Abstract
Objective
We conducted a systematic review to assess the effect of natural language processing (NLP) systems in improving the accuracy and efficiency of eligibility prescreening during the clinical research recruitment process.
Materials and Methods
Guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standards of quality for reporting systematic reviews, a protocol for study eligibility was developed a priori and registered in the PROSPERO database. Using predetermined inclusion criteria, studies published from database inception through February 2021 were identified from 5 databases. The Joanna Briggs Institute Critical Appraisal Checklist for Quasi-experimental Studies was adapted to determine the study quality and the risk of bias of the included articles.
Results
Eleven studies representing 8 unique NLP systems met the inclusion criteria. These studies demonstrated moderate study quality and exhibited heterogeneity in the study design, setting, and intervention type. All 11 studies evaluated the NLP system’s performance for identifying eligible participants; 7 studies evaluated the system’s impact on time efficiency; 4 studies evaluated the system’s impact on workload; and 2 studies evaluated the system’s impact on recruitment.
Discussion
NLP systems in clinical research eligibility prescreening are an understudied but promising field that requires further research to assess its impact on real-world adoption. Future studies should be centered on continuing to develop and evaluate relevant NLP systems to improve enrollment into clinical studies.
Conclusion
Understanding the role of NLP systems in improving eligibility prescreening is critical to the advancement of clinical research recruitment.
Keywords: natural language processing, clinical trial matching, clinical research, eligibility prescreening, cohort identification
INTRODUCTION
Clinical research is crucial for knowledge generation and the development of empirical evidence to improve health outcomes.1 However, clinical research requires robust effort to ensure the enrollment of adequate numbers of study participants needed to reach the study’s statistical power to ensure significant findings.2–4 A major bottleneck in clinical research recruitment is eligibility prescreening—a costly, time-consuming, and inefficient process where the clinical research staff manually reviews the patients’ medical history for demographics and clinical conditions, collating and matching the information, and identifying candidates based on the protocol’s eligibility criteria.5 The goal of eligibility prescreening is to identify all eligible participants and minimize the risk of misclassifying an eligible participant as ineligible.6 Hence, there is a growing effort to advance the automation of eligibility prescreening in clinical research by developing tools to extract structured (eg, demographics, medication list) and unstructured data (eg, narrative clinical notes, imaging results) from the electronic health record (EHR) to identify eligible patients.7–15 However, the inability to extract relevant information from unstructured data poses a significant limitation to accurately identify eligible patients for clinical research studies.16
One way to automate this process is through natural language processing (NLP), which is a subfield of artificial intelligence that enables the computer “to parse, segment, extract, or analyze text data” using a collection of processing algorithms based on statistics, syntactic, and/or semantic rules.17,18 In clinical settings, NLP systems leverage the information available in the EHR to provide data for specialized applications (eg, clinical decision support systems).19 In clinical research eligibility prescreening, an NLP system can extract relevant information from structured and unstructured data types essential to determine whether the patient is potentially eligible to participate in a research protocol.20 The use of NLP to identify eligible patients can prompt investigators when trials are available for patients.21 Despite the potential for NLP systems to enhance the eligibility prescreening process, these approaches are usually experimental, and their efficacy in clinical practice remains inconclusive.10,22,23 The existing evidence on the effect of informatics-driven tools such as NLP systems on clinical research recruitment is focused on computer-aided eligibility prescreening and recruitment systems rather than specifically looking at NLP systems.24,25 It is unclear whether NLP systems in clinical settings improve the recruitment process, as measured through accuracy and efficiency in identifying and enrolling eligible patients.6
Objective
The purpose of this study is to systematically review the literature on the use of NLP systems for eligibility prescreening in clinical research and its impact on the recruitment process. We will report the following aspects of the included studies: (1) study purpose and characteristics, (2) patient cohort and disease domain of the research protocols for eligibility prescreening, (3) NLP system task and evaluation, and (4) effect of NLP system on clinical research recruitment process outcomes (ie, accuracy in eligibility determination, workload efficiency, and impact on recruitment).
MATERIALS AND METHODS
Guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standards of quality for reporting systematic reviews,26 a protocol for study eligibility was developed a priori and registered in the PROSPERO database of systematic reviews (https://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=215071; registration number: CRD42020215071).
Search strategy
We searched 5 databases (PubMed, Embase, Cumulative Index of Nursing and Allied Health Literature [CINAHL], Institute of Electrical and Electronics Engineers [IEEE] Xplore and Association for Computing Machinery [ACM]) to identify studies where an NLP system was used for clinical research eligibility prescreening. The search strategy27 is detailed in Supplementary Materials Tables S1 and S2. We included the term “text mining” to broadly capture systems that extract information from free or unstructured clinical text.28 The search included studies from database inception through February 2021. We broadly defined eligibility prescreening as the process of determining a potential participant’s eligibility to participate in clinical research based on the information in the medical records.6,9,16,29 Inclusion criteria were: (1) quantitative study designs evaluating an NLP system for eligibility prescreening for any disease-specific clinical research (eg, drug trials, observational studies) that includes human participants, (2) eligibility prescreening conducted in an electronic clinical data source (eg, EHR, registry), (3) studies with objective outcomes and measures on the performance of the NLP system, (4) studies comparing the NLP system with a non-NLP approach in eligibility prescreening (ie, studies comparing an NLP system to another NLP system or algorithm were excluded), and (5) full-text article is available. There were no restrictions on searches by publication date, but only English language or those translated to the English language were included. We excluded studies that: (1) evaluated NLP systems intended to identify eligible participants for purposes other than research (eg, clinical procedure), (2) evaluated NLP systems used for different aspects of clinical research other than eligibility prescreening (eg, data collection, analysis), (3) evaluated an algorithm instead of a system because, though NLP is an algorithm, a system is required for a user to carry out the NLP tasks in a real-world setting,30 (4) do not involve patient data to evaluate the NLP system, and (5) involved patient simulations.
