Abstract
Objective
The COVID-19 (coronavirus disease 2019) pandemic response at the Medical University of South Carolina included virtual care visits for patients with suspected severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. The telehealth system used for these visits only exports a text note to integrate with the electronic health record, but structured and coded information about COVID-19 (eg, exposure, risk factors, symptoms) was needed to support clinical care and early research as well as predictive analytics for data-driven patient advising and pooled testing.
Materials and Methods
To capture COVID-19 information from multiple sources, a new data mart and a new natural language processing (NLP) application prototype were developed. The NLP application combined reused components with dictionaries and rules crafted by domain experts. It was deployed as a Web service for hourly processing of new data from patients assessed or treated for COVID-19. The extracted information was then used to develop algorithms predicting SARS-CoV-2 diagnostic test results based on symptoms and exposure information.
Results
The dedicated data mart and NLP application were developed and deployed in a mere 10-day sprint in March 2020. The NLP application was evaluated with good accuracy (85.8% recall and 81.5% precision). The SARS-CoV-2 testing predictive analytics algorithms were configured to provide patients with data-driven COVID-19 testing advices with a sensitivity of 81% to 92% and to enable pooled testing with a negative predictive value of 90% to 91%, reducing the required tests to about 63%.
Conclusions
SARS-CoV-2 testing predictive analytics and NLP successfully enabled data-driven patient advising and pooled testing.
Keywords: medical informatics [L01.313.500], natural language processing (nlp) [L01.224.050.375.580], machine learning [g17.035.250.500], data science [L01.305]
INTRODUCTION
The first coronavirus disease 2019 (COVID-19) case in the United States was confirmed January 21, 2020. Waves of rapid expansion to all 50 U.S. states followed, with about 33 million confirmed cases and 600 000 deaths in the United States as of June 2021.1 Increased severity of the disease has been especially noted with comorbid conditions2 and mortalities as high as 21%.3 One of the key public health measures for controlling the spread of COVID-19 is aggressive testing.4
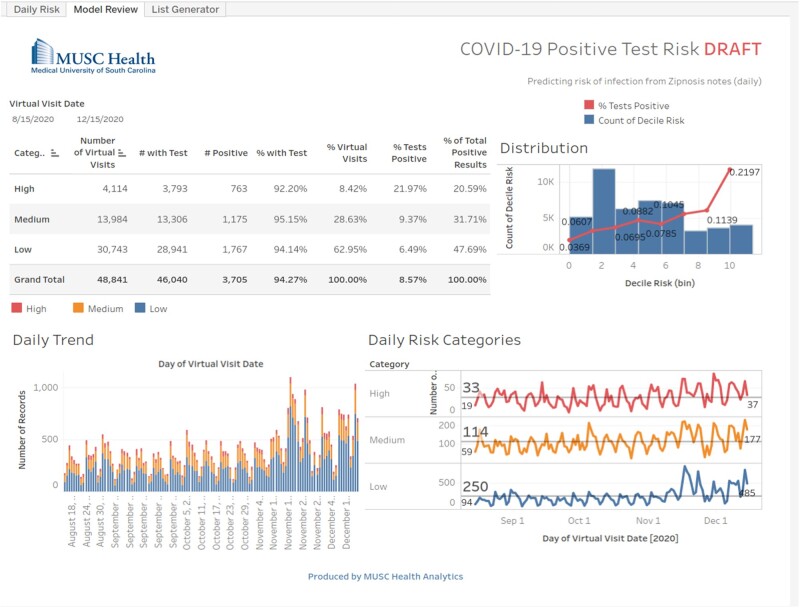
At the Medical University of South Carolina (MUSC) (Charleston, SC), a telehealth system (Zipnosis)5 was implemented in March 2020 as the preferred option for patients interested in COVID-19 testing. Patients would start a virtual visit and answer a questionnaire about their symptoms, COVID-19 exposure and travel history, and brief medical history. The telehealth system then exported an automatically generated summary text note generated from the information entered by the patient.6 This note was the only information available in the electronic health record (EHR). Care management based on some form of COVID-19 dashboard (Figure 1) or decision support capabilities was required, but the unstructured text format of this note made them difficult to deploy. More generally, detailed clinical information is needed to help assess the extent of the pandemic, assess characteristics of the virus and the disease it is causing, and discover and compare supportive or therapeutic approaches and population health measures applied at the patient level. This detailed information is typically found in unstructured text notes in EHR systems or other ancillary systems. Extracting such information manually is costly, not scalable, and far too slow to address current needs. As an effective and scalable approach to extract structured and coded information from unstructured text, natural language processing (NLP) has been used for many years.7 To enable access to structured and coded COVID-19–related information as documented by patients in the telehealth system, a new NLP application (COVID-NLP tool) was rapidly developed and is described in more detail in the present article. Along with the NLP tool, a new database (COVID data mart) was created in March 2020 to combine clinical information from patients assessed or treated for COVID-19 at MUSC. It was progressively enriched with information extracted from the telehealth system and combined with select clinical information from existing patient records at MUSC. It included clinical information from about 220 000 patients as of March 2021 .
Figure 1.
Medical University of South Carolina virtual care visits dashboard sample.
Early success with using information from the telehealth system to predict positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) results8 encouraged further efforts to enhance the accuracy of these predictions and enable applications supporting patient care such as a novel data-driven COVID-19 symptom checker giving patients testing advice according to their predicted test result. This initial effort was based on text analytics and brute force deep learning–based approach applied to the telehealth system notes. Further experiments with NLP and predictive analytics for SARS-CoV-2 test results prediction with various behaviors and optimization experiments followed9,10 and are described in detail subsequently along with applications for data-driven patient advising (symptom checker) and enhanced pooled testing.
