Significance
Overflow metabolism, referred to as the Crabtree effect in yeast, is the seemingly wasteful strategy of using aerobic fermentation instead of the more efficient respiration for energy generation. This allows cells to grow faster at high glucose availability. Although well-studied, the underlying reasons that not all yeasts experience the Crabtree effect while still able to grow at comparable rates as yeasts exhibiting the effect, are not known. We combined physiological and proteome quantification with metabolic modeling to perform a quantitative comparison of four yeasts, two exhibiting and two not exhibiting the Crabtree effect, under glucose excess conditions. Our analyses provide insight into the underlying causes of the Crabtree effect, demonstrating a coupling to adaptations in both metabolism and protein translation.
Keywords: Crabtree effect, yeast, systems biology, proteomics, constraint-based modeling
Abstract
Aerobic fermentation, also referred to as the Crabtree effect in yeast, is a well-studied phenomenon that allows many eukaryal cells to attain higher growth rates at high glucose availability. Not all yeasts exhibit the Crabtree effect, and it is not known why Crabtree-negative yeasts can grow at rates comparable to Crabtree-positive yeasts. Here, we quantitatively compared two Crabtree-positive yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe, and two Crabtree-negative yeasts, Kluyveromyces marxianus and Scheffersomyces stipitis, cultivated under glucose excess conditions. Combining physiological and proteome quantification with genome-scale metabolic modeling, we found that the two groups differ in energy metabolism and translation efficiency. In Crabtree-positive yeasts, the central carbon metabolism flux and proteome allocation favor a glucose utilization strategy minimizing proteome cost as proteins translation parameters, including ribosomal content and/or efficiency, are lower. Crabtree-negative yeasts, however, use a strategy of maximizing ATP yield, accompanied by higher protein translation parameters. Our analyses provide insight into the underlying reasons for the Crabtree effect, demonstrating a coupling to adaptations in both metabolism and protein translation.
Growth is a fundamental property of life, and a main concern of organisms is therefore to absorb and break down nutrients from their surroundings to generate energy in the form of adenosine triphosphate (ATP) and synthesize the building blocks needed to proliferate. Many organisms use glucose as their preferred carbon and energy source. There are two main strategies for metabolizing glucose: 1) complete oxidation of glucose to carbon dioxide and water using the oxygen-consuming respiratory pathway consisting of glycolysis, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation and 2) incomplete oxidation through glycolysis and fermentation resulting in the secretion of by-products. In terms of energy generated per molecule of glucose, complete oxidation of glucose via respiration is about 10 times as efficient as incomplete oxidation through fermentation and has a fivefold-higher biomass yield (1–3). Despite this, the use of fermentative pathways to metabolize glucose at high growth rates is seen in many organisms, ranging from bacteria and yeast to human cells (4–6). This phenomenon is referred to as overflow metabolism and is characterized by high glycolytic fluxes, increased channeling of carbon toward fermentation by-products, and decreased flux through respiration even in the presence of oxygen.
There are numerous theories for the underlying causes of overflow metabolism (7). Pfeiffer et al. (3) proposed that overflow is the result of a trade-off between the rate and yield of ATP synthesis of the two metabolic modes. Recent studies have expanded this idea to include the protein cost, suggesting that overflow metabolism is caused by limited proteome resources. As the protein content of a cell is finite, there is a natural constraint on the proportion of the proteome that can be allocated to metabolism. Although respiration has a high ATP yield, it requires synthesis of components of the TCA cycle and electron transport chain (ETC), as well as an additional mitochondrial machinery for eukaryal cells, while fermentation only requires a few cytosolic enzymes. Therefore, at high growth rates, fermentation is more efficient in terms of the proteome mass needed to produce the same amount of ATP. Indeed, various modeling approaches (1, 7–9) and proteomics measurements (10) have shown that when conditions allow, such as when glucose is present in abundance, cells will engage in overflow metabolism, allowing them to grow at a faster rate. For microorganisms such as bacteria and yeast, this presents an evolutionary advantage that enables cells to outcompete their neighbors.
Among yeasts, overflow metabolism is referred to as the Crabtree effect (11, 12), but it is interesting that not all yeast species exhibit overflow metabolism. Some yeasts can fully oxidize glucose through respiration even at high glucose concentrations (Crabtree-negative yeasts) and still grow at rates comparable with those of Crabtree-positive yeasts. The mechanism of this is unclear. We therefore performed absolute proteome quantification of two Crabtree-positive yeasts, Saccharomyces cerevisiae and Schizosaccharomyces pombe, and two Crabtree-negative yeasts, Kluyveromyces marxianus and Scheffersomyces stipitis, cultivated in batch cultures where glucose is provided in excess. We combined physiological and proteome quantification with genome-scale metabolic modeling (GEM) to quantitatively describe the differences between Crabtree-positive and -negative yeasts on both the proteomic and metabolic levels. We show that the Crabtree effect is coupled to metabolic flux differences in glycolysis, the pentose phosphate pathway (PPP), and respiratory fluxes. We further found that proteome allocation reflects the trade-off between the two distinct glucose utilization strategies in Crabtree-positive and -negative yeasts, with metabolic capacity limitations particularly in the ETC and ATP synthase. Finally, we show that the Crabtree effect is accompanied by differences in ribosomal protein (RP) content and protein translation efficiencies. Taken together, our analyses show that adaptations in both metabolism and protein translation underlie the Crabtree effect in yeasts.
Results
Characterization of the Physiological Differences between Crabtree-Positive and -Negative Yeasts.
