Abstract
A protocol for the rapid detection of fungal DNA in ocular samples, derived from three species, Candida albicans, Aspergillus fumigatus, and Fusarium solani, has been developed. Two novel panfungal primers complementary to 18S rRNA sequences present in all three species were designed. Panfungal PCR was followed by three nested PCRs utilizing species-specific primers. PCR sensitivity ranged from 50 to 100 fg of free DNA and between one and two C. albicans organisms. In addition, we also developed a rapid and reliable DNA extraction protocol. This protocol minimized DNA loss during extraction, whilst removing compounds from vitreous and aqueous fluids that have previously been shown to have inhibitory effects on PCR. Preliminary results obtained after testing the protocol on three patient samples support culture results and medical history. However, one patient was PCR positive but culture negative, suggesting that the sensitivity of this protocol may exceed that of traditional culture techniques. This system, therefore, constitutes an additional protocol that may significantly aid patient management in cases where fungal endophthalmitis is suspected.
Fungal endophthalmitis accounts for 4 to 11% of all cases of culture-proven endophthalmitis (14, 35) and is usually acquired from an endogenous source via hematogenous spread (1, 2, 8, 11, 13, 25). It may, however, also be secondary to intraocular surgery (20, 38), corneal ulceration, or trauma (3, 19, 30). Confirmation of suspected clinical disease currently presents a challenge to the clinician, with difficulty in making a diagnosis frequently delaying treatment. Fungal endophthalmitis may have a classical appearance (e.g., Candida spp. [7]), may present in a similar manner to chronic bacterial endophthalmitis (31, 33), or may coexist with bacterial endophthalmitis (9). The time to diagnosis from onset of symptoms has been reported as varying from 3 days to 4 months (30), during which time bilateral ocular disease may cause severe morbidity, especially in patients who are already debilitated. In cases with a typical history and clinical signs of fungal endophthalmitis, vitreous sampling is implemented. While intraocular samples are often culture negative, cases that respond to antifungal treatment are considered to be infective in origin. Blood cultures are useful but insensitive for the detection of candidemia (with estimates of only 50 to 75% of cases detected), and serological testing remains experimental (10, 36). Isolation of fungi may also be difficult, as coexisting bacteremia may hide fungemia in culture, with bacteria competing with fungi for nutrients in vitro (16, 34).
Amplification of target DNA through PCR with sequence-specific primers is potentially more sensitive and rapid than microbiologic techniques, as a number of constraints are removed. Unlike culture, PCR does not require the presence of viable organisms for success and may be performed even when sample volumes are small. In addition, the sensitivity may be dramatically increased through the use of nested PCR. A number of protocols have already been developed for the detection of bacteria and fungi from a variety of clinical samples, such as blood, serum, and ocular fluids (4, 12, 15, 17, 18, 23, 24, 26, 28, 37, 40). Since these protocols either involve subsequent processing of PCR products or are focused on only one fungal genus, we have developed a PCR-based system for the rapid detection of Candida albicans, Aspergillus fumigatus, and Fusarium solani in ocular samples. This rapid protocol increases the number of samples from which a diagnosis may be made and thereby facilitates effective patient management.
MATERIALS AND METHODS
All reagents used were purchased from Sigma Chemicals (Poole, Dorset, United Kingdom) unless otherwise stated.
Fungal isolates.
Identification was performed by using standard mycological methods of germ tube formation in serum, carbohydrate assimilation tests using the API 20C kit (bioMérieux, Inc., Hazelwood, Mo.), and morphology on cornmeal agar. C. albicans and F. solani were obtained from the National Collection of Pathogenic Fungi (EK3619 and G135, respectively) (PHLS Laboratory, Bristol, United Kingdom). A. fumigatus was derived from the culture of clinical specimens. Organisms were subsequently stored on beads (Mast Diagnostics, Bootle, Merseyside, United Kingdom) at −70°C.
DNA extraction for PCR optimization.
Previously extracted DNA derived from Candida spp. was used (28), while a protocol based on that described by Cenis was used for filamentous fungi (6). Briefly, 500 μl of liquid potato dextrose medium (Difco Laboratories Ltd., Surrey, United Kingdom) was inoculated with fungal hyphal threads and left at room temperature for 72 h. The resulting mycelial mat was pelleted by centrifugation at 13,000 rpm (Eppendorf microcentrifuge 5415c; Eppendorf UK Ltd., Cambridge, United Kingdom) for 5 min and was washed with 500 μl of Tris-EDTA (pH 8.0). The mat was then homogenized by hand in 300 μl of extraction buffer (200 mM Tris-HCl [pH 8.5], 250 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate) for 5 min. One hundred fifty microliters of 3 M sodium acetate (pH 5.2) was added, and the mixture was cooled to −20°C for 10 min. Fungal debris was pelleted by centrifugation at 13,000 rpm for 5 min, the supernatant was transferred to a fresh tube, and an equal volume of isopropanol was added. DNA was then pelleted by centrifugation at 13,000 rpm for 10 min. Excess salt was removed by washing with 70% ethanol, and DNA was resuspended in Tris-EDTA (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Bacterial DNA extraction was performed as described elsewhere (5), while human DNA extraction from 10 ml of whole blood was performed by using a Nucleon BACC2 kit (Vector Laboratories, Ltd., Peterborough, United Kingdom).