BACKGROUND
As part of COVID-19 pandemic response efforts, NLP has been applied mostly to help analyze the large amount of scientific publications focused on COVID-1911–13 or process social media for sentiment analysis14 or misinformation detection.15 Extracting information from clinical text, ie, narrative text notes found in EHR systems, has been another important application of NLP. To extract COVID-19–related information, COVID-19 SignSym built on the CLAMP tool to extract signs and symptoms.16 Another example was developed using the spaCy framework and applied at the Department of Veterans Affairs to detect positively tested patients.17 Both were released in July 2020. At the University of Washington, a new corpus of 1472 clinical notes has been annotated and used to develop an NLP application extracting symptoms and mentions of COVID-19.18
To help manage patient care and resources, various predictive analytics algorithms focused on COVID-19 have been developed at the population level (infections and mortality prediction)19–22 or at the individual patient level to predict mortality and critical illness events (eg, intubation, intensive care unit admission).23 In most cases, existing structured and coded information was used without any NLP application. Information used included demographics, diagnostic codes, specific laboratory test orders (including SARS-CoV-2), and death. Other clinical information has also been used with predictive analytics algorithms to predict SARS-CoV-2 infections. Routine laboratory test results (complete blood count) and patient gender allowed for a sensitivity of 86% to 93% and specificity of 35% to 55% when predicting SARS-CoV-2–positive results.24
No data-driven efforts to predict SARS-CoV-2–positive results using machine learning and information provided by patients have been reported. A data-driven symptom checker (K Health) was used in a recent study, but the knowledge specific to COVID-19 was added separately and based on manually crafted rules instead of trained machine learning algorithms.25
For increased testing efficiency and enabling asymptomatic cases testing, pooled sample testing has been proposed and approved by the Food and Drug Administration for SARS-CoV-2 diagnostic testing since June 2020.26 The Centers for Disease Control and Prevention later published an interim guidance for pooled testing in October 2020.27 Pooled testing involves combining multiple samples and testing with a single diagnostic test but only works when the prevalence of cases (ie, positive results) is low. Pooled testing can be implemented using various strategies and studies demonstrated the absence of reduced sensitivity.28 Strategies include the Dorfman protocol29 and “split pooling,” the latter possibly allowing for a lower number of false negatives.30 No efforts applying predictive analytics to enhance or enable pooled testing have been reported.
MATERIALS AND METHODS
The first version of the COVID-NLP tool, the data mart and all related data extraction, transfer, and loading were developed, tested, and made available for production in about 10 days only, between March 16 and March 26, 2020.
NLP for COVID-19–related information extraction
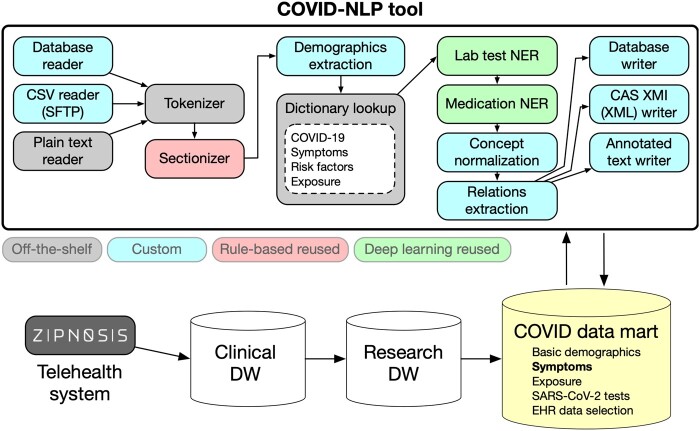
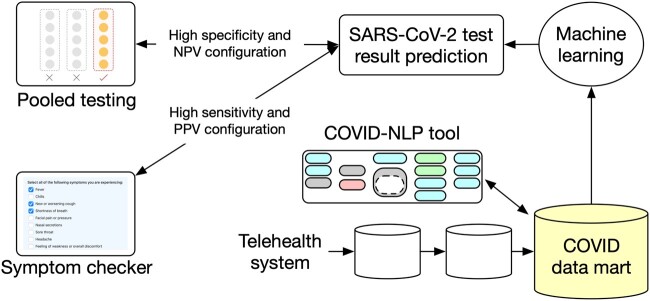
The COVID-NLP tool builds on a standard framework (Apache UIMA)31 and combines components we had developed in past efforts and could reuse (“off-the-shelf”) with components from other local current research (“rule-based” or “deep learning,” not retrained) and a few new custom components (Figure 2). Symptom and comorbidity extraction relies on ConceptMapper with new dictionaries generated using lexgen,32 a tool to automatically create dictionaries from the Unified Medical Language System Metathesaurus. The deep learning components are based on bidirectional long-short term memory sequence labeling models using word tokens as input. Vector representations of words were constructed using fastText embeddings33 pretrained with clinical texts (MIMIC-III [Medical Information Mart for Intensive Care-III]).34 The training set of the 2019 n2c2 challenge was used to train the “laboratory test NER” model. For the “medication NER” component, the 2009 i2b235 and 2018 n2c236 shared tasks corpora were used. All output from the COVID-NLP tool was represented using the Observational Medical Outcomes Partnership common data model.37 Nineteen categories of information were extracted (Table 1). The COVID-NLP tool was deployed on servers to process hourly batches of new text notes from the telehealth system. All system improvements and bug fixes were retroactively reapplied to existing notes to ensure that all derived output had been consistently extracted. A more recent version of the tool is available with open source.38
Figure 2.
