Significance
A central goal in invasion genomics is to identify and determine the mechanisms that underlie the successful colonization, establishment, and subsequent range expansion of invasive populations of nonindigenous species. Using a whole-genome approach, we evaluate the importance of genetic diversity for the successful establishment of nonindigenous species. Our study shows that genetic diversity per se is not the major factor driving invasions, since we observed all possible scenarios with invasive populations showing reduced, similar but also increased, genetic diversity relative to the native population. Using coalescent methods, we reconstruct the demographic history of the invasion and infer the source population of each invasion event, which shows that propagule pressure and multiple introductions play an important role in determining invasion success.
Keywords: nonindigenous species, global change, invasion dynamics, gelatinous zooplankton, Mnemiopsis leidyi
Abstract
Invasion rates have increased in the past 100 y irrespective of international conventions. What characterizes a successful invasion event? And how does genetic diversity translate into invasion success? Employing a whole-genome perspective using one of the most successful marine invasive species world-wide as a model, we resolve temporal invasion dynamics during independent invasion events in Eurasia. We reveal complex regionally independent invasion histories including cases of recurrent translocations, time-limited translocations, and stepping-stone range expansions with severe bottlenecks within the same species. Irrespective of these different invasion dynamics, which lead to contrasting patterns of genetic diversity, all nonindigenous populations are similarly successful. This illustrates that genetic diversity, per se, is not necessarily the driving force behind invasion success. Other factors such as propagule pressure and repeated introductions are an important contribution to facilitate successful invasions. This calls into question the dominant paradigm of the genetic paradox of invasions, i.e., the successful establishment of nonindigenous populations with low levels of genetic diversity.
Spurred by anthropogenic interference, ecosystems around the globe have become increasingly connected, allowing species to establish nonindigenous populations far from their native range (1, 2). While some of these nonindigenous species (NIS) may provide benefits (3), others can cause severe ecological or economic damage, with documented ecosystem impacts, making them invasive (4). Negative impacts range from habitat destruction and disease transmission to displacement or even extinction of native species (5, 6). Though invasive species are on the rise, increasing in both magnitude and extent (7), only a small fraction of NIS appear to form invasive populations (8, 9). Therefore, a major focus in invasion ecology has been on the identification and characterization of traits which contribute to successful invasions (10). For example, recent experimental and modeling results illustrate how selection for fecundity traits can favor population growth in invasive populations (11). However, the mechanisms by which ecological, genetic, and demographic factors interact to confer invasion success of NIS have yet to be fully understood, specifically patterns of genetic diversity during colonization, establishment, and subsequent range expansion.
Initially, populations of NIS are generally small and as a result inbreeding, reduced genetic diversity and lack of adaptation potential elevate NIS extinction risks (12–14). However, a variety of apparently successful invasive species, have led to the formulation of the genetic paradox of invasions (12, 15), which describes the successful establishment of NIS populations with low levels of genetic diversity (13). This hypothesis has recently been challenged (16, 17), with several studies of invasive populations that do not show reduced genetic diversity compared to their source populations (16, 18–20). In contrast, recent whole-genome data analyses have documented a severe reduction of genetic diversity during successful freshwater invasions of marbled crayfish in Madagascar (21). Hence, it remains highly debated how ecological, genetic, and demographic factors interact to confer invasion success, and specifically in the marine realm, how patterns and mechanisms affecting genetic diversity during NIS colonization, establishment, and expansion interplay.
Here we have used the warty comb jelly Mnemiopsis leidyi A. Agassiz, 1865 (Bolinopsidae, Lobata) as a marine invasion genomic model species. Native to the western Atlantic Ocean, it is considered one of the 100 most notorious invasive species (22), which successfully invaded different environments world-wide (23, 24). M. leidyi possess some characteristics of successful invasive species, including short generation times (11, 25), large reproductive capacity (26) and the ability to self-fertilize (27). Most importantly, M. leidyi is one of few invasive marine invertebrate species with a fully sequenced reference genome (28), making it an enticing target to develop the field of invasion genomics.
We conducted whole-genome resequencing of 72 individuals from five populations including two native and three invasive sites to 1) evaluate genetic differentiation and genetic diversity at the whole-genome level during different invasion scenarios and 2) use coalescent methods to reconstruct the invasion routes and past demographic changes in the context of the recent invasion events. We test the paradox of invasions hypothesis by comparing native versus invasive populations that represent spatially and temporally independent marine invasions. Our study closes an important gap in understanding how invasion success relates to genetic diversity and establishes M. leidyi as genomic model organism for invasion ecology.
Results
Genetic Differentiation of M. leidyi Populations at the Whole-Genome Level.
Whole-genome sequencing of 72 M. leidyi individuals from two native range sites in North America and three invasive locations in Europe was conducted (Fig. 1A and SI Appendix, Table S1). Mean sequencing depth per individual was 26.3× with a mean mapping rate of 87.9% (mean mapping quality of 42.8), leading to a set of 1,632,777 single nucleotide polymorphisms (SNPs) after quality filtering. No closely related individuals were sampled (pairwise kinship-coefficients <0.06).
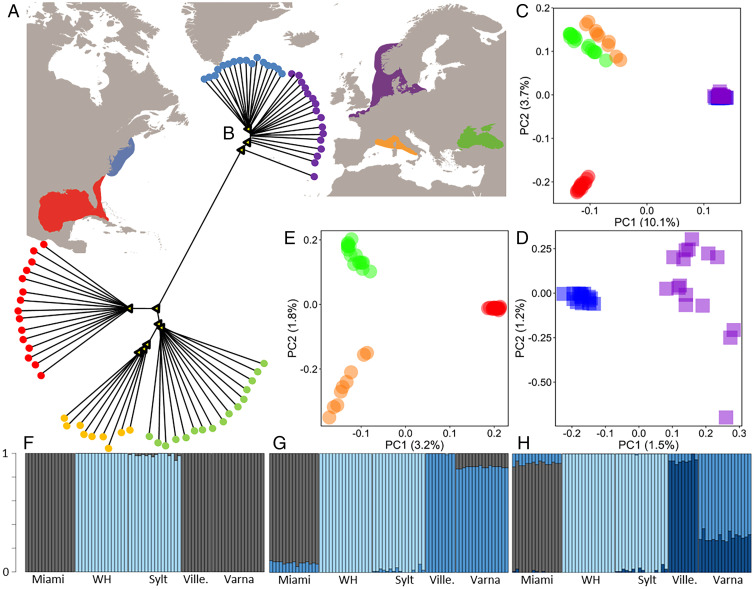
Fig. 1.