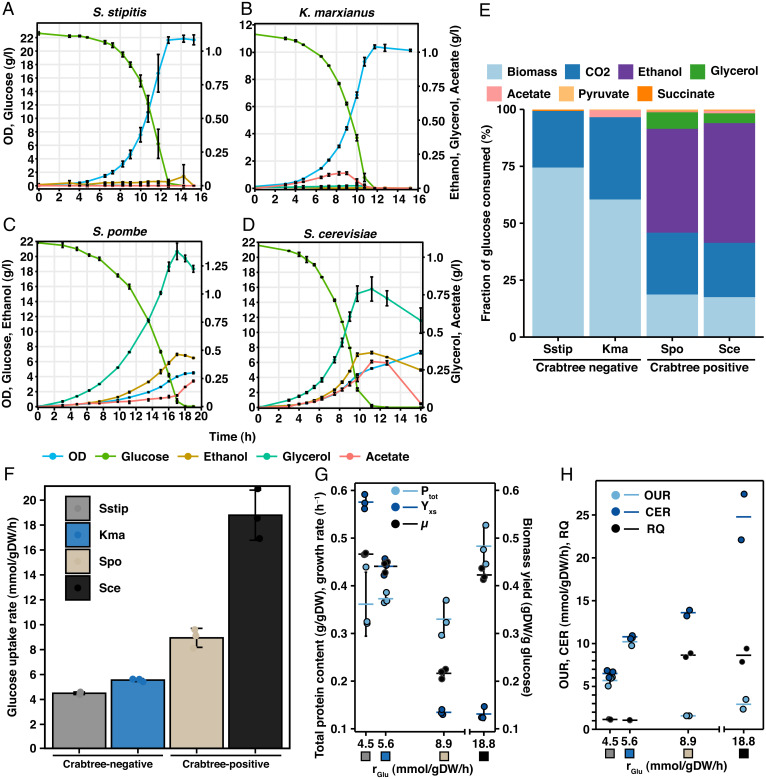
We selected S. cerevisiae and S. pombe as representative Crabtree-positive yeasts, as they are widely used as model eukaryals and have evolved the Crabtree effect independently (13). As Crabtree-negative yeasts we selected K. marxianus and S. stipitis, which have emerged as attractive options for use as cell factories (14, 15). To characterize the physiology of the Crabtree effect, we cultivated all four yeast species in triplicates in bioreactors in minimal media (16) with glucose as the carbon source and measured important physiological parameters, including biomass, exometabolite levels, as well as oxygen uptake and carbon dioxide production (Fig. 1 A–D). Since overflow metabolism in K. marxianus and S. stipitis has been reported to occur in response to limitations in oxygen availability (14, 17), we controlled the dissolved oxygen (DO) levels to always be above 60% (SI Appendix, Fig. S1), and the initial glucose concentration of K. marxianus used was 10 g/L instead of 20 g/L as used for the other species. As seen from the exometabolite profile, the two Crabtree-negative yeasts showed low by-product formation (Fig. 1 A, B, and E and SI Appendix, Fig. S2 A and B). K. marxianus excreted low amounts of acetate, corresponding to <3% of the glucose consumed in line with previous studies of K. marxianus during batch cultivation (17, 18). The Crabtree-positive species secreted higher levels of by-products, with ethanol as the main product, corresponding to 42 and 47% of the glucose consumed for S. pombe and S. cerevisiae, respectively (Fig. 1 C–E). We also observed a large difference in the glucose uptake rate (GUR) between the groups (Fig. 1F and SI Appendix, Table S1). S. pombe and S. cerevisiae had roughly a twofold- and >threefold-higher uptake rate than the Crabtree-negative species, respectively. This is in line with the observation that the GUR plays an important role in overflow metabolism (19).
Fig. 1.
Characterization of physiological differences between Crabtree-positive and -negative yeast. (A–D) Cell density (optical density at 600 nm [OD600]) and extracellular concentrations of glucose, ethanol, glycerol, and acetate during the course of the batch cultivation. Mean values ± SD of biological triplicates are shown. See SI Appendix, Fig. S2 A–D for additional metabolites. (E) The fractional consumption of glucose, C-mmol/C-mol glucose, normalized against the total carbon recovery. Mean values of biological triplicates are shown. The carbon recovery was between 91 and 100% for all four yeasts. (F) GUR. Mean values ± SD of biological triplicates are shown. (G) Protein content (Ptot), biomass yield (Ysx), and growth rate (µ) plotted against GUR. Mean values ± SD and individual values of biological triplicates are shown. Colored boxed represent the organism, as indicated in panel F. (H) OUR, CER, and RQ plotted against GUR. Mean values ± SD and individual values of biological triplicates are shown for S. stipitis and mean values, and individual values for biological duplicates are shown for S. pombe, S. cerevisiae, and K. marxianus. Colored boxes represent the organism, as indicated in panel F. Spo, S. pombe; Sce, S. cerevisiae; Kma, K. marxianus; Sstip, S. stipitis.
The main growth-related parameters are summarized in Fig. 1 G and H. The growth rate measured was similar for S. cerevisiae and the two Crabtree-negative species, 0.42 h−1 compared to 0.44 and 0.47 h−1 for K. marxianus and S. stipitis, respectively, while S. pombe grew at 0.22 h−1. The fermentative metabolism in the Crabtree-positive yeasts was reflected in biomass yields (Ysx) of around 0.1 g dry weight (g DW)/g glucose and respiratory quotients (RQ) around 9. The respiratory metabolism of the Crabtree-negative species was confirmed by RQ values 1.09 and 1.15 for K. marxianus and S. stipitis, respectively, and resulted in biomass yields of 0.44 and 0.58 g DW/g glucose, respectively. Interestingly, we observed differences in the oxygen uptake rate (OUR) and carbon dioxide evolution rate (CER) when comparing the species within each group. For the Crabtree-negative species, S. stipitis showed about a 50% lower OUR and CER than K. marxianus (Fig. 1H). This difference could be due to the presence of respiratory chain complex I in S. stipitis, which results in a higher phosphate-to-oxygen ratio (P/O) and thus a higher ATP yield per mitochondrial NADH oxidized. This is associated with a higher biomass yield for S. stipitis than for K. marxianus. In the Crabtree-positive species, S. cerevisiae had about 2 times higher OUR and CER than S. pombe, indicative of a lower metabolic rate in S. pombe, which is also reflected in the lower growth rate for this species.
The Crabtree Effect Is Coupled to Differences in Glycolytic, PPP, and Respiratory Fluxes.
To further characterize the differences between the Crabtree-positive and negative yeasts, we performed absolute quantification of the proteome. For this we first determined the absolute concentrations of proteins in four individual reference samples, constructed by separately pooling the three replicates for each yeast, through mass spectrometry (MS) using intensity-based absolute quantification (iBAQ) (20) with Proteomics Dynamic Range Standard (UPS2) as internal standard. The absolute concentrations of all proteins in the proteomes were then determined by tandem mass tag (TMT)-based (21) MS using the pooled reference samples as internal references. This resulted in the quantification of 3,925, 3,765, 3,612, and 4,110 proteins in S. cerevisiae, S. pombe, K. marxianus, and S. stipitis, respectively (Datasets S1–S4).