COVID-NLP tool components. COVID-19: coronavirus disease 2019; DW: data warehouse; EHR: electronic health record; NER : named entity recognition; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Table 1.
Clinical information extracted by the COVID-NLP tool
| Demographics and Social History | Medical Risk Factors | Laboratory Tests | Medications | Environment Risk Factors | Structural Components |
|---|---|---|---|---|---|
|
|
|
|
|
|
To assess the accuracy of the COVID-NLP tool, a small random sample of 15 text notes was manually annotated by domain experts. Two experts independently used WebAnno39 to annotate each text note and a third expert adjudicated annotation disagreements. Evaluation metrics included recall (ie, sensitivity), precision (ie, positive predictive value), and the F1-measure with micro- and macro-averaging. The ETUDE tool40 was used to automate all evaluation computations.
SARS-CoV-2 test results prediction
The study population included all patients with virtual care visits at MUSC since April 16, 2020 (date when the telehealth system started including anosmia questions), who had a SARS-CoV-2 diagnostic test within 14 days after the virtual visit. Information extracted by the COVID-NLP tool from the text note generated by the telehealth system was then used for predicting the SARS-CoV-2 test result. Initial efforts used the telehealth system text directly, without information extraction,8 or used simple keywords extraction with regular expressions to extract symptoms.
A separate “curated” dataset was developed at MUSC by clinical experts in the division of Infectious Diseases in March-May 2020. At that time, it included 125 cases (ie, patients with positive SARS-CoV-2 diagnostic testing) and 242 controls (ie, patients with confirmed negative SARS-CoV-2 testing) with detailed clinical information collected by domain experts through manual chart review and patient interviews. This curated dataset was used for validation of our predictive algorithms.
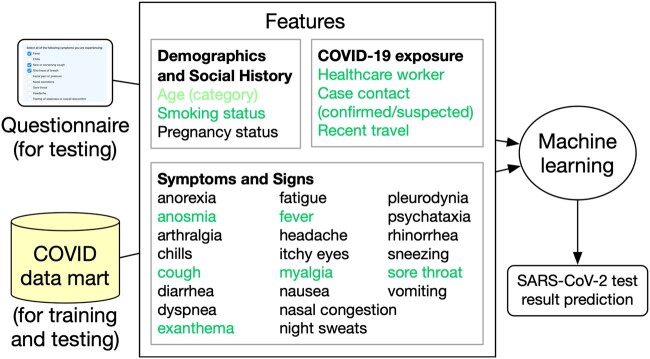
Further efforts to improve predictions were based on information extracted using NLP and started with a comparison of shallow and deep learning algorithms. Support vector machines, decision trees, logistic regression, and artificial neural networks (multilayer perceptron) were compared along with 2 deep learning–based classifiers (convolutional neural network and fastText)41 using word embeddings trained with clinical notes from the MIMIC-III clinical dataset (version 1.4.34 In general, we favored easily explainable, rather than “black-box,” algorithms, following recommendations for explicit and data-driven predictions based on simple, easily understood, and easily applied models.42 The initial features used for prediction included a selection of symptoms, exposure, and other information (Figure 3). The outcome was the SARS-CoV-2 diagnostic test result (positive or negative).
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) predictive analytics algorithm features. COVID-19: coronavirus disease 2019.
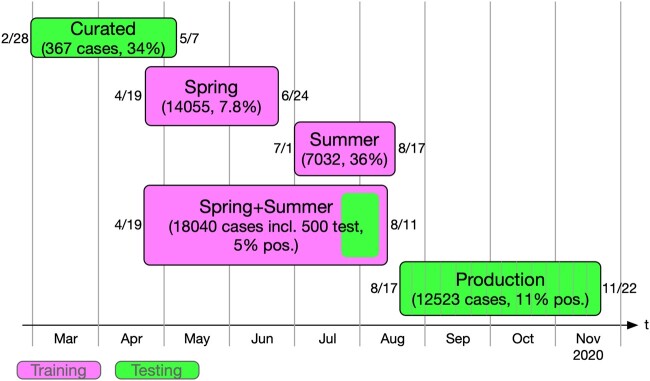
Various datasets were used to train and test our predictive analytics algorithms (Figure 4): a Spring dataset included visits from April 19 to June 24, 2020 (34 597 notes from 14 055 patients; 1101 testing positive and 12 954 negative). We used all positive cases and downsampled to 10% of negative cases. A second Summer dataset was created in response to the drastically changing positivity rates from July 1 to August 17, 2020. We used all positive cases but downsampled to 40% of the negative cases. It included 7032 cases with a positivity rate of 36%. A third Spring+Summer dataset combined visits from April 19 to August 11, 2020, and included a random sample of positive cases to reduce the proportion of positive cases to about 5% (902 from 3569 positives) with a held-out test set of 500 randomly selected cases (30 positive cases and 470 negative cases). The remaining training set included 872 positive cases and 16 668 negative cases. A final Production dataset included visits from August 17 to November 22. This dataset was used to assess live performance on production data and included 12 523 cases (1398 positive and 11 125 negative).
Figure 4.
Datasets used for training and testing predictive analytics algorithms.
