Abstract
A highly reproducible and sensitive real-time detection assay based on TaqMan technology was developed for the detection of hepatitis B virus (HBV) DNA and compared with two commercially available assays. The assay was validated with the Viral Quality Control panel, which also includes EUROHEP HBV DNA standards. This real-time PCR detection system had a dynamic range of 373 to 1010 genome copies per ml and showed an excellent correlation with both the commercial HBV Digene Hybrid Capture II microplate assay (Digene Diagnostics) and the HBV MONITOR assay (Roche Diagnostics). To demonstrate its clinical utility, four chronically HBV-infected patients treated with lamuvidine were monitored using the three different assays. From the results we concluded that this assay is an excellent alternative for monitoring of HBV-infected patients in routine diagnostics and clinical practice, enabling the analysis of a large dynamic range of HBV DNA in a single, undiluted sample.
It has been estimated that worldwide approximately 300 million individuals are chronically infected with hepatitis B virus (HBV). The measurement of HBV DNA in serum has become an important tool to identify individuals with high viral replication, to monitor patients on therapy, and to predict whether antiviral therapy will be successful. With the introduction of new antivirals like lamivudine [(−)2′,3′-dideoxy-3′-thiacytidine], close monitoring of patients has become increasingly important due to the occurrence of antiviral drug-resistant virus strains or the presence of flares after withdrawal from antiviral therapy (6, 11).
Several homemade and commercial molecular assays have been used to quantify the level of HBV DNA in serum samples (3, 7–10, 13, 18). However, due to the lack of standardization and the inability of these assays to quantify the whole dynamic range over which HBV DNA should be measured, different assays and serum dilutions have to be used for adequate monitoring of antiviral therapy. In this paper, we describe the results of the validation of a real-time PCR detection assay, based on TaqMan technology, for the detection of HBV DNA in serum samples. This assay is able to measure the large dynamic range in which HBV DNA can be present in chronically infected patients.
The assay is based on linearity, takes into account intra- and interassay variability, and can be performed in a routine setting. The real-time PCR detection assay is validated using the viral quality control (VQC) HBV DNA panel (CLB, Amsterdam, The Netherlands) and compared with other commercially available quantitative assays (HBV Digene Hybrid Capture II microplate assay [Digene, Gaithersburg, Md.] and HBV MONITOR assay [Roche Diagnostics, Almere, The Netherlands]). Furthermore, to demonstrate its use in clinical practice, four chronically HBV-infected patients were monitored over a period of time in which they received antiviral treatment.
MATERIALS AND METHODS
Patients and clinical samples.
The clinical samples used for this study were well-characterized samples obtained from previous study protocols. Additional samples for the correlation and precision study were routine samples from our Virology Department. Samples requiring dilution, including the VQC standards, were diluted in serum known to be HBV DNA negative. All aliquots were stored frozen at −20°C or a lower temperature until use. For the specificity analysis, samples (kindly provided by the Blood Bank Rotterdam) obtained from 200 healthy blood donors were used.
For a standardized evaluation, we obtained an international reference VQC plasma preparation panel (CLB) containing well-characterized HBV DNA levels including EUROHEP HBV DNA standard A. These samples were tested extensively and contain HBV DNA levels ranging from no HBV DNA to 4.37 × 107 HBV molecules per ml.
DNA extraction method for real-time PCR detection assay.
For the isolation of HBV DNA from serum, the High Pure Viral Nucleic Acid kit (Roche Diagnostics) was used. Briefly, 200 μl of serum was added to 200 μl of a freshly prepared working solution containing 6 M guanidine-HCl, 10 mM urea, 10 mM Tris-HCl, and 20% (vol/vol) Triton X-100 supplemented with 1 μg of poly(A)+ carrier RNA and 800 μg of proteinase K. After incubation for 10 min at 72°C, 100 μl of isopropanol was added and the mixture was transferred onto a High Pure filter tube combined with a collection tube. The filter tube was centrifuged for 1 min at 5,000 × g in a standard tabletop centrifuge at room temperature. After being washed twice with 450 μl of buffer (20 mM NaCl, 2 mM Tris-HCl [pH 7.5] in ethanol), the filter was placed in a new collection tube and 50 μl of RNase- and DNase-free water was added to elute the DNA. This resulted in fourfold concentration of the original input material.
HBV Digene Hybrid Capture II microplate assay.
