Significance
Molecular imaging at the single-molecule level of large and flexible proteins such as monoclonal IgG antibodies is possible by low-energy electron holography after chemically selective sample preparation by native electrospray ion beam deposition (ES-IBD) from native solution conditions. The single-molecule nature of the measurement allows the mapping of the structural variability of the molecules that originates from their intrinsic flexibility and from different adsorption geometries. Additionally, we can distinguish gas-phase–related conformations and conformations induced by the landing of the molecules on the surface. Our results underpin the relation between the gas-phase structure of protein ions created by native electrospray ionization (ESI) and the native protein structure and are of relevance for structural biology applications in the gas phase.
Keywords: low-energy electron holography, single-molecule imaging, native electrospray ion beam deposition
Abstract
Imaging of proteins at the single-molecule level can reveal conformational variability, which is essential for the understanding of biomolecules. To this end, a biologically relevant state of the sample must be retained during both sample preparation and imaging. Native electrospray ionization (ESI) can transfer even the largest protein complexes into the gas phase while preserving their stoichiometry and overall shape. High-resolution imaging of protein structures following native ESI is thus of fundamental interest for establishing the relation between gas phase and solution structure. Taking advantage of low-energy electron holography’s (LEEH) unique capability of imaging individual proteins with subnanometer resolution, we investigate the conformational flexibility of Herceptin, a monoclonal IgG antibody, deposited by native electrospray mass-selected ion beam deposition (ES-IBD) on graphene. Images reconstructed from holograms reveal a large variety of conformers. Some of these conformations can be mapped to the crystallographic structure of IgG, while others suggest that a compact, gas-phase–related conformation, adopted by the molecules during ES-IBD, is retained. We can steer the ratio of those two types of conformations by changing the landing energy of the protein on the single-layer graphene surface. Overall, we show that LEEH can elucidate the conformational heterogeneity of inherently flexible proteins, exemplified here by IgG antibodies, and thereby distinguish gas-phase collapse from rearrangement on surfaces.
Proteins are dynamic objects whose biological function is often tied to structural changes (1–5). Mapping this conformational variability and associated dynamics is one of the major challenges in protein structure determination (6, 7). To this end, high-resolution imaging capable of resolving submolecular detail plays a central role because protein interactions occurring on the submolecular or atomic level are linked to conformational or stoichiometric changes in higher levels of the structural hierarchy; i.e., structural motifs, domains, or entire subunits are influenced (1, 8).
X-ray crystallography, cryogenic electron microscopy, and NMR are the leading techniques for atomically resolved structure determination from protein ensembles (9–13), which, in the case of NMR, includes information about protein dynamics on various timescales (14, 15).
For imaging conformational variability of inherently flexible proteins such as antibodies, single-molecule techniques (16, 17) have the advantage of being able to access the full conformational space given by the flexibility of the protein. Specifically, in the case of antibodies, understanding this flexibility can help in the development of antibody-based therapeutics (18–20). Recent developments toward the imaging of single macromolecules include coherent diffraction with free electron lasers (XFEL) (21–23), scanning probe microscopy (24–26), tomographic methods in electron microscopy (16, 17), and low-energy electron holography (LEEH), the method used here. LEEH (27–29) operates at electron energies of 50 to 150 eV, which results in negligible radiation damage and high contrast. Recently, LEEH has been shown to be capable of imaging nanoscale objects (30) and individual proteins at a spatial resolution in the range of 1 nm (31). To apply an imaging method such as LEEH to a biological question, the sample preparation procedure and imaging method have to maintain a biologically relevant state of the specimen. Simultaneously, LEEH requires ultrapure substrate conditions, which suggests a vacuum-based deposition method. Thus, the ideal sample preparation method for the imaging of proteins with LEEH is native electrospray ion beam deposition (ES-IBD) (31, 32) (see Materials and Methods for details). Native electrospray ionization (ESI) provides protein ions of intact stoichiometry and three-dimensional shape, which has been confirmed by structure-sensitive gas-phase measurements, for instance by ion mobility spectrometry (33–35). Subsequent deposition of these molecular gas-phase ions on surfaces in ultrahigh vacuum (UHV) ( mbar) yields an ultrapure sample, ideal for LEEH microscopy.
Until now, only systems without a high degree of flexibility, i.e., small, compact proteins ([31]) and suspended macromolecules (30, 36, 37), have been successfully imaged by LEEH. Sample preparation based on native ESI, however, is able to deliver mass-selected native protein structures as large as an entire virus (38) or as intricate as a membrane protein receptor complex embedded in membrane mimetics (39, 40) into the gas phase and onto the surface. Deposition under vacuum conditions enables the imaging of large and flexible proteins and protein complexes with LEEH.
Native ES-IBD, however, involves several processes that can potentially alter the protein’s conformation (41): a liquid–gas-phase transfer, the electrospray ionization process, the transfer of the molecular ions into vacuum, and a collision with freestanding graphene upon deposition. Thus, we are presented with two questions: 1) whether LEEH, at the present subnanometer resolution, can elucidate the different conformations assumed by large and flexible proteins following deposition onto freestanding graphene and 2) to what extent the observed protein conformations are influenced by the ES-IBD process, specifically by native ESI, the gas-phase flight, and the subsequent landing on the graphene surface.
