Abstract
Background:
Combined antiretroviral treatment (cART) for HIV infection is highly effective in controlling viral replication. However, it cannot achieve a sterilizing cure. Several strategies have been proposed to achieve a functional cure, some of them based on immune-mediated clearing of persistently infected cells. Here, we aimed at identifying factors related to CD8TC and CD4TC quality before cART initiation that associate with the persistence of CD8TC antiviral response after cART, inflammation levels, and the size of the viral reservoir.
Methods:
Samples from 25 persons living with HIV were obtained before and after (15 months) cART initiation. Phenotype and functionality of bulk and HIV-specific T cells were assayed by flow cytometry ex vivo or after expansion in pre-cART or post-cART samples, respectively. Cell-Associated (CA) HIV DNA (total and integrated) and RNA (unspliced [US] and multiple spliced [MS]) were quantitated by real-time PCR on post-cART samples. Post-cART plasma levels of CXCL10 (IP-10), soluble CD14 (sCD14) and soluble CD163 (sCD163) were measured by ELISA.
Results:
Pre-cART phenotype of CD8TCs and magnitude and phenotype of HIV-specific response correlated with the phenotype and functionality of CD8TCs post-cART. Moreover, the phenotype of the CD8TCs pre-cART correlated with markers of HIV persistence and inflammation post-cART. Finally, exhaustion and differentiation of CD4TCs pre-cART were associated with the composition of the HIV reservoir post-cART and the level of inflammation.
Conclusions:
Overall, this work provides data to help understand and identify parameters that could be used as markers in the development of immune-based functional HIV cure strategies.
Keywords: HIV, persistence, CD8 T cells, CD4 T cells, inflammation, viral reservoir
INTRODUCTION
Human Immunodeficiency Virus (HIV) is a retrovirus that causes a chronic infection, which produces an irreversible and profound deterioration of the immune system, ultimately leading to the development of acquired immunodeficiency syndrome (AIDS) in the vast majority of untreated HIV+ persons [1]. Combination antiretroviral treatment (cART) has drastically improved the life expectancy and quality of life of people living with HIV/AIDS (PLWHA). Simultaneously, cART has an impact on the epidemic dynamics since good coverage of suppressive cART has been proven to lower transmission rates. Finally, cART is available as pre-exposure or post-exposure prophylaxis [2, 3]. Once cART is initiated, viral replication is suppressed and plasma viral load (VL) falls below detectable levels but, if treatment is interrupted, VL rapidly increases as a consequence of viral reactivation from the latent reservoir [4–6]. Consequently, cART alone is not sufficient to eliminate the infection and PLWHA are dependent on a lifelong treatment which still has several limitations in terms of administration, toxicity, and resistance, among others [7, 8]. On the other hand, there are several reports describing cases of PLWHA who had shown long-term control of the infection either spontaneously or induced by an intervention [9–13] indicating that achieving virus control is feasible. Based on this, different strategies have been designed to find a cure for HIV infection [14, 15].
The ability to maintain viral control is multifactorial and involves host features (clinical, genetic, immune, metabolic, etc), as well as viral features (genome integrity, fitness, escape mutations). However, HIV-specific CD8+ T cells (CD8TC) emerge as key factors to achieve control in different models (reviewed in [16]). CD8TC specificity, function, phenotype, and localization are determinants of viral control and disease progression and may determine, at least in part, the success of future HIV cure and prevention strategies.
Our group has provided evidence regarding different qualitative aspects of the CD8TC response (specificity, functionality, and phenotype) that better associate with virus control in patients not receiving cART in an acute infection cohort from Argentina [17–19]. In the same cohort, we also determined that a higher proportion of terminally differentiated CD8TC and increased PD1 expression before cART initiation were correlated with HIV persistence once cART was initiated [20]. In another report, we analyzed the phenotype (memory/effector maturation subsets and PD-1 expression) and function (production of IFN-γ, IL-2, CCL4[MIP-1β], and/or TNF-α; CD1071/b mobilization, and direct and indirect antiviral activity) of in vitro expanded CD8TC from PLWHA receiving cART who initiated treatment either early or late after infection [21]. Results indicated that HIV-specific cells could be selectively stimulated and expanded in vitro, presenting a high proportion of cells able to carry out multiple effector functions. Although cART initiation timing had an impact on the memory/effector differentiation phenotype, expanded cells from both groups had potent antiviral activity. Here, we aimed at extending our previous work by providing a more complete picture that identifies factors related to CD8TC and CD4TC quality before cART initiation that associates with the persistence of CD8TC antiviral response after cART, inflammation levels, and viral reservoir size. For this, distribution of memory sub-populations and PD-1 expression at CD4TC and CD8TC compartments, and polyfunctionality of HIV-specific CD8TCs were evaluated pre-cART. Additionally, the distribution of memory/effector subsets in CD8TC, HIV-specific CD8TC polyfunctionality, cell-associated (CA) HIV DNA and RNA forms, and plasma levels of IP-10, sCD14, and sCD163 were determined in samples obtained post-cART. Then, correlation analyses were performed between pre-cART and post-cART measurements.
METHODS
Study subjects:
Twenty-five participants diagnosed with HIV infection were enrolled as part of the Grupo Argentino de Seroconversión study group [22] and included in this study. Plasma and peripheral blood mononuclear cells (PBMCs) were collected before cART initiation (pre-cART sample, median time from the presumed date of infection=90 days) and after cART initiation (post-cART sample, median= 15 months on cART) (Table 1). This study was reviewed and approved by the institutional review board Comité de Ética Humana, Facultad de Medicina, Universidad de Buenos Aires, Buenos Aires, Argentina. All participants provided written informed consent.
Table 1:
Characteristics of enrolled participants.
| Pre-ART sampleb | Post-ART samplec | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject IDa | Gender | Age | ART regimen | Viral Loadd (HIV RNA copies/ml plasma) | CD4 counte (cells/ml) | CD8 counte (cells/ml) | CD4/CD8 ratio | Time from infectionf (days) (HIV RNA copies/ml plasma) | Viral Loadd (cells/ml) | CD4 counte (cells/ml) | CD8 counte | CD4/CD8 ratio | Time from ART initi-ationg (days) |
| DT1 | F | 34 | 3TC AZT LPV/r | 48526 | 383 | 531 | 0,72 | 1140 | <50 | 562 | 513 | 1,09 | 960 |
| DT2 | M | 34 | TDF 3TC ATV/r | 132317 | 324 | 703 | 0,46 | 630 | <50 | 297 | 571 | 0,52 | 210 |
| DT3 | M | 42 | 3TC ABC EFV | 125716 | 391 | 602 | 1,36 | 210 | <50 | 711 | 1094 | 0,65 | 570 |
| DT4 | M | 25 | NA | 27244 | 465 | 1027 | 0,45 | 330 | <50 | 535 | 723 | 0,74 | 150 |
| DT5 | F | 23 | NA | NA | NA | NA | NA | NA | <40 | 739 | 739 | 1 | 210 |
| DT6 | M | 26 | 3TC AZT EFV | 66929 | 246 | 1047 | 0,24 | 570 | <50 | 501 | 808 | 0,62 | 330 |
| DT7 | M | 45 | 3TCLPV | 15297 | 330 | 780 | 0,42 | 1530 | <50 | 468 | 608 | 0,77 | 150 |
| DT8 | F | 43 | NA | >500000 | 279 | 1085 | 0,26 | 120 | <40 | 528 | 733 | 0,72 | 270 |
| DT9 | M | 42 | NA | 28288 | 184 | 606 | 0,3 | 640 | <40 | 579 | 657 | 0,88 | 450 |
| DT10 | M | 37 | 3TC AZT EFV | NA | NA | NA | NA | NA | <40 | 563 | 780 | 0,72 | 600 |
| DT11 | M | 49 | 3TC AZT EFV | NA | NA | NA | NA | NA | <40 | 1023 | 674 | 1,52 | 1350 |
| DT12 | M | 27 | TDF FTC EFV | 111893 | 629 | 741 | 0,85 | 240 | ND | 696 | 472 | 1,48 | 690 |
| Median DT (IQR25%-75%) | 57728 (27505122260) | 330 (263428) | 741 (6041037) | 0,45 (0,280,79) | 570(225-890) | 563 (508707) | 699 (580770) | 1 (0,671,07) | 390(210 - 668) | ||||
| ET1 | M | 39 | TDF FTC ATV/r | >500000 | 627 | 943 | 0,66 | 30 | <40 | 594 | 668 | 0,89 | 450 |
| ET2 | M | 45 | 3TC AZT EFV | 283441 | 187 | 928 | 0,2 | 90 | <40 | 522 | 596 | 0,88 | 600 |
| ET3 | M | 41 | TDF ATD 3FTC | 36338 | 768 | 649 | 1,18 | 30 | <50 | 1099 | 599 | 1,83 | 330 |
| ET4 | M | 25 | TDF FTC EFV | 17292 | 612 | 1828 | 0,33 | 60 | <50 | 1342 | 1010 | 1,31 | 450 |
| ET5 | M | 52 | TDF 3TC ATV/r | 20106 | 787 | 743 | 1,06 | 60 | <40 | 829 | 491 | 1,69 | 480 |
| ET6 | M | 70 | TDF FTC | >500000 | 421 | 1379 | 0,31 | 60 | <40 | 854 | 452 | 1,89 | 540 |
| ET7 | M | 26 | TDF FTC EFV | 671440 | 435 | 3346 | 0,13 | 30 | <40 | 842 | 646 | 1,3 | 510 |
| ET8 | M | 30 | TDF 3TC ATV/r | 172468 | 345 | 761 | 0,45 | 30 | <40 | 657 | 566 | 1,16 | 360 |
| ET9 | M | 31 | TDF FTC EFV | 26547 | 400 | 571 | 0,7 | 30 | ND | 824 | 413 | 1,99 | 1000 |
| ET10 | M | 35 | NA | 231360 | 330 | 1881 | 0,18 | 60 | <40 | 1080 | 868 | 1,24 | 360 |
| ET11 | M | 30 | NA | 9294958 | 654 | 1570 | 0,42 | 30 | <40 | 1026 | 696 | 1,47 | 360 |
| ET12 | M | 26 | TDF FTC EFV | >500000 | 339 | 4308 | 0,08 | 90 | <40 | 691 | 1044 | 0,66 | 480 |
| ET13 | M | 42 | TDF FTC ATV/r | >500000 | 213 | 1126 | 0,19 | 90 | <40 | 386 | 480 | 0,81 | 360 |
| Median ET (IQR25%-75%) | 172468 (23327477441) | 421 (335640) | 1126 (7521855) | 0,33 (0,190,70) | 60 (30-75) | 829 (6261053) | 599 (489782) | 1,3 (0,891,76) | 450 (360 - 525) | ||||
| Median Total (IQR25%-75%) | 66929 (26896201914) | 387 (313616) | 935 (6891427) | 0,42 (0,230,70) | 90 (30390) | 691 (531848) | 657 (539759) | 1 (0,731,47) | 450 (330 - 585) | ||||
Participants denoted as DT initiated ART after 4 months since the estimated date of infection (DT=Delayed Treatment). Participants denoted as ET initiated ART within 4 months post-infection (ET=Early Treatment). Further analyses were performed using data from all participants without segregation. All study participants were White.
Sample obtained immediately before participants started ART.
Sample obtained on ART.