To ease the predictive analytics algorithm deployment, a feature selection effort using both the weight and Shapley value of each feature resulted in a subset of 6 general risk features and 6 sign or symptom features (top features, in green font in Figure 3).
Predictive analytics models generated values from 0 to 1. Model interpretation required choosing a threshold above which a test would be predicted positive. Different values of this threshold allowed for either high sensitivity (eg, for patient testing advices) or high specificity and negative predictive value (NPV) (eg, for pooled testing) (Figure 5). These 2 versions of the predictive analytics were deployed as Web service. More details about these models are available in Supplementary Appendix 2 and in a public repository.43 For the predictive analytics accuracy evaluation, metrics included sensitivity, specificity, and NPV.
Figure 5.
Coronavirus disease 2019 (COVID-19) predictive analytics general workflow. NPV: negative predictive value; PPV: positive predictive value; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Data-driven personalized COVID-19 advices for patients
To provide patients with data-driven COVID-19 testing and behavior advices, the predictive analytics configuration favoring high sensitivity was preferred. The objective was to ensure that all possible SARS-CoV-2 infections would be identified. The logistic regression predictive analytics algorithm based on simple keywords extraction from text was used for this task. For training and testing, the only source of information was the notes generated by the telehealth system. After testing, the predictive analytics algorithm was deployed as a web application with a simple user interface listing questions about the symptoms patients were experiencing (Python Django application containerized using Docker and deployed as a Microsoft Azure web application for public access.44 The symptom checker was designed to return 3 possible levels of risk for an individual as proposed in Centers for Disease Control and Prevention recommendations: low risk (recommending no action), medium risk (recommending testing, staying home, and caution), and high risk (recommending testing, staying home, and medical help within 24 hours).
Data-driven SARS-CoV-2 pooled testing optimization
When the expected population rate of positive SARS-CoV-2 diagnostic test results would exceed levels typically feasible for efficient pooling, the predictive analytics algorithm should balance the configuration favoring high specificity and NPV. The objective is to pool only those predicted as negative.
Pragmatic evaluation of predictive model thresholds for pooled testing can be achieved by direct estimation of the expected number of tests needed to assess a given number of specimens. Note that this quantity is not a simple function of the traditional machine learning objectives. The number of true negatives should be as large as possible, and the number of false positives should be minimized as well. An objective function metric that combines these 2 constraints would allow for direct evaluation of the predictive model and corresponding tuning parameter and threshold selection. We developed a straightforward Monte Carlo approximation that provides reasonable and practical estimates of the number of tests required based on predicted risk values (details in Supplementary Appendix 1). The simulation directly calculates the number of resulting pools that are positive and the total number of tests needed. The R markdown code for the Monte Carlo estimation procedure is available in a public repository.43
To capture COVID-19-related information for all patients tested for SARS-CoV-2 and not only patients using virtual visits, a new form integrated with the test ordering system was developed and implemented using REDCap (Research Electronic Data Capture)45 and Epic (Epic Systems, Verona, WI) HL7 FHIR (Fast Healthcare Interoperability Resources)46 interfaces. Information captured in the form was then used with the predictive analytics algorithm before placing the order for SARS-CoV-2 testing. A flag was placed on the order identifying whether the specimen could be pooled (if predicted negative) or should be processed individually (if predicted positive). Once the order was released from the EHR system, the flag would be evaluated by the Cloverleaf interface engine47 and the appropriate laboratory test (pool or single specimen testing) would be transmitted to the laboratory information system (Cerner Millennium PathNet; Cerner, North Kansas City, MO). Specimens designated for pooling were placed on the pooling system (Hamilton Microlab STAR Liquid Handling System; Hamilton, Reno, NV) where 5 samples are aliquoted into a single tube for testing (Abbott M2000 RealTime system SARS-CoV-2 assay; Abbott, Chicago, IL). If the pooled specimens result was negative, testing results for all component specimens were reported negative. If the pooled specimen result was positive, the individual component specimens were tested to determine which of the 5 contributing samples was positive.
RESULTS
NLP for COVID-19–related information extraction
When comparing the COVID-NLP tool output with the aforementioned reference standard, we measured an overall micro-averaged F1-measure of 0.725 and macro-average of 0.832 (Table 2). As example of domain portability, the medication NER module achieved a precision of 0.878, recall of 0.682, and F1-measure of 0.768 with medication names. For comparison, the system achieves an F1-measure of 0.927 on the 2018 n2c2 shared task test set, for which it was originally developed.48
Table 2.
COVID-19 information extraction accuracy results
| TP | FP | FN | Precision | Recall | F1-measure | |
|---|---|---|---|---|---|---|
| Environmental risk factors | ||||||
| CloseContact | 25 | 2 | 5 | 0.926 | 0.833 | 0.877 |
| HealthcareWorker | 10 | 4 | 3 | 0.714 | 0.769 | 0.741 |
| Travel | 13 | 2 | 1 | 0.867 | 0.929 | 0.897 |
| Communal setting | 0 | 0 | 0 | – | – | – |
| Demographics and social history | ||||||
| IsPregnant | 6 | 0 | 0 | 1.000 | 1.000 | 1.000 |
| Smokes | 13 | 2 | 0 | 0.867 | 1.000 | 0.929 |
| Gender | 15 | 0 | 0 | 1.000 | 1.000 | 1.000 |
| Weight | 27 | 3 | 0 | 0.900 | 1.000 | 0.947 |
| Height | 0 | 0 | 0 | – | – | – |
| Medical risk factor | ||||||
| Comorbidity, symptom | 319 | 182 | 136 | 0.637 | 0.701 | 0.667 |
| Laboratory tests | ||||||
| LabName | 0 | 0 | 16 | |||
| LabValue | 0 | 0 | 15 | |||
| Medications | ||||||
| MedDosage | 67 | 27 | 34 | 0.713 | 0.663 | 0.687 |
| MedDuration | 16 | 8 | 7 | 0.667 | 0.696 | 0.681 |
| MedFrequency | 55 | 23 | 1 | 0.705 | 0.982 | 0.821 |
| MedName | 137 | 19 | 64 | 0.878 | 0.682 | 0.768 |
| MedRoute | 88 | 35 | 10 | 0.715 | 0.898 | 0.796 |
| micro-average | 791 | 307 | 292 | 0.720 | 0.730 | 0.725 |
| macro-average | 0.815 | 0.858 | 0.832 | |||
COVID-19: coronavirus disease 2019; FN: false negative; FP: false positive; TP: true positive.
