Significance
Plants modulate photosynthesis activity in response to the surrounding environment. It is well known that the redox-responsive protein thioredoxin (Trx) activates photosynthesis-related enzymes in the light. However, the factors involved in deactivating them are not well understood. Recent in vitro experiments suggest that several Trx and Trx-like proteins serve as oxidation factors for Trx-targeted proteins; thus, we examined their functions in vivo. Consequently, we found that f-type Trx and two types of Trx-like proteins, Trx-like 2 and atypical Cys His-rich Trx, were involved in oxidative deactivation of photosynthesis-related enzymes (e.g., fructose-1,6-bisphosphatase, Rubisco activase, and the ATP synthase γ-subunit). Thus, this study reveals the functions of oxidation factors in vivo and elucidates the regulation system for photosynthesis in the dark.
Keywords: redox regulation, oxidation, thioredoxin, thioredoxin-like protein, 2-Cys peroxiredoxin
Abstract
Thioredoxin (Trx) is a protein that mediates the reducing power transfer from the photosynthetic electron transport system to target enzymes in chloroplasts and regulates their activities. Redox regulation governed by Trx is a system that is central to the adaptation of various chloroplast functions to the ever-changing light environment. However, the factors involved in the opposite reaction (i.e., the oxidation of various enzymes) have yet to be revealed. Recently, it has been suggested that Trx and Trx-like proteins could oxidize Trx-targeted proteins in vitro. To elucidate the in vivo function of these proteins as oxidation factors, we generated mutant plant lines deficient in Trx or Trx-like proteins and studied how the proteins are involved in oxidative regulation in chloroplasts. We found that f-type Trx and two types of Trx-like proteins, Trx-like 2 and atypical Cys His-rich Trx (ACHT), seemed to serve as oxidation factors for Trx-targeted proteins, such as fructose-1,6-bisphosphatase, Rubisco activase, and the γ-subunit of ATP synthase. In addition, ACHT was found to be involved in regulating nonphotochemical quenching, which is the mechanism underlying the thermal dissipation of excess light energy. Overall, these results indicate that Trx and Trx-like proteins regulate chloroplast functions in concert by controlling the redox state of various photosynthesis-related proteins in vivo.
Plant chloroplasts have evolved multiple strategies with which to adapt photosynthesis to fluctuating light environments. One such strategy involves the redox regulation of various enzymes that function in photosynthesis reactions. Multiple photosynthesis-related proteins, such as the four Calvin–Benson cycle enzymes (glyceraldehyde-3-phosphate dehydrogenase, fructose-1,6-bisphosphatase [FBPase], sedoheptulose-1,7-bisphosphatase [SBPase], and phosphoribulokinase [PRK]), possess redox-active Cys residues (1, 2). In addition, the γ-subunit of ATP synthase (CF1-γ) and two regulatory proteins associated with Calvin–Benson cycle enzymes, CP12 and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase (RCA), are also redox-regulated (2–4). In the 1970s thioredoxin (Trx) was identified as a reducing power mediator for FBPase and SBPase in chloroplasts (5, 6). In a light-containing environment, reducing power is transferred from the photosynthetic electron transport system to Trx via ferredoxin and ferredoxin-Trx reductase (6). Trx then achieves light-dependent activation of its target enzymes by reducing the disulfide bond on these enzymes.
In chloroplasts, NADPH-Trx reductase C (NTRC) works in parallel with the Trx-dependent system as another redox pathway. NTRC is also a redox-responsive protein containing both NADPH-dependent Trx reductase and Trx domains; these enable NTRC to reduce its target proteins using the reducing power of NADPH (7). NTRC can reduce 2-Cys peroxiredoxin (2-Cys Prx) in addition to several Trx-targeted proteins (8–12). 2-Cys Prx utilizes reducing power to reduce reactive oxygen species such as H2O2 (13). In chloroplasts, NTRC is thought to be a major electron donor for 2-Cys Prx (14) because the reducing power transfer efficiency from NTRC is extremely high compared with that from typical chloroplast Trx proteins (12). Plants deficient in NTRC show severe phenotypes, such as stunted growth, low chlorophyll content, and very high nonphotochemical quenching (NPQ) (7, 11, 12, 14–17). Thus, it is clear that NTRC plays important physiological roles in chloroplasts.
Redox-regulated proteins in the stroma are reduced in the light and then reoxidized in the dark (18, 19). Reoxidation is an important process in plants; for example, we recently showed that the reoxidation of chloroplast NADP-malate dehydrogenase is important for maintaining NADPH homeostasis in chloroplasts, particularly in an environment with fluctuating light (20). Despite the importance of the oxidation process, the proteins involved in target oxidation have yet to be clarified. Recently, Trx-like proteins, such as Trx-like 2 (TrxL2) and atypical Cys His-rich Trx (ACHT), have been suggested as oxidation factors (21–27). These reports were mainly based on the results of in vitro experiments, suggesting that Trx-like proteins transfer the reducing power of Trx-targeted proteins to H2O2 via 2-Cys Prx. However, the functions of these proteins in vivo are not known very well. The so-called common Trxs belonging to f-, m-, x-, y- (or z-?) types were also thought to be the candidate of the oxidation factor. Because it is known that, particularly, Trx-f can oxidize its target proteins under certain conditions in vitro (25, 28), we focused this work on Trx-f.
Target oxidation by ACHT1 and ACHT2, among five ACHT isoforms in Arabidopsis thaliana (29), has been demonstrated in vitro (25). ACHT1 and ACHT2 are broadly conserved in photosynthetic organisms, including green algae, moss, and seed plants (30). Their amino acid sequences and biochemical properties are similar (SI Appendix, Fig. S1A) (25, 29). In addition, ACHT1 and ACHT2 (designated also as Lilium5 and Lilium2, respectively) are predicted to originate from the same ancestral gene (31). Comparison of the expression patterns of ACHT1 and ACHT2 in the database shows that ACHT2 is expressed more than ACHT1, especially in leaves (SI Appendix, Fig. S1B) (32), suggesting that ACHT2 may play a dominant role in A. thaliana leaves.
