ABSTRACT
The prevalence of non-alcoholic fatty liver disease (NAFLD) is constantly increasing, and altered expression of microRNAs (miRNAs) fosters the development and progression of many pathologies, including NAFLD. Therefore, we explored the role of new miRNAs involved in the molecular mechanisms that trigger NAFLD progression and evaluated them as biomarkers for diagnosis. As a NAFLD model, we used apolipoprotein E-deficient mice administered a high-fat diet for 8 or 18 weeks. We demonstrated that insulin resistance and decreased lipogenesis and autophagy observed after 18 weeks on the diet are related to a concerted regulation carried out by miR-26b-5p, miR-34a-5p, miR-149-5p and miR-375-3p. We also propose circulating let-7d-5p and miR-146b-5p as potential biomarkers of early stages of NAFLD. Finally, we confirmed that circulating miR-34a-5p and miR-375-3p are elevated in the late stages of NAFLD and that miR-27b-3p and miR-122-5p are increased with disease progression. Our results reveal a synergistic regulation of key processes in NAFLD development and progression by miRNAs. Further investigation is needed to unravel the roles of these miRNAs for developing new strategies for NAFLD treatment.
This article has an associated First Person interview with the joint first authors of the paper.
KEY WORDS: Non-alcoholic fatty liver disease, MicroRNAs, Lipid metabolism, Insulin resistance, Autophagy, Extracellular vesicles
Summary: Apoe−/− mice administered a high-fat diet represent a model of non-alcoholic fatty liver disease, revealing the synergistic regulation of key processes in disease progression by miRNAs and indicating some miRNAs as biomarkers for diagnosis.
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is considered the most common hepatic disorder in Western countries, where its prevalence is continuously growing (Wree et al., 2013; Wong et al., 2014; McPherson et al., 2015; Rinella and Sanyal, 2016; Younossi et al., 2018). This condition represents the hepatic hallmark of metabolic syndrome, because it is strongly related to obesity, insulin resistance, dyslipidemia and hypertension (Younossi et al., 2018). Hence, owing to the research of recent years and the heterogeneity, experts have considered that the term NAFLD is not accurate enough, and the name metabolic dysfunction-associated fatty liver disease (MAFLD) has been proposed (Eslam et al., 2020; Tilg and Effenberger, 2020). NAFLD is defined by the accumulation of fat in the liver, and it consists of two main stages, the earliest one characterized by benign steatosis in >5% of hepatocytes (fatty liver) that can progress to non-alcoholic steatohepatitis (NASH) [European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) and European Association for the Study of Obesity (EASO), 2016; Arab et al., 2018; Friedman et al., 2018]. The hallmark of this severe stage is hepatocyte ballooning, inflammation and/or fibrosis, eventually resulting in hepatic cirrhosis and, ultimately, hepatocellular carcinoma [Marra et al., 2008; European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) and European Association for the Study of Obesity (EASO), 2016; Friedman et al., 2018; Pierantonelli and Svegliati-Baroni, 2019]. Owing to the high prevalence mentioned above, although hepatitis C is the leading cause of liver transplantation, it is expected that NASH will exceed it in the near future (Wree et al., 2013; Wong et al., 2014; Rinella and Sanyal, 2016). Although some factors involved in the progression from steatosis to NASH are still unknown, the roles of lipotoxicity, oxidative stress and activation of the immune system have been well characterized (Vernon et al., 2011). Considering these data, it is essential to gain deeper knowledge about the molecular mechanisms involved in NAFLD, with the aim of discovering new targets for treatment.
In recent years, microRNAs (miRNAs) have been proposed as novel and interesting tools for this purpose, belonging to an evolutionarily conserved class of short (20-22 nucleotides in length) and single-stranded non-coding RNAs. In mammals, the majority of gene expression is repressed at the post-transcriptional level by miRNAs (Hussain, 2012; Hartig et al., 2015). Particularly, owing to the complementarity between these molecules and their targets, miRNAs are able to associate with the 3′ untranslated region (UTR) of a gene mRNA, leading to mRNA decay and/or translational repression (Huntzinger and Izaurralde, 2011). Consequently, they have a critical role in a huge number of physiological processes, such as cell growth, tissue differentiation and embryonic development. Indeed, their alteration is involved in the onset and progression of pathological states, including obesity, NAFLD and cardiovascular diseases (Hussain, 2012; Hartig et al., 2015). Currently, the major drawback of miRNAs is poor understanding of the mechanisms by which they exert their actions. A concerted effort is needed before manipulation of miRNA expression can be used in disease management worldwide.
Aberrant miRNA expression in both liver and plasma has been described as a main feature of liver diseases in many studies, including NAFLD (Yokota et al., 2001; Madan et al., 2006; Bettaieb et al., 2015). In this sense, extracellular vesicles (EVs), mainly exosomes, are able to vehiculize miRNAs mediating tissue crosstalk while protecting them from degradation by RNAses. Therefore, circulating miRNAs are being considered as potential biomarkers in the diagnosis and prognosis of many pathologies, such as NAFLD, cancer, cardiovascular diseases, obesity and diabetes (Mori et al., 2019).
With this background, we used a mouse deficient in apolipoprotein E (Apoe) and administered a high-fat diet (HFD) as a model of NAFLD. The principal aim of the present work was to determine whether dysregulation of the expression of certain miRNAs could trigger NAFLD development. With this objective in mind, we tried to identify molecular targets of these miRNAs as key proteins involved in fostering disease progression. Finally, we also analyzed EVs in order to find miRNAs that could serve as NAFLD biomarkers.
RESULTS
Evaluation of weight gain and lipid profile
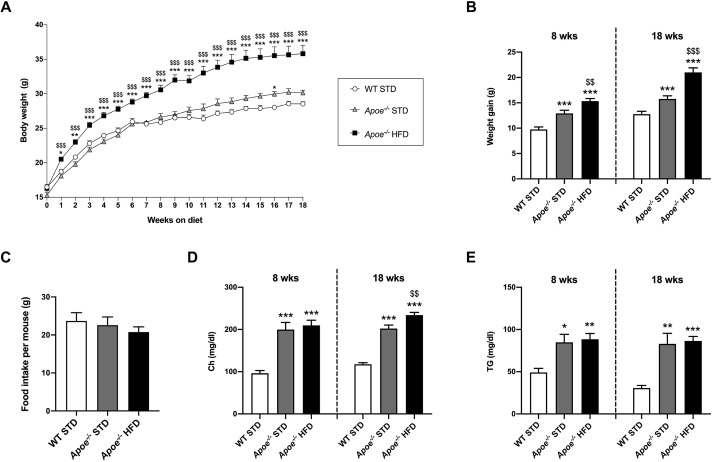
After genotype characterization, wild-type (WT) and apolipoprotein E-deficient (Apoe−/−) mice were administered a standard diet (STD) or HFD for 8 or 18 weeks (Fig. S1). In order to study the diet-associated weight gain, we evaluated body weight (BW) and gain from weaning until euthanasia (Fig. 1A,B), BW at the time of euthanasia, and weights of liver, subcutaneous fat, represented by inguinal white adipose tissue (iWAT) and visceral fat (vWAT), normalized versus BW (Table 1). According to the results, BW, weight gain and adipose depots were significantly increased, while the liver/BW ratio was significantly diminished, in the HFD-fed Apoe−/− versus STD-fed Apoe−/− and WT groups (Fig. 1A,B and Table 1). Additionally, food intake was measured; however, no statistically significant differences were observed when comparing the three groups (Fig. 1C).
Fig. 1.
Characterization of obesity and lipid profile in the mouse model of NAFLD. (A) Graph showing BW progression in STD-fed WT (WT STD; n=20-39), STD-fed Apoe−/− (Apoe−/− STD; n=10-37) and HFD-fed Apoe−/− (Apoe−/− HFD; n=18-38) mice from weaning until the 18th week on the diet. (B) Representation of BW gain from weaning to 8 or 18 weeks of diet in the three experimental groups. (C) Assessment of food intake per mouse and per week (g) in WT STD (n=16), Apoe−/− STD (n=23) and Apoe−/− HFD (n=27) mice. (D) Levels of plasma Ch (mg/dl) in WT STD (n=6-7), Apoe−/− STD (n=5-9) and Apoe−/− HFD (n=8-10) mice after 8 or 18 weeks on the diet. (E) Circulating TG levels (mg/dl) in WT STD (n=7), Apoe−/− STD (n=4-9) and Apoe−/− HFD (n=8-10) mice after 8 or 18 weeks on the diet. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, except for TGs at 8 weeks of diet, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $$P<0.01 and $$$P<0.001 versus Apoe−/− STD mice. BW, body weight; Ch, cholesterol; HFD, high-fat diet; STD, standard diet; TG, triglycerides; WT, wild type.
Table 1.
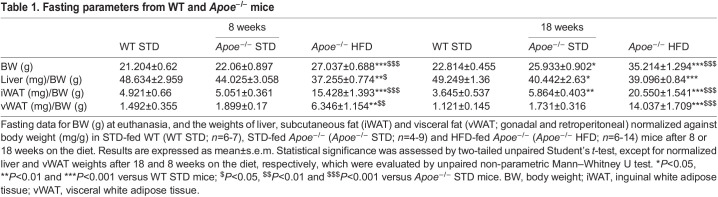
Fasting parameters from WT and Apoe−/− mice
To obtain the plasma lipid profile, cholesterol (Ch) and triglyceride (TG) levels were measured. The circulating levels of Ch and TGs were significantly higher in Apoe−/− mice than in the WT groups (Fig. 1D,E). Furthermore, 18-week HFD-fed Apoe−/− mice had significantly higher plasma Ch levels than STD-fed Apoe−/− mice (Fig. 1D).
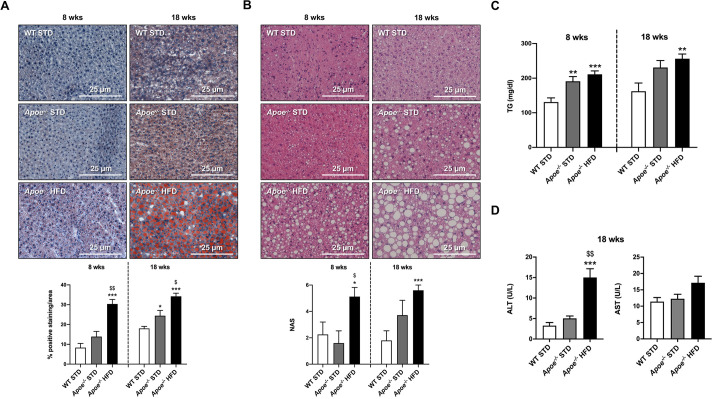
Given that Apoe deficiency induces NAFLD, in order to investigate the liver tissue and possible lipid ectopic depot, we evaluated hepatic lipid accumulation by Oil Red O staining in liver sections from Apoe−/− and WT mice. The results revealed a significant increase in the percentage Oil Red O-positive area in HFD-fed mice compared with STD-fed mice after 8 and 18 weeks on the respective diets. Moreover, in the case of 18-week STD-fed Apoe−/− mice, a statistically significant increase was observed with respect to the STD-fed WT group (Fig. 2A).
Fig. 2.