Here, we demonstrate the imaging of individual Herceptin molecules, a monoclonal IgG antibody, deposited by native ES-IBD on ultraclean freestanding single-layer graphene (SLG). We use a newly constructed LEEH instrument, in combination with our mass-selective ES-IBD method (32), to record and analyze a large number of holograms. LEEH imaging of the deposited proteins reveals a multitude of structures, which can be interpreted as intact antibodies in different conformations. We find that the understanding of both the gas-phase conformation and the landing process are crucial for interpreting the protein conformations observed on the graphene surface. The majority of the molecules appear in a compact conformation, which is presumably due to gas-phase folding about the hinge region in the flexible antibody structure. Other conformations, tied to energy transfer upon landing on the surface, feature more extended hinge regions and can be mapped to the known crystallographic structure of IgG antibodies.
Results and Discussion
IgG antibodies are glycoproteins consisting of four peptide chains (two heavy chains and two light chains) that interconnect to form three subunits, each of which is composed of two peptide chains that form a loop around a cavity. The subunits are arranged in a characteristic Y shape constituted by two antigen-binding Fab subunits and the Fc subunit, which mediates the biological function. The hinge region connecting the subunits exhibits a high degree of flexibility, which allows each of the subunits to move and reorient independently. This flexibility is crucial for antibodies to fulfill their biological role (16, 42) and is reflected by a remarkable variety of possible antibody conformations. Because of this flexibility, the imaging of antibodies poses a substantial challenge for techniques relying on averaging over many molecules (7, 17). Thus, antibodies are ideal candidates for exploring protein flexibility on the level of individual molecules with LEEH.
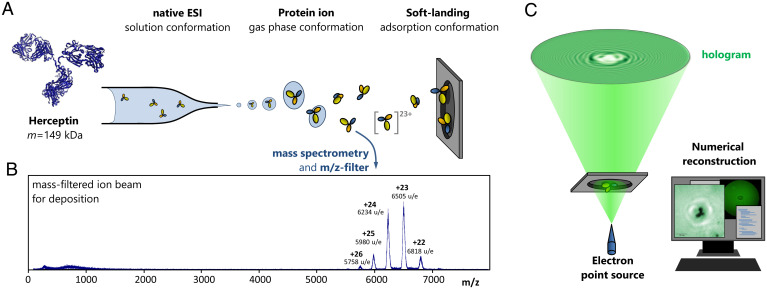
Herceptin molecules (molar mass 149 kDa) are deposited on freestanding single-layer graphene following native ES-IBD and mass-to-charge selection in a time-of-flight mass spectrometer (43) (Fig. 1 A and B). Native ES-IBD inherits accurate control over the chemical identity and protein fold from the use of native ESI (39, 40, 44, 45) and combines it with controlled, gentle deposition of the mass-selected species in UHV (31). Soft landing at low kinetic energies ensures that the molecules remain chemically intact upon impact on the substrate (25, 26, 31, 43, 46–49). Mass spectra recorded before deposition (Fig. 1B) show that the molecules are chemically intact and retain a tertiary structure close to the native conformation (41, 44, 50). We refer to the antibody gas-phase ions as native-like to indicate that changes in environment during the sample preparation process might have induced deviations from the native structure of the proteins in solution, such as side-chain collapse (51) or subunit rearrangement (41).
Fig. 1.
Deposition and imaging workflow for native ES-IBD/LEEH. (A) Scheme of the native ES-IBD process, proceeding from solution through the gas phase to soft-landing deposition. Before deposition, the beam is mass selected to ensure that only the relevant species land on the surface. (B) Mass spectrum of the Herceptin deposition beam after mass selection. The low charge states indicate that the proteins are in native-like conformations. (C) Scheme of the LEEH measuring and reconstruction process.
After deposition, the proteins are investigated by LEEH (Fig. 1C). Images of the proteins are obtained from holograms by numerical reconstruction. We employ a one-step propagation-based reconstruction algorithm (52), which results in a two-dimensional (2D) amplitude image that maps the absorption of the imaged protein (see Materials and Methods for details).
LEEH is a single-molecule imaging method, and hence we can access the entirety of different conformations the antibodies assume on the SLG surface. Some of the conformations show the characteristic Y shape (53, 54) with three distinct subunits known from the crystallographic models (Fig. 2A and see Fig. A–C). Additionally, structures with two distinguishable subunits (see Fig. 4 E–G) and compact structures without substructure (see Fig. 5 C and D) are observed. The correct interpretation of the molecules that do not appear Y-shaped requires an in-depth consideration of the processes occurring during native ES-IBD. These processes include conformational changes occurring in the gas phase, deposition on the surface, and adsorption in different molecular geometries. Before we discuss this in detail, we focus on the subset of antibody molecules presenting Y-shaped structures.
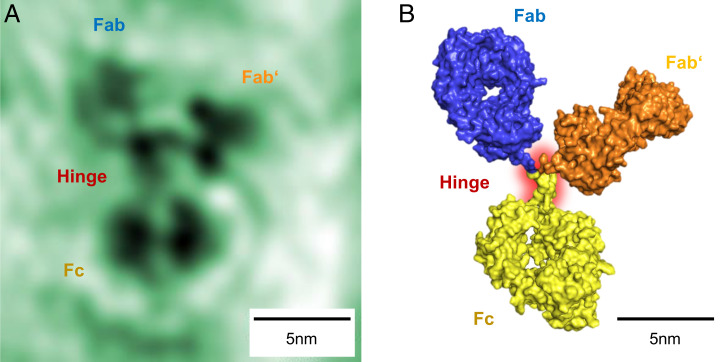
Fig. 2.
Y-shaped conformation. (A) Amplitude reconstruction of an individual antibody molecule. All three subunits and the hinge region are clearly distinguishable. In the hinge region, two strands stem from the bottom subunit identifying this subunit as the Fc subunit. The two Fab subunits (top) are captured in different orientations; on the top left subunit, the cavity in the center of the subunit appears as a bright spot. (B) Model of the antibody conformation observed in A, obtained from the 1IGT crystallographic structure by bond rotation in the hinge region. The sizes of the individual subunits are in close agreement; the slight deviations are likely due to additional degrees of freedom in the hinge region that have not been included in the model.
