ABSTRACT
Protein abnormalities can accelerate aging causing protein misfolding diseases, and various adaptive responses have evolved to relieve proteotoxicity. To trigger these responses, cells must detect the buildup of aberrant proteins. Previously we demonstrated that the Hsp70–Bag3 (HB) complex senses the accumulation of defective ribosomal products, stimulating signaling pathway proteins, such as stress kinases or the Hippo pathway kinase LATS1. Here, we studied how Bag3 regulates the ability for LATS1 to regulate its key downstream target YAP (also known as YAP1). In naïve cells, Bag3 recruited a complex of LATS1, YAP and the scaffold AmotL2, which links LATS1 and YAP. Upon inhibition of the proteasome, AmotL2 dissociated from Bag3, which prevented phosphorylation of YAP by LATS1, and led to consequent nuclear YAP localization together with Bag3. Mutations in Bag3 that enhanced its translocation into nucleus also facilitated nuclear translocation of YAP. Interestingly, Bag3 also controlled YAP nuclear localization in response to cell density, indicating broader roles beyond proteotoxic signaling responses for Bag3 in the regulation of YAP. These data implicate Bag3 as a regulator of Hippo pathway signaling, and suggest mechanisms by which proteotoxic stress signals are propagated.
KEY WORDS: Bag3, Lats1, Proteotoxicity
Summary: We show how the Hsp70–Bag3 module regulates the Hippo pathway. This regulatory mechanism involves control of nuclear translocation of the transcription factor Yap.
INTRODUCTION
In aging and protein conformation disorders, organisms face malfunction or failure of the ubiquitin-proteasome system (UPS), which causes a major buildup of abnormal proteins. Such abnormal species increase the load on the protein degradation and folding machineries (Sherman and Goldberg, 2001). Upon accumulation in the cell, these species can form toxic aggregates, and further exacerbate ageing and facilitate age-related protein conformation disorders (Johnston and Samant, 2021; Sherman and Goldberg, 2001). In response to proteasome insufficiency, cells activate pathways that can relieve the proteotoxic stress, such as aggresome formation or autophagy (Johnston and Samant, 2021). Other pathways activated under these conditions, for examples, those mediated by JNK stress kinases and p38 MAPK proteins, the heat shock response, the unfolded protein response and that mediated by NRF2 , can regulate programmed cell death and other aspects of cell physiology (Bush et al., 1997; Lee and Goldberg, 1998; Meriin et al., 1998; Taguchi et al., 2011). Previously, we demonstrated that a protein complex that includes the heat-shock protein Hsp70 (herein referring to HspA1 and HspA8) and co-chaperone Bag3 (HB module) monitors abnormal polypeptides in the cytosol upon proteasome inhibition and regulates the activities of a series of signaling pathways (Meriin et al., 2018). One such signaling pathway regulated by the HB complex is the Hippo pathway, which plays important roles in integrating cell density or mechanical stress cues. LATS1, which is a major effector kinase of the Hippo pathway, directly interacts with Bag3 in the HB complex (Ulbricht et al., 2013). This interaction is mediated by the WW domain of Bag3 (Ulbricht et al., 2013), and depends on association of the latter with Hsp70 (Meriin et al., 2018).
Major targets of LATS1 include the transcriptional effectors YAP and Taz (also known as YAP1 and WWTR1, respectively) (Zhu et al., 2015), with phosphorylation stabilizing their association with 14-3-3 proteins, which anchor YAP in the cytoplasm and prevent nuclear translocation (Zhu et al., 2015). Accordingly, reduced LATS1 and LATS2 (LATS1/2) activity results in YAP nuclear translocation and transcriptional regulation (Meng et al., 2016). LATS1-dependent YAP phosphorylation is mediated by the scaffold protein AmotL2 (Varelas, 2014). LATS1 has a UBA domain, which binds monoubiquitin on AmotL2 and recruits the latter together with YAP (Kim et al., 2016). Diverse upstream cues have been shown to regulate the LATS kinases, including signals regulating the Mst1 and Mst2 kinases (Mst1/2; also known as STK4 and STK3, respectively) and signals induced by increased cell density (Meng et al., 2016).
The molecular mechanisms by which the HB complex regulates downstream signaling pathways upon proteotoxic stress are poorly characterized. Here, we studied these mechanisms in relation to how the buildup of abnormal polypeptides impacted LATS1-mediated phosphorylation and nuclear translocation of YAP. We demonstrate that under proteotoxic stress, the HB complex regulates the association of LATS1 and YAP with AmotL2, and that Bag3 plays a wide-spread role in regulating YAP nuclear localization.
RESULTS
Proteotoxicity regulates association of AmotL2 with the Hsp70–Bag3 module
Previously, we have shown that the Hsp70–Bag3 (HB) module ‘senses’ accumulation of abnormal polypeptides and triggers signaling events involved in adaptation to proteotoxicity. We had found that the phosphorylation of the Hippo pathway effector YAP by the LATS1 kinase was reduced in response to the buildup of abnormal proteins in a HB-dependent manner (Meriin et al., 2018), which was interesting given that LATS1 directly interacts with Bag3 (Ulbricht et al., 2013).
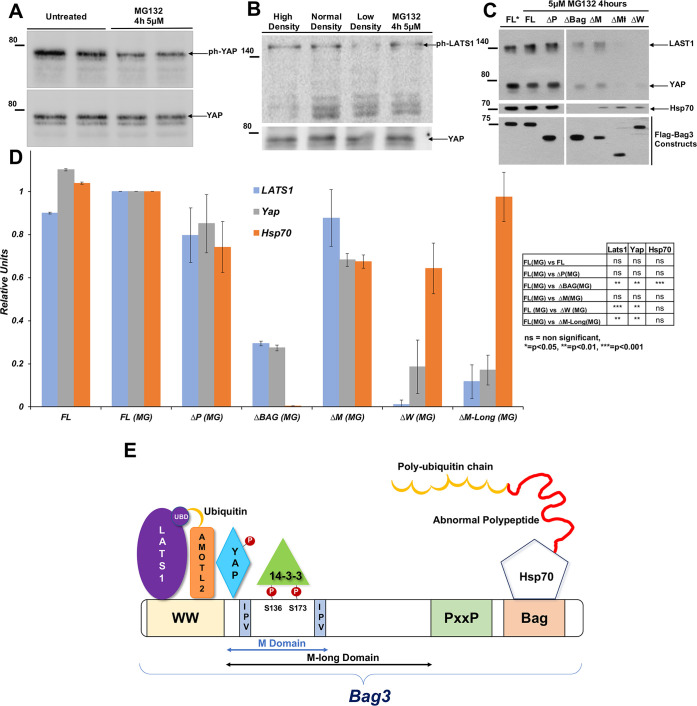
LATS1/2 activity is regulated by a variety of stimuli, which canonically lead to LATS phosphorylation by upstream kinases, such as Mst1/2 (Meng et al., 2016). To investigate how the HB complex regulates LATS1 activity in response to the proteotoxic stress, we measured the levels of LATS1 phosphorylation following the treatment with the proteasome inhibitor MG132, which results in proteotoxic stress, by immunoblotting with antibodies against phospho-S909-LATS1, a modification associated with LATS1 activation (Chan et al., 2005). We observed that incubation of cells with MG132 reduced the phosphorylation of YAP (Meriin et al., 2018) (Fig. 1A), but did not affect the levels of phospho-LATS1 (Fig. 1B). However, shifting cells from a high to low cell density, as expected, reduced phospho-LATS1 levels, suggesting that the regulation of LATS-dependent YAP phosphorylation by proteotoxic stress involves unconventional mechanisms.
Fig. 1.
