Significance
In this study, we found that selecting individuals with extremely severe forms of schizophrenia led to a significantly improved ability to detect disease-associated rare variants. The high prevalence of rare variant risk factors in individuals with severe, extremely treatment-resistant schizophrenia suggests future clinical opportunities for risk prediction, prognostic stratification, and genetic counseling. These findings have implications for the design of future genetic studies in schizophrenia and highlight a strategy to reduce phenotypic heterogeneity and improve gene discovery efforts in other neuropsychiatric disorders.
Keywords: schizophrenia, genomics, rare variants, treatment-resistant schizophrenia
Abstract
Extreme phenotype sequencing has led to the identification of high-impact rare genetic variants for many complex disorders but has not been applied to studies of severe schizophrenia. We sequenced 112 individuals with severe, extremely treatment-resistant schizophrenia, 218 individuals with typical schizophrenia, and 4,929 controls. We compared the burden of rare, damaging missense and loss-of-function variants between severe, extremely treatment-resistant schizophrenia, typical schizophrenia, and controls across mutation intolerant genes. Individuals with severe, extremely treatment-resistant schizophrenia had a high burden of rare loss-of-function (odds ratio, 1.91; 95% CI, 1.39 to 2.63; P = 7.8 × 10−5) and damaging missense variants in intolerant genes (odds ratio, 2.90; 95% CI, 2.02 to 4.15; P = 3.2 × 10−9). A total of 48.2% of individuals with severe, extremely treatment-resistant schizophrenia carried at least one rare, damaging missense or loss-of-function variant in intolerant genes compared to 29.8% of typical schizophrenia individuals (odds ratio, 2.18; 95% CI, 1.33 to 3.60; P = 1.6 × 10−3) and 25.4% of controls (odds ratio, 2.74; 95% CI, 1.85 to 4.06; P = 2.9 × 10−7). Restricting to genes previously associated with schizophrenia risk strengthened the enrichment with 8.9% of individuals with severe, extremely treatment-resistant schizophrenia carrying a damaging missense or loss-of-function variant compared to 2.3% of typical schizophrenia (odds ratio, 5.48; 95% CI, 1.52 to 19.74; P = 0.02) and 1.6% of controls (odds ratio, 5.82; 95% CI, 3.00 to 11.28; P = 2.6 × 10−8). These results demonstrate the power of extreme phenotype case selection in psychiatric genetics and an approach to augment schizophrenia gene discovery efforts.
Schizophrenia affects nearly 1% of the population (1) and has sufficiently high heritability (80%) (2) to suggest that identifying genetic risk factors would provide insights into disease mechanisms. Yet, with one exception (3), definitively linking the hundreds of schizophrenia-associated common variant loci (4) to the pathophysiology of the disorder has been difficult. A more direct path to understanding the molecular basis of schizophrenia would come from the identification of highly penetrant rare variants in single genes, but this approach has so far been challenging. The Schizophrenia Exome Sequencing Meta-Analysis (SCHEMA) consortium recently conducted the largest sequencing study of schizophrenia to date, analyzing over 24,000 cases and identified 10 genes with a statistically significant excess of rare variants (5). While this represents a significant step forward, this yield is low compared to the hundreds of genes discovered in other neuropsychiatric disorders such as developmental delay and autism spectrum disorder (6, 7).
In many complex genetic disorders, individuals with more severe or earlier-onset manifestations are more likely to harbor rare, deleterious variants of large effect (8–11). As a consequence, extreme phenotype sampling—selecting individuals at the extreme ends of the distribution of a phenotypic trait—substantially reduces the sample size needed to identify genetic causes of disease (12). Sequencing studies of extreme phenotype cohorts have successfully identified rare variants of large effect in hypercholesterolemia (10), epilepsy (9), autism (11), childhood-onset schizophrenia (8, 13), and other conditions. We therefore applied this approach in a sequencing study of individuals with severe, extremely treatment-resistant schizophrenia.
Our primary analyses focused on testing for an enrichment of rare loss-of-function and damaging missense variants across “intolerant” genes. Intolerant genes are significantly depleted of functional variation (i.e., missense and loss-of-function) in healthy populations and are under strong negative selection as they are associated with a significant reduction in fecundity (14). Prior studies of schizophrenia have shown that the case-control burden of rare loss-of-function (odds ratio, 1.26) and damaging missense variants (odds ratio, 1.06 to 1.25) is concentrated exclusively in intolerant genes (5, 15). Using an extreme phenotype sequencing approach, we identified the highest burden of rare variants reported in the schizophrenia literature to date, suggesting that careful phenotype selection can augment gene discovery efforts in psychiatric genetics.
Results
Severe, Extremely Treatment-Resistant Schizophrenia Cohort.
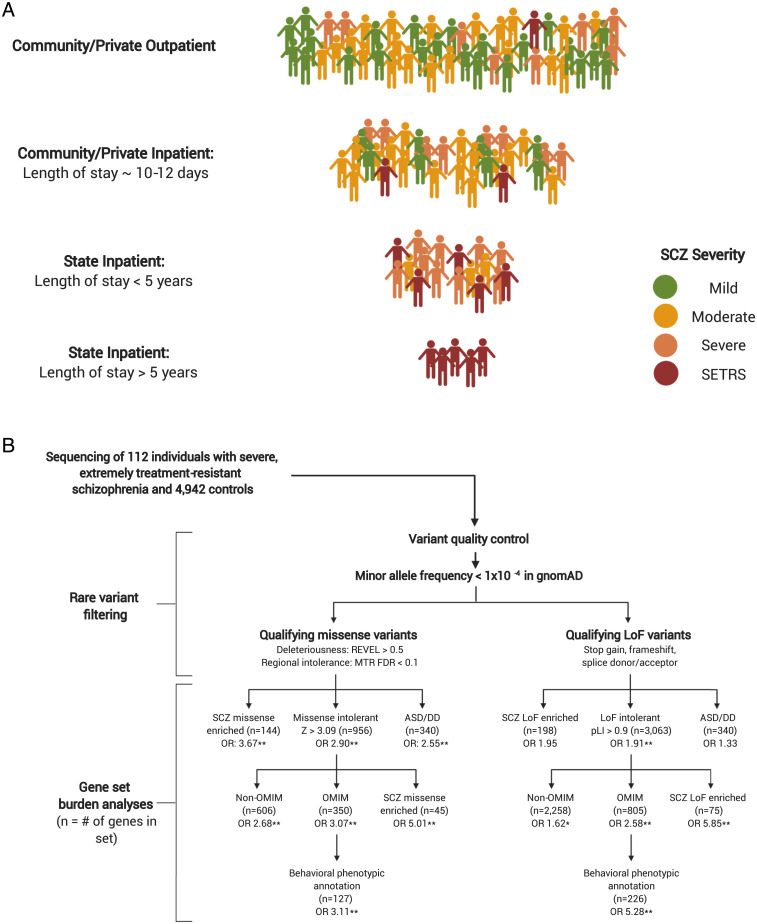
We defined individuals with severe, extremely treatment-resistant schizophrenia as those whose illness severity has required continuous hospitalization for at least 5 y in a long-term New York State inpatient facility (Fig. 1A). Because of the 97% reduction of inpatient psychiatric state hospital beds in the United States since 1955, schizophrenia patients who have remained hospitalized during the process of deinstitutionalization (roughly 1 out of every 500 schizophrenia patients in New York) represent the most severe and treatment-resistant forms of the disease. To be included in the study, we required that the 5 continuous years of hospitalization were due to primary schizophrenia symptoms and not due to discharge refusal or high risk of violence, suicide, or sexual offense. We also excluded individuals who had medical or neurologic disorders that predated the onset of schizophrenia and were the most likely cause of psychosis (see SI Appendix for detailed description of enrollment, phenotypic assessments, and inclusion/exclusion criteria).