Sampling range (A) and population characteristics (B–H) of the comb jelly M. leidyi including native (USA: Miami, red; Woods Hole [WH], green) and invasive (western Eurasia: North Sea, Sylt, purple; Black Sea, Varna, orange; Western Mediterranean, Villefranche, blue) populations with (B) neighbor-joining dendrogram (yellow triangle nodes indicate >95% bootstrap support), principal coordinate analyses (C–E) for invasive and native populations (C), split into northern native (blue) and invasive (purple) (D), southern native (red) and invasive (green and orange) (E) populations and results of admixture analyses (F–H) with most likely scenario of two clusters (F) and alternative scenarios with k = 3 (G) and k = 4 (H).
We explored population structure using several independent methods. First, a neighbor-joining dendrogram clearly separated all southern from northern populations (Fig. 1B). Within the southern group, individuals clustered by sampling location and the native population (Florida, Miami) was more distant compared to the two invasive populations (Black Sea, Varna and Western Mediterranean, Villefranche). Within the northern group, individuals displayed distinctively shorter branch lengths and weaker separation by location compared to the southern invasive range. While all individuals from the native northern population (Woods Hole) clustered together, some individuals from the invasive population (Sylt) were genetically closer to the native population (Woods Hole) than to their invasive counterparts. In a second step, genetic differentiation was assessed by calculating the fixation index (FST) between population pairs (Table 1). Similarly, significant population differentiation was found between the two native populations in North America (Miami and Woods Hole; FST = 0.122). Comparing native versus invasive populations, high FST values were found comparing all southern populations: 0.041 between the native (Miami) and the invasive Black Sea (Varna) population, 0.062 between the native population (Miami) and the invasive Western Mediterranean (Villefranche) population and 0.03 between the two invasive southern populations. Low but significant genetic differentiation was found between the northern native and northern invasive populations (Woods Hole vs. Sylt, FST = 0.014). More restrictive filtering (minimum allele frequency MAF ≤ 0.05) resulted in similar population structure (Table 1).
Table 1.
Pairwise genetic differentiation (FST) between sampling locations
| Miami | Woods Hole | Varna | Villefranche | Sylt | |
| Miami | — | 0.122 | 0.041 | 0.062 | 0.118 |
| Woods Hole | 0.159 | — | 0.118 | 0.207 | 0.014 |
| Varna | 0.059 | 0.154 | — | 0.030 | 0.113 |
| Villefranche | 0.086 | 0.226 | 0.045 | — | 0.196 |
| Sylt | 0.151 | 0.016 | 0.146 | 0.210 | — |
All values tested significantly different from zero (P < 0.001).
Differentiation calculated using 1,632,777 SNPs after filtering for linkage disequilibrium and a 80% call rate (upper diagonal) and using a reduced SNP set of 832,323 SNPs with a minimum allele frequency MAF > 0.05 (lower diagonal).
In accordance with the observed genetic structure, principal component analysis supports the separation of the northern and southern populations (Fig. 1C). The northern native population and the northern invasive population clustered together tightly along both PC1 and PC2. The two invasive southern populations clustered closer to each other compared to the southern native population along PC2, whereas both the native and invasive southern population cluster closely along PC1 (Fig. 1C). Dedicated PCA analyses for northern (Fig. 1D) and southern (Fig. 1E) populations only, show clear geographic population separation. Interestingly, individuals from the northern invasive population (Sylt) display larger intrapopulation differences compared to individuals among the native northern population (Fig. 1D).
We then used STRUCTURE analyses to investigate the membership of each individual among k ancestral populations (k = 2 to k = 4; Fig. 1 F –H). When exploring the overall number of genetic groups present, STRUCTURE analyses suggested a scenario with two clusters (K = 2) as the most likely (see Fig. 1F), grouping both northern versus all southern populations. At k = 3, a further separation of the southern native versus the southern invasive populations was recovered (Fig. 1G), while at k = 4, additional population substructure components were observed among the Black Sea and the Western Mediterranean populations (Fig. 1H) in accordance with the PCA results.
Genetic Diversity at the Whole-Genome Level.
Genetic diversity was further assessed by analyzing nucleotide diversity π (SI Appendix, Table S2). In the native range, the southern population was characterized by a two-fold higher nucleotide diversity (Miami; π = 0.134) relative to the northern population (Woods Hole; π = 0.06). When comparing native versus invasive populations, the northern invasive population showed a 30% increase in nucleotide diversity (Sylt; π = 0.078) relative to Woods Hole, which was statistically significant (P = 0.036). For the southern populations, the invasive population in the Black Sea showed a slight decrease of 2.24% (Varna; π = 0.131) relative to Miami, but differences were not statistically significant (P = 0.718). Comparing the recent range expansion into the Western Mediterranean with the native southern population, an even more pronounced significant decrease of 23% was observed (Villefranche; π = 0.103, P < 0.001). All results were independent of sample size (SI Appendix, Table S2).
Testing Contrasting Colonization Scenarios.