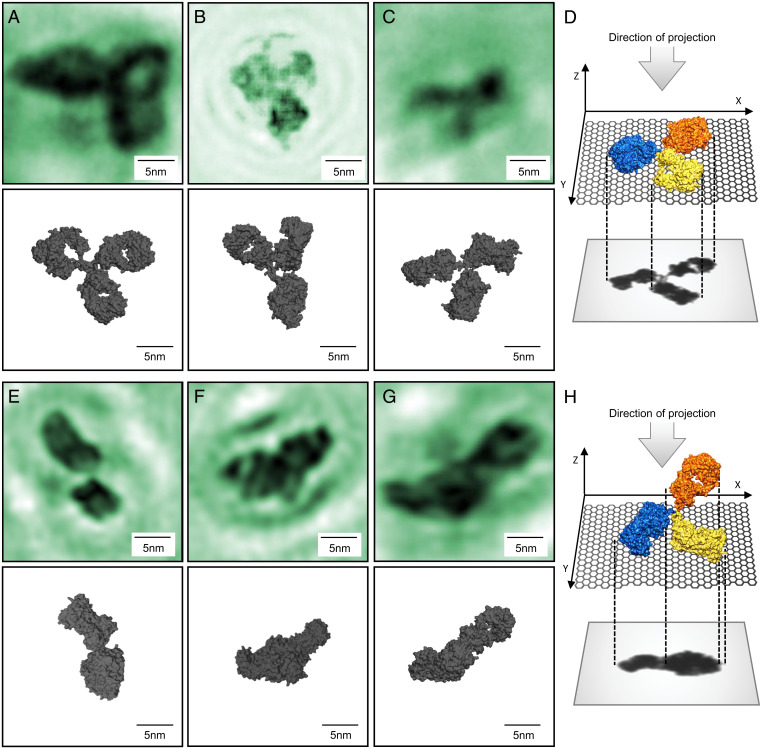
Fig. 4.
Diversity in antibody adsorption geometry. (A–C) Amplitude reconstructions of individual antibody molecules in a flat adsorption geometry, depicted schematically in D, and corresponding conformations obtained from the PDB model. For all three molecules, the three subunits are clearly distinguishable, and the visibility of the hinge region and the cavities of the subunits varies. (A) Hinge region not visible, cavities can be seen on all three subunits. (B) Hinge region partially visible, cavity visible on one subunit (top left). (C) Hinge region partially visible, no cavities can be seen. (E–G) Amplitude reconstructions of individual antibody molecules in a vertical adsorption geometry schematically depicted in H. All three molecules have two clearly distinguishable subunits with the third one partially visible in F. The fact that one of the subunits appears larger in both E and G could indicate a contribution of the third subunit located above the other subunits as sketched in H. All six images correspond to a field of view of .
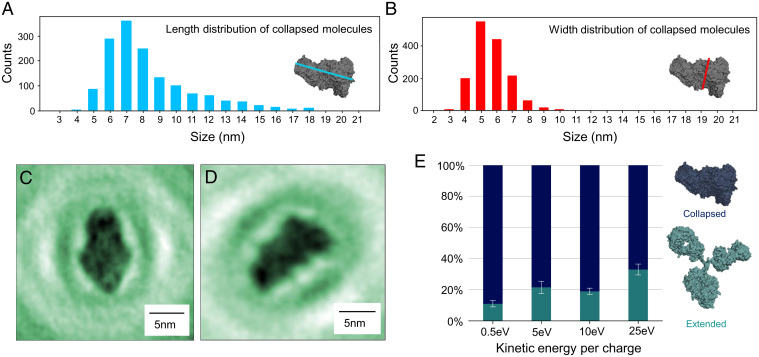
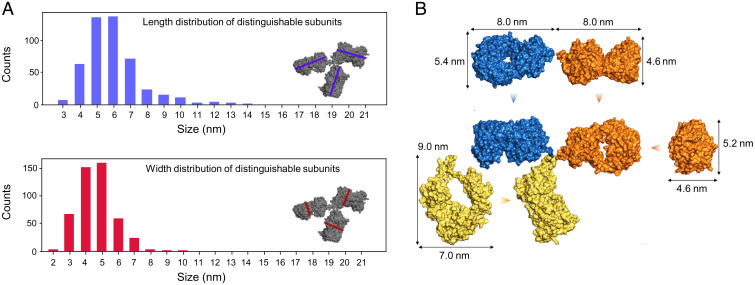
Fig. 5.
Gas-phase–related conformations. (A and B) Length (A) and width (B) distribution of molecules in collapsed conformations. Length and width, longer and shorter distance, respectively, were measured at right angles as indicated by the blue and red lines in A and B, Insets. Comparison with Fig. 3A shows that the collapsed antibody conformations are larger than individual antibody subunits. (C and D) Examples of amplitude reconstructions of molecules in a collapsed conformation. (E) Percentage of extended and collapsed conformations, depending on the landing energy. The amount of collapsed structures increases at lower landing energies, indicating that the collapsed structure is closely related to the gas-phase conformation. For this analysis, a large number of molecules were evaluated at each landing energy: 378 molecules at 0.5 eV, 1,259 molecules at 5 eV, 382 molecules at 10 eV, and 239 molecules at 25 eV.