Fig. 1.
Study and analysis workflow. (A) Approximate distribution of illness severity within the schizophrenia spectrum by system of care (numbers not to scale). Typically, patients who do not respond to treatment after several months in community inpatient facilities are transferred to state-funded inpatient facilities and represent a more severe form of schizophrenia. Patients who require very long stays (>5 y) in state-funded inpatient facilities represent the most severe and treatment-resistant subset of the schizophrenia spectrum, which we define as severe, extremely treatment-resistant schizophrenia (SETRS). SCZ: Schizophrenia. (B) Analytic workflow of 112 individuals with SETRS and 4,929 controls. Qualifying missense and loss-of-function variants are analyzed separately in their respective gene sets. SCZ missense enriched: Genes with prior evidence of enrichment for missense variation in the schizophrenia exome meta-analysis (SCHEMA) study of schizophrenia. SCZ loss-of-function enriched: Genes with prior evidence of enrichment for loss-of-function variation in SCHEMA. ASD/DD: combined gene set of 102 genome-wide significant genes associated with autism spectrum disorder and 299 genome-wide significant genes associated with developmental delay. OMIM: All genes associated with Mendelian disorders in the OMIM database. Non-OMIM: Missense and loss-of-function intolerant genes without a known disease association in OMIM. Behavioral OMIM: The subset of OMIM genes with clinical phenotype annotations including “behavioral” manifestations. *P < 0.05; **FDR < 0.1.
While not an exclusion criterion, we assessed all participants for a history of intellectual disability prior to the onset of psychosis given the potential strong contribution of intellectual disability to rare variant burden in schizophrenia (16). We used a combination of interviews with the participant, collateral information from family members, and the primary clinical team/social workers, highest educational attainment, history of special education, work history, and review of neuropsychological testing records to assess for evidence of premorbid intellectual disability. The results of this review were then discussed with a neuropsychologist (T.E.G.) with extensive experience in cognitive assessments for consensus diagnosis. No participants with severe, extremely treatment-resistant schizophrenia met diagnostic criteria for intellectual disability. The typical schizophrenia comparison cohort was composed of 218 individuals with typical schizophrenia recruited from outpatient and short-term inpatient settings (see SI Appendix for further details).
We conducted whole-genome sequencing on 112 individuals with severe, extremely treatment-resistant schizophrenia who met Diagnostic and Statistical Manual of Mental Disorders 5 (DSM-5) (17) criteria for schizophrenia and compared them to 4,929 predominantly whole-exome–sequenced healthy controls that passed quality control (SI Appendix and Tables S1 and S2). Individuals with severe, extremely treatment-resistant schizophrenia had a mean age of 61.0 y (SD = 9.2), were predominantly male (67.9%), and had an average duration of lifetime state hospitalization of 24.8 y (SD = 12.6) (see Table 1, Methods, and SI Appendix for further details). To ensure that our results were not due to sequencing artifact from comparing genomes to exomes, we also performed a separate genome-only analysis with 4,146 whole-genome sequenced individuals with nonpsychiatric illnesses such as amyotrophic lateral sclerosis, renal disease, and HIV (see SI Appendix and Tables S3 and S4 for details).
Table 1.
Demographic and clinical characteristics of SETRS cohort (n = 112)
| Demographic characteristics | |
| Mean age in years (± SD) | 61.0 (9.2) |
| Sex (%) | |
| Male | 76 (67.9) |
| Female | 36 (32.1) |
| Had children (%) | |
| Male | 4 (5.3) |
| Female | 13 (36.1) |
| Cohort | 17 (15.2) |
| Ancestry (%) | |
| Caucasian | 50 (44.6) |
| African American | 39 (34.8) |
| Admixed | 15 (13.4) |
| Hispanic | 8 (7.2) |
| Highest level of educational attainment (%) | |
| Less than ninth grade | 17 (15.2) |
| 9th to 12th grade | 41 (36.6) |
| High school graduate or equivalency | 27 (24.1) |
| Completed some college | 18 (16.1) |
| Graduated college | 8 (7.1) |
| Unknown | 1 (0.9) |
| Clinical characteristics | |
| Age of onset of psychosis: mean in years (±SD) | |
| Male | 18.3 (3.7) |
| Female | 18.3 (3.5) |
| Cohort | 18.3 (3.7) |
| Average years of state hospitalization (± SD) | 24.8 (12.6) |
Rare Variant Burden across Intolerant Genes in Severe, Extremely Treatment-Resistant Schizophrenia.
Our primary analysis focused on comparing the burden of rare variants in individuals with severe, extremely treatment-resistant schizophrenia to controls in two intolerant gene sets that have been previously shown to be enriched for rare damaging variants in individuals with neuropsychiatric disorders (5, 18, 19): genes with a high probability of being loss-of-function intolerant ((pLI) score > 0.9, n = 3,063 genes) and genes intolerant to missense variation (missense Z score > 3.09, n = 956 genes) (Fig. 1B) (20). To reduce potential confounding due to population stratification, we adopted a stratified analysis in which individuals were clustered based on genetic ancestry. We then performed a burden test in each ancestral cluster separately and generated a combined P value and odds ratio using the Cochran–Mantel–Haenszel test (see Methods for further details; SI Appendix, Figs. S1 and S2). We defined rare variants as those with a minor allele frequency of less than 1 × 10−4 in each represented ancestral population in the nonpsychiatric (“non-neuro”) subset of the Genome Aggregation Database (gnomAD) (20). Recent studies of neuropsychiatric disorders have used this cutoff (21), which ensures that variants are sufficiently rare but also allows for likely inherited variants to be included. Variants were also filtered for a variety of stringent quality control measures to remove likely artifacts (SI Appendix). We filtered missense variants for deleteriousness using the rare exome variant ensemble learner (REVEL) pathogenicity predictor using a cutoff of REVEL > 0.5 (22). The cutoff of 0.5 maximizes sensitivity and specificity for deleteriousness prediction (22) and has been previously used for deleteriousness prediction (18, 23). While the missense Z score represents a gene’s overall intolerance to missense variation, intolerance can vary between genic subregions, as functionally important regions (e.g., key protein domains) are more likely to be depleted of missense variation. We therefore used the missense tolerance ratio tool with the recommended cutoff of a false discovery rate < 0.1 to identify the most intolerant subregions of a gene (24). We categorized missense variants with REVEL > 0.5 and missense tolerance ratio false discovery rate < 0.1 as “qualifying” missense variants. “Qualifying” loss-of-function variants included stop gain, frameshift, or canonical splice donor/acceptor variants that met additional filtering criteria using the loss-of-function transcript effect estimator (LOFTEE) (20) and proportion expressed across transcripts (PEXT) (25) tools to remove variants unlikely to cause a true loss-of-function (SI Appendix).
