Significance
The taste of acids is critical for animal survival since it enables them to differentiate potentially dangerous from nutritious foods. Due to the general requirement for acid taste for survival, we tested the idea that the receptor mechanism functioning in acid taste may be evolutionarily conserved. Here, we demonstrate that mutation of a Drosophila gene, Otopetrin-Like A (OtopLA), encoding a protein distantly related to the recently identified mammalian acid taste receptor, OTOP1, is essential for both the strong repulsion to highly acidic food and mild attraction to low acidity. The mild attraction and strong aversion to acids requires expression of OtopLA in distinct taste neurons in the fly equivalent of the vertebrate tongue.
Keywords: acid taste, sour taste, proton sensor, Drosophila, Otopetrin
Abstract
Receptors for bitter, sugar, and other tastes have been identified in the fruit fly Drosophila melanogaster, while a broadly tuned receptor for the taste of acid has been elusive. Previous work showed that such a receptor was unlikely to be encoded by a gene within one of the two major families of taste receptors in Drosophila, the “gustatory receptors” and “ionotropic receptors.” Here, to identify the acid taste receptor, we tested the contributions of genes encoding proteins distantly related to the mammalian Otopertrin1 (OTOP1) proton channel that functions as a sour receptor in mice. RNA interference (RNAi) knockdown or mutation by CRISPR/Cas9 of one of the genes, Otopetrin-Like A (OtopLA), but not of the others (OtopLB or OtopLC) severely impaired the behavioral rejection to a sweet solution laced with high levels of HCl or carboxylic acids and greatly reduced acid-induced action potentials measured from taste hairs. An isoform of OtopLA that we isolated from the proboscis was sufficient to restore behavioral sensitivity and acid-induced action potential firing in OtopLA mutant flies. At lower concentrations, HCl was attractive to the flies, and this attraction was abolished in the OtopLA mutant. Cell type–specific rescue experiments showed that OtopLA functions in distinct subsets of gustatory receptor neurons for repulsion and attraction to high and low levels of protons, respectively. This work highlights a functional conservation of a sensory receptor in flies and mammals and shows that the same receptor can function in both appetitive and repulsive behaviors.
Humans possess the ability to distinguish among five basic tastes: sweet, bitter, salt, sour, and umami. Interestingly, there is considerable variety in the ability of other mammals to detect these qualities. For example, cats are missing sweet taste (1) and the bottlenose dolphin only detects salt in food (2). Yet the fruit fly, Drosophila melanogaster, responds to a similar repertoire of tastes as humans. This is all the more remarkable given the very distant evolutionary relatedness and the enormous differences in the anatomy of the fly and mammalian taste organs and points to a conserved function of these taste qualities in assessing food quality.
Many of the receptors involved in Drosophila taste have been defined (3, 4). Those that contribute to sweet and bitter tastes have been characterized extensively and are members of the “gustatory receptor” (GR) family (3, 4). GRs are unrelated to the G protein–coupled receptors that function in mammalian sweet and bitter taste (3). Therefore, the abilities of insects and humans to respond to similar repertoires of chemicals such as sweet and bitter tastants have emerged independently.
In mice, the taste of acids depends on a proton-selective channel, Otopetrin1 (OTOP1), which is expressed in type III taste receptor cells (5–7). OTOP1 was first identified based on its essential role in the vestibular systems of the mouse and zebrafish (8–11) and was found to encode a family of proton-selective ion channels functionally conserved from worms to humans (5, 9, 12). In sea urchins, an Otop channel functions in calcifying primary mesenchymal cells by promoting the removal of protons generated during the production of CaCO3 (13). Otop family members are structurally unrelated to other ion channels and are composed of 12 transmembrane segments (12, 14, 15), which assemble as a dimer with no obvious permeation pathway (14, 15). Flies, mice, and human genomes each contain three otop genes, although the fly genes are not direct homologs of the vertebrate genes (9).
In Drosophila, low or moderate levels of some organic acids are attractive and promote feeding, while the same acids at higher concentrations repress food consumption (16, 17). This rejection contributes to survival as it discourages the animals from eating very acidic foods in the environment that can decrease lifespan. Two members of the large family of “ionotropic receptors” (IRs; IR25a and IR76b) function in GR neurons (GRNs) in the legs for sensing carboxylic acids and HCl (18). Mutation of either of these IRs disrupts the preference to lay eggs on acid-containing substrates (18). Flies prefer consuming lactic acid over water, and this preference is mildly reduced in Ir25a mutants (19).
The receptors required for the gustatory rejection of noxious levels of acids have been largely enigmatic. An exception is IR7a, which is needed to suppress feeding on foods laced with acetic acid (17). IR7a is very narrowly tuned, as it does not impair the rejection of foods with HCl or any other carboxylic acid tested. This receptor acts in a subset of GRNs called B GRNs that are also activated by bitter chemicals and certain other aversive compounds (4, 17).
Here, we identified a member of the family of Otop channels that in Drosophila is required for the detection of protons in food. Wild-type flies are strongly repelled by high levels of HCl and mildly attracted to a low level of HCl. We found that these responses depend on the Otopetrin-Like protein (OtopLA), which has a common evolutionarily origin with mammalian OTOP channels. By performing cell type–specific rescue experiments, we found that the strong repulsion and mild attraction to different levels of acids depends on expression of OtopLA in distinct subsets of GRNs.
Results
Silencing OtopLA Reduces Gustatory Repulsion to Acids.
To determine the degree to which acids are repulsive, we conducted proboscis extension response (PER) assays in which we laced an appetitive cue, 100 mM sucrose, with varying concentrations of acids. For these experiments, the flies were food deprived for 24 to 26 h to motivate feeding. The sucrose stimulus is then applied to the major taste organ (the labellum), and the extent to which the animals extend their proboscis is monitored. A full extension is scored as 1.0, a partial extension as 0.5, and no extension as 0. When we offered control flies sucrose only (pH 6.8), virtually all the animals fully extended their proboscis (Fig. 1A).
Fig. 1.
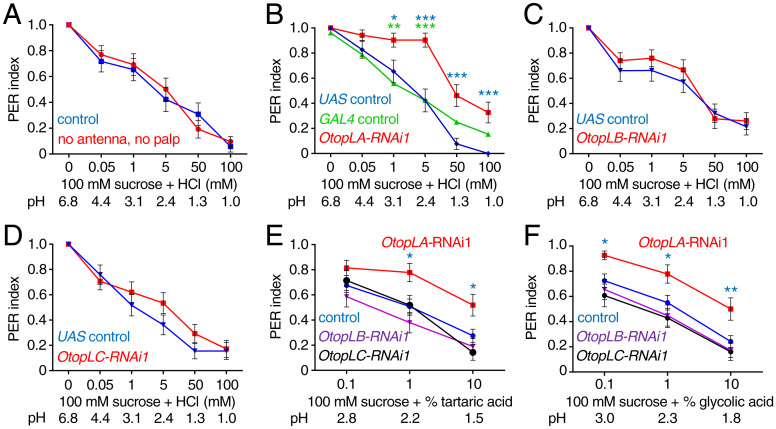
RNAi screen for OtopL genes required for aversion of acid taste. PER assays were performed by touching the proboscis with either 100 mM sucrose alone or sucrose plus the indicated concentrations of HCl (mM) or organic acids (%). The pH values are indicated in each panel. A full extension is scored as 1.0, a partial extension as 0.5, and no extension as 0. All RNAi lines (B–F) were generated by crossing the indicated UAS-RNAi lines to elav-GAL4;UAS-Dcr2 flies. (A) Effects of removing olfactory organs on the PER responses to the indicated concentrations of HCl. Control (w1118) and w1118 flies in which the antenna and maxillary palp were surgically removed (no antenna, no palp). n = 26. (B–F) Effects of RNAi knockdown of different OtopL genes on the PER responses to the indicated acids. (B) Effect of knockdown of OtopLA on the PER to sucrose plus HCl. OtopLA-RNAi1 is UAS-OtopLA-RNAi1 (v104973) crossed to elav-GAL4;UAS-Dcr2 flies (n = 30). The UAS control (n = 26) and GAL4 control (n = 25) were generating by crossing w1118 to either UAS-OtopLA-RNAi1 or elav-GAL4;UAS-Dcr2, respectively. The blue and green asterisks indicate statistically significance differences between OtopLA silenced flies and the UAS and GAL4 controls, respectively. (C) Effect of knockdown of OtopLB on the PER to sucrose plus HCl. OtopLB-RNAi1 is UAS-OtopLB-RNAi1 (v101936) crossed to elav-GAL4;UAS-Dcr2 flies (n = 27). UAS control (n = 28). (D) Effect of knockdown of OtopLC on the PER to sucrose plus HCl. OtopLC-RNAi1 is UAS-OtopLC RNAi (v108591) crossed to elav-GAL4;UAS-Dcr2 flies (n = 29). UAS control (n = 29). (E) Effect of knockdown of OtopL genes on PERs using the indicated concentrations of tartaric acid. OtopLA-RNAi1 (n = 27), OtopLB-RNAi1 (n = 29), and OtopLC-RNAi1 (n = 28). The “control” is elav-GAL4;UAS-Dcr2 flies (n = 65). The blue asterisks indicate significance differences from the control. (F) Effect of knockdown of OtopL genes on PERs using the indicated concentrations of glycolic acid. OtopLA-RNAi1 (n = 27), OtopLB-RNAi1 (n = 29), OtopLC-RNAi1 (n = 28). The “control” is elav-GAL4;UAS-Dcr2 (n = 65). The blue asterisks indicate significance differences from the control. Mann–Whitney u tests. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
When tested with sucrose laced with increasing concentrations of HCl, thereby decreasing the pH from 6.8 to between 4.4 and 1.0, wild-type flies reduced their PER in proportion to the concentration of HCl (Fig. 1A). We obtained similar results using organic acids, such as tartaric acid (SI Appendix, Fig. S1A). Flies sense acids through both taste and smell (4). To assess a potential contribution of smell to the suppression of the PER by HCl, we surgically ablated the two olfactory organs: the antenna and maxillary palp. After removing these organs, addition of HCl and tartaric acid still diminished the PER to the same extend as in intact control flies (Fig. 1A and SI Appendix, Fig. S1A). Thus, olfaction was not needed for the flies to display a distaste for acids.
