Significance
Metabolomics predictive models constructed from high-resolution magic angle spinning (HRMAS) proton magnetic resonance spectroscopy (1H MRS) data measured from 10 μL blood serum of human lung cancer patients collected prior to diagnosis reflect disease status and can be developed into a screening tool to triage patients with suspicious readings for advance imaging tests.
Keywords: magnetic resonance spectroscopy, high-resolution magic angle spinning, metabolomics, human lung cancer, blood serum
Abstract
The current high mortality of human lung cancer stems largely from the lack of feasible, early disease detection tools. An effective test with serum metabolomics predictive models able to suggest patients harboring disease could expedite triage patient to specialized imaging assessment. Here, using a training-validation-testing-cohort design, we establish our high-resolution magic angle spinning (HRMAS) magnetic resonance spectroscopy (MRS)-based metabolomics predictive models to indicate lung cancer presence and patient survival using serum samples collected prior to their disease diagnoses. Studied serum samples were collected from 79 patients before (within 5.0 y) and at lung cancer diagnosis. Disease predictive models were established by comparing serum metabolomic patterns between our training cohorts: patients with lung cancer at time of diagnosis, and matched healthy controls. These predictive models were then applied to evaluate serum samples of our validation and testing cohorts, all collected from patients before their lung cancer diagnosis. Our study found that the predictive model yielded values for prior-to-detection serum samples to be intermediate between values for patients at time of diagnosis and for healthy controls; these intermediate values significantly differed from both groups, with an F1 score = 0.628 for cancer prediction. Furthermore, values from metabolomics predictive model measured from prior-to-diagnosis sera could significantly predict 5-y survival for patients with localized disease.
Lung cancer is currently the leading cause of cancer death in humans (1). Early-stage lung cancer is mostly asymptomatic. This asymptomatic status contributes to delayed diagnoses and results in an overall 5-y survival rate of 19% over all stages. However, the detection of lung cancer at an early stage increases 5-y survival rates to 57% (1). This significant statistical difference demonstrates the importance of lung cancer detection at early and symptomatic stages for improving overall patient survival. The availability of treatments able to achieve improved outcomes with early detection has also increased. All these factors underscore the present urgency to advance early lung cancer detection through effective, widely applicable screening tests.
At present, low-dose spiral computerized tomography (LDCT) is considered the most sensitive imaging tool for detecting small and early-stage lung cancer lesions (2). However, alongside potential reluctance on the part of patients (e.g., views that screenings may be subjectively assessed based on, for example, smoking behavior), logistical and scientific concerns, including costs to the nation and potential radiation hazard, limit LDCT’s use in general populations for widespread screening (2, 3). Such reasonable caution supports the need for a simple, non- or minimally invasive screening test, one without harmful side effects and preferably portable, to provide an alert at suspicious signs of early malignancy. Such a tool could help activate triage to further testing, optimize LDCT’s targeted use for specialized imaging, as for other advanced technologies, and advance total treatment efficiency, cost-effectiveness, and outcomes across the effort to minimize lung cancer–associated mortality.
Molecular biology, cancer genomics, proteomics, and metabolomics, interrelated in their probing and identification of biological processes, offer distinct perspectives of these processes. Genomics, with measured genetic mutations, can suggest the possibility of a disease’s development during an individual’s lifetime, but proteomics and metabolomics measure ongoing bioactivity, with alterations in these bioactivities due to the presence of disease (4). In cancer, metabolomics detects oncological developments by interrogating measurable metabolic profiles from metabolic pathways through global metabolite variations (5).
Here, we report our human lung cancer predictive models for metabolomics screening that we established through study of lung cancer patient blood serum samples collected before and at the time of disease diagnosis and comparisons of patient results with those observed in serum samples obtained from healthy control subjects. Our results demonstrate the potential of magnetic resonance spectroscopy (MRS)-based serum metabolomics predictive models to indicate the presence of lung cancer in asymptomatic patients and its potential for development into an efficient, cost-effective screening tool.
Results
Study Populations.
The current institutional review boards (IRB)–approved study utilized a training-validation-testing-cohort design and included 183 blood serum samples from 79 patients with non–small cell lung cancer (NSCLC) and 79 healthy controls, matched according to gender, age, and smoking status (pack years when available). Written consents were obtained from all patients and control subjects, and detailed study demographics are listed in SI Appendix, Table S1.
Blood serum samples from 25 NSCLC patients (18F [female], 7M [male], Age = 66.9 ± 5.9 y), obtained at the time of their NSCLC diagnosis (AtDx), along with blood samples from 25 controls (Healthy, Age = 66.4 ± 5.9 y, Charlson Health Index [CHI] = 2.8 ± 1.4), were used as the training cohort.
Blood serum samples, collected at 0.5 to 5 y prior to diagnosis (PriorDx, 2.8 ± 1.4 y) from these same 25 NSCLC patients, were used as the validation cohort. Of note, our comparisons used samples obtained from the same patients; however, the validation cohort samples, collected at least half a year before any NSCLC diagnosis, were compared with training cohort samples that were obtained at the time of diagnosis. The comparison showed that validation cohort samples 1) formed a distinctively different group from those samples obtained at diagnosis, and 2) can serve as an ideal agent for NSCLC screening.
Additional blood serum samples, obtained up to 2 y prior to NSCLC diagnosis (0.6 ± 0.5 y) from 54 NSCLC patients (40F, 14M, Age = 64.0 ± 8.9 y), and from 54 matched healthy controls (Age = 62.7 ± 8.4 y, CHI = 1.9 ± 0.9), were used as the testing cohort.
Serum MRS metabolomics predictive models for NSCLC were constructed from the training cohort and validated and tested with the validation and testing cohorts.
Serum MRS of the Training and Validation Cohorts.