Screening, abstraction, appraisal, and analysis
The references of all eligible studies were imported into EndNote X9 (Clarivate Analytics, Philadelphia, PA, USA) and then deduplicated using the Bramer method.31 Two reviewers (BI and CD) independently screened all articles by title and abstract using Covidence (https://www.covidence.org); the reason for exclusion was documented. With discussion to provide consensus, the same reviewers independently assessed all potentially relevant articles in the full-text review to comprehensively determine eligibility for inclusion and searched the reference lists; relevant articles were included for full-text review. The third reviewer (RS) helped resolve any discrepancies. The data collection form32 was piloted by BI and CD and used to manually extract the variables of interest for data synthesis from each included article. These included authors, year of publication, study geographic location, study aim and design, funding source, clinical research study type and disease focus, size and source of the dataset, NLP system characteristics (ie, NLP tasks, terminologies, or ontologies used), comparator, outcomes, and results. The Joanna Briggs Institute Critical Appraisal Checklist for Quasi-experimental Studies33 were adapted to assess the study quality and the risk of bias of the included articles; a question regarding the authors’ conflict of interest was added. Studies earned 1 point for each component that was met; the overall score indicated the quality of the study as low (1–4), moderate (5–7), and high (8–10).34 All studies were described qualitatively. The outcomes of interest (ie, accuracy in eligibility determination, workload efficiency, and impact on recruitment) were described and synthesized by its effect on the clinical research recruitment process.14
RESULTS
Search results
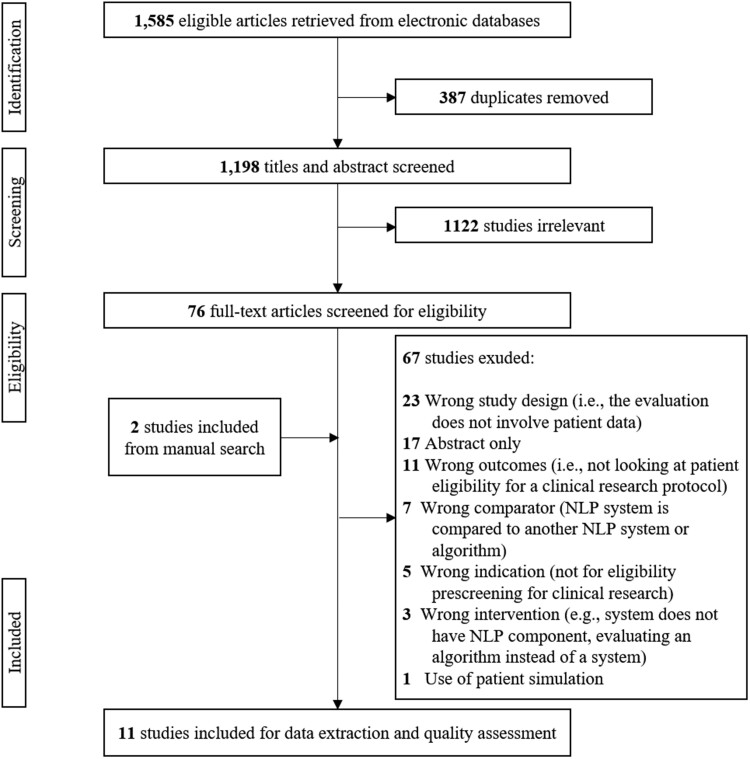
Among the 1585 articles the initial literature search yielded, 1198 studies were included for the title and abstract screening after deduplication. Seventy-six articles were included for full-text review; 2 additional studies were added through manual search. Eleven studies were eligible and were included for data extraction and quality assessment. The reasons for excluding 67 studies are listed in Figure 1. Three studies were excluded due to wrong intervention because the authors compared criteria classifiers for a chatbot,35 query formalism using structured data,36 and classifiers for eligibility criterion.37 Seven studies were excluded because the NLP system being evaluated is compared to another NLP system or algorithm.7,38–43
Figure 1.
PRISMA flowchart for literature search. PRISMA: Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
Description of studies
Supplementary Material Table S3 summarizes the characteristics and findings of the 11 included studies. Eight were retrospective, one-group pretest-posttest design studies comparing the effect of the NLP system on the eligibility prescreening to manual eligibility prescreening of the same cohort of patients.44–51 Three were prospective studies; 2 were posttest-only design where the cohort of patients was prescreened using an NLP system, then the results were manually reviewed to confirm the eligibility determination of the patients who were deemed eligible52,53 and one was a nonequivalent group design study where the recruitment outcome of the prospective cohort of patients was compared to the outcome of the historical controls.54 Eight studies were conducted in the United States, and the other 3 studies were conducted in Australia, the Netherlands, and the United Kingdom.