Drosophila encodes three Otopetrin-Like proteins (OtopLA, OtopLB, and OtopLC), which share a low level of amino acid homology (20.1 to 30.6% identities) to the mouse and human OTOP1 proteins (5, 12). To investigate if any of the three fly OtopLs was required for gustatory aversion toward acids, we used two RNA interference (RNAi) lines for each gene and drove expression with a pan-neuronal driver (elav-GAL4). Both OtopLA RNAi lines (OtopLA-RNAi1 and OtopLA-RNAi2) caused large decreases in repulsion to low pH (Fig. 1B and SI Appendix, Fig. S1B). Knockdown of OtopLA nearly eliminated repulsion to concentrations of HCl between 50 μM and 5 mM (pH 4.4 to 2.4; Fig. 1B and SI Appendix, Fig. S1B). Moreover, we observed attenuation of the aversiveness to concentrations as high as 100 mM HCl (pH 1.0; Fig. 1B and SI Appendix, Fig. S1B). The remaining rejection of sucrose at very low pHs (1.3 and 1.0), which are expected to be tissue damaging, may be due to the nociceptive response to extremely high acidity (20). In contrast to the effects of silencing OtopLA, RNAi knockdown of either OtopLB or OtopLC had no effect on the HCl-induced suppression of the PER (Fig. 1 C and D and SI Appendix, Fig. S1 C and D).
To investigate if OtopLA is also required for sensing carboxylic acids, we repeated the PER assays using sucrose solutions containing varying concentrations of tartaric acid and glycolic acid. The suppression of the PER by these carboxylic acids seen in control flies was significantly attenuated by silencing OtopLA (Fig. 1 E and F and SI Appendix, Fig. S1 E and F). In contrast, knockdown of OtopLB or OtopLC had no impact on the PER under similar conditions (Fig. 1 E and F and SI Appendix, Fig. S1 E and F).
A Proboscis OtopLA Isoform Functions in Acid Sensation.
The OtopLA locus is predicted to be expressed as six messenger RNA (mRNA) isoforms, which encode proteins with 12 transmembrane segments similar to other Otop proteins (5, 12). Five isoforms share a common translation start site (OtopLAc–OtopLAg), and one begins at an alternative site (OtopLAa; SI Appendix, Fig. S2A). Using CRISPR/Cas9, we inserted the LexA and miniwhite transgenes at the translation start site for OtopLAc–OtopLAg to create the OtopLA1 allele (SI Appendix, Fig. S2A). The knock-in deleted 40 base pairs that removed residues 1 to 14 and shifted the reading frame (SI Appendix, Fig. S2A). The deletion also disrupted expression of the mRNAs (SI Appendix, Fig. S2B). We introduced a similar insertion at the start site of OtopLAa, which removed the region encoding amino acids 1 to 18 and introduced a frame shift (SI Appendix, Fig. S2A; OtopLA2).
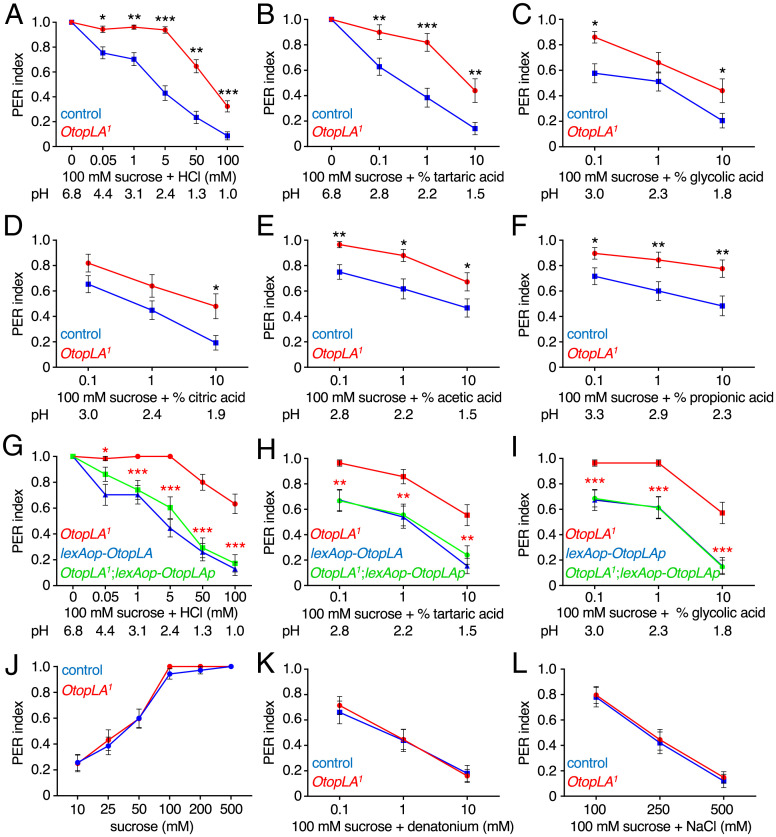
We performed PER assays with 100 mM sucrose mixed with a range of HCl concentrations. The repulsion to HCl was virtually eliminated in OtopLA1 flies at levels up to 5 mM, and there were significant reductions in the PERs at 50 and 100 mM (Fig. 2A). This effect was independent of the concentration of sucrose, as 5 mM HCl suppressed the attraction to concentrations of sucrose ranging from 10 to 500 mM (SI Appendix, Fig. S2C), and the OtopLA1 flies were impaired in HCl repulsion at all concentrations of sucrose tested (SI Appendix, Fig. S2C).
Fig. 2.