Proton MRS of sera (10 μL) without any pretreatment were measured by using our previously described high-resolution magic angle spinning (HRMAS) method (6, 7). Spectra presented as group averages with SDs for the training (patients and controls) and validation (patients only) cohorts are shown in Fig. 1A. From these spectra, 57 spectral regions of interest were determined [reference SI Appendix, Table S2 for region details and their potential contributing metabolites (8)]. Hereafter, we will refer to these spectral regions of interest as “spectral regions” or “regions,” and will discuss metabolites potentially contributing to these regions as needed. We elected to analyze data according to these regions, instead of the metabolites, due to the fact that each region may be contributed to by different metabolites and that each metabolite may also present in different spectral regions.
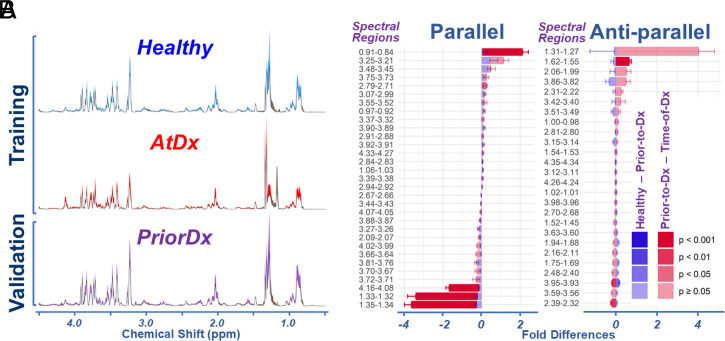
Fig. 1.
Serum HRMAS proton MRS measured from the training and validation cohorts. (A) Averaged spectra with SDs in color shades for each group in the cohorts. (B) Spectral fold differences between Healthy and PriorDx, and between PriorDx and AtDx, for 57 identified spectral regions are plotted as Parallel when the two differences have the same sign or Anti-parallel when they have different signs. The error bars indicate 0.5 SE. The P values of group differences are color coded in the figure.
For the three sample groups—Healthy, PriorDx, and AtDx—two differences were calculated: Healthy versus PriorDx, and PriorDx versus AtDx. The means of these differences, together with their respective SEs (presented as SE/2), are plotted as fold differences in Fig. 1B, with the respective P values of group differences color coded in the figure. If the “above two differences” of a particular spectral region are in the same direction (i.e., have the same sign [+ or −]), they are grouped as “parallel”; if they have the opposite sign, they are “anti-parallel.” A total of 12 regions (∼21%) presented significant differences between the PriorDx and the AtDx groups, with P values less than 0.05; fewer regions presented significant differences between the Healthy and the PriorDx groups.
Establishing Serum NSCLC MRS Metabolomics Predictive Models with the Training and Validation Cohorts.
The data presented are the results obtained from our analyses, conducted according to the described training-validation-testing-cohort design, of samples from the Healthy, PriorDx, and AtDx groups. Of note, the data analyses and model constructions were conducted solely on the training cohort to obtain coefficients and other parameters, means, and SDs. These parameters and coefficients were then applied onto spectral data of the validation and testing cohorts through the calculation processes determined by the training cohort.
To reduce metabolic data dimensions and establish predictive models, principal component analysis (PCA) was performed on the 57 spectral regions of the 50 serum samples in the training cohort. PCA identified 13 principal components (PCs) with >1.0 eigenvalues.
Training cohort P values from either paired or group analyses, as determined by t test or (where appropriate) the Kruskal–Wallis–Wilcoxon (KWW) test, were used to analyze the potential ability of the 13 PCs to differentiate Healthy from AtDx NSCLC patients. Fig. 2A presents these findings in the order of increasing P values. Discrimination between the training cohort’s Healthy and AtDx NSCLC groups was achieved using canonical analysis by varying the numbers of the first PCs in the P value order, beginning with the smallest P values. As would be expected, including more PCs improves the discrimination of these groups by canonical score, as seen by the reduction of resulting P values (Inset in Fig. 2A). We further analyzed the ranking order of the P values for both paired cases and groups and discovered three statuses that included significant P values for PC3 in both rankings, followed by mixed rankings of the next 6 PCs, all with P values smaller than 0.2; the remaining 6 PCs all had P values greater than 0.25 (Fig. 2B). Using a threshold of PC3, plus the 6 mixed-ranking PCs, the first 7 PCs with P values smaller than 0.2 were recruited into the canonical analysis to establish the predictive model. Resulting canonical discriminant scores of the predictive model for these training cohort cases are presented in Fig. 2C (reference SI Appendix, Table S2 for the contributions of each spectral region to the final score).
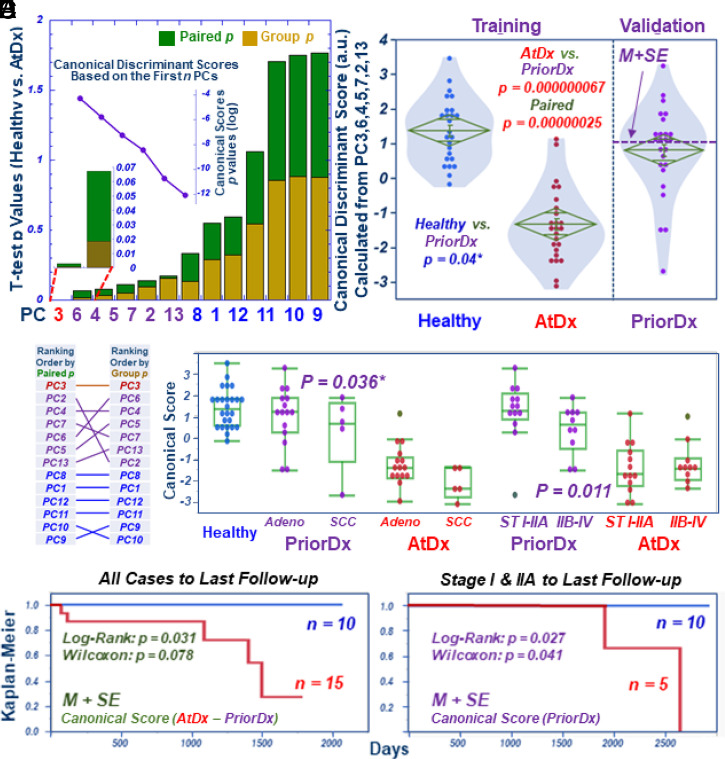
Fig. 2.