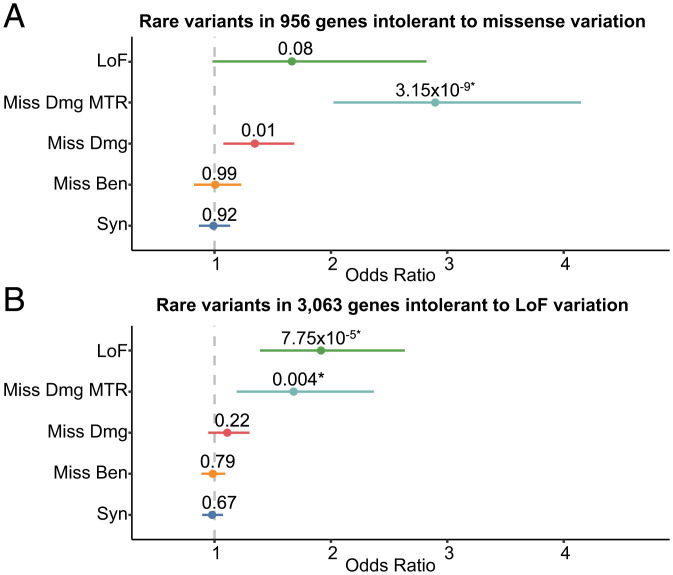
We found that individuals with severe, extremely treatment-resistant schizophrenia have a significantly higher burden of damaging rare missense variants (REVEL > 0.5) in missense-intolerant genes compared to controls (odds ratio, 1.35, 95% CI, 1.08 to 1.69; P = 0.011) (Fig. 2A) (22). Incorporating subgenic intolerance (missense tolerance ratio false discovery rate < 0.1) revealed a significant increase in the burden of damaging missense variants in these genes (odds ratio, 2.90; 95% CI, 2.02 to 4.15; P = 3.2 × 10−9; false discovery rate adjusted P value (FDR P) = 4.4 × 10−8) (Fig. 2A). This signal was absent for benign missense (odds ratio, 1.01; 95% CI, 0.82 to 1.22; P = 0.99) and synonymous variants (odds ratio, 0.99; 95% CI, 0.87 to 1.13; P = 0.92) and remained significant when restricting the analysis to only individuals of European descent (odds ratio, 2.86; 95% CI, 1.81 to 4.52; P = 5.3 × 10−6). These results reveal a substantial role of missense variation in severe schizophrenia with a burden of rare, damaging missense variants concentrated in intolerant subregions of the most missense intolerant genes.
Fig. 2.
The burden of rare variants in missense and loss-of-function intolerant genes in 112 individuals with severe, extremely treatment-resistant schizophrenia and 4,929 healthy controls. (A) Missense intolerant genes were those with missense Z > 3.09 and (B) loss-of-function intolerant genes were those with probability of being loss-of-function intolerant (pLI) score > 0.9 as per gnomAD. Unadjusted two-sided Cochran–Mantel–Haenszel exact P values, effect sizes between severe, extremely treatment-resistant schizophrenia and healthy controls represented as odds ratios, and horizontal bars indicating 95% confidence intervals are shown. LoF: Loss-of-Function variants, Miss Dmg Missense Tolerance Ratio (MTR): Damaging missense variants with REVEL score > 0.5 and missense tolerance ratio false discovery rate < 0.1, Miss Dmg: Damaging missense variants with REVEL score > 0.5, Miss Ben: Benign missense variants with REVEL score < 0.15, Syn: Synonymous variants. *False discovery rate < 0.1.
We next assessed whether individuals with severe, extremely treatment-resistant schizophrenia have a greater burden of loss-of-function variation in loss-of-function intolerant genes compared to controls. We found a significant enrichment of qualifying loss-of-function variants in loss-of-function intolerant genes (pLI > 0.9, odds ratio, 1.91; 95% CI, 1.39 to 2.63; P = 7.8 × 10−5; FDR P = 1.1 × 10−3). Neither synonymous variants (odds ratio, 0.98; 95% CI, 0.89 to 1.07; P = 0.67) nor benign missense variants (odds ratio, 0.98; 95% CI, 0.89 to 1.09, P = 0.79) were enriched in individuals with severe, extremely treatment-resistant schizophrenia for this set of genes. The loss-of-function enrichment remained significant when restricting the analysis to only individuals of European descent (odds ratio, 2.26; 95% CI, 1.52 to 3.36; P = 6.6 × 10−5). Moreover, there was no enrichment of qualifying missense or loss-of-function variants in genes tolerant to missense (missense Z < 1) and loss-of-function variation (pLI < 0.001) and no genome-wide enrichment of synonymous variation between severe, extremely treatment-resistant schizophrenia and controls (SI Appendix, Figs. S3 and S4). Finally, we observed nearly the same qualifying loss-of-function (odds ratio, 1.97; 95% CI, 1.42 to 2.75; P = 6.1 × 10−5) and missense variant (odds ratio, 2.59; 95% CI, 1.77 to 3.78; P = 5.4 × 10−7) burden in our separate genome-only analysis without an increase in benign variation or signal in tolerant genes (SI Appendix, Figs. S5–S9). The lack of enrichment of benign variation either genome-wide or in the intolerant gene sets combined with the rare variant burden being restricted to deleterious variants only in the most intolerant genes (and still present in the European- and genome-only analyses) indicates that the enrichment observed in individuals with severe, extremely treatment-resistant schizophrenia was not due to technical artifact or population substructure. Altogether, these results demonstrate a significant burden of rare variants in severe, extremely treatment-resistant schizophrenia with 48.2% of cases carrying either a qualifying missense or loss-of-function variant in intolerant genes, nearly twice the frequency (25.4%) observed in controls (odds ratio, 2.74; 95% CI, 1.85 to 4.06; P = 2.9 × 10−7).
Mendelian Disease Gene Analysis and Molecular Diagnostic Yield in Severe, Extremely Treatment-Resistant Schizophrenia.
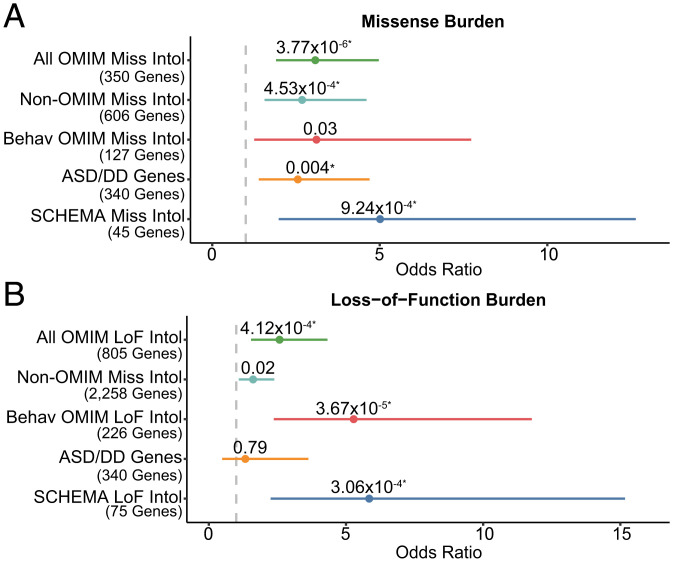
As there are over 60 Mendelian diseases that are known to present with psychosis (26, 27), we aimed to test whether severe, extremely treatment-resistant schizophrenia might represent psychiatric manifestations of known Mendelian diseases. To test this hypothesis at the cohort level, we compared the burden of qualifying missense and loss-of-function variants in the missense (n = 350 genes) and loss-of-function intolerant (n = 805 genes) subsets of 3,509 Mendelian disease genes contained in the Online Mendelian Inheritance in Man (OMIM) database (Fig. 1B and SI Appendix). Individuals with severe, extremely treatment-resistant schizophrenia had a greater burden of both qualifying missense (odds ratio, 3.07; 95% CI, 1.9 to 4.87; P = 3.77 × 10−6, FDR P = 5.3 × 10−5) and loss-of-function variants (odds ratio, 2.58; 95% CI, 1.54 to 4.32; P = 4.1 × 10−4, FDR P = 1.9 × 10−3) in intolerant Mendelian disease genes than controls (Fig. 3 A and B). We then evaluated whether this signal was strengthened in the genes whose OMIM phenotypic entry included annotated behavioral manifestations (e.g., “aggression” or “hallucinations”). The signal for loss-of-function but not missense variants were strengthened by restricting to behaviorally annotated genes, though we were likely underpowered to detect more subtle effects (Fig. 3 A and B). The enrichment in OMIM genes overall indicates that genes involved in Mendelian disorders are associated with risk for severe, extremely treatment-resistant schizophrenia. However even excluding OMIM genes, individuals with severe, extremely treatment-resistant schizophrenia still had more rare variants in missense intolerant genes and loss-of-function intolerant genes than controls (Fig. 3 A and B), indicating that there are likely schizophrenia-associated genes that have not yet been linked to Mendelian diseases.