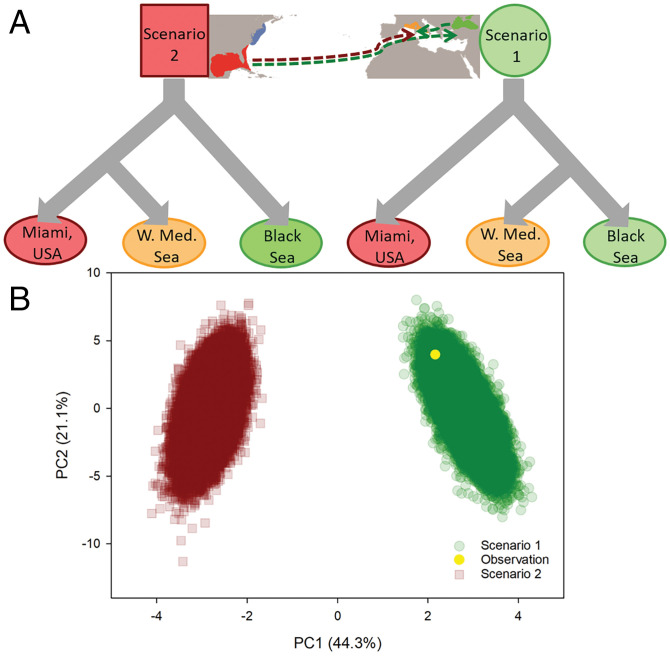
Two different model-testing approaches were then applied to test the likelihood of different invasion routes for M. leidyi into the Western Mediterranean Sea: 1) a stepping-stone model via the Black Sea vs. 2) a direct invasion from the native southern range. First, using a coalescent approach (Fastsimcoal2), a stepping-stone model was preferred over a model with a direct secondary transatlantic invasion into the Western Mediterranean (Fig. 2 and SI Appendix, Table S3). The preferred model suggested current effective population sizes (Ne) of 11,271 (9,323 to 12,043) for the southern native population, 11,606 (10,093 to 13,405) for invasive Black Sea and 844 (773 to 943) for Western Mediterranean population with an estimated mutation rate of 7.6 × 10−8 per nucleotide per generation. When checking for consistency of the estimated mutation rate, a similar mutation rate of 6.1 × 10−8 was obtained for the North Sea invasion (SI Appendix, Table S3). We are aware that these mutation rates are high and further studies are needed to substantiate if ctenophores are characterized by exceptionally high mutation rates, as suggested for mtDNA in ctenophores (29). Second, we compared the two invasion routes using an Approximate Bayesian Computation (ABC) modeling approach. Similarly, the ABC model favored the stepping-stone range expansion model, with comparable effective population size estimates (Fig. 3 and SI Appendix, Table S3).
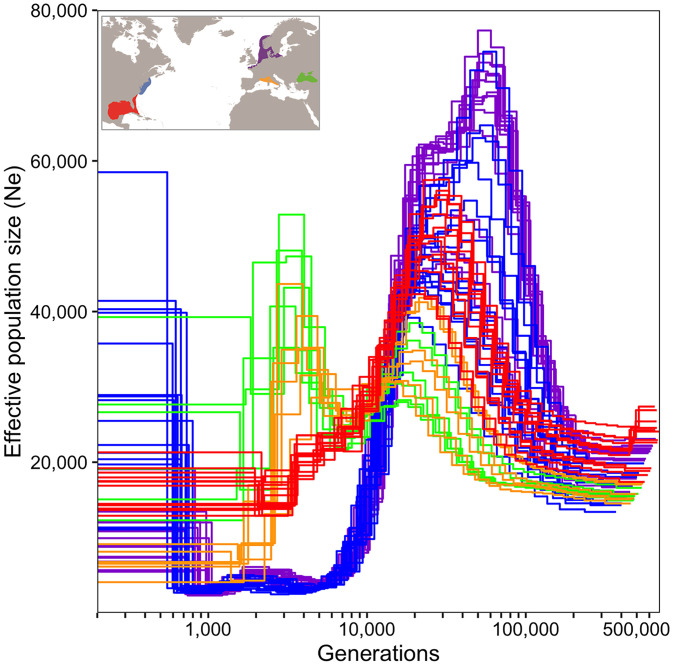
Fig. 2.
Demographic history of M. leidyi for invasive and native populations. PSMC model with estimates of the demographic changes (Ne = effective population size) over time (generations, log scale) for all populations including the native southern (Miami, red, n = 15) and northern (Woods Hole, blue, n = 16) populations as well as the invasive southern (Black Sea, Varna, green, n = 16; Western Mediterranean, Villefranche, orange, n = 9) and northern (North Sea, Sylt, purple, n = 16) populations.
Fig. 3.
Invasion scenario testing of the recent southern expansion. Results from ABC model analyses with (A) general scenario explanation of invasion pathways of the Western Mediterranean Sea via 1) a stepping-stone range expansion from the Black Sea (Scenario 1, green circle) or 2) a direct reintroduction from its native range in the United States (Scenario 2, red squares) and (B) ABC model results for both scenarios where direct observation (yellow point) clearly indicates Scenario 1, with stepping-stone invasion from the Black Sea as the most likely explanation.
Reconstruction of the Demographic History of M. leidyi.
The demographic history of our genomic model species was inferred using the Pairwise Sequentially Markovian Coalescent (PSMC) method. The PSMC plots clearly show that all five populations share ancestry and demographic history for most of the investigated time period, as they converge into a single ancestral population (Fig. 2). At around 200,000 generations ago, the ancestral population showed a protracted period of demographic expansion, likely reflecting an increase in its geographic distribution in the native range. After this period of expansion, the Ne curves of the northern and southern native populations diverged, into two genetically distinct populations in the native range (Fig. 2).
In regard to the invasion events, the Ne curve for the southern invasive Black Sea (Varna) population converged with the Ne curve for the native Florida population (Miami), indicating that the Black Sea population originated from Florida or a closely related southern lineage population (Fig. 2). A large increase in Ne was observed when the curve for the Black Sea splits from Miami, suggestive of a relatively large inoculum during the invasion. The Ne curve for the Western Mediterranean population (Villefranche) was identical to the curve for the Black Sea population up until very recent times, indicating their origin from the Black Sea (Fig. 2). The drop in Ne of Villefranche where the two curves diverge suggests the invasion of the Western Mediterranean Sea by few founder individuals and a large bottleneck. On the other hand, when looking at the Ne curve of the northern invasive population (Fig. 2), it shows a similar pattern to the native northern population, indicating that Sylt originated from Woods Hole or a closely related population from the northern lineage. In recent times, a small population size increase in the northern invasive population is observed, likely indicating the invasions into the North Sea. While timing of events and absolute Ne values are dependent on the mutation rate used (and hence these estimates should be taken with caution) we can be certain of the origin of the invasions and that the Black Sea invasion was proportionally much larger compared the North Sea invasion.