Non-canonical regulation of LATS1/2 activity by Bag3. (A) Reduced phosphorylation of YAP following treatment with MG132. (B) Effect of MG132 and cell density on the phosphorylation of LATS1. MCF10A cells were plated on wells of a 96-well plate (3000 cells for low density, 6500 cells normal density and 19,500 cells for high density). Cells of normal density were also treated with 5 µM MG132 for 4 h. Images in A and B are representative of three experiments. (C) Association of LATS1, YAP and Hsp70 with various deletion mutants of Bag3. Cells were transfected with plasmids encoding normal (full-length) Bag3 and indicated deletion mutants. Cells were either untreated or treated with MG132 for 4 h (MG). Bag3 versions were pulled down using a Co2+-affinity column, and eluates were blotted with Hsp70, YAP and LATS1 antibodies. FL, full-length Bag3; ΔP, deletion of PxxP; ΔBag, deletion of Bag domain; ΔM, deletion of M domain short (see scheme on the lower panel); ΔW, deletion of WW domain; ΔM-long, deletion of M-domain long. (D) Quantification of data in C. The values for LATS1, Hsp70 and YAP were normalized by the content of Bag3 constructs in these samples and expressed as a fraction of association with the FL Bag3. Values for association of the proteins with the FL Bag3 in MG132-teated cells were taken as 1. To the right of the graph statistical significance values for differences between association of proteins with Bag3 deletion constructs and FL Bag3. The statistical values were calculated using the unpaired two-tailed t-test. (E) Hsp70–Bag3 interaction model based on the pulldown data.
To uncover these mechanisms, we tested whether YAP associates with the HB module. His-tagged Bag3 was expressed in HeLa cells and pulled down using a Co2+ column. The presence of the associated YAP and other components in the eluted fraction was detected by immunoblotting with the corresponding antibodies. YAP was clearly associated with Bag3 (Fig. 1C,D). This suggests that phosphorylation of YAP by LATS1/2 might be mediated by association with the HB module and that proteotoxic stress may lead to dissociation of LATS1 or YAP from the HB module. However, we found that MG132 did not affect the association of Bag3 with either LATS1 or YAP (Fig. 1C,D), indicating that the mechanism linking proteotoxic stress with the regulation of YAP phosphorylation by LATS1 does not involve dissociation of either LATS or YAP from the HB complex.
To identify protein interaction regions, we analyzed a series of Bag3 deletion mutants. Cells were transfected with plasmids expressing various Bag3 deletion constructs, including ΔBag, a deletion mutant that lacks the Bag domain, which is essential for interaction with Hsp70 (Merabova et al., 2015; Rauch, et al., 2017), ΔP, a deletion mutant that lacks the PxxP domain, ΔW, a deletion mutant that lacks the WW domain, ΔM-short, a deletion mutant that lacks the region of small heat-shock protein binding sites, and ΔM-long, a deletion mutant that lacks the entire region between the WW and PxxP regions. Cells were treated with the proteasome inhibitor MG132, and Bag3, together with the associated proteins, was isolated using cobalt affinity resin and the complexes were analyzed by immunoblotting. Bag3 complexes isolated from cells transfected with full-length Bag3 were used as control. As reported previously (Takayama et al., 1999), the level of Hsp70 family members was drastically reduced in the ΔBag pulldown compared to the control, indicating specificity and quantification reliability of the analysis (Fig. 1C,D). Deletion of the WW domain completely prevented association of LATS1, and reduced association of YAP with Bag3 (Fig. 1C,D). Of note, association of Bag3 with Hsp70 was not significantly influenced by the deletion. The M-long deletion reduced association of both LATS and YAP. On the other hand, the deletion mutants ΔM-short and ΔP did not significantly affect interaction between Bag3, LATS1 and YAP. These data indicate that the WW domain within Bag3 is critical for association with LATS1 with some additional contribution by the M-domain. Both the WW and M-domains also mediated interaction with YAP (Fig. 1E).
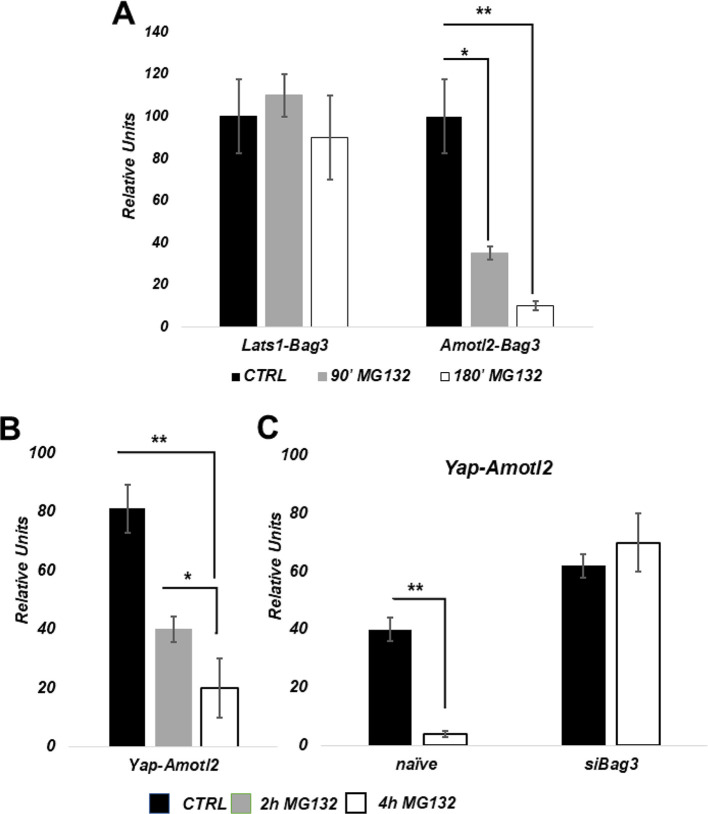
Previous studies have shown that AmotL2 serves as a scaffold that bridges LATS1 and YAP association, which is essential for LATS-dependent phosphorylation of YAP (Chan et al., 2005; Varelas, 2014). Interestingly, association of Amotl2 with Bag3 has been reported in proteomics studies (BAG3 Result Summary|BioGRID), suggesting that Amotl2 might be linked to Bag3-mediated regulation of YAP. To validate the association of Amotl2 and Bag3 we employed a quantitative proximity ligation assay developed in our previous studies (Meriin et al., 2018, and see Materials and Methods). Unlike standard proximity ligation assay, we do not image protein–protein interactions microscopically, but rather evaluate the interactions by qPCR of the ligated oligonucleotide product that is formed only when antibodies against proteins of interest are localized in proximity to each other. We found that this assay is significantly more sensitive and more quantitative, and has several other advantages compared to pulldowns. qPCR detected significant association between Amotl2 and Bag3 in naïve cells, and this interaction was strongly reduced in response to MG132 (Fig. 2A). Under these conditions, association of Amotl2 with YAP was also dramatically reduced, as measure similarly by the quantitative proximity ligation assay (Fig. 2B). Therefore, our observations indicate that, in naïve cells, the LATS–Amotl2–YAP complex associates with the HB module, and that Amotl2 dissociates from this complex upon proteotoxic stress.
Fig. 2.
Effects of proteotoxic stress on the HB–LATS–AmotL2–YAP complex. (A) Effects of proteasome inhibition on the association between LATS1–Bag3 and AmotL2–Bag3 (the interacting proteins are indicated underneath each set) measured by the proximity ligation assay (see Materials and Methods). MCF10A cells were treated with 5 µM MG132 for the indicated time periods, cells were fixed, and the interactions measured. (B) Effects of proteasome inhibition on association between AmotL2 and YAP. (C) Effects of Bag3 depletion on association between AmotL2 and YAP. *P≤0.05; **P≤0.01 (unpaired two-tailed t-test).