Oxidation of target proteins by the TrxL2 isoforms from A. thaliana, namely TrxL2.1 and TrxL2.2, has been demonstrated in vitro (22). TrxL2 genes are also conserved in photosynthetic organisms, such as seed plants, moss, and some green algae, but not in Chlamydomonas reinhardtii (30). Although the amino acid sequences and biochemical properties of TrxL2.1 and TrxL2.2 are similar (SI Appendix, Fig. S2A) (22), their expression patterns are different, and TrxL2.1 is reported to be more expressed than TrxL2.2, particularly in leaves (SI Appendix, Fig. S2B) (32). In addition, TrxL2.1 expression seems to be regulated by the circadian rhythm and the rhythm of temperature change (SI Appendix, Fig. S2C). The expression of TrxL2.1 is more strongly induced before and during light-to-dark transitions than TrxL2.2, suggesting that TrxL2.1 plays a predominant role during these periods.
In the present study, we generated A. thaliana mutant plant lines deficient in Trx-f1 and Trx-f2, TrxL2.1, or ACHT1 and ACHT2, whose target oxidation activities are well studied in vitro, and used these plants to investigate redox state changes in chloroplasts. We found that Trx-f, TrxL2.1, ACHT1, and ACHT2 are involved in the oxidation of FBPase, CF1-γ, and RCA. ACHT2 also seemed to be involved in the regulation of NPQ. Furthermore, the knockout of Trx-like proteins suppressed the impact of NTRC deficiency in plants, suggesting that a connection existed between the NTRC system and Trx-like protein-involving system.
Results
Generation of A. thaliana Mutant Plants Deficient in TrxL2 and ACHT.
To investigate the role of Trx-like proteins in vivo, we first attempted to generate A. thaliana mutant plant lines deficient in TrxL2.1, TrxL2.2, and both ACHT1 and ACHT2, utilizing CRISPR/Cas9-mediated genome editing (33). Consequently, we obtained two TrxL2.1-deficient lines [trxl2.1(1) and trxl2.1(2)] generated with different CRISPR constructs (Fig. 1A and SI Appendix, Fig. S3). We also obtained the acht line, which was intended to be a double-knockout line for ACHT1 and ACHT2 (Fig. 1A and SI Appendix, Fig. S4A). All of the mutants had 1-nt insertions in exon regions that resulted in frameshifts and disruptions of the genes (SI Appendix, Figs. S3 and S4A). Our repeated attempts to obtain a mutant line deficient in TrxL2.2 were unsuccessful (SI Appendix, Fig. S3B).
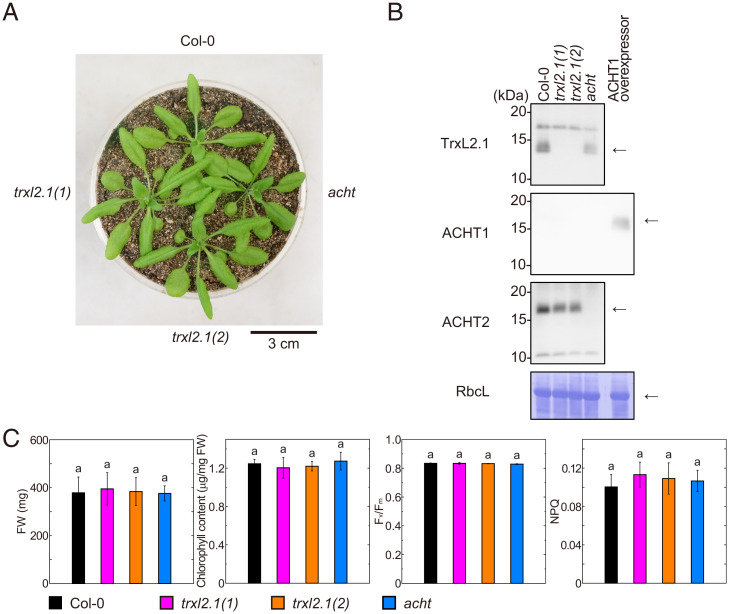
Fig. 1.
Phenotypes of mutant plants deficient in Trx-like proteins. (A) Visible phenotypes of plants deficient in Trx-like proteins. Plants were grown for 4 wk. (B) Confirmation of the knockout of Trx-like proteins by Western blotting. Arrows indicate the proteins of interest. Bands without arrows are nonspecific proteins. Leaf extract from the ACHT1 overexpressor was used as a positive control. Coomassie brilliant blue (CBB)-stained Rubisco large subunit (RbcL) is shown as a loading control. (C) Physiological parameters of plants deficient in Trx-like proteins. Each value is presented as the mean ± SD (n = 5). Different letters indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey honestly significant difference [HSD]).
The deficiency of each protein was confirmed by Western blotting. As shown in Fig. 1B, TrxL2.1 and ACHT2 proteins were absent in the lines for which TrxL2.1 and ACHT2 genes were respectively disrupted; however, the deficiency of ACHT1 was not confirmed by Western blotting. In addition, ACHT1 was detected in an ACHT1 overexpressor line but not in others, including Col-0. The messenger RNA expression level of ACHT1 in rosette leaves in A. thaliana is very low [37- and 15-fold lower than those of Trx-f1 and ACHT2, respectively (32)]. Our protein quantification also showed that less than 0.1 ng of ACHT1 protein and more than 1 ng ACHT2 protein were detected in a 30 μg leaf protein extract (SI Appendix, Fig. S4B). The original low expression level of ACHT1 is likely the reason why we could not detect this protein in both the Col-0 and mutant plants. Taking these findings together with DNA sequencing data, we concluded that the acht line is a double-knockout line for ACHT1 and ACHT2.