Serial dilution of C. albicans for DNA extraction and PCR.
A single overnight colony of C. albicans was spiked into 5 ml of liquid potato dextrose medium (Difco) and left overnight at room temperature. The following serial dilutions were made in water: 101, 102, 103, 104, 504, and 105, in a final volume of 10 ml. Equal volumes (20 μl) were plated out on Sabouraud dextrose agar (Difco) in duplicate for enumeration and were used for DNA extraction and PCR.
DNA extraction from serially diluted C. albicans and spiked normal vitreous fluid.
DNA was extracted with a QIAamp DNA Mini Kit (QIAGEN Ltd., Crowley, West Sussex, United Kingdom). A modified protocol based on that recommended by the manufacturers and that described by Flahaut and coworkers (12) was used. The initial step of spheroplast formation through incubation with zymolase was omitted. This was not detrimental to DNA recovery but reduced loss of free DNA already present, through the omission of the centrifugation and supernatant removal steps. The protocol was as follows: 5 to 100 μl of the sample to be investigated was mixed with 20 μl of proteinase K (QIAGEN) and 180 μl of digestion buffer (30 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate) and was incubated at 56°C for 1 h. Buffer AL (200 μl) (QIAGEN) was then added, and the mixture was heated to 70°C for 10 min. Absolute ethanol (200 μl) and 100 ng of carrier human DNA were subsequently added to each sample. The mixture was added to a QIAamp spin column (QIAGEN) and was centrifuged at 13,000 rpm for 1 min. The column was washed twice with 500 μl of buffer AE (QIAGEN), and DNA was eluted by the addition of 50 μl of preheated buffer AE (QIAGEN) following incubation at 60°C for 5 min. The eluate was reapplied to the column after centrifugation, thereby increasing DNA recovery. Incubation and centrifugation were repeated, with the resulting eluate ready for PCR. Negative controls were 20 μl of sterile water subjected to an identical extraction procedure.
Normal vitreous fluid.
Samples were collected by sterile technique at the time of vitrectomy, during planned surgical procedures. All patients showed no evidence of intraocular infection, inflammation, or medical history of uveitis and/or diabetes mellitus. Vitreous samples were aliquoted in a sterile manner and stored at −20°C. To show that vitreous was not inhibitory to DNA extraction and PCR, 20 μl of normal vitreous was spiked with 2 μl of serially diluted C. albicans. DNA extraction was then performed as described above, while 10 μl of the Candida dilution was plated out in duplicate for enumeration.
Cases of suspected fungal endophthalmitis.
Intraocular sampling is routinely undertaken for all patients with suspected fungal endophthalmitis. The extraocular environment was sterilized with 5% povidone iodine solution prior to surgery. Approximately 100 to 200 μl of aqueous fluid was withdrawn with a 27-gauge (0.33-mm) needle via a limbal paracentesis. Vitreous samples (200 to 400 μl) were taken at the time of three-port pars plana vitrectomy. These samples were transported to the microbiology laboratory for immediate processing.
PCR primer design.
Small-subunit (18S) rRNA sequences from the relevant fungal species were accessed through GenBank. An alignment was constructed by using the Clustal algorithm contained in Megalign, a component of the sequence analysis package Lasergene (DNASTAR Inc., Madison, Wis.). Panfungal primers were chosen from regions that showed high levels of conservation between species, whereas species-specific primers were chosen from divergent regions inside the panfungal amplicon. Panfungal primers used were as follows: Pffor, 5′-AGGGATGTATTTATTAGATAAAAAATCAA-3′, and Pfrev2, 5′-CGCAGTAGTTAGTCTTCAGTAAATC-3′. These generated PCR products of 728, 743, and 744 bp in C. albicans, A. fumigatus, and F. solani, respectively. C. albicans-specific primers used were Cafor2, 5′-GGGAGGTAGTGACAATAAATAAC-3′, and Carev3, 5′-CGTCCCTATTAATCATTACGAT-3′, which produced a 402-bp PCR product. A. fumigatus-specific primers used were Asfufor, 5′-CCAATGCCCTTCGGGGCTCCT-3′ and Asfurev, 5′-CCTGGTTCCCCCCACAG-3′, which produced a 520-bp PCR product. F. solani-specific primers used were Fusofor, 5′-CCAATGCCCTCCGGGGCTAAC-3′, and Fusorev, 5′-GCATAGGCCTGCCTGGCG-3′, which produced a 565-bp PCR product.
PCR parameters.