We previously reported that Bag3 is required for the suppression of YAP phosphorylation in response to proteotoxic stress (Meriin et al., 2018), raising the possibility that Bag3 is required for the dissociation of Amotl2 from LATS–YAP complexes. To test this possibility, we measured interaction between Amotl2 and YAP in the presence and the absence of Bag3. Fig. 2C shows that the association between Amotl2 and YAP was higher in Bag3-depleted cells. Importantly, upon proteasome inhibition, Amotl2–YAP interaction was reduced in control cells, while in Bag3-depleted cells there was no dissociation between Amotl2 and YAP (Fig. 2C). Therefore, Bag3 plays a critical role in the dissociation of Amotl2 from YAP upon proteotoxic stress.
Bag3 facilitates the nuclear translocation of YAP
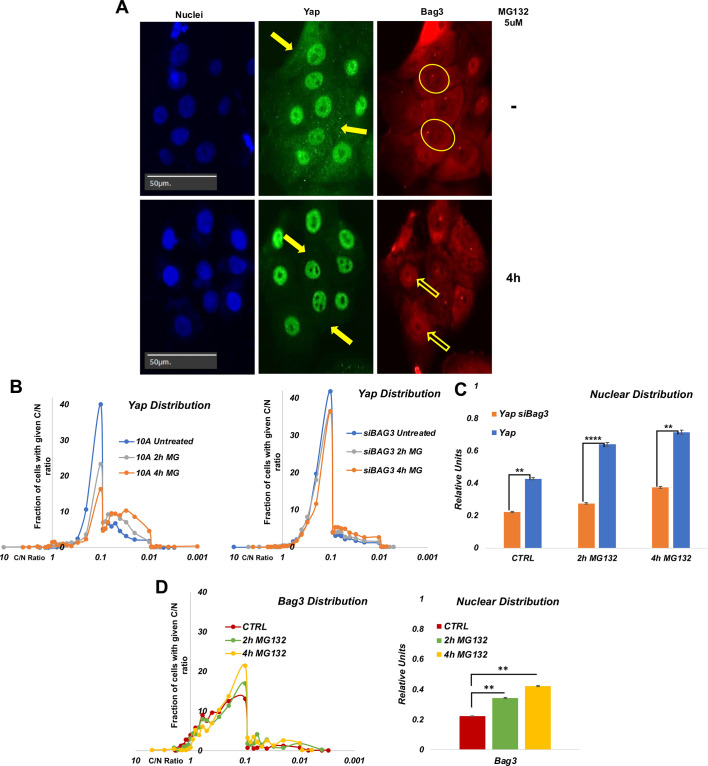
Upon dephosphorylation, YAP is translocated into nucleus and activates transcription of a series of target genes (BAG3 Result Summary|BioGRID; https://thebiogrid.org/114907/summary/homo-sapiens/bag3.html; Zhao et al., 2007). Indeed, we previously demonstrated that YAP-dependent transcription of CTGF was activated in cells exposed to proteasome inhibitor in cells exposed to proteasome inhibitor (Meriin et al., 2018). Given that YAP does not dissociate from Bag3 under the proteotoxic conditions (addition of MG132) (Fig. 1C,D) we hypothesized that YAP remains in the complex with Bag3 upon nuclear translocation, and that Bag3 may play a role in this translocation. To address this possibility, MCF10A cells were seeded on a 96-well plate at ∼60% confluency (medium confluency) then treated with MG132, and nuclear localization of Bag3 and YAP was assessed by immunohistochemistry. It was critical to keep medium confluency in these experiments, since cell density regulates the Hippo pathway (Yu and Guan, 2013) independently of proteotoxicity (see below).
For each condition, nuclear localization of the proteins was quantitatively assessed by analyzing thousands of cells in ∼100 images using the high-content imaging system Hermes (see Materials and Methods). The Hermes software package Athena provided us nuclear and cytoplasmic intensity of fluorescence per each cell. Although a significant fraction of YAP was present in the nucleus in naïve cells, additional nuclear YAP accumulation was observed in response to the proteotoxic treatment (Fig. 3A,B). Importantly, we observed high heterogeneity in nuclear localization of YAP in both naïve and stressed cell populations. Following MG132 treatment, the proportion of cells with a high nuclear content of YAP increased, but even following strong proteasome inhibition, some cells still had predominantly cytoplasmic YAP (Fig. 3B,C). To test whether Bag3 is involved in the nuclear translocation of YAP, MCF10A cells were depleted of Bag3 using siRNA, and nuclear localization of YAP in response to MG132 was assessed as in the above experiment. Indeed, after silencing of Bag3, YAP translocation was reduced despite treatment with MG132 (Fig. 3B,C). Altogether, these data suggest that interaction with Bag3 has an important role in the nuclear localization of YAP.
Fig. 3.
Effects of proteotoxic stress on nuclear localization of Bag3 and YAP. (A) Representative images of localization of YAP and Bag3 in the presence and absence of MG132. Filled arrows indicate the cytoplasmic content of YAP. Circles indicate nuclei with a low content of Bag3. Empty arrows indicate nuclei with a high content of Bag3. Images are representative of three experiments. (B) Distribution of cells in the population according to C/N ratio (cytoplasmic/nuclear localization) of YAP in control cells (left panel) and Bag3-depleted cells (right panel). Cells were treated on 96-well plates and fixed, and localization of YAP was assessed by immunofluorescence. Image acquisition and analysis were performed using the Hermes imaging system. Unlike control culture where MG132 caused redistribution of YAP in a large proportion of cells, in Bag3-depleted cells MG132 does not cause significant YAP redistribution. (C) Quantification of effects of MG132 treatment and Bag3 depletion on nuclear localization of YAP based on experiment in Fig. 3B. (D) Distribution of cells in the population according to C/N ratio of Bag3 in control cells (left panel). The treatments were performed as described in B. Right panel, quantification of effects of MG132 treatment on nuclear localization of Bag3 based on experiment in the left panel. **P≤0.01; ****P≤0.0001 (unpaired two-tailed t-test).
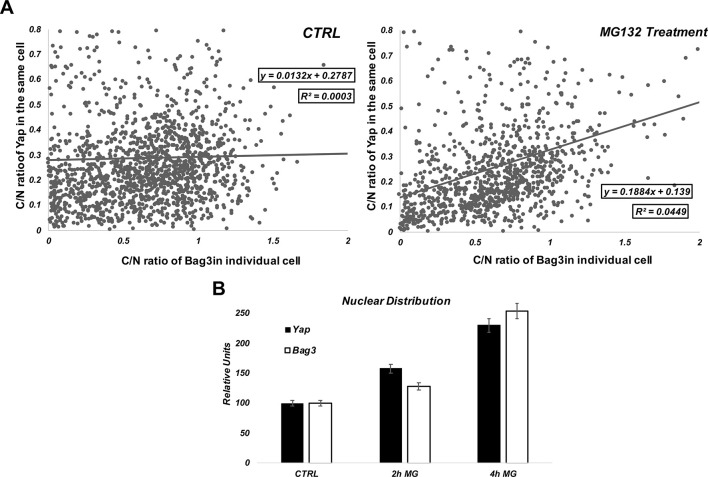
Similar to YAP, Bag3 also demonstrated nuclear translocation in response to MG132 in a population of cells (Fig. 3D). When analyses were performed across the populations, we found that, in MG132-treated cells, there was a positive correlation between the degrees of nuclear localization of YAP and Bag3 (Fig. 4), and such a correlation was not observed in naïve cells. These data suggest that Bag3 and YAP proteins migrate to nucleus together, and that Bag3 is important for nuclear localization of YAP.
Fig. 4.
Correlation between C/N ratio of YAP and C/N ratio of Bag3 in cell population. Cells expressing FLAG-tagged Bag3 were double stained for FLAG and YAP. (A) The C/N ratio (cytoplasmic/nuclear localization) of YAP in each cell was plotted against the C/N ratio of Bag3 in the same cell. In control sample there is no correlation between C/N ratios of Bag3 and YAP in individual cells. On the other hand, upon MG132 treatment, the correlation between nuclear distribution of YAP and Bag3 becomes obvious. (B) Effects of MG132 on C/N distribution of YAP and Bag3. In A, we performed single linear regression using a Person's R with a confidence level of 95% and then calculated R-squared.