We then assessed the impact of the Trx-like proteins on the fresh weight (FW) of plants, chlorophyll content, maximum quantum yield of photosystem II (PSII) (Fv/Fm), and NPQ. NPQ represents the degree of nonphotochemical energy dissipation around PSII. The values of these variables did not differ significantly among plant lines (Fig. 1C), indicating that the deficiency of TrxL2.1 alone or ACHT1 and ACHT2 scarcely affected the photosynthetic performance of the plants in the conditions examined.
Effects of Trx-Like Protein Deficiency on Target Oxidation during Light-to-Dark Transitions.
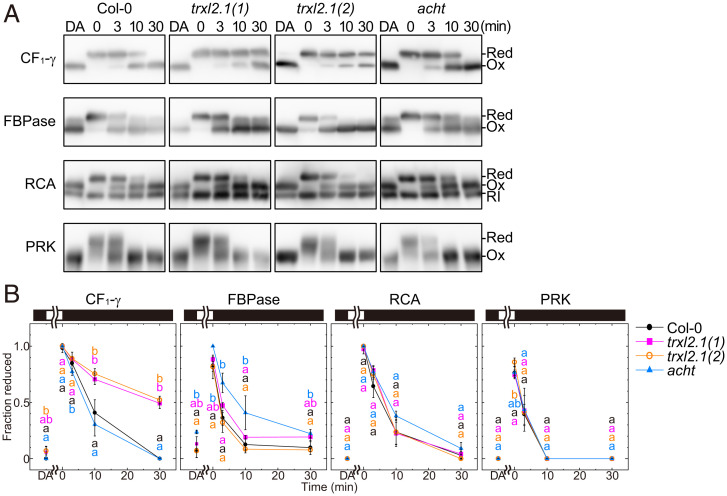
Using the generated mutant lines, we investigated the influence of Trx-like proteins in the oxidation of target proteins during light-to-dark transitions. Dark-adapted plants were illuminated with high-light intensity (700 μmol photons m−2·s−1) and subsequently transferred into the dark. The change in the redox states of Trx-targeted proteins—such as CF1-γ, FBPase, RCA, and PRK—during these transitions was monitored by thiol modification with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS). The oxidation of CF1-γ in the dark was drastically retarded in the trxl2.1(1) and trxl2.1(2) lines but not in the acht line (Fig. 2). In contrast, the oxidation of FBPase was significantly retarded in the acht line (Fig. 2). These results imply that each of the Trx-like proteins is involved in the oxidation of Trx-targeted proteins in chloroplasts with a particular specificity: that is, TrxL2.1 plays a dominant role in CF1-γ oxidation, and ACHT1 and ACHT2 are involved in the FBPase oxidation (Fig. 2B). These findings are consistent with our previous in vitro results, which indicated that ACHT1 and ACHT2 are efficient oxidation factors for FBPase (25). Because the oxidation of FBPase was also observed in the acht line, there might be other oxidation factors for this enzyme in addition to ACHT1 and ACHT2.
Fig. 2.
Oxidation of Trx-targeted proteins in Trx-like protein-deficient plants during light-to-dark transitions. (A) Western blotting. The redox states of Trx-targeted proteins in dark-adapted (DA) plants or plants during light-to-dark transitions were monitored. Abbreviations: Ox, oxidized; Red, reduced; RI, redox-insensitive splicing variant. (B) Redox state dynamics of Trx-targeted proteins. Using the data from A, each band intensity was measured, and the redox state was determined and plotted. Black and white bars represent 0- and 700-μmol photons m−2·s−1 light intensity, respectively. Each value is presented as the mean ± SD [n = 6 (Col-0) or n = 3 (the others)]. Different letters at each time point indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD).
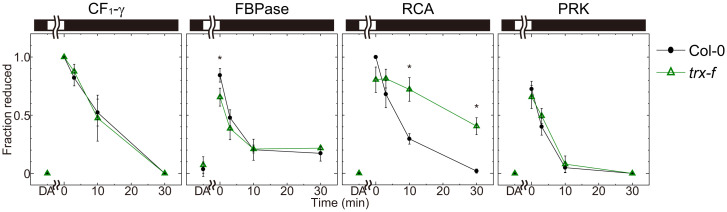
The oxidation of RCA and PRK was not altered significantly in the examined lines (Fig. 2B). Because Trx-f has also been suggested to serve as an oxidation factor (34), we investigated the oxidation of Trx-targeted proteins in a Trx-f–deficient line (trx-f) that we produced in a previous study (35). The oxidation of RCA was significantly impaired in the trx-f line compared with that in the Col-0 line (Fig. 3 and SI Appendix, Fig. S5A), indicating that Trx-f is a dominant oxidation factor for RCA.
Fig. 3.
Oxidation of Trx-targeted proteins in plants deficient in Trx-f. Redox states of Trx-targeted proteins were plotted. For this experiment, plants were preilluminated for 60 min and then transferred into the dark. Using the data from SI Appendix, Fig. S5A, the intensity of each band was measured, and the redox state was determined and plotted. Black and white bars represent 0 and 700 μmol photons m−2·s−1 light intensity, respectively. Each value is presented as the mean ± SD (n = 3). An asterisk indicates a significant difference between the Col-0 and trx-f lines (P < 0.05; Welch’s t test).
Suppression of ntrc Phenotypes through Disruption of Trx-Like Proteins.