Fig. 2A shows a high-resolution image in which the size of the smallest resolved features is 5 Å. The features of the antibody are readily recognized: The molecule resembles the characteristic Y shape of the antibody structure with three clearly distinguishable subunits. The size of the molecule and its subunits match the expected sizes based on X-ray crystallography data of IgG antibodies (53–56). We identify three major subunits, interconnected by a well-resolved hinge region of two discernible peptide chain segments originating from the lower subunit. This feature of the hinge region, along with the lower subunit’s slightly larger dimensions, allows its identification as the Fc subunit. The other two high-contrast regions in the upper part of Fig. 2A are hence the Fab subunits. Beyond the identification of the subunits, further detail can be inferred: The two separate dark areas in the Fc subunit can be interpreted as the two heavy chains constituting this part of the antibody, with the area of low contrast dividing the peptide chains marking the hydrophobic pocket hosting the glycan chains.
The reconstruction of holograms yields 2D images, which can be interpreted as projections of the individual molecules along the optical axis. Thus, different molecular configurations will be mapped to different shapes. Depending on their orientation with respect to the surface, the shape and size of the antibody subunits, as well as the visibility of the cavity, can vary significantly. The experimentally observed shapes can be reproduced by merely rotating the subunits around the hinge region, assuming a flexible hinge and intact, rigid subunits. The best-fitting structure for the molecule presented in Fig. 2A is shown in Fig. 2B (also SI Appendix, Fig. S2). We find that the two identical Fab subunits are captured in different orientations: The right Fab subunit exhibits a compact shape, and the left subunit presents a torus-like shape with an area of low contrast in its center that can be interpreted as the characteristic cavity in the subunit structure. Depending on its orientation, the Fab subunit can indeed appear in a variety of shapes, including both the compact and the torus-shaped forms observed in Fig. 2. The range of this variety, especially regarding subunit size, is depicted in Fig. 3.
Fig. 3.
Subunit size and shape. (A) Size distributions (length and width) of individual distinguishable subunits. Length and width, longer and shorter distance, respectively, were measured at right angles as indicated by the blue and red lines in Insets. (B) Projections of the 1IGT crystallographic structure and its individual subunits. The arrows indicate the direction of projection. Projection directions were chosen to display maximal differences in shape and size expected for subunits of different orientation. Given the flexibility of the hinge region, these projections indicate the range of shapes and sizes the antibody subunits can assume on the surface. Comparison of the experimental distribution in A to the size range obtained in B suggests that the antibody subunits remain intact upon landing on the surface.
The intrinsic flexibility of the hinge region, which is manifest at the single-molecule level as evidenced in Fig. 2, allows the subunits of one molecule to reorient independently. This results in a large number of subunit configurations that map to significantly different appearances of subunits in the reconstructed images of different antibody molecules. The examples of experimentally observed Y-shaped antibody molecules shown in Figs. 2A and 4 A–C display a wide variety of subunit shapes and sizes. The variations observed in the overall size of the molecules can be justified by noting that subunits can not only reorient by rotation around the hinge region within the substrate plane, but also rotate out of this plane, which effectively decreases the apparent distances between subunits. The statistical distribution of the sizes of individual subunits (Fig. 3A) matches the expected size distribution for the subunits of the crystallographic model (55) under different orientations (indicated in Fig. 3B). The agreement of measured and expected size distributions of the individual antibody subunits, as well as the observation of characteristic structural features such as the cavity, suggests that individual antibody subunits, which are in themselves more rigid than the full antibody structure, retain their conformation during the ES-IBD process even when the antibody as a whole changes shape due to the flexibility of the hinge region.
We can reproduce the plethora of experimentally observed structures from the crystallographic model by simulating the flexibility of the hinge region via a stepwise rotation of the and ψ angles of the Gly-236 residues (55) of both the heavy chains by 60∘, sampling different configurations (SI Appendix, Fig. S2). Among them, roughly 400 configurations with three clearly distinguishable subunits can be identified. From each of these structures, many projections are generated for different viewing directions. A visual comparison of these projections with the experimental data allows the identification of suitable matches (Fig. 4 and further detail in SI Appendix, Figs. S2 and S3).
While this model creates many projections that show three distinct subunits, the majority of the projections obtained feature overlapping subunits, and hence fewer than three subunits can be clearly distinguished. Likewise, in the experiment, only 2% of the antibody molecules appear as Y-shaped with three clearly identifiable subunits, which correspond to Y-shaped conformations adsorbed in a flat geometry (Fig. 4D).
In addition, two types of structures can be identified: A total of 20% of the analyzed molecules (1,259 molecules in five separate experiments) lead to images with two distinguishable subunits (Fig. 4 E–G) and around 78% of the observed molecules are compact structures with no discernible substructure, neither subunits nor cavities (Fig. 5 C and D). The structures in which only two subunits can be distinguished can be understood as projections of molecular configurations that are adsorbed on the graphene in a vertical geometry, as schematically depicted in Fig. 4H, with at least one subunit rotated out of the surface plane. In a vertical geometry, the antibody interacts with the graphene only via one or two of its subunits leading to a partial (Fig. 4F) or complete (Fig. 4G) eclipse of the third subunit when viewed as a projection onto the graphene plane. This interpretation is supported by the fact that a third subunit is partially recognizable in some of these structures (Fig. 4F). The subunits observed in these conformations match the intact subunits of the crystallographic model in size and shape (Fig. 3). Additionally, in some images, one of the two subunits appears larger than the other, which is indicative of partially overlapping subunits (for example, Fig. 4E). Because of the flexibility of the molecule, the resulting vertical conformations are diverse in appearance; further examples are given in SI Appendix, Fig. S4. Vertical antibody adsorption geometries, as proposed here, have also been observed using other imaging techniques, e.g., in a combined atomic force microscopy and molecular dynamics investigation, both in water and in air (57).