Discussion
The genetic paradox of invasions (15) describes the successful establishment of invasive populations with low levels of genetic diversity. However, the general applicability of the paradox itself has also been challenged (16, 17), with several reports of invasive populations that do not show reduced genetic diversity as compared to their source populations (19) or even showing increased diversity due to admixture from diverse source regions (18). Importantly, the mechanisms by which low genetic diversity due to founder effects in the invasive range can be circumvented are highly controversial (16, 20). The lack of evidence for strong founder events accompanying marine invasions is possibly driven by high propagule loads (20), multiple introductions (16), admixture from distant source populations (18) or a combination thereof. Furthermore, recurrent introductions can introduce additional genetic diversity over time. However, the diverse patterns of successful invasive populations may also stem from methodological shortcomings. We highlight the value of whole-genome data to gain a full representation of the genomic diversity that is not affected by ascertainment bias via selection of a few representative marker loci. By investigating genetic diversity patterns in contrasting invasion scenarios across western Eurasia at the whole-genome scale and using one of the 100 most successful invasive species world-wide (22), we address the genetic paradox of invasions and discuss our findings in the context of the specific invasion scenarios.
First Invasion Wave: The Black Sea Invasion.
M. leidyi was first recognized as an invasive species in the Black Sea during the 1980s, where it led to a significant ecosystem restructuring (30). During the following four decades, M. leidyi spread through vast areas of southwestern Eurasia (24). Our genome-wide study confirms that the invasive Black Sea population originated from the southern Gulf of Mexico/Florida peninsula (31–34). Given the time period of four decades since this initial invasion event, the substantial genetic distance between native and invasive southern populations observed here can be explained by allopatric divergence in the absence of gene flow. On the other hand, similar levels of genetic diversity were found when comparing the southern native and invasive (Black Sea) populations. These results are in stark contrast to previous studies using a small number of genetic markers, which reported a significant decrease in genetic diversity of M. leidyi in the Black Sea (31–34). Considering the high genetic differentiation observed between southern native and invasive populations and the high nucleotide diversity found, this suggests a single or time-limited invasion event constituted by a significant amount of individuals that transferred the bulk of the genetic diversity of the native population to the Black Sea. This interpretation is further supported by the PSMC plots, which show a large increase in population size for the invasive Black Sea population, coinciding with the time of the first M. leidyi sightings. Such high propagule pressure could be the result of large vessel ballast water discharge containing M. leidyi, especially from oil tankers, which need substantial ballast water during return journeys for stabilization (35). In accordance, the first records of M. leidyi in the Black Sea date back to the early 1980s (23, 24, 36), a period characterized by high shipping traffic between Cuba and the U.S.S.R.’s major Black Sea ports (23, 36), which are in close proximity to M. leidyís first sighting site (36). However, shipping traffic came to an abrupt end in the late 1990s, just before the collapse of the Soviet Union, which could explain the current lack of gene flow and continued divergence between the two populations. Our demographic reconstruction using PSMC shows a rapid expansion of the Black Sea population shortly after its establishment, which can be attributed to the initial lack of predators (36) and in agreement with large blooms observed before the arrival of the nonindigenous predator Beroe ovata (36) followed by a drastic drop in the effective population size.
A large number of initial founders, such as inferred from our PSMC analyses for the Black Sea, is expected to be important for avoiding random drift, inbreeding and provides sufficient standing genetic variation upon which selection can act (37). Also, this increases the likelihood of introducing genotypes preadapted to the new environment (20, 38). This seems particularly relevant for the Black Sea invasion, as native and invasive distribution ranges show contrasting environmental conditions. In native southern areas around Florida, surface salinities over 33‰ are characteristic (39), compared to 18‰ in the western Black Sea (40). While adults of M. leidyi are tolerant to a wide range of salinity values, egg production rates are severely affected by salinity due to reduced fertility (41). Thus, the presence of substantial standing genetic variation in introduced populations can be critical to NIS adaptation to new conditions present in the invasive habitat and determine secondary range expansions into adjacent areas.
Secondary Range Expansion into the Western Mediterranean.
After the establishment of M. leidyi in the Black Sea, it was first recorded in the Western Mediterranean in 2005, with blooms observed along the coasts of Italy, France, and Spain during 2009 (24). The origin of the Western Mediterranean M. leidyi population has remained controversial, either stemming from a secondary introduction from the Black Sea (31, 32, 34) or representing a new direct introduction from its native range (33). By using full genome resequencing data, we recover the origin of the Western Mediterranean invasion as secondary range expansion from the Black Sea.
This secondary range expansion is characterized by a significant reduction in genetic diversity and fits the classical view that invasive populations are often associated with a genetic bottleneck. This result is also supported by the inferred demographic history using coalescent methods, which suggest a small effective population size for the Western Mediterranean population, much lower than the native Florida and the invasive Black Sea populations. Both independent coalescence-based methods lead to the same result of secondary range expansion into the Western Mediterranean by the Black Sea population. Connectivity via ocean currents could be a possible vector seeding animals from the Black Sea via surface water outflow through the Sea of Marmara and Aegean Sea (24). However, the current direction is dominated by an eastward flow for the central Mediterranean Sea (24), hampering direct drift dispersal from the Black Sea to the Western Mediterranean. However, this eastward current is characterized by very low current stability estimates along the continental coast, which could explain the long lag-time (24) of over three decades and the severe genetic bottleneck observed in our study, possibly due to current mediated drift of animals. Irrespective of the mechanisms, we conclude that M. leidyi has successfully invaded the Western Mediterranean Sea, despite a significantly decreased genetic diversity, which supports the genetic paradox of invasions paradigm.
Repeated Invasions into Northern Europe.
Our results confirm that M. leidyi populations in northern Europe originated from the northern native population in the NE coast of North America. We found low but significant levels of genetic differentiation between the native and invasive populations. Notably, within the invasive population, we observe a 30% increase in genetic diversity. This allows us to resolve earlier analyses of genetic diversity in northern European populations that have used small numbers of molecular markers (34, 42). Increased genomic diversity in the invasive range is often accounted for by multiple introductions from distinct source regions leading to admixture of naturally isolated populations (18, 19, 43). However, low genetic differentiation based on pairwise fixation indices, lack of structure and lack of introgression found here suggest source matching without admixture from other isolated populations. Compared to the southern system, the northern invasion appears to be younger but given M. leidyi’s short generation time (11, 25), an initial small founding population should have been expected to diverge from the native population during the past 10 y/250 generations. Specifically, low genetic differentiation coupled with an increase in nucleotide diversity suggests multiple transfers of a few individuals over an extended period of time from slightly different areas within the native range. In accordance, our PSMC results suggest that the northern invasion was not an event of the same magnitude as the invasion of the Black Sea from the Southern native population. Due to the presence of some of the largest ports of the world with vigorous trade activity connected to the NE coast of the United States, Northern Europe is susceptible to repeated invasions of planktonic marine organisms via ballast water discharge (35, 44). Importantly, those Northern European ports are also characterized by very high connectivity with large areas of NW Europe via ocean currents, making recurrent reseedlings of nonnative species very likely (24).
