Significance
The human calcium-sensing receptor maintains a stable concentration of calcium in the blood. Naturally occurring mutations in the receptor are linked to calcium homeostatic disorders including hypercalcemia and hypocalcemia. Modulation of the receptor function can provide therapeutic relief of these conditions. The structures of the calcium-sensing receptor in the absence and presence of various allosteric modulators reveal the mechanism by which these modifiers up- or down-regulate receptor function. We have captured symmetric forms of the homodimeric receptor both at rest and upon stimulation and concluded that receptor activation involves a conformational change at its dimer interface. The structural details of the calcium-sensing receptor in different functional states may assist the design of new therapeutics for diseases related to calcium homeostasis.
Keywords: calcium-sensing receptor, cryo-EM structure, allosteric modulation, activation mechanism, symmetry
Abstract
The human extracellular calcium-sensing (CaS) receptor controls plasma Ca2+ levels and contributes to nutrient-dependent maintenance and metabolism of diverse organs. Allosteric modulation of the CaS receptor corrects disorders of calcium homeostasis. Here, we report the cryogenic-electron microscopy reconstructions of a near–full-length CaS receptor in the absence and presence of allosteric modulators. Activation of the homodimeric CaS receptor requires a break in the transmembrane 6 (TM6) helix of each subunit, which facilitates the formation of a TM6-mediated homodimer interface and expansion of homodimer interactions. This transformation in TM6 occurs without a positive allosteric modulator. Two modulators with opposite functional roles bind to overlapping sites within the transmembrane domain through common interactions, acting to stabilize distinct rotamer conformations of key residues on the TM6 helix. The positive modulator reinforces TM6 distortion and maximizes subunit contact to enhance receptor activity, while the negative modulator strengthens an intact TM6 to dampen receptor function. In both active and inactive states, the receptor displays symmetrical transmembrane conformations that are consistent with its homodimeric assembly.
Critical to the maintenance of Ca2+ homeostasis, the extracellular calcium-sensing (CaS) receptor was the first G protein–coupled receptor (GPCR) discovered to sense ions (1–3). The CaS receptor detects fluctuations in plasma Ca2+ at the parathyroid. In response to increases in Ca2+, it transmits signals to inhibit the release of parathyroid hormone, in turn preventing further rises in Ca2+ concentration (2, 3). In the cortical thick ascending limb of the renal nephron, the CaS receptor is also activated by surges in plasma Ca2+ and responds by inhibiting Ca2+ reabsorption. The excess urinary calcium excretion arising from CaS receptor activation lowers the plasma Ca2+ level. The CaS receptor is implicated in various pathologies associated with hypercalcemia and hypocalcemia (4). It has also been linked to the progression of diseases such as breast and colon cancer, in which the receptor modulates tumor growth (3, 5–7).
The CaS receptor senses a diverse array of extracellular stimuli. During normal function, it activates multiple intracellular signaling pathways involving Gq/11, Gi/o, or G12/13; in tumor cells, it is coupled to Gs (2, 3, 8, 9). In addition to the principal agonist Ca2+, the receptor is directly activated by aromatic l-amino acids (10, 11). Other CaS agonists include various divalent and trivalent cations (12), referred to as type I calcimimetics for mimicking the action of Ca2+ (13).
The activity of the CaS receptor is also subject to allosteric modulation. Positive allosteric modulators (PAMs) are classified as type II calcimimetics for increasing the receptor sensitivity for Ca2+ (12–16). The prototypical PAM molecules share a phenylalkylamine structure, including cinacalcet and NPS R-568 (abbreviated as R-568). Cinacalcet was the first drug described to target a GPCR allosterically, and it is used clinically to treat hyperparathyroidism in patients with chronic kidney diseases (15). Negative allosteric modulators (NAMs) of the CaS receptor are referred to as calcilytics for suppressing the receptor response to Ca2+ (12–16). Synthetic calcilytics such as NPS-2143 and ronacaleret are also structurally related to phenylalkylamines. Recently, inorganic phosphate has been identified as an inhibitor of the receptor (11, 17).
The CaS receptor rests within the class C family of GPCRs and functions as an obligate homodimer. Like other class C GPCRs, each CaS subunit contains a large extracellular domain (ECD) involved in orthosteric ligand binding, a seven-helix transmembrane (TM) domain responsible for G protein coupling, followed by an extended cytoplasmic tail (18–23). The conformations of the CaS ECDs in both the inactive and active states have been determined by X-ray crystallography (11, 24). The ECD structures also revealed how the receptor recognizes various extracellular ligands, including Ca2+, the amino acid l-Trp, and inorganic phosphate. Although the role of amino acids is still under debate (25), recent structural studies of full-length CaS receptor further confirmed that Ca2+ and amino acids cooperate to activate the receptor (26–28).
The TM domain of the CaS receptor harbors the binding sites for PAM and NAM molecules according to previous mutagenesis studies (29–32). Recently reported modulator-bound CaS receptor structures revealed asymmetric TM configurations that are stabilized by PAM molecules binding in different poses within the separate subunits of the homodimer (33). We have determined PAM- and NAM-bound, as well as PAM-free, structures of a near–full-length CaS receptor using cryogenic-electron microscopy (cryo-EM) that display symmetric TM dimers and modulator poses, instead. This finding presents the possibility of receptor activation without requiring asymmetric conformational transition. Our structures also illustrate how distortion of TM6 provides the driving force for receptor activation. Furthermore, the presence of a PAM or NAM stabilizes distinct TM6 helix conformations to promote specific dimer arrangements and differentially modulate receptor function.
Results
Cryo-EM Reconstructions of Multiple Functional States.
We acquired active-state structures of the CaS receptor using protein expressed in mammalian cells and purified in the presence of excess Ca2+, the tryptophan analog cyclomethyltryptophan (TNCA) (24), and the PAM molecule R-568 (34) (Fig. 1 and SI Appendix, Figs. S1, S2, and S3 A–E and Table S1) (35). Our initial global reconstruction in C1 symmetry revealed that the receptor exhibits twofold symmetry in both the ECD and TM regions, but their dimer axes are offset by 4.5° (SI Appendix, Fig. S3F). A similar displacement of the ECD and TM domains has been observed in the active-state structure of full-length CaS receptor from Gallus gallus (27) (SI Appendix, Fig. S3G). Using three-dimensional (3D) variability analysis of the cryo-EM data, we found that the active receptor is in constant motion along two principal dimensions (Movies S1 and S2). Along the first component of motion exhibited by the active receptor, the TM domains of the two subunits oscillate relative to each other while maintaining the symmetry of the TM dimer (Movie S1). Along the second component, the ECD and TM domains flex back and forth about the linker region (Movie S2). This motion underlies the offset of their dimer axes in our C1 reconstruction. On the other hand, the TM domains of the two subunits are not completely synchronous as they bend, but slide vertically relative to each other and thus break the twofold symmetry in the process (Movie S2). Overall, we have captured an active CaS population that is predominantly symmetric. The symmetric TM dimer of our reconstruction is in contrast to the asymmetric arrangement reported in cinacalcet- and evocalcet-bound active CaS receptor structures (33) (SI Appendix, Fig. S3H). In addition, these receptor forms also undergo dynamic motion and appear to adopt a symmetric pose at certain points along their movement trajectory (33).