SARS-CoV-2 diagnostic test results prediction
When assessing the predicted SARS-CoV-2 diagnostic test result accuracy, we found that the simpler models tended to outperform the more complex models. Specifically, we focused on SVM and logistic regression models in the later stages of this work because decision trees, neural networks, and deep neural networks did not reliably outperform the former models (additional results in the Supplementary Appendix).
Table 3 summarizes the accuracy of test result predictions across 3 generations of models. To most accurately reflect production usage, we present average weekly performance metrics for these models tested on the production dataset when the model threshold (used to determine positive vs negative results) was set to optimize accuracy metrics (ie, increase NPV and specificity) or to optimize testing efficiency (ie, reduce the number of tests needed). The logistic regression algorithm trained on the Spring+Summer dataset had a slightly lower NPV at 0.831 than earlier models with more features. Using the Monte Carlo estimation of the expected number of tests needed to determine the most efficient threshold actually raises the NPV to 0.889, making this model more competitive with more feature-rich models. The trade-off for this improved NPV is a large increase in specificity (from about 0.12 to 0.99) and a drop in sensitivity (from about 0.81 to 0.01) when using the efficiency-optimized threshold.
Table 3.
Diagnostic test results prediction accuracy results
| Accuracy Optimized |
Efficiency Optimized |
|||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | NPV | Sensitivity | Specificity | NPV | |
| Testing on production dataset | ||||||
| Spring dataset training (10% negatives; all features) | ||||||
| Logistic regression | 0.397 | 0.805 | 0.914 | 0.199 | 0.926 | 0.903 |
| Support vector machine | – | 0.000 | – | 0.299 | 0.883 | 0.910 |
| Summer dataset training (40% negatives, features without age) | ||||||
| Logistic regression | 0.408 | 0.706 | 0.906 | 0.286 | 0.804 | 0.901 |
| Support vector machine | 0.999 | 0.001 | 0.929 | 0.373 | 0.789 | 0.911 |
| Spring+Summer dataset training (positives subset to reach 5%, top features) | ||||||
| Logistic regression | 0.806 | 0.120 | 0.831 | 0.013 | 0.989 | 0.889 |
| Testing on curated dataset | ||||||
| Spring dataset training (10% negatives; all features) | ||||||
| Logistic regression | 0.736 | 0.744 | 0.845 | 0.576 | 0.967 | 0.815 |
| Support vector machine | 1.000 | 0.302 | 1.000 | 0.520 | 0.979 | 0.798 |
| Summer dataset training (40% negatives, features without age) | ||||||
| Logistic regression | 0.672 | 0.682 | 0.801 | 0.592 | 0.789 | 0.789 |
| Support vector machine | – | 0.000 | – | 0.920 | 0.318 | 0.885 |
| Spring+Summer dataset training (positives subset to reach 5%, top features) | ||||||
| Logistic regression | 0.920 | 0.227 | 0.846 | – | 0.000 | – |
NPV: negative predictive value.
When evaluated against the “curated” dataset, which has fewer possible features for extraction, we see a less severe impact of removing features from the models. The logistic regression algorithm trained on all available features reaches an NPV of 0.8451 and a similar NPV of 0.8462 when trained on the top features only.
Data-driven personalized COVID-19 advices for patients
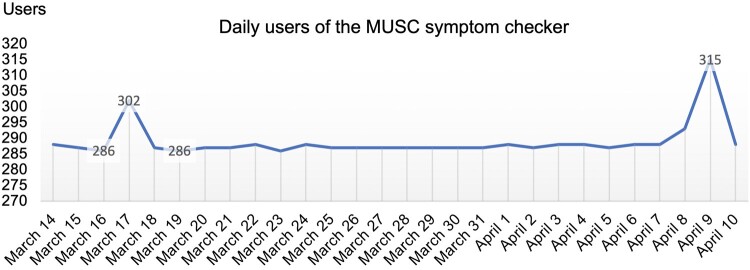
The symptom checker Web application was designed to store only telemetry data on usage, not actual user inputs (although this was proposed for future versions). We therefore monitored its use by patients. As example, the symptom checker was used between 286 and 315 times daily between March 14 and April 10, 2021 (Figure 6).
Figure 6.
Medical University of South Carolina (MUSC) data-driven symptom checker usage sample (daily usage between March 14 and April 10, 2021).
Data-driven SARS-CoV-2 pooled testing optimization
The predictive analytics Web service used for pooled testing also stores only telemetry data on usage, not actual user inputs. It was used between 0 and 125 times daily between March 14 and April 10, 2021.