The HBV Digene Hybrid Capture II microplate assay was performed in accordance with the manufacturer's protocol. Briefly, 30 μl of serum samples, controls, and standards or calibrators ranging from 0.5 to 6,000 pg/ml (equivalent to 1.42 × 105 to 1.7 × 109 HBV DNA copies per ml) were incubated with 30 μl of sodium hydroxide solution (denaturation reagent) for 30 min at 65°C in a 96-well microplate. No additional sample preparation step was needed. After preparation of the probe mixture, 30 μl of RNA HBV probe was added to each well and the plate was incubated for 1 h at 65°C. To capture the DNA-RNA hybrids, 75 μl of each solution in the microplates was transferred to the corresponding well of the anti-RNA-DNA hybrid antibody-coated capture microplate and subsequently shaken at room temperature for 1 h. The hybrid was detected using an anti-hybrid antibody conjugated to alkaline phosphatase and detected with the chemiluminiscent substrate CDP-star with Emerald II.
To enable detection of HBV DNA levels of less than 1.42 × 105 copies per ml, the ultrasensitive format of the assay was used. Briefly, 1-ml serum samples and controls along with 50 μl of precipitation buffer were centrifuged at 33,000 × g for 110 min at 4°C in a Hereaus Stratos Biofuge. The supernatant was discarded, and the precipitated virus was dissolved in 25 μl of diluent. This procedure yields a 30-fold increase in sensitivity and enhances the lower detection limit of the assay to approximately 8,000 HBV copies/ml.
HBV MONITOR assay.
HBV DNA levels were monitored using the Roche Molecular Systems Amplicor HBV MONITOR Assay in accordance with the instructions of the manufacturer. Briefly, we subjected 50 μl of serum to precipitation by polyethylene glycol 8000, followed by sodium hydroxide lysis and neutralization. For PCR, a primer set which amplifies a 104-bp fragment of the precore and core gene was used. During the amplification, internal standard (IS) DNA was coamplified with the target DNA. One of the primers was labeled with biotin. After amplification, two aliquots were pipetted into separate wells of a streptavidin-coated plate. The PCR products of the HBV core gene and the IS were separately hybridized with a dinitrophenyl-labeled HBV- or IS-specific probe. The hybridization products were detected colorimetrically with an antidinitrophenyl-alkaline phosphatase conjugate and para-nitrophenylphosphate substrate.
The amount of HBV DNA was calculated from the ratio of the HBV-specific well to the IS-specific well. The number of copies per ml was calculated from a standard curve prepared from each amplification run, ranging from 0 to 106 HBV copies per ml. The detection limit of the assay, according to the manufacturer, is 400 genome equivalents per ml. The dynamic range of the assay in the format we used was 400 to 4 × 107 genome equivalents per ml.
Real-time PCR detection assay.
The PCR primers and probe used were designed using Primer Express software (PE Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). Amplification was performed in a 50-μl reaction mixture containing 2× TaqMan Universal MasterMix (PE Biosystems), 45 pmol of forward primer (5′-GGA.CCC.CTG.CTC.GTG.TTA.CA-3′, nucleotides 184 to 203), 45 pmol of reverse primer (5′-GAG.AGA.AGT.CCA.CCM.CGA.GTC.TAG.A-3′, nucleotides 273 to 249), 15 pmol of TaqMan probe (5′-FAM-TGT.TGA.CAA.RAA.TCC.TCA.CCA.TAC.CRC.AGA-TAMRA-3′, nucleotides 218 to 247), and 10 μl of isolated DNA. The primers and probe were selected in the pre-S gene of the HBV genome and generated a product of 89 bp.
All isolations and amplification reactions were performed in duplicate. After preparation of the reaction mixtures in 96-well plates, the plates were centrifuged at 1,200 rpm for 1 min in a Rotina 48R swing rotor (Hettig, Tuttlingen, Germany) to remove small air bubbles in the vessels. Amplification and detection were performed with an ABI Prism 7700 Sequence Detection System (PE Biosystems). After incubation for 2 min at 50°C, which enables uracil N′-glycosylase (present in the 2× Universal MasterMix) to inactivate possible contaminating amplicons, incubation for 10 min at 95°C allowed AmpliTaq Gold polymerase to activate and inactivate the uracil N′-glycosylase. The PCR cycling program consisted of 45 two-step cycles of 15 s at 95°C and 60 s at 60°C.