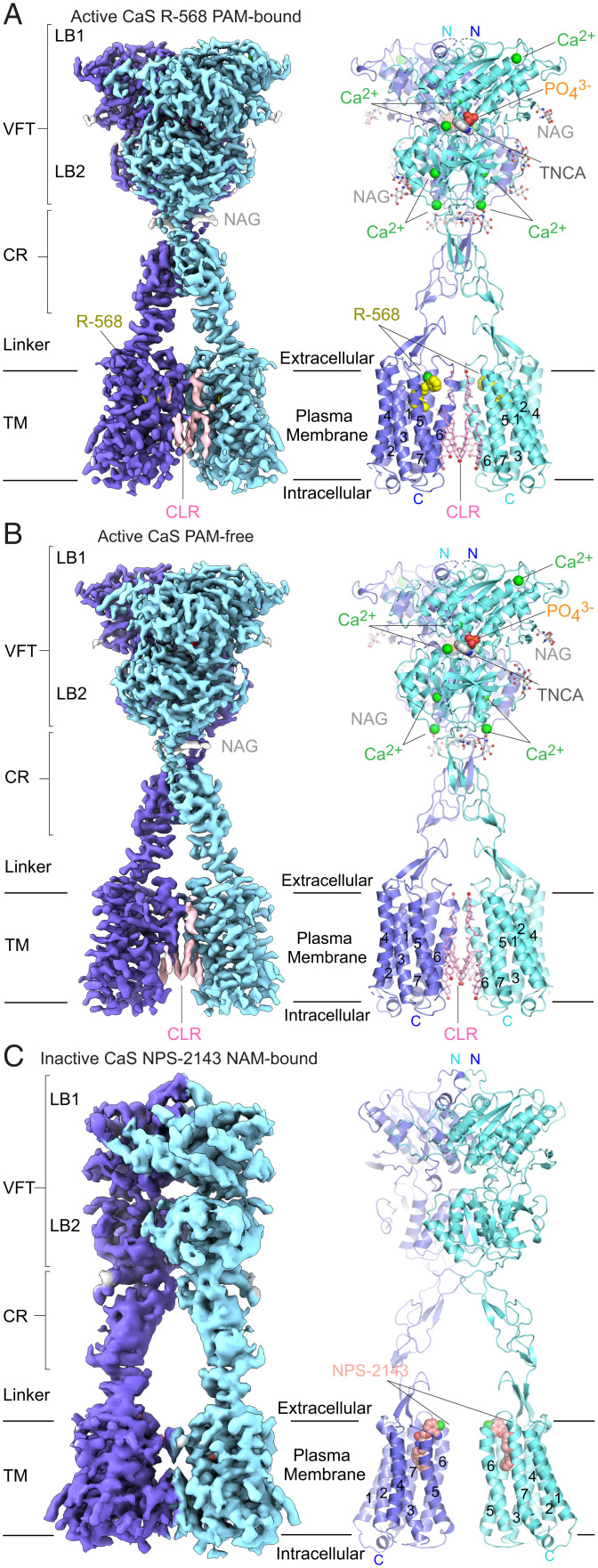
Fig. 1.
Cryo-EM structures of the human CaS receptor. The active structures include the PAM-bound (A) and PAM-free (B) states; the inactive structure is in the NAM-bound form (C). For each structure, the cryo-EM density map (Left) and cartoon representation (Right) are oriented toward the TM–homodimer interface. (Discontinuous densities below a size threshold of 2 voxels were removed from map display.) Subunits are individually colored blue and cyan, and TM helices are numbered. Ligands bound to the active CaS receptor include TNCA (dark gray), Ca2+ (green), PO43− (orange and red), and R-568 in the PAM-bound state (yellow). Additional molecules present are N-linked glycans (NAG; light gray) and CLR (pink). In the inactive state, only the NAM NPS-2143 (salmon) was modeled.
In our C1 reconstruction, we found high-resolution features in the ECD consistent with the estimated overall resolution of 2.8 Å; however, the density for the TM domain lacked most side chains, reflecting its dynamic nature. We took several approaches to improve the density of the TM domain. First, we generated a reconstruction for the TM domain of a single CaS subunit using a combination of local refinement and symmetry-expanded particles, which contain the original particle set and their symmetry mates generated based on the inherent twofold symmetry axis of the global reconstruction. Second, we performed 3D classification of the single TM domain based on the PAM-binding region. We identified two major classes, corresponding to PAM-free (29% of particles) and PAM-bound states (36% of particles), respectively. Furthermore, we observed only one conformation of R-568 in the PAM-bound class. Local refinements of the single TM domains reached 3.4- and 3.5-Å resolution for the PAM-bound and PAM-free classes, respectively (SI Appendix, Fig. S3). This procedure brought out density for nearly all the side chains in the TM domain as well as the allosteric modulator in the PAM-bound class (SI Appendix, Fig. S4 A and B).
For both active-state conformational classes, we then reversed symmetry expansion of the corresponding particle sets and obtained global C1 reconstructions that once more displayed twofold symmetry in both the ECD and TM domains (SI Appendix, Fig. S3 I and J). We proceeded to calculate overall reconstructions imposing C2 symmetry, each achieving 2.7-Å resolution. We generated a composite map for each class by combining the corresponding C2 overall density map with single TM reconstructions aligned to each subunit (SI Appendix, Fig. S3 A–E) (36, 37). Complete receptor structures of the PAM-bound and PAM-free states were then built into these composite reconstructions (38, 39).
In the active state, each CaS subunit features one TNCA, four Ca2+, and one phosphate bound to the ECD (SI Appendix, Fig. S5). Additionally, we observed density for multiple cholesterol or cholesteryl hemisuccinate (CHS) molecules surrounding the TM complex, four of which are located at the subunit interface (Fig. 1 A and B). In the PAM-bound class, one R-568 molecule was also found within the TM domain of each CaS subunit (Fig. 1A). Conversely, modulator-binding sites of the PAM-free class were unoccupied in both subunits.
To capture an inactive-state structure of the CaS receptor, we introduced the NAM NPS-2143, the inhibitory anion sulfate, and ethylene glycol tetraacetic acid (EGTA) to drive the receptor into an inactive conformation (17, 40). We also lowered the pH and increased ionic strength to further stabilize the inactive state (41, 42). Our initial global reconstruction displayed twofold symmetry but was only resolved to 6.0-Å resolution. To improve the density, we separately constructed maps for the ECD and TM domains of a single CaS subunit using symmetry-expanded particles. Local refinement of the individual ECD and TM domains reached 4.0- and 4.2-Å resolution, respectively (SI Appendix, Fig. S6). We assembled an overall reconstruction of the inactive state by combining two single-ECD and two single-TM maps, each aligned to the respective subunit of the global map (Fig. 1C) (43, 44).
We built the inactive CaS receptor structure by fitting the individual domains of the active structure as rigid bodies into the density (45). The overall model was then optimized through molecular dynamics flexible fitting simulation (46). The inactive CaS receptor structure contains one NPS-2143 molecule bound within the TM domain of each subunit (Fig. 1C and SI Appendix, Fig. S4C).
Activation-associated Conformational Changes.
The overall architecture of the homodimeric CaS receptor is composed of two subunits interacting side-by-side while facing opposite directions (Fig. 1). Each subunit is crowned by a Venus flytrap (VFT) module composed of lobe-shaped LB1 and LB2 domains, followed by a cysteine-rich (CR) domain and a peptide linker. Below these elements, the seven-helix TM bundle is found enveloped by a detergent micelle. In addition, protruding extracellular and intracellular loops (ECLs and ICLs) connect adjoining TM helices. A comparison of the inactive and active structures of the CaS receptor reveals several key conformational differences. The changes in the extracellular region are consistent with the paradigm established for the CaS ECD structures (11).
First, in the inactive state, the VFTs of both subunits adopt an open conformation but, in the active state, each closes around the activator TNCA (SI Appendix, Fig. S7 A and B). Our inactive structure is more representative of the ground state than any known inactive CaS structure reporting either an open/closed or closed/closed VFT arrangement (26, 28, 33) since closure of one VFT has been shown to initiate activation in class C GPCRs (47). The closed VFT conformation observed in other inactive CaS receptor structures is likely caused by the presence of ambient amino acids or their analogs (26, 28, 33). The l-Trp analog TNCA is known to remain bound within the VFT despite extensive washing during purification (24). Although the NAM NPS-2143 was also used in some studies to stabilize the inactive structure (28, 33), the combination of low pH, high ionic strength, inhibitory sulfate, and EGTA in our protein preparation might have allowed us to obtain a fully inactive state with an open/open VFT configuration. Relative to the inactive CaS structure, the active form has a VFT that is closed by 18°. The closed conformation of the active VFT is consistent with its anion-binding pattern. Thus, we observed strong density at a known anion-binding site that is important for structural integrity (site 2) (11), and modeled a phosphate at this position (SI Appendix, Fig. S5 C and D). An anion at the adjacent site (site 1) would inhibit receptor activity (11, 17). The absence of a bound anion at site 1 in the active cryo-EM structure is consistent with its active functional state.