Of the 11 studies included, 2 studies evaluated the same NLP system, the International Business Machines (IBM) Watson Clinical Trial Matching44,45; 3 studies evaluated the Automated Clinical Trial Eligibility Screener (ACTES), unnamed in the first 2 studies48,49,54; 1 study evaluated CogStack50; 1 study evaluated Trial Prospector53; and 4 studies evaluated unnamed NLP systems.46,47,51,52 All NLP systems in the included studies extracted patient information from the EHR. Two studies also extracted patient information from study databases.44,45 In addition to patient information extraction, 6 studies used the NLP system to also extract eligibility criteria from research protocols in the institution’s protocol library,46–48 ClinicalTrials.gov,49 or both.44,45
All studies reported the manual review and eligibility determination of a clinician and/or clinical research staff as the gold standard comparator to the NLP system. Five studies also included a historical study log to compliment the gold standard.47,49–51,54 Though there are 6 studies that involve oncology research, only one study used a specialized ontology from the National Cancer Institute Thesaurus for oncology terms.53 Four studies did not mention the terminologies or ontologies used for knowledge representation.44,45,51,52 The disease domain of the varied, and included oncology, pediatric emergency care, cardiology, and critical care. The sample definition varied; 4 studies analyzed the NLP system performance by the patient encounter,48,52–54 and the others analyzed it by unique patient case. Three NLP systems were used to prescreen for one study protocol46,50,51; the remaining were used to prescreen multiple study protocols. A total of 69 research protocols were included in all 11 studies.
Quality assessment
Table 1 details the quality assessment ratings for each included study and Table 2 provides a summary of the quality assessment ratings of all the included studies. Three studies were rated low quality primarily because the gold standard varied across research protocols for prescreening, and the evaluation was done only on the eligible patients the NLP system identified. The majority (n = 6, 54.5%) of the studies only measured the outcome after the intervention (ie, the accuracy of the gold standard was not measured). Only 2 studies performed multiple measurements of the outcome before and after the intervention. A follow-up visit is only applicable to one study involving time-motion study and pre- and postusability survey54; the study was unclear if the follow-up was complete because although the sample size was clearly stated in their methods section, there was no mention of loss to follow-up and the sample size in the “Results” section. Though all studies compared the outcomes of the eligibility prescreening on the same cohort of potential participants before and after using an NLP system, the outcomes of 6 studies were not measured reliably due to unclear eligibility classification and lack of information on the historical study log. Nine studies conducted appropriate statistical analysis; one study only looked at the yield,52 and though it investigated the false positives of the output by manual review, it did not evaluate the patients the system classified as ineligible. Majority of the studies exhibited moderate quality (n = 7, 64%). Overall, with a mean score of 5.3, the included studies demonstrated moderate quality.
Table 1.
Detailed quality assessment ratings of the included studies
| Criteria | Penberthy et al52 | Sahoo et al53 | Ni et al48 | Ni et al49 | Jonnalagadda et al46 | Meystre et al47 |
|---|---|---|---|---|---|---|
| Clear cause (ie, intervention of interest) and effect (ie, outcome of interest) | Yes. | Yes. | Yes. | Yes. | Yes. | Yes. |
| Potential participants included in any comparisons are similar | Yes. | Yes. | Yes. | Yes. | Yes. | Yes. |
| Single group. | Single group. | Single group. | Single group. | Single group. | Single group. | |
| Potential participants included in any comparisons are receiving similar intervention, other than the exposure or intervention of interest | No. | No. | Yes. | No. | Yes. | Unclear. |
| There is variety in studies, time, and clinical research staff as gold standard. | Different clinicians for the comparator. | The potential participants are from the same clinical practice. | The oncologist only reviewed a subset of patients. | The potential participants are from the same clinical practice. | The historical prescreening process as gold standard was not described; different staff may be involved in the determination. | |
| Has control group | No. | No. | No. | No. | No. | No. |
| Has multiple measurements of the outcome both pre- and post-the intervention or exposure | No. | No. | Unclear. | No. | No. | Unclear. |
| Only post. | Only post. | Inter-rater agreement for the gold standard was mentioned but not reported. | Only post. | Only post. | Inter-rater agreement evaluation was only for eligibility criteria | |
| Complete follow-up | N/A. | N/A. | N/A. | N/A. | N/A. | N/A. |
| Outcomes of eligibility prescreening included in any comparisons measured in the same way | Yes. | Unclear. | Yes. | Yes. | No. | Yes. |
| All outcomes for the protocols were reported. | Unclear if there are different physicians across trials. | All outcomes for the protocols were reported. | All outcomes for the protocols were reported. | Potential participants were not manually reviewed before NLP system prescreening; only after. | All outcomes for the protocols were reported. | |