Developing serum metabolomics predictive models for NSCLC detection with PriorDx samples. (A) PCs calculated from the 57 spectral regions of the training cohort differentiate Healthy from AtDx NSCLC sera, with paired (green) or group (brown) P values shown as stacked bars in an increasing order. To construct predictive models, canonical analysis using the first n PCs of the training cohort’s smallest P values shows increased discriminant ability between the two groups in the cohort, that is, reduced P values (Inset curve) were seen with the inclusion of more PCs in calculations. (B) Ranking paired and group P values in the increasing order revealed three statuses: the first rank of PC3 with both significant values; the next six PCs (<0.2) with mixed rankings; and the last six PCs (>0.25). (C) Predictive model of canonical discriminant scores calculated from the first seven PCs (PC3 and PCs with mixed ranking) of the training cohort passively yielded scores for the validation cohort’s PriorDx group, with intermediate values between the Healthy and the AtDx groups; significant differences between PriorDx and each of the other two groups were noted. The dashed line represents the value of mean plus one SE (M + SE), calculated from the validation cohort. * indicates one-sided analysis. (D) Presentation of scores from the model according to NSCLC types and stages. Stages I and IIA are localized NSCLC. (E) After setting mean plus one SE as a threshold, as calculated from score differences of the model between each case’s AtDx and PriorDx, we observed that patients whose score differences were higher than the threshold had significantly better survival rates. (F) For localized Stage I and IIA patients, survival was significantly predicted by using the M + SE threshold defined in B.
Fig. 2C, which compares scores calculated from the model for cases of the Healthy, PriorDx, and AtDx groups in the training and validation cohorts, reveals statistically significant differences between these three groups. The metabolomics values (M ± SD: 0.79 ± 1.30) calculated for the PriorDx group, that is, the validation cohort, is lower than the Healthy (1.36 ± 0.92) but higher than AtDx (−1.36 ± 1.07) groups in the training cohort, with statistical significances of P = 0.04 and 6.7 × 10−8, respectively. Possible covariances, including days between PriorDx and AtDx, patient age for AtDx, gender, and smoking status, were tested, but no significant contribution to the predictive model was observed (SI Appendix, Table S3). Differences among the Healthy, AtDx, and PriorDx groups, sorted according to cancer types and stages, are presented in Fig. 2D. In the figure, localized Stages I and IIA NSCLC, with neither lymph node involvement nor metastasis, are grouped and compared with an advanced-disease group that combined Stages IIB to IV, owing to our study’s limited case numbers for each of these more advanced stages.
We also observed a significantly better survival rate by Kaplan–Meier survival analyses for those patients whose score difference from the model was higher than the threshold (Fig. 2E), calculated by setting a threshold of mean plus one SE (M + SE), as calculated from the score difference of the model between AtDx and PriorDx for each case (i.e., the difference of these two scores for each patient). Furthermore, within the Stage I and IIA group, individual patient survival could be significantly predicted from that patient’s PriorDx blood samples (dashed line in Fig. 2C) if these score values were higher than the M + SE threshold calculated for the validation cohort, as shown in Fig. 2F.
Evaluating Serum NSCLC MRS Metabolomics Predictive Models with Testing Cohort.
The serum metabolomic predictive models, established through a canonical discriminant procedure on the training cohort and examined by the validation cohort, were further evaluated by the testing cohort. Independent from the training and validation cohorts, the testing cohort originally included additional serum samples from 56 NSCLC patients prior to their diagnoses, together with samples from 56 matched controls. Examination of MRS results revealed that among these 56 paired serum samples, two samples in two pairs demonstrated significantly different metabolic profiles; they were deemed outliers by outlier analysis and excluded from further study. Thus, 54 paired serum samples were analyzed by passively following all calculations of the training cohort, as previously described (SI Appendix, Table S1).
Fig. 3A presents resulting values from the model for cases in the testing cohort as compared with those in the training and validation cohorts. When compared with the Healthy group, the PriorDx group in the testing cohort demonstrated the same significant trend of value changes as were seen in the validation cohort.
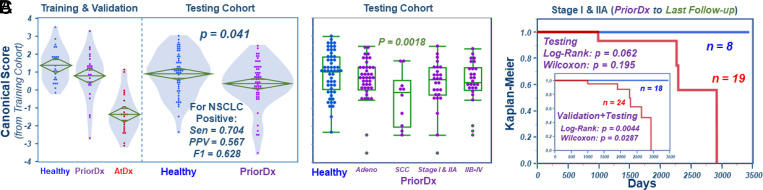
Fig. 3.
Examining serum metabolomics predictive model for NSCLC detection with the testing cohort. (A) Significantly different values from the model, as first obtained for the three serum groups in the training and the validation cohorts, are equally observed for the testing cohort’s Healthy and PriorDx groups. Using the M + SE defined in Fig. 2, the model predicted NSCLC positive presented sensitivity (Sen) = 0.704; specificity = 0.463; positive predictive value (PPV) = 0.567; negative predictive value = 0.610; accuracy = 0.583; and F1 score = 0.628. (B) PriorDx cases, grouped according to NSCLC types and stages for the testing cohort, where squamous cell carcinoma (SCC) is significantly different from Healthy. (C) Kaplan–Meier analysis predictions of overall survival for each of the testing cohort’s localized Stage I and IIA cases. The sum of all localized cases in both the validation and the testing cohorts shows significant Kaplan–Meier survival predictions (Inset), using the M + SE threshold established in Fig. 2C).