Second, a major distinction between the inactive and active structures of the CaS receptor is the extent of homodimer contact. In the inactive state, the two subunits partner through the extracellular LB1 domains exclusively. A large gap is observed between the LB2 and CR domains, with a 50-Å separation between the C termini of the CR domains (SI Appendix, Fig. S7C). In the active state, however, subunit association occurs between all individual domains in the extracellular and TM regions. The gap between the CR domains is decreased to 31 Å as a result of the formation of a new homodimer interface between the LB2 and CR domains of both subunits (SI Appendix, Fig. S7D). The distance between the membrane-proximal domains reflects the conformation sensed by the TM region and is therefore indicative of the functional state. As such, activation of the CaS receptor is correlated with tighter subunit association and, in turn, an expansion of homodimer interactions (Fig. 1). Recently, a nanobody-bound CaS receptor structure has been proposed to represent the fully inactive state because of a larger distance between LB2 domains than in other known natural CaS conformations (28). Since the inactivating nanobody is wedged between the LB2 domains of the two subunits, however, their separation may be artificially expanded due to steric hindrance induced by the nanobody.
The unique homodimer interface formed in the active state requires the presence of additional Ca2+. Specifically, we found ion density at all four Ca2+-binding sites that we previously identified using anomalous diffraction (sites 1 to 4) (11) (SI Appendix, Fig. S5 A and B). The Ca2+ bound to site 4 is important for receptor activation as it bridges the LB2 and CR domains of both subunits to form homodimer interactions in the active state. We also carried out elemental analysis of purified CaS receptor and found that the amount of Ca2+ bound to the active receptor is substantially higher than that found in the inactive state, consistent with the role of Ca2+ as an agonist (SI Appendix, Table S2).
The disparity between inactive and active homodimer interactions continues into the TM region. In the inactive structure, direct TM contact is absent, and the two subunits face each other remotely through parallel surfaces formed by the TM5 and TM6 helices from each subunit (Fig. 2 A and C). At the extracellular end of the TM homodimer, the closest helix pair is that of TM6. Conversely, the neighboring TM5 helices draw nearest at the intracellular end. The TM5–TM6 dimer orientation distinguishes the CaS inactive conformation from other class C receptors including mGlu (48–51) and GABAB (52–56) receptors.
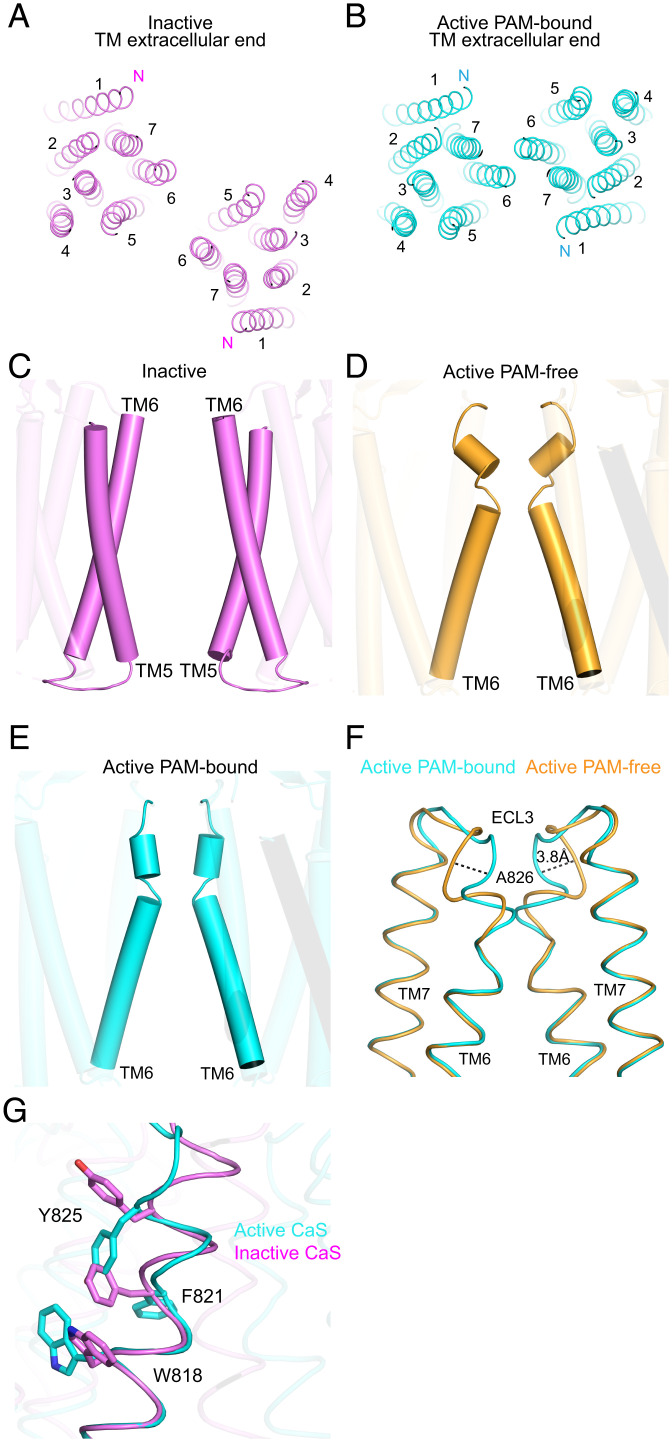
Fig. 2.
Activation-induced conformational changes in the TM. (A and B) TM dimer structures of the NAM-bound inactive (A) and PAM-bound active (B) CaS receptors viewed from the extracellular end. (C–E) Structural elements at the homodimer interface of the NAM-bound inactive (C), PAM-free active (D), and PAM-bound active (E) CaS receptors viewed from the side. (F and G) Alignment of the TM6 helix and ensuing ECL3 region based on residues in the intracellular portion of the TM6 helix (A8046.36 to I8166.48). The diagrams show the comparison between PAM-bound and PAM-free active forms (F) as well as that between the PAM-bound active and NAM-bound inactive states (G).
In our active-state structures, the two subunits associate to form a new homodimer interface through their TM6 helices (Fig. 2 B, D, and E). The pair of TM6 helices start far apart at the intracellular end, drawing closer together as they extend toward the extracellular side. These trajectories continue until direct subunit contacts are formed near the extracellular membrane surface. Furthermore, cholesterol molecules found at the dimer interface facilitate additional interactions between TM6 helices of both subunits (Fig. 1 A and B).
Finally, the inactive and active CaS receptor structures display different relative orientations between ECD and TM domains, which can be described using the intersecting angle between their corresponding dimer interfacial planes. In the inactive state, the homodimer interfacial planes of the membrane-proximal LB2/CR units and TM domains are misaligned by ∼70°. In contrast, the active-state CaS structures feature a 90° difference between these dimer interfacial planes (SI Appendix, Fig. S7E). The increased planar angle between TM and membrane-proximal regions in the active state highlights the importance of ECD-induced mechanical torsion for rotating the TM domains of the receptor into its active conformation. The maintenance of this high torsion requires closure of both VFT modules by way of an amino acid activator, in addition to stabilizing forces of Ca2+ at the LB2 and CR dimer interfaces. In the membrane, the approach of neighboring TM6 helices permits the formation of a new homodimer interface between TM domains of both subunits for receptor activation.
Activation-induced Helix Disruption in TM6.
A prominent feature of the active CaS receptor is an interruption of the helical fold that splits TM6 into two parts at Phe8216.53 (Fig. 2 D and E) as compared with inactive CaS (Fig. 2C). Nearby residue Pro8236.55 facilitates this break and initiates the formation of a second helix. We conclude that the TM6 break is an integral part of the receptor activation mechanism for several reasons. First, the integrity of the entire TM6 helix is maintained in the inactive state. Second, we found this TM6 break both in the absence and presence of a PAM molecule, indicating that the TM6 helix deformation is an inherent part of the activation process. Third, despite the absence of dimer symmetry, the recently reported cinacalcet- and evocalcet-bound active structures demonstrate a similar TM6 distortion (33). Taken together, this suggests that the TM6 break, as opposed to changes in symmetry (33), encapsulates the activation-induced conformational change of the CaS receptor.