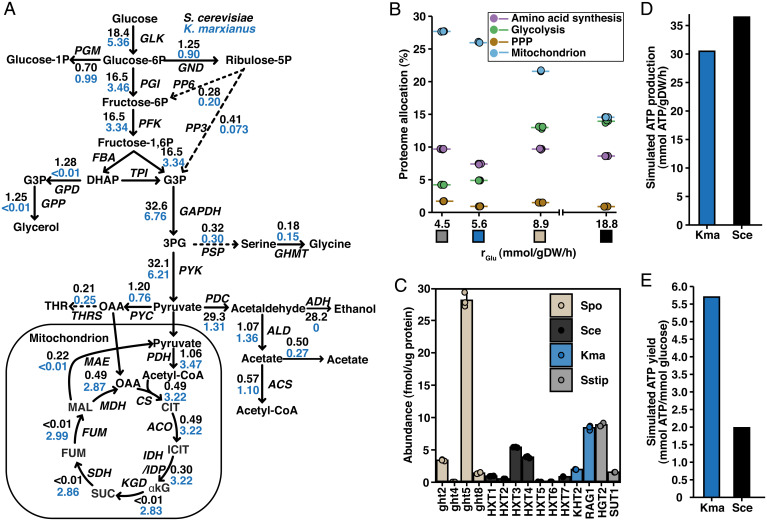
We set out to investigate the metabolic adaptations and mechanisms underlying the Crabtree effect using enzyme-constrained (ec) GEM (9). This method extends the classical flux balance analysis (FBA) (22) approach used for GEMs by incorporating a detailed description of enzymatic demands of metabolic reactions, while also allowing for integration of protein abundance data. We used the GECKO toolbox (23) to generate condition-specific models of two recently published ec models for S. cerevisiae and K. marxianus by constraining them with experimentally measured exchange fluxes, growth rates, and protein levels and used the models to predict the flux distribution under these conditions (Fig. 2A). The model-based investigation was limited to these two models based on their quality and standardized formats used, facilitating comparison of the simulation results (24). In the flux distribution, two main differences were observed. First, we observed a difference in the flux entering the PPP from glucose-6-phosphate (G6P). Although the absolute flux through the PPP in S. cerevisiae was higher, 1.25 compared to 0.9 mmol/gDW/h, the relative fraction of G6P, excluding the loss of G6P to polysaccharide biomass components via glucose-1-phosphate, metabolized in the PPP was 7% compared to 20.5% in K. marxianus. This is in agreement with previously observed differences in flux distributions between Crabtree-positive and -negative yeasts, including S. stipitis (25–27). Second, we observed a large difference in the metabolism of pyruvate. S. cerevisiae had a >threefold-higher flux through glycolysis than K. marxianus, and the majority of the pyruvate was consumed by pyruvate decarboxylase (PDC), followed by production of ethanol and acetate by the alcohol and aldehyde dehydrogenases (ADH and ALD). Only a small fraction of the pyruvate was metabolized by pyruvate dehydrogenase (PDH) and entered the TCA cycle. In K. marxianus, the majority of the pyruvate was metabolized by PDH and entered the TCA cycle. This indicates that the TCA cycle plays a dual role in Crabtree-negative yeasts, serving both as a supply of precursor metabolites and in respiratory metabolism of pyruvate. In S. cerevisiae, the main flux-carrying reactions were the early steps of the cycle, indicating that its main role during rapid growth is to provide precursors for biomass components through synthesis of ɑ-ketoglutarate. The differences in pyruvate metabolism were reflected in the levels of proteins around the pyruvate branching point (SI Appendix, Fig. S3). We observed a much larger abundance of the enzymes involved in fermentation, including PDC, ADH, and ALD in Crabtree-positive yeasts, while the Crabtree-negative yeasts had higher levels of PDH. In S. cerevisiae, PDC has a higher activity than PDH and therefore has been proposed to be an important factor driving fermentation at high glycolytic flux (28). Our data on GUR, protein levels around the pyruvate branch point suggests that at the lower glycolytic flux in the Crabtree-negative yeasts, the capacity of PDH is sufficient to support a fully respiratory dissimilation of pyruvate, indicating that balancing the glycolytic flux is an important aspect in the Crabtree effect.
Fig. 2.
Adaptations in metabolism and proteome allocation reflect the trade-off between glucose utilization and ATP yield. (A) Flux distribution of central carbon metabolism as predicted by FBA using condition-specific enzyme-constrained models, constrained with experimentally measured exchange fluxes and protein levels. (B) Summed proteome allocation to selected processes plotted against GUR. Protein members of each process were defined by GO annotation. Individual values and mean values of biological triplicates are shown. Colored boxes represent the organism, as specified in Fig. 1F. See SI Appendix, Fig. S3 for absolute abundances of the pathways. (C) Protein abundance of glucose transporters measured in this study. Mean values ± SD of biological triplicates are shown. (D) ATP production rate calculated based on FBA simulation results. (E) ATP yield calculated from FBA simulation results. Spo, S. pombe; Sce, S. cerevisiae; Kma, K. marxianus; Sstip, S. stipitis.
Proteome Allocation Reflects the Trade-Off between Glucose Utilization Strategies and ATP Yield.
Next, we analyzed the proteomics data to evaluate how the flux distribution is reflected in the proteome allocation. By summing the individual protein abundances based on GO term annotation (29), we observed differences in the allocation of proteins to mitochondria and glycolysis between the Crabtree-positive and -negative yeasts, while the PPP and amino acid biosynthesis pathways showed similar allocation comparing the two groups (Fig. 2B). Furthermore, we observed no clear differences in the allocation or abundance for all other major biosynthetic processes (SI Appendix, Fig. S4), indicating that the main metabolic differences are in energy metabolism. In the Crabtree-positive species, where the glycolytic flux was high, we observed a higher allocation toward glycolysis, at the expense of allocation of proteins toward mitochondria. Similar trends were also seen in the absolute protein levels of the GO term groups (SI Appendix, Fig. S5), except from S. cerevisiae showing a higher summed abundance of glycolytic proteins than S. pombe, possibly as a result of having a higher protein content (Fig. 1G). This finding indicates that the proteome size is an important factor determining growth rate in Crabtree-positive yeasts.
A major event in the evolution of S. cerevisiae was the whole-genome duplication (WGD) (30, 31), resulting in increased gene dosage of glycolytic proteins (32), which is thought to be a main driver of the Crabtree effect. Indeed, we observed higher protein levels of all glycolytic proteins in both Crabtree-positive species compared to the Crabtree negative (SI Appendix, Fig. S6). No WGD has been detected in S. pombe, and the yeast has thus relied on small-scale duplication events as well as increasing the expression of single enzymes (33). Interestingly, although S. pombe has fewer duplicated glycolytic genes than S. cerevisiae, the single-isoform proteins have a similar abundance to the combined abundance of the S. cerevisiae paralogs, especially in the upper half of glycolysis. Additionally, several studies have shown that glucose transport is the main flux controlling step of glycolysis and thus serve as one of the main factors influencing the Crabtree effect (19, 32, 34). This is further supported by the finding that the Crabtree effect is abolished in a strain of S. cerevisiae with limited glucose uptake capacity (35), but this is also accompanied with slower growth. Apart from the difference in the GUR, our data on hexose transporters (HXTs) show key differences both in the levels and number of active glucose transporters (Fig. 2C). The two Crabtree-negative strains, based on abundance, seem to rely mainly on a single glucose transporter, the main difference being that K. marxianus expresses the low-affinity transporter RAG1, while S. stipitis expresses high-affinity transporter HGT2. Both Crabtree-positive yeasts possess additional isoforms of glucose transporters, a total of 17, of which 6 are expressed under normal conditions in the case of S. cerevisiae and 8 in the case of S. pombe, highlighting the importance of glucose transport in the Crabtree effect (34). However, the two species differ in the distribution of expression of the transporters under the conditions studied. S. cerevisiae distributed the expression among several transporters, with different affinities for glucose, while S. pombe predominantly has high-level expression of its high-affinity transporter ght5.