| Outcomes were measured in a reliable way | No. | No. | Yes. | Yes. | No. | Unclear. |
| Research staff (gold standard) vary by study. | Clinicians (gold standard) vary by study. | Gold standard was complimented by the historical study log. | Domain expert verified NLP system output. | No inter-rater agreement evaluation. | No information on the historical prescreening. | |
| Appropriate statistical analysis was used | No. | Unclear. | Yes. | Yes. | Yes. | Yes. |
| Only evaluated the yield. | Only evaluated the accuracy of the potentially eligible patients. | |||||
| Potential conflict of interest disclosure | No. | Yes. | Yes. | Yes. | Yes. | Yes. |
| SCORE | 3 | 3 | 7 | 6 | 5 | 5 |
| Criteria | Ni et al54 | Alexander et al44 | Beck et al45 | Tissot et al50 | van Dijk et al51 |
|---|---|---|---|---|---|
| Clear cause (ie, intervention of interest) and effect (ie, outcome of interest) | Yes. | Yes. | Yes. | Yes. | Yes. |
| Potential participants included in any comparisons are similar | No. | Yes. | Yes. | Yes. | Yes. |
| Unclear if the historical control was prescreened for the same studies in the weekly aggregation. | Single group. | Single group. | Single group. | Single group. | |
| Potential participants included in any comparisons are receiving similar intervention, other than the exposure or intervention of interest | Unclear. | Yes. | Yes. | No. | No. |
| No information on the comparator historical patients. | The potential participants are from the same observational cohort study. | The potential participants are from the same clinical practice. | Some of the participants were not screened in the original trial. | Different EHR systems were used. | |
| Has control group | Unclear. | No. | No. | No. | No. |
| They have historical controls. | |||||
| Has multiple measurements of the outcome both pre- and post-the intervention or exposure | Yes. | Yes. | No. | No. | Unclear. |
| For time-motion study and usability study. | Inter-rater agreement for the gold standard before the intervention. | Only post. | Only post. | There may be difference in how each clinic prescreened patients. | |
| Complete follow-up | Unclear. | N/A. | N/A. | N/A. | N/A. |
| Participants sample size in the result not mentioned. | |||||
| Outcomes of eligibility prescreening included in any comparisons measured in the same way | Yes. | Yes. | Yes. | Yes. | Yes. |
| All outcomes for the protocols were reported. | All outcomes for the protocols were reported. | All outcomes for the protocols were reported. | All outcomes for the protocols were reported. | All outcomes for the protocols were reported. | |
| Outcomes were measured in a reliable way | No. | Yes. | Yes. | No. | Yes. |
| Lack of information regarding the historical control. | Authors explained why one study has low inter-rater agreement. | Manual reviewers were blinded to the results during manual review and errors were reported. | Only 20 of the 173 patients who were not screened in the gold standard were manually reviewed. | Further investigation was made on the participants that were missed by the system | |
| Appropriate statistical analysis was used | Yes. | Yes. | Yes. | Yes. | Yes. |
| Potential conflict of interest disclosure | Yes. | Yes. | Yes. | Unclear. | Yes. |
| SCORE | 5 | 8 | 7 | 4 | 6 |
EHR: electronic health record; NLP: natural language processing.
Table 2.
Quality assessment of included studies
| Characteristics | Number of studies (%)a | |
|---|---|---|
| Clear cause (ie, intervention of interest) and effect (ie, outcome of interest) | Yes | 11 (100) |
| No | 0 (0) | |
| Unclear | 0 (0) | |
| Potential participants included in any comparisons are similar | Yes | 10 (91) |
| No | 1 (9) | |
| Unclear | 0 (0) | |
| Potential participants included in any comparisons are receiving similar intervention, other than the exposure or intervention of interest | Yes | 4 (36) |
| No | 5 (46) | |
| Unclear | 2 (18) | |
| Has control group | Yes | 0 (0) |
| No | 10 (91) | |
| Unclear | 1 (9) | |
| Has multiple measurements of the outcome both pre- and post-the intervention or exposure | Yes | 2 (18) |
| No | 6 (55) | |
| Unclear | 3 (27) | |
| Complete follow-up or if incomplete, the differences between groups in terms of their follow-up were adequately described and analyzed | Yes | 0 (0) |
| No | 0 (0) | |
| Unclear | 1 (9) | |
| NA | 10 (91) | |
| Outcomes of eligibility prescreening included in any comparisons measured in the same way | Yes | 9 (82) |
| No | 1 (9) | |
| Unclear | 1 (9) | |
| Outcomes were measured in a reliable way | Yes | 5 (45.5) |
| No | 5 (45.5) | |
| Unclear | 1 (9) | |
| Appropriate statistical analysis was used | Yes | 9 (82) |
| No | 1 (9) | |
| Unclear | 1 (9) | |
| Authors disclose the presence or absence of potential conflict of interest | Yes | 9 (82) |
| No | 1 (9) | |
| Unclear | 1 (9) | |
| Study quality rating | Low (1–4) | 3 (27) |
| Moderate (5–7) | 7 (64) | |
| High (8–10) | 1 (9) | |
NA: not applicable.
Out of 11 included studies.
Outcomes
There was heterogeneity in the results and outcome measures of the included studies. Metrics used to evaluate the accuracy of the eligibility determination varied across studies. There was also variability in skills and training on the domain experts as gold standard to compare the impact of an NLP system on time and workload efficiency. Lastly, 2 studies investigated the impact of an NLP system to study recruitment.