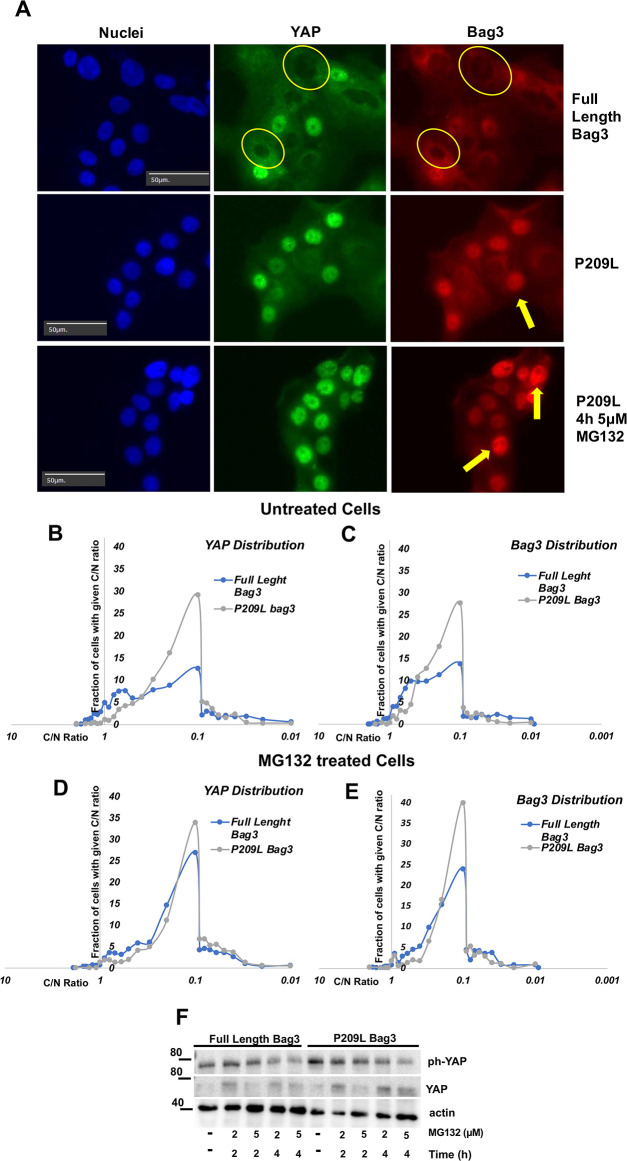
The Bag3 disease mutant Bag3 P209L enhances nuclear translocation of YAP
The P209L mutation in Bag3 causes heritable myopathy (Meister-Broekema et al., 2018) and this mutation has been shown to enhance protein aggregation in cells (Meister-Broekema et al., 2018). The hypothesis of the authors of this work was that the mutation stabilizes abnormal polypeptides in association with the HB complex, and thus promotes their aggregation. Our prior observations indicated that protein aggregation is regulated by LATS, and reversely correlates with phosphorylation of YAP (Meriin et al., 2018). Accordingly, the effects of P209L mutation on aggregation could be associated with effects of Bag3 on the LATS–YAP axis. Therefore, we sought to explore whether this mutation affects nuclear translocation of Bag3 and YAP. MCF10A cells were infected with retroviruses expressing either full-length Bag3 or Bag3 carrying P209L mutation, both of which were resistant to siRNA that targets endogenous Bag3. Cells were seeded at medium density and treated with MG132, and localization of YAP and Bag3 was assessed by immunofluorescence. When endogenous Bag3 was depleted with siRNA, we observed significant enhancement of nuclear localization of P209L recombinant Bag3, compared to that of normal recombinant Bag3 (Fig. 5A). Interestingly, nuclear localization of P209L Bag3 was associated with enhanced nuclear translocation of YAP, both in the absence and in the presence of MG132 (Fig. 5B–E).
Fig. 5.
Distribution of cells in the population according to C/N ratio of YAP and P209L Bag3. (A) Representative image of localization of YAP and Bag3 in the presence and absence of MG132. Arrows indicate nuclei with high content of either YAP or Bag3. Circles indicate nuclei with low content of YAP or Bag3. Images are representative of three experiments. (B) Distribution of cells in the population according to C/N ratio (cytoplasmic/nuclear localization) of YAP in untreated P209L Bag3-expressing cells. (C) Distribution of cells in the population according to C/N ratio of P209L Bag3 in untreated cells. (D) Distribution of cells in the population according to C/N ratio of YAP in P209L Bag3-expressing cells treated with MG132 (4 h, 5 μM). (E) Distribution of cells in the population according to C/N ratio of P209L Bag3 in cells treated with MG132 (4 h, 5 μM). (F) Effect of P209L mutation on phosphorylation of YAP in response to MG132. Cell expressing Bag3 P209L and full length Bag3 were depleted of endogenous Bag3, treated with MG132 at the indicated conditions, and immunoblotted with anti-phospho (ph)-YAP and pan-YAP antibodies. Of note, MG132 treatment seem to increase pan-Yap levels. Nevertheless, p-YAP levels decreased. Image is representative of three experiments. A control for mutant SS-AA Bag3 and P209L Bag3 expression is shown in Fig. S2.
To test whether the P209L mutation affected YAP phosphorylation we compared phospho-YAP levels in cells expressing either full-length Bag3 or P209L Bag3 following endogenous Bag3 depletion. However, the phosphorylation of YAP was not reduced in the presence of P209L compared to wild type Bag3 (Fig. 5F). Therefore, the P209L mutation enhances nuclear translocation of YAP without affecting its phosphorylation, further supporting the role of Bag3–YAP co-translocation in the nuclear localization of the latter.
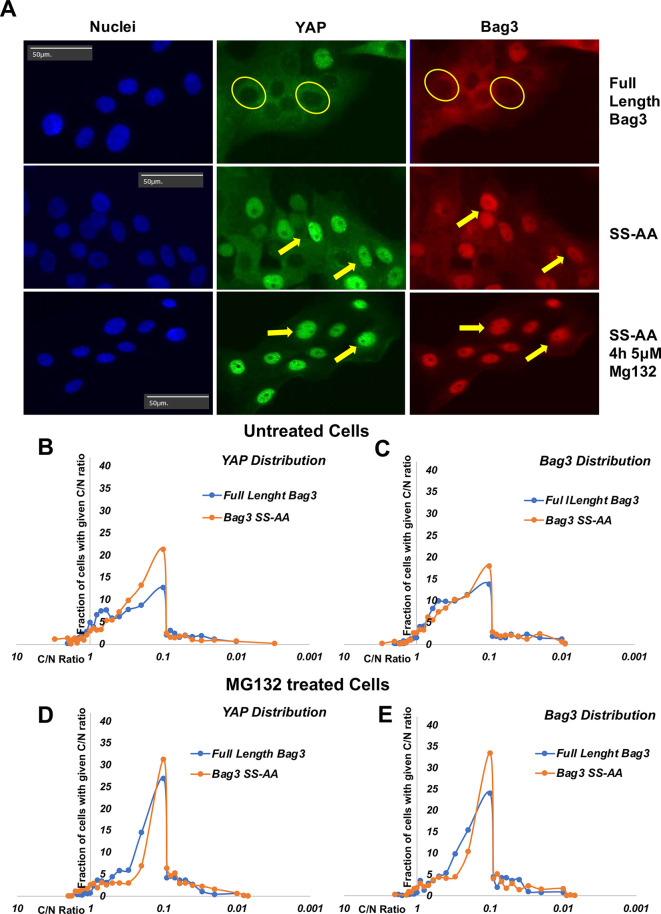
Blocking 14-3-3 protein–Bag3 interaction facilitates nuclear translocation of YAP
Nuclear translocation of YAP is controlled by its association with 14-3-3 proteins (Meng et al., 2016). LATS-dependent phosphorylation of YAP promotes 14-3-3 binding, restricting nuclear YAP translocation. Bag3 also binds several 14-3-3 proteins, with two 14-3-3-binding sites on Bag3 previously identified (Xu et al., 2013). Considering that YAP may co-translocate to nucleus with Bag3, we tested whether 14-3-3 binding to Bag3 affects nuclear translocation of YAP. Accordingly, we created mutations in Bag3 that prevent phosphorylation of both 14-3-3-binding sites S136A and S173A (SS→AA), and thus prevent association of Bag3 with 14-3-3 proteins (Xu et al., 2013). The construct was additionally mutated to make it resistant to siRNA that we use to deplete endogenous Bag3 (see Materials and Methods).