Panfungal and nested PCRs were performed in 50-μl reaction volumes containing PCR buffer (50 mM KCl, 10 mM Tris HCl [pH 8.3], 60 ng of each primer, 200 μm (each) deoxynucleotide triphosphate (Bioline, London, United Kingdom), 2.5 mM magnesium chloride , and 2 U of Taq polymerase (Perkin-Elmer, Buckinghamshire, United Kingdom). Cycling parameters were identical for both panfungal and nested PCRs with the exception of annealing temperatures, which were as follows: panfungal, 58°C; Candida nested, 66°C; Aspergillus nested, 66°C; Fusarium nested, 64.5°C. Cycling conditions consisted of an initial denaturation step at 95°C for 5 min, followed by either 30 or 40 cycles of 95°C for 30 s, annealing temperature for 30 s, and 72°C for 20 s (Genius Thermal Cycler; Techne, Cambridge, United Kingdom). PCR was completed by a final extension at 72°C for 7 min. Panfungal and Fusarium-specific PCRs were performed for 30 cycles, while all others were performed for 40 cycles. After amplification, 1 μl was removed from the panfungal PCR and added to the nested PCR for further amplification.
Electrophoresis and imaging.
PCR products were resolved in a 1% Tris-acetate-EDTA–agarose gel and were visualized with ethidium bromide and ultraviolet illumination. Images were captured and stored by using a UVP gel documentation system (UVP Ltd., Cambridge, United Kingdom).
Sequencing of PCR products.
Four 50-μl PCRs were pooled and resolved in a 1% Tris-acetate-EDTA–agarose gel. The appropriate band was excised from the gel, and DNA was purified by using a Geneclean II kit (BIO101, La Jolla, Calif.). PCR fragments were directly cycle sequenced in both directions with a 377 automated sequencing system (ABI, Warrington, Cheshire, United Kingdom).
RESULTS
PCR primers and specificity.
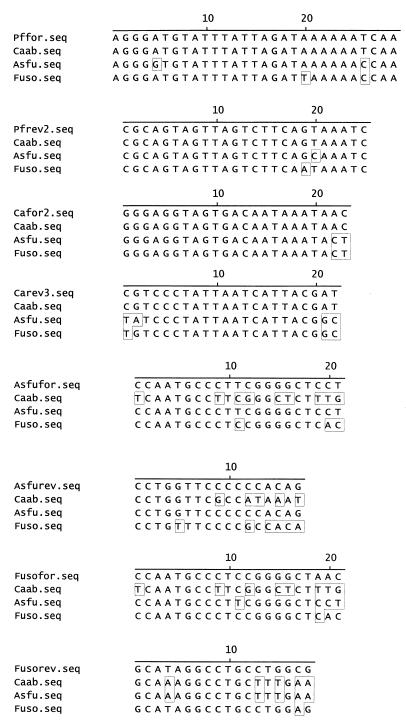
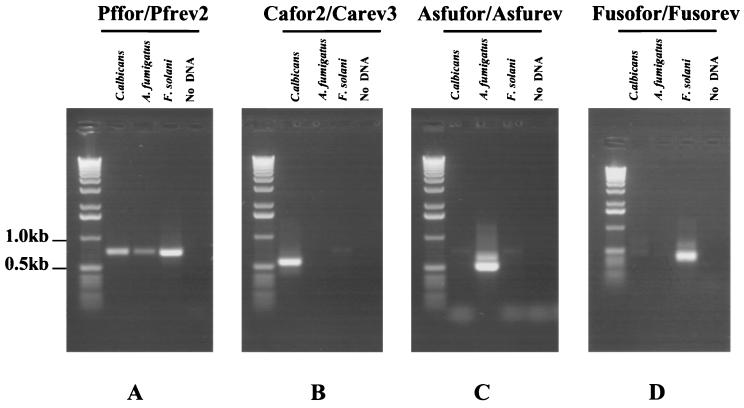
All primers used are shown in Fig. 1, aligned with the relevant sequences from the three fungal species studied. It was not possible to identify long stretches of 18S rRNA sequence that were 100% homologous in all three fungal species. Consequently, the panfungal primers contain a number of mismatches with A. fumigatus and F. solani sequences. However, due to their location within the primers, these mismatches were not problematic and did not significantly decrease the efficiency of PCR amplification from these species. All primers were designed to operate at high annealing temperatures, thereby preventing the coamplification of nonspecific target DNA, including DNA derived from bacteria, humans, and other fungal species. In addition, primer sequences were compared against existing sequences in GenBank to eliminate nonspecific priming. Species-specific primers were chosen with a minimum of two mismatches at the 3′ end when compared to the two nonrelevant species. Due to the similarity between A. fumigatus and F. solani 18S sequences, this was not possible when designing the F. solani-specific reverse primer. Therefore, to reduce the likelihood of Fusofor-Fusorev coamplifying A. fumigatus DNA, two artificial mismatches were introduced towards the 3′ end of the primers. Figure 2 shows results obtained after panfungal and species-specific PCR. Single bands of the correct size were obtained from all three species with Pffor-Pfrev2. In contrast, when DNA derived from human and several bacterial species was used as template, no amplicons were observed (data not shown). Species-specific PCR also resulted in bands of the correct size in only the appropriate species. Occasionally, bands larger than the band of interest were observed in the nested PCR. These corresponded in size to products carried over from the panfungal PCR and also to seminested PCR products. Therefore, they were of no significance and were disregarded.