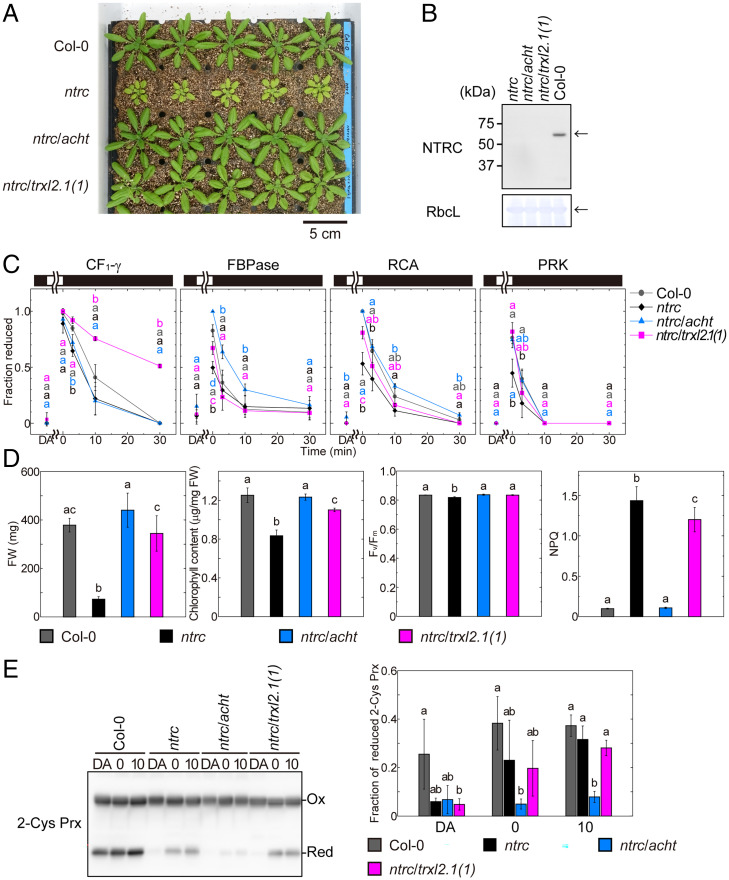
Several reports have previously demonstrated that Trx-like proteins obtain reducing power from reduced-form target proteins and then reduce 2-Cys Prx (21, 22, 24, 25, 27). NTRC is also reported to be an efficient electron donor for 2-Cys Prx (8, 12, 14). Therefore, we crossed an NTRC-deficient line (ntrc) with the acht line (resulting in the ntrc/acht line) or with the trxl2.1(1) line [resulting in the ntrc/trxl2.1(1) line] (Fig. 4 A and B and SI Appendix, Fig. S6) to investigate the relationships among Trx-like proteins, NTRC, and 2-Cys Prx. Although the ntrc line showed severe visible phenotypes (Fig. 4A), as previously reported (7, 11, 12, 14–17), the ntrc/acht and ntrc/trxl2.1(1) plants were visibly indistinguishable from the Col-0 line (Fig. 4A). These results suggest that the visible phenotypes of plants caused by a lack of NTRC are strongly affected by the functions of oxidation factor proteins.
Fig. 4.
Phenotypes of plants deficient in NTRC and Trx-like proteins. (A) Visible phenotypes of plants deficient in NTRC and Trx-like proteins. Plants were grown for 4 wk. (B) Confirmation of NTRC protein deficiency by Western blotting. Arrows indicate the proteins of interest. CBB-stained Rubisco large subunit (RbcL) is shown as a loading control. (C) Redox states of Trx-targeted proteins. Using the data from SI Appendix, Fig. S5B, the intensity of each band was measured, and the redox state was determined and plotted. The same data shown in Fig. 2 was used for Col-0. Black and white bars represent 0 and 700 μmol photons m−2·s−1 light intensity, respectively. Each value is presented as the mean ± SD [n = 6 (Col-0) or n = 3 (the others)]. Different letters at each time point indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD). (D) Physiological parameters. Each value is presented as the mean ± SD (n = 5). Different letters indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD). (E) Redox state of 2-Cys Prx at each time point (DA, 0 min, or 10 min). (Left) The result of Western blotting with anti-2-Cys Prx A antibody is shown. Because oxidized 2-Cys Prx (Ox) forms a homodimer with an intermolecular disulfide bond (or bonds), it is detected at the upper position than the reduced 2-Cys Prx (Red). (Right) By measuring the band intensity, the redox state was determined and is shown as the mean ± SD (n = 3). Different letters at each time point indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD).
We also monitored the redox states of Trx-targeted proteins in these mutant lines during light-to-dark transitions. In the ntrc line, Trx-targeted proteins were relatively oxidized in the light (t = 0) and during light-to-dark transitions when compared with the oxidation observed in the Col-0 line (Fig. 4C and SI Appendix, Fig. S5B), which is consistent with prior reports (36). In the ntrc/trxl2.1(1) and ntrc/acht lines, the redox states of several Trx-targeted proteins in the light were restored to the same level as those detected in the trxl2.1(1) and acht lines, respectively. In particular, CF1-γ and FBPase were in more reduced states in the ntrc/trxl2.1(1) line and the ntrc/acht line, respectively, than the states of these proteins in Col-0 (Fig. 4C).
We also measured four physiological parameters in the mutant lines. As expected from the plant phenotypes shown in Fig. 4A, FW, chlorophyll content, and Fv/Fm did not differ substantially among the ntrc/acht and ntrc/trxl2.1(1) lines and Col-0 (Fig. 4D). Consistent with previously reported results (11, 14, 15), the ntrc line showed very high NPQ (Fig. 4D). This high NPQ was also observed in the ntrc/trxl2.1(1) line, whereas the NPQ in the ntrc/acht line was at the same level as that detected in Col-0 (Fig. 4D). These results indicate that a relationship exists between NTRC and ACHT proteins in NPQ regulation.
A previous study suggested that the ntrc phenotypes are caused by an insufficient supply of reducing power to 2-Cys Prx (14). Therefore, we investigated light-dependent changes in the redox states of 2-Cys Prx in our mutant lines. The redox state of 2-Cys Prx in the ntrc/trxl2.1(1) line was almost identical to that in the ntrc line (Fig. 4E). However, 2-Cys Prx in the ntrc/acht line was more oxidized than 2-Cys Prx in the ntrc line; it was mostly oxidized, even in the light (Fig. 4 E, Right). Thus, we conclude that the recovery of the visible phenotypes, physiological parameters, and redox state of Trx-targeted proteins observed with further loss of oxidation factor proteins in the ntrc line is not due to the recovery of the 2-Cys Prx redox state.
Effects of the ACHT2 Expression Level on Plant Growth and NPQ.