Mutation of OtopLA impairs gustatory repulsion to acids. (A–F) PER indexes obtained using control (w1118) and OtopLA1 flies upon stimulation of the labellum with 100 mM sucrose alone or 100 mM sucrose mixed with the indicated concentrations of acids. (A) Sucrose and HCl. Control, n = 64. OtopLA1, n = 65. (B) Sucrose and tartaric acid. Control, n = 39. OtopLA1, n = 25. (C) Sucrose and glycolic acid. Control, n = 39. OtopLA1, n = 25. (D) Sucrose and citric acid. Control, n = 39. OtopLA1, n = 25. (E) Sucrose and acetic acid. Control, n = 30. OtopLA1, n = 29. (F) Sucrose and proprionic acid. Control, n = 30. OtopLA1, n = 29. (G–I) Testing for rescue of the OtopLA1 defects in acid aversion by expressing the OtopLAp transgene under control of the LexA knocked into OtopLA1. The PERs were assayed from the following lines using the indicated concentrations of acids: 1) OtopLA1, 2) lexAop-OtopLA, and 3) OtopLA1;lexAop-OtopLA (rescue). Asterisks indicate significance differences between OtopLA1 and the rescue flies. (G) HCl. OtopLA1, n = 30. lexAop-OtopLA, n = 27. OtopLA1;lexAop-OtopLA, n = 29. (H) Tartaric acid. OtopLA1, n = 28. lexAop-OtopLA, n = 26. OtopLA1;lexAop-OtopLA, n = 28. (I) Glycolic acid. OtopLA1, n = 28. lexAop-OtopLA, n = 26. OtopLA1;lexAop-OtopLA, n = 28. (J) Testing whether the OtopLA1 mutation impairs the PER response to sucrose. Control (w1118), n = 27. OtopLA1, n = 29. (K) Testing whether the OtopLA1 mutation impairs the PER response to denatonium mixed with 100 mM sucrose. Control (w1118), n = 27. OtopLA1, n = 29. (L) Testing whether the OtopLA1 mutation impairs the PER response to NaCl mixed with 100 mM sucrose. Control (w1118), n = 27. OtopLA1, n = 29. Mann–Whitney U tests. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
We also assessed the responses to different concentrations of HCl using a binary choice assay in microtiter plates. Alternate wells contained either 2 mM sucrose alone or 2 mM sucrose laced with HCl. A 100% preference for 2 mM sucrose alone results in a preference index (PI) of 1.0, while a complete selection of 2 mM sucrose plus HCl yields a PI = −1. An equal preference for both options results in a PI = 0. Control flies were indifferent to 0.005 HCl but showed a modest attraction to 0.01 HCl, which was statistically significant (SI Appendix, Fig. S2D). In contrast, 5 to 100 mM HCl were aversive (SI Appendix, Fig. S2D). We found that the mild attraction to 0.01 mM HCl was eliminated in OtopLA1 flies (SI Appendix, Fig. S2D). In addition, the OtopLA1 mutant exhibited reduced aversion to sucrose mixed with 5 to 100 mM HCl (SI Appendix, Fig. S2D). OtopLA1 flies retained some degree of aversion to acids, even to 5 mM HCl (SI Appendix, Fig. S2D). This is in contrast to the PER results in which repulsion to 5 mM HCl was virtually eliminated (Fig. 2A). The remaining repulsion in the binary feeding assay might be due to a postfeeding effect or to the lower concentration of sucrose that was used (2 mM) than for the PER assays (100 mM). In contrast to the deficit in OtopLA1, the OtopLA2 mutant exhibited a normal rejection of HCl (SI Appendix, Fig. S2E). Consistent with this finding, the OtopLA-RNAi1, which did not target OtopLAa, still disrupted rejection of HCl (Fig. 1B).
To test whether the OtopLA1 flies also show a deficit in rejecting sucrose with organic acids, we conducted PER assays with tartaric acid, glycolic acid, citric acid, acetic acid, and propionic acid. The degree of suppression of the PERs in control flies varies greatly with different carboxylic acids (Fig. 2 B–F) (16, 17, 21). Nevertheless, OtopLA1 flies exhibited decreased behavioral aversion toward each of the organic acids (Fig. 2 B–F).
The preceding analyses indicate that one or more of the five isoforms with the common translational start site (OtopLAc-g; SI Appendix, Fig. S2A) functions in acid taste. Therefore, to identify the key isoform, we isolated mRNA from the fly proboscis and performed RT-PCR using primers that amplify all the isoforms except OtopLAa. We isolated and sequenced eight complementary DNAs (cDNAs) from the proboscis, all of which encoded a 947–amino acid protein (OtopLAp; SI Appendix, Fig. S2A) distinct from those annotated by FlyBase (https://flybase.org/). OtopLAp is most similar to OtopLAg, except for missing residues 204 to 210 in OtopLAg, which are encoded by the small fifth coding exon, and the last four amino acids of the eighth exon (residues 909 to 912; SI Appendix, Fig. S2A). To test whether OtopLAp is sufficient to restore normal acid sensitivity to the OtopLA1 mutant, we generated flies expressing lexAop-OtopLAp and introduced the transgene into the OtopLA1 background so that it would be expressed under control of the LexA knocked into OtopLA1 (SI Appendix, Fig. S2A). The OtopLAp transgene fully restored aversion to HCl, tartaric acid, and glycolic acid (Fig. 2 G–I), demonstrating that OtopLAp is sufficient to confer acid taste.
OtopLA Mutants Display a Narrow Behavioral Deficit in Tasting Low pH.
To evaluate whether OtopLA is specifically required for acid taste, we assayed the PERs to other chemicals. The attraction to sucrose alone was identical between OtopLA1 and the control flies over concentrations ranging from 10 to 500 mM (Fig. 2J). Thus, the reductions in repulsion to sugars mixed with acids did not appear to be due to a change in sugar sensitivity. The aversion to the bitter compound, denatonium, and to high levels of NaCl were also indistinguishable between OtopLA1 and control flies (Fig. 2 K and L). Thus, the OtopLA1 mutation did not cause a broad deficit in gustatory sensation.
OtopLA Mutants Are Impaired in Acid-Induced Action Potentials.
If loss of OtopLA impairs acid sensitivity by disrupting reception in GRNs, then this should cause a decrease in acid-induced action potentials in these sensory neurons. The labellum is decorated with 31 gustatory hairs (sensilla) that fall into three length classes: small (S-type), intermediate (I-type), and large (L-type) (4). A total of 9 out of the 11 S-type sensilla are categorized as either S-a or S-b sensilla on the basis of distinct sensitivities to bitter compounds (22). The two S-c sensilla (S4 and S8) are unresponsive to all bitter chemicals tested (22). The S-b sensilla (S3, S5, and S9) have been reported to be responsive to acids (16). To perform tip recordings, we used the standard electrolyte for this assay (30 mM tricholine citrate, TCC; pH 6.8) because it suppresses water spikes (4, 23). TCC alone induced very few action potentials in S-b sensilla in control flies (Fig. 3 A–C). Consistent with a previous study (16), HCl (pH 4 and pH 2) activated S-b sensilla (Fig. 3 A–D and F) but not S-a (e.g., S6 and S7; SI Appendix, Fig. S3 A and B) or the S-c sensillum (e.g., S4; SI Appendix, Fig. S3C). Also as reported, the I-a type (e.g., I7) are not stimulated by acid (SI Appendix, Fig. S3D), while I-b type of I sensilla (e.g., I8) are responsive (16) (SI Appendix, Fig. S3E).
Fig. 3.