Fig. 3.
Rare variant burden analyses in schizophrenia relevant gene sets. Enrichment of qualifying missense (A) and loss-of-function variants (B) across gene sets of relevance to schizophrenia in individuals with severe, extremely treatment-resistant schizophrenia compared to healthy controls. Unadjusted exact two-sided Cochran–Mantel–Haenszel P values, effect sizes between severe, extremely treatment-resistant schizophrenia and healthy controls represented as odds ratios, and horizontal bars indicating 95% CIs are shown. Gene sets are described in detail in Fig. 1 and SI Appendix. All OMIM: All genes associated with Mendelian disorders in the OMIM database. Non-OMIM: Missense and loss-of-function intolerant genes without a known disease association in OMIM. Behav OMIM: The subset of OMIM genes with clinical phenotype annotations including “behavioral” manifestations. SCHEMA Miss Intol: Genes with prior evidence of enrichment for missense variation in the schizophrenia exome meta-analysis (SCHEMA) study of schizophrenia. SCHEMA LoF Intol: Genes with prior evidence of enrichment for loss-of-function variation in SCHEMA. ASD/DD: combined gene set of 102 genome-wide significant genes associated with autism spectrum disorder and 299 genome-wide significant genes associated with developmental delay. All gene sets ending with “Miss Intol” are restricted to include only those overlapping intolerant genes with a missense Z > 3.09 and those ending in “LoF Intol” overlapping genes with probability of loss-of-function intolerance (pLI) score > 0.9. *FDR < 0.1.
To evaluate the prevalence of Mendelian disorders in severe, extremely treatment-resistant schizophrenia at the individual level, we conducted a clinical genetic diagnostic evaluation of each severe, extremely treatment-resistant schizophrenia genome using the American College of Medical Genetics (ACMG) pathogenicity criteria (28). Molecular diagnostic evaluation of individuals with severe, extremely treatment-resistant schizophrenia revealed two ACMG-defined “likely pathogenic” variants related to a neurodevelopmental or neurologic condition. One participant had a loss-of-function variant in FOXP2, which causes childhood apraxia of speech (29). Of note, FOXP2 is a genome-wide significant autism spectrum disorder risk gene (7) and shows enrichment in the online SCHEMA database with four cases and zero controls with high-confidence loss-of-function variants (5). The second likely pathogenic variant is a loss-of-function variant in WBP11, which has been shown to be an incompletely penetrant cause of vertebral, cardiac, tracheoesophageal, renal, and limb defects and neuropsychiatric manifestations such as attention deficit hyperactivity disorder (30). There is preliminary evidence of enrichment of WBP11 loss-of-function (one case, one control variant, odds ratio: 4.01) and damaging missense variants (two cases, one control variant, odds ratio: 8.03) in SCHEMA. The low diagnostic yield (1.8%) in severe, extremely treatment-resistant schizophrenia is consistent with a prior diagnostic sequencing study of typical schizophrenia and significantly lower than the 31% molecular diagnostic rate for other isolated neurodevelopmental disorders (31).
Given the increased burden of rare variants in OMIM genes in our cohort, one might expect a higher rate of ACMG “pathogenic” diagnoses at the individual level. Closer inspection revealed that loss-of-function variants across OMIM genes in our cohort tended to occur in a heterozygous state, whereas the inheritance model for the Mendelian condition was recessive (SI Appendix, Tables S5–S7). For example, one individual with severe, extremely treatment-resistant schizophrenia had a heterozygous loss-of-function variant in DIS3L2, which in a homozygous loss-of-function state, causes Perlman syndrome, a severe neurodevelopmental disorder with multiple anomalies and autosomal recessive inheritance (32). Interestingly, there is burgeoning evidence that individuals with neuropsychiatric disorders, particularly those with treatment resistance, are enriched for pathogenic Mendelian variants, even in heterozygous “carrier” states (33). This finding is also consistent with recent research that some carriers of rare variants in Mendelian disease genes who were previously thought to be “unaffected” actually present with milder forms of Mendelian disorders (34). These results demonstrate that clinical diagnostic sequencing currently has a low yield for schizophrenia. However, given the substantial burden of rare variants in Mendelian disease genes, these data point toward isolated neuropsychiatric disorders possibly representing an “intermediate” phenotype between health and severe Mendelian disorders. Furthermore, these data combined with the excess of rare variants in intolerant genes suggest a future role for clinical sequencing in severe, extremely treatment-resistant schizophrenia as the number of genome-wide significant schizophrenia–associated genes increases.
Autism Spectrum Disorder and Developmental Delay Genes in Severe, Extremely Treatment-Resistant Schizophrenia.
Because variants in genes conferring risk to autism spectrum disorders and developmental delay have been associated with schizophrenia (5, 35), we tested for a burden of rare variants in a combined gene set of genome-wide significant autism spectrum disorder (7) and developmental delay genes (6). We found no substantial enrichment of qualifying missense (odds ratio, 2.55; 95% CI, 1.39 to 4.69; P = 3.8 × 10−3, FDR P = 0.01) and loss-of-function variants (odds ratio, 1.33; 95% CI, 0.49 to 3.63; P = 0.79, FDR P = 1) in this gene set compared to intolerant genes (Fig. 3 A and B), though we were likely underpowered to detect a smaller enrichment.
Rare Variant Burden in Typical Schizophrenia Risk Genes.
To ascertain whether damaging rare variants carried by individuals with severe, extremely treatment-resistant schizophrenia are relevant to the broader schizophrenia population, we tested for enrichment in the top missense and loss-of-function enriched genes from the SCHEMA consortium, the largest schizophrenia exome sequencing study to date (5). There was a significant enrichment in qualifying missense (odds ratio, 3.67; 95% CI, 1.70 to 7.93; P = 1.4 × 10−3; FDR P = 0.02) but not loss-of-function variants (odds ratio, 1.95; 95% CI, 0.85 to 4.46; P = 0.19; FDR P = 1). Restricting the schizophrenia-associated missense and loss-of-function gene sets to intolerant genes, however, yielded a significant enrichment in both qualifying missense (odds ratio, 5.01, 95% CI, 1.98 to 12.64; P = 9.2 × 10−4, FDR P = 4.3 × 10−3) and loss-of-function variants (odds ratio, 5.85; 95% CI, 2.25 to 15.17; P = 3.06 × 10−4, FDR P = 1.9 × 10−3) (Fig. 3 A and B). In total, 8.9% of individuals with severe, extremely treatment-resistant schizophrenia carry either a qualifying missense or loss-of-function variant in one of these intolerant schizophrenia-associated genes compared to 1.6% controls (odds ratio, 5.82; 95% CI, 3.00 to 11.28; P = 2.6 × 10−8). This observation confirms both the relevance of risk factors in the severe, extremely treatment-resistant schizophrenia population to broader risk of schizophrenia and strongly supports the conclusion that the elevated rate of damaging variants in intolerant genes reported here represent valid risk alleles. Notably, even after removing these SCHEMA genes from our analysis, there remains a strong enrichment of both qualifying missense (odds ratio, 2.68; 95% CI, 1.82 to 3.96; P = 5.8 × 10−7) and loss-of-function (odds ratio, 1.74; 95% CI, 1.24 to 2.44; P = 1.5 × 10−3) among intolerant genes. These results imply that the majority of severe, extremely treatment-resistant schizophrenia risk genes are not among the top risk genes for typical schizophrenia and represent risk genes that are either specific to severe, extremely treatment-resistant schizophrenia or not yet discovered in typical schizophrenia.
