Significance
GPCRs and GPCR pathways are increasingly being implicated in human malignancies, placing them among the most promising cancer drug candidates. Our results reveal enrichment of highly impactful, recurrent GPCR mutations within cancers. We found that cognate mutations in selected class A GPCRs have deleterious effects on signaling function. The results also suggest that olfactory receptors, often considered inconsequential, display a nonrandom mutation pattern in tumors in which they are expressed. These findings support the idea that protein paralogs can act in parallel as members of an onco-group.
Keywords: G protein-coupled receptor (GPCR), cancer, evolutionary action (EA), mutational signatures, β-arrestin
Abstract
G protein-coupled receptors (GPCRs) are the largest family of human proteins. They have a common structure and, signaling through a much smaller set of G proteins, arrestins, and effectors, activate downstream pathways that often modulate hallmark mechanisms of cancer. Because there are many more GPCRs than effectors, mutations in different receptors could perturb signaling similarly so as to favor a tumor. We hypothesized that somatic mutations in tumor samples may not be enriched within a single gene but rather that cognate mutations with similar effects on GPCR function are distributed across many receptors. To test this possibility, we systematically aggregated somatic cancer mutations across class A GPCRs and found a nonrandom distribution of positions with variant amino acid residues. Individual cancer types were enriched for highly impactful, recurrent mutations at selected cognate positions of known functional motifs. We also discovered that no single receptor drives this pattern, but rather multiple receptors contain amino acid substitutions at a few cognate positions. Phenotypic characterization suggests these mutations induce perturbation of G protein activation and/or β-arrestin recruitment. These data suggest that recurrent impactful oncogenic mutations perturb different GPCRs to subvert signaling and promote tumor growth or survival. The possibility that multiple different GPCRs could moonlight as drivers or enablers of a given cancer through mutations located at cognate positions across GPCR paralogs opens a window into cancer mechanisms and potential approaches to therapeutics.
G protein-coupled receptors (GPCRs), the largest class of membrane receptors, with over 800 individual human proteins, are increasingly implicated in human tumorigenesis and cancer progression (1, 2). They play essential and diverse roles in cellular homeostasis (3), and some GPCRs are involved in cancer hallmarks in at least 13 cancer types (4), making them potential targets for anticancer drugs (5). However, the mutational heterogeneity in driver genes often clouds their discovery, due to low recurrent mutation rates and a high number of background mutations (6, 7).
To increase the statistical power of cancer driver gene discovery, one computational approach groups mutations across biologically similar genes based on gene networks, pathways, or protein similarity (8, 9). By accounting for strict comparisons with gene-specific mutational frequencies in noncancerous tissues, the MutSigCV method was able to find genes with a mutational frequency that is greater than random in cancer (10). Such studies recently identified mutated residues in hypermutated GPCR genes near conserved structural motifs in cancer samples (11) and reported preferential disruption of receptors coupled to Gαi/o (12). In a domain-centric approach, cancer variants were found to be enriched in protein families, such as kinases, sharing similar functional and structural domains (13–16). Thus, an emerging possibility, supported by their large number of paralogs, is that cognate mutations in different GPCRs are associated with cancer, as already observed in kinases (16).
The GPCR family is defined by a highly conserved tertiary structure with evolutionarily important motifs mediating their dynamic shifting between various active and inactive states (17, 18). Consequently, signaling is often a tangled, interconnected web with different receptors using overlapping sets of downstream effectors and pathways. Because there are many more GPCRs than effectors, mutations in different GPCRs could perturb signaling similarly so as to favor a tumor. With many of these pathways modulating hallmarks of cancers, we hypothesize that somatic mutations found repeatedly at cognate structural positions across many receptors have a high functional impact, funneling their phenotypic effect into cancer-enabling cellular disruptions. To test this possibility, we create an in silico model that accounts for not only cancer-specific nucleotide substitution frequency, but also innate codon composition of aligned amino acid positions in paralogous proteins, and, in this way, we are able to systematically compute expected mutation rates at individual residue positions across paralogous proteins.
Here, we used sequence homology to identify somatic cancer mutations in 651 class A GPCRs, which include 284 nonolfactory and 367 olfactory GPCRs, occurring at cognate structural positions. In order to discriminate between functionally important mutations and noise contributed by benign and low-impact mutations, we used Evolutionary Action (EA) (19) to quantify the predicted impact of mutations on protein function and the Evolutionary Trace (ET) (20, 21) to identify evolutionarily important positions under significant mutational burden in cancer. While mutation frequencies in most residues are linearly correlated with expected mutational frequencies in cancers, we found that mutations at a few selected positions within the DRY and NPxxY motifs are enriched across class A GPCRs in cancer genomes. These motifs were reported as mutational hotspots in previous studies applying gene- and domain-centric approaches (11, 12). While the agreement between different methods further validated potential oncogenic effects of mutations near DRY and NPxxY motifs, the increased resolution of analysis from genes and protein domains to individual residues enables examination of more GPCRs with rare variants. Our data also showed that olfactory GPCRs have distinct mutational rates and patterns compared to nonolfactory GPCR in cancers, but there are highly mutated positions on both olfactory and nonolfactory GPCRs located at evolutionarily important residues, which are considered to be deleterious. Phenotypic characterization of these mutations indicates severe functional perturbations including loss of cell surface expression and altered G protein and β-arrestin signaling. These data suggest cognate driver mutations may flexibly distribute across protein paralogs to induce functional perturbations to promote disease.
Results
Cancer Somatic Mutations Are Nonrandomly Distributed Throughout the GPCR Structure and Target Select Functional Motifs.
In order to search for mutationally enriched positions within class A GPCRs, we aligned the amino acid sequences of 651 class A GPCRs (284 nonolfactory and 367 olfactory GPCRs) (SI Appendix, Table S1) to define cognate sequence positions. To eliminate possible misalignments, we focused our investigation on the 269 positions that are represented by fewer than 10% of gaps in class A GPCRs multiple sequence alignments (SI Appendix, Table S2). For each receptor, we tallied all the somatic mutations recorded in The Cancer Genome Atlas Research Network (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) to comprehensively characterize changes in 33 human cancer types. Our analysis is restricted to receptors expressed in any cancer cells according to RNA sequencing (RNA-seq) data, using a stringent threshold of fragment per kilobase of transcript per million mapped reads (FPKM) greater than one (SI Appendix, Fig. S1). In comparison with variations from a healthy population [1000 Genome Project (22)], we observed that the distribution of mutations in GPCR genes in TCGA is distinctly different (Fig. 1 A–D). On a residue position basis, the nonsynonymous substitution rate tends to be twice as high for both nonolfactory (median = 0.0023) and olfactory (median = 0.0025) GPCR than for synonymous substitutions (median = 0.0012 for nonolfactory; median =0.0011 for olfactory), and the shapes of distributions differ significantly between nonsynonymous and synonymous substitutions (Kolmogorov–Smirnov test P value of <0.0001 for both olfactory and nonolfactory GPCRs; Fig. 1 A and B). In contrast, in a healthy population, the overall variation rates are much lower than ones in TCGA, while distributions of nonsynonymous and synonymous substitutions are still statistically distinct (Kolmogorov–Smirnov test P value of <0.0001). The nonsynonymous substitution rate is lower than that of the synonymous rate in nonolfactory GPCRs (median = 0.0003 for nonsynonymous; median = 0.0006 for synonymous), but the nonsynonymous rate is still higher in olfactory GPCRs (median = 0.0022 for nonsynonymous; median = 0.0007 for synonymous) (Fig. 1 C and D).
Fig. 1.