Eligibility determination of patient cohort
Although all studies included eligibility prescreening of potential participants, one study did not measure the accuracy of the NLP system’s classification of the patients’ eligibility.54 One study measured the accuracy of the NLP system to determine eligibility by calculating the yield, defined as (1-false positive); where false positive is the “number determined as not eligible on review by research staff divided by the number determined as eligible through the automated screening.”52 Two studies measured the outcome with mean average precision (MAP); one of the studies calculated MAP per research protocol and demonstrated an MAP of 18–35.2% across 3 research protocols,47 while the other study demonstrated a combined MAP of 62.9% for all 13 research protocols.48 Recall (or sensitivity) was reported in 8 studies and ranged from 70.6% to 100%, which collectively represents a total of 43 research protocols. Five studies, collectively including 23 research protocols, reported precision (or positive predictive value) that ranges from 21.6% to 92.8%; of these, 4 studies (a total of 22 research protocols) reported a negative predictive value ranging from 93.7% to 100%. Six studies, with a total of 27 research protocols, reported the NLP system’s specificity ranging from 76% to 99%. Of the 5 studies that measured precision and recall, 3 studies reported an F1-score, which were 67.9%, 86.3%, and 90%. The area under the curve was reported in one study, which was 75.5–83.7 for 3 research protocols. Lastly, 4 studies reported accuracy ranging from 81% to 100% for a total of 30 research protocols. Due to the different outcome measures and variability of the effect, summarizing the magnitude of performance evaluation of the NLP systems in clinical research eligibility prescreening would not be appropriate.
Impact on time and workload efficiency
In the studies that showed benefit to the time and workload needed to identify eligible participants, the outcome measures varied, including the comparison (eg, historical study log, current gold standard) and domain expert used for a gold standard. Hence, a more in-depth summary of the efficiency benefit in terms of time and workload cannot be accurately described. Supplementary Material Table S3 provides further details about the time and workload efficiency outcomes and effects. A study reported that the NLP system took less than 2 min to prescreen 198 patients, while the experienced research coordinator spent 2 weeks (∼80 h) to complete the manual prescreening for one research protocol.46 One study reported a statistically significant reduction of patient screening time by 34% by the research staff using an NLP system compared to manual prescreening, where time was reallocated to work-related activities such as: (1) waiting for sample collection (P = .03), (2) study-related administrative tasks (P = .03), and (3) work-related conversations (P = .006).54 One study looked at efficiency in terms of when the potential participant was identified based on the historical study log; this study showed that out of 203 patients that were historically deemed eligible, 47% (95% CI, 40%, 54%) of the patients were identified by NLP system on the same day as when they were screened; 24% (95% CI, 18%, 30%) were identified a day early, and 23% (95% CI, 17%, 29%) were identified 2 days earlier.39 Lastly, in one study, manual prescreening of 90 patients for 3 research protocols took 110 min; NLP system-assisted eligibility prescreening of the same patients for the same research protocols required 24 min, demonstrating a reduction in the time for screening by 78%.45
Four of the 11 studies measured workload as an outcome, where the workload is defined as the number of patients required to be reviewed to identify all eligible patients.48–51 The workload reduction of the 4 studies ranged from 79.9% to 92%; 2 studies reported statistical significance (P = 1.00E−9 and 8.30E−21). One study reported that, on average, an oncologist would need to manually review 163 patients per research protocol to replicate the historical patient enrollment for each of the 10 research protocols; but with the NLP system, this was reduced to 24 patients per research protocol.49
Impact on recruitment
Two studies investigated the impact of an NLP system on enrollment was evaluated on 6 research protocols used the ACTES and improved the number of patients screened by 11% compared to the historical study log, though only 4 research protocols demonstrated statistical significance.54 It also showed that there was an 11.1% improvement in recruiting eligible patients with the use of the NLP system; however, only one research protocol demonstrated statistical significance. In the time-motion study of the same study, though the NLP system demonstrated that the reduction in prescreening time was redirected to other work-related activities, it did not show significance in the patient contact to introduce the study. Lastly, the enrollment rate of clinicians using Trial Prospector was compared to those who did not and found that there was no difference.53
DISCUSSION
This systematic review provides a detailed overview of the existing studies conducted to examine the effect of NLP systems for clinical research eligibility prescreening. All included studies described the performance of the NLP system in determining the eligibility of a cohort of patients; however, only 9 described the potential benefit of the system in terms of time and workload efficiency for the research team, and only 2 investigated the system’s effectiveness on study enrollment. The limited number of available studies and the variability in outcome measures suggest that this is an understudied area and the effect of an NLP system on the research recruitment process is unclear.
Many studies were excluded that described an NLP system but did not evaluate them in the context of clinical research eligibility prescreening. The initial literature search identified studies where different NLP algorithms were compared that could be used to develop a system for clinical research eligibility prescreening.22,38 It may therefore be that these systems have not yet been developed into completely testable NLP systems, or there may be other obstacles to the evaluation.
Though this systematic review revealed that the NLP system could reach a recall as high as 100%; 2 studies, focusing on breast and lung cancer, demonstrated a recall ranging from 70.6% to 83.3%, which is mainly due to incorrect representation of eligibility criteria and ambiguity in the abbreviations used in the narrative text.44,47 With the goal of minimizing the risk of misclassifying an eligible participant as ineligible during eligibility prescreening, it is better to classify an ineligible participant as eligible and have a clinical research staff further assess the potential eligibility.6 It is imperative for the research team to consider how the rest of the eligible participants can be identified. This may include broadening the eligibility criteria for prescreening to widen the capture of potentially eligible patients and have domain experts review the result.