FIG. 1.
Alignments of primer sequences and corresponding sequences from C. albicans (Caab), A. fumigatus (Asfu), and F. solani (Fuso). Residues that differ from the primer sequence at the top of each alignment are boxed.
FIG. 2.
Results obtained after PCR with (A) Pffor-Pfrev2 and PCR with 1 μl of this with (B) Cafor2-Carev3, (C) Asfufor-Asfurev, and (D) Fusofor-Fusorev. DNA (1 ng) from the following species was used: C. albicans, A. fumigatus, and F. solani. Lane 4 contains no DNA.
In order to confirm that species-specific PCRs were indeed specific, nested PCRs derived from the three species studied were sequenced. Comparison of the sequences obtained with previously published 18S rRNA sequences confirmed the specificity of the primers, since sequences were 100% homologous. In addition, panfungal and species-specific PCRs were set up using 10 ng of DNA from the following bacterial species: Propionibacterium acnes, Enterococcus faecalis, viridans streptococci, Klebsiella pneumoniae, Proteus mirabilis, Haemophilus influenzae, Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa. Positive controls were 10-ng samples of fungal DNA from C. albicans, A. fumigatus, and F. solani. All bacterial PCRs were negative, further supporting the specificity of the system.
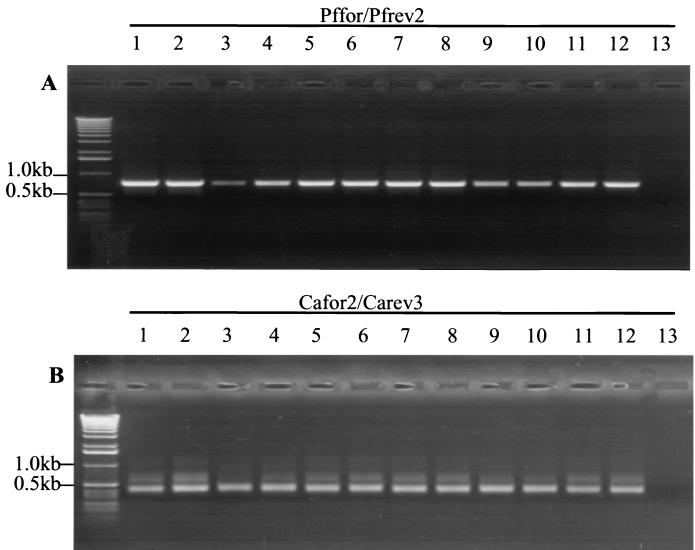
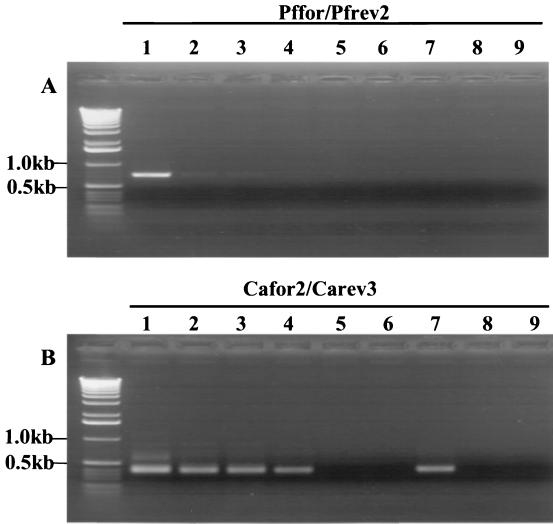
To determine whether panfungal and Candida-specific primers could amplify DNA derived from other Candida species, an alignment of 18S rRNA sequences derived from multiple Candida species was performed. This revealed the conservation of the sequences comprising Pffor-Pfrev2 and Cafor2-Carev3 between species. These were then used to amplify 20-ng samples of DNA from Candida guilliermondii, Candida glabrata, Candida pelliculosa, Candida tropicalis, Candida parapsilosis, and Candida krusei. Results are shown in Fig. 3. As predicted, primary and nested PCR products of the correct size were obtained for all the above-mentioned Candida species.
FIG. 3.
Results obtained after PCR with (A) Pffor-Pfrev2 and PCR with 1 μl of this with (B) Cafor2-Carev3. Ten-nanogram samples of DNA from the following species was used: (lanes 1 and 2) C. guilliermondii, (lanes 3 and 4) C. glabrata, (lanes 5 and 6) C. pelliculosa, (lanes 7 and 8) C. tropicalis, (lanes 9 and 10) C. parapsilosis, (lanes 11 and 12) C. krusei, (lane 13) no DNA.
PCR sensitivity.