Rare Variant Burden Compared to Individuals with Typical Schizophrenia.
We hypothesized that individuals with severe, extremely treatment-resistant schizophrenia would have a higher burden of rare damaging variants in intolerant genes compared to individuals with more typical forms of schizophrenia. Thus, we next compared the burden of rare variants in individuals with severe, extremely treatment-resistant schizophrenia to a cohort of 218 typically ascertained individuals with schizophrenia who had whole-genome or exome sequencing and the same set of controls from the primary analysis (see Online Methods for ascertainment, sequencing, and quality control details). Specifically, we compared the burden of qualifying rare variants per sample across missense, loss-of-function, and SCHEMA missense and loss-of-function intolerant gene sets as previously described.
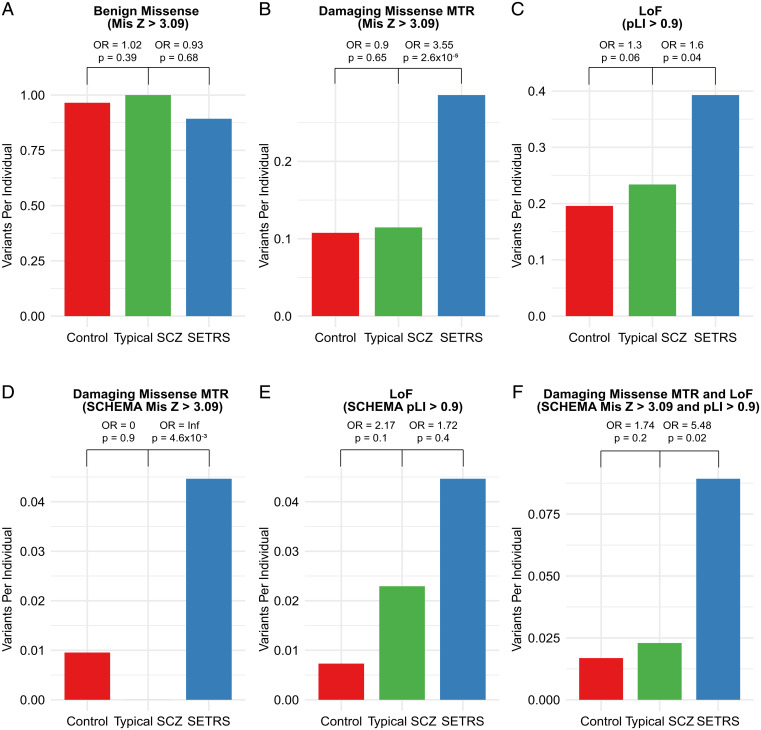
As expected, individuals with typical schizophrenia had an increased burden of loss-of-function variants (odds ratio, 1.30) compared to controls, largely consistent with the recent results from SCHEMA (odds ratio, 1.26) (Fig. 4). The typical schizophrenia cohort did not show an increased burden of regionally intolerant missense variants (odds ratio, 0.90), though this was likely due to the small sample size for a typical schizophrenia cohort and a smaller effect size for most qualifying missense variants in SCHEMA (odds ratio, 1.06) (5). Compared to individuals with typical schizophrenia, individuals with severe, extremely treatment-resistant schizophrenia have a substantially increased burden of qualifying missense and loss-of-function variants in intolerant genes (Fig. 4). Altogether, 48.2% of individuals with severe, extremely treatment-resistant schizophrenia carried at least one qualifying missense or loss-of-function variant in intolerant genes compared to 29.8% of individuals with typical schizophrenia (odds ratio, 2.18; 95% CI, 1.33 to 3.60; P = 1.6 × 10−3) and 25.4% of controls (odds ratio, 2.74; 95% CI, 1.85 to 4.06; P = 2.9 × 10−7).
Fig. 4.
Overall burden of rare variants in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia and control individuals. (A) No difference observed between severe, extremely treatment-resistant schizophrenia, typical schizophrenia, and controls for benign missense variants in missense intolerant genes (Mis Z > 3.09, n = 956 genes). (B) Increased burden of damaging missense MTR variants in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia in missense intolerant genes (Mis Z > 3.09, n = 956 genes). (C) Increased burden of loss-of-function variants in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia in loss-of-function (LoF) intolerant genes (pLI > 0.9, n = 3,063). (D) Increased burden of damaging missense MTR variants in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia in missense intolerant schizophrenia exome meta-analysis (SCHEMA) genes (n = 45 genes). (E) Nonsignificant increase in burden of loss-of-function variants in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia in loss-of-function intolerant SCHEMA genes (n = 75 genes). (F) Increased burden of damaging missense MTR and loss-of-function in severe, extremely treatment-resistant schizophrenia compared to typical schizophrenia in intolerant SCHEMA genes (n = 45 for missense intolerant and n = 75 for loss-of-function intolerant SCHEMA genes). Odds ratios (OR) and P values shown are based on a comparison of typical schizophrenia individuals to controls and severe, extremely treatment-resistant schizophrenia individuals to typical schizophrenia using one-sided Cochran–Mantel–Haenszel test. Benign missense: missense variants with REVEL score < 0.15, Missense damaging MTR: missense variants with REVEL score > 0.5 and missense tolerance ratio false discovery rate < 0.1. SCZ: schizophrenia, SETRS: severe, extremely treatment-resistant schizophrenia.
Notably, this increased burden was also present in the SCHEMA genes that are intolerant to functional variation. Individuals with typical schizophrenia showed an increase in the burden of loss-of-function variants in SCHEMA loss-of-function intolerant genes compared to controls but did not have any qualifying missense variants in SCHEMA missense intolerant genes (Fig. 4). The absence of qualifying missense variants in typical schizophrenia in these genes is likely due to the small size of this sample and the gene set (n = 45 genes) in addition to the stringent deleteriousness and regional intolerance cutoffs.
In comparison, individuals with severe, extremely treatment-resistant schizophrenia have nearly a fourfold increase in the burden of qualifying missense and loss-of-function variants in these intolerant genes previously implicated in schizophrenia risk. Specifically, 8.9% of individuals with severe, extremely treatment-resistant schizophrenia carry a qualifying missense or loss-of-function variant compared to 2.3% of individuals with typical schizophrenia (odds ratio, 5.48; 95% CI, 1.52 to 19.74; P = 0.02). There was no enrichment of benign missense variants between severe, extremely treatment-resistant schizophrenia and typical schizophrenia (odds ratio 0.91). Moreover, there was no evidence of genome-wide synonymous inflation between individuals with severe, extremely treatment-resistant schizophrenia and typical schizophrenia (Mann–Whitney U P = 0.61) and between the typical schizophrenia and control cohorts (Mann–Whitney U P = 0.35) (SI Appendix, Figs. S4 and S9). Lastly, the results of this analysis were replicated in the separate genome-only analysis (SI Appendix, Figs. S11 and S12), further indicating that the enrichment of qualifying variants in severe, extremely treatment-resistant schizophrenia was not due to technical artifact or population substructure.
Gene-Based Rare Variant Collapsing Analysis.
No individual gene reached genome-wide significance in a gene-level burden test (SI Appendix, Fig. S4 and Dataset S1). The top genes from the collapsing analysis (SI Appendix, Table S8) included ZDHHC5 (odds ratio 29.23, 95% CI, 2.27 to 280.56; P = 5.5 × 10−3), which was one of the closest genes to genome-wide significant locus in a schizophrenia genome-wide association study (4) and one of the top five genes from a recent meta-analysis of rare variants in over 14,000 schizophrenia exomes (36). Other top genes from the collapsing analysis included HDAC6 (odds ratio 65.01, 95% CI, 3.39 to 3,721.66; P = 2.6 × 10−3), which has been shown to play an important role in synaptic plasticity and memory (37) and KCNJ2 (odds ratio 65.22, 95% CI, 3.37 to 3,757.49; P = 2.6 × 10−3), which has been associated with Andersen–Tawil syndrome and neuropsychiatric illness (38). While these results are encouraging, larger samples of individuals with severe, extremely treatment-resistant schizophrenia will be required to identify risk genes reaching genome-wide significance.