The differences in strategies of glucose metabolism are further reflected by model simulations of ATP production. The net ATP production flux, calculated by combining the ATP production from glycolysis and respiration and which equals the ATP consumption by the cell, was 36.5 and 30.5 mmol/gDW/h for S. cerevisiae and K. marxianus, respectively (Fig. 2D). However, the ATP yield, defined in mmol ATP/mmol glucose, was threefold higher in K. marxianus (Fig. 2E), reflecting the higher efficiency of respiratory metabolism in terms of ATP yield. These results clearly highlight the difference in the two strategies employed for energy generation. Crabtree-positive yeasts produce ATP by the inefficient pathway of fermentation and compensate the low efficiency by having a large glucose uptake and flux through glycolysis, while the Crabtree-negative yeasts balance the flux through glycolysis and respiration, leading to a high ATP yield. Furthermore, K. marxianus is more efficient in ATP utilization for biomass production than S. cerevisiae, and this could also contribute to the ability to grow fast without overflow metabolism for Crabtree-negative yeasts.
Limitations in the ETC and ATP Synthase Are Characteristic for the Crabtree Effect.
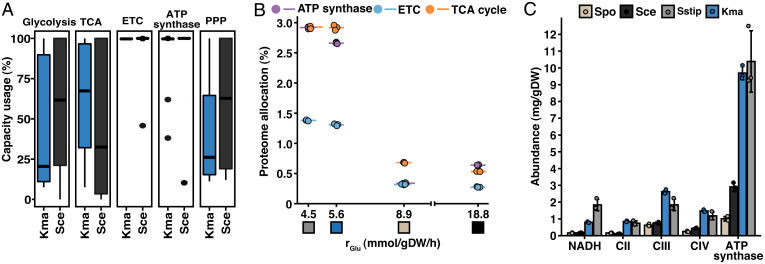
To further characterize the adaptations involved in the Crabtree effect, we calculated the capacity usage, defined as the ratio between the measured enzyme abundances and the enzyme demand as predicted by the ec models of S. cerevisiae and K. marxianus. We observed a higher usage in glycolysis in S. cerevisiae, while K. marxianus had a higher usage in the TCA cycle, in line with the differences seen at the flux level (Fig. 3A). This signifies that the increased fluxes through either of these pathways are not only facilitated by an increased expression but also a higher occupancy of their constituent enzymes. Furthermore, for both yeasts, we observed a full usage of the proteins of the ETC and ATP synthase. This is reasonable, since these proteins are all mitochondrial and present in the mitochondrial inner membrane, and any unused protein in these complexes would result in an inefficient use of mitochondrial membrane space. We also observed a difference in the usage of the enzymes in the PPP, with lower usage in K. marxianus, possibly as a result the lower absolute flux through the PPP. However, when looking at the absolute protein levels, no clear difference was observed (SI Appendix, Fig. S7).
Fig. 3.
Limitations in the ETC and ATP synthase are characteristic for the Crabtree effect. (A) Capacity usage, calculated as the model-predicted enzyme levels divided by the experimentally measured enzyme levels, of the main pathways of central carbon metabolism (CCM). (B) Summed proteome allocation to processes of CCM plotted against GUR. Protein members of each process were defined by GO annotation. Mean values of biological triplicates are shown. Colored boxes represent the organism, as specified in panel C. See SI Appendix, Fig. S7A for absolute abundances of the processes. (C) Absolute protein abundances of the components constituting the ETC and ATP synthase. Mean values ± SD of biological triplicates are shown. See SI Appendix, Fig. S7B for proteome allocation of the components. PPP, pentose phosphate pathway; Spo, S. pombe; Sce, S. cerevisiae; Kma, K. marxianus; Sstip, S. stipitis.
We next analyzed the proteome allocation in mitochondrial energy metabolism, including the TCA cycle, ETC, and ATP synthase, of the four yeasts (Fig. 3B). Here, we observed a higher allocation of all three processes in the Crabtree-negative yeast, where the allocation to the TCA cycle and ATP synthase was both about 3%, while about 1.5% was allocated to the ETC, compared to about 0.5 and 0.25% of the proteome in the Crabtree-positive yeasts. These findings were also reflected in the absolute abundance levels of the processes and individual enzyme complexes (Fig. 3C and SI Appendix, Figs. S8 and S9). For all components of the ETC and ATP synthase, we observed roughly a threefold-higher abundance in the Crabtree-negative compared to the Crabtree-positive yeasts. This clearly underlines the difference in the strategy used for ATP production. Together with the higher allocation of glycolytic proteins at the expense of mitochondrial proteins in Crabtree-positive yeasts, this indicates that the high glycolytic flux causes a saturation of the capacity of NADH oxidation through respiration, causing increased fermentation at high GURs. This is in line with a previous study in S. cerevisiae showing that increasing the capacity of NADH oxidation by expressing an alternative oxidase (AOX), allowing the direct reduction of oxygen from the ubiquinone pool, led to a reduced ethanol formation (36). Interestingly, S. stipitis natively possesses an AOX, STO1. To investigate the role of the AOX, we therefore compared the abundance of the Sto1p to that of complex IV (cytochrome c oxidase). We observed a fivefold-higher median abundance of the subunits of complex IV (SI Appendix, Fig. S10), indicating that the main oxygen-consuming step is cytochrome c oxidase and that the AOX is used to a lower extent under the conditions tested. Furthermore, in S. stipitis, complex I is present. We observed expression of the complex but not the internal NADH dehydrogenase, NDI1, suggesting that NADH oxidation relies on complex I and the external NADH dehydrogenase, NDE1, under the conditions studied. The presence of complex I presents an additional proton-translocating step, which in turn leads to a higher P/O and ATP yield. This could explain the significantly higher biomass yield as well as the lower OUR and CER in S. stipitis compared to K. marxianus (Fig. 1 G and H).
The Crabtree Effect Is Accompanied by Differences in RP Content and Protein Translation Efficiencies.
The Crabtree effect has evolved independently in multiple lineages of yeast, suggesting that there is an evolutionary advantage in switching to fermentation when glucose is present in excess. This advantage is thought to be a higher rate of ATP production and lower proteome cost, which allows Crabtree-positive yeasts to grow at a faster rate (37). However, our data show that the Crabtree-negative yeasts K. marxianus and S. stipitis grow at similar rates as the Crabtree-positive yeast S. cerevisiae, while the Crabtree-positive yeast S. pombe has a significantly lower growth rate (Fig. 1G). This raises the possibility that, under the condition of glucose excess, Crabtree-positive yeasts are subject to additional physiological constraints, which is balanced by the advantage given by high ATP production rate via glucose fermentation. One major candidate for such a constraint is in protein translation. This is evident from an evolutionary perspective: a major evolutionary event in the Saccharomyces lineage, which has a pronounced Crabtree effect, is a WGD event followed by massive gene loss, leading to a retention of about 10% of duplicated genes that have evolved into paralogs (38). Two major groups of the retained paralogs are 1) enzymes in carbohydrate metabolism, which enabled higher glycolytic flux and thereby giving rise to the pronounced Crabtree effect in post-WGD yeasts; and 2) RPs (39). In S. cerevisiae, 59 of 78 RP subunits have paralogs (SI Appendix, Fig. S11 A and B). In S. pombe, which is not in the Saccharomyces lineage but independently evolved to be Crabtree positive, 62 of 78 RP subunits have paralogs (SI Appendix, Fig. S11 A and B). In contrast, K. marxianus and S. stipitis do not have any RP paralogs (SI Appendix, Fig. S11 A and B). The evolution and retention of RP paralogs in Crabtree-positive yeasts suggest that ribosomes in these species may have become more specialized (e.g., translate different subsets of proteins), which may be accompanied by slower kinetics of protein translation. We therefore compared the ribosomal content and efficiency in Crabtree-positive and -negative yeasts.