Mutational burden in class A GPCRs. (A–D) The substitution rate per position is shown as histograms. Nonsynonymous substitution rate for olfactory and nonolfactory GPCRs in pan-cancer, and for olfactory in 1000 Genome Project are significantly right-shifted compared to that of synonymous substitution. In contrast, the synonymous substitution rate is higher than nonsynonymous for nonolfactory GPCRs in the 1000 Genome Project. Kolmogorov–Smirnov distances are (A) 0.725, (B) 0.662, (C) 0.201, and (D) 0.350, and, for all four distribution, P values are less than 0.0001. (E and F) The z score of nonsynonymous substitutions at cognate residue positions. Positions of residues discussed in the text are indicated.
We next investigate the nonsynonymous substitutions rate at cognate residue positions and the listing of residues follows the Ballesteros–Weinstein numbering system (23) in transmembrane regions, and, otherwise, residue numbering is based on the human β2 adrenergic receptor. Residue 3.50 in nonolfactory GPCRs is the most highly mutated in TCGA pan-cancer (z score of 7.20) compared to 1000 Genome cohorts (z score of −0.39) (Fig. 1E). In addition to residue 3.50, other highly mutated positions are 8.51 (z score of 6.01), and 7.50 (z score of 2.92). In olfactory GPCRs, 3.50 is also the most highly mutated position, both in TCGA pan-cancer and 1000 Genome cohorts (z score of 7.26 in TCGA; z score of 5.18 in 1000 Genome) (Fig. 1F), although the subset of receptors mutated at this position is different for each cancer cohort. Among the 56 olfactory receptors having mutations at 3.50 in the TCGA database, 24 are uniquely associated with TCGA (SI Appendix, Figs. S2 and S3). There is a growing appreciation of a role for olfactory receptors in signaling outside of the olfactory system (24), and our data suggest that mutations in subsets of olfactory receptors that are pan-cancer–specific may benefit cancer progression by modulating DRY motif functionality.
To further investigate the structural distribution of synonymous and nonsynonymous mutations, we next mapped separately the 50 most highly mutated positions onto the β2AR inactive structure [Protein Data Bank (PDB) ID code 2RH1 (25)]. In the measurement of structural clustering using selection clustering weight (26), nonolfactory nonsynonymous mutations in TCGA pan-cancer are statistically clustered (z score of 1.7; SI Appendix, Fig. S4) in the structure around functional motifs with 16 residues directly in or within 8 Å of the DRY, NPxxY, or PIF motif. These data suggest that the increased rate of nonsynonymous mutations in cancers is nonrandomly enriched in structure and function.
Biases in Nucleotide Substitution Frequency and Codon Composition Are Insufficient to Explain Mutational Enrichment.
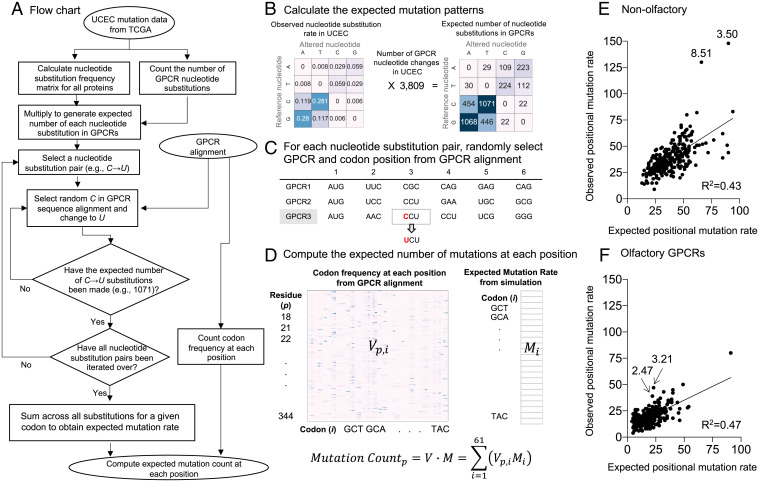
Different nucleotide substitution biases have been observed in different cancer types (27). Therefore, we created an in silico model to test whether the mutation rate at each position could be explained solely by the variation in the nucleotide substitution biases of each cancer type and the codon composition of each alignment position (Fig. 2 A–D; see Materials and Methods). In the model, individual codons experience varied mutation frequencies depending on the nucleotide biases exhibited in different cancers (SI Appendix, Fig. S5). Therefore, some alignment positions are expected to experience increased mutation rates due solely to nucleotide substitutions that are random within the constraints of the tumor substitution frequency matrix.
Fig. 2.
Controlling for nucleotide substitution biases and variable codon mutation expectation. (A) Flowchart of the simulation using UCEC as an example. (B) The observed nucleotide substitution frequencies were tabulated across all proteins for a given cancer type, creating five nucleotide substitution frequency matrices. UCEC is shown here for example. The other cancer types are shown in SI Appendix, Fig. S5. (C) A random GPCR and a random codon within aligned class A GPCRs were chosen, and the codon was mutated following the nucleotide substitution matrix calculated in step B. In this example, GPCR3 and position 3 are randomly selected, and nucleotide C is changed to U. (D) The random simulation in step C results in the expected mutation rate matrix. The codon frequency at each position was then calculated by counting the number of times each codon occurs at every position within the multiple sequence alignment. The expected mutation count at position p is the sum over all codons i of the number of occurrences of that codon at position p (Vp,i) multiplied by the expected mutation rate at a given codon (Mi). (E and F) Expected versus observed pan-cancer positional mutation rate at (E) nonolfactory GPCRs and (F) olfactory GPCRs.
We observed that expected mutation frequency based on the random simulation in the pan-cancer model correlated linearly with the observed mutation frequency both in olfactory (R2 = 0.47) and nonolfactory (R2 = 0.43) GPCRs (Fig. 2 E and F), indicating mutations at most positions are consistent with the random background mutation rate. Strikingly, residues 3.50 and 8.51 in nonolfactory GPCRs (χ2 p value < 0.005, z score > 5.0), and residues 3.21 and 2.47 in olfactory GPCRs (χ2 p value < 0.005, z score > 2.0), were significantly hypermutated compared to the simulation: Residues 3.50 and 8.51 had 58 and 62 more mutations than expected, respectively; and residues 3.21 and 2.47 had 24 and 17 more mutations than expected, respectively (SI Appendix, Table S3). These data indicate that the increased mutation rate at these positions cannot be attributed to codon composition and nucleotide substitution biases.
Nonsynonymous Mutations Display Cancer Type–Specific Patterns of Mutation.
In previous studies, individual cancer types were shown to be driven by type-specific driver mutations (28–30). We therefore repeated the above analyses on all individual cancer types within the TCGA, focusing on the five cancer types with the largest rate of GPCR mutations (UCEC, SKCM, LUAD, COAD, and STAD) (SI Appendix, Fig. S6; TCGA cancer study abbreviations in SI Appendix, Table S4). In nonolfactory GPCRs, residue 3.50 is the most hypermutated in all five cancers (χ2 test P values are, ), followed by residue 8.51 in UCEC and COAD (χ2 test P values are , respectively), and residue 7.42 in UCEC, LUAD, and COAD (χ2 test P values are , respectively). In olfactory GPCRs, on the other hand, residue 3.50 is the most hypermutated in COAD (χ2 test P values is ), and residue 7.50 is the most hypermutated in SKCM and STAD (χ2 test P values are and ). These data suggest that the significant overlap in hypermutated positions may benefit cancer progression through similar cognate functional perturbations in multiple cancer types.
However, we also observed marked differences between individual cancer types. In nonolfactory GPCRs, residue 7.50 is hypomutated in UCEC (chi-squared test P value ), whereas it is hypermutated in SKCM (chi-squared test P value ). In both olfactory and nonolfactory GPCRs, residue 1.49 is hypomutated in UCEC (chi-squared test P value for olfactory and for nonolfactory GPCRs), while being hypermutated in SKCM (chi-squared test P value for olfactory and for nonolfactory GPCRs).