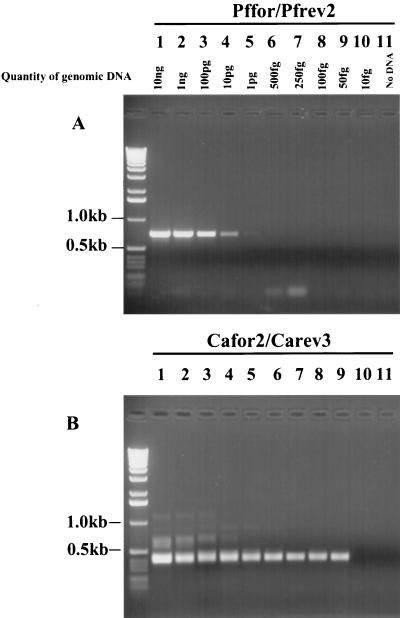
With serially diluted DNA in water, the sensitivity of the panfungal PCR was sufficient to amplify 10 pg of DNA from C. albicans and 100 pg of DNA from A. fumigatus and F. solani. However, after nested PCR, the sensitivity dramatically increased. The lowest amount of DNA consistently amplifiable was 100 fg from A. fumigatus and F. solani (data not shown) and 50 fg of DNA from C. albicans (see Fig. 4). Negative controls processed simultaneously were consistently negative. There is little published data regarding the DNA content of A. fumigatus and F. solani components, hence it is difficult to equate free DNA directly to living organisms. In contrast, the DNA content of a diploid C. albicans cell has been estimated as being approximately 37 fg (32). As the next dilution down from 50 fg was 10 fg, which gave a negative result, it seemed likely that this PCR amplification system could amplify DNA derived from one or two C. albicans cells. To test this theory, PCR was performed on DNA extracted from serially diluted C. albicans. The average numbers of colonies that grew from 20-μl aliquots of various dilutions were as follows: confluent growth per dilution of 102; 38.5 organisms per dilution of 104; 16.5 organisms per dilution of 504; 3 organisms per dilution of 105. PCR results are shown in Fig. 5. DNA was eluted from the QIAamp spin column in a 50-μl volume, 10 μl of which was subsequently used for PCR. This therefore corresponded to DNA derived from the following average number of organisms per dilution: >500 organisms per dilution of 102; 7.7 organisms per dilution of 104; 3.3 organisms per dilution of 504; 0.6 organisms per dilution of 105. A strong band was only obtained after primary PCR for the first dilution, while all dilutions gave positive results for the nested PCR. This indicates that DNA could be successfully extracted, retrieved, and detected from as few as one C. albicans organism.
FIG. 4.
PCR of serially diluted DNA from C. albicans using (A) Pffor-Pfrev2 and PCR with 1 μl of this with (B) Cafor2-Carev3.
FIG. 5.
PCR of DNA extracted from serially diluted C. albicans using (A) Pffor-Pfrev2 and PCR with 1 μl of this with (B) Cafor2-Carev3. DNA used was equivalent to the following average number of organisms: (lane 1) >500, (lane 2) 7.7, (lane 3) 3.3, (lane 4) 0.6. Controls were blank (lanes 5 and 6), and 10 pg of DNA derived from C. albicans (lane 7), A. fumigatus (lane 8) and F. solani (lane 9).
Candida spiked into normal vitreous.
Previous studies have shown that vitreous and aqueous fluids may exhibit inhibitory properties when added directly to PCR (5, 28, 39). In order to prove that DNA extraction using the QIAamp system eliminated this problem, 20-μl volumes of normal vitreous fluid were spiked with 2-μl aliquots of serially diluted C. albicans. The volumes of diluted C. albicans added were small in order to minimize vitreous dilution. DNA extraction and PCR were performed as before. PCR sensitivity in both primary and nested PCRs was identical to that of serially diluted C. albicans alone. No inhibitory PCR effects derived from vitreous fluid were observed (data not shown). It was not possible to test vitreous derived from an eye with intraocular inflammation, even though this material has also been shown to inhibit PCR (27). Such material comprised the actual patient samples that were to be tested for fungal infection, and only small quantities were available. However, the success observed with normal spiked vitreous suggests that PCR inhibitors may also be removed from inflamed vitreous with this system.
Application to patient samples.