However, we further compared the Healthy and PriorDx groups in the training and validation cohorts, respectively, with the Healthy group in the testing cohort. Here, we noted that the latter (M + SD, 0.90 ± 1.18) was closer to the PriorDx group (0.79 ± 1.30) in the validation cohort than to the training cohort’s Healthy group (1.36 ± 0.92). This discrepancy was primarily caused by outliers seen in the testing cohort’s Healthy group. Given that AtDx and PriorDx conditions can alter serum metabolomics status to any degree, regular outlier analyses cannot be applied to them. Instead, outlier analyses can be meaningfully applied to the Healthy groups, as they are considered control samples. Our outlier analyses on both the Healthy groups in the training and testing cohorts showed that no outlier was identified with the Training group. However, all three outlier analysis algorithms (Mahalanobis, Jackknife, and T2) could identify the eight outliers in the testing cohort. After removing these outliers, the resulting M + SD for the Healthy group in the testing cohort changed from the previous 0.90 ± 1.18 to 1.16 ± 0.90, a value closer to that of the Healthy group in the training cohort. Also, the P value between PriorDx in the validation and Healthy in the testing cohorts, as calculated with the KWW test, fell from the previous 0.718 to 0.169.
Similar to that shown in Fig. 2D, when examining PriorDx cases according to NSCLC types and stages, the testing cohort (Fig. 3B) presented the same trends as those noted in the validation cohort (Fig. 2D). The Kaplan–Meier survival analysis for the localized Stage I and IIA cases in the testing cohort, conducted similarly to validation cohort cases shown in Fig. 2F, demonstrated a similar survival predicting trend (Fig. 3C). As stated earlier, samples in the validation cohort were obtained from the same patients as those of the training cohort. Of note, the validation cohort samples, collected at least a half year prior to any NSCLC diagnosis, form a group having similar characteristics to the PriorDx samples of the testing cohort. As neither the validation nor the testing cohorts were involved in construction of the model, we increased our examined case numbers by pooling the data of these two groups, as shown in Fig. 3 C, Inset. A collective examination of all localized cases in the combined cohorts yielded a significantly enhanced Kaplan–Meier survival prediction capability as opposed to that shown by considering either cohort on its own (cf. Fig. 2F).
Examining Serum NSCLC MRS Metabolomics Predictive Model Collectively with the Validation and Testing Cohorts.
We were encouraged by the results in Fig. 3 C, Inset, which showed that larger case numbers could be achieved with pooled data from validation and testing cohorts by considering them as a group. We also noted that the characteristics of PriorDx samples were fundamentally different from the training cohort’s AtDx group, despite the fact that some samples (those of the validation cohort) were from the same patients. We thus further tested their model values collectively by combining validation and testing cohorts, as shown in Fig. 4.
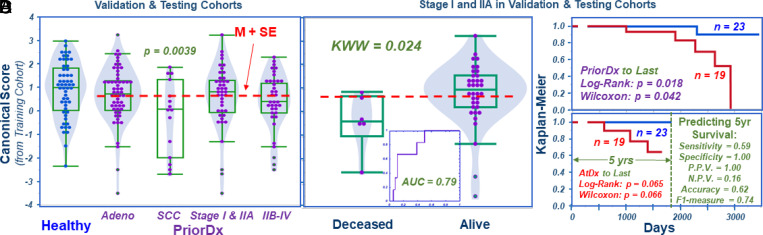
Fig. 4.
Combination of validation and testing cohorts. (A) Values from the predictive model for all cases in the validation and testing cohorts according to NSCLC types and stages, with the threshold M + SE calculated for all PriorDx cases indicated by a dashed line. (B) Values from the predictive model for localized Stage I and IIA cases, according to patient survival status. (C) Using the M + SE threshold defined in A, Kaplan–Meier analysis shows significant survival predictions for PriorDx. (D) Significant Kaplan–Meier survival rates calculated according to the date of patients’ AtDx, with detailed statistical parameters listed in the figure (P.P.V: positive predictive value; N.P.V: negative predictive value). AUC: area under curve; SCC: squamous cell carcinoma.
Fig. 4A presents similar trends that were observed when comparing Healthy cases in the testing cohort with all PriorDx cases in the validation and testing cohorts (subgrouped according to the NSCLC types and stages seen previously for the individual cohorts). As in findings shown in Fig. 3C, Kaplan–Meier survival predictions for all stages I and IIA cases remained significant after recalculating the M + SE threshold from all PriorDx cases in both cohorts. Patient survival status is shown in Fig. 4B and prediction significance in Fig. 4C.
More clinically relevant, Fig. 4D shows that, for these cases, statistically significant Kaplan–Meier survival rates can be predicted by using this threshold and calculating by the patient’s AtDx date, as the detailed statistical parameters listed in Fig. 4D explain.
Probing Significantly Contributing Metabolites toward the Predictive Model of Serum NSCLC MRS Metabolomics.
The presented serum NSCLC MRS metabolomic model, achieved from canonical analyses in Fig. 2C, can be evaluated according to its possibly involved metabolic pathways. The loading factors for each spectral region (presented in SI Appendix, Table S2) represent the product of the combined coefficients from both PCA and canonical analysis and the mean and SD values of the region calculated from the training cohort.