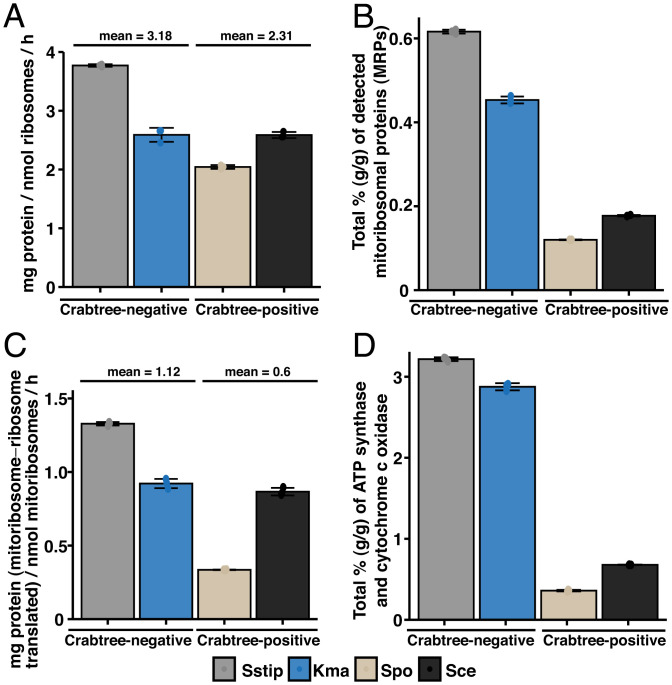
In our proteomics data, we detected 74 to 77% of RPs in S. cerevisiae and S. pombe, respectively, while for K. marxianus and S. stipitis, 97 to 98% of RPs were detected (SI Appendix, Fig. S11A). However, in all species we detected 96 to 99% of RP subunits, wherein a subunit is considered detected if one of the two paralogs is detected (SI Appendix, Fig. S11B). The total RP abundance were 19.5 to 23.6% (g per g total protein) in the faster-growing species (S. cerevisiae, K. marxianus, and S. stipitis),and 16.1% in the slower-growing S. pombe (SI Appendix, Fig. S11C), in line with previous studies showing that RP abundance increases linearly with specific growth rate (40, 41). We then calculated the overall ribosomal translation efficiency as mg total protein per nmol ribosomes per hour and found that S. pombe has less-efficient ribosomes compared to S. stipitis, while K. marxianus and S. cerevisiae exhibited similar efficiencies (Fig. 4A). This indicates that translational efficiency is a factor underlying the Crabtree effect, although data on more yeast species would be required to confirm this. As we have shown that the ETC and ATP synthase are particularly limiting in central carbon metabolism and energy generation (Fig. 3A), we then examined the abundance of mitochondrial ribosomal proteins (MRPs) in our dataset. The number of MRPs in the four yeast species examined ranges between 66 and 77 subunits, and in all four species we detected 90 to 99% of these proteins (SI Appendix, Fig. S11D). Here, we find that S. cerevisiae and S. pombe expressed MRPs at only 28% of the MRP expression levels found in K. marxianus and S. stipitis (Fig. 4B). As MRPs translate key subunits of ATP synthase, cytochrome c oxidase, and mitoribosomes, we then calculated the mitoribosomal protein translation efficiency (mg mitoribosome-translated proteins per nmol mitoribosomes per hour), based on the mean molar abundance of all subunits of these three complexes. Our data showed that the mitoribosome translation efficiency in S. pombe was much lower than that of S. stipitis, while again the efficiencies of K. marxianus and S. cerevisiae were similar (Fig. 4C). These three factors—less-efficient cytoplasmic ribosomes, lower expression of mitoribosomes, and/or less-efficient mitoribosomes—together give rise to a significantly lower expression of ATP synthase and cytochrome c oxidase in both S. cerevisiae and S. pombe, at 17% of the expression levels detected in K. marxianus and S. stipitis (Fig. 4D). Taken together, our data therefore show that the Crabtree effect in yeasts is accompanied by adaptations in metabolism, particularly in the strategies used to produce ATP, as well as adaptations in protein translation, in the efficiency and/or content of ribosomes and mitoribosomes.
Fig. 4.
The Crabtree effect is accompanied by differences in RP content and protein translation efficiencies. (A) Comparison of the overall ribosomal translation efficiency, calculated as milligrams total protein per nanomoles ribosomes per hour. (B) Comparison of the expression levels of MRPs, expressed as percent out of total proteome. (C) Comparison of the mitoribosomal protein translation efficiency, calculated based on the mean molar abundance of all subunits of the mitochondrially expressed protein complexes, including cytochrome c oxidase, ATP synthase, and mitoribosomes. (D) Comparison of the expression levels of cytochrome c oxidase and ATP synthase, expressed in percent of total cellular protein. C, Spo, S. pombe; Sce, S. cerevisiae; Kma, K. marxianus; Sstip, S. stipitis.
Discussion
Here, we quantitatively described the metabolic and proteomic adaptations underlying the Crabtree effect by comparing two Crabtree-positive yeasts, S. cerevisiae and S. pombe, and two Crabtree-negative yeasts, K. marxianus and S. stipitis, cultivated in bioreactors under glucose excess conditions. By combining physiological and proteome quantification with GEM, we found that the Crabtree effect is linked to adaptations in metabolic strategies of glucose utilization, as well as differences in organization and efficiency of both mitochondrial and cytoplasmic translation.
Optimal carbon usage is crucial in order to generate energy, in the form of ATP, and biomass precursors for cell growth. Crabtree-positive yeast produce ATP mainly through the use of glycolysis and fermentation, leading to production of by-products, while Crabtree-negative yeasts fully oxidize glucose to carbon dioxide and water. While the complete oxidation of glucose is more efficient in terms of ATP generated per molecule of glucose (3), it comes at the trade-off of a high protein cost. On the other hand, metabolizing glucose through fermentation comes with the trade-off of a low ATP yield, potentially resulting in a lower growth rate but has a higher efficiency in terms of protein cost (1). To compensate for the low yield, an increased glycolytic flux would be required. Our data confirms that Crabtree-positive yeasts indeed have a higher rate of glucose uptake and glycolysis. We observed roughly a twofold- and threefold-higher uptake rate in S. pombe and S. cerevisiae, respectively, as well as a higher glycolytic flux compared to the Crabtree-negative yeasts. Model simulations further show that fluxes around the pyruvate branch point differ significantly, where S. cerevisiae showed a high flux through PDC and toward ethanol formation, while K. marxianus had a higher flux through the TCA cycle and respiration. Although the total ATP produced was similar between the two yeasts, the molar yield of ATP per glucose was close to threefold higher in K. marxianus. This clearly highlights the trade-off in ATP production using the different metabolic strategies.