Assessment of intrahepatic lipid accumulation and NAS. (A) Representative images of Oil Red O staining to evaluate hepatic-specific lipid content (top; image magnification ×20), and its quantification in WT STD (n=6-7), Apoe−/− STD (n=5-6) and Apoe−/− HFD (n=5-6) mice after 8 or 18 weeks on the diet (bottom). (B) Representative images of H&E staining to perform liver histological analysis (top; image magnification ×20), and quantification of NAS according to liver abnormalities in WT STD (n=4-5), Apoe−/− STD (n=5-7) and Apoe−/− HFD (n=8-10) mice after 8 or 18 weeks on the diet (bottom). (C) Intrahepatic TG quantification (mg/dl) in WT STD (n=6-7), Apoe−/− STD (n=5-6) and Apoe−/− HFD (n=6-8) mice after 8 or 18 weeks on the diet. (D) Quantification of both types of circulating transaminases (ALT, left; AST, right; U/L) in WT STD (n=6-7), Apoe−/− STD (n=7-8) and Apoe−/− HFD (n=9) mice after 18 weeks on the diet. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, except for plasma ALT measurement, which was evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $P<0.05 and $$P<0.01 versus Apoe−/− STD mice. ALT, alanine transaminase; AST, aspartate transaminase; NAS, NAFLD Activity Score; TG, triglycerides.
In a further step, we also checked the hepatic morphological changes through Hematoxylin and Eosin (H&E) staining in other sections. The results were evaluated by a pathologist specializing in hepatic histopathology from Santa Cristina Hospital (Madrid, Spain), using the NAFLD Activity Score (NAS) scoring system as a tool and following the established criteria (Kleiner et al., 2005). These data suggested that 8-week HFD-fed Apoe−/− mice presented a higher NAS with respect to that of the STD-fed groups. However, this increase was only statistically significant versus STD-fed WT mice after 18 weeks on the diet (Fig. 2B). The breakdown of these parameters in steatosis, inflammation and ballooning, together with the mRNA levels of the chemokine monocyte chemoattractant protein-1 (Mcp1), also known as Ccl2, are shown in Fig. S2. Regarding Ccl2 gene expression, levels were significantly higher in the HFD-fed Apoe−/− mice than in the STD-fed Apoe−/− and WT mice (Fig. S2B).
In addition, lipid droplet size was measured, revealing that the lipid droplet size of 8-week HFD-fed Apoe−/− mice was significantly larger than that of STD-fed WT mice. Furthermore, lipid droplet size was markedly increased in HFD-fed Apoe−/− mice after 18 weeks on the diet, compared with that in both groups of STD-fed mice, and this difference was statistically significant. Moreover, the lipid droplet size in STD-fed Apoe−/− mice was significantly larger than that in STD-fed WT mice after 18 weeks on the diet (Fig. S2C).
NASH is characterized not only by the presence of hepatic steatosis and inflammation, but also fibrosis. Thus, we performed Sirius Red staining in liver sections of 18-week-fed mice to determine hepatic fibrosis. Quantification of positive staining revealed the highest levels of fibrosis in 18-week HFD-fed Apoe−/− mice, which were significantly different from those in STD-fed mice (Fig. S2D). The results were also evaluated by the pathologist mentioned above, using the established criteria of fibrosis scoring (Liang et al., 2014). These data suggested that 18-week HFD-fed Apoe−/− mice presented a statistically higher fibrosis score than that of STD-fed WT mice. However, our findings only indicated an initial stage of hepatic fibrosis (Fig. S2D).
Intrahepatic TG concentrations were also measured in our experimental groups after 8 and 18 weeks on the respective diets. Our results showed significantly higher intrahepatic TG concentration in 8-week HFD- and STD-fed Apoe−/− mice than in STD-fed WT mice, but, after 18 weeks on the respective diets, this difference was only statistically significant between the HFD-fed Apoe−/− and STD-fed WT groups (Fig. 2C).
Next, we assessed the extent of hepatocellular injury through the determination of plasma activities of alanine and aspartate transaminases (ALT and AST, respectively) after 18 weeks of diet administration. Regarding ALT, the most specific marker of liver damage, a statistically significant increase was observed in HFD-fed Apoe−/− mice compared with the STD-fed Apoe−/− and WT mice. Concerning plasma AST levels, there were no statistically significant differences among the groups (Fig. 2D).
Lipogenesis impairment as NAFLD progresses
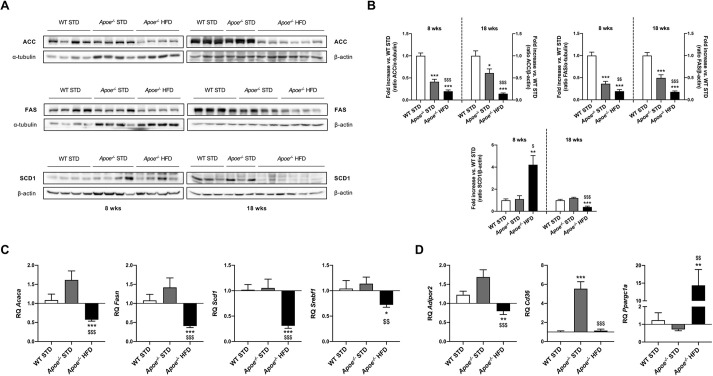
As has been previously verified, hepatic lipid metabolism dysregulation prevails in NAFLD (Arab et al., 2018; Friedman et al., 2018). Expression of key proteins involved in lipid metabolism and NAFLD progression, such as acetyl-CoA carboxylase (ACC; encoded by Acaca), fatty acid synthase (FAS; encoded by Fasn) and stearoyl-CoA desaturase 1 (SCD1), were analyzed by western blotting using α-tubulin or β-actin as normalizers (Fig. 3A,B). Hepatic ACC and FAS expression presented the same profile after 8 and 18 weeks of diet, reducing their levels as NAFLD progressed. The levels of these proteins were significantly reduced in both Apoe−/− groups compared with the WT group, and statistically significant differences were also observed between the two Apoe−/− groups (Fig. 3A,B). Regarding SCD1 expression, statistically significant changes were only observed in the HFD-fed Apoe−/− group, in which expression was significantly higher than in the STD-fed groups after 8 weeks on the diet but significantly lower after 18 weeks on the diet (Fig. 3A,B). Significant decrease in the expression of Acaca, Fasn and Scd1 in Apoe−/− mice after 18 weeks of HFD was also demonstrated by reverse transcription and quantitative PCR (RT-qPCR). In addition, we analyzed the mRNA levels of the upstream regulator of lipogenic gene expression, sterol-regulatory element binding protein-1c (SREBP1c; encoded by Srebf1), after 18 weeks of diet, and we found a statistically significant downregulation in the HFD-fed Apoe−/− group compared with the STD-fed Apoe−/− and WT groups (Fig. 3C).
Fig. 3.
Expression of pivotal proteins and mRNA levels of genes involved in hepatic lipid metabolism. (A) Representative western blot analysis of ACC, FAS and SCD1 in liver homogenates from the six groups studied. α-tubulin or β-actin was used as a loading control. (B) Histograms presenting the protein/α-tubulin or β-actin ratio quantifications of band intensities in WT STD (n=3-4), Apoe−/− STD (n=3-4) and Apoe−/− HFD (n=4-6) mice after 8 or 18 weeks on the diet. (C) mRNA levels of Acaca, Fasn, Scd1 and Srebf1 by RT-qPCR in livers from WT STD (n=5-7), Apoe−/− STD (n=5-8) and Apoe−/− HFD (n=9-14) mice after 18 weeks on the diet. Actb was used as a control. (D) mRNA levels of Adipor2, Cd36 and Ppargc1a by RT-qPCR in livers from WT STD (n=4-5), Apoe−/− STD (n=4-5) and Apoe−/− HFD (n=7-9) mice after 18 weeks on the diet. Actb was used as a control. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, apart from ACC, 8-week-diet FAS and 18-week-diet SCD1 data, and mRNA levels of Cd36 and Ppargc1a, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $P<0.05, $$P<0.01 and $$$P<0.001 versus Apoe−/− STD mice.
Furthermore, mRNA levels of other key genes were analyzed by RT-qPCR in the liver of 18-week-fed mice in order to elucidate the mechanisms involved in the onset of NAFLD. In the case of adiponectin receptor 2 (Adipor2) mRNA, we also observed significantly reduced expression in HFD-fed Apoe−/− mice compared with STD-fed Apoe−/− and WT mice. However, significantly higher levels of cluster of differentiation 36 (Cd36) mRNA expression were observed in STD-fed Apoe−/− mice compared with STD-fed WT mice and HFD-fed Apoe−/− mice, which presented similar levels. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Ppargc1a) mRNA expression was significantly higher in the HFD-fed Apoe−/− mice than in STD-fed Apoe−/− and WT mice (Fig. 3D).
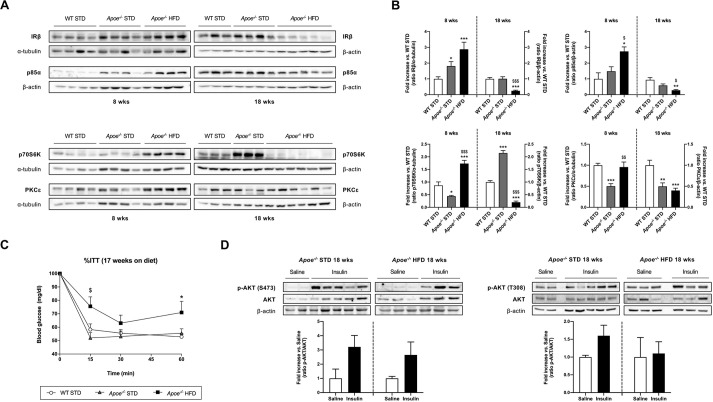
Assessment of diet-induced insulin resistance
One of the main features of NAFLD is hepatic insulin resistance (González-Muniesa et al., 2017). For this reason, the hepatic expression of proteins involved in insulin signaling, such as insulin receptor β subunit (IRβ; also known as INSR), PI3 kinase p85α (p85α; also known as PIK3R1), p70 S6 kinase (p70S6K; also known as RPS6KB2) and protein kinase C epsilon type (PKCε; also known as PRKCE), was analyzed by western blotting (Fig. 4A,B). The expression of IRβ and p85α was markedly changed in the HFD-fed Apoe−/− mice, in which expression was significantly higher than in the STD-fed groups after 8 weeks on the diet but significantly lower after 18 weeks on the diet. IRβ expression was only significantly higher in STD-fed Apoe−/− mice compared with STD-fed WT mice after 8 weeks on the diet (Fig. 4A,B). p70S6K expression in HFD-fed Apoe−/− mice had a similar pattern to that of IRβ and p85α; however, in STD-fed Apoe−/− mice, p70S6K expression was significantly lower than that in STD-fed WT mice after 8 weeks on the diet and significantly higher after 18 weeks on the diet (Fig. 4A,B). The expression of PKCε in STD-fed Apoe−/− mice was similar after 8 and 18 weeks on the diet and significantly lower than that in STD-fed WT mice. In HFD-fed Apoe−/− mice, PKCε expression was significantly higher than that in STD-fed Apoe−/− mice and similar to that in STD-fed WT mice after 8 weeks on the diet, but significantly lower than that in WT mice after 18 weeks on the diet (Fig. 4A,B).
Fig. 4.