Discussion
Using an extreme phenotype sampling approach, we identified the strongest burden of rare missense and loss-of-function variation in schizophrenia patients reported to date. These results have several implications for our understanding of the genetic basis of schizophrenia and future studies in psychiatric genetics. First, this study provides a proof-of-principle of the utility of extreme phenotype case identification and sequencing in psychiatric genetics. Compared to the burden of loss-of-function (odds ratio, 1.30) and damaging missense variants (odds ratio, 0.90) in intolerant genes in our typical schizophrenia cohort, we identified a significantly higher burden of loss-of-function (odds ratio, 1.91) and damaging missense (odds ratio, 2.90) variants in individuals with severe, extremely treatment-resistant schizophrenia compared to controls. Moreover, these damaging rare variants occur in high frequency of individuals with severe, extremely treatment-resistant schizophrenia, with 48.2% carrying at least one qualifying missense or loss-of-function variant in an intolerant single gene compared to 29.8% of individuals with typical schizophrenia and 25.4% of controls. These findings have implications for future gene discovery efforts in schizophrenia and underscore the power of an extreme phenotype approach as we detected a larger and more statistically robust rare variant signal in severe, extremely treatment-resistant schizophrenia despite the typical schizophrenia cohort being nearly twice the sample size.
While the current molecular diagnostic yield based on ACMG criteria was low, the magnitude of the difference between cases and controls in qualifying variants (48.4 versus 25.4%) implies that, as schizophrenia risk genes continue to be identified, disease-associated single-gene variants may be found in ∼20% of individuals with severe, extremely treatment-resistant schizophrenia. This diagnostic rate would be of clinical relevance for an adult-onset disorder like schizophrenia for two reasons. First, employing severe, extremely treatment-resistant schizophrenia criteria could allow for stratification of patients and provide clear indications for sequencing, thus reducing the sample sizes needed for future gene discovery. Second, as early identification and intervention have been shown to modify the course of illness in schizophrenia (39), detection of genomic risk factors for severe, extremely treatment-resistant schizophrenia could identify individuals at risk for severe disease and in need of more intensive or alternative treatments than traditional antipsychotic medications.
Second, our data provide a more complete understanding of the genetic architecture of the schizophrenia spectrum, which now seems to resemble that of autism spectrum disorder and developmental delay: more mildly affected individuals tend to have a stronger polygenic contribution of mostly common variants, whereas severely affected individuals have a larger burden of rare variants in intolerant genes (40–42). Notably, this finding suggests that there may have been an ascertainment bias that led to a relative underrepresentation or even absence of the most severely affected individuals in schizophrenia genetics studies. For example, in childhood-onset disorders such as developmental delay, autism spectrum disorder, and early-onset epilepsies, parents of severely affected individuals are more likely to refer their children for genetic evaluation in research studies or clinical sequencing. The opposite is likely true for schizophrenia. Because it is largely an adult-onset disorder, the most severely affected individuals often do not have legal guardians and are also less likely to participate in research studies precisely due to the severity and nature of their psychotic and cognitive symptoms.
Of note, the increased damaging rare variant signal observed in more severe forms of autism is thought to be driven predominantly by comorbid intellectual disability (19, 42, 43) with little evidence that common measures of autism spectrum disorder severity increase the yield of rare damaging variants (44). The lack of pathogenic variants in known intellectual disability/developmental delay genes combined with our thorough investigation of premorbid intellectual functioning suggests that our rare variant signal is not being driven by undetected intellectual disability. While we cannot rule out the effects of milder premorbid cognitive impairment, prior research shows a smaller role for rare variation in mild intellectual disability and borderline intellectual functioning (41). Long-term hospitalization represents a surrogate marker of disease severity in severe, extremely treatment-resistant schizophrenia that captures a currently undefined component of the phenotype driving the rare variant signal. One potential explanation could be the comparatively early onset of severe, treatment-resistant psychosis and functional decline in individuals with severe, extremely treatment-resistant schizophrenia (mean age of onset 18.3 y) in addition to prodromal social and behavioral abnormalities predating the onset of psychosis. Indeed, the substantially reduced fecundity of the severe, extremely treatment-resistant schizophrenia cohort (only 15% had children) is consistent with the effects of negative selection against high-impact rare variants in intolerant genes and implies that these variants are either of recent origin or de novo. Larger samples will be necessary to characterize what components of the severe, extremely treatment-resistant schizophrenia phenotype are most likely to be driving the rare variant signal observed.
Despite similarities in their genetic architecture, we found that genes and variants conferring risk for severe, extremely treatment-resistant schizophrenia do not significantly overlap with known autism spectrum disorder and developmental delay risk genes, though we were likely underpowered to detect a more subtle enrichment. Our results are consistent with findings from the SCHEMA consortium study (5) in which only 3 and 10 of the top 32 statistically significant risk genes (defined using the more relaxed FDR < 0.05 threshold for this comparison) conferred risk for autism spectrum disorder and developmental delay, respectively. In contrast to pathogenic copy number variants such as the 22q11.2 deletion syndrome, which show pleiotropic effects and increased risk for all three disorders, many single-gene risk variants preferentially confer risk for schizophrenia. This provides an opportunity to study genes such as KDM6B that broadly increase risk for neurodevelopmental disorders and contrast them with genes like CUL1, which are more specific to schizophrenia.
Third, one advantage of identifying large effect coding variants is that they are more tractable for biological studies than modeling the cumulative impact of hundreds of common variants. Studying the functional consequences of large effect variants in model systems will help provide insight into the neurobiology of schizophrenia. Furthermore, we found an unusually high burden of damaging missense variation in severe schizophrenia compared to our typical schizophrenia cohort and previously reported rates of damaging missense variation in schizophrenia. As additional studies confirm and prioritize the most promising candidates, it would be important to investigate the functional consequences of these missense variants and whether some may represent toxic gain-of-function effects that confer risk for a uniquely severe form of schizophrenia.
While severe, extremely treatment-resistant schizophrenia’s genetics overlap with typical schizophrenia-associated genes and biological pathways, most of the intolerant genes carrying qualifying variants in this study have neither been previously linked to schizophrenia nor show enrichment in the SCHEMA database. In the future, studying these severe, extremely treatment-resistant schizophrenia-specific genes and variants may provide insight into the genetic and molecular mechanisms of treatment resistance, increased symptom severity, and the progressive course of illness seen in individuals with severe, extremely treatment-resistant schizophrenia. Additionally, studying disease-associated missense variants would allow for a careful dissection of disease mechanism through altered protein structure and function, providing a unique opportunity to unravel the molecular pathophysiology of schizophrenia.
Given that roughly 30% of individuals with schizophrenia do not respond to antipsychotic treatment (45), understanding the genetic mechanisms of treatment resistance and poor prognosis would be of substantial benefit to all individuals with schizophrenia. Lastly, for all patients with severe, extremely treatment-resistant schizophrenia, traditional antipsychotic treatments are simply ineffective. The high rate of identifiable rare variants (many in druggable genes) in severe, extremely treatment-resistant schizophrenia provides an opportunity to functionally characterize these variants in model systems, paving the way for the development of mechanistically targeted therapeutics for patients in dire need of novel treatments. Altogether, these results suggest that extreme phenotype case identification could augment gene discovery efforts in schizophrenia and other psychiatric disorders and offers potential clinical utility.