To test whether prevention of interaction of Bag3 with 14-3-3 proteins is sufficient to drive YAP to the nucleus, cells were transfected with a plasmid expressing Bag3 with SS→AA mutation, endogenous Bag3 was depleted, and localization of YAP and Bag3 SS→AA was assessed by immunofluorescence. Even in naïve cells, we observed an increased fraction of cells with nuclear YAP and Bag3 SS→AA (Fig. 6A). Treatment with MG132 further increased the translocation of both YAP and Bag3 SS→AA (Fig. 6B–E). Therefore, blocking the interaction between Bag3 and 14-3-3 increases the capability of Bag3 to facilitate translocation of YAP to the nucleus.
Fig. 6.
Distribution of cells in the population according to C/N ratio of YAP and SS→AA Bag3. (A) Representative image of localization of YAP and Bag3 in the presence and absence of MG132. Arrows indicate nuclei with high content of either YAP or Bag3. Circles indicate nuclei with low content of YAP or Bag3. Images are representative of three experiments. (B) Distribution of cells in the population according to C/N ratio of YAP in naïve cells. (C) Distribution of cells in the population according to C/N ratio (cytoplasmic/nuclear localization) of SS→AA Bag3 in naïve cells. (D) Distribution of cells in the population according to C/N ratio of YAP in cells treated with MG132 (4 h, 5 μM). (E) Distribution of cells in the population according to C/N ratio of SS→AA Bag3 in cells treated with MG132 (4 h, 5 μM). A control for mutant SS-AA Bag3 and P209L Bag3 expression is shown in Fig. S2.
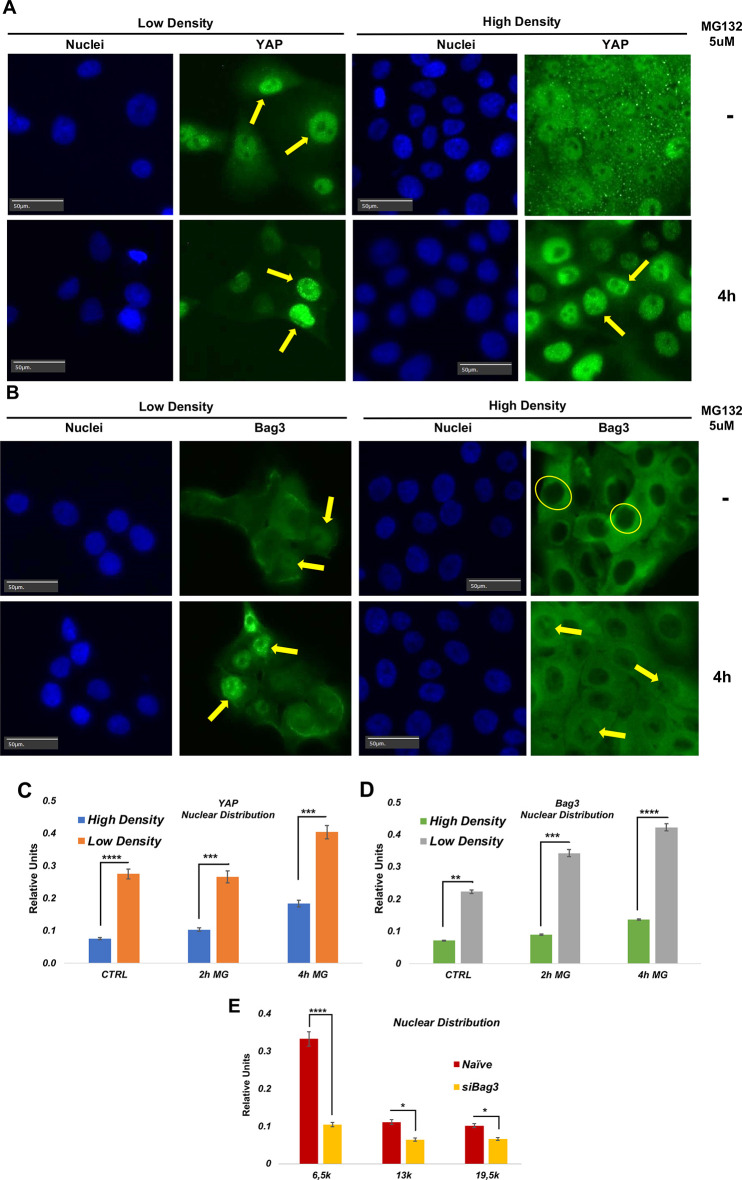
Bag3 regulates YAP nuclear translocation in response to low cell density
Given our observations, we further addressed whether Bag3 might be involved in nuclear localization of YAP in response to stimuli other than proteotoxicity. We tested whether Bag3 influences cell density-regulated control of YAP localization (Yu and Guan, 2013). For this, MCF10A cells were seeded at densities of 20% (low density) and 90% (high density), and cellular localization of YAP in these cells was measured by immunofluorescence. As described previously (Zhao et al., 2007), at high density, most of YAP localized in the cytoplasm, and we observed only a relatively small fraction of cells with nuclear YAP, while at low density the majority of cells demonstrated nuclear localization of YAP (Fig. 7A,B), and YAP remained in the cytoplasm only in a minor fraction of cells. Since Bag3 appeared to co-translocate to nucleus with YAP in response to inhibition of proteasome, we tested whether the cytoplasmic–nuclear distribution of Bag3 was affected by cell density. After seeding MCF10A cells at lower and high density, we assessed localization of Bag3 by immunohistochemistry and performed image analysis as in the above experiments. As with YAP, a significantly lower nuclear localization of Bag3 was observed in cells of the high density culture compared to those of the normal density culture (Fig. 7C,D). Importantly, MG132 treatment led to further redistribution of YAP and Bag3 to the nuclei in a population of cells both at high and low density (Fig. 7A,B for YAP; Fig. 7C,D for Bag3).
Fig. 7.
Distribution of cells in the population according to C/N ratio of YAP and Bag3 in cells seeded at low and high density. Low density corresponds to 3000 cells/well and high density to 19,500 cells/well. (A) Representative image of localization of YAP in low and high density cells in the presence and absence of MG132. Arrows indicate nuclei with high content of either YAP or Bag3. Circles indicate nuclei with low content of YAP or Bag3. (B) Representative image of localization of Bag3 in low and high density cells in the presence and absence of MG132. Arrows indicate nuclei with high content of either Bag3. Circles indicate nuclei with low content of Bag3. (C) YAP C/N ratio (cytoplasmic/nuclear localization) distribution in the population at low and high density. Images in A and B are representative of three experiments. (D) Bag3 C/N ratio distribution in the population at low and high density. E. Distribution of cells in the population according to C/N ratio of YAP upon depletion of Bag3. Effect of depletion of endogenous Bag3 on intracellular distribution of YAP at low and high density. A control for Yap expression in different cell densities and in the absence of Bag3 using siRNA Bag3 is shown in Fig. S4. *P≤0.05; **P≤0.01; ***P≤0.001; ****P≤0.0001 (unpaired two-tailed t-test).