Evaluation of alterations in insulin signaling. (A) Representative gels of western blot analysis of IRβ, p85α, p70S6K and PKCε in liver homogenates from the six groups studied (n=3-6 per group). α-tubulin or β-actin was used as a loading control. (B) Histogram displaying the protein/α-tubulin or β-actin ratio quantification of band intensities in WT STD (n=3-4), Apoe−/− STD (n=3-4) and Apoe−/− HFD (n=4-6) mice after 8 or 18 weeks on the diet. (C) Insulin tolerance test (ITT) in WT STD (n=7), Apoe−/− STD (n=6) and Apoe−/− HFD (n=7) animals after 17 weeks on the diet. (D) Western blot analysis of total AKT and the phosphorylated forms (Ser473, left; Thr308, right) in the liver of mice subjected to in vivo insulin signaling analysis after 18 weeks on the diet. Top: representative gels of Apoe−/− STD and Apoe−/− HFD mice injected with saline (n=2-3) or insulin (n=3-5) before euthanasia. In both cases, β-actin was used as a loading control. Bottom: histogram showing the normalized phospho-AKT (p-AKT)/AKT ratio in Apoe−/− STD (n=2-3) and Apoe−/− HFD (n=3-5) mice. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, apart from IRβ and 8-week-diet PKCε data, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $P<0.05, $$P<0.01 and $$$P<0.001 versus Apoe−/− STD mice.
To evaluate glucose homeostasis, insulin tolerance tests (ITTs) and glucose intolerance tests (GTTs) were performed 17 weeks after weaning (Fig. 4C; Fig. S3). Our results suggested that, 17 weeks after HFD administration, mice developed overt and maintained glucose intolerance and insulin resistance.
Additionally, with the aim of analyzing glucose homeostasis alterations in our experimental conditions, in vivo insulin signaling studies were performed using 18-week STD- and HFD-fed Apoe−/− mice. AKT phosphorylation (Ser473 and Thr308) was also determined in response to insulin in the liver. Our results showed that insulin resistance exhibited by HFD-fed Apoe−/− mice in the ITT experiments could be due to an impairment in AKT phosphorylation (Thr308) because we did not observe an increase in the phosphorylation of this residue after insulin stimulation, hindering the full activation of this kinase (Fig. 4D).
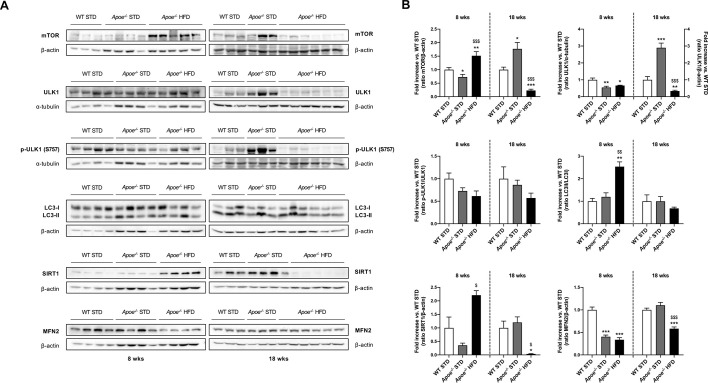
Role of autophagy in NAFLD development
Although autophagy has emerged as a regulatory pathway, its activation state in the development of NAFLD remains unclear. In order to elucidate expression changes in NAFLD, we assessed the protein levels of mammalian target of rapamycin (mTOR), Unc-51-like autophagy-activating kinase 1 (ULK1) and its phosphorylated state, light chain 3 (LC3; also known as MAP1LC3), sirtuin 1 (SIRT1) and mitofusin 2 (MFN2) in liver samples by western blotting (Fig. 5A,B). mTOR expression in the Apoe−/− groups notably differed at 8 and 18 weeks. Although after 8 weeks on the diet, the expression of this protein was significantly decreased in STD-fed Apoe−/− mice compared with STD-fed WT mice, the opposite was observed after 18 weeks of feeding. In the HFD-fed Apoe−/− mice, mTOR expression was significantly higher than that in both STD-fed groups after 8 weeks on the diet and significantly lower after 18 weeks on the diet (Fig. 5A,B). A direct target of mTOR is ULK1, a protein involved in autophagy; its expression presented a similar pattern to that of mTOR, especially after 18 weeks of diet. Quantification showed a statistically significant reduction in ULK1 levels in both Apoe−/− groups compared with the WT group after 8 weeks on the respective diets. We observed that expression of this protein was similar in Apoe−/− HFD mice after 8 and 18 weeks on the diet, but significantly different from that of the STD-fed Apoe−/− mice. In the 18-week STD-fed Apoe−/− group, ULK1 expression was markedly increased compared with that at 8 weeks, and significantly higher than that in the STD-fed WT mice. Regarding the p-ULK1/ULK1 ratio, we observed lower levels in both Apoe−/− groups compared with WT, but these differences were not statistically significant (Fig. 5A,B). LC3 is a molecular marker of autophagy, which, once lipidated with phosphatidylethanolamine, triggers autophagosome formation, as seems to occur in 8-week HFD-fed Apoe−/− mice compared with STD-fed WT mice. Despite western blot analysis results indicating no significant differences between the groups, the data showed that HFD decreased the LC3-II/LC3-I ratio after 18 weeks of diet, suggesting the onset of autophagic flux restriction (Fig. 5A,B). The deacetylase SIRT1 has a key role in the regulation of autophagy by nutrients. Western blot analysis suggested that the higher levels of SIRT1 expression observed in the 8-week HFD-fed Apoe−/− mice compared with the STD-fed mice were only statistically significant with respect to the STD-fed Apoe−/− group, and that expression was significantly lower in this group than in the STD-fed groups after 18 weeks of feeding. In the STD-fed Apoe−/− group, SIRT1 expression was lower than that in the WT group after 8 weeks of diet, but almost the same after 18 weeks (Fig. 5A,B). The mitochondrial outer membrane protein MFN2 showed decreased expression in HFD-fed Apoe−/− mice at 8 and 18 weeks, exhibiting statistically significant differences with respect to STD-fed WT mice and, after 18 weeks of feeding, compared with STD-fed Apoe−/− mice. The STD-fed Apoe−/− group had decreased MFN2 levels after 8 weeks on the diet but the same levels as STD-fed WT mice after 18 weeks on the diet (Fig. 5A,B).
Fig. 5.
Dysregulation of autophagy-related proteins in response to lipid oversupply. (A) Representative gels of western blot analysis of mTOR, phospho-ULK1 (p-ULK1; Ser757), ULK1, LC3-I/II, SIRT1 and MFN2 in liver homogenates from the six groups studied (n=3-6 per group). α-tubulin or β-actin was used as a loading control. (B) Histograms showing the protein/α-tubulin or β-actin, p-ULK1/ULK1 and LC3-II/LC3-I ratio quantification of band intensities in WT STD (n=3-4), Apoe−/− STD (n=3-4) and Apoe−/− HFD (n=4-6) mice after 8 or 18 weeks on the diet. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, with the exception of p-ULK1/ULK1 18-week and mTOR and SIRT1 8- and 18-week data, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $P<0.05, $$P<0.01 and $$$P<0.001 versus Apoe−/− STD mice.
Hepatic alteration in the expression of NAFLD-related miRNAs
Once we had characterized the experimental model, our next objective was to unravel the existing differences in NAFLD-related miRNA expression among the studied groups by RT-qPCR, using miR-191-5p as a normalizer.
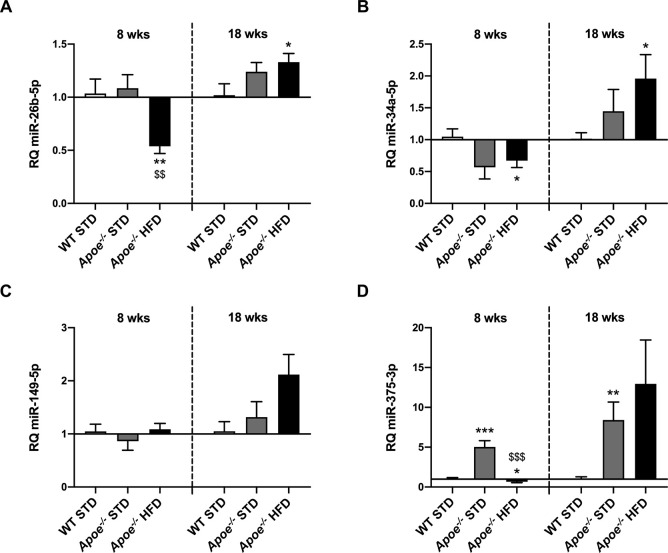
First, in Apoe−/− mice fed a HFD for 8 weeks, we observed significantly lower levels of let-7d-5p (Fig. S4A), miR-22-3p (Fig. S4C), miR-26b-5p (Fig. 6A), miR-122-5p (Fig. S4E), miR-146b-5p (Fig. S4F), miR-181b-5p (Fig. S4G) and miR-194-5p (Fig. S4I) compared with those in STD-fed mice. The profile observed for miR-192-5p was rather similar, but levels in the STD-fed Apoe−/− mice were also significantly lower than those in STD-fed WT mice (Fig. S4H). Levels of miR-27b-3p and miR-34a-5p in HFD-fed Apoe−/− mice were significantly lower than those in STD-fed WT mice (Fig. 6B; Fig. S4D). In addition, significantly higher miR-375-3p levels were observed in STD-fed Apoe−/− mice compared with STD-fed WT mice, and significantly lower levels were observed in HFD-fed Apoe−/− mice compared with both STD-fed groups (Fig. 6D). Additionally, no statistical variation was observed in the levels of miR-15b-5p and miR-149-5p (Fig. 6C; Fig. S4B).
Fig. 6.
RT-qPCR analysis of miRNA expression in the liver. (A-D) Comparison of miR-26b-5p (A), miR-34a-5p (B), miR-149-5p (C) and miR-375-3p (D) expression in WT STD (n=4-7), Apoe−/− STD (n=4-9) and Apoe−/− HFD (n=6-10) mice after 8 or 18 weeks on the diet. miR-191-5p was used as a control. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, with the exception of 18-week miR-149-5p data, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05, **P<0.01 and ***P<0.001 versus WT STD mice; $$P<0.01 and $$$P<0.001 versus Apoe−/− STD mice.
Regarding the 18-week-fed mice, levels of let-7d-5p, miR-22-3p and miR-194-5p tended to be lower, although not significantly, in the Apoe−/− groups than in the WT group (Fig. S4A,C,I). By contrast, miR-149-5p levels were higher, although not significantly, in Apoe−/− HFD mice than in the STD-fed mice (Fig. 6C). In the case of miR-375-3p, expression levels were higher in the two Apoe−/− groups than in the WT group, but a statistically significant difference was only observed between the STD-fed groups (Fig. 6D). Additionally, levels of miR-27b-3p expression were higher in the two Apoe−/− groups than in the WT group, but statistically significant differences were only obtained for the STD-fed Apoe−/− group compared with the WT group (Fig. S4D). For miR-26b-5p and miR-34a-5p, levels of expression were higher in the two Apoe−/− groups than in the WT group, significantly so in the case of HFD-fed Apoe−/− mice compared with STD-fed WT mice (Fig. 6A,B). The well-known hepato-specific miRNA miR-122-5p showed significantly higher expression in STD-fed Apoe−/− mice than in HFD-fed Apoe−/− and STD-fed WT mice (Fig. S4E). Although miR-146b-5p levels tended to be higher in STD-fed Apoe−/− mice and lower in Apoe−/− HFD mice than in STD-fed WT mice, these differences were not statistically significant (Fig. S4F). No significant differences in miR-15b-5p, miR-181b-5p and miR-192-5p expression were observed among the groups (Fig. S4B,G,H).
Overexpression of miR-26b-5p and miR-34a-5p in Huh7 cells results in downregulated expression of some predicted targets
To demonstrate whether some selected miRNAs, such as miR-26b-5p and miR-34a-5p, are involved in regulating the expression of predicted targets, we transfected Huh7 cells with miR-26b-5p and miR-34a-5p miRNA precursors.