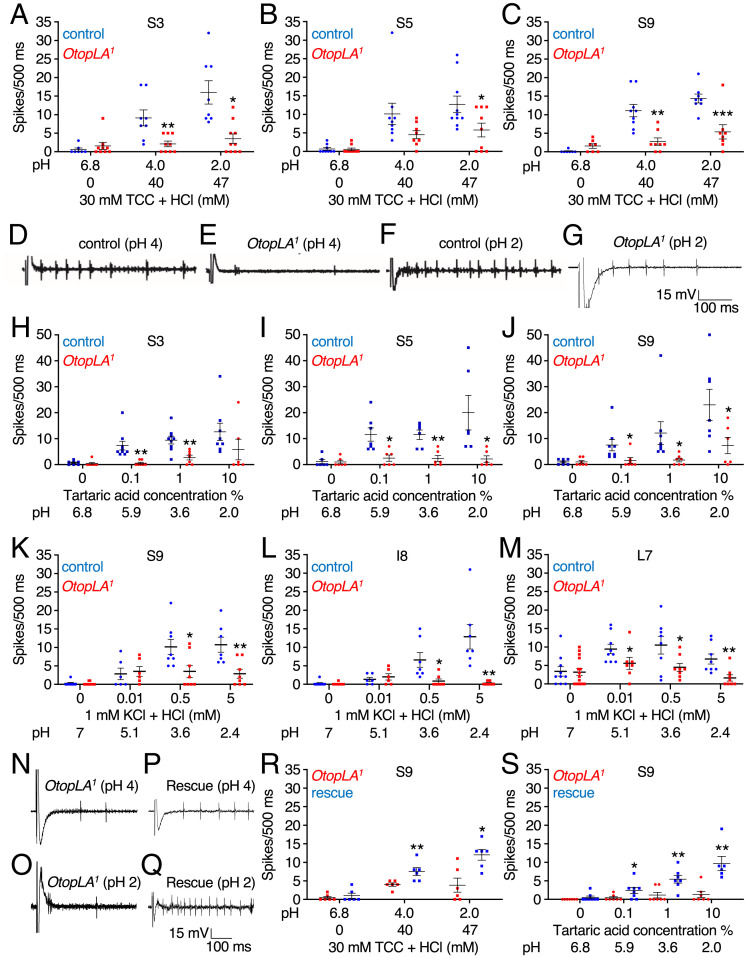
Tip recordings showing that mutation of OtopLA impairs acid-induced action potentials. (A–C) Tip recordings from the indicated S-b class sensilla (22) due to stimulation with HCl at the indicated pH values. Shown are the mean action potentials during the first 500 ms of the recordings with the control (w1118) and OtopLA1. The pH 6.8 solution contained only the electrolyte (30 mM TCC). (A) Responses of S3 sensilla to HCl. Control, n = 7 to 8. OtopLA1, n = 9 to 10. (B) Responses of S5 sensilla to HCl. Control, n = 9 to 10. OtopLA1, n = 8 to 19. (C) Responses of S9 sensilla to HCl. Control, n = 7 to 9. OtopLA1, n = 7 to 8. (D–G) Representative traces for the first 500 ms obtained from S9 sensilla from control (w1118) and OtopLA1 flies during exposure to HCl at pH 4 or 2. (H–J) Tip recordings from the indicated S-b class sensilla stimulated with tartaric acid at the indicated pH values. Shown are the mean action potentials during the first 500 ms of the recordings with the control (w1118) and OtopLA1. The 0% tartaric acid solution consisted only of the electrolyte (30 mM TCC). (H) Responses of S3 sensilla to tartaric acid. Control, n = 7 to 9. OtopLA1, n = 6 to 7. (I) Responses of S5 sensilla to tartaric acid. Control, n = 6 to 7. OtopLA1, n = 6. (J) Responses of S9 sensilla to tartaric acid. Control, n = 7 to 8. OtopLA1, n = 6 to 7. (K–M) Tip recordings from the indicated class of sensilla due to stimulation with HCl at the indicated concentrations and pH values. Shown are the mean action potentials during the first 500 ms of the recordings with the control (w1118) and OtopLA1. The 0% HCl solution contained only the electrolyte (1 mM KCl). (K) Responses of S9 sensilla to HCl. Control, n = 6 to 8. OtopLA1, n = 8 to 12. (L) Responses of I8 sensilla to HCl. Control, n = 6 to 9. OtopLA1, n = 6 to 13. (M) Responses of L7 sensilla to HCl. Control, n = 8 to 9. OtopLA1, n = 8 to 16. (N–Q) Testing for rescue of the deficit in HCl-induced action potentials in OtopLA1 flies expressing the OtopLAp transgene. Representative traces for the first 500 ms obtained from S9 sensilla of OtopLA1 and OtopLA1;lexAop-OtopLAp (rescue) flies upon stimulation with HCl at pH 4 or 2. In all cases, 30 mM TCC was used as the electrolyte. (R) Assaying for changes in mean HCl-induced action potentials in OtopLA1 flies expressing the OtopLAp transgene (rescue). The mean action potentials were during the first 500 ms from S9 sensilla stimulated with HCl at the indicated pHs. OtopLA1, n = 6 and OtopLA1;lexAop-OtopLA (rescue), n = 6. (S) Assaying for changes in mean tartaric acid-induced action potentials in OtopLA1 flies expressing the OtopLAp transgene (rescue). The mean action potentials were during the first 500 ms from S9 sensilla stimulated with tartaric acid at the indicated pH values. We used 30 mM TCC as the electrolyte. OtopLA1, n = 7 and OtopLA1;lexAop-OtopLA (rescue), n = 6 to 8. Student’s unpaired t tests. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
To determine whether the OtopLA1 mutation caused a reduction in acid-induced action potentials, we examined HCl at pH 4 and 2. We found that the frequencies of action potentials were diminished in all three S-b sensilla (Fig. 3 A–G). We recorded from an I-b sensilla (I8) and found that the frequency of acid-induced action potentials was greatly reduced in OtopLA1 flies (SI Appendix, Fig. S3E). We also compared the neuronal excitability of control and OtopLA1 over a range of concentrations of tartaric acid. The s-b and I-b taste sensilla from the mutant flies exhibited large reductions in responses to all levels of tartaric acid (Fig. 3 H–J and SI Appendix, Fig. S3F).
A discrepancy between the PER and tip recordings is that 5 mM HCl suppressed the PER induced by 100 mM sucrose but did not elicit action potentials from any sensilla tested (SI Appendix, Fig. S3G). However, if we add the tip recording electrolyte (30 mM TCC) to the solution used for the PER assays (100 mM sucrose plus 5 mM HCl), the pH increased from 2.3 to 5.9, and there was no suppression by the 5 mM HCl (SI Appendix, Fig. S3H). To circumvent the pH buffering by TCC, we performed additional tip recordings using 1 mM KCl as the electrolyte and tested 0.5 and 5 mM HCl. As before, we observed little or no responses from S-a (SI Appendix, Fig. S3I), S-c (SI Appendix, Fig. S3J), and I-a (SI Appendix, Fig. S3K) sensilla. However, both S-b and I-b sensilla responded to 0.5 and 5 mM HCl (Fig. 3 K and L). In addition, we examined an L-type sensilla (L7) and obtained responses (Fig. 3M). In contrast to the other sensilla, the acid-induced action potential frequency from L7 was lower with 5 mM HCl than with 0.5 mM HCl (Fig. 3M). In OtopLA1 flies, the responses to 0.5 and 5 mM HCl were decreased significantly in all three acid-sensitive sensilla types (Fig. 3 K–M). The OtoplA1 flies also exhibited a slightly lower response to 1 mM KCl alone in L7 sensilla as compared to control flies, although the difference was not significant (P = 0.8; Student’s unpaired t test, Fig. 3M). This could potentially be due to water spikes since unlike for TCC, water spikes are not suppressed by KCl.
To test if OtopLAp is sufficient to restore neuronal sensitivity to the OtopLA1 mutant, we conducted extracellular tip recordings. We created flies harboring the lexAop-OtopLAp transgene and crossed it into the OtopLA1 background so that OtopLAp was expressed under control of the LexA in the cells that endogenously express OtopLAp. We then assayed HCl-induced action potentials by recording from S9 sensilla. We found that introduction of the OtopLAp transgene significantly increased the frequency of HCl-driven action potentials in OtopLA1 flies (Fig. 3 N–R). Introduction of the OtopLAp transgene also increased the low sensitivity to tartaric acid in OtopLA1 S9 sensilla (Fig. 3S).
OtopLA Functions in Multiple GRN Types to Sense Acids.
S-type sensilla house four types of GRNs including A GRNs that are activated by attractive tastants such as sugars and B GRNs that are stimulated by acids and other aversive compounds such as bitter chemicals. Loss of OtopLA did not cause a general deficit in electrical excitability of B GRNs as the frequency of denatonium-induced spikes was the same in control and mutant flies (SI Appendix, Fig. S3 L and M). S-type sensilla also include C GRNs that respond to water, and D GRNs that are activated by cations (4). I-type sensilla harbor just A GRNs and B GRNs, while L-type sensilla house A, C, D, and E (low salt) GRNs (4).
Carboxylic acids have been reported to suppress feeding by activating B GRNs and suppressing the sugar response of A GRNs (16). We focused on citric acid and an L-type sensilla (L7), which houses A but not B GRNs, and found that citric acid suppressed sugar-induced action potentials in a concentration-dependent manner as reported (SI Appendix, Fig. S3N) (16). However, HCl did not suppress the sugar activation of A GRNs even at a pH 2.0 (SI Appendix, Fig. S3O).
To identify the type of GRN that requires OtopLA for sensitivity toward acids, we performed gene silencing in each class of GRNs using cell type–specific GAL4s and the two OtopLA RNAi lines that we used for the global knock down OtopLA (see Silencing OtopLA Reduces Gustatory Repulsion to Acids). We then conducted PER assays on the cell type–specific knockdown strains, focusing on the suppression of 100 mM sucrose appeal by 5 mM HCl. Knockdown of OtopLA in A GRNs (Gr5a-GAL4), C GRNs (ppk28-GAL4), D GRNs (ppk23-GAL4), or E (Ir94a-GAL4) GRNs had no effect (SI Appendix, Fig. S4A). Taste hairs also contain an associated mechanosensory (M) neuron (4). There was no effect resulting from RNAi knockdown of OtopLA in these neurons (nompC-GAL4; SI Appendix, Fig. S4A). However, when we silenced OtopLA in B neurons (Gr66a-GAL4), using either RNAi, the repulsion toward 5 mM HCl was strongly suppressed and the flies showed a PER close to 1 (SI Appendix, Fig. S4A). To further characterize the effect of silencing OtopLA in B neurons, we conducted PER assays with a range of HCl concentrations. The outcome was the same as that obtained with pan-neuronal silencing of OtopLA—there was a general loss of sensitivity toward low pH across a range of concentrations (SI Appendix, Fig. S4B).