To test whether Bag3 plays a role in YAP nuclear localization under these conditions, MCF10A cells were transfected with a siRNA against Bag3, and cellular localization of YAP was evaluated. Importantly, upon depletion of Bag3, we observed a significant redistribution of YAP to the cytoplasm at both low and high cell density, indicating that Bag3 is involved in YAP nuclear localization even in the absence of proteotoxic stress. Bag3 depletion also suppressed additional redistribution of YAP to the nucleus at both high and low density (Fig. 7E). Interestingly, previously, we showed that in the absence of proteotoxicity Bag3 does not affect YAP phosphorylation (Meriin et al., 2018). Therefore, although activity of LATS1/2 in response to changes of cell density is Bag3 independent, nuclear localization of YAP under these conditions is Bag3 dependent. These data indicate that the role of Bag3 in regulation of YAP translocation is not limited to proteotoxicity, and appears to be more general.
DISCUSSION
The main question addressed in this work was how Bag3 transmits the signal about proteotoxicity to downstream pathways. As an example, we chose LATS1/2–YAP axis because we knew that LATS can directly associate with Bag3, and this association is mediated by WW domain of the latter. Unlike regulation of LATS in response to more conventional stimuli, that is, a change in cell density, proteotoxicity did not appear to affect phosphorylation of LATS mediated by upstream signals. We interestingly observe that, in addition to associating with LATS1, Bag3 also associated with YAP. This interaction involved both the WW and M-domains of Bag3. Accordingly, our observations suggest that LATS1 and YAP bind to Bag3 in a relative proximity to each other. Proteasome inhibition did not change the association of either LATS1 or YAP with Bag3, leading us to uncover that AmotL2, a scaffold protein that bridges LATS1 and YAP interactions, also associates with Bag3 in naïve cells. Importantly, in response to proteasome inhibition, AmotL2 readily dissociates, suggesting disrupted interaction between LATS1 and YAP that leads to reduced YAP phosphorylation. The dissociation of AmotL2 was dependent on the presence of Bag3, suggesting the major role of the latter in this regulation. Therefore, our observations suggest that proteotoxicity does not affect the activity of the LATS1 kinase, but rather the availability of YAP as a substrate.
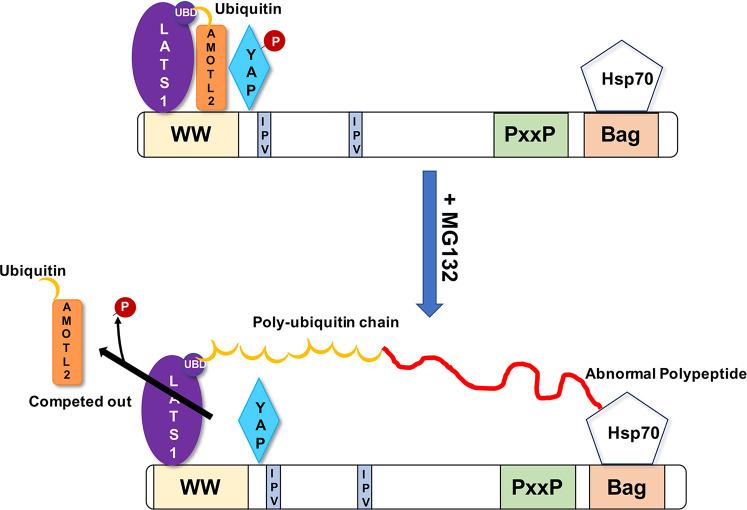
Interestingly, LATS1 carries a ubiquitin-binding UBA domain. It was previously demonstrated that this domain is critical for association of LATS with the scaffold AmotL2 (Varelas, 2014). Indeed, AmotL2 is monoubiquitylated, and this ubiquitin directly interacts with the UBA domain of LATS1/2. Removal of this ubiquitin or mutation in the UBA domain prevents association of LATS1/2 with AmotL2, which blocks phosphorylation of YAP (Kim et al., 2016). The presence of these structural elements suggests the mechanism of regulation of YAP phosphorylation in response to proteotoxic stress. We propose a model in which, in naive cells, the entire LATS1/2–AmotL2–YAP complex interacts with Bag3 (Fig. 8). Upon the buildup of abnormal proteins, the ubiquitylated species would be recruited to the proximity of Bag3 by Hsp70. Such ubiquitylated species would start to compete with monoubiquitin of AmotL2 for binding of the UBA domain of LATS1/2 (Fig. 8). Such a competition could therefore remove AmotL2 from the complex with LATS, Bag3 and YAP, which would disrupt the interaction between LATS1/2 and YAP, and thus suppress YAP phosphorylation.
Fig. 8.
A model of the mechanism of regulation of AmotL2 dissociation from HB complex upon proteotoxic stress. Recruitment of poly-ubiquitylated proteins to Hsp70–Bag3 competitively inhibits association of monoubiquitin on AmotL2 with the UBA domain of LATS. This leads to dissociation of AmotL2 and disruption of interaction between LATS and YAP.
An important surprise was that YAP remained associated with Bag3 even after proteotoxic stress, that is, likely following its dephosphorylation. Previously it was shown that YAP dephosphorylation leads to its dissociation from 14-3-3 proteins, resulting in nuclear translocation and activation of transcription (Meng et al., 2016). The fact that YAP remains associated with Bag3 after proteotoxic stress suggested that it can translocate into the nucleus in the complex with Bag3. This in turn led to a hypothesis that Bag3 may be involved in nuclear translocation of YAP. Indeed, we observed correlations between nuclear translocation of YAP and Bag3. Cells with higher nuclear localization of Bag3 also had higher nuclear localization of YAP. This correlation was seen only in cells following proteotoxic stress, but not in naïve cells. Furthermore, depletion of Bag3 significantly reduced nuclear translocation of YAP. However, more direct indication of involvement of Bag3 in nuclear translocation of YAP came from the analysis of Bag3 mutants. We analyzed the P209L mutant, which causes cardiomyopathy in humans and mouse model (Meister-Broekema et al., 2018). It appears that this mutation significantly enhances nuclear translocation of Bag3, both in naïve and stressed cells. Importantly, it also significantly enhanced nuclear translocation of YAP, further highlighting the link between these phenomena. In another approach to test the role of Bag3 in nuclear translocation of YAP, we mutated 14-3-3-binding sites on Bag3. It was previously shown that Bag3 can interact with several 14-3-3 proteins, and these interactions play an important role in Bag3 function in aggresome formation (Xu et al., 2013). Since interaction with 14-3-3 proteins typically prevents nuclear translocation of proteins (Mackintosh, 2004), we introduced S>A mutations into two 14-3-3 binding sites of Bag3, which indeed facilitated Bag3 nuclear translocation. YAP also translocated into nucleus more readily with this mutant Bag3. Therefore, it appears that YAP translocation into nucleus is regulated not only by 14-3-3 protein binding to YAP itself, but also by 14-3-3 protein binding to Bag3. It is likely that upon dissociation of 14-3-3 proteins from both YAP and Bag3, the complex moves into the nucleus, where YAP can start transcription of the target genes.
Interestingly, whereas LATS interacts with Bag3 via the WW domain, non-receptor tyrosine kinases Src and Yes associate with Bag3 via the PxxP motif (Mackintosh, 2004; Xu et al., 2013). Previously, LATS was shown to regulate nuclear translocation of YAP via its phosphorylation at position S127, while Src and Yes phosphorylate YAP at position Y357, which inhibits its nuclear export (Sugihara et al., 2018). Therefore, association of YAP with Bag3 provides both facilitation of nuclear translocation of YAP and suppression of nuclear export, which could further enhance the transcriptional activity of YAP. It is possible that Bag3 may have a more general role in nuclear localization of transcription factors with which it interacts.