This difference in metabolism is also reflected on the proteome level. Along with the higher glycolytic flux, we observed higher levels of glycolytic proteins in both Crabtree-positive yeasts, while the levels of protein in the TCA cycle, ETC, and ATP synthase were higher in the Crabtree-negative yeasts. This indicates that the increased glycolytic flux and the associated increase in protein levels comes at the expense of respiratory proteins, as also seen in our data comparing the distribution of proteome resources. This is in line with the glucose repression of respiration resulting from a regulatory rewiring associated with the evolution of aerobic fermentation (42–44).
The Crabtree effect evolved independently in S. cerevisiae and S. pombe (13). A main driver in the evolution of the Crabtree effect in S. cerevisiae is the WGD, including the duplication of 6 of 13 glycolytic enzymes and an increase in the number of HXTs, resulting in increased glycolytic flux (32, 34). In S. pombe, no WGD has been observed, and fewer glycolytic paralogs are present (33). Interestingly, although the glucose influx in S. pombe is about one-half of that of S. cerevisiae, some single glycolytic enzymes in the early stages of glycolysis show similar levels to the summed abundance of the S. cerevisiae isozymes. Enzymes in the later stages showed lower abundance, indicating a limitation in the second half of glycolysis. A potential explanation is the lower activity of PYK, an important regulator of glycolytic flux (45), in S. pombe resulting from a mutation rendering it insensitive to stimulation by fructose bisphosphate (46). Furthermore, we observed a difference in HXTs. S. cerevisiae and S. pombe contain 17 and 8 HXTs, respectively, compared to 2 in the Crabtree-negative yeasts. S. pombe and the Crabtree-negative yeasts expressed mainly a single HXT, while S. cerevisiae distributed expression among multiple paralogs. The distribution of flux over multiple paralogs of HXTs and glycolytic proteins indicates that S. cerevisiae has optimized its metabolism for fast utilization of glucose, while the other yeasts have a more constrained glucose uptake.
In addition to maintaining paralogous glycolytic enzymes and HXTs, Crabtree-positive yeasts have also evolved to contain paralogs for many RPs. The resulting increase in gene dosage of RP has been linked to an increase in fermentative capacity (47). RP genes are highly transcribed, accounting for 50% of RNA polymerase II transcription (48). Doubling the gene dosage of 76 to 77% of RPs in Crabtree-positive yeasts means that mature ribosomes are likely to be highly diverse, with some studies estimating that the diversity of ribosomes could outnumber the actual ribosomal content of the cell (49). This high diversity has the benefit of allowing ribosomal activities to be finely controlled but would necessarily mean that the overall protein translation kinetics becomes slower. Here, our analyses show that the cytosolic translational efficiency was indeed lower in certain Crabtree-positive species, which would exacerbate the proteomic limitations in fast-growing cells and thereby favoring the use of fermentative pathways to metabolize glucose when conditions allow. Furthermore, we found that the allocation of MRPs was significantly lower in the Crabtree-positive species, possibly explained by the loss of regulatory elements coupling the expression of MRPs and RPs following the WGD (50). We also found that the efficiency of mitochondrial translation was lower in certain Crabtree-positive species. These factors give rise to a significantly lower expression of ATP synthase and cytochrome c oxidase and highlight the significance of adaptations in the content and efficiency of ribosomes in the Crabtree effect.
In conclusion, this study provides insight into the underlying mechanisms giving rise to the Crabtree effect in terms of adaptations in metabolism and protein translation, supporting that a trade-off between the rate and yield of ATP generation, as well as ribosomal specialization and translation efficiency, are important factors in the evolution of the Crabtree effect. Furthermore, the proteomic datasets generated herein, as well as our modeling approach, together provide a framework for analyzing multispecies omics data to understand the process of species diversification, an ongoing challenge in systems and synthetic biology.
Methods
Culture Conditions.
The yeasts used in this study were S. cerevisiae CEN.PK113-7D (MATa, MAL2-8c, SUC2), S. pombe (ATCC24843), K. marxianus CBS6556, and S. stipitis (ATCC58376). Cells were stored in aliquoted glycerol stocks at −80 °C. Batch cultures were carried out in DASGIP 1-L bioreactors with off-gas analysis, pH-control, temperature control, and DO sensors. A working volume of 700 mL, temperature of 30 °C, pH of 5, controlled by addition of 2 M KOH and 2 M HCl, and a starting inoculum of optical density 0.1 was used. The initial aeration and agitation was set to 50 sLh−1 and 800 rpm, respectively, and a DO was kept above 60%, by increasing the stirring rate. The basis of all cultures was a minimal medium that contained 20 g/L glucose (10 g/L for K. marxianus), 5 g/L (NH4)2SO4, 3 g/L KH2PO4, 0.5 g/L MgSO4⋅7H2O, 1 mL/L trace metal solution, 1 mL/L vitamin solution, and 0.1 mL/L antifoam 204 (Sigma-Aldrich). The trace metal solution consisted of 19 g/L Na2EDTA⋅2H2O (disodium ethylenediaminetetraacetate dihydrate), 4.5 g/L ZnSO4⋅7H2O, 1 g/L MnCl2⋅4H2O, 0.3 g/L CoCl2⋅6H2O, 0.3 g/L CuSO4⋅5H2O, 0.4 g/L Na2MoO4⋅2H2O, 4.5 g/L CaCl2⋅2H2O, 3 g/L FeSO4⋅7H2O, 1 g/L H3BO3, and 0.1 g/L potassium iodide. The vitamin solution consisted of 0.05 g/L d-Biotin, 0.2 g/L 4-aminobenzoic acid, 1g/L nicotinic acid, 1 g/L D-Pantothenic acid hemicalcium salt, 1 g/L pyridoxine-HCl, 1 g/L thiamine-HCl, 25 g/L, and myo-inositol.
Sampling from Bioreactor.
For all sampling timepoints, the dead volume of the sampling port was removed prior to sampling. Determination of dry cell weight concentration was performed by vacuum filtering the samples using preweighed 0.45-µm filter membrane (Sartorius Biolabs) followed by drying the filters by microwaving at 125 W for 15 min and storing the filters in a vacuum desiccator for at least 3 d. Sampling for analysis of exometabolites was performed by filtering the sample through 0.45-µm nylon filters. The cell-free samples were stored at −20 °C until analysis. For proteomics, samples were centrifuged at <0 °C for 5 min. Cell pellets were washed with phosphate-buffered saline, flash frozen in liquid nitrogen, and stored at −80 °C until analysis.
Statistical and Data Analysis.
The analysis of all data was performed based on three biological replicates, except for calculations of the OUR, CER, and the RQ, where data from two biological replicates were used for S. pombe, S. cerevisiae, and K. marxianus.
Exometabolite Analysis.