We show that all scenarios with increase, decrease and maintenance of genetic diversity can occur within the same nonindigenous species, and that all three scenarios can lead to the successful establishment forming invasive populations. This contradicts the general applicability of the genetic paradox of invasions. Specifically, we find 1) a single invasion by a large number of propagules from the southern native population of M. leidyi so that most of the genetic diversity was retained in the Black Sea, 2) repeated introductions from the northern native population of M. leidyi by a small number of propagules leading to an increased genetic diversity in the North Sea, and finally 3) a secondary stepping-stone invasion by a small number of propagules from the invasive Black Sea population that accounted for the significant loss in genetic diversity in the Western Mediterranean. Despite the different modes of invasion events that accounted for the origins of these three independent invasion events in western Eurasia, all populations appear to be equally successful and are regarded as invasive with tangible impacts at the ecosystem level. Thus our results suggest that genetic diversity per se is not the major driving force behind invasions. Propagule pressure and multiple introductions also play an important role in determining invasion success.
As perspective of relevance for management actions, our whole-genome data highlight that in the case of northern Europe, despite international conventions in place such as UNEP’s Barcelona Convention, Convention on Biological Diversity or the IMO Ballast Water Management convention to halt species introductions, continued seeding of M. leidyi from the native range has persisted and is likely associated with repeated ballast water introductions from large transatlantic shipping vessels. This suggests that other marine invaders could experience similar temporal invasion dynamics as outlined here, which is of importance to understanding NIS establishment success, NIS range expansion and the potential for new invasions of nonnative species in Northern Europe. Future studies could consider impacts of selection along native environmental gradients (e.g., ref. 45) or use molecular informed niche modeling (46) to understand fine scale population differences, but our results demonstrate that whole-genome sequencing methods (invasion genomics) are powerful to describe the invasion dynamics and detect recurrent invasions. This may allow for the relatively rapid assessment of NIS and the development of management policies, for example, directed at regulation of ballast water release in areas associated with repeated NIS invasions. The North Sea is particularly vulnerable to large input of species from other regions (44) and will benefit from management actions aimed at restricting NIS introductions and their potential subsequent successful establishment. Invasion genomic tools outlined here will allow for detecting risk areas and are thereby important to halt species introductions in the marine environment.
Materials and Methods
Study Organism and Invasion History.
The warty comb jelly M. leidyi, native to the Eastern Americas, was first sighted outside its native range in the Black Sea during the early 1980s (23, 24, 36). From there it expanded into the Sea of Azov, the Sea of Marmara, the Eastern Mediterranean and Caspian Sea and most lately, the Western Mediterranean Sea (24). In a second invasion wave, it was recorded in Northern Europe and expanded from the North Sea (2005) into the central Baltic Sea and was recorded all along the western Norwegian coast until 63.4°N (24). Previous genetic studies, using few markers (COI, CYT-B, microsatellites), confirmed two source regions for the southern and northern invasions, respectively (31–34, 47). The southern invasion had been shown to originate from the Gulf of Mexico area, whereas the northern invasion was sourced from the NE coast of the United States (31–34, 47). So far, the origin of the Western Mediterranean invasion was less certain and direct colonization from the native range (33) as well as a stepping-stone invasion from the Black Sea were proposed (32). The species possesses some of the common characteristics of successful invasive species, including short generation times (11, 25), high growth rates (11), large reproductive capacity (26), high predation rate (48) and wide environmental tolerance (23).
Sampling.
Sampling sites were chosen based on major population structures of M. leidyi (34). A total of 72 M. leidyi individuals were collected from five distinct geographic locations: two in the native range on the East coast of the United States, Florida, Miami (n = 15) and Woods Hole (n = 16) in New England; as well as three sites across the invasive range with Varna (n = 16) in the Western Black Sea, Villefranche-sur-Mer (n = 9) in the Western Mediterranean Sea and Sylt (n = 16) in the North Sea (Fig. 1 and SI Appendix, Table S1, Figs. S1, S2). All specimens were collected during June/July 2016 in surface waters, either from shore, diving/snorkeling or from small vessels operated nearshore. Specimens were preserved individually, frozen below −40 °C or lyophilized until further processing. For lyophilizations, live specimens were briefly rinsed in freshwater to minimize salt carryover, transferred to an airtight container, prefrozen at −80 °C for at least 2 h and placed in the freeze dryer for 48 h.
DNA Extraction and Resequencing.
High-molecular-weight genomic DNA was extracted from ctenophore tissue using a modified CTAB/Chloroform protocol (49), with subsequent quality control via electrophoresis on a 0.8% agarose gel. Whole-genome resequencing was conducted at the Max-Planck Institute for Evolutionary Biology in Plön (Germany). Individually indexed, paired-end libraries with an insert size of 550 bp were constructed using the Illumina TruSeq Nano DNA HT Library Preparation Kit (Illumina). An Agilent 2100 Bioanalyzer System (Agilent Technologies) was used to perform quality control of the libraries. Libraries were sequenced in 2 × 150 bp paired-end configuration on the Illumina NextSEq. 500 System (Illumina). The Bcl to FastQ conversion was performed using BCL2FASTQ version 2.17.1.4 (Illumina).
Data Filtering and SNP Calling.