The helix break facilitates the formation of a TM6–TM6 homodimer interface. In the PAM-bound active CaS structure, the break bends the TM6 helix so that its extracellular fragment runs nearly parallel to its counterpart in the opposing subunit (Fig. 2E). This kinked conformation permits the entire extracellular portion of the TM6 pair to interact along the helical path. In the PAM-free active structure, the extracellular end of TM6 and ensuing ECL3 bend toward the interior of the TM bundle (3.8-Å Cα shift in A826) (Fig. 2F), appearing to fill the space that is occupied by the modulator in the PAM-bound active structure. As a result, the curvature of the bend is more pronounced in the absence of the PAM, and the pair of TM6 helices move slightly apart after the kink. This results in more sparse contacts in the PAM-free than PAM-bound conformation. Therefore, the presence of the PAM serves to increase homodimer interactions between TM bundles, thereby enhancing receptor activity.
The distortion of TM6 prevents steric clash between rearranged TM domains in the active conformation. Indeed, if we align each subunit of the inactive TM domain onto the active dimer based on the intracellular fragment of TM6, the extracellular end of TM6 and following ECL3 from the two subunits would collide (SI Appendix, Fig. S7 F–I). Therefore, the intact TM6 found in the inactive receptor would not be compatible with the TM6–TM6 dimer orientation found in the active conformation. Although a TM6-centered dimer arrangement is common to the active state of all known class C GPCR structures (52–54, 56), the formation of a dimer interface that is dependent on a distinct break in TM6 is novel, having only been observed in the CaS receptor. This is more akin to G protein–bound monomeric class B GPCRs, which also display a disrupted TM6 (57).
The partial unwinding of the TM6 helix in the active state results in substantial conformational changes in its residues near the extracellular membrane surface (Fig. 2G). First, relative to its counterpart in the inactive state, Trp8186.50 undergoes a rotamer switch from lining the exterior to filling the interior of the TM domain. Second, a dramatic change occurs at Phe8216.53, the breakpoint of TM6. In the inactive state, Phe8216.53 points toward the core of the TM bundle while its backbone atoms form part of a helix turn. In the active state, Phe8216.53 is diverted outward to face the dimer interfacial area. Finally, Tyr8256.57 also changes its rotamer conformation. In the inactive state, however, the side chain of Tyr8256.57 aims upward to the extracellular membrane surface. In the active state, Tyr8256.57 is directed downward, filling the void left by the side chain of Phe8216.53. The packing of Trp8186.50 and Tyr8256.57 along one side of the TM6 helix further reinforces the bend at Phe8216.53 as it protrudes into the external space. In summary, the bending of TM6 allows the formation of a new homodimer interface between TM domains of both subunits for receptor activation. This provides the transmembrane molecular switching mechanism for receptor activation in the absence and presence of PAMs.
Transmission of Extracellular Signals to the TM.
Within each CaS receptor subunit, communication of conformational signals from the VFT to the TM is facilitated by the CR domain and linker, as well as the ECL2 running parallel to the linker (SI Appendix, Fig. S8 A–C). The CR domain is a rigid scaffold held together by four intradomain disulfide bonds (SI Appendix, Fig. S1). The relative orientation between the CR and LB2 domains is fixed through an interdomain disulfide bridge, allowing the CR domain to transduce and amplify the movement of the LB2 domain.
Each CaS receptor subunit possesses a bonded network that seamlessly integrates the CR domain, linker, and ECL2 components as a joint entity (SI Appendix, Fig. S8 A–C). These interactions are largely conserved across different activation states. This allows the joint entity to serve as a mechanical junction between the VFT and TM domain to enable communication of extracellular signals across the membrane. The interfacial contacts within this junction can be dissected into two main categories.
The first set of interactions involves a hydrogen-bonding network between the peptide linker and a β-hairpin formed by ECL2, and the contacts are mostly mediated by backbone atoms (SI Appendix, Fig. S8B). The linker–ECL2 interaction serves to strengthen the otherwise unsupported peptide linker.
A second set is an intercalated stacking interaction among all three components (SI Appendix, Fig. S8C). This is mediated mostly by the linker residue Lys601, which is positioned between Trp590 of the CR domain and Ile761 of ECL2 for nonpolar interactions. Mutating any of these three residues to shorten or eliminate their side chains was harmful to signal propagation, as the receptor sensitivity toward Ca2+ is reduced equivalently in each case (SI Appendix, Fig. S8D). On the other hand, replacing neighboring residues in the CR domain (Asp587, Asp588) and ECL2 (Arg752, Gln754) with alanine had little effect on receptor function, suggesting these residues play a supportive role to assist signal transmission (SI Appendix, Fig. S8D).
We also examined the functional effect of introducing mutations at the apex of ECL2. We found that simultaneously replacing the two ECL2 residues at the tip with glycines (D758G/E759G) enacts only minor functional change in the receptor. This finding is consistent with the structural observation that the ECL2 apex primarily forms contacts with the linker and CR domain through main-chain atoms. Taken together, the combined interaction among the CR domain, linker, and ECL2 affords the junction sufficient rigidity to transform the conformational changes in the VFT into a rotation of the TM domain. This reorientation of the TM domain induces TM6 breakage for the formation of a stable active-state interface.
Active TM Homodimer Interface.
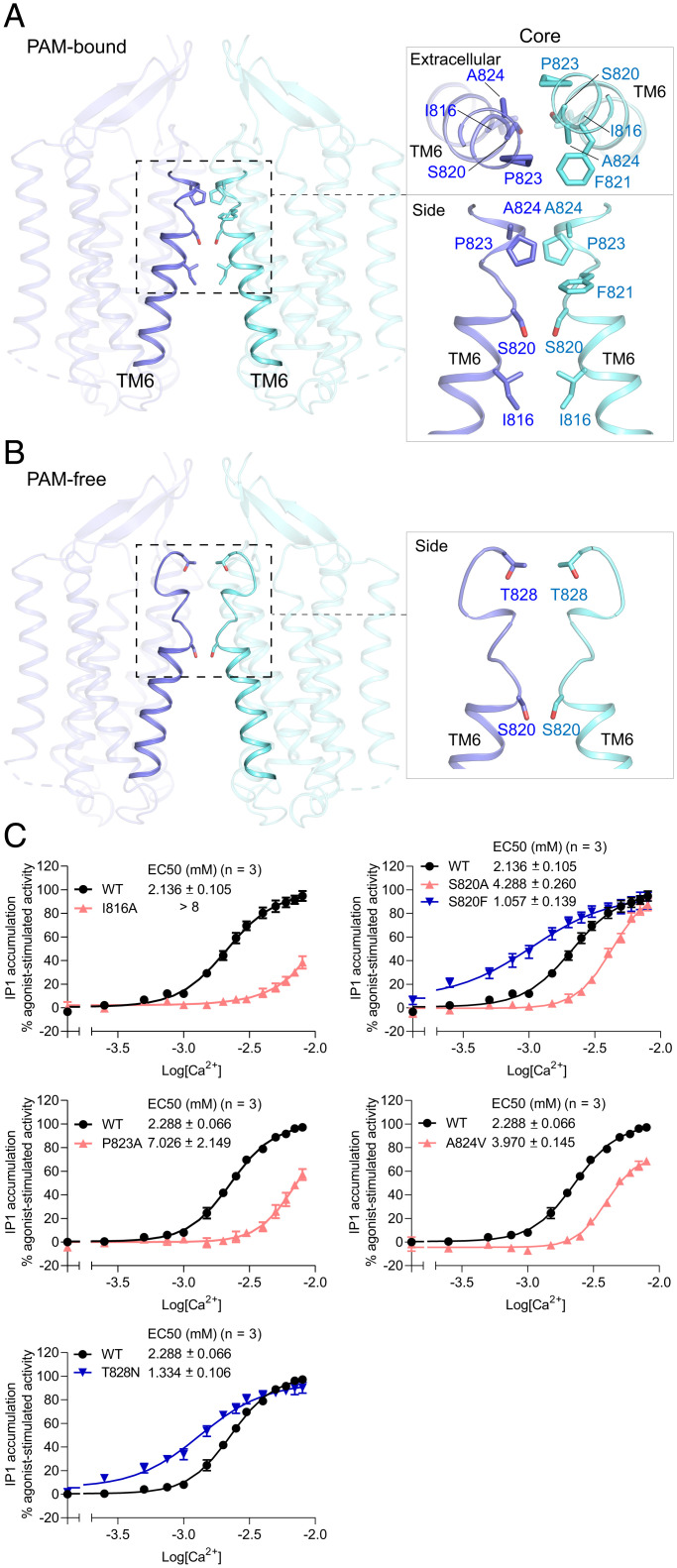
The homodimer interactions between CaS TM domains are most extensive in the presence of PAM, consistent with the role of PAM in stabilizing the active conformation (Fig. 3 A and B). In the active CaS receptor structures, direct homodimer contacts are formed by the extracellular part of TM6 from both subunits. The interacting residues are found on one side along the helical path of TM6 and include Ser8206.52 and Thr828 in the PAM-free state, but are expanded to include Ile8166.48, Phe8216.53, Pro8236.55, and Ala8246.56 in the PAM-bound form (Fig. 3A).