We observed that miR-34a-5p overexpression downregulates FAS, MFN2 and mTOR protein levels. In addition, miR-34a-5p transfection was validated by assessing the downregulation of SIRT1, a widely known target of this miRNA (Fig. S5).
We performed similar experiments overexpressing miR-26b-5p in order to validate our conclusions. In these experiments, we found a significant decrease in FAS expression (Fig. S5).
EV miRNAs as potential NAFLD biomarkers
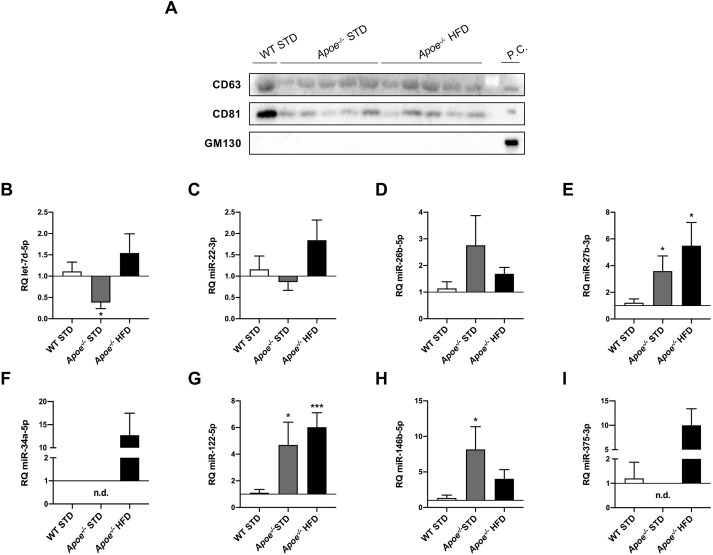
After demonstrating liver injury generated in the Apoe−/− model, we proceeded to isolate miRNAs contained in plasma EVs from 18-week-fed mice in order to examine, by RT-qPCR, whether some of the previous selected miRNAs were secreted into EVs and could be used as biomarkers of NAFLD. To validate EV isolation, CD63 and CD81, two of the most frequently used markers of exosomes, were analyzed by western blotting. GM130 (also known as GOLGA2) was used as a negative control because it is a cis-Golgi matrix protein. Our results suggest that the isolated EVs were enriched in exosomes (Fig. 7A).
Fig. 7.
RT-qPCR analysis of miRNA expression in circulating extracellular vesicles. (A) Western blot analysis of markers to identify exosomes, such as CD63 and CD81, and GM130 as a cis-Golgi matrix protein used as a negative control, in WT STD (n=1), Apoe−/− STD (n=5) and Apoe−/− HFD (n=5) mice after 18 weeks on the diet. Huh7 cells were used as a positive control. (B-I) Comparison of let-7d-5p (B), miR-22-3p (C), miR-26b-5p (D), miR-27b-3p (E), miR-34a-5p (F), miR-122-5p (G), miR-146b-5p (H) and miR-375-3p (I) expression in WT STD (n=4-7), Apoe−/− STD (n=3-6) and Apoe−/− HFD (n=4-7) mice after 18 weeks on the diet. miR-16-5p was used as a control. Results are expressed as mean±s.e.m. Statistical significance was assessed by two-tailed unpaired Student's t-test, with the exception of miR-22-3p data, which were evaluated by unpaired non-parametric Mann–Whitney U test. *P<0.05 and ***P<0.001 versus WT STD mice. No statistics could be performed for miR-34a-5p and miR-375-3p data owing to undetermined values obtained in most WT STD and Apoe−/− STD liver samples. n.d., non-detected; P.C., positive control.
The STD-fed Apoe−/− mice had significantly lower levels of let-7d-5p in plasma EVs compared with the STD-fed WT group (Fig. 7B). miR-22-3p contained in EVs tended to be higher, although not significantly, in HFD-fed Apoe−/− mice compared with the STD-fed group (Fig. 7C). The EV-contained miR-26b-5p levels tended to be higher, although not significantly, in the STD-fed Apoe−/− mice compared with the other groups (Fig. 7D). The content of miR-27b-3p and miR-122-5p in EVs was significantly higher in Apoe−/− mice, particularly HFD-fed Apoe−/− mice, than in STD-fed WT mice (Fig. 7E,G). The plasma miR-34a-5p and miR-375-3p levels presented similar profiles. In both cases, we observed a tendency for higher levels, although not significantly, in the HFD-fed Apoe−/− mice compared with the STD-fed groups and undetectable levels in the STD-fed Apoe−/− group (Fig. 7F,I). In 18-week-fed mice, EV-contained miR-146b-5p levels were higher in Apoe−/− mice than in WT mice, significantly so in the case of the STD-fed Apoe−/− group (Fig. 7H).
DISCUSSION
Although the Apoe−/− murine model has been widely used in atherosclerosis research, in this study, we confirmed that these mice are also prone to developing hepatic steatosis and serve as a model for NAFLD, mimicking the major characteristics of this disease, as has been reported (Schierwagen et al., 2015; Hansen et al., 2017; Nascimento et al., 2020).
For instance, the abnormal amount of lipids in the liver, especially in HFD-fed mice, was caused by lipid level dysregulation as a consequence of insulin resistance, which is probably due to a defect in mitochondrial lipid oxidation (Saltiel and Kahn, 2001; Stumvoll et al., 2005; Muoio and Newgard, 2008; Castro et al., 2014). From a clinical perspective, if we consider the results obtained after the double-blind hepatic examination, it could be said that HFD administration accelerated disease progression without changes in food intake between the groups. The lipid droplet size itself represents an essential parameter indicating the processes used for cellular TG catabolism, especially under nutrient oversupply. Two TG disposal pathways exist in hepatocytes, lipolysis and lipophagy, which degrade large and small lipid droplets, respectively (Schott et al., 2019). Although NAS did not differ between the two experimental Apoe−/− HFD groups, lipid droplets were obviously bigger after 18 weeks of feeding. Furthermore, the hepatic fibrosis increased with phenotype severity. Thus, the observed fibrosis was in a preliminary stage.
Intriguingly, Cd36 expression seemed to be highest in STD-fed Apoe−/− mouse livers. Although CD36 is weakly expressed in hepatocytes, it makes sense that accumulated lipid overload upregulates its expression, reinforcing clinical studies that have described a CD36 content increase in NAFLD patients (García-Monzón et al., 2014; Rada et al., 2020). However, Cd36 mRNA levels in HFD-fed Apoe−/− mice were not augmented as expected but were similar to those in STD-fed WT mice. It should be pointed out that Cd36 deletion increases Ccl2 transcription in hepatocytes without affecting hepatic lipid uptake (Zhong et al., 2017). The chemokine CCL2 is produced in adipose tissue and triggers the development of insulin resistance and steatosis (Xu et al., 2015), two of the major characteristics present in the HFD-fed Apoe−/− mice, in which its expression was significantly increased. Therefore, Cd36 deficiency might contribute to the progression of non-alcoholic fatty liver to NASH, which is managed by inflammation and subsequent cytokine and chemokine production.
ACC catalyzes the rate-limiting step of de novo lipogenesis and regulates fatty acid β-oxidation in hepatocytes. In our mouse model, the HFD-fed Apoe−/− group showed dramatically reduced ACC expression. Many reports have demonstrated that a decrease in Acaca expression indicates steatosis amelioration (Donnelly et al., 2005; Li et al., 2018; Lopez-Pastor et al., 2019). However, in accordance with our findings, Chow et al. observed that the phenotype of their liver-specific ACC1/2 double knockout mice promoted hepatic TG accumulation and fatty acid oxidation repression, thus preserving fat storage in this tissue by a compensatory pathway (Chow et al., 2014). Liver SCD1 protein levels were significantly higher in HFD-fed Apoe−/− mice than in STD-fed mice after 8 weeks on the diet, but significantly lower after 18 weeks. Previous reports suggested that SCD1 downregulation aggravates hepatocyte lipoapoptosis and liver injury (Cazanave and Gores, 2010; Fernández Gianotti et al., 2013). Two HFD-fed rat models displayed a similar downregulation of Scd1 mRNA and protein levels, suggesting that SCD oligomer formation can alter enzyme activity and function, thus aggravating NAFLD (Fernández Gianotti et al., 2013). Regarding the global downregulation of the lipogenic enzymes in our 18-week HFD-fed Apoe−/− mice, Luo et al. reported that these effects can be related to high-fat feeding (Luo et al., 2012). After feeding, SREBP1c is activated by insulin signaling, upregulating lipogenic genes. However, our HFD-fed mice presented insulin resistance, avoiding this upregulation. A reduction in SREBP1c and, subsequently, lipogenic genes was described in liver-specific insulin receptor knockout mice, in agreement with our findings (Haas et al., 2012).
A pivotal and common feature of metabolic disease is insulin resistance, and the overwhelming effect of a HFD has been associated with increased prevalence of this impairment, as our outcomes show. In the liver, the suppression of glucose production and stimulation of lipogenesis and glycogen synthesis are insulin-mediated processes, the dysregulation of which affects both insulin secretion and action, producing elevation in fasting and postprandial glucose and lipid levels (Michael et al., 2000; Saltiel and Kahn, 2001). Regarding this, although increased expression of IRβ was found in 8-week HFD-fed Apoe−/− mouse livers, suggesting an initial compensatory mechanism against HFD-induced insulin resistance, this increase was no longer observed after 18 weeks on the diet. A study based on partial deletion of hepatic IR in mice fed a HFD observed an amelioration of hepatic steatosis (Merry et al., 2020); therefore, the decrease observed in our HFD-fed Apoe−/− mice could serve as an attempt to avoid NAFLD progression. Indeed, under pathophysiological and nutrient excess conditions, the induction of endoplasmic reticulum stress degrades IR by autophagy-dependent processes, contributing to obesity-induced insulin resistance in peripheral tissues (Zhou et al., 2009). In the same way, we also observed a reduction in several proteins involved in insulin signaling in 18-week HFD-fed mice, contributing towards insulin resistance. Upon hepatic insulin resistance, the content of diacylglycerol (DAG) leads to PKCε activation, which in turn plays a major role in binding to the IR and repressing its tyrosine kinase activity by Thr1160 phosphorylation (Monetti et al., 2007; Jornayvaz et al., 2011; Petersen et al., 2016; Lyu et al., 2020). Certainly, our findings revealed decreased PKCε expression, but this did not correlate with a reduction in insulin resistance. In a previous study, mice with Prkce ablation and 16 weeks of fat feeding did not exhibit reversed diet-induced inhibition of insulin signaling in the liver; thereby, PKCε-independent mechanisms might maintain long-term insulin resistance (Raddatz et al., 2011). Bearing in mind in vivo insulin signaling studies and the aforementioned insulin resistance observations, western blot analysis of AKT activation failed to show statistically significant differences between the 18-week STD- and HFD-fed Apoe−/− groups, verifying insulin signaling impairment. As has previously been described, insulin-induced AKT activation requires phosphorylation of both Ser473 and Thr308 sites to result in synergistic and full activation, and these phosphorylations are not dependent on each other (Alessi et al., 1996; Taniguchi et al., 2006).