Versant HIV-1 RNA 3.0 assay (Siemens. Lower and upper detection limits are 50 and 500,000 RNA copies/ml, respectively) or Abbott Real Time HIV-1 assay (Abbott Park, Lower and upper detection limits are 40 and 107 RNA copies/ml, respectively. ND: Non-detected).
Single platform flow cytometry (BD Biosciences, USA).
Relative to the presumed date of infection.
Time from the moment of cART initiation to sample obtaining.
F: Female. M: Male. NA= Data not available.
Drugs = 3TC: Lamivudine; AZT: Zidovudine; LPV/r: Lopinavir/ritonavir; TDF: Tenofovir; ATV/r: Atazanavir/ritonavir; ABC: Abacavir; EFV: Efavirenz; FTC: Emtricitabine
Samples:
We collected 40 ml of whole blood. Upon centrifugation, plasma was recovered, and stored at −80C. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation (Amersham, Sweden) and cryopreserved in liquid nitrogen for subsequent functional assays. Plasma viral load (VL) was determined by branched-DNA, Versant HIV-1 RNA 3.0 assay (Siemens Healthcare, UK) or by Abbott Real Time HIV-1 assay (Abbott Park, IL) depending on kit availability.
CD4+ and CD8+ T-cell counts were determined by flow cytometry (FACSCalibur; BD Biosciences, USA). Cellular immune activation was evaluated as the percentage of CD38- and/or HLA-DR-expressing CD4+ and CD8+ T cells by flow cytometry.
Ex vivo phenotype and functionality of bulk and HIV-specific T-cells by flow cytometry:
Pre-cART samples were thawed, rested for 2 hours, stimulated with HIV peptide pools for 5 hours and then stained as described previously by our group [21]. Peptide pools spanning Nef (127 peptides) or p24 (128 peptides) were constructed using peptides from the potential T-cell epitope (PTE) peptide panels obtained from the NIH AIDS Reagent Program [23]. For stimulation, peptide pools were used at 2 μg/mL together with costimulatory antibodies (anti-CD28 and anti-CD49d; 1 μg/mL; BD Biosciences), monensin (Golgistop, 0.7 μL/mL; BD Biosciences) and brefeldin A (10 μg/mL; BD Biosciences). A DMSO condition was included to account for background. To evaluate polyfunctionality, anti-CD107a/b-FITC antibodies (BD Biosciences) were also added to identify degranulating cells. After stimulation, cells were further stained with Zombie NIR™ Fixable Viability Kit (Biolegend, USA), and the following conjugated antibodies: anti-CD14-V450, anti-CD19-V450, anti-CD3-BV786, anti-CD8-APC, and anti-CD4-BV650 (BD Biosciences). Then, cells were permeabilized (Permeabilization Wash Buffer, Biolegend), fixed (Fixation Buffer, Biolegend), and subsequently stained using anti-IL-2–PerCP-Cy5.5, anti-TNF-α-PECy7, anti-IFN-γ-BV711, and anti-CCL4-PE conjugated antibodies (BD Biosciences).
In parallel, T-cell memory phenotype was studied as also described by our group [21]. Cells were briefly stimulated as described above and afterwards stained with Zombie NIR™ Fixable Viability Kit plus the following conjugated antibodies, anti-CCR7-Alexa700, anti-PD-1-PE, anti-CD3-BV786, anti-CD8-APC, anti-CD4-BV650, anti-CD14-V450, anti-CD19-V450, anti-CD45RO-Per-CPCy5.5, and anti-CD95-PE-CF594 (BD Biosciences, USA). Following permeabilization, cells were fixed and stained with anti-IL-2, anti-TNF-α, and anti-IFN-γ antibodies, all of them conjugated to FITC (BD Biosciences) to identify specific cells regardless of function.
Flow cytometry data acquisition was performed on a 3-laser 14-color BD FACSAria FUSION flow cytometer using the BD FACSDiva v 8.0.1 software (BD Biosciences). Instrument settings and fluorescence compensation were performed using unstained samples and single-stained BD CompBeads (BD Bioscience). Isotype controls, consisting of stimulated cells stained with conjugated antibodies to CD14, CD19, CD3, CD4, and CD8 plus the isotype controls corresponding to the CCR7, CD45RO, CD95, PD-1, and/or the corresponding intracellular marker were performed for each individual in order to set negative populations accurately.
Acquired data was analyzed using FlowJo v10 (Data Analysis Software, LLC). Gating strategy was performed as described before [21]. First, single cells were selected in a forward scatter area (FSC-A) vs FSC-Height plot. Then, dead cells were excluded based on Zombie NIR™ fluorescence, and monocytes and B lymphocytes were also excluded according to CD14 and CD19 staining. Subsequently, the lymphocyte population was selected in an FSC-A vs side scatter (SSC) plot. Samples with at least 100,000 events in the lymphocyte gate were included in subsequent analyses. Finally, CD3+ CD8+ (or CD4+) cells were gated in CD3-vs-CD8 (or CD4) dot plots.
To study T-cell polyfunctionality, CD107a/b, IFN-γ, IL-2, CCL4, or TNF-α plots were constructed on the CD8+ and CD4+ populations and a Boolean gate platform was used to create all possible combinations. Data presented correspond to background-subtracted results using the DMSO plus CD28/CD49d control. This was performed on a cytokine-subset-by-cytokine-subset basis, ie, subtracting the result from this condition for a given cytokine subset to the same subset of a peptide-stimulated condition. One standard deviation (SDs) above background was set as the threshold for determining positive responses. Values below this threshold were set at 0.
The distribution of the different T-cell phenotype subsets was studied in bulk and HIV-specific CD8+ T cells and bulk CD4+ T cells. HIV-specific CD8+ T cells were identified in a CD8 vs FITC plot (CD107a/b, IFN-γ, IL-2, CCL4, and TNF-α). A positive cytokine response was defined as at least twice the background value, >0.05% after subtraction of background and at least 1,000 events. This criterion was established to minimize the possibility of error due to a low number of events when further subdividing these cells into the different memory subsets. Central memory T cells (TCM, CCR7+/CD45RO+), effector memory T cells (TEM, CCR7−/CD45RO+), and terminal effector T cells (TTE, CCR7−/CD45RO−) were identified. Within the CD45RO−CCR7+ cells naive T-cells (Tnaive, CCR7+/CD45RO−/CD95−) and stem-cell memory T-cells (TSCM, CCR7+/CD45RO−/CD95+) were gated. Additionally, PD-1 expression was studied on bulk and memory subpopulations. Based on the knowledge that HIV-specific CD8+ T-cell frequency decays significantly following cART initiation, an in vitro cell expansion protocol was implemented as reported previously by our group [21]. Phenotype and polyfunctionality were subsequently studied on expanded cells.
Due to limitations in sample availability, bulk CD8TC and CD4 TC responses at pre-cART samples were evaluated in 22 and 23 participants, respectively. In all participants except 1, replicates could be made, adding a total of 43 and 45 responses, respectively. Bulk CD8TC parameters at post-cART were evaluated in the 25 participants of the cohort, all of them in duplicate except for 3. This adds a total of 47 responses. The phenotypes of HIV-specific CD8TC responses were evaluated in 18 individuals pre-cART and in 23 individuals post-cART. When the quantity of sample allowed it, replicates were performed with different antigens in the stimulation (Nef and p24 peptide pools). Thus, a total of 35 responses were obtained pre-cART and 43 responses were obtained post-cART. CD8 TC polyfunctionality was evaluated in 24 and 23 pre-cART and post-cART samples, respectively. When possible, samples were tested in replicate against Nef and p24 peptide pools adding a total of 42 and 46 responses, respectively.
Additionally, antiviral activity of expanded CD8+ T cells was evaluated using the Viral Inhibition Assay (VIA) and the VITAL assay as described previously by our group [21]. Briefly, the VIA protocol was initially published by Sáez-Cirión et al [24] and it assesses the capacity of expanded CD8TC to inhibit viral replication in primary autologous CD4TCs, accounting for both cytolytic and non-cytolytic mechanisms. Here, target cells (consisting of autologous CD4TCs purified after PBMC treatment with CD3/8 bi-specific antibodies) were infected and co-cultured at 1:1 ratio with purified expanded Nef-specific or p24-specific CD8+ T cells (effectors). At day 7, p24 antigen was quantified in cell culture supernatants by ELISA. CD8TC anti-HIV suppressive capacity was calculated as the log10 of the percentage of p24 antigen loss when CD8TCs were present in the culture compared to infected CD4TC controls without effectors. On the other hand, the VITAL assay was adapted from the publication by Hermans et al [25], and it assesses direct HIV-specific cell-mediated cytotoxicity.
Briefly, CD4+ target cells (prepared as described above) were either labeled with CFSE or PKH-26 red fluorescent cell linker. CFSE-labeled cells were loaded with Nef or p24 peptides and then combined with PKH-26-labeled cells. Nef- or p24-specific effector cells were added at different target-to-effector (T:E) ratios (1:1, 1:5, 1:10, and 1:1 adjusted to proportion of specific CD8+ T-cell effectors). Following overnight incubation, cells were stained with Zombie Viability Kit and anti-CD3-PECy7, anti-CD4-PerCP, and anti-CD8-BV510 antibodies, and analyzed by flow cytometry. Adjusted survival was calculated as the mean percentage of CFSE+ events with added effector vs the condition with no effectors. Finally, the percentage of specific lysis was calculated using the equation % specific lysis= 100 - % adjusted survival. Further details can be found in Salido et al [21]. The VIA assay could be performed in 16 post-cART samples. In all cases, effector cells were prepared in parallel with Nef and p24 peptide pools resulting in 2 sets of effectors per sample that were tested separately. This resulted in 32 VIA values for the whole set of samples. Moreover, these 2 sets of effectors were obtained for the VITAL assay. However, this assay requires a larger number of cells and thus the number of samples tested and replicates were smaller: 1:1 relation: 15 samples, 30 responses; 1:5: 12 samples, 21 responses; 1:10: 10 samples, 15 responses.
Quantitative real-time PCR for Cell-Associated (CA) HIV RNA and DNA:
Cell-associated (CA) HIV DNA (total and integrated) and RNA (unspliced [US]) and multiple spliced [MS]) were quantitated by real-time PCR on post-cART samples. Total HIV DNA PCR measures all forms of viral DNA present in the cell (integrated and non-integrated forms), while HIV-integrated DNA PCR amplifies integrated provirus exclusively. The CA US-RNA form represents the genomic RNA (result of genuine HIV transcription) but can also represent host-HIV read-through transcripts. However, the latter transcripts have been shown to contribute poorly to the bulk of HIV RNA. The CA MS-RNA transcripts are more closely related to advanced stages in the cycle of viral replication and production of infectious particles [26].