Read quality was initially assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Raw sequence data were trimmed using Trimmomatic v0.36 (50), with a minimum Phred score of 15. After removing adapter sequences and low quality bases, paired and unpaired reads were aligned to the M. leidyi reference genome (28) using the mem algorithm in BWA v0.7.15 (51). A trimmed version of the genome was used only containing scaffolds longer than 10 kb, which reduced the number of scaffolds from 5,100 to 1,254 while retaining 94.9% of bases. Sequence alignments were preprocessed using Picard v2.6.0 (Broad Institute 2018 https://broadinstitute.github.io/picard/) which included merging (MergeSamFiles), coordinate sorting (SortSam) and identifying/removing duplicate reads arising during library amplification (MarkDuplicates). Subsequently, variant calling was performed with the mpileup and call commands in BCFtools v1.9 (52). We excluded sites for which read depth was less than 10 or more than 100, sites for which quality was below 20 and sites in proximity to indels. Finally, variants were filtered using vcftools v0.1.14 (53) so that only biallelic SNPs with a Phred quality score above 13 were retained. Furthermore, only loci genotyped in more than 80% of individuals were retained. For some of the analyses, an additional minor allele frequency (MAF) filter was implemented, removing sites with MAF below 0.05.
Genetic Diversity and Population Structure.
Genetic diversity and population structure analyses were conducted using filtered SNP datasets following linkage disequilibrium-based variant pruning implemented in PLINK version 1.90b3.42 (54), which produces a pruned subset of markers in approximate linkage equilibrium. The indep-pairwise option was used with a window size in variant count of 100, a variant count of 5 to switch window at the end of each step and a r2 threshold of 0.5. Prior to the analysis, PCAngsd version 0.95 (55) was used to detect potentially related individuals based on the kinship matrix. Levels of genetic diversity across populations were assessed using Nei and Li (56) nucleotide diversity (π) calculated using vcftools v0.1.14 (53). π was calculated for each population using the entire dataset with all samples and additionally with a resized dataset in which all populations were randomly subsampled to the largest common sample size of n = 9. SEs and confidence intervals for π were obtained by performing a bootstrap function with 1,000 replicates using the boot package in R.
Standardized genetic differentiation statistics between population pairs were calculated using vcftools version 0.1.14 (53), in accordance with Weir and Cockerham (57). All genotype data were used to conduct a Principal Component Analysis (PCA) in order to visualize population structure using smartPCA from the Eigensoft package (58). Population structure was visualized by means of a neighbor-joining dendrogram constructed using the BIONJ algorithm (59). Bootstrap support was calculated with the aboot function in the software package poppr v2.3.0 (60). In order to assess the significance of pairwise FST values, we carried out bootstrapping over loci to generate a CI around the observed FST. P values were calculated using a one-sample T-test for each pairwise FST. Population structure was further investigated using the Bayesian assignment approach implemented in STRUCTURE (61), a model based clustering algorithm that infers the most likely number of groups (K) in the data. The analysis was performed with K = 1 to 5, assuming an admixture model, correlated allele frequencies and without population priors. A burn-in of 10,000 steps followed by 100,000 additional Markov Chain Monte Carlo iterations were performed. For each K, 10 independent runs were conducted to check the consistency of results. The most likely K was inferred using the method of Evanno et al. (62), which measures the steepest increase of the ad hoc statistic ΔK based on the rate of change in the log probability of data between successive K values.
Testing Alternative Invasion Routes.
We used two different model-testing approaches to identify the most likely invasion routes of M. leidyi into western Eurasia. In particular, we tested whether the invasion occurred following 1) a stepping-stone scenario where M. leidyi was first introduced from the native southern range to the Black Sea and subsequently expanded into the Western Mediterranean versus 2) a scenario in which the Western Mediterranean population was directly seeded from the native southern population. First, the two scenarios were simulated and compared using Fastsimcoal2 (63), which infers demographic parameters from the site frequency spectrum of the populations using a coalescent simulation framework. The joint allele frequency spectra (JAFS) for all populations were directly generated from the final variant calls using the scripts in https://github.com/shenglin-liu/vcf2sfs. Wide priors for population sizes were used while mutation rates were constrained to vary between 1 × 10−7 – 1 × 10−9. As calibration points, we used the time since the first reported observations of substantial amounts of animals in the invasive range (1985 for the Black Sea and 2009 for the Western Mediterranean) and a generation time of two weeks (25), based on laboratory experiments. For each demographic model, 100 independent estimations with different initial parameter values were run. Results for the estimation with the highest likelihood were reported. The best fitting demographic model was identified using Akaike's Information Criterion (AIC) score (64). In order to check for consistency of the mutation rate obtained, we also ran Fastsimcoal2 using the North Sea invasion data (SI Appendix, Table S3). Second, we applied an ABC approach implemented in DIYABC version 2.1 (65) to evaluate the relative likelihood of the two invasion scenarios described above. The analysis was conducted using a subset of 960 informative SNPs after filtering for a 100% call rate and only selecting one SNP per scaffold. Estimates of contemporary effective population sizes, derived from PSMC (see below), and divergence times (same as historical events in Fastsimcoal2) were used to parameterize the models. A total of 1 million data sets were simulated for each invasion scenario. The similarity between observed and simulated data sets was assessed using the following statistics: mean genetic diversity, FST and Nei’s genetic distance between sample pairs. Model validity was checked by conducting a PCA of the summary statistics for all simulated data sets under the contrasting scenarios to confirm if the observed data overlapped with the distribution of simulated data sets. The 10,000 (1%) simulated data sets with the best fit to the observed data were used to obtain posterior distributions of the model parameters.
Demographic Analyses Using PSMC.
Coalescent methods can be applied to infer past population dynamics and demographic changes over large timescales with high resolution. The demographic history of M. leidyi was inferred using the PSMC method (66), which can extract demographic information about a given population/species from the distribution of heterozygous sites across the genome. For each individual, a consensus sequence was generated using BCFtools v1.9 (52), following the approach of Nadachowska-Brzyska et al. (67). First, variants were called with the mpileup and call commands in BCFtools using the M. leidyi genome as reference. Same filtering criteria as outlined above were applied. Additionally, mitochondrial regions were excluded. Subsequently, the consensus command in BCFtools was used to incorporate all variants to create a consensus sequence for each individual. The resulting vcf file was converted to the psmc input format with vcfutils.pl (distributed with BCFtools). Next, the consensus sequence was divided into nonoverlapping 10 bp bins, with bins scored as either heterozygous if there was at least one heterozygote nucleotide position in the bin or homozygous. PSMC v0.6.5 (66) was run with the total number of iterations set to 25, T max (-t) set to 15, initial mutation/recombination ratio (-r) set to 5 and time bin parameter (-p) set to “4 + 25*2 + 4+6”.