While the dimensions of the species with two distinguishable subunits are congruent with out-of-plane antibody geometries, the compact structures do not match the projections obtained from Y-shaped antibody conformations. The dimensions of the latter structures are larger in comparison with the length and width distribution expected of the individual subunits (Fig. 5 A and B); hence these structures cannot be individual subunits. This suggests that the compact conformations are intact antibodies whose subunits are in significant overlap, which results in the lack of observable substructure.
To explain the origin of the different classes of antibody structures (structures with distinguishable subunits and compact structures) observed on the surface and how they relate to the antibody’s native conformation, we need to consider the influence of the entire sample preparation process of ES-IBD on the protein structure. The conformation of a flexible protein can change at several points in this process: during ionization, gas-phase flight, collision with the graphene surface, and due to adsorption interaction. Mass spectrometry and ion mobility measurements have shown that antibodies neither unfold nor fragment in the gas phase; i.e., their collision cross-sections are in the range expected for native-like protein structures and the peaks observed in the mass spectra correspond to the intact protein (41, 58, 59). We additionally verified that the structures we observe are tied to native-like conformations by comparing them to structures obtained by deliberately denaturing the proteins in solution, which leads to a significant change in both the mass spectrum and the size distribution of the molecules observed on the surface (SI Appendix, Fig. S5). However, ion mobility measurements along with corresponding molecular dynamics simulations suggest that the tertiary structure of flexible proteins collapses onto itself in the gas phase during native ES-IBD (41, 60–62). This is reflected in the collision cross-section (CCS) measurements: The CCS of the gas-phase antibody measured by ion mobility () (41) is significantly smaller than the CCS calculated for extended structures (e.g., for the Protein Data Bank structure 1IGT) (55). In the case of antibodies, the highly flexible hinge region plays a key role since it allows the subunits to come into close contact (41, 60). Compact structures without distinguishable subunits observed on the surface can hence be interpreted as chemically intact antibodies that have collapsed in the gas phase and retained a compact conformation upon landing on the surface. To substantiate this interpretation, we compared the sizes of three different collapsed antibody structures simulated by Hansen et al. (41) that match the reduced CCS to our experimental data. These models yield a length range of 6 to 12 nm and a width range of 5 to 9 nm, which is in good agreement with the experimental size distributions found here (Fig. 5 A and B).
Despite the strong indication that antibody molecules collapse in the gas phase, we observe a significant population of molecules with an extended hinge region that fit the Y-shaped antibody structures known from crystallography, both in flat and vertical geometries. There are two possible explanations for the occurrence of these extended structures on the surface. On the one hand, the extended conformations could already be present in the gas phase and retained upon landing on the surface. On the other hand, these structures could result from the landing process. Since both ion mobility measurements and the corresponding simulations (41) suggest that extended gas-phase conformations are very unlikely, the former process would not significantly contribute to the number of extended structures on the surface. This is further confirmed by the fact that collision cross-sections calculated from the models associated with the extended structures (Fig. 4) using the software tool IMPACT (63) yield CCSs in the range of , which is significantly larger than the experimentally observed values (41) and correlates more closely with the values calculated from the crystallographic model.
Generally, the landing of a three-dimensional flexible object has to be considered as a transition from a gas-phase conformation into an adsorption conformation (25). This transition is characterized by the conversion of translational kinetic energy upon landing (46, 64); hence, kinetic energy transfer is a defining factor regarding possible structures on the surface. Charge transfer and adsorption interaction, between molecule and surface, also influence the adsorption conformations. Since we do not observe molecular motion or diffusion during the measurement, we can exclude thermal energy as a factor that influences the surface conformations. The occurrence of extended antibody conformations on the surface can thus be related to the dissipation of kinetic energy during the landing process or the adsorption interaction, which can convert collapsed molecules into extended conformations. Whether the transition from a collapsed into an extended structure occurs depends on the orientation of the molecule upon impact and strongly on its kinetic energy (46, 65). The molecules are slowed down to landing energies of less than 5 eV per charge, and hence the kinetic energy at impact is around 110 to 130 eV per molecule at charge states between +22 and +26. This translates to ∼10 meV per atom, which is less than the thermal energy at room temperature () and hence far from a reactive collision regime (65). However, this energy is mostly transferred into soft vibrational modes of the molecule as well as into the deformation of the surface. As a consequence, major conformational changes coupled to these soft modes can occur even at low collision energies, while changes in primary and secondary structure are unlikely. For a collapsed antibody, soft modes can be the rotations of the subunit around the hinge region. These processes favor extended surface conformations with increasing deposition energy (46). Experimentally, we have observed a dependence of the conformation of the antibody molecules on deposition energy. The percentage of extended structures with distinguishable subunits increases from at the lowest landing energy of 0.5 eV per charge to at a landing energy of 5 to 10 eV per charge and to at 25 eV per charge (Fig. 5E). This means that more molecules present collapsed adsorption conformations when deposited at very low landing energy.
Considering that even high-energy protein–surface collisions of 500 eV per charge, as used in surface-induced dissociation (SID) (66, 67), yield folded subunits recoiling from the surface, we can conclude that the antibody subunits remain intact upon landing at the energies used in our experiments. Thus, our observation of the dependence of the distribution of antibody appearance on the deposition energy further underpins the idea that the compact structures observed are indeed collapsed and that both extended and collapsed conformations are chemically intact antibody molecules with folded subunits.