To examine expression of OtopLA in the labellum, we used two approaches. We first took advantage of the LexA reporter knocked into the OtopLA1 allele (SI Appendix, Fig. S2A) to drive expression of lexAop-mCherry. Although the mCherry signal was modest, it appeared to label GRNs associated with both taste pegs (SI Appendix, Fig. S4C), which are flat sensilla situated between the pseudotrachea (24), and taste hairs (SI Appendix, Fig. S4H). However, when we used the poxneuro70 (poxn70) mutation to eliminate all gustatory hairs without effecting peg GRNs (25), we found that neither 5 mM HCl nor 1% tartaric acid was effective in suppressing the attraction to sucrose (SI Appendix, Fig. S4D). The poxn70 mutation only affects gustatory hairs and not olfactory hairs (26, 27). Thus, our results highlight a role for taste hairs in acid-induced repulsion. Furthermore, surgical removal of the olfactory organs in poxn70 did not result in a further reduction in acid-induced repulsion (SI Appendix, Fig. S4D). When we inactivated peg GRNs only (57F03-GAL4and UAS-kir2.1) (28), there was a normal acid-induced suppression of the sugar response (SI Appendix, Fig. S4E). Thus, the peg GRNs appear to have little if any role in acid-induced taste responses.
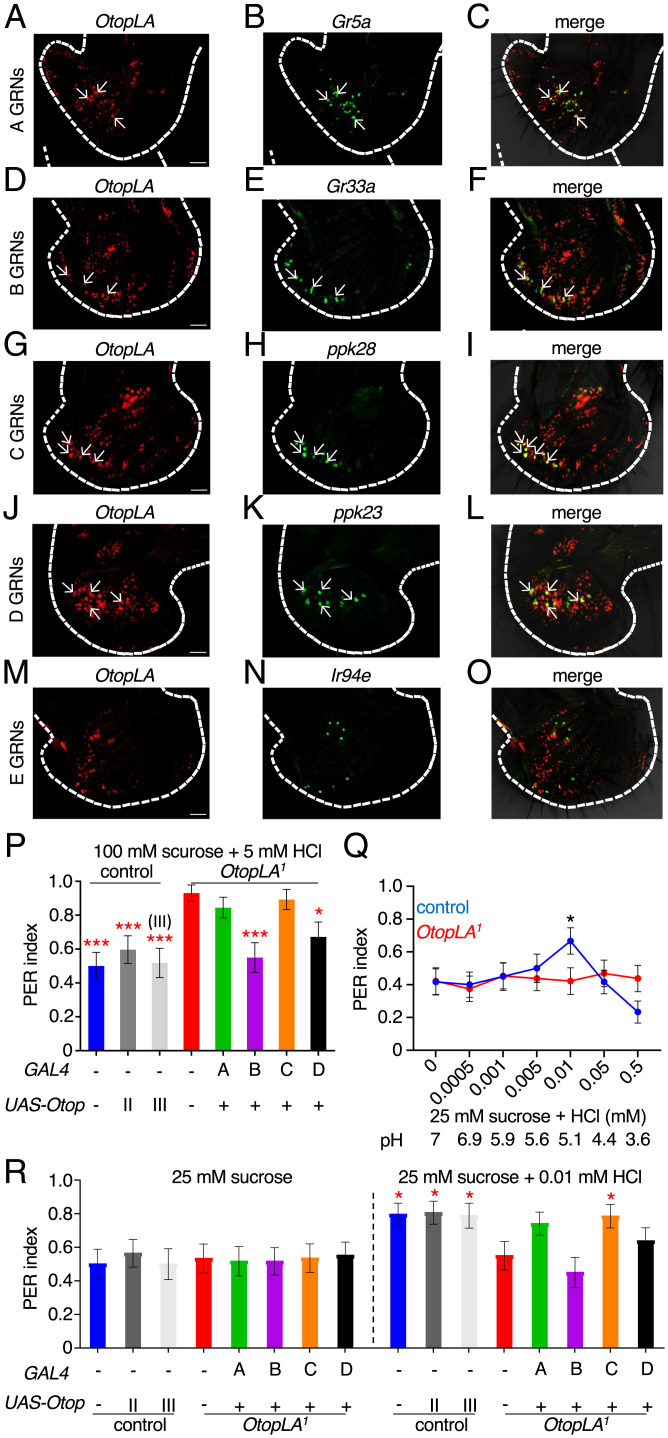
Because the reporter signals were weak, in order to more accurately determine which type of neurons express OtopLA, we also performed in situ hybridizations. When we hybridized the labellum with the OtopLA probe alone, we observed signals in control but not OtopLA1 labella, confirming the specificity of the probe (SI Appendix, Fig. S4 F and G). Note that the labeling of any given labellum was not complete due to incomplete penetration of the probe. Next, we performed double in situ hybridization experiments using markers for the five different GRN subsets (A to E; Table 1). ∼50% of the OtopLA-positive neurons were positive for markers for A to D GRNs, and none were positive for the marker for E GRNs (Fig. 4 A–O and Table 1). The remaining neurons may be peg GRNs. However, as described in the previous paragraph, peg GRNs do not contribute significantly to acid taste.
Table 1.
Overlap of OtopLA neurons with different GRN subtypes
| GRN | Former name | GRN maker | GRNs labeled by marker | GRNs labeled by OtopLA | OtopLA+ GRNs labeled by marker | % OtopLA+ GRNs labeled by marker |
| A | sweet | Gr5a | 24.0 ± 1.6 | 75.6 ± 3.3 | 5.6 ± 0.3 | 7.4 |
| B | bitter | Gr33a | 18.6 ± 1.3 | 83.0 ± 3.5 | 5.0 ± 0.5 | 6.0 |
| C | water | ppk28 | 16.0 ± 0.6 | 66.0 ± 3.0 | 11.6 ± 0.3 | 17.5 |
| D | cation | ppk23 | 16.6 ± 1.2 | 71.6 ± 4.3 | 14.2 ± 0.6 | 19.7 |
| E | low salt | Ir94a | 9.3 ± 0.6 | 70.0 ± 4.6 | 0.0 | 0 |
GRNs expressing OtopLA and markers for A to E classes of GRNs. These classes of GRNs were defined previously (4). Double in situ hybridizations were performed using RNAscope. Mean numbers of GRNs in proboscises (n ≥ 3) expressing OtopLA and the indicated A to E markers are shown. The average numbers of GRNs expressing OtopLA varied in different double-labeling experiments due to variations in probe penetration are shown.
Fig. 4.
OtopLA functions in multiple GRNs for detection of acids. (A–O) Images of in situ hybridizations of OtopLA and the indicated reporters in labella from w1118 flies. In each row, the Left panel indicates the distribution of OtopLA transcripts (red). The Middle panel shows the markers for A–E GRNs (green). The Right panel shows the merge of the OtopLA RNAs and marker RNAs. (P) PER assays to investigate the effects of cell-specific rescue of OtopLA function to acid taste in OtopLA1 flies. The assays were performed using 100 mM sucrose plus 5 mM HCl. Genotypes tested: control (w1118), control with UAS-OtopLAp (chromosome II or III insertion), OtopLA1, and OtopLAp expressed in an OtopLA1 background using the Gr5a-GAL4 (A neurons), Gr66a-GAL4 (B neurons), ppk28-GAL4 (C neurons), and the ppk23-GAL4 (D neurons). Control, n = 27. UAS-OtopLAp (chromosome II), n = 31. UAS-OtopLAp (chromosome III), n = 26. OtopLA1, n = 29. OtopLAp rescued in: A neurons, n = 29. B neurons, n = 30. C neurons, n = 28. D neurons, n = 29. (Q) PER assays using 25 mM sucrose and the indicated concentrations of HCl. The pH values of the solutions are indicated. Control (w1118), n = 30. OtopLA1, n = 33. (R) PER assays to investigate the effects of cell-specific rescue of OtopLA function in restoring attraction to 0.01 mM HCl. Genotypes tested: (w1118), control with UAS-OtopLAp (chromosome II or III insertion), OtopLA1, and OtopLAp expressed in OtopLA1 background using Gr5a-GAL4 (A neurons), Gr66a-GAL4 (B neurons), ppk28-GAL4 (C neurons), and ppk23-GAL4 (D neurons). PER indices were obtained by testing either with 25 mM sucrose alone or 25 mM sucrose mixed with 0.01 mM HCl. Asterisks indicate statistically significant differences from OtopLA1 flies tested with 25 mM sucrose laced with 0.01 mM HCl. Control, n = 27. UAS-OtopLAp (chromosome II), n = 31. UAS-OtopLAp (chromosome III), n = 26. OtopLA1, n = 29. OtopLAp rescued in: A neurons, n = 29. B neurons, n = 30. C neurons, n = 28. D neurons, n = 29 (Scale bars, 10 μm). Mann–Whitney U tests. Error bars, SEMs. *P < 0.05, **P < 0.01, and ***P < 0.001.