MATERIALS AND METHODS
Reagents and antibodies
MG132 was purchased from A2S; 36% formaldehyde was from Sigma. Antibodies against YAP (14074S, 1:1000), phospho-YAP (S127) (4911S, 1:1000), LATS1 (3477S, 1:1000), phospho-LATS1 (S909) (9157S, 1:1000) and β-actin (3700S, 1:1000), and DyLight™ 554–phalloidin were from Cell Signaling Technology; anti-BAG3 (10599-1-AP, 1:1000) was from ProteinTech; anti-puromycin (MABE343, 1:500) was from Millipore; anti-FLAG DYKDDDDK was from Thermo Fisher Scientific; Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (H+L) and Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H+L) were from Jackson ImmunoResearch. Donkey anti-rabbit-IgG (711-005-152) and Donkey anti-mouse-IgG (715-005-150) secondary antibodies were purchase from Jackson ImmunoResearch Inc. and conjugated with a pair of oligonucleotides from ActivSignal Inc. (Watertown, MA, USA). The linker for the ligation reaction was provided by ActivSignal Inc. To measure Hsp70, we used a mixture of antibodies (ADI-SPA-822-D, 1:1000) against HspA1 and HspA8 from Enzo Life Sciences.
Cell culture
MCF10A (a human breast epithelial cell line; ATCC CRL-10317) and MCF10A cells expressing Flag-tagged Bag3, LATS and Amotl cells (these cell were prepared and checked in the laboratory of Professor Michael Y Sherman at Boston University), were grown in DMEM/F-12 50/50 medium (Biological Industries, 01-170-1A) supplemented with 5% horse serum (Biological Industries, 04-004-1A), 20 ng/ml epidermal growth factor (PeproTech, AF-100-15-1000), 0.5 μg/ml hydrocortisone (Sigma-Aldrich, H0888), 10 μg/ml human insulin (Sigma-Aldrich, 19278) and 100 ng/ml cholera toxin (Sigma-Aldrich, C8052); all cultures were supplemented with 1× (from a 100 × solution) L-glutamine (Gibco, 25030-024), as well 0.1 mg/ml penicillin and streptomycin (Biological Industries, 03-031-1B) and grown at 37°C with 5% CO2.
For siRNA transfection we used Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific) and followed the manufacturer's protocol. For a well on a 35 mm dish, we mixed 9 μl of the reagent with 3 μl of 10 μM siRNA in 150 μl of OptiMEM and added the mixture to 1500 μl of a cell suspension in the well. After 48 h the transfection was stopped and the cells were plated for an experiment conducted in the same day. We used the following siGENOME siRNAs purchased from Dharmacon: standard personalized sequence Bag3 5′-GCAAAGAGGUGGAUUCUUU-3′ or 3′UTR Bag3 5′-GCCAUAGGAAUAUCUGUAUUU-3′ for normal silencing or for the siRNA-resistant constructs.
PLA based on a modified activsignal protocol
The scheme to show the idea underlying this method is shown in Fig. S1. MCF10A cells transfected with FLAG-tagged Amotl2 were treated with 5 µM MG132 for 90 min, 2 h, 180 min or 4 h, fixed with 4% formaldehyde and permeabilized with 0.2% Triton X-100 on a 96-well plastic plate. The samples were left in buffer B provided by ActivSignal at 4°C overnight, with all following steps conducted at room temperature. The samples were incubated for 2 h with a pair of primary antibodies diluted in the buffer B. After 3× PBS with 0.05% Tween 20 (PBST) washes, consecutive 1-h incubations with each of secondary antibodies in the buffer B were performed. After 2× PBST followed by 2×10 mM Tris-HCl pH 8.0, 1 mM EDTA (TE) washes, a ligation mixture including a linker oligonucleotide (50 nM) and T4 DNA ligase (5 U/ml) was dispensed to the wells, and the reaction was conducted for 30 min. The wells were washed (1× PBST and 2× TE), 50 μl of TE was added to each well, and the ligated conjugates were extracted by 30-min incubation of the plate at 95°C. The eluates were assessed by qPCR using PerfeCTa SYBR® Green FastMix ROX (QuantaBio) according to the manufacturer's protocol (Fig. S1).
Plasmids and infection
We used pcDNA3.1-based plasmids used for overexpression of N-terminally FLAG-tagged Bag3 or its ΔC, ΔP, ΔW deletion mutants, which have been described previously (Colvin et al., 2014). Additionally, we constructed a ΔM mutant missing a stretch from amino acids 86–214. For expression in MCF10A cells, we cloned some of these constructs in retroviral vector pBabe (puro). For pulldown experiments we replaced FLAG on N-terminus of these constructs with the four following sequences connected by short linkers: FLAG-tag, S-tag, SBP-tag and (His)9-tag.
FLAG-tag mutated BAG3 constructs were made by site-directed mutagenesis of WT Bag3 in a retroviral vector pBABE using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) following the manufacturer's protocol.
Cell lysis and analysis
MCF10A cells in 35 mm or 60 mm dish were lysed with lysis buffer (40 mM HEPES, pH 7.5, 50 mM KCl, 1% Triton X-100, 2 mM dithiothreitol, 1 mM Na3VO4, 50 mM β-glycerophosphate, 50 mM NaF, 5 mM EDTA, 5 mM EGTA), and supplemented with proteasome inhibitor cocktail (Sigma) and PMSF before the use. Samples were adjusted to have equal concentration of total protein and subjected to PAAG electrophoresis followed by immunoblotting as described previously (Magi and Liberatori, 2005).
For pull-down analysis of Bag3-associated proteins, for each condition, transfected HeLa (ATCC CCL-2) cells from 2 of 100 mm dishes were washed with DPBS, fixed with 1.2% formaldehyde for 10 min at room temperature, then Tris-HCl (pH 7.4) was added to 50 mM final concentration which was followed by a wash with 50 mM Tris-HCl (pH 7.4) in DPBS. All following steps were performed at 4°C. The cells were lysed in: DPBS (Corning) supplemented with 30 mM NaCl, 10 mM Hepes (pH 7.4), 1.5 mM MgCl2, 0.5% Triton X-100, 5% glycerol, 10 mM imidazole, 1 mM PMSF and protease and phosphatase inhibitor cocktails (Sigma P8849 and 4906845001, respectively). The lysates were passed three times through a syringe (21G needle), clarified by centrifugation for 7 min at 16,000 g. The supernatants were adjusted to have equal concentration of total protein and loaded on 15 μl of HisPur Cobalt Resin (Pierce, Thermo Fisher Scientific). After incubation for 40 min, the flow through was allowed to pass through the beads twice more, and the beads were washed five times with DPBS (Corning) supplemented with 146 mM NaCl, 20 mM Tris-HCl (pH 8.0), 0.5% Triton X-100, 5% glycerol and 15 mM imidazole. The His-tagged Bag3 along with associated proteins was eluted with 300 mM imidazole in 50 mM Na(PO4) (pH 6.8) and 300 mM NaCl.
Microscopy
Microscopy was performed using two different cells line, MCF10A and MCF10A Flag–BAG3. The cells were fixed with a solution of 4% formaldehyde in PBS at room temperature for 5 min, then they were washed once with PBS and after they were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Consecutive, the cells were blocked for 1 h in blocking solution [5% BSA (w/v) in PBST] at room temperature and rinsed three times and washed three times for 5 min with PBST.
Next, a solution of primary antibodies (1:200) in blocking solution were added to the cells, which were incubated overnight at 4°C. After, the cells were washed rinsed three times and washed six times for 5 min with PBST. A solution of secondary antibodies (1:500) in blocking solution was added to cells and left to incubate for 1 h at room temperature, after which the cells were washed as previously but for seven times. DAPI was added at the concentration of 1:5000.