CD4+ T cells were isolated from frozen PBMCs using an immunomagnetic selection kit (BD Bioscience, USA) and only 90% purity samples (determined by flow cytometry) were assayed. DNA and RNA were extracted using a proteinase K lysis protocol or a commercial kit (PureLink RNA Mini Kit, Invitrogen, USA) respectively, then both were quantified and stored at −80°C until use. Total and integrated HIV DNA was quantitated following the highly sensitive real-time nested PCR-based assay developed by Chomont et al [27] with modifications. Briefly, cell lysates were used as input in a pre-PCR step in which the total or integrated HIV DNA was pre-amplified together with the CD3 gene (2 copies per cell) in the same reaction, in triplicate. Pre-amplified products were diluted and used as input in a second amplification reaction (HIV or CD3) in which each form was quantified by real-time PCR. The frequency of cells harboring each molecular form was calculated from the ratio of HIV copy number/2 x (CD3 copy number). For quantification of US and MS-RNA forms, Pasternak's protocol was used [28] with modifications. Briefly, a hemi-nested PCR was performed with 16 cycles of amplification followed by a second amplification round of quantitative real time PCR. Then US and MS-RNA copy numbers were standardized to cellular equivalents using a 18s RNA real time PCR (Invitrogen, USA). Each sample was assayed in quadruplicate and a control with no reverse transcriptase was used to reveal DNA contamination. Quantitative real time PCR assays were run for 40 cycles. For both HIV DNA and RNA PCRs, the LLODs were 1 copy per well. If there was amplification but it was not quantified (below the LLOD), an arbitrary value of 0.5 copies was assigned. If no amplification occurred at all, the value was set to 0 copies. Due to sample availability, these assays were performed in the following number of samples: Total HIV DNA: 17 samples, Integrated HIV DNA: 5 samples, CA HIV US-RNA: 18 samples, and CA HIV MS-RNA: 12 samples.
Plasma Soluble factors:
The levels of CXCL10 (IP-10, BD Biosciences), soluble CD14 (sCD14) and soluble CD163 (sCD163) (Thermo Fisher Scientific) were measured in post-cART plasma samples. Due to sample availability, 9, 23, and 22 samples were used to evaluate IP-10, sCD14, and sCD163 respectively.
Data analysis:
Data was expressed as median values with interquartile ranges (25% to 75%, IQ25-75) and analyzed by nonparametric methods using GraphPad Prism v7.0 software, unless otherwise stated. Correlation analyses were performed using the Spearman's rank test. In this case, P-values were adjusted for multiple comparisons using a false discovery rate (FDR) procedure, according to the Benjamini and Hochberg method, using the GraphPad Prism 7 software (Graph-Pad Software Inc., San Diego, CA, USA). Distribution analysis of CD4+ and CD8+ T-cell subsets in pre- and post-cART samples was performed with SPICE 6.0 software (https://niaid.github.io/spice/). All tests were considered significant when the P-value was <0.05. Adjusted P-values for correlation analyses were considered significant when <0.1.
RESULTS
Cohort description and experimental determinations:
Twenty-five participants from the Grupo Argentino de Seroconversión study group were selected. Samples were obtained immediately before cART initiation (pre-cART sample) and at a median of 15 months after cART initiation (post-cART samples). Paired pre- and post-cART samples were obtained for all participants but 3. A detailed description of participants is shown in Table 1. As expected, all participants had detectable viral load pre-cART and reached levels below the limit of detection post-cART.
Median CD8TC count significantly diminished while median CD4TC count and CD4/CD8 ratio significantly increased post-cART (Wilcoxon's test P<0.0001, P=0.0001, and P<0.0001, respectively).
At the pre-cART samples, the following determinations were performed: i) distribution of memory subpopulations at bulk CD4TC and CD8TC compartments; ii) HIV-specific CD8TC polyfunctionality and memory/effector differentiation profile; iii) PD-1 expression in bulk and memory/effector subpopulations of CD4TC, CD8TC, and HIV-specific cells. In post-cART samples, CA HIV DNA and RNA forms as well as plasma levels of IP-10, sCD14, and sCD163 were determined ex vivo. Also, the distribution of memory/effector subsets in bulk and HIV-specific CD8TC and HIV-specific CD8TC polyfunctionality were determined after cell expansion in vitro as performed previously [21]. Results for these determinations are shown in supplemental figures 1 and 2. Overall, it could be observed that the pre-cART distribution of naive and memory/effector T-cell subsets, both at the CD4TC and CD8TC compartments, behaved as expected according to previous publications [18, 29] (Supplemental Figure 1A). Since it is known that the frequency of HIV-specific T cells decays significantly following cART initiation, cells from post-cART samples were expanded as reported previously [21]. No immune parameter was evaluated directly ex vivo in post-cART samples. During the expansion process, CD8TC are preferentially expanded over CD4TCs. This could be due to suboptimal culture conditions for CD4TCs proliferation, out-competition by CD8TCs, underrepresentation of MHC-II-restricted compared to MHC-I-restricted peptides, or for other reasons. Thus, CD4TC were not evaluated at post-cART samples.
Both bulk and HIV-specific CD8TC post-cART also showed the anticipated phenotype [21] with a noticeable enrichment of effector memory (TEM) at the specific compartment (Supplemental Figure 1B). HIV-specific pre-cART CD8TC were dominated by monofunctional cells. The most frequent functions were CD107A/B mobilization (degranulation) and TNF-α production followed by IFN-γ, Mip1β (CCL4), and IL-2 production. On the other hand, the most frequent functions in expanded CD8TC from post-cART samples were IFN-γ, Mip1β (CCL4), and TNF-α production followed by degranulation and, to a lesser extent, IL-2 production. In this group, a significantly higher proportion of polyfunctional cells able to carry out multiple effector functions were found (permutation test P<0.0001, SPICE software; Supplemental Figure 1C). In line with this, expanded CD8TC were able to exert significant cytotoxic activity as measured by the VITAL assay, ie, specific direct lysis of HIV-peptide-loaded target CD4TCs by the autologous HIV-specific expanded CD8TC (Supplemental Figure 1D). Also, 12 out of 25 participants (52%) had detectable HIV-specific CD4TC responses pre-cART. Positive responses showed a median of 0.56% (IQR25-75=0.42%-0.86%, data not shown).
Pre-cART PD-1 showed a significant expression when analyzed in bulk CD4TC and CD8TC (Supplemental Figure 1E, left and right panel, respectively), especially in those individuals with delayed treatment initiation. When its expression was studied in the different subpopulations, it was concentrated within TEM and central memory T cells (TCM). Regarding viral reservoir composition post-cART, total HIV DNA was more readily measured than integrated DNA, while US-RNA was more abundant than MS-RNA (Supplemental Figure 2A). Plus, US-RNA to total or integrated DNA ratios were calculated in order to account for transcriptional activity per infected cell; and MS- to US-RNA ratio to illustrate the stage in viral genome transcription, ie, lower MS/US-RNA reflects higher frequency of cells in the later stages of the viral replication cycle [26] (Supplemental Figure 2B). Finally, plasma levels of IP-10, sCD14, and sCD163 were measured. These molecules have been proposed as markers of persistent inflammation in HIV infection since their levels remain elevated despite years of cART, compared to healthy donors [30–37]. Both IP-10, sCD14, and sCD163 were detected in all the samples tested. Moreover, IP-10 and sCD14 medians were considerably elevated compared to values corresponding to healthy donors found in the bibliography (Supplemental Figure 2C) [30–34, 38–41].
Correlation analysis showed a relationship between pre- and post-cART CD8TC response quality (preserved distribution of memory subsets, functionality, and lower PD-1 expression), while CD4TC phenotype (distribution of memory subsets and PD-1 expression) pre-cART was better associated with the level of HIV persistence and inflammation post-cART.
As a first approach on how pre-cART CD8TC and CD4TC phenotype and function relate to the parameters evaluated post-cART, correlation analyses were performed and heat-maps were prepared to better visualize results (Supplemental Figure 3). The rows and columns correspond to determinations performed at the pre-cART and post-cART samples, respectively. Also, the estimated time of infection to sampling and the pre-cART VL value were included as row variables. In this manner, we were able to identify patterns of consistent correlations that provide support to the ideas presented in our hypothesis:
i) The phenotype of CD8TC pre-cART influences the functionality of CD8TC post-cART:
First, we observed that the percentages of pre-cART bulk CD8+ Tnaive cells correlated inversely with the proportions of post-cART bulk CD8+ TCM, TEM cells and with the levels of CD8+ T-cell arrest (calculated as the ratio TEM/[TEM+TTE] as described previously [18]), while it correlated directly with the proportions of bulk CD8+ TTE cells (Figure 1A). The percentage of HIV-specific CD8TC pre-ART evaluated ex vivo by flow cytometry correlated directly with the capacity of expanded CD8TC from post-cART samples to mediate direct cytolytic antiviral function measured by the VITAL assay (Figure 1B). In addition, the phenotype of the HIV-specific CD8+ T cells pre-cART was related to cell functionality post-cART: higher percentages of CD8+ stem-cell memory T cells (TSCM) and TTE cells correlated inversely with proportions of monofunctional HIV-specific CD8+ T cells post-cART and correlated directly with higher proportions of polyfunctional (4- and 5-function cells for TSCM and 5-function cells for TTE) post-cART. Conversely, higher percentages of CD8+ TEM cells and higher arrest correlated directly with higher frequencies of monofunctional cells (Figure 1C) and correlated inversely with frequencies of polyfunctional HIV-specific CD8+ T cells post-cART.
Figure 1:
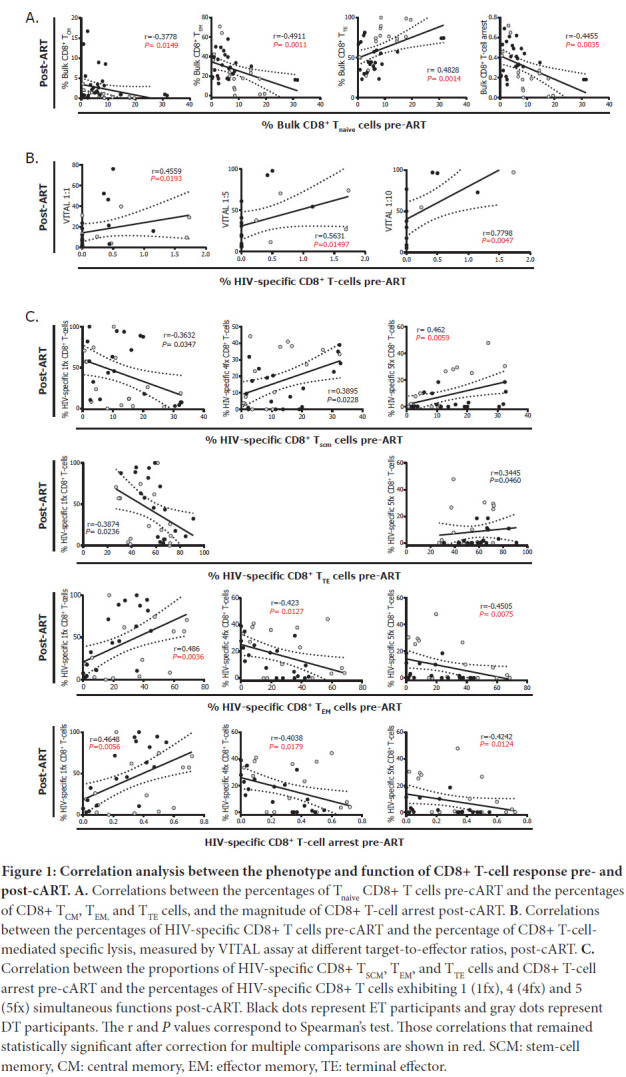
Correlation analysis between the phenotype and function of CD8+ T-cell response pre- and post-cART. A. Correlations between the percentages of Tnaive CD8+ T cells pre-cART and the percentages of CD8+ TCM, TEM, and TTE cells, and the magnitude of CD8+ T-cell arrest post-cART. B. Correlations between the percentages of HIV-specific CD8+ T cells pre-cART and the percentage of CD8+ T-cell-mediated specific lysis, measured by VITAL assay at different target-to-effector ratios, post-cART. C. Correlation between the proportions of HIV-specific CD8+ TSCM, TEM, and TTE cells and CD8+ T-cell arrest pre-cART and the percentages of HIV-specific CD8+ T cells exhibiting 1 (1fx), 4 (4fx) and 5 (5fx) simultaneous functions post-cART. Black dots represent ET participants and gray dots represent DT participants. The r and P values correspond to Spearman's test. Those correlations that remained statistically significant after correction for multiple comparisons are shown in red. SCM: stem-cell memory, CM: central memory, EM: effector memory, TE: terminal effector.
ii) An exhausted phenotype on CD8+ T cells pre-cART also relates to HIV persistence and inflammation post-cART:
The percentages of CD8+ T cells expressing PD-1 and PD-1high pre-cART were correlated directly with higher levels of total HIV DNA (Figure 2A). Higher percentages of CD8+ TEM cells, lower percentages of CD8+ TTE cells and higher CD8+ T-cell arrest were related to lower levels of HIV US-RNA and lower US-RNA/total DNA ratios post-cART (Figure 2B). Finally, frequencies of CD8+ TEM cells, CD8+ T–cell arrest, and the frequency of CD8+ T-cells expressing PD-1 correlated directly with the levels of plasma IP-10 post-cART (Figure 2C).