Extracellular glucose, ethanol, glycerol, pyruvate, succinate, and acetate were quantified using an ultimate 3000 high-performance liquid chromatography system (Thermo Fisher) equipped with a BioRad HPX-87H column (BioRad) and a refractive index detector, with 5 mM H2SO4 as the elution buffer at a flow rate of 0.6 mL ⋅ min−1, and an oven temperature of 45 °C.
Gas Analysis.
The oxygen and carbon dioxide concentrations in the exhaust gas were measured in real time using a GA4 gas analyzer (DasGip). The OUR at each time point was calculated, correcting for the dilution by carbon dioxide production, according to Eq. 1:
| [1] |
where F is the gas flow, is the fraction of oxygen in the air entering the reactor, is the fraction of oxygen in the gas exiting the reactor, is the combined fraction of nitrogen and argon in air, is the combined fraction of nitrogen and argon exiting the reactor, and Vm is the molar volume at 25 °C (24.465).The total amount of moles of oxygen consumed until each time point was calculated as the area under the OUR curve, and the resulting values were plotted against the biomass concentration (g cell dry weight). The specific OUR was calculated by taking the slope of the linear trendline and multiplying by the growth rate. The CER was calculated according to Eq. 2:
| [2] |
where and are the fraction of carbon dioxide in the gas entering and exiting the reactor, respectively. Further calculations were performed using the same approach as for the specific OUR.
Quantitative Proteome Measurements.
The liquid chromatography-MS (LC-MS) experiments were carried out on an Orbitrap Fusion or Orbitrap Fusion Lumos mass spectrometer interfaced with an Easy-nLC1200 nanoflow LC systems (all from Thermo Fisher Scientific). Peptide and protein identification was performed using Proteome Discoverer version 2.4 (Thermo Fisher Scientific) with Mascot version 2.5.1 (Matrix Science) as a database search engine.
The global relative protein quantification between the samples was performed via the modified filter-aided sample preparation (FASP) method (51), which included the two-stage digestion with trypsin in 0.5% sodium deoxycholate/50 mM triethylammonium bicarbonate buffer and labeling with the TMT 10plex isobaric reagents (Thermo Fisher Scientific) according to the manufacturer's instructions. The polled reference sample for each species was prepared from aliquots of the individual replicates and processed alongside the individual samples. The combined TMT-labeled set was prefractionated into 40 primary fractions on via basic-pH reversed-phase chromatography (bRP-LC) on an XBridge BEH C18 column (3.5 μm, 3.0 × 150 mm, Waters Corporation) at pH 10 using a Dionex Ultimate 3000 ultra high-performance liquid chromatography system (Thermo Fisher Scientific). The most abundant peptide precursors were selected in a data-dependent manner, collision-induced dissociation MS2 spectra for peptide identification were recorded in the ion trap, the 10 most abundant fragment ions were isolated, fragmented using the higher-energy collision dissociation, and the MS3 spectra for reporter ion quantification were recorded in the Orbitrap.
For the label-free quantification, an aliquot of 25 µg of each of the the pooled reference samples was spiked with 5.3 µg UPS2 Proteomics Dynamic Range Standard (Sigma-Aldrich), digested using the FASP protocol as described in the previous paragraph, and separated into eight fractions using a Pierce high-pH reversed-phase spin column kit (Thermo Fisher Scientific). Each peptide fraction was analyzed in three replicates, with the MS1 spectra recorded at 120,000 resolution and the data-dependent MS2 spectra recorded in the ion trap. The label-free data were processed using the Minora feature detection node in Proteome Discoverer version 2.4, and the intensity values of the three injection replicates were averaged. A modified iBAQ (20) approach was used to estimate the absolute protein concentration in the pooled reference sample. Protein intensity was divided by the number of identified peptides to yield the iBAQ intensity for each protein. Linear regression was calculated for the relationship between the known log10-transformed iBAQ abundance and the measured log10-transformed protein amounts of the spiked UPS2 standard proteins. The resulting regression model and measured iBAQ abundances of the proteins in the pooled reference sample were used to estimate their protein amounts.
The detailed experimental procedures, LC-MS, and data processing parameters are described in SI Appendix, Supplementary Methods.
For determination of total protein content subsequent to the proteomics analysis, proteins were extracted by boiling cell pellets in a lysis buffer containing 1M NaOH and 10% sodium dodecyl sulfate for 10 min. Supernatant was collected and protein concentration was measured using the Pierce bichionic acid assay Protein Assay Kit (ThermoFisher 23225) and normalized to the cell dry weight.
Ortholog Prediction with OrthoFinder and GO Annotation.
To identify orthologous protein among the four yeasts analyzed in this study, and to facilitate the assignment of GO terms for analyzing the proteomics data, proteome-wide homology matching was performed using OrthoFinder (52). The results were processed and classified into single-copy and multicopy orthologs. Single-copy orthologs were defined as having a single ortholog in each of the four species, while multicopy orthologs were defined as having multiple protein orthologs in one or more of the species. To assign GO terms to all proteins of the four yeasts, GO term annotation was first downloaded for S. cerevisiae using the biomaRt R package (53). This annotation was then used to annotate the proteins of S. pombe, K. marxianus, and S. stipitis based on the protein orthology. The GO annotations were used to divide protein into groups by function used for analysis throughout the study.
FBA.
The ec consensus yeast metabolic model version 8.3.4 and the eciSM966 K. marxianus models used in this study were obtained from a GitHub repository hosted within the group (https://github.com/SysBioChalmers/ecModels) (23). Condition-specific models were created by incorporating and constraining the models with experimental measurements of protein content, metabolite exchange rates, and metabolic enzyme abundances using the GECKO toolbox (23). This included scaling the coefficient of protein pseudoreaction to equal the measured protein content, as well as scaling the carbohydrate coefficient to maintain an equivalent amount of mass in the biomass pseudoreaction (9). Additionally, the growth-associated maintenance, which largely reflects the polymerization costs of proteins and carbohydrates, was recalculated as described previously (9). MATLAB R2018b (MathWorks Inc.) with Gurobi solver (Gurobi Optimizer) in the RAVEN toolbox (54) was used for simulations. For the FBA simulations, the total protein pool exchange pseudoreaction was set as an objective function to minimize, and the problem was solved using the solveLP function available in the RAVEN toolbox. In ecModel simulations, each enzyme has an exchange reaction that serves as a proxy for the enzyme requirements and has the unit mmol gDW−1. The capacity usage of individual enzymes, representing the fraction of available enzyme that is in fact used for catalysis, was calculated as the in silico–predicted enzyme exchange flux divided by the in vivo–measured enzyme abundance.
Supplementary Material
Acknowledgments
We thank Egor Vorontsov at the Proteomics Core Facility of Sahlgrenska Academy at the University of Gothenburg for conducting the proteomic analysis. The Proteomics Core Facility is grateful to Inga-Britt and Arne Lundbergs Forskningsstiftlese for the donation of the Orbitrap Fusion Tribrid MS instrument. This research was supported by funding from the Novo Nordisk Foundation and the Knut and Alice Wallenberg Foundation. Open access funding is provided by Chalmers University of Technology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112836118/-/DCSupplemental.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (55) partner repository with the dataset identifier PXD026313. Processed quantitative proteomics data are available in Datasets S1–S4. All code and data associated with this study have been deposited to GitHub and are available at https://github.com/SysBioChalmers/ComparativeStudyCrabtreeEffect (56) or through Zenodo at https://doi.org/10.5281/zenodo.5004293.