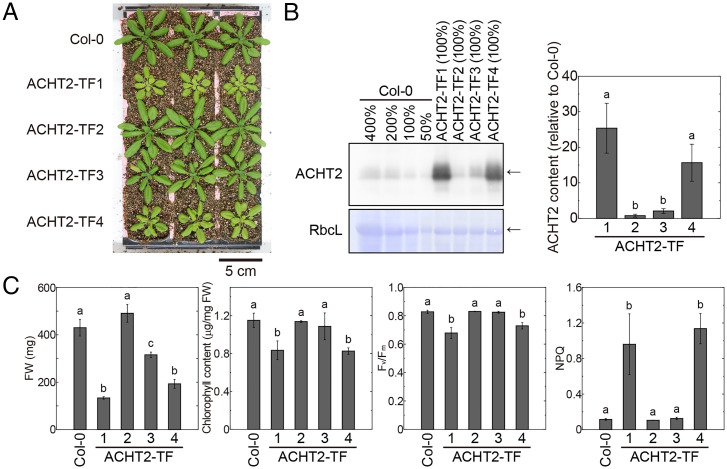
Because ACHT proteins are suggested to be involved in ntrc phenotypes, particularly in the extent of NPQ (Fig. 4), we further investigated the role of ACHT2 in chloroplasts. We transformed the Col-0 line with a vector for ACHT2 overexpression and obtained four mutant lines in which ACHT2 was expressed at various levels (Fig. 5 A and B): ACHT2-TF1, 25-fold; ACHT2-TF2, 0.8-fold; ACHT2-TF3, 2-fold; and ACHT2-TF4, 16-fold (“TF” denotes transformed). The highest expressing lines, ACHT2-TF1 and ACHT2-TF4, showed high NPQ as well as stunted growth, lower chlorophyll content, and lower Fv/Fm (Fig. 5 A and C).
Fig. 5.
Phenotypes of plants with various expression levels of ACHT2. (A) Visible phenotypes of plants with various expression levels of ACHT2. Plants were grown for 4 wk. (B) Expression levels of ACHT2. (Left) The results of Western blotting with anti-ACHT2 antibody are shown. Values (%) indicate the loaded amounts of leaf extract proteins. Arrows indicate the proteins of interest. CBB-stained Rubisco large subunit (RbcL) is shown as a loading control. (Right) To compare the ACHT content more accurately, the data obtained from diluted samples (see SI Appendix, Fig. S7) were used for band intensity measurements. ACHT2 content for each mutant line relative to that of the Col-0 line was calculated and is shown as the mean ± SD (n = 3). Different letters indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD). (C) Physiological parameters. Each value is presented as the mean ± SD (n = 3). Different letters indicate significant differences among plant lines (P < 0.05; one-way ANOVA and Tukey HSD).
Discussion
In this study, we investigated the physiological impacts of Trx and Trx-like proteins as oxidation factors in chloroplasts. Trx-like proteins (i.e., TrxL2.1, ACHT1, and ACHT2, as well as a typical Trx, Trx-f) were found to be involved in target oxidation with different target specificities (Figs. 2B and 3). These results are consistent with those of previous biochemical studies (22, 25, 34, 37).
We demonstrated that TrxL2.1 is involved in the oxidation of CF1-γ and that ACHT1 and ACHT2 are involved in the oxidation of FBPase (Fig. 2B). In contrast, Trx-f appeared to be involved in the oxidation of RCA (Fig. 3). Recent studies have suggested that 2-Cys Prx serves as a major electron sink for the oxidation of Trx-targeted proteins and that oxidation factors transfer reducing power from Trx-targeted proteins to 2-Cys Prx (22, 28, 36). The reduction efficiency of 2-Cys Prx promoted by typical Trx isoforms, such as Trx-f, is also known to be low compared with that promoted by NTRC or Trx-like proteins (12, 25, 29). Thus, Trx-f proteins may not be able to efficiently transfer the reducing power from their targets directly to 2-Cys Prx, particularly when they are competing with Trx-like proteins. One feasible and previously suggested (34) pathway for Trx-mediated oxidation is the transfer of reducing power by Trx to other chloroplast Prx proteins, such as PrxQ. This pathway is possible because chloroplast Trx can reduce PrxQ in vitro in a relatively efficiently manner (25, 35, 38); however, a deficiency of PrxQ alone does not influence target oxidation in vivo (28). Hence, the Trx-mediated oxidation pathway in vivo requires further investigation in future research.