For preparation of an external standard containing noninfectious material, HBV DNA (3.2 kb) was cloned into vector pBR322 (kindly provided by R. Heijtink, Erasmus University, Rotterdam, The Netherlands) and transformed into INVαF′ One Shot Escherichia coli bacteria (InVitrogen, Leek, The Netherlands). Plasmid DNA was isolated on the Vistra Labstation (Amersham Pharmacia Biotech, Roosendaal, The Netherlands). For standardization of the real-time PCR detection assay, a standard curve of the plasmid ranging from 10 million to 670 copies/ml and a run control of 30,000 copies/ml (CLB) were included in each run. Validation of the plasmid controls was done with the VQC panel (CLB).
Data analysis and statistics.
Analysis of raw data was done with the Sequence Detector V1.6.3 software (PE Biosystems). Data were collected at the annealing step (60°C) of every cycle, and the threshold cycle (Ct) for each sample was calculated by determining the point at which the fluorescence exceeded the threshold limit, which was set at 0.04 U. The standard curve was calculated automatically by plotting the Ct values against each standard of known concentration and calculation of the linear regression line of this curve. Calculation of the correlation coefficient (r2) was done for each run, and the minimal value was 0.98. Sample copy numbers were calculated by interpolation of the experimentally determined standard curve. In order to determine the extent to which the data obtained were in agreement, the data were also analyzed as described by Bland and Altman (1), whose method is based on comparison of the differences between measurements for the same sample by plotting of the differences against the average. All other statistical analysis was performed using the functions of the SPSS (version 8.0) software.
RESULTS
Precision study.
Inter- and intra-assay variability was determined by isolating one-third-log serial dilutions of a serum sample ranging from 5.49 × 109 to 3.82 × 102 HBV DNA copies per ml. Interassay variability was determined by isolating quadruplicates of the dilution series on three consecutive days (n = 12), while intra-assay variability was evaluated by running quadruplicates of one isolation on one plate (n = 4). Table 1 shows the mean Ct values, the percent coefficient of variation (%CV), and the amount of input HBV DNA within a run and between separate runs. There is no statistically significant difference between the %CV of interassay variability (mean, 1.85) and the %CV of intra-assay variability (mean, 1.65) (P = 0.761).
TABLE 1.
Study of intra- and interassay variability of the HBV DNA real-time PCR detection assay to determine its precision
| Sample | Amt of input HBV DNA (no. of copies/ml) | Mean Ct (%CV)
|
|
|---|---|---|---|
| Intra-assaya | Interassayb | ||
| V15 | 3.82 × 102 | 43.97 (4.67) | 44.06 (3.40) |
| V14 | 1.03 × 103 | 40.49 (7.62) | 40.79 (5.71) |
| V13 | 1.15 × 103 | 39.04 (5.12) | 37.90 (1.65) |
| V12 | 3.44 × 103 | 37.33 (1.34) | 36.87 (1.50) |
| V11 | 3.10 × 104 | 34.64 (0.70) | 34.83 (1.01) |
| V10 | 9.30 × 104 | 33.22 (0.43) | 33.61 (1.11) |
| V9 | 2.79 × 105 | 31.82 (1.11) | 31.46 (1.55) |
| V8 | 8.37 × 105 | 29.76 (0.84) | 30.02 (0.84) |
| V7 | 2.51 × 106 | 28.35 (0.78) | 28.35 (0.94) |
| V6 | 7.53 × 106 | 26.86 (0.60) | 26.76 (1.14) |
| V5 | 2.26 × 107 | 24.48 (0.35) | 24.93 (1.27) |
| V4 | 6.78 × 107 | 23.25 (0.12) | 23.35 (1.76) |
| V3 | 2.03 × 108 | 21.73 (0.54) | 21.79 (1.27) |
| V2 | 6.10 × 108 | 20.23 (0.53) | 20.40 (1.61) |
| V1 | 1.83 × 109 | 19.21 (0.52) | 18.92 (1.73) |
| V0 | 5.49 × 109 | 17.62 (1.20) | 17.38 (3.05) |
Mean %CV, 1.65; n = 4.
Mean %CV, 1.85; n = 12.
Specificity.
In order to determine the specificity of the real-time PCR detection assay, 200 anti-hepatitis B surface antigen-negative EDTA-plasma samples were analyzed. None of the samples showed false-positive reactions in duplicate. Four samples had to be reisolated and retested because of inconsistency between the duplicates but showed no reactivity in the second analysis.