This systematic review also revealed the potential for NLP systems’ promising success in excluding patients who did not meet the eligibility criteria. Achieving an NPV of 100%, an NLP system demonstrated that the patients who were classified as ineligible are truly ineligible. Even though the system may misclassify ineligible patients as eligible, the system is not filtering out eligible patients because of misclassifying them as ineligible. Notably, research staff would need to ascertain that the patients identified by the system are eligible before recruitment. However, depending on the number of patients to be prescreened and/or protocols for prescreening, clinical research staff availability, and study design, research teams may opt for a system that is more precise in identifying eligible patients and redirect effort of the research staff to recruitment.
Optimizing recruitment process with an NLP system
All 11 studies acknowledged that the time and effort needed for the research staff to manually go through complex medical records to identify patients who are eligible for a research study impedes recruitment into research studies.55 Seven studies assessed the impact of the NLP system in the time spent for eligibility prescreening reported a substantial decrease in time. Workload reduction when using an NLP system was significantly high, and it can potentially increase as the number of protocols and/or patients increase. Further, the domain experts for the gold standard in these studies vary in their expertise (eg, research coordinator, physician). Though the findings are promising, it is important to note that NLP systems are embedded in a social-organizational environment, and their effects can vary in different settings and users.56 Assessing the adoption of the NLP systems in real-world clinical research settings is crucial.57 It is also recognized that the integration of the system into the EHR could potentially improve the research team’s efficiency in their recruitment effort. It is unclear that the time and effort saved in eligibility prescreening translates to increased enrollment.
As mentioned, the NLP systems in the included studies were used to extract patient information from the clinical data source and eligibility criteria from the research protocol. One common problem identified in both information extraction tasks is ambiguity. For the patient information extraction, imperfection of the NLP system in understanding language semantics (eg, inability to recognize “ started four days ago” for a criterion indicating “more than 72 hours”) and syntax (eg, abbreviations in the clinical text) cause false-positive recommendations.44,49 This also poses a challenge in recognizing the temporality of the eligibility criteria during the prescreening. Further, the limitations of the EHR, such as lack of information, can extend its impact on the utilization of an NLP system.16,45 For eligibility criteria extraction, the underspecified requirements in the criteria (eg, not defining “severe” in “severe disease”) or the specific requirement of information not readily available in the EHR such as patient’s living situation (eg, “ study partner availability”) warrants validation from the research team.46 The 3 studies that used the NLP system to extract eligibility criteria from the research protocol did not evaluate the accuracy of the system’s retrieval of eligibility criteria due to the needed human intervention in ensuring that the criteria were accurate to measure the study outcome (ie, eligibility determination). The NLP systems of the other 7 studies require manual entry or an expert-generated algorithm of the eligibility criteria. The capability of an NLP system to extract eligibility criteria warrants further investigation because NLP-driven tools developed for this task show promising results of accuracy.41,58
Limitations
To the best of our knowledge, this is the first study that has synthesized the effects of NLP systems for clinical research eligibility prescreening. An earlier study reviewed the use of NLP systems for clinical care.59 However, we were unable to conduct a meta-analysis to assess the effectiveness of NLP systems for clinical research eligibility prescreening due to the diversity in outcome measures and the varying methods for evaluating the study design. Second, although the literature search was conducted in multiple databases, it is possible that relevant studies were missed because they may be indexed elsewhere. Grey literature was not included in the literature search, which may have limited the inclusion of pilot studies or studies with negative findings. In addition, the use of retrospective data, which accounted for the bulk of the included studies, may lead to conclusions that differ from the current state of adoption. Finally, studies across a decade, several continents, and various settings were captured, and such variability in time and space complicates the ability of the findings to inform practice in any one clinical research setting.
CONCLUSION
This systematic review highlights the evidence related to NLP systems for clinical research eligibility prescreening. Studies had wide heterogeneity in their study setting, design, and outcome measures. Despite limitations, 3 considerations in the evaluation and adoption of NLP systems for clinical research eligibility prescreening were identified: (1) study design and magnitude of workload are important factors in deciding on the preferred comprehensiveness of the system, (2) the role of the NLP system is to optimize the recruitment process, not to replace domain expertise, and (3) determination of the eligibility criteria in the prescreening process needs expert validation. The findings underscore the need for real-world evaluation of an NLP system to fully understand its strengths and limitations as it is adopted by the research team for eligibility prescreening. Future research should focus on continuing to develop applicable NLP systems that ultimately impact clinical enrollment outcomes. Understanding the role of NLP systems in improving eligibility prescreening is critical to the advancement of recruitment effort optimization in clinical research.
FUNDING
Research reported in this publication was supported by the National Institute of Nursing Research grant numbers T32 NR007969 (PI: Bakken), P30 NR016587 (PI: Bakken), and K24NR018621 (PI: RS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
BI completed the search, deduplicated the retrieved studies, and drafted the manuscript. BI and CD reviewed and screened the searched studies for inclusion, and manually extracted the data; RS helped resolve conflicts to attain consensus. CD, CW, and RS contributed to final edits and approval for publication.