For parameter conversion we used a generation time of 2 wk (25) and an average mutation rate of 6.85 × 10−8 per nucleotide per generation obtained from analyses of the northern and southern invasion events, respectively (Fastsimcoal2 results see SI Appendix, Table S3). The rate was estimated using invasion data as we were interested in examining relatively recent divergent times (68). However, since we conducted the PSMC analyses using resequencing data, which is not as informative as high-coverage sequencing methods (67), we use the PSMC plots to infer the origin of the different invasions and the relative size of the invasions but the timing of the population splits can only be properly estimated with high-coverage sequencing data (i.e., de novo genome sequencing). Besides knowledge of the mutation rate, the main underlying assumptions of PSMC analyses are: 1) populations are panmictic and there is no substructuring within populations and 2) sequences are evolving neutrally. PSMC can be sensitive to violations of the underlying assumptions (69). High population structure could result in distorted effective population size (Ne) changes over time, while natural selection could lead to an observed drop in Ne (69). As invasive populations are likely not in equilibrium during part of their invasion history and might be impacted by nonrandom mating, inbreeding due to founder effects and natural selection, this can affect estimates of absolute coalescence times and Ne estimates (69, 70). In this study, no genetic differences were found within populations and all populations were panmictic without signs of inbreeding. The potential impact of selection should be considered as some of the observed drops in Ne might be partially due to selective effects. Combining different approaches and bearing model assumptions and uncertainties about absolute face values in mind, coalescent methods provide a powerful tool of inferring demographic histories from genomic data (70), also for invasion biology.
Supplementary Material
Acknowledgments
This study was financed by VILLUM FONDEN, Denmark with Grant ID 25512 (to C.J.), the Danish Council for Independent Research and the European Commission—Marie-Curie Program with the DFF-MOBILEX mobility Grant No. DFF-1325-00102B (to C.J.), the CRC1182 Origin and Function of Metaorganisms and by Cluster of Excellence 80 “The Future Ocean,” which is funded within the framework of the Excellence Initiative by the Deutsche Forschungsgemeinschaft (DFG) on behalf of the German federal and state governments with Grant ID CP1539 (to C.J., co-PI T.B.H.R., to support M.T.L.). F.L. is supported by the Institut Universitaire de France. We thank Oscar Pueblo and David Duchêne for discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116211118/-/DCSupplemental.
Data Availability
All sequencing data have been deposited on GenBank under the NCBI BioProject accession number: PRJNA689379 (71). All code is provided and available at Zenodo under DOI: 10.5281/zenodo.5668363.
References
- 1.Cariton J. T., Geller J. B., Ecological roulette: The global transport of nonindigenous marine organisms. Science 261, 78–82 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Wonham M. J., et al. , Going to the source: Role of the invasion pathway in determining potential invaders. Mar. Ecol. Prog. Ser. 215, 1–12 (2001). [Google Scholar]
- 3.Schlaepfer M. A., Do non-native species contribute to biodiversity? PLoS Biol. 16, e2005568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simberloff D., et al. , Impacts of biological invasions: What’s what and the way forward. Trends Ecol. Evol. 28, 58–66 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Bax N., et al. , Marine invasive alien species: A threat to global biodiversity. Mar. Policy 27, 313–323 (2003). [Google Scholar]
- 6.Molnar J. L., Gamboa R. L., Revenga C., Spalding M. D., Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 6, 485–492 (2008). [Google Scholar]
- 7.Seebens H., et al. , No saturation in the accumulation of alien species worldwide. Nat. Commun. 8, 14435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson M., Fitter A., The varying success of invaders. Ecology 77, 1661–1666 (1996). [Google Scholar]
- 9.Mack R. N., et al. , Biotic invasions: Causes, epidemiology, global consequences and control. Ecol. Appl. 10, 689–710 (2000). [Google Scholar]
- 10.Parker J. D., et al. , Do invasive species perform better in their new ranges? Ecology 94, 985–994 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Jaspers C., Marty L., Kiørboe T., Selection for life-history traits to maximize population growth in an invasive marine species. Glob. Change Biol. 24, 1164–1174 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Allendorf F. W., Lundquist L. L., Introduction: Population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30 (2003). [Google Scholar]
- 13.Dlugosch K. M., Parker I. M., Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Gaither M. R., Toonen R. J., Bowen B. W., Coming out of the starting blocks: Extended lag time rearranges genetic diversity in introduced marine fishes of Hawai’i. Proc. R. Soc. Lond. Ser. B-Biol. Sci. 279, 3948–3957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sax D. F., Brown J. H., The paradox of invasion. Glob. Ecol. Biogeogr. 9, 363–371 (2000). [Google Scholar]
- 16.Roman J., Darling J. A., Paradox lost: Genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Estoup A., et al. , Is there a genetic paradox of biological invasions? Annu. Rev. Ecol. Evol. Syst. 47, 51–72 (2016). [Google Scholar]
- 18.Facon B., Pointier J. P., Jarne P., Sarda V., David P., High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr. Biol. 18, 363–367 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Kolbe J. J., et al. , Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lockwood J. L., Cassey P., Blackburn T., The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 20, 223–228 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Gutekunst J., et al. , Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2, 567–573 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Lowe S., Browne M., Boudjelas S., De Poorter M., 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database (Hollands Printing Ltd., 2000). [Google Scholar]
- 23.Costello J. H., et al. , Transitions of Mnemiopsis leidyi (Ctenophora: Lobata) from a native to an exotic species: A review. Hydrobiologia 690, 21–46 (2012). [Google Scholar]
- 24.Jaspers C., et al. , Ocean current connectivity propelling the secondary spread of a marine invasive comb jelly across western Eurasia. Glob. Ecol. Biogeogr. 27, 814–827 (2018). [Google Scholar]
- 25.Baker L. D., Reeve M. R., Laboratory culture of the lobate ctenophore Mnemiopsis mccradyi with notes on feeding and fecundity. Mar. Biol. 26, 57–62 (1974). [Google Scholar]