Fig. 3.
TM dimer interface of the active CaS receptor. (A and B) TM dimer interface formed by TM6 of both subunits in the PAM-bound (A) and PAM-free (B) states. Magnified panels depict direct contacting residues viewed from the extracellular end or the side. (C) Functional analysis of the TM dimer interface. Dose-dependent Ca2+-stimulated IP1 accumulation was measured in cells transiently expressing either WT or mutant CaS receptor bearing one of the selected substitutions at the core TM interface. Cell-surface expression levels of the I816A, S820A, S820F, P823A, A824V, and T828N mutants were 78, 71, 91, 100, 69, and 93% of WT level, respectively. Data points represent the average ± SEM of multiple experiments (n), each with triplicate measurements.
Among these interfacial residues, we have identified both loss-of-function and gain-of-function mutations that corroborate our structural model (Fig. 3C). Specifically, we found that replacing Ile8166.48 with an alanine (I816A) markedly reduced receptor sensitivity toward Ca2+ and the maximal receptor response upon Ca2+ stimulation. In addition, I816T and I816V are naturally occurring inactivating mutations that result in familial hypocalciuric hypercalcemia (FHH) (58). Similarly, S820A, P823A, and A824V are all inactivating disease mutations (58, 59) that are detrimental to receptor function. Our functional data are in agreement with the dimer interface configuration where these residues contribute to subunit association. Furthermore, Pro8236.55 may provide a vulnerable point along TM6 for helix disruption. The P823A mutant suffers substantial functional loss, possibly due to the reduced propensity for TM6 to break in the absence of a proline residue near its extracellular end.
On the other hand, S820F is an activating disease mutation that results in autosomal dominant hypocalcemia (6). The sharp increase in Ca2+ potency of the S820F mutant indicates more extensive contacts are formed by the phenylalanine residues, possibly through aromatic stacking interactions with neighboring Phe8216.53. Finally, although Thr828 of ECL3 only contributes to dimer interactions in the PAM-free state, substituting Thr828 with asparagine could facilitate new polar interactions with Ser8276.59. T828N is a known activating disease mutation that causes distinctly higher receptor sensitivity toward Ca2+ (6).
In addition to direct contacts, the CaS subunits also interact through cholesterol molecules (SI Appendix, Fig. S9A). Near both the extracellular and intracellular membrane surfaces, we found density at the dimer interface that we identified as cholesterols (CLR1, 2, 4, and 6), although CHSs are also possible (Fig. 1 A and B).
On the extracellular side, cholesterol-mediated interactions involve TM7 elements that surround the direct TM6 dimer interface (SI Appendix, Fig. S9A). Residues from TM6 such as Phe8216.53 and Pro8236.55 also participate in these peripheral dimer interactions, which are auxiliary to the formation of a TM6-centered homodimer interface.
On the intracellular side, homodimer interactions are almost exclusively mediated by cholesterol molecules (CLR4 and CLR6) as the TM6 helices are far apart in this region (SI Appendix, Fig. S9A). The only exception occurs midway along TM6, where the pair of Ile8166.48 side chains pack against each other and form indirect dimer contacts through cholesterols. Other TM6 residues that projected into the interfacial area include Phe8096.41, Leu8126.44, and Ile8136.45. We found that alanine substitution for each individual residue, as well as leucine substitution of Phe8096.41, showed a distinct reduction in Ca2+ potency and efficacy (SI Appendix, Fig. S9B). Among these, F809L is an inactivating disease mutation that causes FHH (6). These observations suggest that cholesterol may play an essential role in receptor activation by stabilizing the TM6–TM6 dimer interface. Indeed, we found that cholesterol depletion has deleterious effects on the overall functioning of the receptor, as it lowers receptor basal activity as well as the efficacy and potency of Ca2+ (SI Appendix, Fig. S9B). The CaS receptor has been previously identified in cholesterol-rich membranes in parathyroid cells (60).
PAM Binding.
In the PAM-bound structure of active CaS receptor, we found density for one R-568 molecule within the TM domain of each subunit. The PAM-binding site is located near the extracellular membrane surface and is framed by residues from the TM2, TM3, TM5, TM6, and TM7 helices. The PAM-binding pocket has a lateral opening between TM5 and TM6, through which the entire bound R-568 is accessible (Fig. 4 A and B).
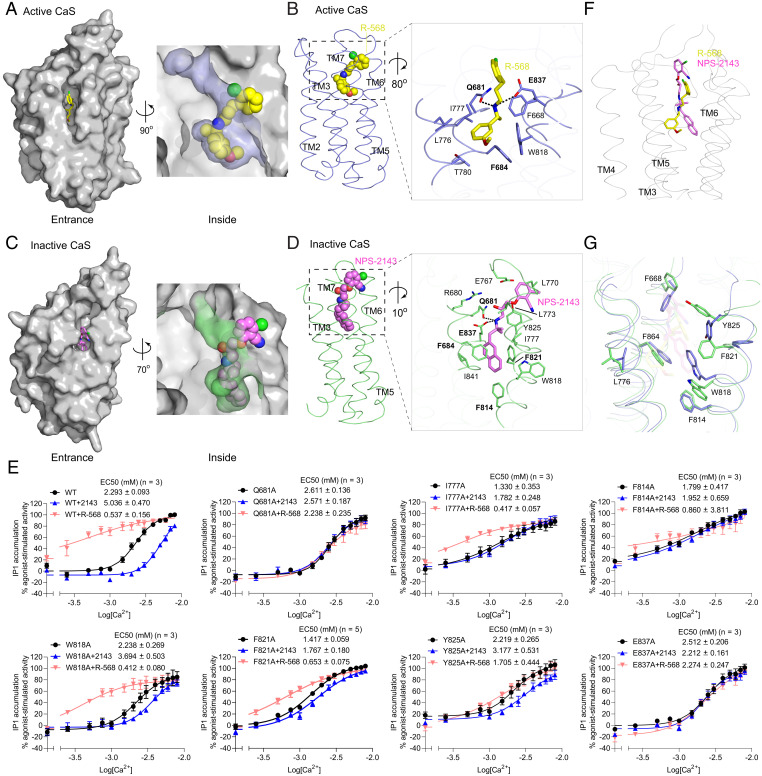
Fig. 4.