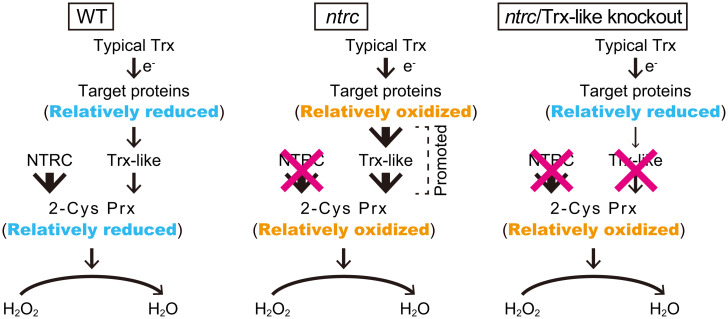
Here, we clarified the relationships among NTRC, 2-Cys Prx, and Trx-like proteins using mutant lines deficient in NTRC and Trx-like proteins. The important role of 2-Cys Prx in linking the NTRC system to the Trx system has already been reported in a previous study (14). Based on the present and previous results (14, 26), we propose a model for the NTRC and Trx systems connected by 2-Cys Prx (Fig. 6), which is explained as follows: 1) NTRC is an effective electron donor for 2-Cys Prx (8, 12, 14); 2) when compared to typical Trx proteins, Trx-like proteins efficiently transfer reducing power to 2-Cys Prx (22, 25, 29); and 3) Trx-like proteins serve as oxidation factors for target proteins in vitro (22, 24, 25, 27) and in vivo (Fig. 2). In the ntrc line, Trx-targeted proteins and 2-Cys Prx were in more oxidized states than they were in the Col-0 line under light conditions (Fig. 4 C and E) (t = 0), which is also consistent with previous findings (12, 14, 36). The relatively oxidized state of 2-Cys Prx in the ntrc line must receive reducing power from Trx-like proteins more strongly (Fig. 6; model for ntrc). Consequently, the target proteins must become more oxidized by the stronger flow of the reducing power to Trx-like proteins. The resultant oxidized states of the target proteins are suggested to cause ntrc phenotypes, such as stunted growth (14). We found that knockout of Trx-like proteins in the ntrc background, namely ntrc/trxl2.1(1) and ntrc/acht, restored the redox states of several target proteins to the levels of those in trxl2.1(1) and acht, respectively (Fig. 4C), even though 2-Cys Prx was similarly or more oxidized in these lines relative to its oxidation in the ntrc line (Fig. 4E). This was probably due to the impairment of the Trx-like protein-mediated electron flow from target proteins to 2-Cys Prx (Fig. 6; model for ntrc/Trx-like knockout). Therefore, we speculate that Trx-like proteins are major redox mediators between target proteins and 2-Cys Prx in the WT plant. In fact, 2-Cys Prx in the acht line was in more oxidized state than that in the Col-0 line during light-to-dark transitions (SI Appendix, Fig. S8). The knockdown of 2-Cys Prx is also known to suppress the ntrc phenotypes (14), indicating that the decreased amount of 2-Cys Prx limits the electron flow from Trx-like proteins to 2-Cys Prx, which in turn leads to impaired oxidation of target proteins. In contrast, the further knockout of typical Trx proteins in the ntrc background is known to accelerate ntrc phenotypes and the oxidation of target proteins (14, 15, 39). These results, which are in contrast to those of ntrc/Trx-like knockouts, imply that Trx proteins are not major mediators of electron transfer between targets and 2-Cys Prx. Taking these findings together, we suggest that Trx-like proteins—such as ACHT1, ACHT2, and TrxL2.1—must mediate electron transfer from Trx-targeted proteins to 2-Cys Prx in WT plants. ACHT appeared to play an important role in 2-Cys Prx reduction during light-to-dark transitions (SI Appendix, Fig. S8), whereas NTRC seems to be a major electron donor for 2-Cys Prx in the dark-adapted state and may also work during light-to-dark transitions.
Fig. 6.
Model of the redox cascade in chloroplasts under light conditions. A model for the 2-Cys Prx-linked NTRC and Trx systems, inspired by previous studies (14, 26), is shown. The size of arrows reflects the rates of electron flow.
Importantly, we found a relationship between redox-responsive proteins, particularly ACHT proteins, and the extent of NPQ (Figs. 4D and 5C). As shown in Fig. 4D, high NPQ in the ntrc line was suppressed by the lack of ACHT1 and ACHT2 (in the ntrc/acht line) but not by the lack of TrxL2.1 [in the ntrc/trxl2.1(1) line]. In addition, the plant lines that highly expressed ACHT2 showed high NPQ (Fig. 5C) similar to the ntrc line. A previous study also showed that overexpression of ACHT1 leads to high NPQ (40). Therefore, ACHT1 and ACHT2 may function in NPQ regulation. Although many studies have investigated the mechanism of NPQ induction and its relationship with the redox state (11, 41–45), the functional elements of NPQ regulation and redox regulation are not yet fully understood. Nevertheless, the results of the present study demonstrate the relevance of these systems.
In conclusion, we observed the oxidation of Trx-targeted proteins by Trx and Trx-like proteins in vivo. However, as in the case of PRK oxidation (Figs. 2B and 3), the oxidation of all Trx-targeted proteins cannot be explained only by the Trx and Trx-like proteins studied here, and other Trx or ACHT isoforms may be involved. Moreover, the effects of Trx-like proteins on growth and physiological parameters are not yet clear. One explanation could be functional redundancy among Trx proteins and Trx-like proteins, as many isoforms are localized in chloroplasts (30, 46). Further studies, especially focusing on other oxidation factors and their redundancy, are therefore needed to clarify the oxidative regulation pathways and to understand the entire redox regulation system in chloroplasts.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana Col-0 was used as a WT line. The transfer DNA (T-DNA) insertion mutant line, trx-f, which is deficient in Trx-f1 and Trx-f2 and was designated as “trxf1-1 trxf2” in our previous report (35), was also used here. In addition, we used another T-DNA insertion mutant line, ntrc, which is deficient in NTRC and was designated as “ntrc-1” in our previous report (12). T-DNA insertion lines for TrxL2.2., SALK_036628C and SALK_045620C, were tested but we could not confirm them as knockout by Western blotting. Plants were grown under a 16-h light (60 μmol photons m−2·s−1)/8-h dark cycle at 22 °C for 4 wk before they were used for each experiment.
CRISPR/Cas9-Mediated Genome Editing.
A plasmid for CRISPR/Cas9-mediated genome editing was constructed according to Hahn et al. (33) using the pFH6_new vector, pUB-Cas9 vector, and primers shown in SI Appendix, Table S1. To simultaneously target two different sites in the plant genome, two single-guide RNA expression cassettes were integrated into one plasmid using the primers FH41, FH42, FH254, and FH255 (SI Appendix, Fig. S3A and Table S1). Plant transformation and screening were conducted as described by Hahn et al. (33). The trxl2.1(1), trxl2.1(2), and acht lines were generated using the Col-0 line as a background.
Protein Extraction from Plant Leaves.
Rosette leaves of A. thaliana grown for 4 wk were excised and immediately immersed in liquid nitrogen. The leaves were powdered with a pestle and then homogenized with HES buffer [25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-NaOH (pH 7.5), 1 mM ethylenediamine-N,N,N′,N′-tetraacetic acid, and 2% (wt/vol) sodium dodecyl sulfate (SDS)]. The sample was incubated (at room temperature for 30 min and then 95 °C for 5 min) and centrifuged (at 20,400 × g and 20 °C for 15 min). The protein concentration of the supernatant was determined using a BCA Protein Assay Kit (Pierce). After adding SDS sample buffer with 2-mercaptoethanol to the supernatant, the sample was subjected to SDS/PAGE followed by Western blotting.