As seen throughout the study and as NAFLD progresses, abrupt changes occur in the expression of different proteins involved in either glucose or lipid metabolism. These changes can be triggered by many mediators, including miRNAs. Moreover, these molecules are recognized as potential biomarkers of NAFLD. Indeed, comparing NAFLD patients and healthy individuals, miR-375 is upregulated in pathologic serum samples (Pirola et al., 2015; Lei et al., 2018). Although miR-375 function in NAFLD has still not been elucidated, our results revealed a noteworthy increase in 18-week HFD-fed Apoe−/− mice and Adipor2 downregulation, as previously reported (Lei et al., 2018). This result is of importance as Adipor2 acts as a regulator of glucose and lipid metabolism through modulating the peroxisome proliferator-activated receptor alpha (PPARα) pathway (Lei et al., 2018). As other authors have checked in vivo and in vitro (Choi et al., 2013; Fu and Kemper, 2016; Xu et al., 2021b), another miRNA that regulates this gene is miR-34a, which is also overexpressed in our model. This miRNA is altered in NAFLD and other related diseases, such as obesity and atherosclerosis. Its expression is triggered by lipids and aggravates NAFLD and NASH, either by impairing lipid accumulation, promoting oxidative stress or fostering hepatocyte apoptosis (Lee et al., 2010; Castro et al., 2013; Choi et al., 2013; Zhang et al., 2020; Xu et al., 2021a,b). The regulation of Adipor2 by miR-34a induces ectopic deposition of lipids and decreases mitochondrial content by resistin action through the AMP-activated protein kinase (AMPK)/PPARα pathway (Wen et al., 2018). These findings and our database research suggest that Adipor2 expression can be modulated by miR-149-5p. The prominent hepatic increase in Ppargc1a mRNA in the HFD-fed group might be due to a compensatory response to enhance mitochondrial biogenesis and function in a steatosis environment, foster fatty acid oxidation, and reduce TG accumulation and secretion (Morris et al., 2012). Furthermore, it has been described that miR-34a decreases expression of SREBP1c as well as that of its downstream targets, ACC, FAS and SCD1 (Wang et al., 2020), which would explain our findings based on their downregulated expression after 18 weeks of HFD feeding. In the same way, our results suggest that overexpression of miR-34a-5p in cultured hepatocytes induced evident FAS downregulation. Database analysis revealed that these three enzymes were not only targets of miR-34a-5p, but also miR-149-5p, as our results confirmed. Although the role of miR-149 in the liver remains unclear, it is elevated in a HFD-induced NAFLD mouse model and fibroblast growth factor-21 (FGF-21) was identified as its target (Xiao et al., 2016; Chen et al., 2020). In addition, miRNA–target interaction (MTI) databases predict Fasn as a miR-26b-5p and miR-375-3p target, which is consistent with the outcomes obtained after 18 weeks of feeding and with our in vitro experiments on miR-26b-5p overexpression. Concerning insulin signaling, IR and p85α are targets of some assessed miRNAs, but, according to our data, only miR-149-5p or miR-375-3p regulation would explain the decrease in expression observed after 18 weeks on the HFD and, as a consequence, the insulin resistance. Indeed, miR-149-5p was upregulated in the liver of HFD-induced hepatic insulin resistance mice (Zhao et al., 2019), and miR-375 is highly expressed in pancreatic islets involved in insulin secretion and glucose homeostasis (Poy et al., 2004, 2009). Thus, according to our results and database research, p85α expression can also be regulated by miR-34a-5p, while PKCε is a target of miR-34a-5p and miR-375-3p. Moreover, p70S6K might be regulated by miR-26b-5p and miR-34a-5p in 18-week HFD-fed Apoe−/− mice.
Autophagy is a quality control program that can not only degrade abnormal subcellular organelles and misfolded proteins, but also lipid droplets, especially in the early stage of steatosis, alleviating disease progression. However, its inhibition occurs in the late stage of NAFLD progression (Ding et al., 2020; Lee et al., 2020; Lu et al., 2020). For this reason, although research on autophagy in NAFLD is still scarce, evidence suggests that its dysfunction is closely related to this liver disease. Consistent with what is mentioned above, our results indicated increased LC3 lipidation in 8-week HFD-fed Apoe−/− mice and, therefore, autophagic flux activation as observed in the early stages of NAFLD. However, after 18 weeks of feeding, ULK1 expression and LC3 lipidation were decreased in the livers from HFD-fed Apoe−/− mice, suggesting autophagy inhibition, a feature previously described in advanced stages of NAFLD (Ding et al., 2020; Lee et al., 2020; Lu et al., 2020). Lee et al. attributed this phenomenon to long-term HFD feeding (Lee et al., 2020). Similar outcomes were obtained for SIRT1 expression, another regulator of autophagy through autophagy-related (ATG) protein modulation. Our findings showed a reduction in SIRT1 expression after 18 weeks of HFD feeding, as occurred in a HFD-induced NAFLD rat model (Deng et al., 2007). In liver diseases, mitochondrial dysfunction triggers oxidative stress, inflammation and lipotoxicity. Activation of the SIRT1/PGC-1α pathway effectively induces mitophagy, which is involved in mitochondrial dysfunction resolution (Jiang et al., 2021). Aside from autophagy, liver-specific SIRT1 knockout mice challenged with HFD developed hepatic steatosis, inflammation and endoplasmic reticulum stress. As a consequence of SIRT1 loss, PPARα signaling failed, decreasing fatty acid oxidation and PPARα target gene expression, including Cd36 expression, as our aforementioned findings show (Purushotham et al., 2009). Another hallmark of NAFLD is morphological alterations of mitochondria. Mitofusin proteins are located at the outer mitochondrial membrane and control fusion events, maintaining a proper energetic and metabolic cellular performance. In particular, MFN2 has a direct role in lipid transfer, especially phosphatidylserine, between mitochondria and endoplasmic reticulum, prompting progression of liver diseases if depleted, as our findings in Apoe−/− HFD mice showed (Hernández-Alvarez et al., 2019; Joaquim and Escobar-Henriques, 2020). Another consequence of a decrease in MFN2 is impairment of glucose, pyruvate and fatty acid oxidation. Under excessive lipid intake, this loss is also connected to mitophagy defects, leading to the accumulation of dysfunctional mitochondria, unmetabolized fatty acid overload in mitochondria and, ultimately, steatosis (Joaquim and Escobar-Henriques, 2020).
Next, we assessed the role of miRNAs in the regulation of proteins involved in autophagy. MTI databases predict that miR-34a-5p could be targeting mTOR and ULK1 expression, which is aligned with our results. It is also likely that autophagy regulation occurs elsewhere in the signaling pathway, owing to long-term presence of nutrients. After 8 weeks of HFD administration, our results revealed upregulated SIRT1, which has been reported as beneficial upon HFD exposure. The effects described in a previous study were the prevention of hepatosteatosis targeting SREBP1c, the induction of antioxidant enzymes, the protection against inflammation and diet-induced glucose intolerance (Pfluger et al., 2008). The upregulation and later downregulation of SIRT1 expression found in our HFD mouse model are consistent with miR-34a-5p expression differences, and several works have described the deacetylase as a miR-34a-5p target (Ding et al., 2015; Wang et al., 2020). Indeed, Choi et al. also reported that the elevation of hepatic miR-34a is related to the severity of NAFLD and reduced SIRT1 in the liver on account of targeting nicotinamide phosphoribosyltransferase (NAMPT) and, consequently, impairing NAD biosynthesis (Choi et al., 2013). In addition to miR-34a-5p, we have found that SIRT1 is also a target of miR-26b-5p, and its protein expression might have corresponded with a synergistic effect, although miR-26b-5p has not yet been described as a NAFLD mediator, unlike its family member, miR-26a (Ali et al., 2018; Xu et al., 2021a,b). Finally, we hypothesized that the lower levels of MFN2 in HFD-fed Apoe−/− mice could be due to miR-149-5p and/or miR-34a-5p regulation. Although MTI databases predicted the regulation of MFN2 by miR-149-5p, this has not yet been described. Conversely, in palmitic acid-induced muscle cell dysfunction, miR-34a-5p overexpression downregulated MFN2 expression (Simão et al., 2019). We found the same outcome in our miR-34a-5p overexpression assays in cultured hepatocytes.
Recently, EVs have gained attention owing to their potential role in disease development as a mechanism of cell–cell communication. Within their cargoes, miRNAs have emerged as promising non-invasive biomarkers for the diagnosis of many diseases, including NAFLD (Povero et al., 2014; Tan et al., 2014; Garcia-Martinez et al., 2020; Srinivas et al., 2021). According to the literature and our data, miR-34a and miR-375 may be potential biomarkers of NAFLD due to the marked increase in expression observed in the plasma of HFD-fed mice, as has been previously described in the serum of NAFLD patients (Yamada et al., 2013; Pirola et al., 2015). Furthermore, the higher plasma levels of miR-27b-3p and miR-122-5p, the more severe the disease. Hence, these two circulating miRNAs might serve as NAFLD-stage markers, supporting previous publications (Yamada et al., 2013; Tan et al., 2014). Finally, we propose circulating let-7d-5p and miR-146b-5p as biomarkers of early stages of NAFLD because both are only significantly altered in the STD-fed Apoe−/− group.
In summary, NAFLD is a multifactorial complex disease with limited diagnostic and treatment tools, which threatens a large part of the world’s population. Increasing knowledge of miRNAs and their performance and/or regulation has allowed the investigation of the mechanisms involved in obesity-associated metabolic diseases, including NAFLD. Considering the research on miRNA expression levels and MTI databases in NAFLD mice administered a HFD, we suggest that decreases in lipogenesis-related proteins, such as ACC, FAS and SCD1, are due to miR-34a-5p and miR-149-5p regulation. In relation to insulin resistance, they could be modulated by miR-34a-5p, miR-149-5p and miR-375-3p, targeting IR and p85α. We have also confirmed the pivotal role of autophagy in NAFLD, which is negatively impaired because miR-34a-5p induced a decrease in mTOR, ULK1, SIRT1 and MFN2 expression. Regarding circulating miRNAs, the initial stages of NAFLD might be diagnosed by measuring let-7d-5p and miR-146b-5p levels; the later stages are associated with notable increases in circulating miR-34a-5p and miR-375-3p levels. As disease progresses, miR-27b-3p and miR-122-5p plasma levels increase. The findings of this study have been summarized in Fig S7. Further investigation will be necessary to shed light on the interplay between miRNAs and their targets, as well as elucidate their participation in NAFLD progression and their potential use as biomarkers.
MATERIALS AND METHODS
Mice and diets
WT and Apoe−/− male mice, both of a C57BL/6J genetic background, were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and maintained on a 12 h light/dark cycle at room temperature. Animals were fed ad libitum with a STD (5.3% calories from fat; D.Rod18.H07, LASvendi GmbH, Soest, Germany) or HFD (60% calories from fat; TD 06414, Envigo Teklad, Barcelona, Spain) from weaning until euthanasia at 8 or 18 weeks. For each period of study, three groups (5-14 mice/group) were established: STD-fed WT, STD-fed Apoe−/− and HFD-fed Apoe−/−. Sacrifice of 16 h-fasted mice was performed after 8 or 18 weeks on the respective diets. All animal experimentation was conducted in accordance with the accepted standards of animal use approved by the Complutense University of Madrid Committee.
In vivo insulin signaling studies
In order to study the hepatic insulin signaling in physiological conditions, in vivo insulin signaling assays were performed. Fasted mice were intraperitoneally injected with 1 U/kg BW of insulin glulisine (Apidra SoloStar, Sanofi, Paris, France). After 10 min, mice were sacrificed and harvested tissues were immediately frozen in liquid nitrogen. Insulin signaling was assessed by western blot analysis of phospho-AKT (Ser473 and Thr308) in liver homogenates.