Conclusion
We have shown the potential for nondestructive imaging of highly flexible proteins at subnanometer resolution using LEEH. Our investigation of individual antibody molecules evidences a large structural variability, both of the molecules’ subunit orientation and of the overall molecular geometry, as expected for such a highly flexible protein. This large array of conformations includes the antibody structures known from other imaging methods.
Furthermore, we found that the sample preparation process plays a crucial role in shaping the molecular conformations on the surface. While the deposited proteins retain their primary, their secondary, and a native-like tertiary structure, both the transition from solution into the gas phase and the landing on a surface can induce conformation alterations, such as a collapse of the molecules onto themselves in the gas phase and the recovering of extended conformations due to surface collisions. This specifically allows us to image proteins that retain structures reminiscent of the gas-phase conformations and to compare them to known crystallographic structures, which is an important consideration for the many applications of structural biology in the gas phase (68). Furthermore, our study allows us to assess the conformational changes induced by the surface interaction.
Overall, we have shown that LEEH can already serve as a complementary method to other imaging techniques in elucidating flexible protein structures. Future research toward single-molecule protein structure determination will have to focus on an improved resolution and full three-dimensional reconstruction as well as on controlling the protein conformation during the landing process.
Materials and Methods
Low-Energy Electron Holography
Our LEEH microscope is set up in an in-line holography geometry as originally suggested by Gabor (69). The sample is illuminated by low-energy electrons (50 to 150 eV) that are emitted by a sharp tungsten tip brought in close distance (ca. 200 to 500 nm) to the graphene. The pattern emerging from the interference between the wave scattered by the proteins and the unscattered reference wave forms the hologram, which appears on the fluorescent screen of a microchannel plate detector (Fig. 1C) and is reconstructed numerically. By tuning the tip–sample distance, it is possible to adjust the magnification factor (up to magnifications of 107 for the system presented here), permitting the acquisition of both survey images (SI Appendix, Fig. S1) and holograms of individual molecules. Our LEEH microscope operates at a base pressure of mbar and under room temperature conditions. The tungsten tip emitters are prepared by electrochemical etching followed by annealing in UHV.
LEEH requires an atomically clean, electron-transparent substrate. Thus, the proteins are deposited by native ES-IBD in UHV on SLG. SLG is the substrate of choice for LEEH imaging because it is transparent for electrons even at the low electron energies used in LEEH. Furthermore, graphene is conductive, which is essential for distortion-free imaging as it provides an equipotential surface at the sample and reduces possible charging effects. Additionally, the interaction of SLG with proteins is mediated by weak van der Waals forces minimizing potential structural alterations of the proteins due to substrate interactions (70).
Preparation of Ultraclean Graphene
Single-layer graphene can be fabricated and maintained in an ultraclean state. We follow the protocol described in ref. 71: Flakes of poly(methyl methacrylate) (PMMA) coated graphene are fished on transmission electron microscopy grids (500-nm holes, plasma cleaned and sputter coated) and then heated up to remove the PMMA and transferred into vacuum. The cleanliness of the graphene is ascertained by LEEH before molecules are deposited.
Protein Preparation
The antibodies are stored in buffer (Tris-buffered saline solution, pH 7.6), which is exchanged with 200 mM ammonium acetate via a biospinning process. The concentration of the spray solution was 0.5 mg/mL.
Native ESI and Preparative Mass Spectrometry
Native ESI is used to bring the antibody molecules into the gas phase from a buffered solution, using low ionization voltages (1 to 1.5 kV) applied to very sharp metal-coated glass emitters to avoid unfolding of the molecules during the ionization process. The temperature at the air–vacuum interface is kept at 70 ∘C and minimal voltage offsets for activation with background gas collisions are used. This process yields protein ions of native-like conformation, which are guided into vacuum, where an ion beam of defined kinetic energy is formed. A quadrupole mass filter and a time-of-flight mass spectrometer are used to characterize and select molecules whose m/z ratio corresponds to the native-like species for deposition (Fig. 1B). The m/z filter range ( to u/e) used for deposition corresponds to folded antibody structures of low charge states ( to + 26) as established by ion mobility measurements (41). Deposition is carried out in UHV ( mbar). The landing energy of the proteins can be tuned by applying a retarding voltage to the sample. The coverage is controlled via a current measurement at the sample to guarantee the deposition of isolated proteins. After the deposition, the sample is transferred to the LEEH microscope using a UHV suitcase ( mbar). This method ensures the high chemical purity of the sample required for LEEH characterization. First, it provides the conditions for generating high-resolution holograms by creating a well-defined environment for the individual proteins that is free of additional scatterers. Second, the chemical control over the deposited species, achieved by preparative mass spectrometry, facilitates the interpretation of the data due to the unambiguous chemical identity of the deposited proteins.
Reconstruction
The method of reconstruction used here is a propagation-based method that stems from a wave-optical approach to holography (52, 72). In the case of reconstruction, the propagation of the hologram multiplied by the reference wave from the detector plane to the image plane is expressed in integral form, yielding a Kirchhoff–Fresnel diffraction integral of the form
with , where U(x, y) approximates the wave scattered by the object (exit wave). This integral has the form of a convolution:
where S is a propagation function.
The convolution theorem allows us to carry out the analysis in Fourier space. The exit wave can hence be expressed as
The amplitude reconstructions shown in this work are obtained by taking the absolute value of the complex-valued exit wave.
To simplify things further, we make use of the fact that the opening angle of the beam is small, which allows us to use the paraxial approximation to linearize the square root appearing in the propagation function.
Supplementary Material
Acknowledgments
We thank Dr. Argyris Politis and Kjetil Hansen for providing us with their simulated models of collapsed antibodies and Dr. Jean-Nicolas Longchamp for his support in constructing our LEEH microscope.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. V.W. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112651118/-/DCSupplemental.