Figure 2:
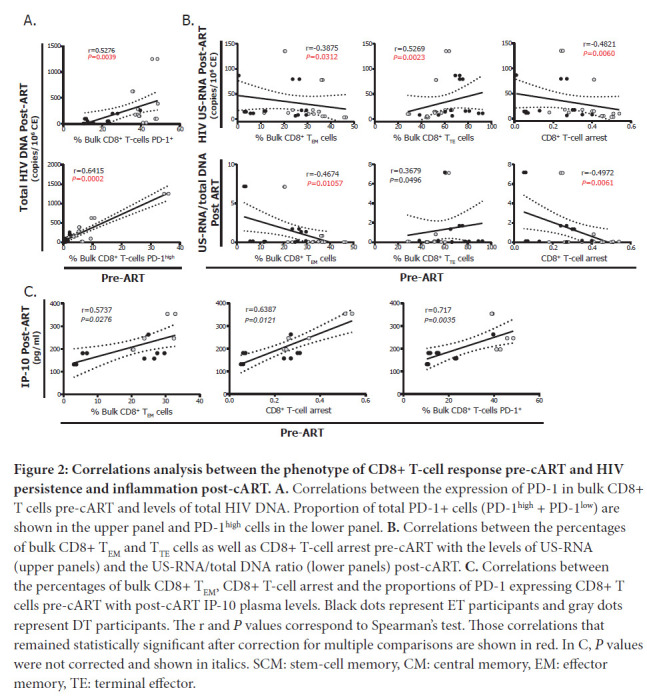
Correlations analysis between the phenotype of CD8+ T-cell response pre-cART and HIV persistence and inflammation post-cART. A. Correlations between the expression of PD-1 in bulk CD8+ T cells pre-cART and levels of total HIV DNA. Proportion of total PD-1+ cells (PD-1high + PD-1low) are shown in the upper panel and PD-1high cells in the lower panel. B. Correlations between the percentages of bulk CD8+ TEM and TTE cells as well as CD8+ T-cell arrest pre-cART with the levels of US-RNA (upper panels) and the US-RNA/total DNA ratio (lower panels) post-cART. C. Correlations between the percentages of bulk CD8+ TEM, CD8+ T-cell arrest and the proportions of PD-1 expressing CD8+ T cells pre-cART with post-cART IP-10 plasma levels. Black dots represent ET participants and gray dots represent DT participants. The r and P values correspond to Spearman's test. Those correlations that remained statistically significant after correction for multiple comparisons are shown in red. In C, P values were not corrected and shown in italics. SCM: stem-cell memory, CM: central memory, EM: effector memory, TE: terminal effector.
iii) Exhaustion and differentiation of CD4+ T cells pre-cART determine the composition of the HIV reservoir post-cART and also the level of inflammation:
The frequency of PD-1high CD4+ T cells pre-cART correlated directly with the levels of total HIV DNA and inversely with US-RNA and the US-RNA/total DNA ratio post-cART (Figure 3A). Pre-cART percentages of CD4+ Tnaive cells correlated directly with the levels of total HIV DNA, and MS-RNA/US-RNA ratios post-cART (Figure 3B). Similarly, pre-cART percentages of CD4+ TCM cells correlated directly with the levels of total and integrated HIV DNA, and inversely with the levels of HIV US-RNA as wells as US-RNA/total DNA and US-RNA/integrated DNA ratios (Figure 3C). Finally, the percentages of terminally differentiated CD4+ T cells pre-cART correlated inversely with the total and integrated HIV DNA, and directly with the levels of US-RNA and the US-RNA/total DNA ratio (Figure 3D). Exactly opposite correlations were found with the magnitude of CD4+ T-cell differentiation arrest (Figure 3E). These results prompted us to study the relation of PD-1 expression within each of these CD4+ T-cell subpopulations and HIV persistence.
Figure 3:
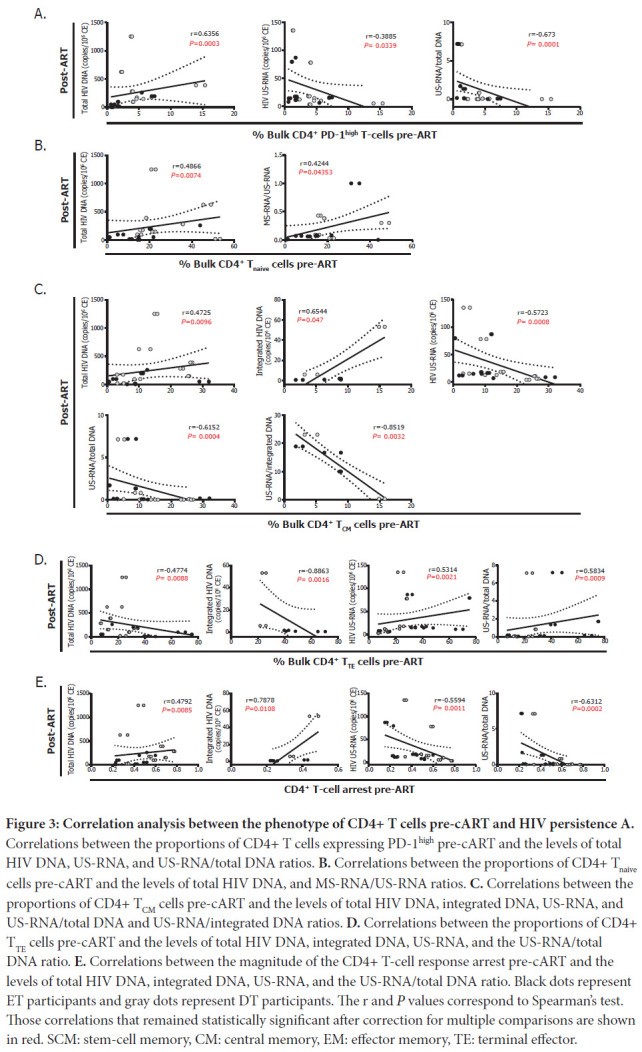
Correlation analysis between the phenotype of CD4+ T cells pre-cART and HIV persistence A. Correlations between the proportions of CD4+ T cells expressing PD-1high pre-cART and the levels of total HIV DNA, US-RNA, and US-RNA/total DNA ratios. B. Correlations between the proportions of CD4+ Tnaive cells pre-cART and the levels of total HIV DNA, and MS-RNA/US-RNA ratios. C. Correlations between the proportions of CD4+ TCM cells pre-cART and the levels of total HIV DNA, integrated DNA, US-RNA, and US-RNA/total DNA and US-RNA/integrated DNA ratios. D. Correlations between the proportions of CD4+ TTE cells pre-cART and the levels of total HIV DNA, integrated DNA, US-RNA, and the US-RNA/total DNA ratio. E. Correlations between the magnitude of the CD4+ T-cell response arrest pre-cART and the levels of total HIV DNA, integrated DNA, US-RNA, and the US-RNA/total DNA ratio. Black dots represent ET participants and gray dots represent DT participants. The r and P values correspond to Spearman's test. Those correlations that remained statistically significant after correction for multiple comparisons are shown in red. SCM: stem-cell memory, CM: central memory, EM: effector memory, TE: terminal effector.
First, higher proportions of CD8+ TCM, TEM and TTE cells expressing PD-1 pre-cART were correlated with higher levels of total HIV DNA post-cART (Figure 4A), suggesting an association between an exhausted CD8+ T-cell response and a higher HIV reservoir post-cART. On the other hand, higher proportions of CD4+ TCM PD-1+ pre-cART correlated with lower levels of US-RNA and US-RNA/total DNA ratios post-cART while higher proportions of CD4+ TEM PD-1+ pre-cART correlated with higher levels of total DNA, lower US-RNA levels, and lower US-RNA/total DNA ratios post-cART. In turn, proportions of CD4+ TTE PD-1+ pre-cART correlated inversely with US-RNA/total DNA ratios (Figure 4B). Overall, these results indicate that HIV may persist in diverse CD4+ T-cell subsets with different efficiency. Pre-cART expression of PD-1 on CD4+ T cells seems to be a marker of lower HIV transcriptional activity, particularly in TCM and TEM cells.
Figure 4:
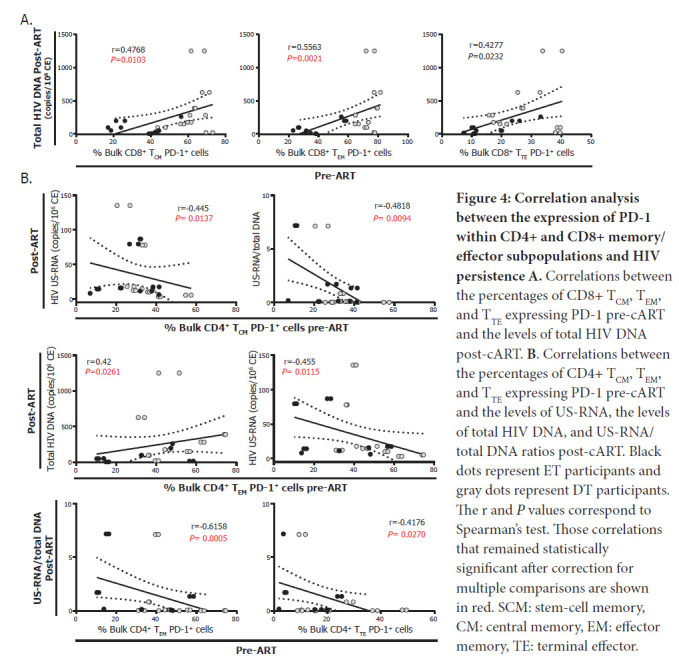
Correlation analysis between the expression of PD-1 within CD4+ and CD8+ memory/effector subpopulations and HIV persistence A. Correlations between the percentages of CD8+ TCM, TEM, and TTE expressing PD-1 pre-cART and the levels of total HIV DNA post-cART. B. Correlations between the percentages of CD4+ TCM, TEM, and TTE expressing PD-1 pre-cART and the levels of US-RNA, the levels of total HIV DNA, and US-RNA/total DNA ratios post-cART. Black dots represent ET participants and gray dots represent DT participants. The r and P values correspond to Spearman's test. Those correlations that remained statistically significant after correction for multiple comparisons are shown in red. SCM: stem-cell memory, CM: central memory, EM: effector memory, TE: terminal effector.