The pooling efficiency or reduction in testing was simulated for the Spring+Summer training dataset and the Production dataset. The threshold that minimized the total number of tests (ie, efficiency optimized) is compared against the threshold determined to maximize accuracy metrics like NPV on the training corpus (ie, accuracy optimized) (Table 4). Testing efficiency optimization is measured as the ratio of expected number of tests simulated (50 Monte Carlo simulations) to the number of subjects tested. A lower number is better. As seen in Table 4, the logistic regression algorithm trained on the Spring+Summer corpus requires roughly 97% of the total number of tests with the accuracy-optimized threshold and only 64% of the total number of tests with the efficiency-optimized threshold.
Table 4.
Pooled testing efficiency prediction results (number of tests divided by the number of subjects tested)
| Training Datasets |
Production Dataset |
|||
|---|---|---|---|---|
| Accuracy Optimized | Efficiency Optimized | Accuracy Optimized | Efficiency Optimized | |
| Spring dataset training (10% negatives; all features) | ||||
| Logistic regression | 0.995 | 0.766 | 0.656 | 0.636 |
| Support vector machine | 1.000 | 0.757 | 1.000 | 0.631 |
| Summer dataset training (40% negatives, features without age) | ||||
| Logistic regression | 0.970 | 0.779 | 0.720 | 0.689 |
| Support vector machine | 0.999 | 0.793 | 0.999 | 0.668 |
| Spring+Summer dataset training (positives subset to reach 5%, top features) | ||||
| Logistic regression | 0.957 | 0.807 | 0.973 | 0.643 |
DISCUSSION
Other efforts to analyze symptoms associated with COVID-19 concluded that anosmia, ageusia, and fever best discriminated positive COVID-19 cases from negative cases.49 The symptoms contributing the most to SARS-CoV-2–positive test results prediction include sore throat, cough, anosmia, fever, myalgia, and exanthema according to our experiments. The first 4 helped include cases and the last 2 helped exclude cases. Several studies examined the typical symptomatology of COVID-19 and the symptoms reported with a sensitivity of at least 50% in at least 1 study include cough, sore throat, fever, myalgia or arthralgia, fatigue, and headache.50
Our current production model has eliminated certain original features due to unintuitive performance or poor upstream data. For instance, ages were binned into 3 categories: <18, 18-64, and 65+. Young patients (<18 years of age) using Zipnosis were largely sicker than older patients (65+ years of age), who were more likely to seek routine care. This implicit sampling bias resulted in young patients receiving a higher risk score when all else was held equal. The curated dataset included only a subset of the features our models were trained on. This difference probably caused the lower accuracy observed with the curated dataset. We have so far ignored temporal considerations in our modeling. Future work will need to address changes in positivity rates at the population level over time and monitor model drift, for instance.
Errors analysis
When evaluating the COVID-NLP tool information extraction accuracy, the worst-performing categories included laboratory test names and values, comorbidities, and exposure risk due to close contact. Understandably, every laboratory test annotated in our evaluation corpus happened to be a body temperature, which was not captured by the reused pretrained laboratory test NER system. Our comorbidity dictionary included respiratory diseases but missed the commonly mentioned concepts of asthma and viral infection. This evaluation also uncovered a mismatch between the tokenized output fed to ConceptMapper for term alignment and how the terms were tokenized in the dictionaries themselves, which resulted in a high false negative rate for any terms followed by specific punctuation in the note. Symptom and comorbidity extraction also suffered from this apparent bug with a precision of 0.701 and a recall of only 0.637. Most false negative errors in this category were due to errant punctuation in the tokenization of the target concept. Finally, the close contact exposure risk details were extracted by a simple regular expression designed to match the Zipnosis note output, the template for which has occasionally changed throughout our development cycle. All of these errors will be addressed in future releases of the COVID-NLP tool.
Study limitations
Our small sample size for the NLP-based information extraction reference corpus was made worse by a preprocessing error that caused 5 of the notes to be duplicated in place of a different set of 5 that we intended to annotate. What we designed to be a 20-note corpus was effectively shrunk to 15 notes.
The COVID-NLP tool was initially only developed for notes from the telehealth system. Current and future efforts to enhance this NLP tool include adapting it to a large variety of clinical notes as found in EHR systems.
The efforts described here only used data from a specific time period. Changes in the COVID-19 clinical presentation, prevalence, and response will probably affect the accuracy of the trained algorithms. To assess and address these possible temporal shifts, we plan to regularly verify the accuracy of the NLP tool, the predictive analytics, and their application for pooled testing. Significant accuracy alterations will be followed by retraining and retesting efforts.
CONCLUSION
In summary, the rapid development and deployment of a dedicated data mart and NLP application enabled access to structured and coded information from patients tested and treated for COVID-19 at MUSC. This information was then successfully used to predict the result of SARS-CoV-2 diagnostic tests, then further used with different configurations to either support patient advices or enable more efficient pooled testing. Both were integrated with clinical care systems and demonstrate possible applications of NLP and predictive analytics to support patient advising and clinical care.
FUNDING
This work was supported by the Patient-Centered Outcomes Research Institute (contract ME-2018C3-14549) and by the SmartState Program (Translational Biomedical Informatics Chair Endowment).