Metabolic tests
GTTs and ITTs were performed as we have previously described (Lopez-Pastor et al., 2019). Assays of total Ch and total TGs (Spinreact, Girona, Spain), as well as transaminase levels (Spinreact, Girona, Spain), were performed with plasma samples obtained from 16 h-fasted mice. According to Triglyceride Colorimetric Assay Kit (Cayman Chemical, Ann Arbor, MI, USA) instructions, intrahepatic TGs were measured in liver samples.
EV extraction
Briefly, plasma was previously clarified by two sequential 20-min centrifugations at 2000 g and 10,000 g. Then, EVs were pelleted with Total Exosome Precipitation Reagent (Invitrogen, Carlsbad, CA, USA). Pelleted vesicles were resuspended in Exosome Resuspension Buffer (Invitrogen) for further processing.
Cell culture
Huh7 cells were kindly supplied by Dr González-Rodríguez (Instituto de Investigación Sanitaria Princesa, Madrid, Spain). They were maintained in 75 cm2 flasks (Thermo Fisher Scientific, Waltham, MA, USA) in high-glucose Dulbecco's modified Eagle medium (Cytiva, Marlborough, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), 100 U/ml penicillin-streptomycin (Gibco) and MycoZap (Lonza, Basel, Switzerland).
Transfection with miR-26b-5p and miR-34a-5p miRNA precursors
Huh7 cells were seeded in 9.6 cm2 six-well plates in complete medium (1×105 cells per well). After 24 h, growth medium was replaced with fresh medium. Cells were then transfected with a miR-26b-5p and miR-34a-5p precursor (PM12899 and PM11030, respectively) (Ambion, Austin, TX, USA) using INTERFERin® (Polyplus Transfection, Strasbourg, France) as the transfection reagent. Once transfection conditions were established, final concentrations in the wells were 10 nM or 20 nM for the miR-26b-5p precursor, and 5 nM or 10 nM for the miR-34a-5p precursor, in 1 ml final volume. After 6 h, cells were supplemented with an additional milliliter of complete medium. The medium was discarded 96 h after transfection and cells were washed with cold phosphate-buffered saline (PBS) (Gibco).
PCR
Apoe−/− mice were genotyped by PCR with DNA AmpliGel Master Mix (Biotools, Madrid, Spain) as previously described (Gómez-Hernández et al., 2013). A 155-bp band was obtained for the WT mice and a 245-bp band for the Apoe−/− mice.
Isolation of RNA and miRNAs
According to the manufacturer's protocol, RNA was isolated from liver tissue and cultured cells with a miRVana miRNA Isolation Kit (Invitrogen), and RNA extraction from EVs was carried out with a Total Exosome RNA & Protein Isolation Kit (Invitrogen). In both cases, miRNAs and long RNAs were obtained in separate fractions.
RT-qPCR
Complementary DNA (cDNA) was synthesized by a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) for mRNA analysis. cDNA from miRNAs was obtained using a TaqMan™ Advanced miRNA cDNA Synthesis Kit (Applied Biosystems). Analysis of gene expression (Acaca, Adipor2, Cd36, Fasn, Ppargc1a, Scd1) was performed with the corresponding primers using PowerUp™ SYBR® Green Master Mix (Applied Biosystems). Analysis of Ccl2 and miRNA expression (let-7d-5p, miR-15b-5p, miR-22-3p, miR-26b-5p, miR-27b-3p, miR-34a-5p, miR-122-5p, miR-146b-5p, miR-149-5p, miR-181b-5p, miR-192-5p, miR-194-5p and miR-375-3p) was performed with TaqMan probes for the corresponding genes using TaqMan™ Fast Advanced Master Mix (Applied Biosystems). All RT-qPCR experiments were performed in an ABI Prism 7900HT Thermal Cycler (Applied Biosystems). The relative abundance of mRNAs or miRNAs was calculated using an endogenous reference gene (Gapdh for conventional TaqMan®, miR-191-5p for TaqMan® from miRNAs, miR-16-5p for TaqMan® from miRNAs of EVs, and Actb for SYBR®). SYBR Green primer sequences and TaqMan probes are shown in Tables S1 and S2, respectively. The results were calculated using the 2−ΔΔCq method.
Western blotting
Western blot analyses were performed on liver homogenates and cultured cells as previously described (Escribano et al., 2003). The primary antibodies used are shown in Table S3 and all of them were diluted in TTBS. Rabbit and mouse primary antibodies were immunodetected using horseradish peroxidase-conjugated anti-rabbit (NA931V; 1:4000 in TTBS) or anti-mouse secondary antibody (NA934V; 1:5000 in TTBS) (GE Healthcare, Buckinghamshire, UK), respectively. When possible, phospho-proteins and their total expression were detected in the same gel, using RestoreTM Western Blot Stripping Buffer (Thermo Fisher Scientific) as per the manufacturer's instructions, blocking the membrane again before the incubation with the other antibody. Loading was normalized by α-tubulin or β-actin. The representative gels that share the same housekeeper were as follows: (1) at 8 weeks, ACC and p70S6K; (2) at 8 weeks, p85α, SIRT1 and MFN2; (3) at 8 weeks, PKCε, ULK1 and p-ULK1; (4) at 18 weeks, ACC and p-ULK1; (5) at 18 weeks, FAS, IRβ, p85α and SIRT1; (6) at 18 weeks, SCD1 and LC3. The band intensities were quantified using ImageJ v1.52k software (http://rsb.info.nih.gov/ij).
Histological characterization of hepatic lesions
A section of liver samples was included in Tissue-Tek® optimum cutting temperature (OCT) compound (Sakura Finetek, Alphen aan den Rijn, The Netherlands), and later in liquid nitrogen for freezing, and another one was maintained in formalin to be included in paraffin. Each OCT-embedded liver block was serially sectioned (7 μm) with a cryostat (CM1510 S, Leica, Wetzlar, Germany) to perform the Oil Red O staining using standard techniques to quantify lipid accumulation. Paraffin-embedded livers were cross-sectioned into 4-μm-thick specimens at 5-mm intervals, dewaxed, and rehydrated to be stained with H&E to analyze the architecture or Sirius Red to measure the hepatic fibrosis. Images of sections of H&E, Sirius Red and Oil Red O were acquired using an inverted Eclipse TE300 microscope coupled to a Digital Sight DS-U2 camera (Nikon, Tokyo, Japan). Quantifications for images of Oil Red O staining were performed using IP Win32 v4.5 software (Acromag, Wixom, MI, USA). Lipid droplet size was quantified using ImageJ v1.52k software, and quantification for images of Sirius Red stains was carried out using Fiji software (NIH, Bethesda, MD, USA).
The assessment of NAS in liver samples was performed as we have previously described (Lopez-Pastor et al., 2019). Assessment of fibrosis stage was carried out by an experienced liver pathologist using the system for mouse models validated by Liang et al., which defined fibrosis staging as: 0, none; 1, perisinusoidal and/or pericentral; 2, incomplete central/central bridging fibrosis; 3, complete central/central bridging fibrosis (Liang et al., 2014).
In silico analysis of miRNAs and targets involved in NAFLD
The identification of the different miRNAs that were analyzed in the study was carried out through an exhaustive search for keywords and different publications in PubMed. Once the miRNAs of interest were chosen, the interaction between them and their mRNA targets was evaluated by searching prediction databases such as miRWalk v3.0, TargetScan Mouse v7.2, miRDB v5.0 and miRTarBase v6.0. The data collected from these databases were analyzed, and only those targets that appeared in two or more databases were considered as possible targets. The choice was also made considering that these targets were key proteins in the signaling pathways that can be altered in NAFLD such as lipogenesis, insulin signaling and autophagy. A diagram showing the MTIs during NAFLD progression that are proposed throughout the main text is supplied in Fig. S6. This representation was made using Cytoscape 3.8.2. software (Shannon et al., 2003).
Statistical analysis
Data are presented as mean±s.e.m. Normality of these variables was tested with Shapiro–Wilk test. Differences were assessed using unpaired two-tailed Student's t-tests and unpaired non-parametric Mann–Whitney U tests, as appropriate. P<0.05 was considered statistically significant. The software used for the analyses was Prism v8.0 (GraphPad Software Inc., San Diego, CA, USA).
Supplementary Material
Acknowledgements
The authors thank Alejandro Alonso Eugenio for his invaluable support in the preparation of Fig. S7.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.R.L.-P., J.I.-M., T.G.-I., A.G.-H., Ó.E.; Methodology: A.R.L.-P., J.I.-M., T.G.-I., P.G.-L., A.G.-H., Ó.E.; Investigation: A.R.L.-P., J.I.-M., T.G.-I., P.G.-L., Á.G.-R., C.G.-M., M.V.d.C., L.E., A.G.-H., Ó.E.; Resources: Á.G.-R., C.G.-M., M.V.d.C., L.E.; Data curation: A.R.L.-P.; Writing - original draft: A.R.L.-P.; Writing - review & editing: J.I.-M., A.G.-H., Ó.E.; Supervision: A.G.-H., Ó.E.; Project administration: A.G.-H., Ó.E.; Funding acquisition: A.G.-H., Ó.E.
Funding
This work was supported by grants from Ministerio de Ciencia, Innovación y Universidades (RTI-2018-095098-B100 to A.G-H. and Ó.E.) and Santander-Universidad Complutense de Madrid (PR75/18-21572 to Ó.E.). J.I.-M. and P.G.-L. were funded by Consejería de Ciencia, Universidades e Innovación from Comunidad de Madrid and the European Social Fund.