Measurement of Growth and Photosynthetic Parameters.
Using plants grown for 4 wk before being dark-adapted for 8 h, Fv/Fm and NPQ were measured with a Dual-PAM-100 spectrometer (Walz). NPQ was determined after illumination for 3 min with 70 μmol photons m−2·s−1 light intensity. After the measurement of these parameters, the FW of the plant above ground and chlorophyll content in rosette leaves were measured. Leaves were frozen in liquid nitrogen and then chlorophyll was extracted with 80% (vol/vol) acetone. Using spectrophotometry, chlorophyll content was determined as the sum of the contents of chlorophyll a and b according to previously published methods (22, 47).
Monitoring the Redox States of Proteins in Plant Leaves.
The redox states of proteins in plant leaves were determined using plants grown for 4 wk and methods described in a previous report (48) with some modifications. Dark-adapted (8 h) plants were illuminated with 700 μmol photons m−2·s−1 light intensity for 30 min (or 60 min for the experiment shown in Fig. 3) and subsequently transferred into the dark. Rosette leaves were excised at each indicated time point and proteins were extracted as described above except using HES buffer with 3 mM AMS and SDS sample buffer that did not contain 2-mercaptoethanol. Samples were subjected to SDS/PAGE and Western blotting followed by band intensity measurement via ImageJ software. To determine the redox state of PRK, 6 M urea was added to a gel for SDS/PAGE.
Overexpression of ACHT1 or ACHT2 in Plants.
First, a kanamycin-resistance cassette of the pRI 201-AN vector (Takara) was replaced by a hygromycin-resistance cassette. Full-length ACHT1 (At4g26160.1) or ACHT2 (At4g29670.1)-coding region was inserted into the vector using the primers shown in SI Appendix, Table S1. Col-0 plants were then transformed with the resulting vector via the Agrobacterium-mediated floral dip method.
Protein Expression and Purification.
Using the vectors prepared in our previous study (25), ACHT1 and ACHT2 (described as ACHT2a in our previous study) were expressed in Escherichia coli. Proteins were purified as described in the aforementioned study (25), except that they were purified without dithiothreitol. Using pET-23c vector (Novagen), cDNA from A. thaliana, and primers shown in SI Appendix, Table S1, an expression plasmid for C-terminally His-tagged PRK was constructed. E. coli strain Rosetta(DE3)pLysS was transformed with this plasmid, and the expressed protein was purified by nickel-nitrilotriacetic acid affinity chromatography with nickel-nitrilotriacetic acid-agarose (Qiagen) and the buffer containing 8 M urea, 25 mM Tris⋅HCl (pH 8.0), and 0 to 200 mM imidazole.
Preparation of Antibodies.
Antibodies against TrxL2.1, CF1-γ, FBPase, NTRC, and 2-Cys Prx A were prepared as described previously (12, 18, 19, 22). Anti-RCA antibody is commercially available (catalog no. AS10-700, Agrisera). Antibodies against ACHT1, ACHT2, and PRK were prepared using each recombinant protein as the antigen.
Supplementary Material
Acknowledgments
We thank the Biomaterial Analysis Center at the Tokyo Institute of Technology for supporting the DNA sequencing and Suzukake-dai Material Analysis Division, Technical Department, Tokyo Institute of Technology, for mass spectrometry analysis. This work was supported by JSPS KAKENHI Grants 16H06556 and 21H02502 (to T.H.), and Dynamic Alliance for Open Innovation Bridging Human, Environment, and Materials.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114952118/-/DCSupplemental.
Data Availability
The sequences of TrxL2.1 (At5g06690.1), TrxL2.2 (At5g04260.1), ACHT1 (At4g26160.1), ACHT2 (At4g29670.1), Trx-f1 (At3g02730.1), Trx-f2 (At5g16400.1), and NTRC (At2g41680.1) genes are available from the Arabidopsis Information Resource database (https://www.arabidopsis.org/). All other study data are included in the main text and SI Appendix.