Since RNAi knockdown of OtopLA in B GRNs disrupted acid repulsion, we examined the overlap of OtopLA with B GRNs in more detail using the LexA introduced into OtopLA1 to drive expression of lexAop-GFP. When we labeled the B GRNs with UAS-td-tomato using the Gr66a-GAL4, we found that a subset of the green fluorescent protein (GFP)-positive GRNs (OtopLA) were also labeled by td-Tomato, although the percentage (∼12%) was higher than obtained using in situ hybridizations (SI Appendix, Fig. S4 H–J). Since ∼50% of the OtopLA-positive neurons were peg GRNs, ∼25% of the OtopLA-positive neurons in taste sensilla are labeled B GRNs. This raises the possibility that OtopLA might play a role in GRNs other than in the B GRNs.
To further investigate the cell-type requirement for OtopLA for acid repulsion, we performed cell type–specific rescue experiments with UAS-OtopLAp transgenic flies. Since OtopLA is expressed in A, B, C, and D GRNs, we tested the effects of introducing OtopLAp in each of these four classes of GRNs. We found that the impairment in acid repulsion exhibited by OtpLA1 flies (due to adding 5 mM HCl to 100 mM sucrose) was rescued by expression of OtopLAp in B neurons (Fig. 4P). Introduction of OtoplAp in D GRNs also increased the PER in response to the addition of 5 mM HCl but did not fully rescue the phenotype (Fig. 4P). In contrast, expression of OtopLAp in either A or C GRNs did not restore aversion to the 5 mM HCl (Fig. 4P). These data indicate that OtopLAp functions in both B and D GRNs for acid repulsion.
In addition to B and D neurons, OtopLA is also expressed in A and C GRNs, which function in sensing appetitive tastants (4). Therefore, we investigated the possibility that OtopLA also contributes to the attraction to low level of protons. However, we did not observe significant attraction to most low concentrations of HCl (Fig. 4Q), except for a modest increase in the PER to 0.01 mM HCl (pH 5.1; Fig. 4Q). This was consistent with the results obtained from the binary choice assays (SI Appendix, Fig. 2D). The attraction to HCl was eliminated in the OtopLA1 mutant (Fig. 4Q). Expression of OtopLAp in C GRNs restored the attraction to 0.01 mM HCl (Fig. 4R). Introduction of OtopLA function in A GRNs also caused an increase in the PER, although it fell below the threshold for significance (P values: 0.07 versus 25 mM sucrose alone; 0.09, versus OtopLA1 with 25 mM sucrose and 0.01 mM HCl; Mann–Whitney U test, Fig. 4R). Expression of OtopLAp in B and D neurons had no significant effect on the PER with 0.01 mM HCl (Fig. 4R).
Discussion
The functional conservation of the Otop channels for acid taste in flies is striking given that chemosensory receptors tend to vary greatly in flies and mammals (3), which diverged ∼800 million y ago. In contrast to the Otop channels, the two major families of fly receptors (GRs and IRs), which function in tasting sugars, bitter compounds, acetic acid, amino acids, polyamines, N, N-diethyl-meta-toluamide (DEET), CO2, and other tastants are not present in mammals (4, 29). The retention of Otop channels for acid taste in flies and mice is remarkable since the gross anatomies of the gustatory systems are very different (3). In addition, the taste receptor cells in flies are neurons, while they are modified epithelial cells in mammals (3).
The conserved role for Otop proteins for acid taste in flies and mammals (5–7) cannot be explained by greater selective pressure for maintaining a receptor for a mineral (e.g., H+) versus organic molecules since other minerals (Ca2+ and Na+) are sensed in flies through IRs, which are not present in mammals (30–32). Thus, the retention of Otop channels for acid taste in flies and mammals underscores the very strong selection for this acid sensor for animal survival. Otop-related proteins are encoded in many distantly related terrestrial and aquatic vertebrates ranging from the platypus to frogs and pufferfish, as well as ancient invertebrates such as worms and insect disease vectors, including Aedes aegypti (9, 12). Thus, despite the considerable diversity of most chemosensory receptors, it is plausible that Otop channels endow a large proportion of the animal kingdom with acid taste.
A question concerns the cellular mechanism through which the sensation of protons is detected. OtopLA is expressed in the four classes of GRNs in taste hairs (A to D). The B and D GRNs respond to aversive tastants (B, bitter, high Na+ etc; D, Ca2+, high Na+, K+), while the A and C GRNs are activated by chemicals that stimulate consumption (A, sugars, low Na+ fatty acids, etc; C, water). Our data indicate that both B and D GRNs contribute to acid repulsion but that B GRNs comprise the major class required for acid repulsion, while D GRNs are the minor class. In support of this conclusion, RNAi knockdown of B but not D GRNs impaired acid repulsion. In addition, we fully rescued the OtopLA1 mutant phenotype by expression of the OtopLAp transgene in B GRNs but only partially rescued the deficit by expression of OtopLAp in D GRNs. In addition, our data indicate that both A and C GRNs contribute to the modest attraction to 0.01 HCl in wild-type flies. This attraction is eliminated in the OtopLA1 mutant. The C GRNs may be more important, as expression of the OtopLAp transgene in A GRNs reduced the impairment in the mutant, but the suppression of the phenotype fell below the threshold for statistical significance.
It has been reported that acids cause repulsion of sugary foods by direct activation of B GRNs and suppression of sugar-induced activation of A GRNs (16). This previous study focused on behavioral responses to carboxylic acids, and we repeated this finding for citric acid. However, at the cellular level, when we decreased the pH of sucrose, we did not observe reduced sucrose-induced action potentials. Thus, we conclude that protons do not suppress A GRNs. Rather, we suggest that A GRNs are inhibited by certain organic anion moieties of carboxylic acids. A mechanism by which the activities of both A GRNs and B GRNs are affected by carboxylic acids, but only B GRNs by protons could provide a coding mechanism for differentiating between protons and carboxylic acids.
Following submission of the initial version of this manuscript, another group also reported a role for OtopLA in acid taste in Drosophila (33). These researchers found that OtopLA is required for attractive responses to low concentrations of acids, as did we, but not for aversive responses to higher concentrations of acids. However, even in wild-type controls, they did not observe significant repulsion until the pH was reduced to high levels (≤2) that may be damaging to cells. At these very low pHs, the nociceptive response is likely to have a major contribution to avoidance. Mi et al. also reported that the flies exhibited a much higher level of attraction to acids than what we have observed (33). The differences in levels of attraction and repulsion might be due to variations in fly food between laboratories, the precise ages of the flies, hours of starvation, or a combination of these factors. Nevertheless, although the level of acid attraction differs, both studies find that the deficit in attraction in OtopLA mutants can be suppressed by expression of a wild-type transgene in A GRNs. In addition, we found that this phenotype is suppressed by expression of the OtopLA rescue transgene in C GRNs.
Another difference between our report and that of Mi et al. (33) is that they reported that expression of OtopLAa in human embryonic kidney 293 (HEK293) cells led to the appearance of inward currents in response to acid stimuli (pH range 6 to 3). We previously demonstrated that both vertebrate and invertebrate Otop proteins form proton channels (5). However, we did not observe any acid-induced currents using stimuli as low as pH 3.0 for either OtopLAp or OtopLAa (FBgn0259994) expressed in either HEK293 cells or Xenopus oocytes, even though we detected surface expression when the channels were tagged with GFP. There are several possible reasons why the Drosophila OtopLA channel did not generate functional currents in either cell type. One possibility is that the native system provides factors or binding partners necessary to gate the channels. We note that OtopLA is the only one of the Drosophila Otop channels to have a large extracellular domain between transmembrane domains 5 and 6, which might bind ligands or proteins.
Together, our data point to a complex role of the OtopLA channel in the gustatory system of Drosophila, where it is expressed in multiple types of sensory cells and can mediate both attractive and aversive responses. Interestingly, humans also find acids appetitive at low concentrations and aversive at higher concentrations. The elucidation of the cellular and molecular mechanism of acid-sensing that we describe here can serve as the basis for further understanding as to how animals assign valence to stimuli that vary only in intensity.
Materials and Methods
Drosophila Stocks.