Validation of the real-time PCR detection assay.
We first validated the HBV plasmid standard curve of the real-time PCR detection assay using the VQC panel, containing both HBV DNA-negative samples and well-characterized samples ranging from 4.27 × 107 to 113 copies per ml.
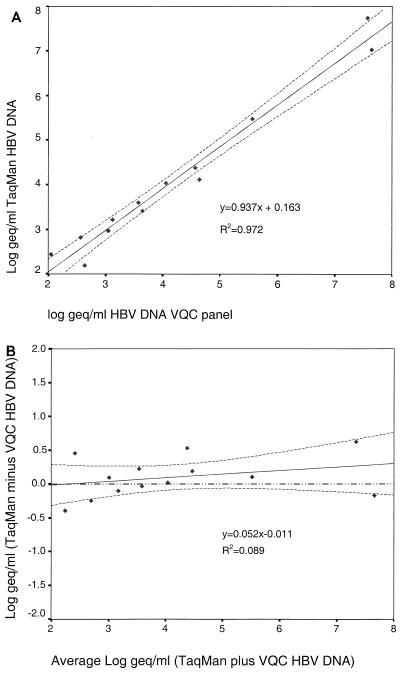
The log10 theoretical HBV DNA concentrations were compared to the log10 HBV DNA concentrations determined by the real-time PCR detection assay, which were calculated through interpolation of the Ct values from the standard curve. Regression analysis showed that the slope approached 1.0 (95% confidence interval, 0.83 to 1.04) and the y intercept approached 0 (95% confidence interval, −0.30 to 0.64; Fig. 1A). The correlation coefficient (r2 = 0.089) and high P value (P = 0.610) of the Bland-and-Altman comparison indicated no significant difference between the values obtained (Fig. 1B).
FIG. 1.
Linear (A) and Bland-and-Altman (B) comparisons of the real-time HBV DNA PCR detection assay and the DNA concentration expected from the VQC panel. Plotted are the calculated regression line (solid line) of these values and the 95% confidence intervals of the mean (broken lines). In panel B, an additional reference line (y = 0; broken-and-dotted line) is plotted to indicated that the data are well within the 95% confidence interval of the expected results.
The VQC sample with a viral HBV copy number of 373/ml still generated a detectable signal in the real-time PCR detection assay, while the sample with 113 copies/ml could not be detected in the format used.
Correlation between the HBV real-time PCR detection assay and commercially available HBV DNA assays.
The HBV DNA real-time PCR detection assay was compared with two commercially available assays to determine its usefulness for a clinical virology laboratory. For routine diagnostics, we selected sera for which the HBV DNA value was determined using the Digene Hybrid Capture II microplate assay or the Roche HBV MONITOR assay.
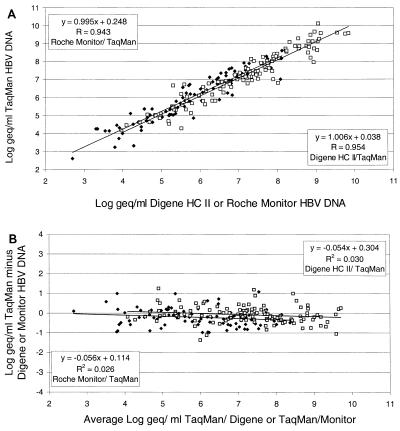
One hundred eighteen HBV DNA-positive samples were tested in both the Digene assay and the real-time PCR detection assay. These samples were within the range of the Digene assay in the standard or sensitive format (range, 8 × 103 to 1.7 × 109 copies of HBV DNA per ml). The log values of all samples were plotted, and regression analysis showed a slope of 1.004 (95% confidence interval, 0.944 to 1.064), while the y intercept approached 0 (0.038, 95% confidence interval, −0.576 to 0.314). Bland-and-Altman analysis furthermore indicated that the slope did not significantly differ from 0 (−0.053, 95% confidence interval, −0.112 to 0.005) and the y intercept approached 0 (0.304, 95% confidence interval, −0.119 to 0.730), indicating no difference in detection between the Digene Hybrid Capture II microplate assay and the real-time PCR detection assay for HBV.