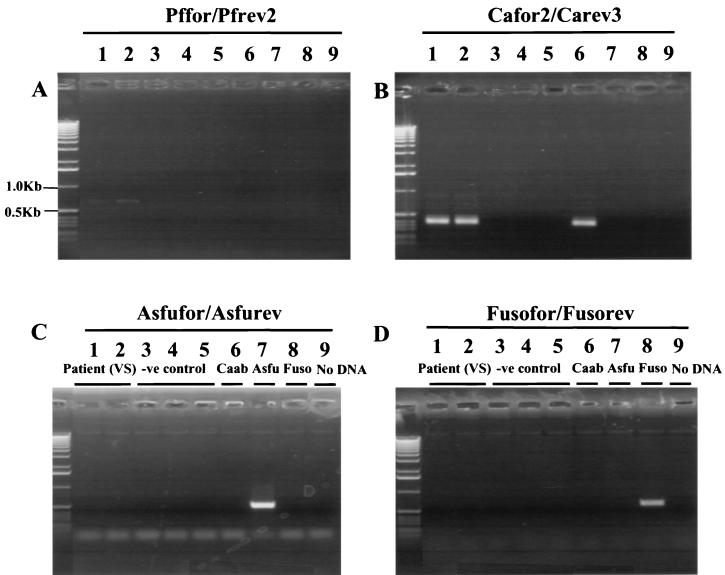
In order to test the viability of the system when applied to clinical samples, ocular material from three patients with suspected fungal or bacterial endophthalmitis were processed. These were 50-μl samples of vitreous derived from patients 1 and 2 and an empty syringe that had previously contained vitreous from patient 3. The latter was rinsed out with 50 μl of sterile water and processed as usual. All patient PCRs were performed in duplicate, and positive Candida results were obtained from both patient 1 (see Fig. 6) and patient 3. This was concordant with their medical history. Patient 1 was a known intravenous drug abuser who presented with clinical signs of fungal endophthalmitis in one eye. Patient 3 had undergone major abdominal surgery, had developed multiple postoperative complications, and was treated with broad-spectrum antibiotics for 5 weeks prior to presentation with bilateral ocular signs typical of fungal endophthalmitis. Both patients responded to treatment with antifungal agents. However, patient 3 was culture positive for C. albicans while patient 1 was culture negative. Patient 2 was PCR negative for all fungal species tested, which was also concordant with medical records. He was an alcoholic with insulin-dependent diabetes mellitus who presented with ocular signs 3 months after initial presentation with osteomyelitis and septicemia. He was successfully treated and responded well to antibiotic therapy for coagulase-negative staphylococci isolated from blood cultures. The patient represented once after treatment with ocular signs suggestive of intraocular infection in one eye. Patient 2 was culture positive for coagulase-negative staphylococci and responded to antibiotic therapy.
FIG. 6.
PCR of eluate derived from vitreous of patient 1 using (A) Pffor-Pfrev2 and PCR with 1 μl of this with (B) Cafor2-Carev3, (C) Asfufor-Asfurev, and (D) Fusofor-Fusorev. Lane contents are as follows: (lanes 1 and 2) 20 μl of eluate from patient 1, (lanes 3 to 5) negative control, (lane 6) 10 pg of C. albicans DNA, (lane 7) 10 pg of A. fumigatus DNA, (lane 8) 10 pg of F. solani DNA, and (lane 9) no DNA.
DISCUSSION
In this report, we describe a rapid, sensitive, and reliable PCR system for the detection and identification of three fungal genera in ocular samples. Panfungal primers were designed by using 18S rRNA gene sequences from C. albicans, A. fumigatus, and F. solani. Ribosomal genes were chosen as targets for amplification, as they are highly conserved genes that exist as multiple copies in the fungal genome (22, 29). This was reflected in the sensitivity of the system, which required 100 fg of DNA from Aspergillus and Fusarium and 50 fg of Candida DNA. When tested on serially diluted Candida cells, as few as two organisms were detected in repeated experiments. The system was not tested on serially diluted Aspergillus or Fusarium fungal material, due to difficulties regarding cellular quantification. However, subsequent analysis of an A. fumigatus-culture-positive patient yielded favorable results (manuscript in preparation).
A major consideration when using this system is the avoidance of contamination. A single PCR amplification may generate 1012 identical amplicons, which in turn may serve as template in subsequent reactions (21). For this reason and to avoid carryover, PCR products were handled in a separate room in which PCR mixtures were set up. In addition, separate pipettes were used for setting up PCRs, aliquoting nested PCR products, and further manipulating PCR products. Potential fungal contaminants derived from airborne sources were also considered; hence, all DNA extractions were performed in a biosafety hood. We encountered contamination in some commercially available laboratory reagents, as has also been described by other researchers (21). In particular, buffer ATL included in the QIAamp DNA Mini Kit was found to be contaminated with Aspergillus matter such as spores or DNA, as negative controls were repeatedly positive. When ATL was replaced with our own digestion buffer, the problem was resolved. It is therefore essential that negative controls are processed at every step to exclude false positives when using this technique.
This system of fungal DNA detection could be expanded to identify individual species within the Candida genus. We demonstrated that PCR amplification with primers Pffor-Pfrev2 and Cafor2-Carev3 allowed the detection of not just C. albicans, but also DNA derived from several other Candida species. These species could potentially be differentiated through the use of restriction enzyme digestion of the PCR products, the efficacy of which has already been demonstrated in a number of previously published protocols (26, 28). Candida speciation would be an important aid to effective patient treatment, facilitating the application of species-specific antifungal therapy, thereby avoiding problems of drug resistance.