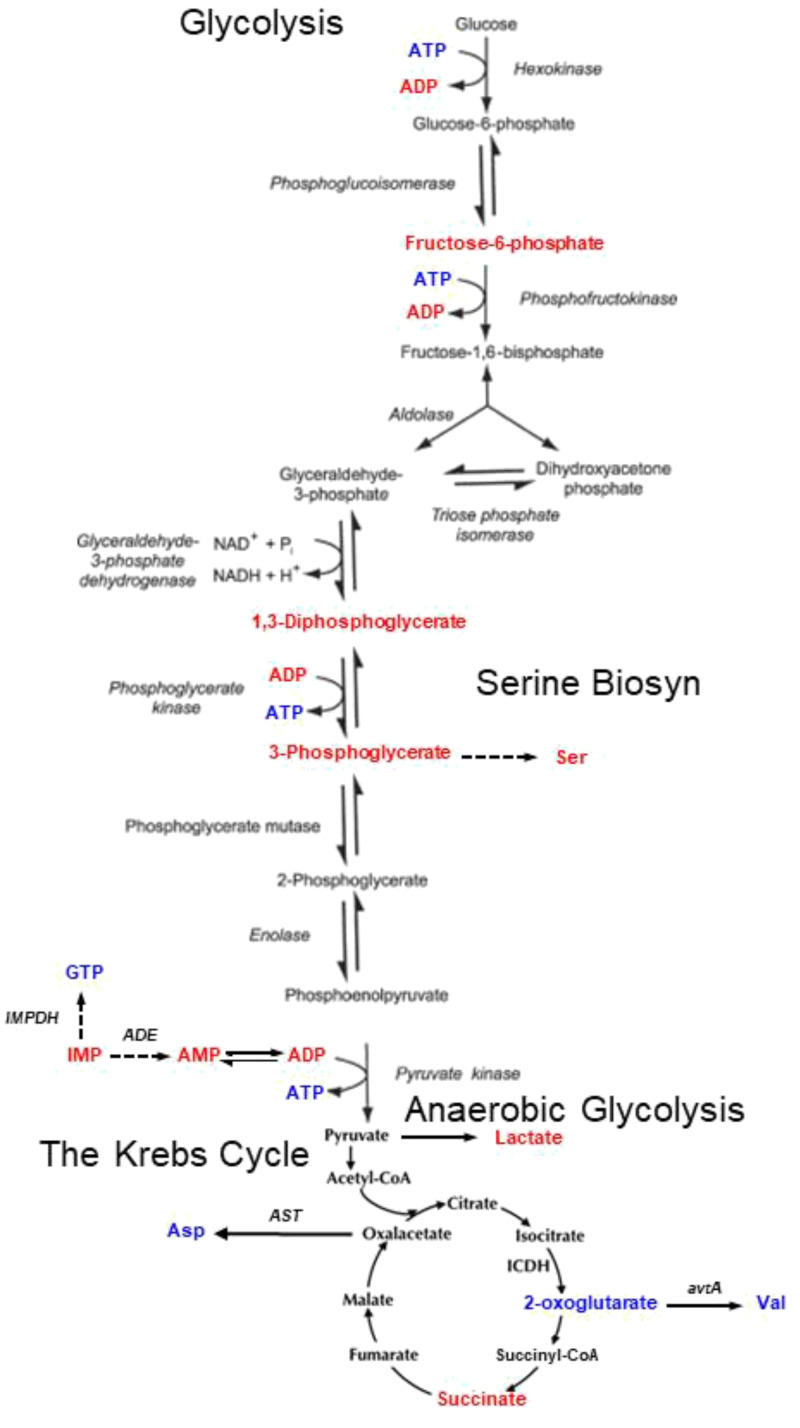
As shown in Fig. 2C, we examined the top 50% of positively contributing regions (which correspond with Healthy cases) and the bottom 50% the most negatively contributing regions (NSCLC cases). Potential metabolites that unidirectionally contributed either to the top 50% for positively contributing regions, or to the bottom 50% for the most negatively contributing regions, are presented in Table 1 [metabolites that might contribute to both lists were removed (8)]. Their associated spectral regions, together with their mean and SDs measured for the Healthy and AtDx groups in the training cohort, are listed in Table 2. As summarized in Fig. 5, examination of the potential involvement of these metabolites in a number of metabolic pathways identified glycolysis, anaerobic glycolysis, the Krebs cycle, etc., as possibly altered metabolic pathways.
Table 1.
Potential major contributing metabolites toward healthy and NSCLC identifications
| Healthy | NSCLC | |
| Nucleotides | ATP | ADP |
| GTP | AMP | |
| IMP | ||
| Nucleosides, nucleobases, and derivatives | 1,7-Dimethyl-xanthine | Caffeine |
| Vitamins, coenzymes | NADP | |
| Sugar phosphates | Diphospho-glycerate | |
| Fructose-6-phosphate | ||
| 3-Phosphoglycerate | ||
| Organic acids | Oxoglutarate | |
| Standard amino acids | Asp | Suc |
| Tyr | Lac | |
| Val | Met | |
| Trp | ||
| Methylated amino acids | Betaine | Dimethyl-proline |
| Other amino acid derivatives | Carnosine | |
| Cr | ||
| Tau | ||
| Carnitines | Carnitine | Acetyl-carnitine |
| Antioxidant | Ergothioneine |
Table 2.
Potential major contributing metabolites and their represented spectral regions measured from the training cohort (mean ± SD)
| Spectral regions | Possible metabolites | Healthy | NSCLC |
| 4.33–4.27 | ATP,GDP,GPC,GTP,Malate,NADP,Thr | 0.0056 ± 0.0018 | 0.0048 ± 0.0023 |
| 3.75–3.73 | Ala,Arg,Citrulline,G6P,Glc,Gln,Glu,Glycerate,GSSG,Leu,Lys | 0.0245 ± 0.0056 | 0.0218 ± 0.0073 |
| 3.37–3.32 | 1,5-AG,1,7-Di-xan,GSSG,Pro | 0.0060 ± 0.0024 | 0.0049 ± 0.0027 |
| 3.25–3.21 | 1,5-AG,Arg,Betaine,Carnitine,Carnosine,Glc,His,m-Ino,Phe,Tau | 0.0865 ± 0.0215 | 0.0749 ± 0.0179 |
| 3.07–2.99 | 2-OG,Carnosine,Cr,Creatinine,Lys,MH,Orn,Tyr | 0.0184 ± 0.0041 | 0.0166 ± 0.0040 |
| 2.94–2.92 | Asn,GSSG | 0.0023 ± 0.0010 | 0.0020 ± 0.0010 |
| 2.91–2.88 | To be determined | 0.0027 ± 0.0012 | 0.0017 ± 0.0014 |
| 2.79–2.71 | Asp,Carnosine | 0.0118 ± 0.0036 | 0.0093 ± 0.0031 |
| 2.06–1.99 | Glu,Ile,Pro | 0.0534 ± 0.0145 | 0.0487 ± 0.0124 |
| 1.94–1.88 | Arg,Citrulline,Ile,Lys,Orn,Pro | 0.0080 ± 0.0018 | 0.0078 ± 0.0023 |
| 1.75–1.69 | Arg,Leu,Lys,Orn | 0.0094 ± 0.0023 | 0.0094 ± 0.0020 |
| 1.62–1.55 | Arg,Citrulline | 0.0121 ± 0.0063 | 0.0066 ± 0.0046 |
| 1.52–1.45 | Ala,Citrulline,Ile,Lys | 0.0165 ± 0.0027 | 0.0171 ± 0.0037 |
| 0.97–0.92 | Ile,Leu,Val | 0.0253 ± 0.0051 | 0.0241 ± 0.0054 |
| 0.91–0.84 | Ile | 0.1100 ± 0.0189 | 0.0886 ± 0.0192 |
| 4.16–4.08 | ADP,F6P,GDP,Glycerate,Lac,Pro | 0.0138 ± 0.0036 | 0.0308 ± 0.0153 |
| 4.07–4.05 | ADP,Creatinine,DPG,G6P,m-Ino,Trp | 0.0025 ± 0.0009 | 0.0031 ± 0.0019 |
| 4.02–3.99 | 1,5-AG,3PG,AMP,Asn,Caffeine,DPG,G6P,His,IMP,Phe | 0.0029 ± 0.0025 | 0.0051 ± 0.0042 |
| 3.81–3.76 | Ala,Arg,Citrulline,Glc,Gln,Glu,Glycerate,GSSG,Lys,Orn | 0.0342 ± 0.0082 | 0.0367 ± 0.0071 |
| 3.70–3.67 | 1,5-AG,F6P,G6P,Glc,GPC,Leu,MH | 0.0107 ± 0.0055 | 0.0132 ± 0.0067 |
| 3.66–3.64 | 1,5-AG,F6P,GPC,Ile | 0.0071 ± 0.0033 | 0.0096 ± 0.0041 |
| 3.55–3.52 | F6P,G6P,Glc,m-Ino | 0.0227 ± 0.0044 | 0.0210 ± 0.0079 |