SUPPLEMENTARY MATERIAL
Supplementary materials are available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
References
- 1. Lenfant C. Clinical research to clinical practice—lost in translation? N Engl J Med 2003; 349 (9): 868–74. [DOI] [PubMed] [Google Scholar]
- 2. Carlisle B, Kimmelman J, Ramsay T, et al. Unsuccessful trial accrual and human subjects protections: an empirical analysis of recently closed trials. Clin Trials 2015; 12 (1): 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gul RB, Ali PA. Clinical trials: the challenge of recruitment and retention of participants. J Clin Nurs 2010; 19 (1–2): 227–33. [DOI] [PubMed] [Google Scholar]
- 4. Lamberti MJ, Mathias A, Myles JE, et al. Evaluating the impact of patient recruitment and retention practices. Drug Inf J 2012; 46 (5): 573–80. [Google Scholar]
- 5. Penberthy LT, Dahman BA, Petkov VI, et al. Effort required in eligibility screening for clinical trials. J Oncol Pract 2012; 8 (6): 365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thadani SR, Weng C, Bigger JT, et al. Electronic screening improves efficiency in clinical trial recruitment. J Am Med Inform Assoc 2009; 16 (6): 869–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuggia M, Campillo-Gimenez B, Bouzille G, et al. Automatic selection of clinical trials based on a semantic web approach. Stud Health Technol Inform 2015; 216: 564–8. [PubMed] [Google Scholar]
- 8. Harrer S, Shah P, Antony B, et al. Artificial intelligence for clinical trial design. Trends Pharmacol Sci 2019; 40 (8): 577–91. [DOI] [PubMed] [Google Scholar]
- 9. Weng C. Optimizing clinical research participant selection with informatics. Trends Pharmacol Sci 2015; 36 (11): 706–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vydiswaran VGV, Strayhorn A, Zhao X, et al. Hybrid bag of approaches to characterize selection criteria for cohort identification. J Am Med Inform Assoc 2019; 26 (11): 1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Butte AJ, Weinstein DA, Kohane IS. Enrolling patients into clinical trials faster using RealTime Recruiting. Proc AMIA Symp 2000; 111–5. [PMC free article] [PubMed] [Google Scholar]
- 12. Ding J, Erdal S, Borlawsky T, . et al. The design of a pre-encounter clinical trial screening tool: ASAP. AMIA Annu Symp Proc 2008; 931. [PubMed] [Google Scholar]
- 13. Pressler TR, Yen PY, Ding J, et al. Computational challenges and human factors influencing the design and use of clinical research participant eligibility pre-screening tools. BMC Med Inform Decis Mak 2012; 12: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmickl CN, Li M, Li G, et al. The accuracy and efficiency of electronic screening for recruitment into a clinical trial on COPD. Respir Med 2011; 105 (10): 1501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Treweek S, Pearson E, Smith N, et al. Desktop software to identify patients eligible for recruitment into a clinical trial: using SARMA to recruit to the ROAD feasibility trial. Inform Prim Care 2010; 18 (1): 51–8. [DOI] [PubMed] [Google Scholar]
- 16. Butler A, Wei W, Yuan C, et al. The data gap in the EHR for clinical research eligibility screening. AMIA Jt Summits Transl Sci Proc 2018; 2017: 320–9. [PMC free article] [PubMed] [Google Scholar]
- 17. Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011; 18 (5): 544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dreisbach C, Koleck TA, Bourne PE, et al. A systematic review of natural language processing and text mining of symptoms from electronic patient-authored text data. Int J Med Inform 2019; 125: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000; 270–4. [PMC free article] [PubMed] [Google Scholar]
- 20. Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995; 122 (9): 681–8. [DOI] [PubMed] [Google Scholar]
- 21. Fogel DB. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp Clin Trials Commun 2018; 11: 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Gu Y, Ji X, et al. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J Am Med Inform Assoc 2019; 26 (11): 1218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu ST, Sohn S, Ravikumar KE, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol 2013; 111 (5): 364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cuggia M, Besana P, Glasspool D. Comparing semi-automatic systems for recruitment of patients to clinical trials. Int J Med Inform 2011; 80 (6): 371–88. [DOI] [PubMed] [Google Scholar]
- 25. Köpcke F, Prokosch HU. Employing computers for the recruitment into clinical trials: a comprehensive systematic review. J Med Internet Res 2014; 16 (7): e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moher D, Liberati A, Tetzlaff J, et al. ; PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med 2009; 6 (7): e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richardson WS, Wilson MC, Nishikawa J, et al. The well-built clinical question: a key to evidence-based decisions. ACP J Club 1995; 123 (3): A12–3. [PubMed] [Google Scholar]
- 28. Kao A, Poteet SR. Natural Language Processing and Text Mining. London: Springer-Verlag London Limited; 2007. [Google Scholar]
- 29. McCaffrey N, Fazekas B, Cutri N, et al. How accurately do consecutive cohort audits predict phase III multisite clinical trial recruitment in palliative care? J Pain Symptom Manage 2016; 51 (4): 748–55. [DOI] [PubMed] [Google Scholar]
- 30. Jones K, Galliers J. Evaluating Natural Language Processing Systems: An Analysis and Review. Berlin, Heidelberg: Springer-Verlag; 1995. [Google Scholar]