PAM- and NAM-binding sites of the CaS receptor. (A and C) Molecular surfaces of the active (A) and inactive (C) CaS TM domain, showing the binding pockets for the PAM R-568 (yellow) (A) and the NAM NPS-2143 (magenta) (C). (B and D) Binding modes of R-568 (B) and NPS-2143 (D), with the magnified views showing the specific contacts between the CaS receptor and each modulator. Hydrogen bonds are denoted by dotted lines. (E) Functional analysis of the modulator-binding sites. Dose-dependent Ca2+-stimulated IP1 accumulation was measured in cells transiently expressing either WT or mutant CaS receptor carrying one of the selected substitutions at the modulator-binding sites. The functional effect of each mutation was also assessed in the presence of 1 µM R-568 or 300 nM NPS-2143. Cell-surface expression levels of the Q681A, I777A, F814A, W818A, F821A, Y825A, and E837A mutants were 102, 89, 92, 89, 97, 96, and 53% of WT levels, respectively. Data points represent the average ± SEM of multiple experiments (n), each with triplicate measurements. (F) Comparison of R-568 and NPS-2143 binding poses. The Cα trace of the PAM-bound active CaS TM domain is shown in gray, and the superimposed R-568 (yellow) and NPS-2143 (magenta) are in stick models. The inactive CaS TM domain was overlapped onto the active form to bring their bound modulators into superposition. (G) Comparison of key residue conformations in the PAM- and NAM-binding sites of the CaS receptor. The TM domains of the active (blue) and inactive (green) CaS receptor are aligned.
The PAM R-568 structure consists of an aromatic ring on each end connected through a linker. When bound to the CaS receptor, R-568 adopts a crescent shape, which differs from the extended or bent conformation observed for the larger PAM molecules cinacalcet and evocalcet (27, 33) (SI Appendix, Fig. S10A). The methoxyphenyl group at one end of R-568 is anchored in the interior of the pocket while the chlorophenyl ring at the other end protrudes out toward the lateral opening. In addition, our observation that R-568 adopted a single conformation is evidence that specific PAM molecules can bind to the receptor through symmetric binding modes.
The linker of R-568 contains an amine group that is responsible for securing the modulator inside the central cavity of the TM bundle. The linker is bent by a ridge inside the pocket that is formed by Gln6813.33 of TM3 and Glu8377.32 of TM7. Both of these residues form hydrogen bonds with the amine group of R-568 to fasten the PAM molecule to the receptor (Fig. 4B and SI Appendix, Fig. S10B). Consistent with our structural observation, we found that an alanine mutation of either Gln6813.33 (Q681A) or Glu8377.32 (E837A) completely abolished the modulating effect of R-568 (Fig. 4E). Previous mutagenesis studies also indicate that Glu8377.32 is critical for the functional affinity of a variety of CaS modulators, including R-568, cinacalcet, and AC-265347 (29, 32). In addition, the linker of R-568 makes hydrophobic contact with Phe6682.56, a residue known to participate in the binding of phenylalkylamine modulators (29, 32).
The methoxyphenyl ring of R-568 occupies a pocket that is lined by the aromatic residues Phe6843.36 and Trp8186.50 on one side, and the aliphatic Leu7765.43 and Ile7775.44 on the other. Among these, Phe6843.36 is primarily responsible for anchoring the methoxyphenyl moiety through π-stacking interactions (Fig. 4B and SI Appendix, Fig. S10B). Extensive mutagenesis studies have established Phe6843.36 as an important binding residue of phenylalkylamine modulators including R-568 (29, 30, 32). The interactions between the methoxyphenyl group and other residues including Trp8186.50 are more distant, and mutations of these residues have minimal impact on CaS receptor function (32) (Fig. 4E). On the other hand, the chlorophenyl ring of R-568 is positioned at the entrance of the modulator-binding cavity. Its partially broken density suggests that the chlorophenyl end of the PAM molecule has a relatively flexible conformation (SI Appendix, Fig. S4A).
The overall binding mode of the PAM R-568 is compatible with the deformed TM6 helix conformation found in the active-state CaS receptor. Its methoxyphenyl group engages the interior-facing Trp8186.50. Its pose runs along the face of TM6 formed by Trp8186.50 and Tyr8256.57 side chains, holding these residues in place to maintain the TM6 kink. Furthermore, the presence of R-568 propels the extracellular end of TM6 toward the dimer interfacial area to increase subunit interactions (Fig. 2 D and F). Therefore, the PAM R-568 enhances agonist potency by stabilizing the TM6–TM6 dimer interface of the activated CaS receptor.
NAM Binding.
In the inactive-state structure of the CaS receptor, we observed partial density for an NPS-2143 molecule within the TM domain of each subunit (SI Appendix, Fig. S4C). The NAM-binding site is situated at a similar location as the PAM-binding site and is defined by residues from the TM3, TM5, TM6, and TM7 helices. The NAM site can be accessed both vertically from the extracellular milieu and laterally through the gap between TM5 and TM6 (Fig. 4 C and D). However, the lateral entrance to the site is partially sealed near the bottom so that the bound NPS-2143 molecule is not fully accessible.
NPS-2143 is also composed of two aromatic groups connected by a linker. When bound to the CaS receptor, the NPS-2143 molecule assumes an extended and relatively straight pose, with its naphthalene moiety buried inside the pocket and the chlorobenzonitrile group exposed outside but not visible in the density map (Fig. 4D and SI Appendix, Fig. S4C). The relatively open environment surrounding the chlorobenzonitrile moiety may have contributed to its flexibility. The disordered chlorobenzonitrile group has been shown to adopt a bent pose in the presence of l-Trp and Ca2+ in a recently reported CaS structure (27, 33).
The naphthalene moiety of NPS-2143 is anchored within a narrow pocket by a cluster of aromatic residues. It rests directly on top of Phe8146.46 and is sandwiched by two additional aromatic residues, Phe6843.36 and Phe8216.53, in π-stacking interactions (Fig. 4D and SI Appendix, Fig. S10C). While Phe6843.36 contributes to the binding affinity of phenylalkylamine modulators of the CaS receptor in general (32), both Phe8146.46 and Phe8216.53 are selectively involved in the recognition of the NAM NPS-2143. Introducing an alanine mutation at either Phe8146.46 (F814A) or Phe8216.53 (F821A) completely abolished the modulating effect of NPS-2143 (Fig. 4E). Other residues surrounding the naphthalene ring include Trp8186.50 and Ile8417.36, but their contacts are less extensive. Functional assessment of the W818A mutation demonstrated limited effect on the potency of NPS-2143 (32), consistent with its minor contribution to NAM binding (Fig. 4E).
The NAM NPS-2143 is also secured to the receptor by interactions involving its linker. As the linker extends toward the extracellular membrane surface, it traverses a path lined by a mixture of hydrophobic and polar residues. First, the amine group of NPS-2143 forms hydrogen bonds with both Gln6813.33 and Glu8377.32 to hold the NAM in place (Fig. 4D). Consistent with our structural observation, the modulating effect of NPS-2143 on receptor activity is completely abolished by the Q681A and E837A mutations (29, 32). Second, the hydroxyl group of NPS-2143 is within hydrogen-bonding distance of both Arg6803.32 and Gln6813.33. The mutation R680A has been demonstrated previously to substantially decrease NPS-2143 potency (32). Finally, the linker forms hydrophobic contacts with residues along its path, including Ile7775.44 and Phe8216.53. Individually replacing Phe8216.53 and Ile7775.44 with an alanine resulted in substantial loss of NPS-2143 efficacy (Fig. 4E). Although the chlorobenzonitrile ring and its neighboring phenoxy group are not visible in the density map, they are poised to interact with Tyr8256.57 and potentially Glu767 of ECL2 to make additional contributions to NAM binding affinity.
Binding of the NAM NPS-2143 stabilizes an intact TM6 conformation. It forms extensive interactions with an inward-facing Phe8216.53, while its naphthalene group precludes Trp8186.50 from adopting a rotamer conformation that occupies the core of the TM domain. Thus, NPS-2143 reduces agonist potency by strengthening TM6 helicity that is sterically incompatible with activation.
Discussion
Allosteric Modulation Mechanism.
Our structures revealed how allosteric modulators modify CaS receptor activity by stabilizing distinct functional states. The PAM R-568 and NAM NPS-2143 bind to overlapping sites within the TM domain of each CaS subunit. The modulator-binding positions are near the extracellular membrane surface and similar to the orthosteric ligand-binding site of class A and B GPCRs (61), the modulator-binding site of the class C receptor mGlu (48–50, 62, 63), as well as the phospholipid-binding sites of the GABAB receptor (54, 55). However, they are distinct from the PAM-binding site of the GABAB receptor, which is located at the dimer interface (52, 53, 56). CaS allosteric modulators share some common binding motifs with one another but also display distinct receptor interactions.