Finally, percentages of pre-cART CD4+ Tnaive and TCM cells correlated directly with plasma levels of IP-10 and sCD163 post-cART while proportions of pre-cART CD4+ TTE cells showed inverse correlations (Figure 5). In turn, percentages of pre-cART CD4+ PD-1+ T cells correlated directly with both markers.
Figure 5:
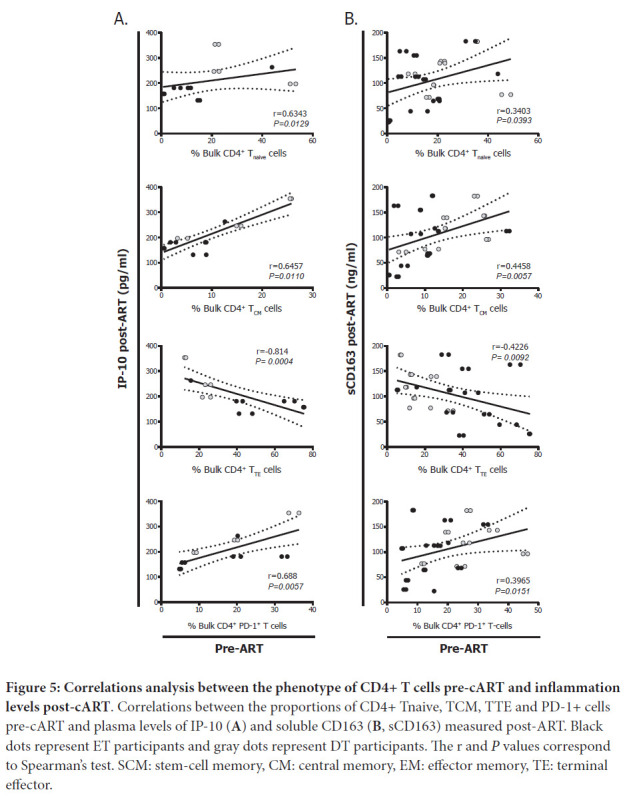
Correlations analysis between the phenotype of CD4+ T cells pre-cART and inflammation levels post-cART. Correlations between the proportions of CD4+ Tnaive, TCM, TTE and PD-1+ cells pre-cART and plasma levels of IP-10 (A) and soluble CD163 (B, sCD163) measured post-ART. Black dots represent ET participants and gray dots represent DT participants. The r and P values correspond to Spearman's test. SCM: stem-cell memory, CM: central memory, EM: effector memory, TE: terminal effector.
DISCUSSION
HIV-specific CD8+ T-cell responses are believed to be a very important factor in achieving sustained long-term viral control as demonstrated in non-human primate models and elite controllers [16]. Since responses elicited in most PLWHA are sub-effective, developing strategies to promote de novo responses or to modulate existing memory immunity in order to resemble the characteristics found in HIV controllers may pave the way to cure the infection. Here, we extended previous studies regarding the association of function and phenotype of CD4+ and CD8+ T cells pre-cART with the immune function and viral markers that persist after cART instauration. It was found that i) the phenotype of CD8+ T cells pre-cART as well as the magnitude and pheno-type of HIV-specific response might be associated with the phenotype and functionality of CD8+ T cells post-cART; ii) the phenotype of the CD8+ T cells pre-cART correlated with markers of HIV persistence and inflammation post-cART; iii) exhaustion and differentiation of CD4+ T cells pre-cART were associated with the composition of the HIV reservoir post-cART and the level of inflammation (Figure 6).
Figure 6:
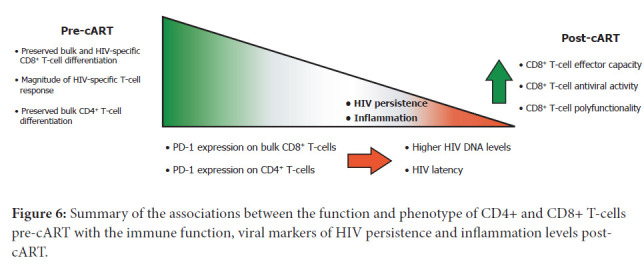
Summary of the associations between the function and phenotype of CD4+ and CD8+ T-cells pre-cART with the immune function, viral markers of HIV persistence and inflammation levels post-cART.
During the natural course of infection, HIV-specific CD8+ T cells play a central role in the control of viral replication, particularly during acute infection [16, 42, 43]. Bulk and HIV-specific CD8+ T cells suffer early alterations in their frequency, differentiation, activation, and functionality that are progressively accentuated, as antigen stimulation persists in untreated infection [17, 18, 44, 45]. As PLWHA initiate cART, some of these alterations revert but CD8 T-cell optimal function is not fully restored [42, 46–54]. As more CD8+ T-cell-based vaccine or CD8+ T-cell dependent cure strategies are being proposed and developed, it is critical to understand and identify the limitations and pitfalls of the immune response that persists in people receiving cART, and to generate tools in order to boost its antiviral function. Here, we show that the magnitude of the damage reached pre-cART associates with the level of dysfunction that remains when receiving cART. This is illustrated by correlations found between percentages of bulk CD8+ Tnaive cells pre-cART and the level of CD8TC arrest post-cART (Figure 1A), the level of CD8TC arrest in the HIV-specific CD8+ T-cell compartment pre-cART and decreased functionality post-cART (Figure 1B-C), and the level of CD8TC arrest and exhaustion of the bulk compartment pre-cART with the levels of inflammation when receiving cART (Figure 2C). It is worth noting that the level of CD8TC arrest as calculated here is an indicator of maturation blockade and skewed differentiation in the T-cell compartment in HIV infection, and it has been correlated with markers of disease progression [18, 46, 55–57]. Thus, these CD8+ T-cell parameters, evaluated immediately before cART initiation, could serve as markers of CD8+ T-cell functionality post-cART, could determine if an individual is eligible for a given intervention, or could be useful to predict the success of a CD8+ T-cell-based strategy or even the need of tailored interventions. In addition, we report that percentages of exhausted bulk CD8+ T cells as well as the level of differentiation arrest in this compartment correlated with the size of the HIV DNA reservoir during cART and the activity of this reservoir, respectively (Figure 2A-B). This recapitulates and extends our previous finding [20] where similar results were obtained in an independent cohort. Two hypotheses arise; it could be first postulated that an early preservation of the CD8+ T-cell compartment limits viral spreading resulting in a smaller reservoir. It is worth noting that the correlations obtained referred to the bulk compartment, not the HIV-specific compartment. This could be illustrating the global alteration that this cell population undergoes in the context of HIV infection. On the other hand, it is known that CD8+ T cells have a role in controlling viral replication during cART although it is neither complete nor efficient since VL inevitably re-emerges upon cART interruption [12, 16, 20, 29, 58–61]. Thus, pre-cART CD8+ T-cell response quality might be standing as a surrogate marker for CD8+ T-cell response quality during cART in these correlations. Recent advances indicate that viral proteins were found to be expressed from latently infected CD4+ T cells, even from those harboring defective HIV proviruses, and that these cells are subjected to immune surveillance [62, 63]. Very recently, Stevenson et al [64] showed that long-term CD8+ T-cell responses during cART are maintained by viral protein-mediated cell triggering. This indicates that CD8+ T-cell response during cART contributes to the dynamics of reservoir maintenance, and warrants more research to understand the molecular biology of HIV persistence during cART and its interaction with the CD8+ T-cell response in order to expand cure and vaccine research.
The population of CD4+ T cells is very heterogeneous and susceptibility to HIV infection and replication activity depends on their differentiation, activation status, functionality, phenotype, and even antigen specificity, among other factors [65]. In treated participants, frequency of HIV persistence varies across different memory subsets. While CD4+ Tnaive cells and TSCM are known to contribute to viral persistence, TCM and TEM are the key contributors to the viral reservoir based not only on their own capacity to self-renew and proliferate but also because they show the highest frequency of infected cells [66, 67] and the highest frequencies of intact and inducible proviruses [68–72], respectively. Here, correlations were observed between the proportions of different CD4+ T-cell subsets pre-cART and either the magnitude of the HIV reservoir measured by total or integrated DNA or the transcriptional activity of that reservoir measured as levels of US-RNA or MS-RNA or HIV RNA/DNA ratios. Levels of HIV DNA post-cART (total and/or integrated) were associated with higher proportions of CD4+ TNaive cells, CD4+ TCM cells, higher CD4+ T-cell arrest, and lower proportions of CD4+ TTE cells pre-cART. However, exactly opposite correlations were found when measuring the transcriptional activity (RNA forms) of the reservoir (Figure 3). This is consistent with the idea that CD4+ TCM cells contribute highly to the pool of HIV-infected cells that persist after cART, but that they harbor virus which rather keeps in a transcriptionally silent state. Then, HIV transcription activity augments as memory cells transition to an activated effector phenotype [65]. In addition to the cellular differentiation state, the expression and coexpression of certain immune checkpoint molecules on CD4+ T-cells (PD-1, TIGIT, LAG-3, CTLA4, among others) have been associated with different states of HIV persistence [73–76].
In particular, we have previously shown that the magnitude of PD-1 expression both at the CD8+ and CD4+ T-cell compartments, evaluated early after infection, was associated with the levels of subsequent viral persistence on cART [20]. Here, we show that although pre-cART expression of PD-1 on CD4+ T-cells associates directly with the frequency of infected cells on cART, higher frequencies of CD4+ PD-1+ TCM, TEM and TTE cells correlated with lower HIV transcription during cART measured as the level of US-RNA or US-RNA/total DNA ratio. This finding is in line with previous evidence suggesting that PD-1 (and other immune checkpoints) are markers of transcriptionally-silent HIV-infected cells during cART [73, 74, 76]. Moreover, blockade of immune checkpoints (more specifically PD-1) was shown to rescue HIV transcription in these cells, and that it could even be proposed as a latency reversal agent [77]. So far, most cure interventions have been proposed to start once PLWHA are receiving suppressive cART. However, it may also be possible that targeting PD-1 at cART initiation could also be beneficial, leading to a smaller HIV reservoir post-cART by reversing HIV latency and increasing CD8+ T-cell function against infected cells earlier.
This is a descriptive study seeking correlations between immune and viral parameters associated with HIV infection. We performed a comprehensive study where multiple immunological and virological parameters were evaluated in paired pre- and post-cART samples. Although our approach is only descriptive, many of the associations that were raised in the analyses are in line with findings from studies involving mechanistical or interventional approaches. This provides reciprocal support to the findings and enhances the evidence in the field. As discussed above, several pre-cART parameters identified here have the potential to act as biomarkers of post-cART immune function and viral persistence, and, hypothetically, could also serve to predict the outcome of an intervention or even indicate whether the intervention is adequate for a given person. Furthermore, it is possible to postulate that the moment of cART initiation may also represent a moment of opportunity to intervene.
This study has several limitations. First, limited sample availability resulted in discrepancies in the number of parameters evaluated in each sample. The second limitation was the need to use a strong and prolonged stimulation protocol for T-cell expansion in samples obtained during cART. Although it is a useful tool to evidence the HIV-specific response post-cART, it could also introduce modifications into the cell phenotype and function. Third, cell phenotype was only studied by means of memory differentiation markers and PD-1 expression. Deeper analysis including other immune checkpoint molecules, co-stimulatory molecules, and survival markers could provide additional and relevant information. Similarly, inclusion of other functional markers such as the production of perforins and granzymes, which have been already described as key components of CD8+ T-cell antiviral function, were not measured.