Hypermutated Positions Target Evolutionarily Important Residues with Mutations Predicted to Be Highly Impactful.
Using ET, we next quantified the functional and structural importance of the top 10 ranked hypermutated positions compared with the hypomutated positions (SI Appendix, Figs. S7 and S8) in the five cancer types with the highest GPCR mutation rates: UCEC, SKCM, LUAD, COAD, and STAD. ET uses phylogenetic lineage analysis to identify correlations between evolutionary changes in genotype and changes in the phylogenetic tree representing divergences in function (20, 31, 32). Genotypic changes correlating with larger phenotypic divergence are considered more important than positions correlating to smaller evolutionary divergences (21). Missense hypermutated and hypomutated positions were enriched for evolutionarily important residues as we would expect, given the essential roles of GPCRs (Fig. 3A).
Fig. 3.
Distribution of TCGA mutations within human class A GPCRs. (A) ET score distribution of hypomutated and hypermutated positions in comparison with ET scores for all positions in class A GPCRs (Mann–Whitney test *P value < 0.05; **P value < 0.005). (B and C) Distribution of predicted mutational impact (EA scores) of the hypermutated positions in comparison with overall class A GPCR mutation EA scores from 1000 Genome Project (healthy population) in (B) nonolfactory and (C) olfactory GPCRs. The red bar indicates median of the distribution. (D) Top hypermutated positions are visualized in the β2AR protein structure (PDB ID code 2RH1). Sphere size corresponds to mutation frequency, and sphere color represents ET score. (E) Primary G protein coupling specificity of GPCRs mutated at the five selected positions (1.40, 3.50, 7.42, 7.50, and 8.51), compared to all class A GPCRs. G protein coupling data are from GPCRdb (34).
Given that mutations may have varying impacts on protein function, we used EA to predict functional impacts quantitatively (19). EA predicts the impact of a mutation on protein function by combining the ET importance of each position and the severity of the substitution, quantified using substitution log odds across evolution, where scores range from 0 (benign) to 100 (drastic functional perturbation) (33). Among the top 10 hypermutated positions in UCEC, SKCM, LUAD, COAD, and STAD, four positions in nonolfactory GPCRs (3.50, 7.42, 7.50, and 8.51) show significant bias toward highly impactful mutations (Mann–Whitney p value < 0.0001) in comparison with the distribution of EA scores of mutations from a healthy population (Fig. 3 B and C and SI Appendix, Fig. S9). Although hypermutated positions in olfactory GPCRs have less overlap between cancers, EA scores for mutations at positions 1.49, 3.50, and 7.50 are highly biased toward drastic functional perturbations (Fig. 3C and SI Appendix, Fig. S9). To investigate the structural distribution of those hypermutated positions, we mapped them onto the β2AR inactive structure, PDB ID code 2RH1 (25) (Fig. 3D). Strikingly, 1.49, 3.50, 7.42, 7.50, and 8.51 are all located near or within known functional motifs: 3.50 within the DRY motif, 7.50 within NPxxY motif, and 7.42 proximal to the toggle switch. Notably, 1.49 and 7.50, while distant in sequence space, are structurally proximal to NPxxY, suggesting a nonrandom enrichment in structure and function. Taken together, mutational patterns and rates are distinct between olfactory and nonolfactory GPCRs, but the hypermutated and hypomutated positions were enriched for evolutionarily important residues for both olfactory and nonolfactory GPCRs. Overall, these data show class A GPCRs in cancer samples are enriched for recurrent, nonrandom, high-impact mutations, occurring at evolutionarily important positions at or near known functional motifs.
We next investigated whether the GPCR mutations in cancer are preferentially associated with specific Gα proteins. The primary G protein association is only available for a subset of GPCRs (256 out of 284 class A nonolfactory GPCRs) from GPCRdb (34), and olfactory receptors were excluded in this analysis because they preferentially use the olfactory-specific Gαolf in their native cell types (35). We observed that the primary Gα coupling profile of GPCRs mutated at 1.49, 3.50, 7.42, 7.50, and 8.51 is similar to the overall preference for primary Gα protein couplings in class A nonolfactory GPCRs expressed in cancer (Fig. 3E and SI Appendix, Fig. S10).
Loss of Function Phenotypes of Mutant GPCRs.
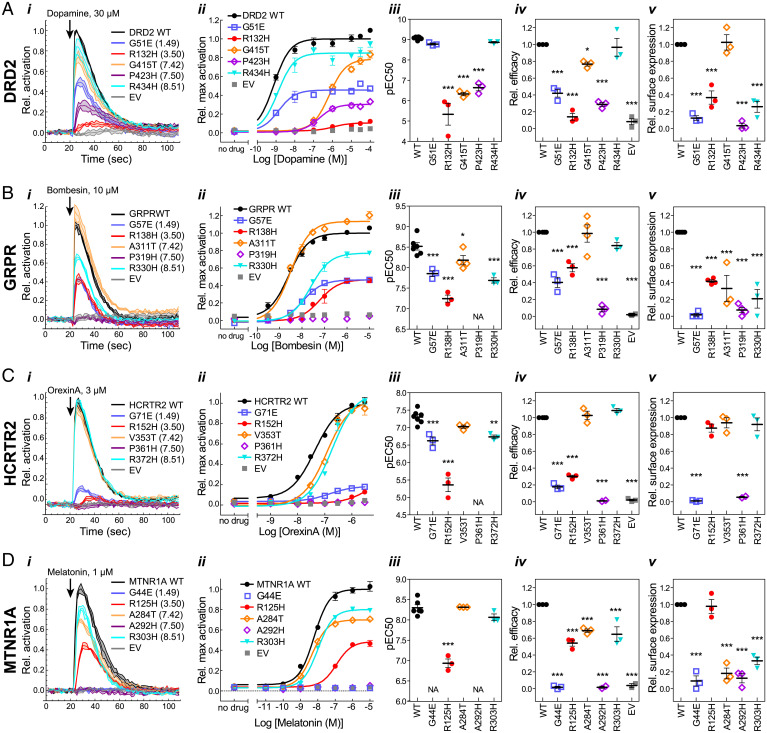
To investigate the functional consequences of substitutions at hypermutated positions with high EA scores, we assayed plasma membrane localization, G protein activation, and β-arrestin recruitment in mutants of four GPCRs. The five identified positions (1.49, 3.50, 7.42, 7.50, and 8.51) were mutated to match the most frequent substitutions found at those positions (SI Appendix, Fig. S11). Diverse receptors were chosen: dopamine D2 receptor (DRD2), gastrin-releasing peptide receptor (bombesin receptor 2; GRPR), hypocretin receptor 2 (orexin receptor type 2; HCRTR2), and melatonin receptor 1A (MTNR1A). All four receptors were represented in the TCGA dataset of mutations at the experimentally tested positions, although none were mutated at all of the positions (SI Appendix, Table S5).
Using transiently transfected cells loaded with a Ca2+ indicator dye, we measured the ability of mutant receptors to mobilize Ca2+ release from internal stores via endogenous Gαq, or, in the case of Gαi/o coupled DRD2 and MTNR1A, via cotransfected chimeric Gαqo (36) (Fig. 4). We measured β-arrestin recruitment using the Tango assay (Fig. 5), in which recruitment of an arrestin–TEV protease fusion protein mediates cleavage of a transcription factor fused to the GPCR, leading to luciferase reporter expression (37). The GPCRs in both assays were modified by the addition of an N-terminal signal peptide and FLAG tag, and the Tango assay construct was additionally fused at the C terminus to the Vasopressin 2 receptor tail and a transcription factor. Plasma membrane expression of the two different protein constructs was assayed in the respective cell lines by labeling with FLAG antibody in nonpermeabilizing conditions.
Fig. 4.