R-568 and NPS-2143 have related phenylalkylamine structures with a secondary amine group located inside the linker. This amine moiety is where the paths of the two modulators intersect when their respective TM domains are aligned (Fig. 4F). Each modulator forms hydrogen bonds with both Gln6813.33 and Glu8377.32 through its amine, demonstrating that it plays a critical role in securing both compounds within the TM bundle. The importance of Glu8377.32 for binding phenylalkylamine modulators has been emphasized previously (29, 32). In this study, we identified Gln6813.33 as an equal partner of Glu8377.32 for the general recognition of CaS modulators.
The PAM R-568 and NAM NPS-2143 compounds of the CaS receptor have opposing functional roles as a result of divergence in their receptor interactions. First, their binding sites exhibit multiple variations involving residues such as Phe6843.36 and Phe8146.46 in the TM core to accommodate the different size and shape of the modulators (Fig. 4G). NPS-2143 is longer and extends slightly deeper into the TM bundle than R-568 to engage Phe8146.46 directly. R-568 traverses the TM cavity diagonally in a pose that is slanted from the nearly vertical NPS-2143 molecule. As a result, the methoxyphenyl ring of R-568 is lodged further inside the core of the TM bundle than the naphthalene group of NPS-2143. The different binding positions of these end groups are partially accommodated by conformational changes in Phe6843.36.
Second, two rotamer switches involving Trp8186.50 and Phe8216.53 further determine the specificity of the bound modulator. The binding mode of R-568 leaves sufficient space for Trp8186.50 to adopt a TM core–facing rotamer. In contrast, the bulky naphthalene moiety of NPS-2143 would only be compatible with a Trp8186.50 conformer that is pointed away from the TM central cavity. In addition, the R-568 pose reinforces the TM6 kink with the pivoting residue Phe8216.53 directed toward the subunit interface. However, the naphthalene group of NPS-2143 forms extensive interaction with a TM core–facing Phe8216.53 to stabilize an uninterrupted TM6 helix. Consistent with the structural data, our mutational analysis showed that F821A selectively impacts NPS-2143–mediated down-regulation while it has little effect on R-568 function.
Taken together, the PAM R-568 enhances receptor activity by stabilizing a distortion of the TM6 helix and maximizing TM6-mediated homodimer interactions in the active conformation. On the other hand, the NAM NPS-2143 reinforces the integrity of the TM6 helix in the inactive state to diminish agonist-induced receptor response.
Symmetric Receptor Activation.
Recent active-state structures of the CaS receptor bound to the larger PAM molecules cinacalcet and evocalcet display marked asymmetry in the TM dimer arrangement and in the PAM pose. This is taken to suggest that G protein activation requires receptor asymmetry (33). Here we find within the limits of our resolution that the active receptor is symmetric in the TM region whether PAM-free or PAM-bound with the smaller R-568 compound. In particular, our PAM-free structure contains only native ligands, suggesting that the receptor is likely symmetric when activated under physiological conditions. In agreement with our findings, the completely full-length structures of human and G. gallus CaS receptor are symmetric regardless of activation state (26, 27). Conformational dynamics of the CaS receptor shows that it can fluctuate between symmetric and asymmetric orientations. However, such a shift might have been permitted by the detergent micelle environment that provides more freedom of movement than the cell membrane. Within a lipid bilayer, translation of one TM domain by about half a helix turn relative to the other as reported for the asymmetric CaS structures (33) would be energetically unfavorable, even taking membrane curvature into consideration. Systematic comparison of our inactive and active structures demonstrates that the critical development in activation-associated transmembrane reorientation arises from a helix-breaking event in TM6. This permits TM bundle rotation and direct apposition of neighboring TM6 helices at the dimer interface. The presence of TM symmetry in our inactive structure as well as in both our PAM-free and PAM-bound active structures leads to the conclusion that while asymmetry may be relevant to receptor activation in the presence of certain modulators, native ligands and other allosteric modifiers drive activation through a mechanism that does not require receptor asymmetry. Ultimately, a G protein–coupled CaS receptor complex structure is needed to determine whether a symmetric or asymmetric dimer is the relevant activated species.
Materials and Methods
The experimental procedures including protein expression and purification, cryo-EM sample preparation, data collection and image processing, model building and refinement, cell-based functional assay, as well as elemental analysis are described in detail in SI Appendix. Briefly, the human CaS receptor (residues 1 to 870) was expressed recombinantly in baculovirus-infected human embryonic kidney (HEK) 293 GnTI− cells (64). Active and inactive forms of the receptor were captured during protein purification using different ligands. The active receptor was stabilized with the CaS activators Ca2+ and TNCA, as well as the PAM R-568. The inactive receptor was obtained in the presence of the NAM NPS-2143.
Purified CaS receptor samples were vitrified and imaged on a Titan Krios transmission electron microscope. Cryo-EM data processing was performed using a combination of cryoSPARC (65) and Relion (66) software. The active CaS receptor dataset yielded two classes, corresponding to PAM-bound and PAM-free conformations, respectively. The inactive receptor reconstruction contained a bound NAM molecule within each subunit. Structural models were built in Coot (67) and refined in Phenix (68).
The function of wild-type (WT) and mutant CaS receptor was analyzed in HEK293 T/17 cells by measuring Ca2+-induced inositol phosphate (IP) accumulation. A homogeneous time-resolved fluorescence IP-One Terbium Kit was used to quantify the production of IP.
Elemental analysis of the purified CaS receptor was conducted using inductively coupled plasma spectrometry. The concentrations of magnesium, aluminum, calcium, manganese, iron, cobalt, nickel, copper, zinc, strontium, lead, cadmium, and barium in the protein samples were measured.
Supplementary Material
Acknowledgments
We thank Dr. Richard Henderson for early-stage cryo-EM investigation and critical reading of the manuscript; Dr. Charles S. Zuker for advice and financial support; Dr. Matthias Quick for access to a PHERAstar FS plate reader; Drs. Alexander Sobolevsky, Kei Saotome, and Erhu Cao for the gift of BacMam vectors; and Dr. Ed Twomey for advice. Y.Z.T. was sponsored by the National University of Singapore and Agency for Science, Technology and Research Singapore. Titan Krios data collection was performed at the Columbia University Cryo-Electron Microscopy Center, as well as at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy, supported by grants from the Simons Foundation (SF349247) and the NIH (GM103310). Detergent screening was performed at the Center on Membrane Protein Production and Analysis, supported by NIH grants (P41GM116799 and R01GM107462 to W.A.H.). This work was supported by NIH grants (R35GM141871 to Q.R.F. and R01GM29169 to J.F.).
Footnotes
Reviewers: E.I., University of California, Berkeley; and P.S., Monash University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115849118/-/DCSupplemental.
Data Availability
The cryo-EM density maps reported in this paper have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-25143 (36), EMD-25144 (37), and EMD-25145 (44). Atomic coordinates of the CaS receptor structures have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under ID codes 7SIL (38), 7SIM (39), and 7SIN (45). The raw micrographs used to calculate the density have been deposited in the Electron Microscopy Public Image Archive under accession numbers EMPIAR-10834 (35) and EMPIAR-10835 (43).
Change History
January 22, 2022: The byline has been updated.