Fourth, pre-cART and post-cART samples are not homogeneous in terms of time from infection and time receiving cART, respectively. Moreover, DT and ET groups differ significantly in time from infection to sampling and pre-cART VL values. Indeed, it is well known that these 2 variables play a significant role in immune function preservation, inflammation, immune-mediated damage, and composition and size of the viral reservoir. In this study, global associations between pre- and post-cART parameters are described without pretending to imply a cause-effect relationship nor suggest a direct underlying mechanism that could account for such associations. Whether the time from infection to sampling or the pre-cART VL values influence these associations could not be discounted. A proper study with an adequate sample size should be designed to rule out this possibility and exclude confounders. On the other hand, immune profiling of CD4TC was not performed in post-ART samples due to technical constraints and lack of enough biological material. Certainly, this could have represented an added value to the study and deserves to be analyzed in a subsequent study. Finally, PCR-based assays aimed at quantifying HIV DNA or RNA have been extensively used to study the viral reservoir in PLWHA receiving cART. However, these techniques fail to discriminate between intact vs defective or inducible vs latent proviruses. This may overestimate the size of the replication-competent virus pool, and the results may not necessarily correlate with the dynamics of the truly inducible, replication-competent viral reservoir. Our results should be extended by including other measurements such as the qVOA, TILDA (Tat/rev Induced Limiting Dilution Assay), and IPDA (intact proviral DNA assay) assays [78].
Overall, combined and parallel analysis of CD8+ and CD4+ T-cell quality before cART together with immune and viral factors during cART represent an important progress in order to understand HIV persistence. Boosting and/or modulation of CD8+ T-cell responses in individuals receiving cART is postulated as essential in HIV cure strategies. Here, we observed that pre-cART quality in CD8+ T-cell response could predict the level of functionality of CD8+ T-cell response during cART. Similarly, proportions of the different CD4+ T-cell memory/effector subsets and the expression of exhaustion markers within each subset could act as determinants not only of the level but also of the activity of the viral reservoir. Advances made in the HIV persistence field over the last few years hold promise for the development of HIV functional cure strategies. Within these strategies, the immune environment will be a key determinant of success. This work provides data to help understand and identify parameters that could be used as markers for subsequent interventions.
ACKNOWLEDGMENTS
Authors especially acknowledge study participants for agreeing to collaborate in this study and to provide blood samples. We thank Mrs. Sabrina Azzolina for technical help during sample processing and Mr. Sergio Mazzini for language assistance during manuscript preparation.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to report.
SUPPLEMENTARY DATA
Supplementary materials are available at the Pathogens and Immunity website. Supplementary data may be provided by the authors to benefit the reader. Supplementary data are not copyedited and are the sole responsibility of the authors. Questions or comments related to supplementary materials should be addressed to the corresponding author.
FUNDING SOURCES
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Grant # PICT2016-1636 to GT and Grant #PICT2018-1856 to YG); and the Universidad de Buenos Aires (UBACyT2017, Grant # 20020160100008BA) to GT. The funders had no role in study design, data collection and interpretation, or the decision to submit the manuscript for publication.
REFERENCES
- 1.Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384(9939):258–71. Epub 2014/June/09. doi: 10.1016/S0140-6736(14)60164-1. PubMed PMID: 24907868. [DOI] [PubMed] [Google Scholar]
- 2.Eisinger RW, Dieffenbach CW, Fauci AS. HIV Viral Load and Transmissibility of HIV Infection: Undetectable Equals Untransmittable. JAMA. 2019;321(5):451–2. Epub 2019/January/11. doi: 10.1001/jama.2018.21167. PubMed PMID: 30629090. [DOI] [PubMed] [Google Scholar]
- 3.Hull M, Lange J, Montaner JS. Treatment as prevention–where next? Curr HIV/AIDS Rep. 2014;11(4):496–504. Epub 2014/November/12. doi: 10.1007/s11904-014-0237-5. PubMed PMID: 25384357; PMCID: PMC4268430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, Miller M, Vella S, Schmitz JE, Ahlers J, Richman DD, Sekaly RP. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet. 2013;381(9883):2109–17. Epub 2013/April/02. doi: 10.1016/S0140-6736(13)60104-X. PubMed PMID: 23541541; PMCID: PMC3815451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewin SR, Rouzioux C. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? AIDS. 2011;25(7):885–97. Epub 2011/March/23. doi: 10.1097/QAD.0b013e3283467041. PubMed PMID: 21422987. [DOI] [PubMed] [Google Scholar]
- 6.Davey RT, Jr., Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96(26):15109–14. Epub 1999/December/28. doi: 10.1073/pnas.96.26.15109. PubMed PMID: 10611346; PMCID: PMC24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39(4):633–45. Epub 2013/October/22. doi: 10.1016/j.immuni.2013.10.001. PubMed PMID: 24138880; PMCID: PMC4012895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zicari S, Sessa L, Cotugno N, Ruggiero A, Morrocchi E, Concato C, Rocca S, Zangari P, Manno EC, Palma P. Immune Activation, Inflammation, and Non-AIDS Co-Morbidities in HIV-Infected Patients under Long-Term ART. Viruses. 2019;11(3). Epub 2019/March/02. doi: 10.3390/v11030200. PubMed PMID: 30818749; PMCID: PMC6466530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RK, Peppa D, Hill AL, Galvez C, Salgado M, Pace M, McCoy LE, Griffith SA, Thornhill J, Alrubayyi A, Huyveneers LEP, Nastouli E, Grant P, Edwards SG, Innes AJ, Frater J, Nijhuis M, Wensing AMJ, Martinez-Picado J, Olavarria E. Evidence for HIV-1 cure after CCR5Delta32/Delta32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV. 2020;7(5):e340–e7. Epub 2020/March/15. doi: 10.1016/S2352-3018(20)30069-2. PubMed PMID: 32169158; PMCID: PMC7606918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang OO, Cumberland WG, Escobar R, Liao D, Chew KW. Demographics and natural history of HIV-1-infected spontaneous controllers of viremia. AIDS. 2017;31(8):1091–8. Epub 2017/March/17. doi: 10.1097/QAD.0000000000001443. PubMed PMID: 28301422; PMCID: PMC5657480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang C, Lian X, Gao C, Sun X, Einkauf KB, Chevalier JM, Chen SMY, Hua S, Rhee B, Chang K, Blackmer JE, Osborn M, Peluso MJ, Hoh R, Somsouk M, Milush J, Bertagnolli LN, Sweet SE, Varriale JA, Burbelo PD, Chun TW, Laird GM, Serrao E, Engelman AN, Carrington M, Siliciano RF, Siliciano JM, Deeks SG, Walker BD, Lichterfeld M, Yu XG. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature. 2020;585(7824):261–7. Epub 2020/August/28. doi: 10.1038/s41586-020-2651-8. PubMed PMID: 32848246; PMCID: PMC7837306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, Group AVS. Post-treatment HIV-1 controllers with a long-term viro-logical remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9(3):e1003211. Epub 2013/March/22. doi: 10.1371/journal.ppat.1003211. PubMed PMID: 23516360; PMCID: PMC3597518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–8. Epub 2009/February/14. doi: 10.1056/NEJMoa0802905. PubMed PMID: 19213682. [DOI] [PubMed] [Google Scholar]
- 14.Pitman MC, Lau JSY, McMahon JH, Lewin SR. Barriers and strategies to achieve a cure for HIV. Lancet HIV. 2018;5(6):e317–e28. Epub 2018/June/13. doi: 10.1016/S2352-3018(18)30039-0. PubMed PMID: 29893245; PMCID: PMC6559798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, Chomont N, Douek D, Lifson JD, Lo YR, Kuritzkes D, Margolis D, Mellors J, Persaud D, Tucker JD, Barre-Sinoussi F, International ASTaCWG, Alter G, Auerbach J, Autran B, Barouch DH, Behrens G, Cavazzana M, Chen Z, Cohen EA, Corbelli GM, Eholie S, Eyal N, Fidler S, Garcia L, Grossman C, Henderson G, Henrich TJ, Jefferys R, Kiem HP, McCune J, Moodley K, Newman PA, Nijhuis M, Nsubuga MS, Ott M, Palmer S, Richman D, Saez-Cirion A, Sharp M, Siliciano J, Silvestri G, Singh J, Spire B, Taylor J, Tolstrup M, Valente S, van Lunzen J, Walensky R, Wilson I, Zack J. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med. 2016;22(8):839–50. Epub 2016/July/12. doi: 10.1038/nm.4108. PubMed PMID: 27400264; PMCID: PMC5322797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins DR, Gaiha GD, Walker BD. CD8(+) T cells in HIV control, cure and prevention. Nat Rev Immunol. 2020;20(8):471–82. Epub 2020/February/14. doi: 10.1038/s41577-020-0274-9. PubMed PMID: 32051540; PMCID: PMC7222980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turk G, Ghiglione Y, Falivene J, Socias ME, Laufer N, Coloccini RS, Rodriguez AM, Ruiz MJ, Pando MA, Giavedoni LD, Cahn P, Sued O, Salomon H, Gherardi MM. Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8+ T-cell antiviral activity and correlates with preservation of the CD4+ T-cell compartment. J Virol. 2013;87(13):7445–62. Epub 2013/April/26. doi: 10.1128/JVI.00865-13. PubMed PMID: 23616666; PMCID: PMC3700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghiglione Y, Falivene J, Ruiz MJ, Laufer N, Socias ME, Cahn P, Giavedoni L, Sued O, Gherardi MM, Salomon H, Turk G. Early skewed distribution of total and HIV-specific CD8+ T-cell memory phenotypes during primary HIV infection is related to reduced antiviral activity and faster disease progression. PLoS One. 2014;9(8):e104235. Epub 2014/August/06. doi: 10.1371/journal.pone.0104235. PubMed PMID: 25093660; PMCID: PMC4122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falivene J, Ghiglione Y, Laufer N, Socias ME, Holgado MP, Ruiz MJ, Maeto C, Figueroa MI, Giavedoni LD, Cahn P, Salomon H, Sued O, Turk G, Gherardi MM. Th17 and Th17/Treg ratio at early HIV infection associate with protective HIV-specific CD8(+) T-cell responses and disease progression. Sci Rep. 2015;5:11511. Epub 2015/June/24. doi: 10.1038/srep11511. PubMed PMID: 26099972; PMCID: PMC4477236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghiglione Y, Trifone C, Salido J, Rhodes A, Ruiz MJ, Polo ML, Salomon H, Laufer N, Sued O, Lewin SR, Turk G. PD-1 Expression in HIV-Specific CD8+ T cells Before Antiretroviral Therapy Is Associated With HIV Persistence. J Acquir Immune Defic Syndr. 2019;80(1):1–6. Epub 2018/November/07. doi: 10.1097/QAI.0000000000001887. PubMed PMID: 30399040. [DOI] [PubMed] [Google Scholar]