G protein activation by (A) DRD2, (B) GRPR, (C) HCRTR2, and (D) MTNR1A mutants. (i) Representative time courses showing G protein activation at a single ligand concentration. Lines indicate means ± SD of technical replicates at each timepoint. (ii) Representative dose–response curves generated from maximum activation values from data as in i. Points indicate means ± SEM of technical replicates. (iii–v) EC50 (iii) and efficacy (Emax) (iv) were determined from dose–response curve fits, and surface expression (v) was measured by cell ELISA, as described in Materials and Methods. Points represent means from independent experiments, and error bars show SEM. Mutants were compared to WT using one-way ANOVA and Dunnett’s posttest (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). Mutants for which an EC50 could not be determined are indicated by “NA.” For mutants which did not reach a plateau in the dose–response curves, EC50 and efficacy determinations are estimates and represent lower limits. Error bars that are not visible are smaller than the associated symbol.
Fig. 5.
β-arrestin recruitment activity of (A) DRD2, (B) GRPR, (C) HCRTR2, and (D) MTNR1A mutants. (i) Representative Tango assay dose–response curves. Points indicate means ± SEM of technical replicates. EC50 (ii) and efficacy (iii) were determined from dose–response curve fits, and surface expression (iv) was measured by cell ELISA (DRD2, HCRTR2) or immunofluorescence microscopy (GRPR, MTNR1A), as described in Materials and Methods. Points represent means from independent experiments, and error bars show SEM. Mutants were compared to WT using one-way ANOVA and Dunnett’s posttest, except where only one mutant was analyzed, in which case a t test was used (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). For mutants which did not reach a plateau in the dose–response curves, EC50 and efficacy determinations are estimates and represent lower limits.
In both functional assays (G protein activation and β-arrestin recruitment), most of the mutants exhibited reduced potency, efficacy, or both, where the losses of functional activity are consistent with high EA scores. The 7.50H mutation, located in the NPxxY motif, severely compromised activity in all receptors, such that an effective concentration, 50% (EC50) could not be measured in most cases. This loss of function is likely explained by misfolding and/or mislocalization, as surface expression was severely reduced or not detectable for all of the 7.50H mutants, with the exception of the MTNR1A Tango construct. The 1.49E mutation, also located near the NPxxY motif, led to very low or undetectable surface expression in most mutants. Despite the low surface expression of 1.49E mutants, G protein activation was measurable, albeit with significantly reduced potency and/or efficacy, indicating the relative insensitivity of the enzyme-linked immunosorbent assay (ELISA) for detecting surface expression. These results are consistent with roles for residues in the vicinity of the NPxxY motif in folding and/or localization, as well as in potency of agonist-induced G protein activation. The 3.50H mutation, located in the DRY motif, led to significantly reduced potency and efficacy for G protein activation in all receptors, including HCRTR2 and MTNR1A, in which surface expression was not significantly different from wild type (WT). In contrast, the 3.50H mutation led to reduced potency, but not efficacy, for β-arrestin recruitment, with the exception of the GRPR mutant which had severely reduced surface expression and no activity.
In general, 7.42T and 8.51H were the least deleterious mutations with respect to G protein activation. The substitution 8.51H exhibited near-normal potency and/or efficacy in all four receptors, while 7.42T exhibited near-normal potency and/or efficacy in all but DRD2, despite reduced surface expression in some cases. However, in the β-arrestin recruitment assay, the DRD2 and GRPR 8.51H mutants had no detectable activity or surface expression, suggesting defects in folding or stability due to this mutation that are exacerbated by the large fusion protein required for the Tango assay.
In contrast to the loss of function phenotypes observed with most mutants, the DRD2 1.49E mutant had striking constitutive β-arrestin recruitment activity (Fig. 5A). Although dopamine dose-dependent β-arrestin recruitment was impaired, activity was near WT efficacy in the absence of ligand. The Ca2+ mobilization assay for G protein activation does not report constitutive activity. However, cells transfected with the DRD2 1.49E mutant tended to detach from wells during the course of the assay, suggesting that this mutant may be deleterious for cell health. We also observed detachment of cells transfected with HCRTR2 1.49E and 7.50H mutants in some G protein activation experiments.
In the case of GRPR and MTNR1A constructs for β-arrestin recruitment, surface expression was not detectable in ELISA, even for WT. Therefore, as a complementary and more sensitive method, we labeled cells in nonpermeabilizing conditions and imaged by confocal microscopy (Fig. 5 B and D and SI Appendix, Figs. S12–S15). Results for DRD2 and HCRTR2 are similar to those obtained with ELISA, and surface expression was measurable for GRPR and MTNR1A. Further, labeling of total protein in permeabilizing conditions was detected at similar levels for all mutants, indicating that all mutants were expressed, but some had defects in folding and/or trafficking that resulted in impaired surface expression. Specific labeling of cell boundaries in nonpermeabilizing conditions, clearly distinguishable from the labeling pattern in permeabilizing conditions, further validates the specificity of ELISA for detecting cell surface epitopes.
Discussion
We present a combined genomic and structural analysis of class A GPCR mutations in cancers. The observation of an increased rate of nonsynonymous mutations compared to synonymous mutations, per residue position (Fig. 1), is consistent with previous studies showing an increased ratio of nonsynonymous to synonymous (dN/dS) mutations in cancer cells compared to germline variants from a healthy population (38, 39). We observed that the rate of nonsynonymous mutations was nonrandomly distributed in the GPCR structure, and that certain cognate residue positions were enriched for nonsynonymous mutations across class A GPCRs in pan-cancer compared with mutations from a healthy population (Figs. 2 and 3). Furthermore, the predicted impacts of mutations at enriched residue positions in cancers were significantly more severe than those of overall mutations in a healthy population (Fig. 3 and SI Appendix, Fig. S9). These results suggest the increased rate of nonsynonymous mutations in cancers in comparison with healthy population is nonrandomly distributed in GPCR structure, and those recurrently observed mutations at cognate positions in paralogous GPCRs could share similar phenotypic consequences in cells, since sequence comparisons of class A receptors make apparent that all share evolutionary and structurally conserved functional motifs (40).
No attempt is made in this study to address germline mutations that may predispose to cancer. Rather, our goal has been to contrast statistical properties of common inherited variations in 1000 Genomes with impactful and cancer-specific somatic mutations, and to identify positions across a set of paralogous genes that are hypermutated in tumors, and therefore candidates for positions at which mutations favor cancer progression.
Although the role of olfactory GPCRs in cancer has not been well studied, overexpression of certain olfactory receptors has been reported in various cancers including skin, breast, prostate, colorectal, lung, and bladder (41–46). Our results show that a large number of olfactory receptors are expressed in cancer samples (SI Appendix, Figs. S1 and S3). Despite similar nonsynonymous mutation positional rates between TCGA pan-cancer and 1000 Genomes, olfactory mutations in cancers are more often located at evolutionarily important residues, which are considered to be deleterious (Figs. 1 and 4 and SI Appendix, Fig. S4). This result suggests that nonsynonymous mutations on olfactory GPCRs are enriched at highly impactful and cognate positions across paralogues proteins, and thus may have functional roles in cancer tissues.
Unlike nonolfactory GPCRs, numerous healthy individuals have loss of function variants in olfactory GPCRs, including mutations at residue 3.50, and different subsets of inactivated olfactory GPCRs play important roles in smell sensitivity and preferences in humans (47). Although olfactory GPCRs typically couple to the heterotrimeric G protein subunit Gαolf, they may also couple to Gαs and Gα15 (48, 49), mediating adenylate cyclase and phospholipase C activation. In cancers, Gαolf is expressed in all 33 cancer types but at low levels, while expression of Gα15 is relatively highly expressed on LAML, HNSC, CESC, THYM, BLCA, and LUSC (SI Appendix, Figs. S16 and S17) Indeed, activation of olfactory receptors in cancer cell lines, such as OR10H1 in bladder cancer (46), OR51E2 in prostate cancer (45), OR2AT4 in leukemia (50), and OR2J3 in non-small-cell lung cancer (43), inhibits cell proliferation and induces apoptosis mediated by adenylate cyclase. While more experiments are required to measure potential oncogenic effects from individual olfactory GPCRs, our finding of the unique subsets of olfactory receptor mutations in cancer tissues suggests that these receptors might contribute to cancer progression by modulating DRY motif functionality.