References
- 1.Brown E. M., et al. , Cloning and characterization of an extracellular Ca(2+)-sensing receptor from bovine parathyroid. Nature 366, 575–580 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Hofer A. M., Brown E. M., Extracellular calcium sensing and signalling. Nat. Rev. Mol. Cell Biol. 4, 530–538 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Leach K., et al. , International Union of Basic and Clinical Pharmacology. CVIII. Calcium-sensing receptor nomenclature, pharmacology, and function. Pharmacol. Rev. 72, 558–604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannan F. M., Kallay E., Chang W., Brandi M. L., Thakker R. V., The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat. Rev. Endocrinol. 15, 33–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown E. M., Clinical lessons from the calcium-sensing receptor. Nat. Clin. Pract. Endocrinol. Metab. 3, 122–133 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Hendy G. N., Guarnieri V., Canaff L., Calcium-sensing receptor and associated diseases. Prog. Mol. Biol. Transl. Sci. 89, 31–95 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Ward B. K., Magno A. L., Walsh J. P., Ratajczak T., The role of the calcium-sensing receptor in human disease. Clin. Biochem. 45, 943–953 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Conigrave A. D., Ward D. T., Calcium-sensing receptor (CaSR): Pharmacological properties and signaling pathways. Best Pract. Res. Clin. Endocrinol. Metab. 27, 315–331 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Magno A. L., Ward B. K., Ratajczak T., The calcium-sensing receptor: A molecular perspective. Endocr. Rev. 32, 3–30 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Conigrave A. D., Quinn S. J., Brown E. M., L-amino acid sensing by the extracellular Ca2+-sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 4814–4819 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng Y., et al. , Structural mechanism of ligand activation in human calcium-sensing receptor. eLife 5, e13662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saidak Z., Brazier M., Kamel S., Mentaverri R., Agonists and allosteric modulators of the calcium-sensing receptor and their therapeutic applications. Mol. Pharmacol. 76, 1131–1144 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Nemeth E. F., Allosteric modulators of the extracellular calcium receptor. Drug Discov. Today Technol. 10, e277–e284 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Conn P. J., Christopoulos A., Lindsley C. W., Allosteric modulators of GPCRs: A novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 8, 41–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemeth E. F., Shoback D., Calcimimetic and calcilytic drugs for treating bone and mineral-related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 27, 373–384 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Urwyler S., Allosteric modulation of family C G-protein-coupled receptors: From molecular insights to therapeutic perspectives. Pharmacol. Rev. 63, 59–126 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Centeno P. P., et al. , Phosphate acts directly on the calcium-sensing receptor to stimulate parathyroid hormone secretion. Nat. Commun. 10, 4693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai M., Trivedi S., Brown E. M., Dimerization of the extracellular calcium-sensing receptor (CaR) on the cell surface of CaR-transfected HEK293 cells. J. Biol. Chem. 273, 23605–23610 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Pidasheva S., et al. , Calcium-sensing receptor dimerizes in the endoplasmic reticulum: Biochemical and biophysical characterization of CASR mutants retained intracellularly. Hum. Mol. Genet. 15, 2200–2209 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Pin J. P., et al. , The activation mechanism of class-C G-protein coupled receptors. Biol. Cell 96, 335–342 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Ray K., et al. , Identification of the cysteine residues in the amino-terminal extracellular domain of the human Ca(2+) receptor critical for dimerization. Implications for function of monomeric Ca(2+) receptor. J. Biol. Chem. 274, 27642–27650 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Ward D. T., Brown E. M., Harris H. W., Disulfide bonds in the extracellular calcium-polyvalent cation-sensing receptor correlate with dimer formation and its response to divalent cations in vitro. J. Biol. Chem. 273, 14476–14483 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z., Sun S., Quinn S. J., Brown E. M., Bai M., The extracellular calcium-sensing receptor dimerizes through multiple types of intermolecular interactions. J. Biol. Chem. 276, 5316–5322 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Zhang C., et al. , Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci. Adv. 2, e1600241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H., et al. , Illuminating the allosteric modulation of the calcium-sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 117, 21711–21722 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling S., et al. , Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca2+ ions and L-tryptophan. Cell Res. 31, 383–394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen T., et al. , Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor. Sci. Adv. 7, eabg1483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., et al. , Structural insights into the activation of human calcium-sensing receptor. eLife 10, e68578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen S. E., Gether U., Bräuner-Osborne H., Investigating the molecular mechanism of positive and negative allosteric modulators in the calcium-sensing receptor dimer. Sci. Rep. 7, 46355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josephs T. M., et al. , Negative allosteric modulators of the human calcium-sensing receptor bind to overlapping and distinct sites within the 7-transmembrane domain. Br. J. Pharmacol. 177, 1917–1930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller A. N., et al. , Identification of global and ligand-specific calcium sensing receptor activation mechanisms. Mol. Pharmacol. 93, 619–630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leach K., et al. , Towards a structural understanding of allosteric drugs at the human calcium-sensing receptor. Cell Res. 26, 574–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y., et al. , Asymmetric activation of the calcium-sensing receptor homodimer. Nature 595, 455–459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemeth E. F., et al. , Calcimimetics with potent and selective activity on the parathyroid calcium receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 4040–4045 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J., et al. , Structures of positive allosteric modulator-bound and unbound active human calcium-sensing receptor. Electron Microscopy Public Image Archive. https://www.ebi.ac.uk/empiar/EMPIAR-10834. Deposited 25 October 2021.
- 36.Park J., et al. , Structure of positive allosteric modulator-bound active human calcium-sensing receptor. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-25143. Deposited 15 October 2021.
- 37.Park J., et al. , Structure of positive allosteric modulator-free active human calcium-sensing receptor. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-25144. Deposited 15 October 2021.
- 38.Park J., et al. , Structure of positive allosteric modulator-bound active human calcium-sensing receptor. Protein Data Bank. https://www.rcsb.org/structure/7SIL. Deposited 15 October 2021.
- 39.Park J., et al. , Structure of positive allosteric modulator-free active human calcium-sensing receptor. Protein Data Bank. https://www.rcsb.org/structure/7SIM. Deposited 15 October 2021.
- 40.Nemeth E. F., et al. , Calcilytic compounds: Potent and selective Ca2+ receptor antagonists that stimulate secretion of parathyroid hormone. J. Pharmacol. Exp. Ther. 299, 323–331 (2001). [PubMed] [Google Scholar]
- 41.Quinn S. J., Bai M., Brown E. M., pH sensing by the calcium-sensing receptor. J. Biol. Chem. 279, 37241–37249 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Quinn S. J., et al. , Sodium and ionic strength sensing by the calcium receptor. J. Biol. Chem. 273, 19579–19586 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Park J., et al. , Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor. Electron Microscopy Public Image Archive. https://www.ebi.ac.uk/empiar/EMPIAR-10835. Deposited 25 October 2021.
- 44.Park J., et al. , Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-25145. Deposited 15 October 2021.
- 45.Park J., et al. , Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor. Protein Data Bank. https://www.rcsb.org/structure/7SIN. Deposited 15 October 2021.
- 46.Kidmose R. T., et al. , Namdinator—Automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kniazeff J., et al. , Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Koehl A., et al. , Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du J., et al. , Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594, 589–593 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Lin S., et al. , Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature 594, 583–588 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Seven A. B., et al. , G-protein activation by a metabotropic glutamate receptor. Nature 595, 450–454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y., Jeong E., Jeong J. H., Kim Y., Cho Y., Structural basis for activation of the heterodimeric GABAB receptor. J. Mol. Biol. 432, 5966–5984 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Mao C., et al. , Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 30, 564–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papasergi-Scott M. M., et al. , Structures of metabotropic GABAB receptor. Nature 584, 310–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J., et al. , Structure of human GABAB receptor in an inactive state. Nature 584, 304–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaye H., et al. , Structural basis of the activation of a metabotropic GABA receptor. Nature 584, 298–303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hilger D., et al. , Structural insights into differences in G protein activation by family A and family B GPCRs. Science 369, eaba3373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorvin C. M., Molecular and clinical insights from studies of calcium-sensing receptor mutations. J. Mol. Endocrinol. 63, R1–R16 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Gorvin C. M., et al. , Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias. Hum. Mol. Genet. 27, 3720–3733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kifor O., Diaz R., Butters R., Kifor I., Brown E. M., The calcium-sensing receptor is localized in caveolin-rich plasma membrane domains of bovine parathyroid cells. J. Biol. Chem. 273, 21708–21713 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Hilger D., Masureel M., Kobilka B. K., Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doré A. S., et al. , Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Wu H., et al. , Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves P. J., Callewaert N., Contreras R., Khorana H. G., Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps reported in this paper have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-25143 (36), EMD-25144 (37), and EMD-25145 (44). Atomic coordinates of the CaS receptor structures have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under ID codes 7SIL (38), 7SIM (39), and 7SIN (45). The raw micrographs used to calculate the density have been deposited in the Electron Microscopy Public Image Archive under accession numbers EMPIAR-10834 (35) and EMPIAR-10835 (43).