Fluorescence microscopy was performed with WiScan® Hermes High Content Imaging System (IDEA Bio-Medical, Rehovot, Israel). The cells were plated in a black 96-well plate with transparent bottom. 52 images per well were automatically acquired, which corresponds to thousands of cells per sample. The images were taken at room temperature using 20× objective, utilizing mainly the red (Ex. 560/32, Em. 607/36), blue (Ex. 390/22, Em. 440/40) and green (Ex. 485/25, Em. 525/30) channels. Image quantification was performed with the WiSoft® Athena software Translocation Application (IDEA Bio-medical, Rehovot, Israel). The software application performs automated, multiplexed image analysis by processing the blue fluorescence channel for nuclei detection using watershed segmentation which was calibrated with parameters that allow separation between adjacent objects; the red channel for detection of cytoplasm using the nucleus as a seed in a seeded watershed analysis, and the green channel to quantify for the amount of protein in each compartment. Throughout different experiments these parameters were kept equal to maintain homogeneity during the analysis. The program allows adjusting the following parameters: Nucleus Smooth, Nucleus Background Subtraction, Nucleus Intensity Threshold, Nucleus Max Patch Size, Nucleus Maximum Area, Nucleus Minimum Area, Cell Smooth, Cell Background Subtraction and Cell Intensity Threshold (Mlcochova et al., 2020, 4; Rasaiyaah et al., 2013). These parameters were adjusted in each experiment (which had different object densities and fluorescence) to obtain the best object recognition and the best separation between the objects. An example of masking is given in Fig. S3.
Supplementary Material
Acknowledgements
We thank our colleagues Santosh Kumar, Valid Gahramanov and Shivani Patel for their assistance and support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.B., A.B.M., J.Y., X.V., Z.-X.J.X., M.Y.S.; Methodology: I.A.; Investigation: S.B., A.B.M., J.Y.; Writing - original draft: M.Y.S.; Writing - review & editing: S.B., X.V., M.Y.S.; Supervision: M.Y.S.; Project administration: M.Y.S.; Funding acquisition: M.Y.S.
Funding
This work was supported by the National Institutes of Health (NIH) grant R01 800000163 and Israel Science Foundation grants ISF-1444/18 and ISF-2465/18. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.259107.
References
- Bush, K. T., Goldberg, A. L. and Nigam, S. K. (1997). Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272, 9086-9092. 10.1074/jbc.272.14.9086 [DOI] [PubMed] [Google Scholar]
- Chan, E. H. Y., Nousiainen, M., Chalamalasetty, R. B., Schäfer, A., Nigg, E. A. and Silljé, H. H. W. (2005). The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076-2086. 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Colvin, T. A., Gabai, V. L., Gong, J., Calderwood, S. K., Li, H., Gummuluru, S., Matchuk, O. N., Smirnova, S. G., Orlova, N. V., Zamulaeva, I. A.et al. (2014). Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 74, 4731-4740. 10.1158/0008-5472.CAN-14-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, H. E. and Samant, R. S. (2021). Alternative systems for misfolded protein clearance: life beyond the proteasome. FEBS J. 288, 4464-4487. 10.1111/febs.15617 [DOI] [PubMed] [Google Scholar]
- Kim, M., Kim, M., Park, S.-J., Lee, C. and Lim, D.-S. (2016). Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 17, 64-78. 10.15252/embr.201540809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. H. and Goldberg, A. L. (1998). Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 30-38. 10.1128/MCB.18.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh, C. (2004). Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 381, 329-342. 10.1042/BJ20031332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi, B. and Liberatori, S. (2005). Immunoblotting techniques. In Immunochemical Protocols (ed. Burns R.), vol. 295, pp. 99-120. Humana Press. 10.1385/1-59259-873-0:227 [DOI] [Google Scholar]
- Meister-Broekema, M., Freilich, R., Jagadeesan, C., Rauch, J. N., Bengoechea, R., Motley, W. W., Kuiper, E. F. E., Minoia, M., Furtado, G. V., van Waarde, M. A. W. H.et al. (2018). Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks. Nat. Commun. 9, 5342. 10.1038/s41467-018-07718-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z., Moroishi, T. and Guan, K.-L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1-17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabova, N., Sariyer, I. K., Saribas, A. S., Knezevic, T., Gordon, J., Turco, M. C., Rosati, A., Weaver, M., Landry, J. and Khalili, K. (2015). WW domain of BAG3 is required for the induction of autophagy in glioma cells. J. Cell Physiol. 230, 831-841. 10.1002/jcp.24811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin, A. B., Gabai, V. L., Yaglom, J., Shifrin, V. I. and Sherman, M. Y. (1998). Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 273, 6373-6379. 10.1074/jbc.273.11.6373 [DOI] [PubMed] [Google Scholar]
- Meriin, A. B., Narayanan, A., Meng, L., Alexandrov, I., Varelas, X., Cissé, I. I. and Sherman, M. Y. (2018). Hsp70-Bag3 complex is a hub for proteotoxicity-induced signaling that controls protein aggregation. Proc. Natl. Acad. Sci. USA 115, E7043-E7052. 10.1073/pnas.1803130115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova, P., Winstone, H., Zuliani-Alvarez, L. and Gupta, R. K. (2020). TLR4-mediated pathway triggers interferon-independent G0 arrest and antiviral SAMHD1 activity in macrophages. Cell Rep. 30, 3972-3980.e5. 10.1016/j.celrep.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah, J., Tan, C. P., Fletcher, A. J., Price, A. J., Blondeau, C., Hilditch, L., Jacques, D. A., Selwood, D. L., James, L. C., Noursadeghi, M.et al. (2013). HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402-405. 10.1038/nature12769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, J. N., Tse, E., Freilich, R., Mok, S. A., Makley, L. N., Southworth, D. R. and Gestwicki, J. E. (2017). BAG3 is a modular, scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins. J. Mol. Biol. 429, 128-141. 10.1016/j.jmb.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, M. Y. and Goldberg, A. L. (2001). Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29, 15-32. 10.1016/S0896-6273(01)00177-5 [DOI] [PubMed] [Google Scholar]
- Sugihara, T., Werneburg, N. W., Hernandez, M. C., Yang, L., Kabashima, A., Hirsova, P., Yohanathan, L., Sosa, C., Truty, M. J., Vasmatzis, G.et al. (2018). YAP tyrosine phosphorylation and nuclear localization in cholangiocarcinoma cells are regulated by LCK and independent of LATS activity. Mol. Cancer Res. 16, 1556-1567. 10.1158/1541-7786.MCR-18-0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, K., Motohashi, H. and Yamamoto, M. (2011). Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123-140. 10.1111/j.1365-2443.2010.01473.x [DOI] [PubMed] [Google Scholar]
- Takayama, S., Xie, Z. and Reed, J. C. (1999). An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274, 781-786. 10.1074/jbc.274.2.781 [DOI] [PubMed] [Google Scholar]
- Ulbricht, A., Eppler, F. J., Tapia, V. E., van der Ven, P. F. M., Hampe, N., Hersch, N., Vakeel, P., Stadel, D., Haas, A., Saftig, P.et al. (2013). Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430-435. 10.1016/j.cub.2013.01.064 [DOI] [PubMed] [Google Scholar]
- Varelas, X. (2014). The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141, 1614-1626. 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Xu, Z., Graham, K., Foote, M., Liang, F., Rizkallah, R., Hurt, M., Wang, Y., Wu, Y. and Zhou, Y. (2013). 14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes. J. Cell Sci. 126, 4173-4186. 10.1242/jcs.126102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.-X. and Guan, K.-L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27, 355-371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., Xie, J., Ikenoue, T., Yu, J., Li, L.et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Li, L. and Zhao, B. (2015). The regulation and function of YAP transcription co-activator. Acta Biochim. Biophys. Sin. 47, 16-28. 10.1093/abbs/gmu110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.