Flies were reared at 22 to 25 °C in standard media. Control flies were w1118 unless otherwise mentioned. The following lines were obtained from the Drosophila Bloomington Stock Center: w1118 (BL 5905), nompC-GAL4 (BL 50131), Ir94e-GAL4 (BL 60725), elav-GAL4;UAS-Dcr2 (BL 25750), Gr5a-GAL4 (BL 57592), lexAop-GFP (BL 32207), lexAop-mCherry (BL52271),UAS-tdTomato (BL 36328), poxn70 (BL 60688), UAS-kir2.1 (BL 6596), and 57F03-GAL4 (BL 46386). The following lines were obtained from the Vienna Drosophila RNAi Center: OtopLA-RNAi1 (v104973), OtopLA-RNAi2 (v100847), OtopLB-RNAi1 (v101936), OtopLB-RNAi2 (v3452), OtopLC-RNAi1 (v47248), and OtopLC-RNAi2 (v19613). The following lines were obtained from other investigators: ppk23-Gal4 (34), ppk28-GAL4 (35), and Gr66a-GAL4 (36).
Chemicals.
Sucrose (84097), denatonium (D5765), tartaric acid (251380), glycolic acid (124737), propionic acid (81910), acetic acid (A6283), and citric acid (C2404) were obtained from Sigma. NaCl (S271-3) and HCl (A144-212) were obtained from Fisher Scientific. For PER assays, water was used as the solvent. Tastants were dissolved in 30 mM TCC (Sigma, T0252) for the extracellular tip recordings.
Generation of OtopLA1 and OtopLA2 Mutant Flies.
To create the OtopLA1 and OtopLA2 mutants, we used CRISPR/Cas9. To generate OtopLA1, we replaced 40 base pairs at the beginning of the coding sequence of OtopLA with the LexA and miniwhite (w+) genes (SI Appendix, Fig. S2A). The deletion removed the first 14 amino acids and also changed the reading frame. The guide RNAs for creating this line were: guide RNA1: gcgaggggaaaggaatgcagcgg and guide RNA2: tgagatgcgcgaaagattactgg. The PCR primers used to generate the 5′ and 3′ homology arms were: forward primer for 5′ arm: gacgcataccaaacggtaccaatgctcccgatttgctggctagc, reverse primer for 5′ arm: ttttgattgctagcggtacctcctttcccctcgctcagc, forward primer for 3′ arm: ctaggcgcgcccatatgtactggaccagccgcggg, and reverse primer for 3′ arm: gacaagccgaacatatgggtgggcaggactcgtg
To generate OtopLA2, we replaced the first 52 base pairs at the beginning of the coding sequence of OtopLAa with the LexA and miniwhite (w+) genes (SI Appendix, Fig. S2A ). The deletion removed the first 18 amino acids and changed the reading frame. The guide RNAs for creating this line were: guide RNA1: acggaaacggaaacaatgggcgg and guide RNA2: cgtcgagggcggggacaatatgg. The PCR primers used to generate the 5′ and 3′ homology arms were: forward primer for 5′ arm: gacgcataccaaacggtacccagggcccgtttcagtt, reverse primer for 5′ arm: ttttgattgctagcggtacctgtttccgtttccgtttcggtct, forward primer for 3′ arm: ctaggcgcgcccatatgatatggccaccttgccgg, and reverse primer for 3′ arm: gacaagccgaacatatgaaccccagacagtgtgcag
The plasmids to create OtopLA1 and OtopLA2 were injected by Bestgene into the vas-cas9 background (BL 51324). After confirming the mutations by PCR (forward primer: agtcgttgccaggatagacg, reverse primer: catcgtggggaactcatcat) and DNA sequencing, we outcrossed both mutant lines to the w1118 background for six generations.
Generation of lexAop-OtopLAp Transgenic Flies.
We dissected 50 w1118 proboscises under liquid N2, and extracted RNA using the Qiagen RNeasy mini kit (74104). cDNAs were synthesized from freshly extracted mRNA using SuperScript IV Mastermix with the ezDNase enzyme (Invitrogen 11766050). The entire coding sequence of the OtopLAp isoform was PCR amplified (34 cycles at an annealing temperature 56 °C) from freshly synthesized cDNAs using the following primers: forward primer: gcggccgcggctcgagaaaggaatgcagcggtgt, and reverse primer: acaaagatcctctagattactccagacgt. The OtopLAp cDNA was cloned between the XhoI and XbaI sites of the lexAop vector (Addgene 26224) (37). The plasmid was injected by Bestgene into the attP2 background (BL 8622), which has an attP docking site (68A4) on the third chromosome to create lexAop-OtopLA transgenic flies. The transgenic line was verified by PCR.
Generation of UAS-OtopLAp Transgenic Flies.
The OtopLAp isoform was amplified from lexAop-OtopLAp plasmid using the following primers: forward primer: agggaattgggaattcaaaggaatgcagcggtgtcc and reverse primer: acaaagatcctctagattactccagacgtgccttgtaggt. The OtopLAp cDNA was cloned between the XbaI and EcoRI sites of the pUASTattB vector. The plasmid was injected by Bestgene into two different attP2 backgrounds (BL 8622), which have attP docking sites on the second and third (site 68A4) chromosomes, respectively, to create the UAS-OtopLA transgenic flies. The transgenic lines were verified by PCR.
PER Assays.
To perform PER assays (38), we collected flies that were 0 to 2 d old and aged them in vials containing 10 males and 10 females until they were 5 to 7 d old. The flies were starved on water-soaked Kimwipes for 24 to 26 h prior to the experiments. The flies were fitted within P-200 tips truncated in a manner to allow only the head to protrude outside. The other end of the tip was sealed with clay. Paper wicks were used to present the taste stimuli to the flies. We made contact for ∼2 seconds and scored the fly’s response over the following 7 s. The flies were satiated with water at the beginning of the experiment and between consecutive stimuli. In addition, the flies were tested with 100 mM sucrose both at the beginning and the end of the experiments. Only those flies that gave a positive response in both cases were considered. After every positive response, flies were tested with water to determine whether the PER was due solely to the tastants. Flies that continued to drink water for ≥1 min were discarded. Complete proboscis extensions were scored as 1, while partial extensions were scored as 0.5. A score of 0 indicated no extension. The mean score obtained from all individual flies tested was calculated as the PER index. PER experiments were done blinded except in cases where there were differences in eye colors between the lines that precluded us from doing so.
Binary Choice Assay.
The binary food choice assay was performed as described (30), with slight modifications. Briefly, 40 to 60 flies (5 to 7 d old) were starved for 24 to 26 h in vials containing two Kimwipes soaked in 6 mL water. The assays were conducted in 72-well microwell mini trays (Thermo Fisher Scientific, 15461158) with the two food alternatives in alternate wells. Both foods contained 0.75% agarose, either red (sulforhodamine B, 0.1 mg/mL; Sigma S9012) or blue (Brilliant Blue FCF, 0.1875 mg/mL; Wako, 027-12842) food dyes and either 2 mM sucrose alone or 2 mM sucrose mixed with a range of HCl concentrations. The foods were loaded into the wells using a repeating pipettor. The flies were anesthetized using CO2 and introduced into the plates. The assays were conducted in a dark humid chamber at 25 °C for 90 min. Subsequently, the plates were transferred to −20 °C for ≥1 h to kill the flies. The flies were then viewed under a stereomicroscope to determine their food preferences based on the color of their abdomens: red, blue, or purple. Flies that ate both foods would have purple abdomens. We observed robust feeding with 80 to 100% of the flies having colored abdomen across all experiments. Their PIs were calculated using the following formula:
Extracellular Tip Recordings.
Extracellular tip recordings were conducted as previously described (39) using 7- to 10-d-old flies. The tastants were dissolved in either 30 mM TCC or 1 mM KCl (Fisher Scientific, P217-500), which served as the electrolyte. The taste solutions were back-filled into recording electrodes (World Precision Instruments, 1B150F-3). Action potentials induced by the tastants were amplified and digitalized using IDAC-4 data acquisition software. Autospike software (Syntech) was used to visualize and manually count the spikes. Neuronal responses were quantified by counting the number of spikes in the first 500 ms following contact with the stimulus.
Immunostaining.
Proboscises were dissected in 1% phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 30 min. They were then washed with PBST (PBS+0.3% Triton X-100) for 1 h (three washes of 20 min each), and blocked using normal goat serum for 1 h. The whole-mount tissues were stained in primary antibodies in blocking buffer for 3 to 4 d, washed with PBST for 1 h and subsequently stained using secondary antibodies in blocking buffer for 2 to 3 d. The tissues were mounted on glass slides with Vectashield mounting media, and images were acquired using a Zeiss LSM 700 confocal microscope. Primary antibodies: anti-GFP (chicken 1:20; ThermoFisher Scientific, A10262) and anti-DsRed (rabbit, 1:50, Takara Bio#632496). Secondary antibodies: Alexa Fluor 488 conjugated goat anti-chicken (Thermo Fisher Scientific, A-11039) and Alexa Fluor 568 conjugated goat anti-rabbit (Thermo Fisher Scientific, A-11036).
mRNA Hybridizations.