- 26.Jaspers C., Costello J. H., Colin S. P., Carbon content of Mnemiopsis leidyi eggs and specific egg production rates in northern Europe. J. Plankton Res. 37, 11–15 (2015). [Google Scholar]
- 27.Jaspers C., Møller E. F., Kiørboe T., Reproduction rates under variable food conditions and starvation in Mnemiopsis leidyi: Significance for the invasion success of a ctenophore. J. Plankton Res. 37, 1011–1018 (2015). [Google Scholar]
- 28.Ryan J. F., et al. ; NISC Comparative Sequencing Program, The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pett W., et al. , Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA 22, 130–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kideys A. E., Ecology. Fall and rise of the Black Sea ecosystem. Science 297, 1482–1484 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Reusch T. B. H., Bolte S., Sparwel M., Moss A. G., Javidpour J., Microsatellites reveal origin and genetic diversity of Eurasian invasions by one of the world’s most notorious marine invader, Mnemiopsis leidyi (Ctenophora). Mol. Ecol. 19, 2690–2699 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Bolte S., et al. , Population genetics of the invasive ctenophore Mnemiopsis leidyi in Europe reveal source-sink dynamics and secondary dispersal to the Mediterranean Sea. Mar. Ecol. Prog. Ser. 485, 25–36 (2013). [Google Scholar]
- 33.Ghabooli S., et al. , Invasion pathway of the ctenophore Mnemiopsis leidyi in the Mediterranean Sea. PLoS One 8, e81067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayha K. M., et al. , Worldwide phylogeography of the invasive ctenophore Mnemiopsis leidyi (Ctenophora) based on nuclear and mitochondrial DNA data. Biol. Invasions 17, 827–850 (2015). [Google Scholar]
- 35.Kaluza P., Kölzsch A., Gastner M. T., Blasius B., The complex network of global cargo ship movements. J. R. Soc. Interface 7, 1093–1103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell J. E., Shiganova T. A., Decker M. B., Houde E. D., The ctenophore Mnemiopsis in native and exotic habitats: U.S. estuaries versus the Black Sea basin. Hydrobiologia 451, 145–176 (2001). [Google Scholar]
- 37.Barrett R. D. H., Schluter D., Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Simberloff D., The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102 (2009). [Google Scholar]
- 39.Kelble C. R., et al. , Salinity patterns of Florida Bay. Estuar. Coast. Shelf Sci. 71, 318–334 (2007). [Google Scholar]
- 40.Stanev E. V., On the mechanism of the Black Sea circulation. Earth Sci. Rev. 28, 285–319 (1990). [Google Scholar]
- 41.Jaspers C., Møller L. F., Kiørboe T., Salinity gradient of the Baltic Sea limits the reproduction and population expansion of the newly invaded comb jelly Mnemiopsis leidyi. PLoS One 6, e24065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verwimp C., et al. , Population genomic structure of the gelatinous zooplankton species Mnemiopsis leidyi in its nonindigenous range in the North Sea. Ecol. Evol. 10, 11–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darling J. A., Genetic studies of aquatic biological invasions: Closing the gap between research and management. Biol. Invasions 17, 951–971 (2015). [Google Scholar]
- 44.Seebens H., Gastner M. T., Blasius B., The risk of marine bioinvasion caused by global shipping. Ecol. Lett. 16, 782–790 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Wilder A. P., Palumbi S. R., Conover D. O., Therkildsen N. O., Footprints of local adaptation span hundreds of linked genes in the Atlantic silverside genome. Evol. Lett. 4, 430–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikeda D. H., et al. , Genetically informed ecological niche models improve climate change predictions. Glob. Change Biol. 23, 164–176 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Ghabooli S., et al. , Multiple introductions and invasion pathways for the invasive ctenophore Mnemiopsis leidyi in Eurasia. Biol. Invasions 13, 679–690 (2010). [Google Scholar]
- 48.Colin S. P., Costello J. H., Hansson L. J., Titelman J., Dabiri J. O., Stealth predation and the predatory success of the invasive ctenophore Mnemiopsis leidyi. Proc. Natl. Acad. Sci. U.S.A. 107, 17223–17227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ausubel F. M., Current Protocols in Molecular Biology Core (John Wiley and Sons, 2003). [Google Scholar]
- 50.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danecek P., et al. ; 1000 Genomes Project Analysis Group, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meisner J., Albrechtsen A., Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nei M., Li W. H., Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weir B. S., Cockerham C. C., Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gascuel O., BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Kamvar Z. N., Tabima J. F., Grunwald N. J., Poppr 1-0-3: An R package for genetic analysis of populations with clonal, partially clonal and/or sexual reproduction. PeerJ 2, e281 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pritchard J. K., Stephens M., Donnelly P., Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evanno G., Regnaut S., Goudet J., Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V. C., Foll M., Robust demographic inference from genomic and SNP data. PLoS Genet. 9, e1003905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akaike H., Stochastic theory of minimal realization. IEEE Trans. Automat. Contr. 19, 667–674 (1974). [Google Scholar]
- 65.Cornuet J. M., et al. , DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nadachowska-Brzyska K., Li C., Smeds L., Zhang G., Ellegren H., Temporal dynamics of avian populations during pleistocene revealed by whole-genome sequences. Curr. Biol. 25, 1375–1380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duchêne S., Lanfear R., Ho S. Y., The impact of calibration and clock-model choice on molecular estimates of divergence times. Mol. Phylogenet. Evol. 78, 277–289 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Mazet O., Rodríguez W., Grusea S., Boitard S., Chikhi L., On the importance of being structured: Instantaneous coalescence rates and human evolution—Lessons for ancestral population size inference? Heredity 116, 362–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mather N., Traves S. M., Ho S. Y. W., A practical introduction to sequentially Markovian coalescent methods for estimating demographic history from genomic data. Ecol. Evol. 10, 579–589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaspers C., et al. , Whole-genome resequencing data to article Invasion genomics uncover contrasting scenarios of genetic diversity in a widespread marine invader. Genbank. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA689379. Deposited 1 April 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited on GenBank under the NCBI BioProject accession number: PRJNA689379 (71). All code is provided and available at Zenodo under DOI: 10.5281/zenodo.5668363.