| 3.15–3.14 | Citrulline,Ergothi,His,MH | 0.0011 ± 0.0011 | 0.0016 ± 0.0011 |
| 2.39–2.32 | Gln,Glu,Malate,Pro,Suc | 0.0077 ± 0.0015 | 0.0094 ± 0.0039 |
| 2.16–2.11 | ALC,Gln,Glu,GSSG,Met | 0.0150 ± 0.0025 | 0.0158 ± 0.0020 |
| 2.09–2.07 | Dm-pro,Gln,Glu,Met,Pro | 0.0092 ± 0.0032 | 0.0106 ± 0.0027 |
| 1.35–1.34 | To be determined | 0.0220 ± 0.0077 | 0.0586 ± 0.0321 |
| 1.33–1.32 | Lac,Thr | 0.0384 ± 0.0102 | 0.0727 ± 0.0336 |
Abbreviations: 1,5-AG, 1,5-Anhydroglucitol; 1,7-Di-xan, 1,7-Dimethyl-xanthine; 2-OG, 2-Oxoglutarate; 3PG, 3-Phosphoglycerate; ADP, Adenosine diphosphate; ALC, Acetyl-carnitine; AMP, Adenosine monophosphate; Arg, Arginine; Asn, Asparagine; Asp, Aspartate; ATP, Adenosine triphosphate; Cit, Citrate; Cr, Creatine; Dm-pro, Dimethyl-proline; DPG, Diphospho-glycerate; ErgoThi, Ergothioneine; F6P, Fructose-6-phosphate; G3P, Glyceraldehyde-3-phosphate; G6P, Glucose-6-phosphate; GDP, Guanosine diphosphate; Glc, Glucose; Gln, Glutamine; Glu, Glutamate; GPC, Glycerophosphocholine; GSSG, Glutathione disulfide; GTP, Guanosine triphosphate; His, Histidine; Ile, Isoleucine; IMP, Inosine monophosphate; Lac, Lactate; Leu, Leucine; Lys, Lysine; Met, Methionine; MH, Methyl-histidine; m-Ino, myo-Inositol; NAD, Nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; Orn, Ornithine; Phe, Phenylalanine; Pro, Proline; Ser, Serine; Suc, Succinate; Tau, Taurine; Thr, Threonine; Trp, Tryptophan; Tyr, Tyrosine; Val, Valine.
Fig. 5.
Metabolic pathways potentially altered by NSCLC. Red letters identify metabolites associated with NSCLC, while blue letters emphasize metabolites related to Healthy controls.
Discussion
A recognized need exists for effective NSCLC screening tests to expedite the discovery of early, asymptomatic lung cancer and facilitate prompt treatments that can reduce associated mortality. The development and clinical implementation of these screening tests—ideally low cost, portable, and with minimal side effects—will permit expeditious relay of suspicious readings so as to triage patients toward further evaluation by imaging tests, such as low-dose CT.
Blood has been considered an ideal target for developing tests that could reveal NSCLC presentations. All cardiac output passes through the lungs, with 20% of blood in them at any given time, delivering nutrients and removing the products of biological reactions. The presence of NSCLC, with its altered physiology and pathology, can cause changes in the blood metabolites produced or consumed by cancer cells in the lungs. Blood metabolomic profiles can reflect these ongoing biological activities at the time of sampling. Accurately measured as a metabolomic target, blood could thus prove valuable as a messenger of physiological and pathological conditions in the lungs. However, because blood circulates throughout the entire body, it can potentially be affected by environmental factors, such as diet. Although blood collection protocols require overnight fasting, potential confounding factors from individuals can present. Fortunately, our use of a PCA approach to construct the predictive models will render any alteration seen only from a single studied subject of little significance to PCA overall loading factors, unless that alteration presents as an extreme outlier with a value hundreds of times higher than the average values. Here, for the 57 regions of the training cohort, our analyses of the ratios of the maximum value over the mean for each region presented the maximum ratio as 9.5, with a mean and an SD of 2.4 ± 1.3.
