T. M., Cloning and heterologous expression of NADPH-cytochrome P450 reductases from the Papaveraceae . Arch. Biochem. Biophys. 348, 369–377 (1997). [DOI] [PubMed] [Google Scholar]
- 30. Ikezawa N., Iwasa K., Sato F., Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C–C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821 (2008). [DOI] [PubMed] [Google Scholar]
- 31. Hawkins K. M., Smolke C. D., Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae . Nat. Chem. Biol. 4, 564–573 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farrow S. C., Hagel J. M., Beaudoin G. A., Burns D. C., Facchini P. J., Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 11, 728–732 (2015). [DOI] [PubMed] [Google Scholar]
- 33. Winzer T., et al. , Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349, 309–312 (2015). [DOI] [PubMed] [Google Scholar]
- 34. Facchini P. J., Farrow S. C., Beaudoin G. A. W., “Compositions and methods for making (R)-reticuline and precursors thereof.” WO2015081437A1 (2015).
- 35. Smolke C. D., Wells D. H., “Engineered benzylisoquinoline alkaloid epimerases and methods of producing benzylisoquinoline alkaloids.” WO2019028390A1 (2020).
- 36. Xiao M., et al. , Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 166, 122–134 (2013). [DOI] [PubMed] [Google Scholar]
- 37. Li Y., Smolke C. D., Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 7, 12137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ogiu K., Morita M., Curare-like action of magnocurarine isolated from Magnolia obovata . Jpn. J. Pharmacol. 2, 89–96 (1953). [DOI] [PubMed] [Google Scholar]
- 39. Weng T. C., et al. , Inhibitory effects of armepavine against hepatic fibrosis in rats. J. Biomed. Sci. 16, 78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menéndez-Perdomo I. M., Facchini P. J., Isolation and characterization of two O-methyltransferases involved in benzylisoquinoline alkaloid biosynthesis in sacred lotus (Nelumbo nucifera). J. Biol. Chem. 295, 1598–1612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He S. M., et al. , Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in coptis species. Front. Plant Sci. 9, 731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Z., et al. , Dauricine induces apoptosis, inhibits proliferation and invasion through inhibiting NF-kappaB signaling pathway in colon cancer cells. J. Cell. Physiol. 225, 266–275 (2010). [DOI] [PubMed] [Google Scholar]
- 43. Stadler R., Loeffler S., Cassels B. K., Zenk M. H., Bisbenzylisoquinoline biosynthesis in Berberis stolonifera cell cultures. Phytochemistry 27, 2557–2565 (1988). [Google Scholar]
- 44. Gu M., et al. , Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza . Biochem. Biophys. Res. Commun. 511, 813–819 (2019). [DOI] [PubMed] [Google Scholar]
- 45. Nishimura K., Horii S., Tanahashi T., Sugimoto Y., Yamada J., Synthesis and pharmacological activity of alkaloids from embryo of lotus, Nelumbo nucifera . Chem. Pharm. Bull. (Tokyo) 61, 59–68 (2013). [DOI] [PubMed] [Google Scholar]
- 46. Uche F. I., et al. , In vivo efficacy and metabolism of the antimalarial cycleanine and improved in vitro antiplasmodial activity of semisynthetic analogues. Antimicrob. Agents Chemother. 65, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Srinivasan P., Smolke C. D., Engineering cellular metabolite transport for biosynthesis of computationally predicted tropane alkaloid derivatives in yeast. Proc. Natl. Acad. Sci. U.S.A. 118, e2104460118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goujon M., et al. , A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang S., Li W., Liu S., Xu J., RaptorX-Property: A web server for protein structure property prediction. Nucleic Acids Res. 44, W430–W435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bomati E. K., Austin M. B., Bowman M. E., Dixon R. A., Noel J. P., Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J. Biol. Chem. 280, 30496–30503 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this work are available within the paper and its SI Appendix . Source data for the figures in this paper and the SI Appendix are provided in Dataset S1.