In a clinical setting, this system could potentially provide reliable results within 6.5 h of receiving a sample. Samples were tested from three patients who clearly demonstrated the range and complexity of the clinical problem. Results obtained demonstrate the successful use of PCR in this setting and suggest that this technique is more sensitive than culture for Candida detection. Furthermore, organisms do not have to be viable when sampled, which is in contrast to the requirements of microbiological protocols. This system overcomes problems previously reported with PCR from ocular fluids (5, 28, 39). It therefore constitutes an additional protocol, which, along with traditional culture techniques, may significantly aid patient management in cases where fungal endophthalmitis is suspected.
ACKNOWLEDGMENTS
E.E.M.J., N.M.C., and S.C. were supported by Oclyx, Ltd. P.A. was supported by Fight for Sight. N.O. was supported by Wellcome Vision Research Fellowship no. 045203 and locally organized research funds from Moorfields Eye Hospital no. 221 and 271.
REFERENCES
- 1.Aguilar G L, Blumenkrantz M S, Egbert P R, McCulley J P. Candida endophthalmitis after intravenous drug abuse. Arch Ophthalmol. 1979;97:96–100. doi: 10.1001/archopht.1979.01020010036008. [DOI] [PubMed] [Google Scholar]
- 2.Bodey G P. Candidiasis in cancer patients. Am J Med. 1984;77(Suppl. 4D):13. [PubMed] [Google Scholar]
- 3.Brod R D, Flynn H W, Jr, Clarkson J G, Pflugfelder S C, Culbertson W W, Miller D. Endogenous Candida endophthalmitis. Management without intravenous amphotericin B. Ophthalmology. 1990;97:666–672. doi: 10.1016/s0161-6420(90)32547-2. [DOI] [PubMed] [Google Scholar]
- 4.Candian U, Hofelein C, Luthy J. Polymerase chain reaction with additional primers allows identification of amplified DNA and recognition of specific alleles. Mol Cell Probes. 1992;6:13–19. doi: 10.1016/0890-8508(92)90066-7. [DOI] [PubMed] [Google Scholar]
- 5.Carroll N M, Jaeger E E M, Choudhury S, Dunlop A A S, Matheson M M, Adamson P, Okhravi N, Lightman S. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol. 2000;38:1753–1757. doi: 10.1128/jcm.38.5.1753-1757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cenis J L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chignell A H. Endogenous candida endophthalmitis. J R Soc Med. 1992;85:721–724. doi: 10.1177/014107689208501204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clift R A. Candidiasis in the transplant patient. Am J Med. 1984;77(Suppl. 4D):34. [PubMed] [Google Scholar]
- 9.Cusumano A, Busin M, Spitznas M. Mycotic infection of the capsular bag in postoperative endophthalmitis. J Cataract Refract Surg. 1991;17:503–505. doi: 10.1016/s0886-3350(13)80859-0. [DOI] [PubMed] [Google Scholar]
- 10.Denning D W, Evans E G, Kibbler C C, Richardson M D, Roberts M M, Rogers T R, Warnock D W, Warren R E. Guidelines for the investigation of invasive fungal infections in haematological malignancy and solid organ transplantation. British Society for Medical Mycology. Eur J Clin Microbiol Infect Dis. 1997;16:424–436. doi: 10.1007/BF02471906. [DOI] [PubMed] [Google Scholar]
- 11.Edwards J E, Jr, Foos R Y, Montgomerie J Z, Guze L B. Ocular manifestations of Candida septicemia: review of seventy-six cases of hematogenous Candida endophthalmitis. Medicine. 1974;53:47–75. doi: 10.1097/00005792-197401000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Flahaut M, Sanglard D, Monod M, Billie J, Rossier M. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin J R, Petit T H, Fishman L S, Foos R Y. Candida endophthalmitis: a clinical and pathologic study of 21 cases. Arch Ophthalmol. 1973;89:450–456. doi: 10.1001/archopht.1973.01000040452002. [DOI] [PubMed] [Google Scholar]
- 14.Hassan I J, Macgowan A P, Cook S D. Endophthalmitis at the Bristol Eye Hospital: an 11-year review of 47 patients. J Hosp Infect. 1992;22:271–278. doi: 10.1016/0195-6701(92)90012-b. [DOI] [PubMed] [Google Scholar]
- 15.Haynes K A, Westerneng T J. Rapid identification of Candida albicans, C. glabrata, C. parapsilosis and C. krusei by species-specific PCR of large subunit ribosomal DNA. J Med Microbiol. 1996;44:390–396. doi: 10.1099/00222615-44-5-390. [DOI] [PubMed] [Google Scholar]