Methods
Participant Recruitment.
Participants with severe, extremely treatment-resistant schizophrenia in this study were recruited between December 2017 and July 2019 from four New York state–funded inpatient facilities: Pilgrim Psychiatric Center, Manhattan Psychiatric Center, Creedmoor Psychiatric Center, and Rockland Psychiatric Center. These hospitals provide care for chronically and severely ill patients, most of whom are diagnosed with schizophrenia spectrum disorders. This study was approved by the New York State Psychiatric Institute/Columbia University Medical Center Institutional Review Board under protocol No. 7312. Informed consent was obtained from all participants or their legally authorized representatives if the participant lacked capacity to consent for themselves.
A total of 132 of the typical schizophrenia participants had whole-genome sequencing completed as part of the Genomic Psychiatry Cohort with data downloaded from dbGaP accession: phs001020.v2.p1. Recruitment details have been previously described in detail (46). A total of 68 participants were enrolled as part of an ongoing whole-genome sequencing in psychiatric genetics at Massachusetts General and McLean Hospitals in Boston. A total of 30 of the typical schizophrenia participants were sequenced as part of previously published study, which describes recruitment and ascertainment details (47).
Ancestral Clustering.
To account for ancestral heterogeneity in each of the cohorts, we used the Louvain method for community detection on the first six principal components (PCs) generated using FLASHPCA version 2.0 to identify genetically determined clusters of geographic ancestry as previously described (18, 48). We identified probabilities of geographic ancestry (African, East Asian, European, Latino, Middle Eastern, and South Asian) using a neural network pretrained on samples with known geographic ancestry. We assigned geographic ancestry to participants with a probability greater than 95%, and those who did not reach this cutoff were labeled “Admixed.” We then performed Uniform Manifold Approximation and Projection on the first six PCs to visualize the intersection of cluster membership and geographic ancestry assigned by the neural network (SI Appendix, Figs. S1, S2, S5, and S6). Clusters with at least eight cases were retained, and four clusters were used in the severe, extremely treatment-resistant schizophrenia and typical schizophrenia analyses. Each case-control ancestral cluster was analyzed separately, and an exact two-sided Cochran–Mantel–Haenszel test was used to evaluate the rare variant burden across all clusters in both the single-gene and gene-set burden analyses.
Gene-Set Burden Analyses.
We compared the burden of synonymous, benign missense, damaging missense, qualifying missense, and qualifying loss-of-function variants between severe, extremely treatment-resistant schizophrenia cases, typical schizophrenia, and controls across gene sets of interest (see SI Appendix for details regarding gene set curation). Specifically, we tallied the total number of qualifying variants in cases and controls in each cluster for a given gene set and assessed the significance of this difference across all clusters using a two-sided Cochran–Mantel–Haenszel test with effect sizes represented as odds ratios. The full list of qualifying missense and loss-of-function variants in intolerant genes are shown in SI Appendix, Table S9.
In missense and loss-of-function intolerant genes and intolerant SCHEMA genes, we also calculated the odds of having at least one qualifying missense or loss-of-function variant. Specifically, we created an indicator variable for cases and controls if they had either a qualifying missense or loss-of-function variant (in their respective intolerant gene set) and assessed the significance of this difference using a two-sided Cochran–Mantel–Haenszel test with effects represented as odds ratios. We accounted for multiple comparisons using the Benjamini–Hochberg false discovery rate. Specifically, we accounted for each test of qualifying missense and loss-of-function variants across the fourteen gene sets shown in Fig. 1B. All analyses were performed in RStudio (version 1.2.1335).
Gene-Based Rare Variant Collapsing Analysis.
We performed a gene-based collapsing analysis as previously described (49, 50). Briefly, we used, rare-variant collapsing analysis to aggregate the full catalog of qualifying loss-of-function and missense variants per gene. Qualifying loss-of-function variants included stop gain, frameshift, or canonical splice donor/acceptor variants that passed quality control. Qualifying missense variants for gene-based collapsing were those that passed quality control and rarity filtering and were damaging (REVEL > 0.5). We did not include regional missense intolerance in our gene-based collapsing as this would leave too few variants for analysis. We compared the case-control burden of qualifying variants for each gene by assigning an indicator variable to each participant with a value of 1 if a qualifying variant was present and 0 if no qualifying variant was present. These results are summarized in a gene-by-participant matrix for each cluster. We then extracted the number of affected cases and controls with and without a qualifying variant on a per gene basis and combined the results across clusters using a two-sided Cochran–Mantel–Haenszel test while controlling for cluster membership.
We conducted single-gene–based collapsing analysis with the following models using a minor allele frequency cutoff < 1 × 10−4: damaging missense (REVEL > 0.5), loss-of-function, and combined damaging missense and loss-of-function (SI Appendix, Fig. S4). Furthermore, for both the severe, extremely treatment-resistant schizophrenia and typical schizophrenia cohorts, we used a collapsing analysis that considered only synonymous variation as a neutral model to estimate the degree of inflation due to population substructure or technical artifact. In both the severe, extremely treatment-resistant schizophrenia and typical schizophrenia cohorts (and in the genome-only analysis), there was no evidence of genome-wide synonymous variant inflation when compared to controls (SI Appendix, Figs. S4, S9, S10, and S12). The top 10 ranked genes from each model can be found in SI Appendix (Table S8), and the complete results can be found in Dataset S1. We used a Bonferroni multiplicity-adjusted significance threshold of P < 8.9 × 10−7 (0.05 / [18,650 genes × 3 nonsynonymous models]) for the single-gene collapsing analysis.
Mendelian Disease Gene Molecular Diagnostic Evaluation.
Briefly, our diagnostic approach prioritizes variants that have previously been reported as pathogenic/likely pathogenic affect the same amino acid as a known pathogenic variant or are loss-of-function in a known haploinsufficient gene. Our pipeline incorporates curated data from ClinVar, ClinGen, Human Gene Mutation Database, and OMIM to annotate all variants previously reported as pathogenic/likely pathogenic (see SI Appendix and Tables S5 and S6 for details).
Supplementary Material
Acknowledgments
We thank the participants and their families for their time, effort, and participation in the study. We thank the staff at Pilgrim Psychiatric Center, Creedmoor Psychiatric Center, Manhattan Psychiatric Center, and Rockland Psychiatric Center for their assistance and effort in the study. We acknowledge the National Institute of Mental Health (NIMH) for their funding contribution. We acknowledge Dr. Slavé Petrovski, Dr. Gundula Povysil, and Dr. Tess Pottinger for thoughtful discussions regarding the preparation of the manuscript. Funding: The collection and sequencing of the severe, extremely treatment-resistant schizophrenia cohort was funded by the NIMH; (T32MH018870), (K23MH121669), Chapman Perelman Foundation (GT006595), Pisetsky Foundation, and Leon Levy Foundation. J.E.M. is funded by the NIH (TL1TR001875). The collection and sequencing of the typical schizophrenia cohort was funded by NIMH (5U01MH105670); (5U01MH401136); (R01MH049487); (R01MH31340); (R01MH071523); the Brain and Behavior Foundation, the Sidney R. Baer, Jr. Foundation, the Ellison Foundation, Anonymous Foundation, the Carmela and Menachem Abraham Fund, Team Daniel, and the Thostenson Family. The Genomic Psychiatry Cohort sample ascertainment was supported by NIH Grant Nos. MH085548 and MH085542 and the Stanley Center for Psychiatric Research at the Broad Institute. The whole-genome sequencing of the Genomic Psychiatry Cohort study was supported by NHGRI Grant No. 5U54HG003067, a philanthropic gift to the Stanley Center for Psychiatric Research, and NIH Grant Nos. MH086873 and MH085548, as well as, U01MH105653 (Boehnke), U01MH105573 (Patos), and U01MH105641(McCarroll). Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Competing interest statement: D.B.G. is a founder of and holds equity in Q State Biosciences and Praxis Therapeutics; holds equity in Apostle Inc., and serves as a consultant to AstraZeneca, Gilead Sciences, GoldFinch Bio and Gossamer Bio. J.A.L. does not accept any personal financial remuneration for consulting, speaking, or research activities from any pharmaceutical, biotechnology, or medical device companies. He receives funding and medication supplies for investigator-initiated research from Denovo, Taisho, and Cerevel, and company sponsored phase II, III, and IV studies from Alkermes, Sunovion, and Boehringer Ingelheim, which does not contribute to his compensation. He is a consultant or advisory board member of Intracellular Therapies, Takeda, Karuna, Pear Therapeutics, Systems-1, and Psychogenics for which he receives no remuneration. He is a paid consultant for Signant Health, a clinical research technology and services organization, and holds a patent from Repligen that yields no royalties. R.S.D. is a paid consultant for AstraZeneca. X.W. is a cofounder and stock owner at Waypoint Bio. The other authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112560118/-/DCSupplemental.