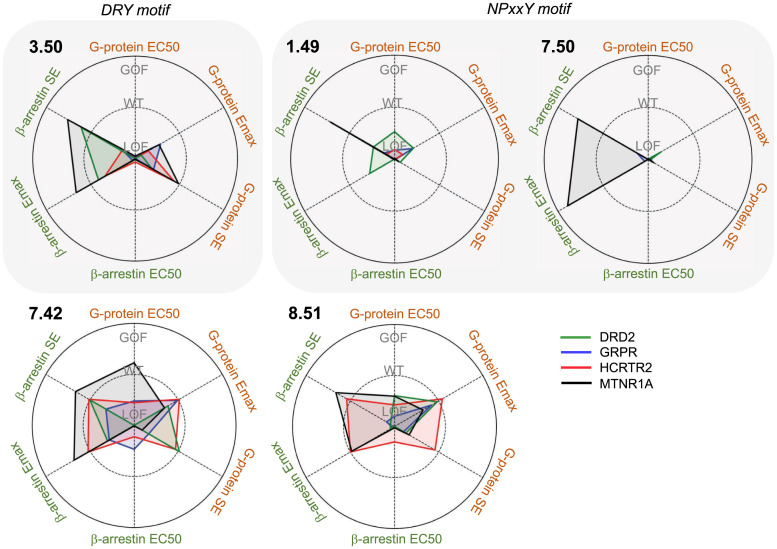
Repeated and independent testing of identical mutations in different nonolfactory GPCRs has revealed that they often lead to conserved functional perturbations. For instance, mutations at 3.50 and 7.50 are associated with many different diseases, including cancers, and have been previously characterized in multiple receptors. Mutation at 3.50 in AVPR2 (51, 52), ADRA1B (53), GPR33 (54), HRH2 (55), and CCR5 (56) result in a complete loss of G protein–dependent signaling with severely decreased cell surface expression and can also trigger constitutive β-arrestin–mediated desensitization. A recent study revealed mutually exclusive mutational patterns between inactivating DRY mutations in GPCR coupled to adenylyl cyclase inhibitor Gαi and activating mutations in adenylyl cyclase activator Gαs (12). Likewise, mutations to 7.50 within CHRM3 (57) and V2R (51) severely impair or eliminate G protein–dependent signaling and cell surface expression. In addition, GPCR signaling through β-arrestins has been reported to serve physiological roles within cancer development, including cell proliferation and migration, in addition to controlling antiapoptotic pathways (58). Consistent with these observations, we observed hypermutated positions located at DRY and NPxxY motifs and significant loss of function in 3.50 and 7.50 mutants of DRD2, GRPR, HCRTR2, and MTNR1A (Figs. 5 and 6). Furthermore, our residue-specific mutational enrichment analysis enabled us to identify additional hypermutated positions at residues 1.49, located close to 7.50 in the NPxxY motif, 7.42, and 8.51. Mutations at these positions were also detrimental (Fig. 6). The receptors tested were selected primarily as convenient and diverse examples of class A GPCR, but there is evidence for their involvement in cancers. Melatonin receptors have been reported to play oncostatic roles by stimulating apoptosis and inhibiting angiogenesis and metastasis (59, 60). Epidemiological studies show the deficiency of melatonin raises risk in ovarian cancer (61), breast cancer (62), and prostate cancer (63), and adjuvant therapy with melatonin has been reported to lead to substantial improvements in solid tumors (64) and colorectal cancer (65). Dopamine D2 receptors contribute to preventing tumor growth by inhibiting vascular endothelia growth factor (VEFG)-mediated angiogenic activities (66), and D2 agonists are reported to inhibit tumor progression in lung cancer (67). A proapoptotic role of orexin receptors has been shown to significantly reduce tumor growths in colon cancers, liver metastases, and glioblastoma (68–70). Given that receptors' function is to sense the environment and regulate cells’ behavior in cancers, the altered responses in G protein activation and/or β-arrestin recruitment caused by hypermutations at cognate positions could alter responses to the environment and cause dramatic changes, ultimately leading to cancers.
Fig. 6.
Summary of mutation profiles at each position. The axes show fold change of measured parameters EC50, Emax, and surface expression (SE), relative to WT (Mut/WT), annotating WT = 1, GOF > 1, and LOF < 1 on the graphs. Values that are closer to the origin represent more severe loss of function (LOF), while values toward the outermost ring indicate more gain of function (GOF).
The present study demonstrated that somatic mutations in tumor samples are distributed across many different class A GPCRs and enriched near functional motifs and have similar effects on GPCR functions. Our results provide insight into flexible use of GPCR paralogs as a functional onco-group and suggest strategies for future studies to discover new therapeutic targets and common downstream pathways shared between GPCRs in specific cancer types. Our in silico tool provides an approach to compute expected mutation rates in cognate positions, allowing identification of mutational hotspots, while taking into account variation in nucleotide substitution biases of each cancer type and the codon composition of each alignment position, without excluding genes with rare variants. Although our current study focuses on class A GPCRs, this tool is widely applicable to different protein families.
Materials and Methods
Sequence Alignment and Position Selection.
A list of 651 class A GPCR was obtained from International Union of Basic and Clinical Pharmacology/British Pharmacological Society (IUPHAR/BPS, https://www.guidetopharmacology.org). The primary sequence for each GPCR was obtained from Uniprot (https://www.uniprot.org) in FASTA format. The sequences were aligned using PROMAL3D (71) using default parameters provided on the web interface. Only alignment positions present (amino acid mapped to that location) in greater than 90% of all GPCR sequences were used for subsequent analyses. This resulted in 269 “high-coverage” positions. Positions below this threshold demonstrated too much variation for confident alignment. In order to eventually visualize the mutation pattern, we further mapped the 269 positions to ADRB2 and the corresponding structure (PDB ID code 2RH1).
GPCR Mutation Acquisition and Mapping.
TCGA has over 10,000 samples from 33 cancer types including ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, UCS, and UVM (SI Appendix, Table S4). Seven different mutation calling algorithm results were combined by using the MC3 pipeline (72), and only proteins whose expression levels were above a threshold of one FPKM in the RNA-seq data were included. Mutations occurring within class A GPCRs were mapped onto the GPCR alignment, for each cancer type within the TCGA, to identify mutations occurring at identical positions across all GPCRs. Both synonymous and nonsynonymous mutations were mapped in this way. Only mutations occurring at the 269 high-coverage positions were considered for further analysis. In cases with multiple mutated isoforms of the same protein, we used the sequence position of the primary isoform of the protein (as annotated by Uniprot). Frameshift, stop mutations (nonsense mutations), and INDELS (insertion and/or deletion mutations) were beyond the scope of this analysis.
In Silico Simulation of GPCR Mutation Patterns.
-
1)
We retrieved the nucleotide sequence of class A GPCRs from University of California Santa Cruz (UCSC) Genome Browser (https://genome.ucsc.edu/) using Human nucleotide RefSeq ID (starting with “NM_”), ensuring that the annotated sequence matched the primary amino acid sequence listed within Uniprot. From these sequences, for each GPCR, the codon triplets corresponding to the 269 high-coverage amino acid alignment positions were obtained. In this way, we calculated how frequently each of the 61 nonstop codons occurred at each “high-coverage” position.