The HRMAS MRS method that we developed for intact tissue metabolomics is an effective tool for conducting blood serum analyses aimed at detecting changes in blood metabolomic profiles occasioned by cancer cells. Although sera are liquids, they contain proteins and other macromolecules that can prevent acquisition of this information by high-resolution serum MRS, particularly in limited samples (<100 μL). By using HRMAS, high-resolution MRS can be measured with very small amounts of serum (10 μL), without any need of sample pretreatment and with the possibility of serum metabolite quantification. However, during HRMAS measurements, a number of NMR parameters could still affect the measured spectral resolutions and thus metabolic intensities or metabolomic profiles. For instance, the higher the NMR field strength used, the better the metabolic spectral resolution one may achieve and, hence, the more accurate the identification of metabolites. Intrinsic NMR properties of the measured biological materials, such as metabolite relaxations rates and T1 or T2 for the measured spin-lattice and spin-spin relaxation times, respectively, can also significantly alter measured results. In the current study, a recycle time of 5 s was used throughout, as this is considered long enough to allow complete spin-lattice relaxations (normally presented as five times their T1s). The Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence with a total T2 filter time of 20 ms was used to remove most spectral broadening caused by contaminating signals from the probe background and from macromolecules in the samples, without significantly attenuating cellular metabolites. These parameters are important to consider during the discovery of disease-specific metabolomic profiles; however, the profiles’ use under the exact same conditions of measurement is crucial.
Our previous investigation of human lung cancer by MRS, conducted on paired tissue and blood serum samples from patients of different disease stages, showed the potential of MRS-based metabolomics in differentiating cancer types and stages of diseases, as well as in estimating overall survival rates for patients (6, 7). In addition to demonstrating these capabilities for our cancer metabolomics approach, our study provided proof of concept for the ability of blood serum metabolomics to detect lung cancer. However, this earlier study’s tested blood samples all had been collected at the time of our patient cohort’s lung cancer diagnoses. Thus, the feasible use of blood serum metabolomics as a screening tool for asymptomatic stage disease could not be evaluated in that work.
In this study, we measured serum MRS metabolomic profile values in samples collected from lung cancer patients prior to their lung cancer diagnoses. Our findings demonstrate the potential for development of serum lung cancer metabolomics into sensitive and specific predictive models that can be implemented as an early detection lung cancer screening tool. We further showed that serum MRS metabolomic profiles present thresholds, based on the biological activities they reflect, that allow NSCLC patient survival status to be predicted, of potential use in guiding clinical strategies and treatment decisions. Our design, which compared blood samples collected prior to NSCLC diagnosis with those obtained at time of diagnosis for the same patients, represents the present study’s strength and rigor; however, caution is important. The two sample groups—the training and validation cohorts—composed of samples collected from the same patient and the degree of independence between these groups rely on the time gap between the two respective collections. In this study, the gap differentiating collection of the training and validation cohort samples was at least 6 mo.
Our analyses of serum MRS metabolomics also highlighted certain metabolic pathways, shown in Table 1 and Fig. 5, including glycolysis and the Krebs cycle, both known in cancer development and progression. In addition to the metabolite changes that we observed (highlighted in Fig. 5), studies in the literature correlate several other metabolites with malignant diseases (shown in Table 1). For instance, a significant association between higher betaine intake and lower lung cancer risk has been reported (9). Carnosine, due to its antitumourigenic effects (10), has been considered as an antineoplastic therapeutic and is proposed for use to reduce lung injury caused by radiation therapy (11). The role of carnitine in cancer metabolism has also recently been reported, including in a recent review (12).
These metabolites, and their potential as deduced from reports in the literature, may indeed be reflected in our observed NSCLC metabolomic profiles and do agree with common understanding of certain metabolic pathways. However, the present untargeted metabolomic study cannot prove that each of these metabolites is indeed present in the serum samples examined. Instead, the results we observed encourage more targeted studies of serum NSCLC to better characterize the MRS metabolomic profiles we observed toward their development and implementation in clinical NSCLC screening procedures.
The current study has a number of limitations. Above all, our study faced the intrinsic challenges of any retrospective study with predetermined criteria: here, the availability of serum samples collected from the same NSCLC patients both prior to and at the time of their diagnosis. Despite screening of tens of thousands of subjects and cross-referencing two well-established human serum resources, the Boston Lung Cancer Study (BLCS) Repository and Mass General Brigham (MGB) BioBank, which contain tens of thousands of human sera, the cases available for the current study were ultimately limited to only 25 patients. This small patient population prevented studies of subgroups, such as cancer types, stages, or treatment differences, as well as detailed studies of the time gaps between PriorDx and AtDx samples aimed at better understanding of the time courses of metabolomic alterations prior to patients’ NSCLC diagnoses.
However, as previously emphasized, the current rigorous work conducted with human serum samples—obtained from the same patient, prior to and at the time of NSCLC diagnosis—does serve as proof of concept and presents a method for establishing serum predictive models that, with further development, may usefully function as lung cancer screening tools. We do not suggest that the metabolomic parameters observed in the limited number of patient samples that comprise our study represent clinically implementable NSCLC screening profiles. Rather, our findings encourage more retrospective and prospective studies toward this aim, conducted in collaboration with lung cancer biorepositories, such as that of the American College of Radiology Imaging Network–The National Lung Screening Trial (ACRIN-NLST) Biospecimen Repository. Using the concept demonstrated in this report, future, large-scale studies can be conducted beyond the purview of any single center, whatever its resources. Specifically, given the proof of concept here established, these studies can compare blood samples obtained from patients PriorDx from other patients at the time of diagnosis and from healthy controls to study the previously described subgroups, with metabolomic profile measures tested to eliminate potential confounding factors. Furthermore, the strategy of deducing metabolomic profiles useful for disease screening by comparing the metabolomics of biofluids obtained at the time of a diagnosis with those obtained prior to the diagnosis can be adopted for other medical evaluations, such as for neurodegenerative diseases.
Conclusion
Blood metabolomics measured with HRMAS MRS on 10-μL serum samples collected from patients prior to their clinical NSCLC diagnosis may indicate the existence of early, asymptomatic lung cancer, as well as predict patient overall 5-y survival. Further prospective studies, guided by the results demonstrated in this report, are needed to validate the use of blood metabolomics models as NSCLC early screening tools in clinical evaluation so as to initiate the triaging of high-risk patients to advanced imaging tests for early-stage diagnosis.