To compare the HBV MONITOR assay with the HBV DNA real-time PCR detection assay, 93 serum samples were tested in both assays. These samples were within the range of the HBV MONITOR assay. Regression analysis showed a slope of 0.995 (95% confidence interval, 0.921 to 1.069) and that the y intercept approached 0 (0.247; 95% confidence interval, −0.195 to 0.690). Bland-and-Altman analysis indicated that both the slope (−0.056, 95% confidence interval, −0.128 to 0.016) and the y intercept (0.114, 95% confidence interval, −0.325 to 0.553) did not significantly differ from 0, indicating no difference between the assays. A summary of the results is given in Fig. 2A (direct comparison) and B (Bland-and-Altman analysis).
FIG. 2.
Correlation of the Digene Hybrid Capture (HC) II microplate assay and the Roche HBV MONITOR assay with the HBV DNA real-time PCR detection assay. The log10 DNA concentrations were calculated for a set of 211 randomly selected clinical samples, 118 of which were detected by the Digene Hybrid Capture II microplate assay (A). Panel B shows the results of Bland-and-Altman analysis performed on both comparisons.
Clinical monitoring.
To further demonstrate the clinical utility of the HBV DNA real-time PCR detection assay, four lamuvidine-treated chronically HBV-infected patients were monitored with all three assays, the Digene Hybrid Capture II microplate assay, the Roche MONITOR assay, and the HBV DNA real-time PCR detection assay. For both the Digene Hybrid Capture II microplate assay and the Roche HBV MONITOR assay, the appropriate dilutions were made in order to enable accurate determination of HBV DNA in the range of the assays. In Fig. 3, the calculated log10 HBV DNA levels (copies per ml) measured with the three different assays are plotted. All four patients showed a rapid decline in HBV DNA during treatment with lamuvidine, and there was a good correlation of the three different assays.
FIG. 3.
Clinical courses of four chronically HBV-infected patients during lamuvidine treatment (A to D). The calculated log10 DNA concentrations (y axis) are plotted against time after start of therapy (hours). The patients were monitored with the HBV Digene Hybrid Capture II microplate assay (□), the HBV MONITOR assay (◊), and HBV real-time PCR detection assay (▴).
DISCUSSION
In this paper, we have presented a quantitative assay based on the TaqMan real-time PCR detection methodology for the detection of HBV DNA. Furthermore, we have demonstrated this detection system to be highly reproducible, with no statistically significant difference in variability, as well as to have the ability to detect HBV DNA between approximately 373 and 10 billion copies per ml without any further sample dilution or concentration. The low variability of this assay makes a twofold decrease or increase in the serum HBV DNA level already significant. The high reproducibility and linearity of the assay described proved to be comparable to those of two commercially available assays.
More than a decade has passed since the introduction and use of PCR technology for the detection of HBV DNA in serum. Although different commercial assays have become available enabling an easy-to-use platform, there is still progress to be made, since the detection range of these assays was shown to be limited. Only the Digene Hybrid Capture II microplate assay was able to detect HBV DNA over a wide range (8 × 103 to 1.7 × 109 copies per ml), although in the lower range a sample concentration step needs to be performed. The Roche HBV MONITOR assay is used for quantitative HBV detection in the lower range of HBV DNA detection in patients (400 to 4 × 107 copies/ml). The high variation of the HBV MONITOR assay within the lower detection range, which can even be more than 100%, is the main disadvantage of this assay (13). The real-time PCR detection assay described here had a good correlation with both the Digene Hybrid Capture II microplate assay and the Roche HBV MONITOR assay and could be used for the monitoring of patients undergoing antiviral treatment. Regression analysis indicated that the standardization based on the VQC panel resulted in a large dynamic range and that data from all of the assays were interchangeable. Since it is usually not known in which range the HBV DNA level of a given sample is to be expected, especially when patients are receiving antiviral treatment, repeated testing is often necessary (13). With the real-time detection assay described here, this problem is largely overcome. Furthermore, since the introduction of TaqMan technology, more cumbersome techniques for PCR quantitation seem to have become obsolete, due to the ability of this technology to quantify HBV DNA rather easily, without the need for the further PCR amplicon processing steps usually required to detect specific sequences (5).