- 21.Salido J, Ruiz MJ, Trifone C, Figueroa MI, Caruso MP, Gherardi MM, Sued O, Salomon H, Laufer N, Ghiglione Y, Turk G. Phenotype, Polyfunctionality, and Antiviral Activity of in vitro Stimulated CD8(+) T-Cells From HIV(+) Subjects Who Initiated cART at Different Time-Points After Acute Infection. Front Immunol. 2018;9:2443. Epub 2018/November/09. doi: 10.3389/fimmu.2018.02443. PubMed PMID: 30405632; PMCID: PMC6205955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Socias ME, Sued O, Laufer N, Lazaro ME, Mingrone H, Pryluka D, Remondegui C, Figueroa MI, Cesar C, Gun A, Turk G, Bouzas MB, Kavasery R, Krolewiecki A, Perez H, Salomon H, Cahn P, Grupo Argentino de Seroconversion Study G. Acute retroviral syndrome and high baseline viral load are predictors of rapid HIV progression among untreated Argentinean seroconverters. J Int AIDS Soc. 2011;14:40. Epub 2011/August/13. doi: 10.1186/1758-2652-14-40. PubMed PMID: 21831310; PMCID: PMC3179691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra U, Li F, Nolin J, Allison M, Zhao H, Mullins JI, Self S, McElrath MJ. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J Virol. 2007;81(10):5225–37. Epub 2007/March/03. doi: 10.1128/JVI.02564-06. PubMed PMID: 17329342; PMCID: PMC1900243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez-Cirion A, Shin SY, Versmisse P, Barre-Sinoussi F, Pancino G. Ex vivo T cell-based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses. Nat Protoc. 2010;5(6):1033–41. Epub 2010/June/12. doi: 10.1038/nprot.2010.73. PubMed PMID: 20539279. [DOI] [PubMed] [Google Scholar]
- 25.Hermans IF, Silk JD, Yang J, Palmowski MJ, Gileadi U, McCarthy C, Salio M, Ronchese F, Cerundolo V. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J Immunol Methods. 2004;285(1):25–40. Epub 2004/February/12. doi: 10.1016/j.jim.2003.10.017. PubMed PMID: 14871532. [DOI] [PubMed] [Google Scholar]
- 26.Pasternak AO, Berkhout B. What do we measure when we measure cell-associated HIV RNA. Retrovirology. 2018;15(1):13. Epub 2018/January/31. doi: 10.1186/s12977-018-0397-2. PubMed PMID: 29378657; PMCID: PMC5789533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, McNulty A, Ramgopal M, Michael N, Kim JH, Ananworanich J, Chomont N. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol. 2014;88(21):12385–96. Epub 2014/August/15. doi: 10.1128/JVI.00609-14. PubMed PMID: 25122785; PMCID: PMC4248919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, Lukashov VV. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46(7):2206–11. Epub 2008/May/09. doi: 10.1128/JCM.00055-08. PubMed PMID: 18463204; PMCID: PMC2446885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, Piovoso M, Shaw A, Dalmau J, Zangger N, Martinez-Picado J, Zurakowski R, Yu XG, Telenti A, Walker BD, Rosenberg ES, Lichterfeld M. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol. 2014;88(17):10056–65. Epub 2014/June/27. doi: 10.1128/JVI.01046-14. PubMed PMID: 24965451; PMCID: PMC4136362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdo TH, Lo J, Abbara S, Wei J, DeLelys ME, Preffer F, Rosenberg ES, Williams KC, Grinspoon S. Soluble CD163, a novel marker of activated macrophages, is elevated and associated with noncalcified coronary plaque in HIV-infected patients. J Infect Dis. 2011;204(8):1227–36. Epub 2011/September/16. doi: 10.1093/infdis/jir520. PubMed PMID: 21917896; PMCID: PMC3203384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JC, Zhang X, Karki M, Chi YY, Wallet SM, Rudy BJ, Nichols SL, Good-enow MM, Sleasman JW. Soluble CD14, CD163, and CD27 biomarkers distinguish ART-suppressed youth living with HIV from healthy controls. J Leukoc Biol. 2018;103(4):671–80. Epub 2018/January/30. doi: 10.1002/JLB.3A0717-294RR. PubMed PMID: 29377283; PMCID: PMC6528780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novelli S, Lecuroux C, Goujard C, Reynes J, Villemant A, Blum L, Essat A, Avettand-Fenoel V, Launay O, Molina JM, Bourgeois C, Meyer L. Persistence of monocyte activation under treatment in people followed since acute HIV-1 infection relative to participants at high or low risk of HIV infection. EBioMedicine. 2020;62:103129. Epub 2020/November/29. doi: 10.1016/j.ebiom.2020.103129. PubMed PMID: 33248370; PMCID: PMC7704414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC, Group ISS. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–90. Epub 2011/January/22. doi: 10.1093/infdis/jiq118. PubMed PMID: 21252259; PMCID: PMC3071127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burdo TH, Lentz MR, Autissier P, Krishnan A, Halpern E, Letendre S, Rosenberg ES, Ellis RJ, Williams KC. Soluble CD163 made by monocyte/macrophages is a novel marker of HIV activity in early and chronic infection prior to and after anti-retroviral therapy. J Infect Dis. 2011;204(1):154–63. Epub 2011/June/02. doi: 10.1093/infdis/jir214. PubMed PMID: 21628670; PMCID: PMC3105035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel N, Boufassa F, Lecuroux C, Saez-Cirion A, Bourgeois C, Dunyach-Remy C, Goujard C, Rouzioux C, Meyer L, Pancino G, Venet A, Lambotte O, Group ACCS. Elevated IP10 levels are associated with immune activation and low CD4(+) T-cell counts in HIV controller patients. AIDS. 2014;28(4):467–76. Epub 2014/January/01. doi: 10.1097/QAD.0000000000000174. PubMed PMID: 24378753. [DOI] [PubMed] [Google Scholar]
- 36.Wada NI, Jacobson LP, Margolick JB, Breen EC, Macatangay B, Penugonda S, Martinez-Maza O, Bream JH. The effect of HAART-induced HIV suppression on circulating markers of inflammation and immune activation. AIDS. 2015;29(4):463–71. Epub 2015/January/30. doi: 10.1097/QAD.0000000000000545. PubMed PMID: 25630041; PMCID: PMC4311407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castillo-Mancilla JR, Brown TT, Palella FJ, Jr., Macatangay BJC, Breen EC, Jacobson LP, Wada NI. Partial Normalization of Biomarkers of Inflammation and Immune Activation Among Virally Suppressed Men With HIV Infection and High ART Adherence. Open Forum Infect Dis. 2020;7(4):ofaa099. Epub 2020/April/24. doi: 10.1093/ofid/ofaa099. PubMed PMID: 32322603; PMCID: PMC7162619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liovat AS, Rey-Cuille MA, Lecuroux C, Jacquelin B, Girault I, Petitjean G, Zitoun Y, Venet A, Barre-Sinoussi F, Lebon P, Meyer L, Sinet M, Muller-Trutwin M. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS One. 2012;7(10):e46143. Epub 2012/October/12. doi: 10.1371/journal.pone.0046143. PubMed PMID: 23056251; PMCID: PMC3462744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toiyama Y, Fujikawa H, Kawamura M, Matsushita K, Saigusa S, Tanaka K, Inoue Y, Uchida K, Mohri Y, Kusunoki M. Evaluation of CXCL10 as a novel serum marker for predicting liver metastasis and prognosis in colorectal cancer. Int J Oncol. 2012;40(2):560–6. Epub 2011/November/01. doi: 10.3892/ijo.2011.1247. PubMed PMID: 22038159. [DOI] [PubMed] [Google Scholar]
- 40.Gomez-Rial J, Curras-Tuala MJ, Rivero-Calle I, Gomez-Carballa A, Cebey-Lopez M, Rodriguez-Tenreiro C, Dacosta-Urbieta A, Rivero-Velasco C, Rodriguez-Nunez N, Trastoy-Pena R, Rodriguez-Garcia J, Salas A, Martinon-Torres F. Increased Serum Levels of sCD14 and sCD163 Indicate a Preponderant Role for Monocytes in COVID-19 Immunopathology. Front Immunol. 2020;11:560381. Epub 2020/October/20. doi: 10.3389/fimmu.2020.560381. PubMed PMID: 33072099; PMCID: PMC7538662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotsch F, Romero R, Friel L, Kusanovic JP, Espinoza J, Erez O, Than NG, Mittal P, Edwin S, Yoon BH, Kim CJ, Mazaki-Tovi S, Chaiworapongsa T, Hassan SS. CXCL10/IP-10: a missing link between inflammation and anti-angiogenesis in preeclampsia? J Matern Fetal Neonatal Med. 2007;20(11):777–92. Epub 2007/October/19. doi: 10.1080/14767050701483298. PubMed PMID: 17943641; PMCID: PMC2396489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perdomo-Celis F, Taborda NA, Rugeles MT. CD8(+) T-Cell Response to HIV Infection in the Era of Antiretroviral Therapy. Front Immunol. 2019;10:1896. Epub 2019/August/27. doi: 10.3389/fimmu.2019.01896. PubMed PMID: 31447862; PMCID: PMC6697065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren JA, Clutton G, Goonetilleke N. Harnessing CD8(+) T Cells Under HIV Antiretroviral Therapy. Front Immunol. 2019;10:291. Epub 2019/March/14. doi: 10.3389/fimmu.2019.00291. PubMed PMID: 30863403; PMCID: PMC6400228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones RB, Walker BD. HIV-specific CD8(+) T cells and HIV eradication. J Clin Invest. 2016;126(2):455–63. Epub 2016/January/06. doi: 10.1172/JCI80566. PubMed PMID: 26731469; PMCID: PMC4731167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott AB, Koup RA. CD8(+) T cells in preventing HIV infection and disease. AIDS. 2012;26(10):1281–92. Epub 2012/March/24. doi: 10.1097/QAD.0b013e328353bcaf. PubMed PMID: 22441256. [DOI] [PubMed] [Google Scholar]