Data Availability
All data needed to evaluate the conclusion in the paper are present in the paper or SI Appendix.
References
- 1.Kumar S., Ma B., Tsai C. J., Wolfson H., Nussinov R., Folding funnels and conformational transitions via hinge-bending motions. Cell Biochem. Biophys. 31, 141–164 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Keskin O., Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: A case study of antibodies. BMC Struct. Biol. 7, 31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobbins S. E., Lesk V. I., Sternberg M. J., Insights into protein flexibility: The relationship between normal modes and conformational change upon protein-protein docking. Proc. Natl. Acad. Sci. U.S.A. 105, 10390–10395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber R., Bennett W. S. Jr., . Functional significance of flexibility in proteins. Biopolymers 22, 261–279 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Schneider S. W., et al., Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc. Natl. Acad. Sci. U.S.A. 104, 7899–7903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianfrocco M. A., Kellogg E. H., What could go wrong? A practical guide to single-particle cryo-EM: From biochemistry to atomic models. J. Chem. Inf. Model. 60, 2458–2469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaeser R. M., How good can cryo-EM become? Nat. Methods 13, 28–32 (2016). [DOI] [PubMed] [Google Scholar]
- 8.MacGillivray R. T., et al., Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry 37, 7919–7928 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Nakane T., et al., Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonic S., Vénien-Bryan C., Protein structure determination by electron cryo-microscopy. Curr. Opin. Pharmacol. 9, 636–642 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Frank J., Single-particle reconstruction of biological macromolecules in electron microscopy–30 years. Q. Rev. Biophys. 42, 139–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartesaghi A., et al., 2.2 Åresolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins M. K., Lea S. M., On the state of crystallography at the dawn of the electron microscopy revolution. Curr. Opin. Struct. Biol. 46, 95–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittermaier A. K., Kay L. E., Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 34, 601–611 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Chao F. A., Byrd R. A., Protein dynamics revealed by NMR relaxation methods. Emerg. Top. Life Sci. 2, 93–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandin S., Öfverstedt L. G., Wikström A. C., Wrange O., Skoglund U., Structure and flexibility of individual immunoglobulin G molecules in solution. Structure 12, 409–415 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., et al., 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci. Rep. 5, 9803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al Qaraghuli M. M., Kubiak-Ossowska K., Ferro V. A., Mulheran P. A., Antibody-protein binding and conformational changes: Identifying allosteric signalling pathways to engineer a better effector response. Sci. Rep. 10, 13696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y., Yu D., Mayani M., Mussa N., Li Z. J., Monoclonal antibody higher order structure analysis by high throughput protein conformational array. MAbs 10, 397–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokarn Y., et al., Biophysical techniques for characterizing the higher order structure and interactions of monoclonal antibodies. ACS Symp. Ser. 1201, 285–327 (2015). [Google Scholar]
- 21.Rose M., et al., Single-particle imaging without symmetry constraints at an X-ray free-electron laser. IUCrJ 5, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurta R. P., et al., Correlations in scattered X-ray laser pulses reveal nanoscale structural features of viruses. Phys. Rev. Lett. 119, 158102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayyer K., et al., Perspectives for imaging single protein molecules with the present design of the European XFEL. Struct. Dyn. 2, 041702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X., et al., Imaging single glycans. Nature 582, 375–378 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Deng Z., et al., A close look at proteins: Submolecular resolution of two- and three-dimensionally folded cytochrome c at surfaces. Nano Lett. 12, 2452–2458 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Rauschenbach S., et al., Two-dimensional folding of polypeptides into molecular nanostructures at surfaces. ACS Nano 11, 2420–2427 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Fink H. W., Stocker W., Schmid H., Holography with low-energy electrons. Phys. Rev. Lett. 65, 1204–1206 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Fink H. W., Stocker W., Schmid H., Coherent point source electron beams. J. Vac. Sci. Technol. B 8, 1323 (1990). [Google Scholar]
- 29.Kreuzer H. J., Nakamura K., Wierzbicki A., Fink H. W., Schmid H., Theory of the point source electron microscope. Ultramicroscopy 45, 381–403 (1992). [Google Scholar]
- 30.Latychevskaia T., Escher C., Andregg W., Andregg M., Fink H. W., Direct visualization of charge transport in suspended (or free-standing) DNA strands by low-energy electron microscopy. Sci. Rep. 9, 8889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longchamp J. N., et al., Imaging proteins at the single-molecule level. Proc. Natl. Acad. Sci. U.S.A. 114, 1474–1479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauschenbach S., Ternes M., Harnau L., Kern K., Mass spectrometry as a preparative tool for the surface science of large molecules. Annu. Rev. Anal. Chem. 9, 473–498 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Uetrecht C., Rose R. J., van Duijn E., Lorenzen K., Heck A. J. R., Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 (2010). [DOI] [PubMed] [Google Scholar]
- 34.van Duijn E., Barendregt A., Synowsky S., Versluis C., Heck A. J. R., Chaperonin complexes monitored by ion mobility mass spectrometry. J. Am. Chem. Soc. 131, 1452–1459 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Bohrer B. C., Merenbloom S. I., Koeniger S. L., Hilderbrand A. E., Clemmer D. E., Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. 1, 293–327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fink H. W., Schmid H., Ermantraut E., Schulz T., Electron holography of individual DNA molecules. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 14, 2168 (1997). [Google Scholar]