- 16.Hockey L J, Fujita N K, Gibson T R, Rotrosen D, Montgomerie J Z, Edwards J E., Jr Detection of fungemia obscured by concomitant bacteremia: in vitro and in vivo studies. J Clin Microbiol. 1982;16:1080–1085. doi: 10.1128/jcm.16.6.1080-1085.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes A R, Cannon R D, Shepherd M G, Jenkinson H F. Detection of Candida albicans and other yeasts in blood by PCR. J Clin Microbiol. 1994;32:228–231. doi: 10.1128/jcm.32.1.228-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hue F, Huerre M, Rouffault M A, de Bievre C. Specific detection of Fusarium species in blood and tissues by a PCR technique. J Clin Microbiol. 1999;37:2434–2438. doi: 10.1128/jcm.37.8.2434-2438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes D S, Hill R J. Infectious endophthalmitis after cataract surgery. Br J Ophthalmol. 1994;78:227–232. doi: 10.1136/bjo.78.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloess P M, Stutling R D, Waring G, Wilson L A. Bacterial and fungal endophthalmitis after penetrating keratoplasty. Am J Ophthalmol. 1993;115:309–316. doi: 10.1016/s0002-9394(14)73580-9. . (Erratum, 115:548.) [DOI] [PubMed] [Google Scholar]
- 21.Loeffler J, Hebart H, Bialek R, Hagmeyer L, Schmidt D, Serey F, Hartmann M, Eucker J, Einsele H. Contaminations occurring in fungal PCR assays. J Clin Microbiol. 1999;37:1200–1202. doi: 10.1128/jcm.37.4.1200-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maleska R, Clark-Walker G D. Yeasts have a four-fold variation in ribosomal DNA copy number. Yeast. 1993;9:53–58. doi: 10.1002/yea.320090107. [DOI] [PubMed] [Google Scholar]
- 23.Mannarelli B M, Kurtzman C P. Rapid identification of Candida albicans and other human pathogenic yeasts by using short oligonucleotides in a PCR. J Clin Microbiol. 1998;36:1634–1641. doi: 10.1128/jcm.36.6.1634-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyakawa Y, Mabuchi T, Fukazawa Y. New methods for detection of Candida albicans in human blood by polymerase chain reaction. J Clin Microbiol. 1993;31:3344–3347. doi: 10.1128/jcm.31.12.3344-3347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomerie J Z, Edwards J E., Jr Association of infection due to Candida albicans with intravenous hyperalimentation. J Infect Dis. 1978;137:197–201. doi: 10.1093/infdis/137.2.197. [DOI] [PubMed] [Google Scholar]
- 26.Morace G, Sanguinetti M, Posteraro B, Cascio G L, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol. 1997;35:667–672. doi: 10.1128/jcm.35.3.667-672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okhravi, N., P. Adamson, N. Carroll, A. Dunlop, M. M. Matheson, H. M. A. Towler, and S. Lightman. PCR-based evidence of bacterial involvement in eyes with suspected intraocular infections. Investig. Ophthalmol. Vis. Sci., in press. [PubMed]
- 28.Okhravi N, Adamson P, Mant R, Matheson M M, Midgley G, Towler H M A, Lightman S. Polymerase chain reaction and restriction fragment length polymorphism mediated detection and speciation of Candida spp. causing intraocular infection. Investig Ophthalmol Vis Sci. 1998;39:859–866. [PubMed] [Google Scholar]
- 29.Olsen G J, Woese C R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 30.Pflugfelder S C, Flynn H W, Jr, Zwickey T A, Forster R K, Tsiligianni A, Culbertson W W, Mandelbaum S. Exogenous fungal endophthalmitis. Ophthalmology. 1988;95:19–30. doi: 10.1016/s0161-6420(88)33229-x. [DOI] [PubMed] [Google Scholar]
- 31.Rao N A, Nerenberg A V, Forster D J. Torulopsis candida (Candida famata) endophthalmitis simulating Propionibacterium acnes syndrome. Arch Ophthalmol. 1991;109:1718–1721. doi: 10.1001/archopht.1991.01080120102037. [DOI] [PubMed] [Google Scholar]
- 32.Riggsby W S, Torres-Bauza L J, Wills J W, Townes T M. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmid S, Martenet A C, Oelz O. Candida endophthalmitis: clinical presentation, treatment and outcome in 23 patients. Infection. 1991;19:21–24. doi: 10.1007/BF01643753. [DOI] [PubMed] [Google Scholar]
- 34.Shin J H, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrader S K, Band J D, Lauter C B, Murphey P. The clinical spectrum of endophthalmitis: incidence, predisposing factors, and features influencing outcome. J Infect Dis. 1990;162:115–120. doi: 10.1093/infdis/162.1.115. [DOI] [PubMed] [Google Scholar]
- 36.Swerdloff J N, Filler S G, Edwards J E., Jr Severe candidial infections in neutropenic patients. Clin Infect Dis. 1993;17(Suppl. 2):S457–S467. doi: 10.1093/clinids/17.supplement_2.s457. [DOI] [PubMed] [Google Scholar]
- 37.van Burik J, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenkel H, Rummelt V, Knorr H, Naumann G O. Chronic postoperative endophthalmitis following cataract extraction and intraocular lens implantation. Report on nine patients. Ger J Ophthalmol. 1993;2:419–425. [PubMed] [Google Scholar]
- 39.Wiedbrauk D I, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]