References
- 1.Buchanan B. B., Role of light in the regulation of chloroplast enzymes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 31, 341–374 (1980). [Google Scholar]
- 2.Michelet L., et al. , Redox regulation of the Calvin-Benson cycle: Something old, something new. Front Plant Sci 4, 470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N., Portis A. R. Jr., Mechanism of light regulation of Rubisco: A specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc. Natl. Acad. Sci. U.S.A. 96, 9438–9443 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marri L., et al. , In vitro characterization of Arabidopsis CP12 isoforms reveals common biochemical and molecular properties. J. Plant Physiol. 167, 939–950 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Schurmann P., Wolosiuk R. A., Breazeale V. D., Buchanan B. B., 2 proteins function in regulation of photosynthetic Co2 assimilation in chloroplasts. Nature 263, 257–258 (1976). [Google Scholar]
- 6.Wolosiuk R. A., Buchanan B. B., Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 266, 565–567 (1977). [Google Scholar]
- 7.Serrato A. J., Pérez-Ruiz J. M., Spínola M. C., Cejudo F. J., A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279, 43821–43827 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Ruiz J. M., et al. , Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18, 2356–2368 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalska J., Zauber H., Buchanan B. B., Cejudo F. J., Geigenberger P., NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc. Natl. Acad. Sci. U.S.A. 106, 9908–9913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Ruiz J. M., Guinea M., Puerto-Galán L., Cejudo F. J., NADPH thioredoxin reductase C is involved in redox regulation of the Mg-chelatase I subunit in Arabidopsis thaliana chloroplasts. Mol. Plant 7, 1252–1255 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Naranjo B., et al. , The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ. 39, 804–822 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Yoshida K., Hisabori T., Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. U.S.A. 113, E3967–E3976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebthal M., Maynard D., Dietz K. J., Peroxiredoxins and redox signaling in plants. Antioxid. Redox Signal. 28, 609–624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Ruiz J. M., Naranjo B., Ojeda V., Guinea M., Cejudo F. J., NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. U.S.A. 114, 12069–12074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thormählen I., et al. , Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiol. 169, 1766–1786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikkanen L., Toivola J., Rintamäki E., Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant Cell Environ. 39, 1691–1705 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Nikkanen L., Rintamäki E., Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 476, 1159–1172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konno H., et al. , Thiol modulation of the chloroplast ATP synthase is dependent on the energization of thylakoid membranes. Plant Cell Physiol. 53, 626–634 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Yoshida K., Matsuoka Y., Hara S., Konno H., Hisabori T., Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol. 55, 1415–1425 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Yokochi Y., et al. , Redox regulation of NADP-malate dehydrogenase is vital for land plants under fluctuating light environment. Proc. Natl. Acad. Sci. U.S.A. 118, e2016903118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eliyahu E., Rog I., Inbal D., Danon A., ACHT4-driven oxidation of APS1 attenuates starch synthesis under low light intensity in Arabidopsis plants. Proc. Natl. Acad. Sci. U.S.A. 112, 12876–12881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K., Hara A., Sugiura K., Fukaya Y., Hisabori T., Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 115, E8296–E8304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cejudo F. J., Ojeda V., Delgado-Requerey V., González M., Pérez-Ruiz J. M., Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front Plant Sci 10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida K., Uchikoshi E., Hara S., Hisabori T., Thioredoxin-like2/2-Cys peroxiredoxin redox cascade acts as oxidative activator of glucose-6-phosphate dehydrogenase in chloroplasts. Biochem. J. 476, 1781–1790 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Yokochi Y., et al. , Impact of key residues within chloroplast thioredoxin-f on recognition for reduction and oxidation of target proteins. J. Biol. Chem. 294, 17437–17450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida K., Yokochi Y., Hisabori T., New light on chloroplast redox regulation: Molecular mechanism of protein thiol oxidation. Front Plant Sci. 10, 1534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida K., Hisabori T., Biochemical basis for redox regulation of chloroplast-localized phosphofructokinase from Arabidopsis thaliana. Plant Cell Physiol. 62, 401–410 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Ojeda V., Pérez-Ruiz J. M., Cejudo F. J., 2-Cys peroxiredoxins participate in the oxidation of chloroplast enzymes in the dark. Mol. Plant 11, 1377–1388 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Dangoor I., Peled-Zehavi H., Levitan A., Pasand O., Danon A., A small family of chloroplast atypical thioredoxins. Plant Physiol. 149, 1240–1250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chibani K., Wingsle G., Jacquot J. P., Gelhaye E., Rouhier N., Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol. Plant 2, 308–322 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Meyer Y., Reichheld J. P., Vignols F., Thioredoxins in Arabidopsis and other plants. Photosynth. Res. 86, 419–433 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Belin C., et al. , A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ. 38, 299–314 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Hahn F., Eisenhut M., Mantegazza O., Weber A. P. M., Generation of targeted knockout mutants in Arabidopsis thaliana using CRISPR/Cas9. Bio Protoc. 7, e2384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaseghi M. J., et al. , The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. eLife 7, e38194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida K., Hara S., Hisabori T., Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J. Biol. Chem. 290, 14278–14288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojeda V., Jiménez-López J., Romero-Campero F. J., Cejudo F. J., Pérez-Ruiz J. M., A chloroplast redox relay adapts plastid metabolism to light and affects cytosolic protein quality control. Plant Physiol. 187, 88–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiura K., et al. , The thioredoxin (Trx) redox state sensor protein can visualize Trx activities in the light/dark response in chloroplasts. J. Biol. Chem. 294, 12091–12098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collin V., et al. , Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 136, 4088–4095 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ojeda V., Pérez-Ruiz J. M., Cejudo F. J., The NADPH-dependent thioredoxin reductase C-2-Cys peroxiredoxin redox system modulates the activity of thioredoxin x in Arabidopsis chloroplasts. Plant Cell Physiol. 59, 2155–2164 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Dangoor I., Peled-Zehavi H., Wittenberg G., Danon A., A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24, 1894–1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks M. D., Sylak-Glassman E. J., Fleming G. R., Niyogi K. K., A thioredoxin-like/β-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, E2733–E2740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., et al. , Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol. 163, 1710–1728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simionato D., et al. , Protein redox regulation in the thylakoid lumen: The importance of disulfide bonds for violaxanthin de-epoxidase. FEBS Lett. 589, 919–923 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Malnoë A., et al. , The plastid lipocalin LCNP is required for sustained photoprotective energy dissipation in arabidopsis. Plant Cell 30, 196–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikkanen L., Guinea Diaz M., Toivola J., Tiwari A., Rintamäki E., Multilevel regulation of non-photochemical quenching and state transitions by chloroplast NADPH-dependent thioredoxin reductase. Physiol. Plant. 166, 211–225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer Y., Belin C., Delorme-Hinoux V., Reichheld J. P., Riondet C., Thioredoxin and glutaredoxin systems in plants: Molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Porra R. J., Thompson W. A., Kriedemann P. E., Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-B extracted with 4 different solvents—Verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989). [Google Scholar]
- 48.Yoshida K., Hisabori T., Simple method to determine protein redox state in Arabidopsis thaliana. Bio Protoc. 9, e3250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences of TrxL2.1 (At5g06690.1), TrxL2.2 (At5g04260.1), ACHT1 (At4g26160.1), ACHT2 (At4g29670.1), Trx-f1 (At3g02730.1), Trx-f2 (At5g16400.1), and NTRC (At2g41680.1) genes are available from the Arabidopsis Information Resource database (https://www.arabidopsis.org/). All other study data are included in the main text and SI Appendix.