To localize mRNAs in proboscis whole-mount tissue, we adapted RNAscope using primers to OtopLA, Gr5a, Gr33a, ppk28, ppk23, and Ir94e, which were designed by Advanced Cell Diagnostics (ACD). A total of ∼10 proboscises were dissected in PBS, placed in 1.5-mL Eppendorf tubes, washed once with PBS, and fixed in 4% paraformaldehyde in 1 mL PBS for 16 h at 4 °C. The tissue was then immersed in a series of 10, 20, and 30% sucrose, each time allowing the tissue to sink the bottom of the tube. Then, the tissue was washed in PBS, refixed in 4% paraformaldehyde in PBS at room temperature for 10 min and washed for 3 × 5 min in PBS. The samples were then dehydrated in a series of 50, 75, and 100% ethanol in PBS. The ethanol was removed completely, and the tissue was air-dried at room temperature for 30 min. The tissue was treated with 3% hydrogen peroxide in PBS for 10 min to inactivate endogenous peroxidase activity and incubated in 50 μL RNAscope Protease III for 30 min. Hybridizations with 100 μL each of the OtopLA and marker probes were performed overnight at 40 °C in ACD HybEZ Hybridization System (110VAC) (ACD cat. No. 321461). The tissue was washed in an RNAscope wash buffer (ACD cat. no. 310091) for 3 × 2 min. The tissue was then incubated in a series of amplifier solutions (Amp’s) provided in the RNAscope Multiplex fluorescent V2 assay kit (ACD cat. no. 323100) according to the manufacturer’s instructions. The tissue was incubated in 100 μL Amp1 for 2 h at 40 °C, in Amp2 for 2 h at 40 °C, Amp3 for 1 h at 40 °C, and C1 for 2 h at 40 °C. Between each step the tissue was washed for 5 × 3 min with wash buffer at room temperature. For fluorescent labeling, a working Opal dye solution was made fresh using a 1:500 ratio of Opal dye (Akoya Biosciences) to TSA buffer. 150 μL of working solution was added to each tube containing 10 proboscises and incubated at 40 °C for 2 h, washed one in a wash buffer and mounted in Vectashield.
Quantification and Statistical Analysis.
Descriptions, results, and sample sizes of each test are provided in the figure legends. All replicates were biological replicates using different flies. Data for all quantitative experiments were collected on at least three different days. For the PER behavioral experiments each “n” represents an individual fly. Based on our experience and common practices in this field, we used a sample size of n ≥ 25 trials for each genotype or treatment for the PER assays. Each “n” for the tip recording experiments represents an analysis of a single, independent fly (n > 6). GraphPad Prism 9 software or Microsoft Excel was used for statistical tests. We used Student’s unpaired t tests for parametric tests (in Microsoft Excel) and Mann–Whitney U test for nonparametric tests. Sample sizes were determined based on previous publications and are cited in the figure legends. In all graphs, error bars indicate the SEM. We set the significance level, a = 0.05. Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Deafness and other Communication Disorders (NIDCD) Grant R01-DC007864 to C.M. and NIDCD Grant R01-DC013741 and National Institute of General Medical Sciences Grant R01-GM131234 to E.R.L. We thank Bochuan Teng and Ziyu Liang for contributions to patch-clamp recordings of HEK293 cells and Kevin Chyung Jackson Walker and Josh Kaplan for expert technical support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110641118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Beauchamp G. K., Maller O., Rodgers J. G., Flavor preferences in cats (Felis catus and Panthera sp.). J. Comp. Physiol. Psychol. 91, 1118–1127 (1977). [Google Scholar]
- 2.Feng P., Zheng J., Rossiter S. J., Wang D., Zhao H., Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liman E. R., Zhang Y. V., Montell C., Peripheral coding of taste. Neuron 81, 984–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montell C., Drosophila sensory receptors-a set of molecular Swiss Army Knives. Genetics 217, 1–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu Y. H., et al. , An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B.. Teng, et al. , Cellular and Neural Responses to Sour Stimuli Require the Proton Channel Otop1. Curr. Biol. 29, 3647–3656.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. Zhang, et al. , Sour Sensing from the Tongue to the Brain. Cell 179, 392–402.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Hughes I., et al. , Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev. Biol. 276, 391–402 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurle B., et al. , Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum. Mol. Genet. 12, 777–789 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Söllner C., Schwarz H., Geisler R., Nicolson T., Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev. Genes Evol. 214, 582–590 (2004). [DOI] [PubMed] [Google Scholar]
- 11.de Caprona M. D., Beisel K. W., Nichols D. H., Fritzsch B., Partial behavioral compensation is revealed in balance tasked mutant mice lacking otoconia. Brain Res. Bull. 64, 289–301 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Hughes I., et al. , NISC Comparative Sequencing Program, Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members. BMC Evol. Biol. 8, 41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang W. W., et al. , An otopetrin family proton channel promotes cellular acid efflux critical for biomineralization in a marine calcifier. Proc. Natl. Acad. Sci. U.S.A. 118, 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q., Zeng W., She J., Bai X. C., Jiang Y., Structural and functional characterization of an otopetrin family proton channel. eLife 8, e46710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saotome K., et al. , Structures of the otopetrin proton channels Otop1 and Otop3. Nat. Struct. Mol. Biol. 26, 518–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlu S., Wisotsky Z., Medina A., Dahanukar A., Acid sensing by sweet and bitter taste neurons in Drosophila melanogaster. Nat. Commun. 4, 2042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimal S., et al. , Mechanism of acetic acid gustatory repulsion in Drosophila. Cell Rep. 26, 1432–1442.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Amrein H., Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr. Biol. 27, 2741–2750.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanley M., Ghosh B., Weiss Z. F., Christiaanse J., Gordon M. D., Mechanisms of lactic acid gustatory attraction in Drosophila. Curr. Biol. 31, 3525–3537.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Lee C. H., Chen C. C., Roles of ASICs in nociception and proprioception. Adv. Exp. Med. Biol. 1099, 37–47 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Devineni A. V., Sun B., Zhukovskaya A., Axel R., Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. eLife 8, e47677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss L. A., Dahanukar A., Kwon J. Y., Banerjee D., Carlson J. R., The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delventhal R., Kiely A., Carlson J. R., Electrophysiological recording from Drosophila labellar taste sensilla. J. Vis. Exp. 84, e51355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk R., Bleiser-Avivi N., Atidia J., Labellar taste organs of Drosophila melanogaster. J. Morphol. 150, 327–341 (1976). [DOI] [PubMed] [Google Scholar]
- 25.LeDue E. E., Chen Y. C., Jung A. Y., Dahanukar A., Gordon M. D., Pharyngeal sense organs drive robust sugar consumption in Drosophila. Nat. Commun. 6, 6667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y. D., Dahanukar A., Molecular and cellular organization of taste neurons in adult Drosophila pharynx. Cell Rep. 21, 2978–2991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y. D., Park S. J., Ja W. W., Dahanukar A., Using Pox-Neuro (Poxn) mutants in Drosophila gustation research: A double-edged sword. Front. Cell. Neurosci. 12, 382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steck K., et al. , Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. eLife 7, e31625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson H. M., Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64, 227–242 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Lee Y., Poudel S., Kim Y., Thakur D., Montell C., Calcium taste avoidance in Drosophila. Neuron 97, 67–74.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y. V., Ni J., Montell C., The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaeger A. H., et al. , A complex peripheral code for salt taste in Drosophila. eLife 7, e37167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi T., Mack J. O., Lee C. M., Zhang Y. V., Molecular and cellular basis of acid taste sensation in Drosophila. Nat. Commun. 12, 3730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toda H., Zhao X., Dickson B. J., The Drosophila female aphrodisiac pheromone activates ppk23(+) sensory neurons to elicit male courtship behavior. Cell Rep. 1, 599–607 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Cameron P., Hiroi M., Ngai J., Scott K., The molecular basis for water taste in Drosophila. Nature 465, 91–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunipace L., Meister S., McNealy C., Amrein H., Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11, 822–835 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Pfeiffer B. D., et al. , Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiraiwa T., Carlson J. R., Proboscis extension response (PER) assay in Drosophila. J. Vis. Exp. 193, 193 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon S. J., Köttgen M., Jiao Y., Xu H., Montell C., A taste receptor required for the caffeine response in vivo. Curr. Biol. 16, 1812–1817 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.