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception of the work or analysis and interpretation of data. SMM and JO led the development of the telehealth system notes capture and COVID-19 data mart. SMM and PMH conceived the COVID-19 NLP tool and led its development. PMH, YK, and MD were responsible for most development work. The SARS-CoV-2 test result prediction algorithms were evaluated and implemented by MD, PMH, and YK. MD implemented the COVID-19 dashboard and Web application for patient advising. AVA developed the Monte Carlo estimation for pooled testing optimization and evaluated it with PMH. JM was responsible for the pooled testing infrastructure implementation. SMM oversaw the REDCap questionnaire for pooled testing development. All authors drafted the work or revised it critically. SMM drafted the initial manuscript. PMH, YK, JO, and AVA provided critical revision of the manuscript. All authors gave final approval of the version to be published. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online
Supplementary Material
ACKNOWLEDGMENTS
We thank Katie Kirchoff for her work building and maintaining the COVID data mart and Dr Scott Curry for offering access to the curated dataset. We also thank Hamilton Baker, Matthew Case, and Michael Kopscik for their help annotating clinical notes.
COMPETING INTERESTS STATEMENT
The authors have no competing interests to declare.
DATA AVAILABILITY STATEMENT
The data underlying this article cannot be shared publicly due to patient healthcare data privacy protection requirements. The COVID-NLP tool software code is available with an open source license in a GitHub public repository (https://github.com/MUSC-TBIC). The predictive analytics algorithms and Monte Carlo estimation details and markdown code are available in the same GitHub public repository and in this publication online supplementary material.
REFERENCES
- 1.Johns Hopkins University Center for Systems Science and Engineering. COVID-19 dashboard. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed June 1, 2021.
- 2. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020; 323 (11): 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fried M, Crawford J, Mospan A, et al. Patient characteristics and outcomes of 11 721 patients with coronavirus disease 2019 (COVID-19) hospitalized across the United States. Clin Infect Dis 2021; 72 (10): e558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parodi S, Liu V. From containment to mitigation of COVID-19 in the US. JAMA 2020; 323 (15): 1441–2. [DOI] [PubMed] [Google Scholar]
- 5.Zipnosis. https://www.zipnosis.com/our-solution/. Accessed June 1, 2021.
- 6. Ford D, Harvey J, McElligott J, et al. Leveraging health system telehealth and informatics infrastructure to create a continuum of services for COVID-19 screening, testing, and treatment. J Am Med Inform Assoc 2020; 27 (12): 1871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008; 128–44. [PubMed] [Google Scholar]
- 8. Obeid JS, Davis M, Turner M, Meystre SM, Heider P, Lenert L. An AI approach to COVID-19 infection risk assessment in virtual visits: a case report. J Am Med Inform Assoc 2020; 27 (8): 1321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meystre S, Kim Y, Heider P. COVID-19 information extraction rapid deployment using natural language processing and machine learning. In: AMIA NLP WG Pre-Symposium; 2020.
- 10. Meystre SM, Heider P, Kim Y. COVID-19 diagnostic testing prediction using natural language processing to power a data-driven symptom checker. In: AMIA Summits Translational Science Proceedings; Virtual (online); 2021.
- 11. Wang LL, Lo K, Chandrasekhar Y, et al. CORD-19: The COVID-19 open research dataset. arXiv, doi: http://arxiv.org/abs/2004.10706, 10 Jul 2020, preprint: not peer reviewed.
- 12.COVID-19 open research dataset challenge (CORD-19). https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge. Accessed June 1, 2021.
- 13. Verspoor K, Cohen KB, Dredze M, et al. Introduction to the 1st Workshop on Natural Language Processing for COVID-19 at ACL 2020. In: Proceedings of the 1st Workshop NLP COVID-19 ACL 2020; Virtual (online); 2020.
- 14. Kruspe A, Haberle M, Kuhn I, Zhu XX. Cross-language sentiment analysis of European Twitter messages during the COVID-19 pandemic. arXiv, doi: https://arxiv.org/abs/2008.12172, 27 Aug 2020, preprint: not peer reviewed.
- 15. Serrano JCM, Papakyriakopoulos O, Hegelich S. NLP-based feature extraction for the detection of COVID-19 misinformation videos on YouTube. 2020. https://openreview.net/pdf?id=M4wgkxaPcyj. Accessed June 1, 2021.
- 16. Wang J, Pham HA, Manion F, Rouhizadeh M, Zhang Y. COVID-19 SignSym: A fast adaptation of general clinical NLP tools to identify and normalize COVID-19 signs and symptoms to OMOP common data model. arXiv, doi: https://arxiv.org/pdf/2007.10286.pdf, 13 Jul 2020, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 17. Chapman AB, Peterson KS, Turano A, Box TL, Wallace KS, Jones M. A natural language processing system for national COVID-19 surveillance in the US Department of Veterans Affairs. In: Proceedings of the 1st Workshop NLP COVID-19 ACL 2020; Virtual (online); 2020
- 18. Lybarger K, Ostendorf M, Thompson M, Yetisgen M. Extracting COVID-19 diagnoses and symptoms from clinical text: a new annotated corpus and neural event extraction framework. In: AMIA NLP WG Pre-Symposium; Virtual (online); 2020: 10. [DOI] [PMC free article] [PubMed]
- 19. Challener DW, Dowdy SC, O'Horo JC. Analytics and prediction modeling during the COVID-19 pandemic. Mayo Clin Proc 2020; 95 (9S): S8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iwendi C, Bashir AK, Peshkar A, et al. COVID-19 patient health prediction using boosted random forest algorithm. Front Public Health 2020; 8: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fokas AS, Dikaios N, Kastis GA. Mathematical models and deep learning for predicting the number of individuals reported to be infected with SARS-CoV-2. J R Soc Interface 2020; 17 (169): 20200494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jewell N, Lewnard J, Jewell B. Predictive mathematical models of the COVID-19 pandemic: underlying principles and value of projections. JAMA 2020; 323 (19): 1893–4. [DOI] [PubMed] [Google Scholar]
- 23. Vaid A, Somani S, Russak AJ, et al. Machine learning to predict mortality and critical events in a cohort of patients with COVID-19 in New York City: model development and validation. J Med Internet Res 2020; 22 (11): e24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joshi RP, Pejaver V, Hammarlund NE, et al. A predictive tool for identification of SARS-CoV-2 PCR-negative emergency department patients using routine test results. J Clin Virol 2020; 129: 104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perlman A, Vodonos Zilberg A, Bak P, et al. Characteristics and symptoms of app users seeking COVID-19–related digital health information and remote services: retrospective cohort study. J Med Internet Res 2020; 22 (10): e23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Food and Drug Administration. Pooled sample testing and screening testing for COVID-19. 2020. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/pooled-sample-testing-and-screening-testing-covid-19. Accessed June 1, 2021.