References
- 1.Chen Y., Nielsen J., Energy metabolism controls phenotypes by protein efficiency and allocation. Proc. Natl. Acad. Sci. U.S.A. 116, 17592–17597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeiffer T., Morley A., An evolutionary perspective on the Crabtree effect. Front. Mol. Biosci. 1, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeiffer T., Schuster S., Bonhoeffer S., Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Vemuri G. N., Altman E., Sangurdekar D. P., Khodursky A. B., Eiteman M. A., Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 72, 3653–3661 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dijken J. P., Weusthuis R. A., Pronk J. T., Kinetics of growth and sugar consumption in yeasts. Antonie van Leeuwenhoek 63, 343–352 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molenaar D., van Berlo R., de Ridder D., Teusink B., Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5, 323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson A., Nielsen J., Metabolic trade-offs in yeast are caused by F1F0-ATP synthase. Sci. Rep. 6, 22264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez B. J., et al. , Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basan M., et al. , Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crabtree H. G., Observations on the carbohydrate metabolism of tumours. Biochem. J. 23, 536–545 (1929). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Deken R. H., The Crabtree effect: A regulatory system in yeast. J. Gen. Microbiol. 44, 149–156 (1966). [DOI] [PubMed] [Google Scholar]
- 13.Hagman A., Säll T., Compagno C., Piskur J., Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One 8, e68734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffries T. W., Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17, 320–326 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Lane M. M., Morrissey J. P., Kluyveromyces marxianus: A yeast emerging from its sister’s shadow. Fungal Biol. Rev. 24, 17–26 (2010). [Google Scholar]
- 16.Verduyn C., Postma E., Scheffers W. A., Van Dijken J. P., Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Fonseca G. G., de Carvalho N. M. B., Gombert A. K., Growth of the yeast Kluyveromyces marxianus CBS 6556 on different sugar combinations as sole carbon and energy source. Appl. Microbiol. Biotechnol. 97, 5055–5067 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Fonseca G. G., Gombert A. K., Heinzle E., Wittmann C., Physiology of the yeast Kluyveromyces marxianus during batch and chemostat cultures with glucose as the sole carbon source. FEMS Yeast Res. 7, 422–435 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Elbing K., et al. , Role of hexose transport in control of glycolytic flux in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70, 5323–5330 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Gallien S., et al. , Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteomics 11, 1709–1723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orth J. D., Thiele I., Palsson B. Ø., What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domenzain I., et al. , Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0. bioRxiv [Preprint] (2021). 10.1101/2021.03.05.433259 (Accessed 12 May 2021). [DOI] [PMC free article] [PubMed]
- 24.Domenzain I., Li F., Kerkhoven E. J., Siewers V., Evaluating accessibility, usability and interoperability of genome-scale metabolic models for diverse yeasts species. FEMS Yeast Res. 21, foab002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papini M., Nookaew I., Uhlén M., Nielsen J., Scheffersomyces stipitis: A comparative systems biology study with the Crabtree positive yeast Saccharomyces cerevisiae. Microb. Cell Fact. 11, 136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiaux J., et al. , Metabolic-flux profiling of the yeasts Saccharomyces cerevisiae and Pichia stipitis. Eukaryot. Cell 2, 170–180 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blank L. M., Lehmbeck F., Sauer U., Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 5, 545–558 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Pronk J. T., Yde Steensma H., Van Dijken J. P., Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12, 1607–1633 (1996). [DOI] [PubMed] [Google Scholar]
- 29.The Gene Ontology Consortium, The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47 (D1), D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfe K. H., Shields D. C., Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Kellis M., Birren B. W., Lander E. S., Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Conant G. C., Wolfe K. H., Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 3, 129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhind N., et al. , Comparative functional genomics of the fission yeasts. Science 332, 930–936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Z., Li W.-H., Expansion of hexose transporter genes was associated with the evolution of aerobic fermentation in yeasts. Mol. Biol. Evol. 28, 131–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otterstedt K., et al. , Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 5, 532–537 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vemuri G. N., Eiteman M. A., McEwen J. E., Olsson L., Nielsen J., Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 2402–2407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Hoek P., Van Dijken J. P., Pronk J. T., Effect of specific growth rate on fermentative capacity of baker’s yeast. Appl. Environ. Microbiol. 64, 4226–4233 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne K. P., Wolfe K. H., The yeast gene order browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15, 1456–1461 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoighe C., Wolfe K. H., Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 2, 548–554 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Metzl-Raz E., et al. , Principles of cellular resource allocation revealed by condition-dependent proteome profiling. eLife 6, e28034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu R., Vorontsov E., Sihlbom C., Nielsen J., Quantifying absolute gene expression profiles reveals distinct regulation of central carbon metabolism genes in yeast. eLife 10, e6572222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gancedo J. M., Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassiliadis D., Wong K. H., Andrianopoulos A., Monahan B. J., A genome-wide analysis of carbon catabolite repression in Schizosaccharomyces pombe. BMC Genomics 20, 251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Z., Li W.-H., The evolution of aerobic fermentation in Schizosaccharomyces pombe was associated with regulatory reprogramming but not nucleosome reorganization. Mol. Biol. Evol. 28, 1407–1413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Blum J. E., Thalacker-Mercer A., Gu Z., Impact of the whole genome duplication event on PYK activity and effects of a PYK1 mutation on metabolism in S. cerevisiae. Front. Mol. Biosci. 8, 656461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamrad S., et al. , Pyruvate kinase variant of fission yeast tunes carbon metabolism, cell regulation, growth and stress resistance. Mol. Syst. Biol. 16, e9270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullis A., et al. , Parallel concerted evolution of ribosomal protein genes in fungi and its adaptive significance. Mol. Biol. Evol. 37, 455–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner J. R., The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Yu R., et al. , Nitrogen limitation reveals large reserves in metabolic and translational capacities of yeast. Nat. Commun. 11, 1881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ihmels J., et al. , Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309, 938–940 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Wiśniewski J. R., Zougman A., Nagaraj N., Mann M., Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durinck S., Spellman P. T., Birney E., Huber W., Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H., et al. , RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLOS Comput. Biol. 14, e1006541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malina C., et al. , Comparative study Crabtree effect. GitHub. https://github.com/SysBioChalmers/ComparativeStudyCrabtreeEffect. Deposited 21 June 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (55) partner repository with the dataset identifier PXD026313. Processed quantitative proteomics data are available in Datasets S1–S4. All code and data associated with this study have been deposited to GitHub and are available at https://github.com/SysBioChalmers/ComparativeStudyCrabtreeEffect (56) or through Zenodo at https://doi.org/10.5281/zenodo.5004293.