It has often been suggested that in-house PCR assays suffer from problems with standardization, false positivity, or contamination, making them unsuitable for routine clinical diagnostic use (14, 16). Quality control programs have indeed indicated that this is a problem (2, 15, 17). It should, however, be realized that for both commercial and in-house assays, sample preparation is still the most difficult step to perform and to automate. Handling of samples and opening of tubes during sample preparation should be carried out under good laboratory practice principles in order to limit the problem of contamination. The complete abolishment of postamplification handling is an additional factor in limiting contamination. Our data further indicate that by using two independent isolates for one sample in our assay, the problem of false positivity is more easily observed and therefore controlled. Furthermore, participation in external quality control programs, as well as the continuous use of well-standardized validation panels, should further guarantee laboratory performance. Available standardized materials, like universal amplification mixtures, primers, and probes, for real-time PCR detection assays leave the input DNA as the only unknown factor. A further point of attention remains the possible inhibition of the amplification reaction by unknown components in the clinical sample. Inclusion of internal markers and selection of appropriate sample preparation devices and methods should limit this problem. We have, for instance, shown that certain sample preparation methods are better than others at removing the inhibitory effect of heparin (12).
Collectively, our data show that HBV DNA quantitation with the real-time PCR detection technique resulted in a sensitive and highly reproducible assay, the results of which correlated well with those obtained with commercially available assays while offering the advantage of a wide dynamic range.
REFERENCES
- 1.Bland J M, Altman D G. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 2.Bootman J S, Kitchin P A. An international collaborative study to assess a set of reference reagents for HIV-1 PCR. J Virol Methods. 1992;37:23–41. doi: 10.1016/0166-0934(92)90018-9. [DOI] [PubMed] [Google Scholar]
- 3.Butterworth L A, Prior S L, Buda P J, Faoagali J L, Cooksley W G. Comparison of four methods for quantitative measurement of hepatitis B viral DNA. J Hepatol. 1996;24:686–691. doi: 10.1016/s0168-8278(96)80264-9. [DOI] [PubMed] [Google Scholar]
- 4.Heermann K-H, Gerlich W H, Chudy M, Schaefer S, Thomssen R The Eurohep Pathobiology Group. Quantitative detection of hepatitis B virus DNA in two international reference plasma preparations. J Clin Microbiol. 1999;37:68–73. doi: 10.1128/jcm.37.1.68-73.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honkoop P, de Man R A, Niesters H G. Quantitative assessment of hepatitis B virus DNA during a 24-week course of lamivudine therapy. Ann Intern Med. 1998;128:697. doi: 10.7326/0003-4819-128-8-199804150-00028. [DOI] [PubMed] [Google Scholar]
- 6.Honkoop P, Niesters H G, de Man R A, Osterhaus A D, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 7.Janssen H L, Schoenmaker-Weber Y A, Kruining H, Schalm S W, Heijtink R A. Quantitative assessment of hepatitis B virus DNA in chronic hepatitis B: comparison of two solution hybridization assays. J Med Virol. 1993;40:307–312. doi: 10.1002/jmv.1890400408. [DOI] [PubMed] [Google Scholar]
- 8.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stunzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 9.Krajden M, Minor J, Cork L, Comanor L. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J Viral Hepatol. 1998;5:415–422. doi: 10.1046/j.1365-2893.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- 10.Lai V C H, Guan R, Wood M L, Lo S K, Yuen M F, Lai C L. Nucleic acid-based cross-linking assay for detection and quantification of hepatitis B virus DNA. J Clin Microbiol. 1999;37:161–164. doi: 10.1128/jcm.37.1.161-164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 12.Niesters H G, van Esser J, Fries E, Wolthers K C, Cornelissen J, Osterhaus A D. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol. 2000;38:712–715. doi: 10.1128/jcm.38.2.712-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noborg U, Gusdal A, Pisa E K, Hedrum A, Lindh M. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J Clin Microbiol. 1999;37:2793–2797. doi: 10.1128/jcm.37.9.2793-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persing D H. Polymerase chain reaction: trenches to benches. J Clin Microbiol. 1991;29:1281–1285. doi: 10.1128/jcm.29.7.1281-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quint W G V, Heijtink R A, Schirm J, Gerlich W H, Niesters H G M. Reliability of methods for hepatitis B virus DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sia I G, Patel R. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin Microbiol Rev. 2000;13:83–121. doi: 10.1128/cmr.13.1.83-121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaaijer H L, Cuypers H T, Reesink H W, Winkel I N, Gerken G, Lelie P N. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]
- 18.Zaaijer H L, ter Borg F, Cuypers H T, Hermus M C, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]