-
2)
The nucleotide substitution frequency across all proteins and all patients for each cancer type was tabulated from TCGA database. For internal consistency, INDELS and nonsense mutations were ignored in this calculation. This process resulted in a 4 × 4 nucleotide substitution frequency matrix for each cancer type.
-
3)
Using this nucleotide frequency matrix, we computed the expected codon substitution rate for all 61 nonstop codons. Specifically, given the number of observed GPCR mutations, N, in each cancer type, we calculated how often we would expect to see each nucleotide substitution, by multiplying Mrate by N to obtain Mcount. For each nucleotide substitution pair, from i to j, in Mcount, we simulated Mcount (i,j) number of mutations by selecting a random codon position in a random codon in a random protein where the original position was nucleotide i and subsequently substituted that position to nucleotide j. This was repeated for all i, j pairs to quantify how frequently each codon was mutated and how often each codon transitioned to the other 61 codons, resulting in the codon × codon mutation frequency Ccount. The sum across the rows of Ccount provides an array, P, of the expected mutation count for each original codon for that cancer type.
-
4)
The expected mutation count at each position is therefore the sum of the normalized mutation count for each codon at that position. If we expect 100 mutations at codon Ci, we normalized this value by dividing by the total number of occurrences of Ci across all GPCRs, resulting in an array of the number of mutations per codon occurrence, Crate.
Code is available at https://github.com/LichtargeLab.
Scoring Protein Position Importance and Predicting the Impact of Mutations.
We used the Evolutionary Trace Webserver (http://lichtargelab.org/software/traceview) to quantify the importance of each sequence position. Using the input sequence of each GPCR in FASTA format, ET performs a blast search of homologous proteins, removes redundant and noisy sequences, aligns protein sequences, and then computes a score for each position quantifying the correlation of changes in genotype to those in phenotype. For this analysis, we used the default parameters available on the webserver.
To quantify the predicted impact a mutation would have on protein function, we used EA. EA is a first-principles–based equation that calculates mutation impact by multiplying the evolutionary importance of the sequence position (ET) by substitution log odds which are measurements for the magnitude of the substitution (19). Mutations are scored from 0, no impact, to 100, highly impactful on protein function. This technique is purely sequence based, utilizing evolutionary patterns and data that can further incorporate secondary structure for improved prediction.
Enrichment of Analysis of EA, ET, and Structural Positions and Motifs.
Enrichments of EA and ET scores were determined using a Mann–Whitney test to measure the level of distribution shift against mutations from a healthy population. Explicitly, each mutation’s EA score or ET score was appended to an array. This array distribution was then compared against the null set. In the case of EA, the null set consisted of EA scores for class A GPCR variants in a healthy population (1000 Genome Project). The null set for ET was all 269 positions. To identify proximal residues (8 Å) to the GPCR functional motifs, the three-dimensional molecular structure of the β2-adrenergic receptor (gene name B2AR, PDB ID code 2RH1) was used to measure interresidue distance from atomic x, y, z coordinates.
Enrichment of a Single G Protein Signaling Pathway.
To measure whether GPCR mutations preferentially occurred within GPCRs which signal through a specific G protein, we determined the primary G protein for each GPCR as reported in GPCRdb (https://gpcrdb.org) (34). For this analysis, we have ignored olfactory receptors, as their signaling pathways in the absence of Golf are not fully understood. Using these data, we were able to determine how many class A GPCRs signaled through each of the primary G proteins.
Cell Lines and Growth Conditions.
FreeStyle 293-F cells (Gibco) were purchased from Thermo Fisher and maintained in FreeStyle media (Gibco) supplemented with 2% fetal bovine serum (FBS) (Sigma). The HTLA cell line, which is a HEK293 line stably expressing a tetracycline transactivator (tTA)-dependent luciferase reporter and βarrestin2 fused to a TEV protease, was a gift from Bryan Roth, University of North Carolina, Chapel Hill, NC. HTLA cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 10% FBS, 2 μg/mL puromycin, and 100 μg/mL hygromycin B. For Tango assays, HTLA cells were seeded in media without antibiotics. All cells were grown in a 37 °C humidified incubator with 5% CO2.
DNA Constructs.
Tango constructs containing human GRPR, HCRTR2, DRD2, or MTNR1A with an N-terminal hemaglutinin signal sequence (73) (HAss) followed by a FLAG tag, and C-terminally fused to the vasopressin 2 receptor tail and TEV protease cleavage site followed by tTA in a pCDNA3.1 backbone (37), were obtained from Addgene (a gift from Bryan Roth; Addgene plasmids 66394, 66399, 66269, and 66443). Mutants were constructed from Tango constructs by site-directed mutagenesis using the Quickchange method. For Ca2+ mobilization assays, WT and mutant GPCRs, along with the N-terminal HAss and FLAG tag, were PCR amplified from Tango constructs, adding a stop codon, and cloned into pCDNA3.1. GPCR-containing open reading frames were confirmed in all constructs by Sanger sequencing. Empty pCDNA3.1 was used as the empty vector (EV) control for both Tango and Ca2+ mobilization assays. Chimeric G protein Gαqo consisting of mouse Gαq with the last five amino acids swapped with those from Gαo and an HA tag replacing Gαq residues 125 to 130, allowing Gi/o coupled receptors to engage the Gq pathway (36), was a gift from Bruce Conklin, University of California, San Francisco, CA, and was subcloned into pCDNA3.1.
Ligands.
Dopamine hydrochloride was purchased from Sigma, and freshly prepared immediately before use. The peptides Bombesin and Orexin-A were from Cayman Chemical, and stock solutions were prepared in water and stored at −80 °C. Melatonin was purchased from Cayman Chemical or Tocris Bioscience, and stock solutions were prepared in dimethyl sulfoxide (DMSO); experimental samples contained a final concentration of 0.1% DMSO or less.
Ca2+ Mobilization Assay for G Protein Activation.
Black clear-bottom poly-D-Lysine–coated 96-well plates (Corning BioCoat) were seeded with 40,000 HEK 293-F cells per well. The next day, cells were transfected with 50 ng per well pCDNA3.1-HAss-FLAG-GPCR constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For DRD2 and MTNR1A, cells were cotransfected with 150 ng of chimeric Gαqo. Approximately 38 h to 46 h posttransfection, Ca2+ mobilization assays were performed essentially as described (74). Cells were washed once with KRH buffer (120 mM NaCl, 4.7 mM KCl, 2.2 mM CaCl2, 10 mM Hepes, 1.2 mM KH2PO4, 1.2 mM MgSO4, 1.8 g/L glucose, pH 7.4, supplemented with 1 mM probenecid), and loaded with 2.7 μM Fluo-4 AM (Invitrogen) in KRH with 0.01% Pluronic F-127 (Biotium) for 1 h at room temperature in the dark. Cells were washed once with KRH; then 120 μL of KRH was added to each well. The assay plate and drug plate containing 4× drug dilutions in KRH were preincubated for 10 min in a plate reader (Flex Station 3, Molecular Devices) preheated to 37 °C. Measurements were acquired every ∼1.6 s (Ex/Em 488/520); 40 μL of 4× drug were added after 20 s. For DRD2, incubations and dopamine dilutions were done in KRH supplemented with 1 mM ascorbic acid.
Tango Assay for β-Arrestin Recruitment.