- 31. Bramer WM, Giustini D, de Jonge GB, et al. De-duplication of database search results for systematic reviews in EndNote. J Med Libr Assoc 2016; 104 (3): 240–3. Erratum in: J Med Libr Assoc 2017; 105 (1): 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. 2nd ed. Chichester: Wiley-Blackwell; 2019: 703. [Google Scholar]
- 33. Porritt K, Gomersall J, Lockwood C. JBI’s systematic reviews: study selection and critical appraisal. Am J Nurs 2014; 114 (6): 47–52. [DOI] [PubMed] [Google Scholar]
- 34. Beauchemin M, Gradilla M, Baik D, et al. A multi-step usability evaluation of a self-management app to support medication adherence in persons living with HIV. Int J Med Inform 2019; 122: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chuan CH, Morgan S. Creating and evaluating chatbots as eligibility assistants for clinical trials: an active deep learning approach towards user-centered classification. ACM Trans Comput Healthcare 2021; 2 (1): 1–19. [Google Scholar]
- 36. Baader F, Borgwardt S, Forkel W. Patient selection for clinical trials using temporalized ontology-mediated query answering. In: Companion Proceedings of the Web Conference 2018. Lyon: International World Wide Web Conferences Steering Committee; 2018: 1069–74. [Google Scholar]
- 37. Spasic I, Krzeminski D, Corcoran P, et al. Cohort selection for clinical trials from longitudinal patient records: text mining approach. JMIR Med Inform 2019; 7 (4): e15980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hassanzadeh H, Karimi S, Nguyen A. Matching patients to clinical trials using semantically enriched document representation. J Biomed Inform 2020; 105: 103406. [DOI] [PubMed] [Google Scholar]
- 39. Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl Sci Proc 2013; 2013: 149–53. [PMC free article] [PubMed] [Google Scholar]
- 40. Segura-Bedmar I, Raez P. Cohort selection for clinical trials using deep learning models. J Am Med Inform Assoc 2019; 26 (11): 1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang X, Xiao C, Glass LM, et al. DeepEnroll: patient-trial matching with deep embedding and entailment prediction. In: Proceedings of The Web Conference 2020. Taipei: Association for Computing Machinery; 2020: 1029–37. [Google Scholar]
- 42. Xiong Y, Shi X, Chen S, et al. Cohort selection for clinical trials using hierarchical neural network. J Am Med Inform Assoc 2019; 26 (11): 1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao J, Xiao C, Glass LM, et al. COMPOSE: Cross-modal pseudo-siamese network for patient trial matching. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. San Francisco, CA: : Association for Computing Machinery: Virtual Event; 2020: 803–12. [Google Scholar]
- 44. Alexander M, Solomon B, Ball DL, et al. Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients. JAMIA Open 2020; 3 (2): 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beck JT, Rammage M, Jackson GP, et al. Artificial intelligence tool for optimizing eligibility screening for clinical trials in a large community cancer center. JCO Clin Cancer Inform 2020; 4: 50–9. [DOI] [PubMed] [Google Scholar]
- 46. Jonnalagadda SR, Adupa AK, Garg RP, et al. Text mining of the electronic health record: An information extraction approach for automated identification and subphenotyping of HFpEF patients for clinical trials. J Cardiovasc Transl Res 2017; 10 (3): 313–21. [DOI] [PubMed] [Google Scholar]
- 47. Meystre SM, Heider PM, Kim Y, et al. Automatic trial eligibility surveillance based on unstructured clinical data. Int J Med Inform 2019; 129: 13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ni Y, Kennebeck S, Dexheimer JW, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc 2015; 22 (1): 166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak 2015; 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tissot HC, Shah AD, Brealey D, et al. Natural language processing for mimicking clinical trial recruitment in critical care: a semi-automated simulation based on the LeoPARDS trial. IEEE J Biomed Health Inform 2020; 24 (10): 2950–9. [DOI] [PubMed] [Google Scholar]
- 51. van Dijk WB, Fiolet ATL, Schuit E, et al. Text-mining in electronic healthcare records can be used as efficient tool for screening and data collection in cardiovascular trials: a multicenter validation study. J Clin Epidemiol 2021; 132: 97–105. [DOI] [PubMed] [Google Scholar]
- 52. Penberthy L, Brown R, Puma F, et al. Automated matching software for clinical trials eligibility: measuring efficiency and flexibility. Contemp Clin Trials 2010; 31 (3): 207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sahoo SS, Tao S, Parchman A, et al. Trial prospector: matching patients with cancer research studies using an automated and scalable approach. Cancer Inform 2014; 13: 157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ni Y, Bermudez M, Kennebeck S, et al. A real-time automated patient screening system for clinical trials eligibility in an emergency department: design and evaluation. JMIR Med Inform 2019; 7 (3): e14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen L, Grant J, Cheung WY, et al. Screening intervention to identify eligible patients and improve accrual to phase II-IV oncology clinical trials. JOP 2013; 9 (4): e174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Salahshour Rad M, Nilashi M, Mohamed Dahlan H. Information technology adoption: a review of the literature and classification. Univ Access Inf Soc 2018; 17 (2): 361–90. [Google Scholar]
- 57. England I, Stewart D, Walker S. Information technology adoption in health care: when organisations and technology collide. Aust Health Rev 2000; 23 (3): 176–85. [DOI] [PubMed] [Google Scholar]
- 58. Yuan C, Ryan PB, Ta C, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. J Am Med Inform Assoc 2019; 26 (4): 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.