References
- Alessi, D. R., Andjelkovic, M., Caudwell, B., Cron, P., Morrice, N., Cohen, P. and Hemmings, B. A. (1996). Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541-6551. 10.1002/j.1460-2075.1996.tb01045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, O., Darwish, H. A., Eldeib, K. M. and Abdel Azim, S. A. (2018). miR-26a potentially contributes to the regulation of fatty acid and sterol metabolism in vitro human HepG2 cell model of nonalcoholic fatty liver disease. Oxid. Med. Cell. Long. 2018, 8515343. 10.1155/2018/8515343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab, J. P., Arrese, M. and Trauner, M. (2018). Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annu. Rev. Pathol. Mech.Dis. 13, 321-350. 10.1146/annurev-pathol-020117-043617 [DOI] [PubMed] [Google Scholar]
- Bettaieb, A., Jiang, J. X., Sasaki, Y., Chao, T.-I., Kiss, Z., Chen, X., Tian, J., Katsuyama, M., Yabe-Nishimura, C., Xi, Y.et al. (2015). Hepatocyte nicotinamide adenine dinucleotide phosphate reduced oxidase 4 regulates stress signaling, fibrosis, and insulin sensitivity during development of steatohepatitis in mice. Gastroenterology 149, 468-480.e10. 10.1053/j.gastro.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, R. E., Ferreira, D. M. S., Afonso, M. B., Borralho, P. M., Machado, M. V., Cortez-Pinto, H. and Rodrigues, C. M. P. (2013). miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 58, 119-125. 10.1016/j.jhep.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Castro, A. V. B., Kolka, C. M., Kim, S. P. and Bergman, R. N. (2014). Obesity, insulin resistance and comorbidities? Mechanisms of association. Arq. Bras. Endocrinol. Metabol. 58, 600-609. 10.1590/0004-2730000003223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave, S. C. and Gores, G. J. (2010). Mechanisms and clinical implications of hepatocyte lipoapoptosis. Clin. Lipidol. 5, 71-85. 10.2217/clp.09.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Chen, D., Yang, H., Wang, X., Wang, J. and Xu, C. (2020). Uric acid induced hepatocytes lipid accumulation through regulation of miR-149-5p/FGF21 axis. BMC Gastroenterol. 20, 39. 10.1186/s12876-020-01189-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.-E., Fu, T., Seok, S., Kim, D.-H., Yu, E., Lee, K.-W., Kang, Y., Li, X., Kemper, B. and Kemper, J. K. (2013). Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 12, 1062-1072. 10.1111/acel.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, J. D. Y., Lawrence, R. T., Healy, M. E., Dominy, J. E., Liao, J. A., Breen, D. S., Byrne, F. L., Kenwood, B. M., Lackner, C., Okutsu, S.et al. (2014). Genetic inhibition of hepatic acetyl-CoA carboxylase activity increases liver fat and alters global protein acetylation. Mol. Metab. 3, 419-431. 10.1016/j.molmet.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.-Q., Chen, L.-L. and Li, N.-X. (2007). The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 27, 708-715. 10.1111/j.1478-3231.2007.01497.x [DOI] [PubMed] [Google Scholar]
- Ding, J., Li, M., Wan, X., Jin, X., Chen, S., Yu, C. and Li, Y. (2015). Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 5, 13729. 10.1038/srep13729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, H., Ge, G., Tseng, Y., Ma, Y., Zhang, J. and Liu, J. (2020). Hepatic autophagy fluctuates during the development of non-alcoholic fatty liver disease. Ann. Hepatol. 19, 516-522. 10.1016/j.aohep.2020.06.001 [DOI] [PubMed] [Google Scholar]
- Donnelly, K. L., Smith, C. I., Schwarzenberg, S. J., Jessurun, J., Boldt, M. D. and Parks, E. J. (2005). Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 115, 1343-1351. 10.1172/JCI23621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano, O., Fernández-Moreno, M. D., Zueco, J. A., Menor, C., Fueyo, J., Ropero, R. M., Diaz-Laviada, I., Román, I. D. and Guijarro, L. G. (2003). Insulin receptor substrate-4 signaling in quiescent rat hepatocytes and in regenerating rat liver. Hepatology 37, 1461-1469. 10.1053/jhep.2003.50245 [DOI] [PubMed] [Google Scholar]
- Eslam, M., Sanyal, A. J., George, J., Sanyal, A., Neuschwander-Tetri, B., Tiribelli, C., Kleiner, D. E., Brunt, E., Bugianesi, E., Yki-Järvinen, H.et al. (2020). MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 158, 1999-2014.e1. 10.1053/j.gastro.2019.11.312 [DOI] [PubMed] [Google Scholar]
- European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) and European Association for the Study of Obesity (EASO) (2016). EASL–EASD–EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 64, 1388-1402. 10.1016/j.jhep.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Fernández Gianotti, T., Burgueño, A., Gonzales Mansilla, N., Pirola, C. J. and Sookoian, S. (2013). Fatty liver is associated with transcriptional downregulation of stearoyl-CoA desaturase and impaired protein dimerization. PLoS ONE 8, e76912. 10.1371/journal.pone.0076912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. and Sanyal, A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908-922. 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, T. and Kemper, J. K. (2016). MicroRNA-34a and impaired FGF19/21 signaling in obesity. Vitam. Horm. 101, 175-196. 10.1016/bs.vh.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez, I., Alen, R., Rada, P. and Valverde, A. M. (2020). Insights into extracellular vesicles as biomarker of NAFLD pathogenesis. Front. Med. 7, 395. 10.3389/fmed.2020.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Monzón, C., Lo Iacono, O., Crespo, J., Romero-Gómez, M., García-Samaniego, J., Fernández-Bermejo, M., Domínguez-Díez, A., Rodríguez de Cía, J., Sáez, A., Porrero, J. L.et al. (2014). Increased soluble CD36 is linked to advanced steatosis in nonalcoholic fatty liver disease. Eur. J. Clin. Invest. 44, 65-73. 10.1111/eci.12192 [DOI] [PubMed] [Google Scholar]
- Gómez-Hernández, A., Escribano, Ó., Perdomo, L., Otero, Y. F., García-Gómez, G., Fernández, S., Beneit, N. and Benito, M. (2013). Implication of insulin receptor A isoform and IRA/IGF-IR hybrid receptors in the aortic vascular smooth muscle cell proliferation: role of TNF-α and IGF-II. Endocrinology 154, 2352-2364. 10.1210/en.2012-2161 [DOI] [PubMed] [Google Scholar]
- González-Muniesa, P., Mártinez-González, M.-A., Hu, F. B., Després, J.-P., Matsuzawa, Y., Loos, R. J. F., Moreno, L. A., Bray, G. A. and Martinez, J. A. (2017). Obesity. Nat. Rev. Dis. Primers 3, 17034. 10.1038/nrdp.2017.34 [DOI] [PubMed] [Google Scholar]
- Haas, J. T., Miao, J., Chanda, D., Wang, Y., Zhao, E., Haas, M. E., Hirschey, M., Vaitheesvaran, B., Farese, R. V., Kurland, I. J.et al. (2012). Hepatic insulin signaling is required for obesity-dependent expression of SREBP-1c mRNA but not for feeding-dependent expression. Cell Metab. 15, 873-884. 10.1016/j.cmet.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, H. H., Feigh, M., Veidal, S. S., Rigbolt, K. T., Vrang, N. and Fosgerau, K. (2017). Mouse models of nonalcoholic steatohepatitis in preclinical drug development. Drug Discov. Today 22, 1707-1718. 10.1016/j.drudis.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Hartig, S. M., Hamilton, M. P., Bader, D. A. and Mcguire, S. E. (2015). The miRNA interactome in metabolic homeostasis. Trends Endocrinol. Metab. 26, 733-745. 10.1016/j.tem.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Alvarez, M. I., Sebastián, D., Vives, S., Ivanova, S., Bartoccioni, P., Kakimoto, P., Plana, N., Veiga, S. R., Hernández, V., Vasconcelos, N.et al. (2019). Deficient endoplasmic reticulum-mitochondrial phosphatidylserine transfer causes liver disease. Cell 177, 881-895.e17. 10.1016/j.cell.2019.04.010 [DOI] [PubMed] [Google Scholar]
- Huntzinger, E. and Izaurralde, E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99-110. 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- Hussain, M. U. (2012). Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell Tissue Res. 349, 405-413. 10.1007/s00441-012-1438-0 [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Chen, D., Gong, Q., Xu, Q., Pan, D., Lu, F. and Tang, Q. (2021). Elucidation of SIRT-1/PGC-1α-associated mitochondrial dysfunction and autophagy in nonalcoholic fatty liver disease. Lipids Health Dis. 20, 40. 10.1186/s12944-021-01461-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joaquim, M. and Escobar-Henriques, M. (2020). Role of mitofusins and mitophagy in life or death decisions. Front. Cell Dev. Biol. 8, 572182. 10.3389/fcell.2020.572182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jornayvaz, F. R., Birkenfeld, A. L., Jurczak, M. J., Kanda, S., Guigni, B. A., Jiang, D. C., Zhang, D., Lee, H.-Y., Samuel, V. T. and Shulman, G. I. (2011). Hepatic insulin resistance in mice with hepatic overexpression of diacylglycerol acyltransferase 2. Proc. Natl. Acad.Sci. USA 108, 5748-5752. 10.1073/pnas.1103451108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner, D. E., Brunt, E. M., Van Natta, M., Behling, C., Contos, M. J., Cummings, O. W., Ferrell, L. D., Liu, Y.-C., Torbenson, M. S., Unalp-Arida, A.et al. (2005). Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313-1321. 10.1002/hep.20701 [DOI] [PubMed] [Google Scholar]
- Lee, J., Padhye, A., Sharma, A., Song, G., Miao, J., Mo, Y.-Y., Wang, L. and Kemper, J. K. (2010). A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 285, 12604-12611. 10.1074/jbc.M109.094524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.-H., Park, S.-H., Ahn, J., Hong, S. P., Lee, E., Jang, Y.-J., Ha, T.-Y., Huh, Y. H., Ha, S.-Y., Jeon, T.-I.et al. (2020). Mir214-3p and Hnf4a/Hnf4α reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy 17, 2415-2431. 10.1080/15548627.2020.1827779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L., Zhou, C., Yang, X. and Li, L. (2018). Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 45, 819-831. 10.1111/1440-1681.12940 [DOI] [PubMed] [Google Scholar]
- Li, M., Xu, C., Shi, J., Ding, J., Wan, X., Chen, D., Gao, J., Li, C., Zhang, J., Lin, Y.et al. (2018). Fatty acids promote fatty liver disease via the dysregulation of 3-mercaptopyruvate sulfurtransferase/hydrogen sulfide pathway. Gut 67, 2169-2180. 10.1136/gutjnl-2017-313778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, W., Menke, A. L., Driessen, A., Koek, G. H., Lindeman, J. H., Stoop, R., Havekes, L. M., Kleemann, R. and Van Den Hoek, A. M. (2014). Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS ONE 9, e115922. 10.1371/journal.pone.0115922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pastor, A. R., Gomez-Hernandez, A., Diaz-Castroverde, S., Gonzalez-Aseguinolaza, G., Gonzalez-Rodriguez, A., Garcia, G., Fernandez, S., Escribano, O. and Benito, M. (2019). Liver-specific insulin receptor isoform A expression enhances hepatic glucose uptake and ameliorates liver steatosis in a mouse model of diet-induced obesity. Dis. Models Mech. 12, dmm036186. 10.1242/dmm.036186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., Mei, J., Yang, J., Wu, Z., Liu, J., Miao, P., Chen, Y., Wen, Z., Zhao, Z., Kong, H.et al. (2020). ApoE deficiency promotes non-alcoholic fatty liver disease in mice via impeding AMPK/mTOR mediated autophagy. Life Sci. 252, 117601. 10.1016/j.lfs.2020.117601 [DOI] [PubMed] [Google Scholar]
- Luo, M., Mengos, A. E., Stubblefield, T. M. and Mandarino, L. J. (2012). High fat diet-induced changes in hepatic protein abundance in mice. J. Proteomics Bioinform. 5, 60-66. 10.4172/jpb.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, K., Zhang, Y., Zhang, D., Kahn, M., Ter Horst, K. W., Rodrigues, M. R. S., Gaspar, R. C., Hirabara, S. M., Luukkonen, P. K., Lee, S.et al. (2020). A membrane-bound diacylglycerol species induces PKCε-mediated hepatic insulin resistance. Cell Metab. 32, 654-664.e5. 10.1016/j.cmet.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan, K., Bhardwaj, P., Thareja, S., Gupta, S. D. and Saraya, A. (2006). Oxidant stress and antioxidant status among patients with nonalcoholic fatty liver disease (NAFLD). J. Clin. Gastroenterol. 40, 930-935. 10.1097/01.mcg.0000212608.59090.08 [DOI] [PubMed] [Google Scholar]