Tango assays were performed as described (37), with modifications. Clear-bottom or opaque poly-D-Lysine–coated 96-well plates (Corning BioCoat) were seeded with 35,000 to 40,000 HTLA cells per well. The next day, cells were transfected with 50 ng per well (DRD2, GRPR, and HCRTR2) or 1 ng per well (MTNR1A) Tango constructs (HAss-FLAG-GPCR-V2tail-TEV-tTA) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Approximately 24 h posttransfection, the media was changed to 100 μL of DMEM with 1% dialyzed FBS (Omega Scientific), and, ∼42 h to 43 h posttransfection, 50 μL of 3× drug diluted in the same media were added to each well. For DRD2, dopamine was prepared in media with 1 mM ascorbic acid. Approximately 20 h after drug addition, media was removed, and 80 μL of BrightGlo luciferase substrate (Promega) diluted 10-fold in 10 mM Hepes pH 7.3 were added to each well, and luminescence (all wavelengths) was read 3 min to 5 min later (Flex Station 3, Molecular Devices).
Functional Assay Data Analysis.
All functional assays were performed with three technical replicates for each condition, as well as WT controls on the same plate on at least three separate days. For Ca2+ assays, data from each well were baseline corrected by subtracting the average of the first 12 points (∼18 s to 19 s), and the maximum of baseline-corrected data between ∼20 and 60 s was extracted, with custom scripts in Mathematica v.12 (Wolfram). Maximum responses from triplicate wells were fit with sigmoidal dose–response curves using Prism v.5 (GraphPad). For Tango assays, raw luminescence values were fit with sigmoidal dose–response curves. Wells with vehicle only (no drug) were included in the curve fits by assigning them a drug concentration of 10−13 M (three to four orders of magnitude lower than the lowest drug concentration), except for MTNR1A, where 10−14 was used. Log (EC50) values obtained from the curve fits are reported; for mutants with no response to ligand or a nonconverging curve fit, no EC50 values could be determined. Efficacy was calculated as the top plateau minus the bottom plateau of the curve fit and divided by the corresponding WT control from the same plate. For mutants with no response to ligand or a nonconverging curve fit, technical replicates were averaged, and the efficacy was calculated as the maximum drug-induced response minus the no-drug value, and divided by WT. For display only, time courses and dose–response curves from multiple plates are shown together by normalizing as follows: Time courses are shown normalized to the maximum of the WT control from the same plate, and dose–response curves are shown normalized to the top plateau of the curve fit for WT from the same plate.
Cell ELISA for Surface Expression.
The ELISA procedure was adapted from Sung et al. (73). HEK293F cells in 96-well poly-D-Lysine–coated plates were transfected with 50 ng per well Ca2+ assay constructs, and fixed ∼40 h to 45 h later. HTLA cells transfected with 50 ng per well Tango constructs were changed to DMEM+1% dialyzed FBS media ∼24 h later and fixed 1 d to 2 d after that. Cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, then washed with PBS and blocked in PBSA (PBS with 1% bovine serum albumin) for 15 min, followed by incubation with FLAG antibody (rabbit monoclonal, Cell Signaling #14793) diluted 1:1,000 in PBSA for 1 h. Cells were washed and incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch) 1:1,000 (0.8 μg/mL antibody) in PBSA for 30 min. Cells were washed again, 50 μL of SuperSignal Pico substrate (Thermo) were added to each well, and luminescence (all wavelengths) was read immediately. ELISAs were performed with five or six technical replicates for each mutant, along with WT and EV controls on the same plate. Replicates were averaged, and relative surface expression values were calculated by subtracting the EV value and then dividing by WT from the same plate. In some cases, negative values were obtained—these were set at zero.
Immunofluorescence Microscopy and Image Processing.
HTLA cells on poly-D-lysine–coated coverslips in 24-well plates were transfected with 400 ng of Tango assay constructs, along with 200 ng of pCDNA3.1-EGFP to identify transfected cells. Two days posttransfection, cells were fixed, blocked, and labeled with FLAG antibody in nonpermeabilizing conditions as described above for ELISAs. Coverslips were then incubated with Alexa-555–conjugated donkey anti-rabbit secondary antibody (Invitrogen), 2 μg/mL, in PBSA for 30 min, washed, and mounted with Prolong Diamond (Invitrogen). For analysis of total protein in permeabilizing conditions, PBSA was supplemented with 1% Triton X-100 for blocking and antibody steps.
Cells were imaged with a Zeiss LSM-710 confocal microscope using a 63× oil immersion objective (Zeiss, Plan-Apochromat 63×/1.4 Oil DIC M27) with Immersol 518F immersion oil (Zeiss). Single optical sections in Alexa 555 and EGFP channels were acquired sequentially with 561- and 488-nm lasers, with laser power and gain adjusted to avoid saturated pixels. For each GPCR, WT and mutant constructs were imaged with identical settings. Three images were acquired for each coverslip, total intensity in the Alexa 555 channel was measured for each image using Mathematica (Wolfram), and values for mutants were normalized by dividing by the values for their respective WT control.
Supplementary Material
Acknowledgments
This work was supported by the NIH Grant (GM066099 and AG061105) and the Robert A. Welch Foundation Grant (Q0035).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113373118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix and our code is publicly accessible on GitHub (https://github.com/LichtargeLab/Recurrent-High-Impact-Mutations-on-Class-A-GPCRs.git).
References
- 1.Bar-Shavit R., et al. , G protein-coupled receptors in cancer. Int. J. Mol. Sci. 17, 1320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lappano R., Maggiolini M., GPCRs and cancer. Acta Pharmacol. Sin. 33, 351–362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wettschureck N., Offermanns S., Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Dorsam R. T., Gutkind J. S., G-protein-coupled receptors and cancer. Nat. Rev. Cancer 7, 79–94 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Nieto Gutierrez A., McDonald P. H., GPCRs: Emerging anti-cancer drug targets. Cell. Signal. 41, 65–74 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Greenman C., et al. , Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratton M. R., Campbell P. J., Futreal P. A., The cancer genome. Nature 458, 719–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandin F., Clay P., Upfal E., Raphael B. J., Discovery of mutated subnetworks associated with clinical data in cancer. Pac. Symp. Biocomput. 2012, 55–66 (2012). [PubMed] [Google Scholar]
- 9.Jia P., Zhao Z., VarWalker: Personalized mutation network analysis of putative cancer genes from next-generation sequencing data. PLoS Comput. Biol. 10, e1003460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence M. S., et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu V., et al. , Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 294, 11062–11086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raimondi F., et al. , Rare, functional, somatic variants in gene families linked to cancer genes: GPCR signaling as a paradigm. Oncogene 38, 6491–6506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller M. L., et al. , Pan-cancer analysis of mutation hotspots in protein domains. Cell Syst. 1, 197–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson T. A., Gauran I. I. M., Park J., Park D., Kann M. G., Oncodomains: A protein domain-centric framework for analyzing rare variants in tumor samples. PLoS Comput. Biol. 13, e1005428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F., et al. , Protein domain-level landscape of cancer-type-specific somatic mutations. PLoS Comput. Biol. 11, e1004147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallion J., Wilkins A. D., Lichtarge O., Human kinases display mutational hotspots at cognate positions within cancer. Pac. Symp. Biocomput. 22, 414–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D., Zhao Q., Wu B., Structural studies of G Protein-coupled receptors. Mol. Cells 38, 836–842 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tehan B. G., Bortolato A., Blaney F. E., Weir M. P., Mason J. S., Unifying family A GPCR theories of activation. Pharmacol. Ther. 143, 51–60 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Katsonis P., Lichtarge O., A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 24, 2050–2058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkins A. D., Bachman B. J., Erdin S., Lichtarge O., The use of evolutionary patterns in protein annotation. Curr. Opin. Struct. Biol. 22, 316–325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtarge O., Bourne H. R., Cohen F. E., An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 257, 342–358 (1996). [DOI] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium, et al. , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballesteros J. A., Weinstein H., “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors” in Methods in Neurosciences, Sealfon S. C., Ed. (Academic, 1995), pp. 366–428. [Google Scholar]