Data Availability
Summary statistics for case and control qualifying variants are provided in Dataset S1. All code for the Analysis Tool for Annotated Variants (ATAV) is publicly available: https://github.com/nickzren/atav.
References
- 1.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1545–1602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan P. F., Kendler K. S., Neale M. C., Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Sekar A., et al. , Schizophrenia Working Group of the Psychiatric Genomics Consortium, Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh T., et al. , Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv [Preprint] (2020). 10.1101/2020.09.18.20192815. Accessed 14 October 2020. [DOI]
- 6.Kaplanis J., et al. , Deciphering Developmental Disorders Study, Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satterstrom F. K., et al. , Autism Sequencing Consortium; iPSYCH-Broad Consortium, Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn K., et al. , High rate of disease-related copy number variations in childhood onset schizophrenia. Mol. Psychiatry 19, 568–572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen A. S., et al. , Epi4K Consortium; Epilepsy Phenome/Genome Project, De novo mutations in epileptic encephalopathies. Nature 501, 217–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotowski I. K., et al. , A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 78, 410–422 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner T. N., et al. , Loss of δ-catenin function in severe autism. Nature 520, 51–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnett I. J., Lee S., Lin X., Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet. Epidemiol. 37, 142–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambalavanan A., et al. , De novo variants in sporadic cases of childhood onset schizophrenia. Eur. J. Hum. Genet. 24, 944–948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner E. J., et al. , Sex-biased reduction in reproductive success drives selective constraint on human genes. bioRxiv [Preprint] (2020). 10.1101/2020.05.26.116111. Accessed 10 January 2021. [DOI]
- 15.Gulsuner S., et al. , Genetics of schizophrenia in the South African Xhosa. Science 367, 569–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh T., et al. , INTERVAL Study; UK10K Consortium, The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 49, 1167–1173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.American Psychiatric Association, Diagnostic and Statistical Manual of Mental Disorders: DSM-5 (American Psychiatric Association, Washington, D.C., ed. 5, 2013. [Google Scholar]
- 18.Collaborative E.; Epi25 Collaborative. Electronic address: jm4279@cumc.columbia.edu; Epi25 Collaborative, Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals. Am. J. Hum. Genet. 108, 965–982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satterstrom F. K., et al. , iPSYCH-Broad Consortium, Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 22, 1961–1965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski K. J., et al. , Genome Aggregation Database Consortium, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halvorsen M., et al. , Exome sequencing in obsessive-compulsive disorder reveals a burden of rare damaging coding variants. Nat. Neurosci. 24, 1071–1076 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Ioannidis N. M., et al. , REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh R., Oak N., Plon S. E., Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 18, 225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silk M., Petrovski S., Ascher D. B., MTR-Viewer: Identifying regions within genes under purifying selection. Nucleic Acids Res. 47 (W1), W121–W126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummings B. B., et al. , Genome Aggregation Database Production Team; Genome Aggregation Database Consortium, Transcript expression-aware annotation improves rare variant interpretation. Nature 581, 452–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamin S., Lauterbach M. D., Stanislawski A. L., Congenital and acquired disorders presenting as psychosis in children and young adults. Child Adolesc. Psychiatr. Clin. N. Am. 22, 581–608 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Lauterbach M. D., Stanislawski-Zygaj A. L., Benjamin S., The differential diagnosis of childhood- and young adult-onset disorders that include psychosis. J. Neuropsychiatry Clin. Neurosci. 20, 409–418 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Richards S., et al. , ACMG Laboratory Quality Assurance Committee, Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDermot K. D., et al. , Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 76, 1074–1080 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin E. M. M. A., et al. , Heterozygous loss of WBP11 function causes multiple congenital defects in humans and mice. Hum. Mol. Genet. 29, 3662–3678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S., et al. , NDD Exome Scoping Review Work Group, Meta-analysis and multidisciplinary consensus statement: Exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 21, 2413–2421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astuti D., et al. , Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat. Genet. 44, 277–284 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Sriretnakumar V., Harripaul R., Vincent J. B., Kennedy J. L., So J., Enrichment of pathogenic variants in genes associated with inborn errors of metabolism in psychiatric populations. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 180, 46–54 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Bastarache L., et al. , Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science 359, 1233–1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genovese G., et al. , Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci. 19, 1433–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lescai F., et al. , Meta-analysis of Scandinavian schizophrenia exomes. bioRxiv [Preprint] (2019). 10.1101/836957. Accessed 10 December 2020. [DOI]
- 37.Perry S., Kiragasi B., Dickman D., Ray A., The role of histone deacetylase 6 in synaptic plasticity and memory. Cell Rep. 18, 1337–1345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan H. F., et al. , A novel neuropsychiatric phenotype of KCNJ2 mutation in one Taiwanese family with Andersen-Tawil syndrome. J. Hum. Genet. 55, 186–188 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Stone W. S., et al. , Association between the duration of untreated psychosis and selective cognitive performance in community-dwelling individuals with chronic untreated schizophrenia in Rural China. JAMA Psychiatry 77, 1116–1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson E. B., et al. , Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc. Natl. Acad. Sci. U.S.A. 111, 15161–15165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichenberg A., et al. , Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proc. Natl. Acad. Sci. U.S.A. 113, 1098–1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo H., et al. , University of Washington Center for Mendelian Genomics, Genome sequencing identifies multiple deleterious variants in autism patients with more severe phenotypes. Genet. Med. 21, 1611–1620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers S. M., et al. , Insufficient evidence for “autism-specific” genes. Am. J. Hum. Genet. 106, 587–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bishop S. L., et al. , Identification of developmental and behavioral markers associated with genetic abnormalities in autism spectrum disorder. Am. J. Psychiatry 174, 576–585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kane J., Honigfeld G., Singer J., Meltzer H., Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Arch. Gen. Psychiatry 45, 789–796 (1988). [DOI] [PubMed] [Google Scholar]
- 46.Pato M. T., et al. , Genomic Psychiatry Cohort Consortium, The genomic psychiatry cohort: Partners in discovery. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B, 306–312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Need A. C., et al. , Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. Am. J. Hum. Genet. 91, 303–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron-Christie S., et al. , Exome-based rare-variant analyses in CKD. J. Am. Soc. Nephrol. 30, 1109–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrovski S., et al. , An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 196, 82–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Povysil G., et al. , Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat. Rev. Genet. 20, 747–759 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for case and control qualifying variants are provided in Dataset S1. All code for the Analysis Tool for Annotated Variants (ATAV) is publicly available: https://github.com/nickzren/atav.