- Marra, F., Gastaldelli, A., Svegliati Baroni, G., Tell, G. and Tiribelli, C. (2008). Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol. Med. 14, 72-81. 10.1016/j.molmed.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Mcpherson, S., Hardy, T., Henderson, E., Burt, A. D., Day, C. P. and Anstee, Q. M. (2015). Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J. Hepatol. 62, 1148-1155. 10.1016/j.jhep.2014.11.034 [DOI] [PubMed] [Google Scholar]
- Merry, T. L., Hedges, C. P., Masson, S. W., Laube, B., Pöhlmann, D., Wueest, S., Walsh, M. E., Arnold, M., Langhans, W., Konrad, D.et al. (2020). Partial impairment of insulin receptor expression mimics fasting to prevent diet-induced fatty liver disease. Nat. Commun. 11, 2080. 10.1038/s41467-020-15623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, M. D., Kulkarni, R. N., Postic, C., Previs, S. F., Shulman, G. I., Magnuson, M. A. and Kahn, C. R. (2000). Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87-97. 10.1016/S1097-2765(05)00015-8 [DOI] [PubMed] [Google Scholar]
- Monetti, M., Levin, M. C., Watt, M. J., Sajan, M. P., Marmor, S., Hubbard, B. K., Stevens, R. D., Bain, J. R., Newgard, C. B., Farese, R. V.et al. (2007). Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 6, 69-78. 10.1016/j.cmet.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B. and Kahn, C. R. (2019). Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 30, 656-673. 10.1016/j.cmet.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, E. M., Meers, G. M. E., Booth, F. W., Fritsche, K. L., Hardin, C. D., Thyfault, J. P. and Ibdah, J. A. (2012). PGC-1α overexpression results in increased hepatic fatty acid oxidation with reduced triacylglycerol accumulation and secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G979-G992. 10.1152/ajpgi.00169.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio, D. M. and Newgard, C. B. (2008). Molecular and metabolic mechanisms of insulin resistance and β-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9, 193-205. 10.1038/nrm2327 [DOI] [PubMed] [Google Scholar]
- Nascimento, J. C. R., Matos, G. A., Pereira, L. C., Mourão, A. E. C. C. B., Sampaio, A. M., Oriá, R. B. and Toniutto, P. (2020). Impact of apolipoprotein E genetic polymorphisms on liver disease: an essential review. Ann. Hepatol. 19, 24-30. 10.1016/j.aohep.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Petersen, M. C., Madiraju, A. K., Gassaway, B. M., Marcel, M., Nasiri, A. R., Butrico, G., Marcucci, M. J., Zhang, D., Abulizi, A., Zhang, X.-M.et al. (2016). Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J. Clin. Invest. 126, 4361-4371. 10.1172/JCI86013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger, P. T., Herranz, D., Velasco-Miguel, S., Serrano, M. and Tschop, M. H. (2008). Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl Acad. Sci. USA 105, 9793-9798. 10.1073/pnas.0802917105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierantonelli, I. and Svegliati-Baroni, G. (2019). Nonalcoholic fatty liver disease: basic pathogenetic mechanisms in the progression from NAFLD to NASH. Transplantation 103, e1-e13. 10.1097/TP.0000000000002480 [DOI] [PubMed] [Google Scholar]
- Pirola, C. J., Fernández Gianotti, T., Castaño, G. O., Mallardi, P., San Martino, J., Mora Gonzalez Lopez Ledesma, M., Flichman, D., Mirshahi, F., Sanyal, A. J. and Sookoian, S. (2015). Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 64, 800-812. 10.1136/gutjnl-2014-306996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero, D., Eguchi, A., Li, H., Johnson, C. D., Papouchado, B. G., Wree, A., Messer, K. and Feldstein, A. E. (2014). Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS ONE 9, e113651. 10.1371/journal.pone.0113651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P. E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P.et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226-230. 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- Poy, M. N., Hausser, J., Trajkovski, M., Braun, M., Collins, S., Rorsman, P., Zavolan, M. and Stoffel, M. (2009). miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl Acad. Sci. USA 106, 5813-5818. 10.1073/pnas.0810550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham, A., Schug, T. T., Xu, Q., Surapureddi, S., Guo, X. and Li, X. (2009). Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327-338. 10.1016/j.cmet.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada, P., González-Rodríguez, Á., García-Monzón, C. and Valverde, Á. M. (2020). Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis. 11, 802. 10.1038/s41419-020-03003-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz, K., Turner, N., Frangioudakis, G., Liao, B. M., Pedersen, D. J., Cantley, J., Wilks, D., Preston, E., Hegarty, B. D., Leitges, M.et al. (2011). Time-dependent effects of Prkce deletion on glucose homeostasis and hepatic lipid metabolism on dietary lipid oversupply in mice. Diabetologia 54, 1447-1456. 10.1007/s00125-011-2073-0 [DOI] [PubMed] [Google Scholar]
- Rinella, M. E. and Sanyal, A. J. (2016). Management of NAFLD: A stage-based approach. Nat. Rev. Gastroenterol. Hepatol. 13, 196-205. 10.1038/nrgastro.2016.3 [DOI] [PubMed] [Google Scholar]
- Saltiel, A. R. and Kahn, C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799-806. 10.1038/414799a [DOI] [PubMed] [Google Scholar]
- Schierwagen, R., Maybüchen, L., Zimmer, S., Hittatiya, K., Bäck, C., Klein, S., Uschner, F. E., Reul, W., Boor, P., Nickenig, G.et al. (2015). Seven weeks of Western diet in apolipoprotein-E-deficient mice induce metabolic syndrome and non-alcoholic steatohepatitis with liver fibrosis. Sci. Rep. 5, 12931. 10.1038/srep12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, M. B., Weller, S. G., Schulze, R. J., Krueger, E. W., Drizyte-Miller, K., Casey, C. A. and Mcniven, M. A. (2019). Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 218, 3320-3335. 10.1083/jcb.201803153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. and Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão, A. L., Afonso, M. B., Rodrigues, P. M., Gama-Carvalho, M., Machado, M. V., Cortez-Pinto, H., Rodrigues, C. M. P. and Castro, R. E. (2019). Skeletal muscle miR-34a/SIRT1:AMPK axis is activated in experimental and human non-alcoholic steatohepatitis. J. Mol. Med. 97, 1113-1126. 10.1007/s00109-019-01796-8 [DOI] [PubMed] [Google Scholar]
- Srinivas, A. N., Suresh, D., Santhekadur, P. K., Suvarna, D. and Kumar, D. P. (2021). Extracellular vesicles as inflammatory drivers in NAFLD. Front. Immunol. 11, 627424. 10.3389/fimmu.2020.627424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumvoll, M., Goldstein, B. J. and Van Haeften, T. W. (2005). Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365, 1333-1346. 10.1016/S0140-6736(05)61032-X [DOI] [PubMed] [Google Scholar]
- Tan, Y., Ge, G., Pan, T., Wen, D. and Gan, J. (2014). A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 9, e105192. 10.1371/journal.pone.0105192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, C. M., Emanuelli, B. and Kahn, C. R. (2006). Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85-96. 10.1038/nrm1837 [DOI] [PubMed] [Google Scholar]
- Tilg, H. and Effenberger, M. (2020). From NAFLD to MAFLD: when pathophysiology succeeds. Nat. Rev. Gastroenterol. Hepatol. 17, 387-388. 10.1038/s41575-020-0316-6 [DOI] [PubMed] [Google Scholar]
- Vernon, G., Baranova, A. and Younossi, Z. M. (2011). Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment. Pharmacol. Ther. 34, 274-285. 10.1111/j.1365-2036.2011.04724.x [DOI] [PubMed] [Google Scholar]
- Wang, L., Sun, M., Cao, Y., Ma, L., Shen, Y., Velikanova, A. A., Li, X., Sun, C. and Zhao, Y. (2020). miR-34a regulates lipid metabolism by targeting SIRT1 in non-alcoholic fatty liver disease with iron overload. Arch. Biochem. Biophys.. 695, 108642. 10.1016/j.abb.2020.108642 [DOI] [PubMed] [Google Scholar]
- Wen, F., An, C., Wu, X., Yang, Y., Xu, J., Liu, Y., Wang, C., Nie, L., Fang, H. and Yang, Z. (2018). MiR-34a regulates mitochondrial content and fat ectopic deposition induced by resistin through the AMPK/PPARα pathway in HepG2 cells. Int. J. Biochem. Cell Biol. 94, 133-145. 10.1016/j.biocel.2017.11.008 [DOI] [PubMed] [Google Scholar]
- Wong, R. J., Cheung, R. and Ahmed, A. (2014). Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology 59, 2188-2195. 10.1002/hep.26986 [DOI] [PubMed] [Google Scholar]
- Wree, A., Broderick, L., Canbay, A., Hoffman, H. M. and Feldstein, A. E. (2013). From NAFLD to NASH to cirrhosis—new insights into disease mechanisms. Nature Reviews Gastroenterology and Hepatology 10, 627-636. 10.1038/nrgastro.2013.149 [DOI] [PubMed] [Google Scholar]
- Xiao, J., Lv, D., Zhao, Y., Chen, X., Song, M., Liu, J., Bei, Y., Wang, F., Yang, W. and Yang, C. (2016). miR-149 controls non-alcoholic fatty liver by targeting FGF-21. J. Cell. Mol. Med. 20, 1603-1608. 10.1111/jcmm.12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Kitade, H., Ni, Y. and Ota, T. (2015). Roles of chemokines and chemokine receptors in obesity-associated insulin resistance and nonalcoholic fatty liver disease. Biomolecules 5, 1563-1579. 10.3390/biom5031563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Tian, Y., Tang, D., Zou, S., Liu, G., Song, J., Zhang, G., Du, X., Huang, W., He, B.et al. (2021a). An endoplasmic reticulum stress-microRNA-26a feedback circuit in NAFLD. Hepatology 73, 1327-1345. 10.1002/hep.31428 [DOI] [PubMed] [Google Scholar]
- Xu, Y., Zhu, Y., Hu, S., Pan, X., Bawa, F. C., Wang, H. H., Wang, D. Q.-H., Yin, L. and Zhang, Y. (2021b). Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol. Metab. 51, 101244. 10.1016/j.molmet.2021.101244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, K., Ichino, N., Ando, Y., Sawada, A., Osakabe, K., Sugimoto, K., Ohashi, K., Teradaira, R., Inoue, T.et al. (2013). Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta 424, 99-103. 10.1016/j.cca.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Yokota, S., Oda, T. and Fahimi, H. D. (2001). The role of 15-lipoxygenase in disruption of the peroxisomal membrane and in programmed degradation of peroxisomes in normal rat liver. J. Histochem. Cytochem. 49, 613-621. 10.1177/002215540104900508 [DOI] [PubMed] [Google Scholar]
- Younossi, Z., Anstee, Q. M., Marietti, M., Hardy, T., Henry, L., Eslam, M., George, J. and Bugianesi, E. (2018). Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11-20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Wu, K., Bo, T., Zhou, L., Gao, L., Zhou, X. and Chen, W. (2020). Integrated microRNA and proteome analysis reveal a regulatory module in hepatic lipid metabolism disorders in mice with subclinical hypothyroidism. Exp. Ther. Med. 19, 897-906. 10.3892/etm.2019.8281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Chen, Z., Zhou, Z., Li, Y., Wang, Y., Zhou, Z., Lu, H., Sun, C. and Chu, X. (2019). High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with high-fat diet-induced hepatic insulin resistance in mice. Genes Nutr. 14, 6. 10.1186/s12263-019-0630-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S., Zhao, L., Wang, Y., Zhang, C., Liu, J., Wang, P., Zhou, W., Yang, P., Varghese, Z., Moorhead, J. F.et al. (2017). Cluster of differentiation 36 deficiency aggravates macrophage infiltration and hepatic inflammation by upregulating monocyte chemotactic protein-1 expression of hepatocytes through histone deacetylase 2-dependent pathway. Antioxid Redox Signal. 27, 201-214. 10.1089/ars.2016.6808 [DOI] [PubMed] [Google Scholar]
- Zhou, L., Zhang, J., Fang, Q., Liu, M., Liu, X., Jia, W., Dong, L. Q. and Liu, F. (2009). Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol. Pharmacol. 76, 596-603. 10.1124/mol.109.057067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.