- 24.Lee S.-J., Depoortere I., Hatt H., Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 18, 116–138 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen S. G. F., et al. , Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihalek I., Reš I., Yao H., Lichtarge O., Combining inference from evolution and geometric probability in protein structure evaluation. J. Mol. Biol. 331, 263–279 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Alexandrov L. B., et al. , Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood L. D., et al. , The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Parmigiani G., et al. , Validity of models for predicting BRCA1 and BRCA2 mutations. Ann. Intern. Med. 147, 441–450 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson I. R., Takahashi K., Futreal P. A., Chin L., Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet. 14, 703–718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson S. M., et al. , Elucidation of G-protein and β-arrestin functional selectivity at the dopamine D2 receptor. Proc. Natl. Acad. Sci. U.S.A. 112, 7097–7102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erdin S., Ward R. M., Venner E., Lichtarge O., Evolutionary trace annotation of protein function in the structural proteome. J. Mol. Biol. 396, 1451–1473 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsonis P., et al. , Single nucleotide variations: Biological impact and theoretical interpretation. Protein Sci. 23, 1650–1666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooistra A. J., et al. , GPCRdb in 2021: Integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones D. T., Reed R. R., Golf: An olfactory neuron specific-G protein involved in odorant signal transduction. Science 244, 790–795 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Conklin B. R., Farfel Z., Lustig K. D., Julius D., Bourne H. R., Substitution of three amino acids switches receptor specificity of Gq α to that of Gi α. Nature 363, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Kroeze W. K., et al. , PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostrow S. L., Barshir R., DeGregori J., Yeger-Lotem E., Hershberg R., Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 10, e1004239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenman C., Wooster R., Futreal P. A., Stratton M. R., Easton D. F., Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madabushi S., et al. , Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J. Biol. Chem. 279, 8126–8132 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Masjedi S., Zwiebel L. J., Giorgio T. D., Olfactory receptor gene abundance in invasive breast carcinoma. Sci. Rep. 9, 13736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelis L., Jovancevic N., Bechara F. G., Neuhaus E. M., Hatt H., Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp. Dermatol. 26, 569–576 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Kalbe B., et al. , Helional-induced activation of human olfactory receptor 2J3 promotes apoptosis and inhibits proliferation in a non-small-cell lung cancer cell line. Eur. J. Cell Biol. 96, 34–46 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Weber L., et al. , Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLoS One 12, e0172491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhaus E. M., et al. , Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 284, 16218–16225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber L., et al. , Characterization of the olfactory receptor OR10H1 in human urinary bladder cancer. Front. Physiol. 9, 456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raimondi F., et al. , Genetic variants affecting equivalent protein family positions reflect human diversity. Sci. Rep. 7, 12771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Touhara K., Deorphanizing vertebrate olfactory receptors: Recent advances in odorant-response assays. Neurochem. Int. 51, 132–139 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Kato A., Touhara K., Mammalian olfactory receptors: Pharmacology, G protein coupling and desensitization. Cell. Mol. Life Sci. 66, 3743–3753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manteniotis S., et al. , Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell Death Discov. 2, 15070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal W., Antaramian A., Gilbert S., Birnbaumer M., Nephrogenic diabetes insipidus. A V2 vasopressin receptor unable to stimulate adenylyl cyclase. J. Biol. Chem. 268, 13030–13033 (1993). [PubMed] [Google Scholar]
- 52.Barak L. S., Oakley R. H., Laporte S. A., Caron M. G., Constitutive arrestin-mediated desensitization of a human vasopressin receptor mutant associated with nephrogenic diabetes insipidus. Proc. Natl. Acad. Sci. U.S.A. 98, 93–98 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheer A., Fanelli F., Costa T., de Benedetti P. G., Cotecchia S., Constitutively active mutants of the a1B-adrenergic receptor: Role of highly conserved polar amino acids in receptor activation. EMBO J. 15, 3566–3578 (1996). [PMC free article] [PubMed] [Google Scholar]
- 54.Römpler H., Yu H.-T., Arnold A., Orth A., Schöneberg T., Functional consequences of naturally occurring DRY motif variants in the mammalian chemoattractant receptor GPR33. Genomics 87, 724–732 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Alewijnse A. E., et al. , The effect of mutations in the DRY motif on the constitutive activity and structural instability of the histamine H(2) receptor. Mol. Pharmacol. 57, 890–898 (2000). [PubMed] [Google Scholar]
- 56.Lagane B., et al. , Mutation of the DRY motif reveals different structural requirements for the CC chemokine receptor 5-mediated signaling and receptor endocytosis. Mol. Pharmacol. 67, 1966–1976 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Wess J., Nanavati S., Vogel Z., Maggio R., Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor. EMBO J. 12, 331–338 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu S., et al. , Involvement of β-arrestins in cancer progression. Mol. Biol. Rep. 40, 1065–1071 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Li Y., et al. , Melatonin for the prevention and treatment of cancer. Oncotarget 8, 39896–39921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sainz R. M., et al. , Melatonin and cell death: Differential actions on apoptosis in normal and cancer cells. Cell. Mol. Life Sci. 60, 1407–1426 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao M., et al. , The reduction in circulating melatonin level may contribute to the pathogenesis of ovarian cancer: A retrospective study. J. Cancer 7, 831–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basler M., et al. , Urinary excretion of melatonin and association with breast cancer: Meta-analysis and review of the literature. Breast Care (Basel) 9, 182–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sigurdardottir L. G., et al. , Urinary melatonin levels, sleep disruption, and risk of prostate cancer in elderly men. Eur. Urol. 67, 191–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y. M., et al. , The efficacy and safety of melatonin in concurrent chemotherapy or radiotherapy for solid tumors: A meta-analysis of randomized controlled trials. Cancer Chemother. Pharmacol. 69, 1213–1220 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Gil-Martín E., Egea J., Reiter R. J., Romero A., The emergence of melatonin in oncology: Focus on colorectal cancer. Med. Res. Rev. 39, 2239–2285 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Basu S., et al. , The neurotransmitter dopamine inhibits angiogenesis induced by vascular permeability factor/vascular endothelial growth factor. Nat. Med. 7, 569–574 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Hoeppner L. H., et al. , Dopamine D2 receptor agonists inhibit lung cancer progression by reducing angiogenesis and tumor infiltrating myeloid derived suppressor cells. Mol. Oncol. 9, 270–281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voisin T., et al. , Aberrant expression of OX1 receptors for orexins in colon cancers and liver metastases: An openable gate to apoptosis. Cancer Res. 71, 3341–3351 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Xu T.-R., Yang Y., Ward R., Gao L., Liu Y., Orexin receptors: Multi-functional therapeutic targets for sleeping disorders, eating disorders, drug addiction, cancers and other physiological disorders. Cell. Signal. 25, 2413–2423 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Biegańska K., Sokołowska P., Jöhren O., Zawilska J. B., Orexin A., Orexin A suppresses the growth of rat C6 glioma cells via a caspase-dependent mechanism. J. Mol. Neurosci. 48, 706–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pei J., Kim B.-H., Grishin N. V., PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ellrott K., et al. , MC3 Working Group; Cancer Genome Atlas Research Network, Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sung Y.-M., Wilkins A. D., Rodriguez G. J., Wensel T. G., Lichtarge O., Intramolecular allosteric communication in dopamine D2 receptor revealed by evolutionary amino acid covariation. Proc. Natl. Acad. Sci. U.S.A. 113, 3539–3544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.G. J. Rodriguez, R. Yao, O. Lichtarge, T. G. Wensel, Evolution-guided discovery and recoding of allosteric pathway specificity determinants in psychoactive bioamine receptors. Proc. Natl. Acad. Sci. U.S.A 107, 7787–7792 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix and our code is publicly accessible on GitHub (https://github.com/LichtargeLab/Recurrent-High-Impact-Mutations-on-Class-A-GPCRs.git).