- 27.Centers for Disease Control and Prevention. Interim guidance for use of pooling procedures in SARS-CoV-2 diagnostic, screening, and surveillance testing. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/pooling-procedures.html. Accessed June 1, 2021.
- 28. Lim KL, Johari NA, Wong ST, et al. A novel strategy for community screening of SARS-CoV-2 (COVID-19): sample pooling method. PLoS One 2020;15(8):e0238417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dorfman R. The detection of defective members of large populations. Ann Math Statist 1943; 14 (4): 436–40. [Google Scholar]
- 30. Litvak E, Dentzer S, Pagano M. The right kind of pooled testing for the novel coronavirus: first, do no harm. Am J Public Health 2020; 110 (12): 1772–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apache. UIMA (Unstructured Information Management Architecture). https:// uima.apache.org. 2008. Accessed June 1, 2021.
- 32. Heider P. Lexicon generation tools. https://github.com/MUSC-TBIC/lexicon-tools [Google Scholar]
- 33. Joulin A, Grave E, Bojanowski P, Mikolov T. Bag of tricks for efficient text classification. In: Proceedings of the 15th Conference of the European Chapter of the Association of Computational Linguist Vol 2 Short Papers. Stroudsburg, PA: Association for Computational Linguistics; 2017: 427–31.
- 34. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 17 (5): 514–8. PMID:20819854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inf Assoc 2020; 27 (1): 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reich C, Ryan P, Belenkaya R, Natarajan K, Blacketer C. OMOP Common Data Model. https://github.com/OHDSI/CommonDataModel/wiki. Accessed June 1, 2021.
- 38. Meystre S. DECOVRI (Data extraction for COVID-19 related information). 2021. https://github.com/MUSC-TBIC/decovri. Accessed June 1, 2021. [DOI] [PubMed]
- 39. de Castilho RE, Mujdricza-Maydt E, Yimam SM, et al. A web-based tool for the integrated annotation of semantic and syntactic structures. In: Proceedings of the Workshop on Language Technology Resources and Tools for Digital Humanities; 2016: 76–84.
- 40. Heider P, Accetta J-K, Meystre SM. ETUDE for Easy and Efficient NLP Application Evaluation. In: AMIA NLP-WG Pre-Symposium; San Francisco, CA; 2018.
- 41. Joulin A, Grave E, Bojanowski P, Mikolov T. Bag of tricks for efficient text classification. arXiv, doi: http://arxiv.org/abs/1607.01759, 9 Aug 2016, preprint: not peer reviewed.
- 42. Kent DM, Paulus JK, Sharp RR, Hajizadeh N. When predictions are used to allocate scarce health care resources: three considerations for models in the era of Covid-19. Diagn Progn Res 2020; 4 (1): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Article addenda for natural language processing enabling COVID-19 predictive analytics to support data-driven patient advising and pooled testing. 2021. https://github.com/MUSC-TBIC/article-addenda/tree/stable/Meystre-etal_2021_NLP-Enabling-COVID-19-Predictive-Analytics. Accessed June 1, 2021. [DOI] [PMC free article] [PubMed]
- 44.MUSC COVID-19 symptom checker. 2020. https://musc-covid19-symptom-risk.azurewebsites.net. Accessed June 1, 2021.
- 45. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.HL7. Welcome to FHIR. https://www.hl7.org/fhir/. Accessed June 1, 2021.
- 47.Infor. Cloverleaf integration suite. https://www.infor.com/products/cloverleaf. Accessed June 1, 2021.
- 48. Kim Y, Meystre SM. Ensemble method-based extraction of medication and related information from clinical texts. J Am Med Inform Assoc 2020; 27 (1): 31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dreyer NA, Reynolds M, DeFilippo Mack C, et al. Self-reported symptoms from exposure to Covid-19 provide support to clinical diagnosis, triage and prognosis: An exploratory analysis. Travel Med Infect Dis 2020; 38: 101909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Struyf T, Deeks JJ, Dinnes J, et al. ; Cochrane COVID-19 Diagnostic Test Accuracy Group. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 disease. Cochrane Database Syst Rev 2020; 7: CD013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to patient healthcare data privacy protection requirements. The COVID-NLP tool software code is available with an open source license in a GitHub public repository (https://github.com/MUSC-TBIC). The predictive analytics algorithms and Monte Carlo estimation details and markdown code are available in the same GitHub public repository and in this publication online supplementary material.