Materials and Methods
Study Design.
Human serum samples.
This study was approved by the MGB Human Research IRB (Protocol 2009P000982), and all research was performed in accordance with relevant guidelines and regulations. To identify cases for the training and validation cohorts, that is, serum samples from NSCLC patients at the time of their diagnosis versus prior-to-diagnosis samples for the same patients, we first surveyed the record of all serum samples from more than 58,600 individuals that were stored at MGB BioBank; we identified 153 cases for whom sera collected at ∼0.5 to 5 y prior to NSCLC diagnoses were available. We then cross-referenced these 153 cases with records of the Harvard/MGH Lung Cancer Susceptibility Study (also known as BLCS) Repository, which contains about 10,000 serum samples obtained from lung cancer patients at time of diagnosis, and discovered 25 NSCLC patients on both lists. For these 25 patients, we acquired prior-to-diagnosis serum samples from the MGB BioBank and time-of-NSCLC-diagnosis serum samples from the BLCS Repository. Serum samples from another 54 NSCLC patients, obtained PriorDx, served as the testing cohort and were obtained from the MGB BioBank. Informed consent was obtained from patients and healthy controls, after explanation of the nature and possible consequences of the study and prior to banking samples collected following overnight fasting, in accordance with standard protocols of the BLCS and the MGB BioBank. The NSCLC patients were matched to healthy controls according to age, gender, and smoking habit. Healthy controls were selected from 14,906 potential candidates by using the highest possible CHI, predicting 10-y survival, and without any history or current malignant neoplasm or metabolic disorders. Samples were grouped into and treated as training, validation, and testing cohorts as previously described.
MRS.
Samples were stored at −80 °C until analysis. HRMAS proton MRS measurements were performed using our previously developed method on a Bruker Avance 600 MHz spectrometer. Measurements were conducted at 4 °C with a spin rate of 3,600 ± 2 Hz and a CPMG sequence (36 180° pulses with a total 20-ms T2 filtering time), with and without continuous-wave water suppression during the 5-s recycle delay. A total of 10 μL untreated serum was placed in a 4-mm Kel-F zirconia rotor, with 2 μL D2O added for field locking. MRS spectra were processed using a laboratory developed MATLAB-based program, and peak intensities from 4.5 to 0.5 parts per million (ppm) were curve fit. Relative intensity values were obtained by normalizing peak intensities by the total spectral intensity between 4.5 to 0.5 ppm. Resulting values smaller than 1% of the median of all curve fit values were considered noise and eliminated. Spectral regions were defined by regions that had at least 70% of training cohort samples showing a detectable value, resulting in 57 regions of interest.
Statistical Analysis.
Statistical analyses were performed using JMP Pro-14 and MATLAB 2017a. Univariate statistical tests included Student’s t test (for spectral regions with normal distribution according to Shapiro–Wilk W test) or Mann–Whitney–Wilcoxon test (for spectral regions with nonnormal distributions) for binary comparisons. Multivariate analyses included PCA and canonical correlation analysis. Associations between canonical correlation scores and survival were assessed using Kaplan–Meier survival curves and log-rank tests. Except where noted and explained, two-sided testing was used.
Supplementary Material
Acknowledgments
Funding was provided by NIH Grants CA141139, CA243255, AG070257, and OD023406, and by the A.A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital.
Footnotes
Author contributions: T.A.S., M.J.L., D.C.C., and L.L.C. designed research; T.A.S., M.J.L., L.A.V., and L.L.C. performed research; T.A.S., M.J.L., Y.B., M.R.C., L.A.V., P.H., J.N., M.T., M.A., M.A.M.-K., D.C.C., and L.L.C. analyzed data; T.A.S., M.J.L., Y.B., M.R.C., L.A.V., P.H., J.N., M.T., M.A.M.-K., D.C.C., and L.L.C. wrote the paper; and L.L.C. acquired funding.
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110633118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Raw data will be available at Metabolomics Workbench DataTrack ID 2950 (https://www.metabolomicsworkbench.org/data/show_mwtabfile.php?F=LeoCheng_20211128_062842_mwtab.txt).
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Oudkerk M., Liu S., Heuvelmans M. A., Walter J. E., Field J. K., Lung cancer LDCT screening and mortality reduction—Evidence, pitfalls and future perspectives. Nat. Rev. Clin. Oncol. 18, 135–151 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Toumazis I., Bastani M., Han S. S., Plevritis S. K., Risk-based lung cancer screening: A systematic review. Lung Cancer 147, 154–186 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Cheng L. L., Pohl U., “The role of NMR-based metabolomics in cancer” in The Handbook of Metabonomics and Metabolomics, Lindon J. C., Nicholls J. K., Holmes E., Eds. (Elsevier, Amsterdam, 2007), pp. 345–374. [Google Scholar]
- 5.Vandergrift L. A., et al. , Metabolomic prediction of human prostate cancer aggressiveness: Magnetic resonance spectroscopy of histologically benign tissue. Sci. Rep. 8, 4997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan K. W., et al. , Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue-serum pairs. Lung Cancer 68, 44–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berker Y., et al. , Magnetic resonance spectroscopy-based metabolomic biomarkers for typing, staging, and survival estimation of early-stage human lung cancer. Sci. Rep. 9, 10319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaleckis R., Murakami I., Takada J., Kondoh H., Yanagida M., Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. U.S.A. 113, 4252–4259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying J., et al. , Associations between dietary intake of choline and betaine and lung cancer risk. PLoS One 8, e54561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaunitz F., Hipkiss A. R., Carnosine and cancer: A perspective. Amino Acids 43, 135–142 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Guney Y., et al. , Carnosine may reduce lung injury caused by radiation therapy. Med. Hypotheses 66, 957–959 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Melone M. A. B., et al. , The carnitine system and cancer metabolic plasticity. Cell Death Dis. 9, 228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Raw data will be available at Metabolomics Workbench DataTrack ID 2950 (https://www.metabolomicsworkbench.org/data/show_mwtabfile.php?F=LeoCheng_20211128_062842_mwtab.txt).