- 37.Stevens G. B., et al., Individual filamentous phage imaged by electron holography. Eur. Biophys. J. 40, 1197–1201 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Snijder J., Rose R. J., Veesler D., Johnson J. E., Heck A. J., Studying 18 MDa virus assemblies with native mass spectrometry. Angew. Chem. Int. Ed. Engl. 52, 4020–4023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urner L. H., et al., Modular detergents tailor the purification and structural analysis of membrane proteins including G-protein coupled receptors. Nat. Commun. 11, 564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen H. Y., et al., Ligand binding to a G protein-coupled receptor captured in a mass spectrometer. Sci. Adv. 3, e1701016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen K., et al., A mass-spectrometry-based modelling workflow for accurate prediction of IgG antibody conformations in the gas phase. Angew. Chem. Int. Ed. Engl. 57, 17194–17199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janeway C. J., Travers P., Walport M., Shlomchik M., “The structure of a typical antibody molecule” in Immunobiology: The Immune System in Health and Disease, Schanck D. et al., Eds. (Garland Publishing, 2001), chap. 3, pp. 93–122. [Google Scholar]
- 43.Rauschenbach S., et al., Electrospray ion beam deposition of clusters and biomolecules. Small 2, 540–547 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Heck A. J., Native mass spectrometry: A bridge between interactomics and structural biology. Nat. Methods 5, 927–933 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Kuhlen L., et al., Structure of the core of the type iii secretion system export apparatus. Nat. Struct. Mol. Biol. 25, 583–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anggara K., et al., Exploring the molecular conformation space by soft molecule-surface collision. J. Am. Chem. Soc. 142, 21420–21427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laskin J., Wang P., Hadjar O., Soft-landing of peptide ions onto self-assembled monolayer surfaces: An overview. Phys. Chem. Chem. Phys. 10, 1079–1090 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Johnson G. E., Gunaratne D., Laskin J., Soft- and reactive landing of ions onto surfaces: Concepts and applications. Mass Spectrom. Rev. 35, 439–479 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Ouyang Z., et al., Preparing protein microarrays by soft-landing of mass-selected ions. Science 301, 1351–1354 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Hopper J. T., et al., Detergent-free mass spectrometry of membrane protein complexes. Nat. Methods 10, 1206–1208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warnke S., von Helden G., Pagel K., Protein structure in the gas phase: The influence of side-chain microsolvation. J. Am. Chem. Soc. 135, 1177–1180 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Latychevskaia T., Fink H. W., Practical algorithms for simulation and reconstruction of digital in-line holograms. Appl. Opt. 54, 2424–2434 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Harris L. J., Larson S. B., Hasel K. W., McPherson A., Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Harris L. J., Skaletsky E., McPherson A., Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Harris L. J., Larson S.B., Hasel K. W., McPherson A., Data from “Structure of immunoglobulin.” Protein Data Bank. https://www.rcsb.org/structure/1IGT. Accessed 9 December 2021.
- 56.Harris L. J., McPherson A., Data from “Structure of immunoglobulin.” Protein Data Bank. https://www.rcsb.org/structure/1IGY. Accessed 9 December 2021.
- 57.Vilhena J. G., et al., Adsorption orientations and immunological recognition of antibodies on graphene. Nanoscale 8, 13463–13475 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Parsons T. B., et al., Optimal synthetic glycosylation of a therapeutic antibody. Angew. Chem. Int. Ed. Engl. 55, 2361–2367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upton R., et al., Hybrid mass spectrometry methods reveal lot-to-lot differences and delineate the effects of glycosylation on the tertiary structure of HerceptinⓇ. Chem. Sci. (Camb.) 10, 2811–2820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Devine P. W. A., et al., Investigating the structural compaction of biomolecules upon transition to the gas-phase using ESI-TWIMS-MS. J. Am. Soc. Mass Spectrom. 28, 1855–1862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pacholarz K. J., et al., Dynamics of intact immunoglobulin—G explored by drift-tube ion-mobility mass spectrometry and molecular modeling. Angew. Chem. Int. Ed. Engl. 53, 7765–7769 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Porrini M., et al., Compaction of duplex nucleic acids upon native electrospray mass spectrometry. ACS Cent. Sci. 3, 454–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marklund E. G., Degiacomi M. T., Robinson C. V., Baldwin A. J., Benesch J. L., Collision cross sections for structural proteomics. Structure 23, 791–799 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Rinke G., et al., Active conformation control of unfolded proteins by hyperthermal collision with a metal surface. Nano Lett. 14, 5609–5615 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Krumbein L., et al., Fast molecular compression by a hyperthermal collision gives bond-selective mechanochemistry. Phys. Rev. Lett. 126, 056001 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Harvey S. R., et al., Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. Proc. Natl. Acad. Sci. U.S.A. 116, 8143–8148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seffernick J. T., Harvey S. R., Wysocki V. H., Lindert S., Predicting protein complex structure from surface-induced dissociation mass spectrometry data. ACS Cent. Sci. 5, 1330–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barth M., Schmidt C., Native mass spectrometry–A valuable tool in structural biology. J. Mass Spectrom. 55, e4578 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Gabor D., A new microscopic principle. Nature 161, 777–778 (1948). [DOI] [PubMed] [Google Scholar]
- 70.Nair R. R., et al., Graphene as a transparent conductive support for studying biological molecules by transmission electron microscopy. Appl. Phys. Lett. 97, 153102 (2010). [Google Scholar]
- 71.Longchamp J. N., Escher C., Fink H. W., Ultraclean freestanding graphene by platinum-metal catalysis. J. Vac. Sci. Technol. B 31, 020605 (2013). [Google Scholar]
- 72.Born M., et al., Principles of Optics (Cambridge University Press, 1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusion in the paper are present in the paper or SI Appendix.