- 46.Emu B, Moretto WJ, Hoh R, Krone M, Martin JN, Nixon DF, Deeks SG, McCune JM. Composition and function of T cell subpopulations are slow to change despite effective antiretroviral treatment of HIV disease. PLoS One. 2014;9(1):e85613. Epub 2014/January/28. doi: 10.1371/journal.pone.0085613. PubMed PMID: 24465619; PMCID: PMC3897457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam S, Sung J, Cruz C, Castillo-Caro P, Ngo M, Garrido C, Kuruc J, Archin N, Rooney C, Margolis D, Bollard C. Broadly-specific cytotoxic T cells targeting multiple HIV antigens are expanded from HIV+ patients: implications for immunotherapy. Mol Ther. 2015;23(2):387–95. Epub 2014/November/05. doi: 10.1038/mt.2014.207. PubMed PMID: 25366030; PMCID: PMC4445615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehr M, Cahenzli J, Haas A, Price DA, Gostick E, Huber M, Karrer U, Oxenius A. Emergence of polyfunctional CD8+ T cells after prolonged suppression of human immunodeficiency virus replication by antiretroviral therapy. J Virol. 2008;82(7):3391–404. Epub 2008/January/18. doi: 10.1128/JVI.02383-07. PubMed PMID: 18199637; PMCID: PMC2268491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weekes MP, Wills MR, Sissons JG, Carmichael AJ. Large HIV-specific CD8 cytotoxic T-lymphocyte (CTL) clones reduce their overall size but maintain high frequencies of memory CTL following highly active antiretroviral therapy. Immunology. 2006;118(1):25–38. Epub 2006/April/25. doi: 10.1111/j.1365-2567.2006.02334.x. PubMed PMID: 16630020; PMCID: PMC1782266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Migueles SA, Weeks KA, Nou E, Berkley AM, Rood JE, Osborne CM, Hallahan CW, Cogliano-Shutta NA, Metcalf JA, McLaughlin M, Kwan R, Mican JM, Davey RT, Jr., Connors M. Defective human immunodeficiency virus-specific CD8+ T-cell poly-functionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol. 2009;83(22):11876–89. Epub 2009/September/04. doi: 10.1128/JVI.01153-09. PubMed PMID: 19726501; PMCID: PMC2772718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogg GS, Jin X, Bonhoeffer S, Moss P, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Hurley A, Markowitz M, Ho DD, McMichael AJ, Nixon DF. Decay kinetics of human immunodeficiency virus-specific effector cytotoxic T lymphocytes after combination antiretroviral therapy. J Virol. 1999;73(1):797–800. Epub 1998/December/16. doi: 10.1128/JVI.73.1.797-800.1999. PubMed PMID: 9847391; PMCID: PMC103892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conrad JA, Ramalingam RK, Duncan CB, Smith RM, Wei J, Barnett L, Simons BC, Lorey SL, Kalams SA. Antiretroviral therapy reduces the magnitude and T cell receptor repertoire diversity of HIV-specific T cell responses without changing T cell clonotype dominance. J Virol. 2012;86(8):4213–21. Epub 2012/January/20. doi: 10.1128/JVI.06000-11. PubMed PMID: 22258246; PMCID: PMC3318638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casazza JP, Betts MR, Picker LJ, Koup RA. Decay kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J Virol. 2001;75(14):6508–16. Epub 2001/June/20. doi: 10.1128/JVI.75.14.6508-6516.2001. PubMed PMID: 11413318; PMCID: PMC114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalams SA, Goulder PJ, Shea AK, Jones NG, Trocha AK, Ogg GS, Walker BD. Levels of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte effector and memory responses decline after suppression of viremia with highly active antiretroviral therapy. J Virol. 1999;73(8):6721–8. Epub 1999/July/10. doi: 10.1128/JVI.73.8.6721-6728.1999. PubMed PMID: 10400770; PMCID: PMC112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahnke YD, Song K, Sauer MM, Nason MC, Giret MT, Carvalho KI, Costa PR, Roederer M, Kallas EG. Early immunologic and virologic predictors of clinical HIV-1 disease progression. AIDS. 2013;27(5):697–706. Epub 2012/December/06. doi: 10.1097/QAD.0b013e32835ce2e9. PubMed PMID: 23211771. [DOI] [PubMed] [Google Scholar]
- 56.Burgers WA, Riou C, Mlotshwa M, Maenetje P, de Assis Rosa D, Brenchley J, Mlisana K, Douek DC, Koup R, Roederer M, de Bruyn G, Karim SA, Williamson C, Gray CM, Team CAIS. Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J Immunol. 2009;182(8):4751–61. Epub 2009/April/04. doi: 10.4049/jimmunol.0803801. PubMed PMID: 19342652; PMCID: PMC2792921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, Forster R, Rowland-Jones S, Sekaly RP, McMichael AJ, Pantaleo G. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410(6824):106–11. Epub 2001/March/10. doi: 10.1038/35065118. PubMed PMID: 11242051. [DOI] [PubMed] [Google Scholar]
- 58.Yue FY, Cohen JC, Ho M, Rahman A, Liu J, Mujib S, Saiyed A, Hundal S, Khozin A, Bonner P, Liu D, Benko E, Kovacs C, Ostrowski M. HIV-Specific Granzyme B-Secreting but Not Gamma Interferon-Secreting T Cells Are Associated with Reduced Viral Reservoirs in Early HIV Infection. J Virol. 2017;91(8). Epub 2017/February/10. doi: 10.1128/JVI.02233-16. PubMed PMID: 28179527; PMCID: PMC5375660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas AS, Jones KL, Gandhi RT, McMahon DK, Cyktor JC, Chan D, Huang SH, Truong R, Bosque A, Macedo AB, Kovacs C, Benko E, Eron JJ, Bosch RJ, Lalama CM, Simmens S, Walker BD, Mellors JW, Jones RB. T-cell responses targeting HIV Nef uniquely correlate with infected cell frequencies after long-term antiretroviral therapy. PLoS Pathog. 2017;13(9):e1006629. Epub 2017/September/21. doi: 10.1371/journal.ppat.1006629. PubMed PMID: 28931091; PMCID: PMC5624641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takata H, Buranapraditkun S, Kessing C, Fletcher JL, Muir R, Tardif V, Cartwright P, Vandergeeten C, Bakeman W, Nichols CN, Pinyakorn S, Hansasuta P, Kroon E, Chalermchai T, O'Connell R, Kim J, Phanuphak N, Robb ML, Michael NL, Chomont N, Haddad EK, Ananworanich J, Trautmann L, Rv254/Search, the RVSSG . Delayed differentiation of potent effector CD8(+) T cells reducing viremia and reservoir seeding in acute HIV infection. Sci Transl Med. 2017;9(377). Epub 2017/February/17. doi: 10.1126/scitranslmed.aag1809. PubMed PMID: 28202771; PMCID: PMC5678930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G. CD8(+) Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy. Immunity. 2016;45(3):656–68. Epub 2016/September/23. doi: 10.1016/j.immuni.2016.08.018. PubMed PMID: 27653601; PMCID: PMC5087330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, Capoferri AA, Beg SA, Huang SH, Karandish S, Hao H, Halper-Stromberg E, Yong PC, Kovacs C, Benko E, Siliciano RF, Ho YC. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe. 2017;21(4):494–506 e4. Epub 2017/April/14. doi: 10.1016/j.chom.2017.03.008. PubMed PMID: 28407485; PMCID: PMC5433942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imamichi H, Smith M, Adelsberger JW, Izumi T, Scrimieri F, Sherman BT, Rehm CA, Imamichi T, Pau A, Catalfamo M, Fauci AS, Lane HC. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci U S A. 2020;117(7):3704–10. Epub 2020/February/08. doi: 10.1073/pnas.1917876117. PubMed PMID: 32029589; PMCID: PMC7035625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevenson EM, Ward AR, Truong R, Thomas AS, Huang SH, Dilling TR, Terry S, Bui JK, Mota TM, Danesh A, Lee GQ, Gramatica A, Khadka P, Alberto WDC, Gandhi RT, McMahon DK, Lalama CM, Bosch RJ, Macatangay B, Cyktor JC, Eron JJ, Mellors JW, Jones RB, Team ACTGA. HIV-specific T cell responses reflect substantive in vivo interactions with antigen despite long-term therapy. JCI Insight. 2021;6(3). Epub 2021/January/06. doi: 10.1172/jci.insight.142640. PubMed PMID: 33400687; PMCID: PMC7934865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fromentin R, Chomont N. HIV persistence in subsets of CD4+ T cells: 50 shades of reservoirs. Semin Immunol. 2020:101438. Epub 2020/December/05. doi: 10.1016/j.smim.2020.101438. PubMed PMID: 33272901. [DOI] [PMC free article] [PubMed]
- 66.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol. 2014;88(24):14070–7. Epub 2014/September/26. doi: 10.1128/JVI.01900-14. PubMed PMID: 25253353; PMCID: PMC4249150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. Epub 2009/June/23. doi: 10.1038/nm.1972. PubMed PMID: 19543283; PMCID: PMC2859814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wonderlich ER, Subramanian K, Cox B, Wiegand A, Lackman-Smith C, Bale MJ, Stone M, Hoh R, Kearney MF, Maldarelli F, Deeks SG, Busch MP, Ptak RG, Kulpa DA. Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals. PLoS Pathog. 2019;15(10):e1008074. Epub 2019/October/15. doi: 10.1371/journal.ppat.1008074. PubMed PMID: 31609991; PMCID: PMC6812841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, Chomont N. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. 2019;15(2):e1007619. Epub 2019/February/28. doi: 10.1371/journal.ppat.1007619. PubMed PMID: 30811499; PMCID: PMC6411230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grau-Exposito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocana I, Ribera E, Torrella A, Planas B, Badia R, Castellvi J, Falco V, Crespo M, Buzon MJ. A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4(+) T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. mBio. 2017;8(4). Epub 2017/July/13. doi: 10.1128/mBio.00876-17. PubMed PMID: 28698276; PMCID: PMC5513707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants. Cell Rep. 2017;21(3):813–22. Epub 2017/October/19. doi: 10.1016/j.celrep.2017.09.081. PubMed PMID: 29045846; PMCID: PMC5960642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kulpa DA, Talla A, Brehm JH, Ribeiro SP, Yuan S, Bebin-Blackwell AG, Miller M, Barnard R, Deeks SG, Hazuda D, Chomont N, Sekaly RP. Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4(+) T Cells. J Virol. 2019;93(24). Epub 2019/October/04. doi: 10.1128/JVI.00969-19. PubMed PMID: 31578289; PMCID: PMC6880164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eller MA, Hong T, Creegan M, Nau ME, Sanders-Buell E, Slike BM, Krebs SJ, Ratto-Kim S, McElrath MJ, Katabira ET, Bolton DL, Michael NL, Robb ML, Tovanabutra S, Baeten JM, Sandberg JK. Activated PD-1+ CD4+ T cells represent a short-lived part of the viral reservoir and predict poor immunologic recovery upon initiation of ART. AIDS. 2020;34(2):197–202. Epub 2019/November/26. doi: 10.1097/QAD.0000000000002432. PubMed PMID: 31764072. [DOI] [PubMed] [Google Scholar]
- 74.McGary CS, Deleage C, Harper J, Micci L, Ribeiro SP, Paganini S, Kuri-Cervantes L, Benne C, Ryan ES, Balderas R, Jean S, Easley K, Marconi V, Silvestri G, Estes JD, Sekaly RP, Paiardini M. CTLA-4(+)PD-1(-) Memory CD4(+) T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques. Immunity. 2017;47(4):776–88 e5. Epub 2017/October/19. doi: 10.1016/j.immuni.2017.09.018. PubMed PMID: 29045906; PMCID: PMC5679306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, Palmer S, Fromentin R, Chomont N, Sekaly RP, Cameron PU, Lewin SR. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS. 2018;32(11):1491–7. Epub 2018/May/11. doi: 10.1097/QAD.0000000000001849. PubMed PMID: 29746296; PMCID: PMC6026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sekaly RP, Chomont N. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog. 2016;12(7):e1005761. Epub 2016/July/16. doi: 10.1371/journal.ppat.1005761. PubMed PMID: 27415008; PMCID: PMC4944956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, Hoh R, Deeks SG, Hazuda DJ, Lewin SR, Routy JP, Sekaly RP, Chomont N. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat Commun. 2019;10(1):814. Epub 2019/February/20. doi: 10.1038/s41467-019-08798-7. PubMed PMID: 30778080; PMCID: PMC6379401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, Chomont N, Bar KJ, Yu XG, Lichterfeld M, Alcami J, Hazuda D, Bush-man F, Siliciano JD, Betts MR, Spivak AM, Planelles V, Hahn BH, Smith DM, Ho YC, Buzon MJ, Gaebler C, Paiardini M, Li Q, Estes JD, Hope TJ, Kostman J, Mounzer K, Caskey M, Fox L, Frank I, Riley JL, Tebas P, Montaner LJ, infection B-HDCtCH- . Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med. 2020;26(9):1339–50. Epub 2020/September/09. doi: 10.1038/s41591-020-1022-1. PubMed PMID: 32895573; PMCID: PMC7703694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


