Abstract
We have used high-density DNA microarrays to provide an analysis of gene regulation during the mammalian cell cycle and the role of E2F in this process. Cell cycle analysis was facilitated by a combined examination of gene control in serum-stimulated fibroblasts and cells synchronized at G1/S by hydroxyurea block that were then released to proceed through the cell cycle. The latter approach (G1/S synchronization) is critical for rigorously maintaining cell synchrony for unambiguous analysis of gene regulation in later stages of the cell cycle. Analysis of these samples identified seven distinct clusters of genes that exhibit unique patterns of expression. Genes tend to cluster within these groups based on common function and the time during the cell cycle that the activity is required. Placed in this context, the analysis of genes induced by E2F proteins identified genes or expressed sequence tags not previously described as regulated by E2F proteins; surprisingly, many of these encode proteins known to function during mitosis. A comparison of the E2F-induced genes with the patterns of cell growth-regulated gene expression revealed that virtually all of the E2F-induced genes are found in only two of the cell cycle clusters; one group was regulated at G1/S, and the second group, which included the mitotic activities, was regulated at G2. The activation of the G2 genes suggests a broader role for E2F in the control of both DNA replication and mitotic activities.
Rapid progress has been made in the understanding of regulatory pathways that govern the transition of cells from a quiescent state into a cell cycle. Such studies have highlighted the critical role of the signaling pathway that involves the accumulation of D cyclin/cdk4 activity leading to the phosphorylation of the retinoblastoma protein, which then allows an accumulation of E2F transcription activity (21, 24). A variety of experiments have demonstrated the role of E2F proteins in the control of expression of genes important for DNA replication as well as further cell cycle progression (5, 18). In particular, E2F activity is responsible for the activation of genes encoding DNA replication proteins, enzymes responsible for deoxynucleotide biosynthesis, proteins that assemble to form functional origin complexes, and kinases that are involved in the activation of initiation.
Although much has been learned from these studies of E2F transcription control, important questions remain. For one, the scope of the gene-regulatory control by E2F proteins has not been addressed. In large part, the identification of target genes has followed from the initial studies of the DNA tumor virus oncoproteins, such as adenovirus E1A and simian virus 40 T antigen; previous work demonstrated that these proteins were capable of inducing quiescent cells to enter S phase, and associated with this induction was an activation of various genes encoding DNA replication activities (17). This activity coincides with an ability to inactivate the Rb tumor suppressor protein and thus allow an accumulation of E2F proteins. Analysis of promoters for genes such as DNA polymerase α, thymidine kinase, and others revealed the presence of E2F binding sites that were shown to be critical for the normal control of expression of these genes. As additional DNA replication genes have been identified, including those encoding proteins that recognize and establish a functional origin of replication, the majority have been shown to be targets for E2F control. As such, it now appears that a primary role of the G1 cdk/Rb/E2F pathway is the control of genes that allow cells to enter S phase and begin DNA replication.
Despite these advances, the study of E2F gene control has been incremental, following from preconceived views of the role of the Rb/E2F pathway in cell proliferation. As one approach to better understanding the full extent of gene expression under the control of the Rb/E2F pathway, not influenced by the bias of previous work, we have analyzed the expression of a large number of genes using high-density DNA microarrays. The strength of this approach lies in the ability to assay a very large number of potential targets in an unbiased manner—no presumptions are made about the nature of the pathway(s) that might be affected or regulated by E2F activities. For these experiments, we have made use of Affymetrix GeneChip DNA microarrays that contain murine gene sequences and expressed sequence tags (ESTs) and then assayed the profile of gene expression following expression of E2F proteins in quiescent cells.
At the same time, and to serve as a basis for comparison with the E2F-induced genes, we have also profiled the pattern of gene expression changes that occur as cells are initially stimulated to proliferate as well as when cells cycle in the presence of growth factors. We find that many of the E2F-induced genes are normally regulated at G1/S of the cell cycle, consistent with previous studies. Strikingly, however, we also find that a substantial number of the E2F-induced genes are normally regulated at G2 of the cell cycle, suggesting a role for E2F activity in initiating a cascade of gene control during the cell cycle.
MATERIALS AND METHODS
Cells and viruses.
The mouse embryo fibroblast (MEF) cell line 2r15 was established from a wild-type 13.5-day embryo essentially as described (20). MEFs were grown in Dulbecco's modified Eagle's medium (DMEM) containing 15% fetal bovine serum (FBS). To bring cells to quiescence for the serum stimulation experiment, nearly confluent cells were split 1:5 and incubated overnight in DMEM containing 15% FBS. The medium was replaced with DMEM containing 0.2% FBS, and the cells were cultured for 30 h. These quiescent cells were stimulated by adding FBS at the final concentration of 15%. To bring cells to quiescence for the hydroxyurea (HU) experiment, almost-confluent cells were split 1:2 and incubated for 48 to 60 h in DMEM containing 15% FBS. Cells became quiescent due to contact inhibition during this period. These quiescent cells were released to grow by splitting 1:5 in DMEM containing 15% FBS. Three hours after splitting, HU was added to the medium at a final concentration of 0.5 mM, and cells were incubated for a further 18 h. Cells were washed twice with DMEM and refed with DMEM containing 15% FBS to release them from HU block. Cell synchrony in both experiments was assessed by flow cytometry (22).
The methods for preparation and determining the titer of viruses have been described (19). For infection with recombinant adenoviruses, 2r15 cells were brought to quiescence by serum starvation, and virus infection was carried out as described (13). Following infection, cells were cultured in DMEM containing 0.2% FBS for 18 h before harvesting for further treatment. Recombinant adenoviruses expressing E2F1 or E2F2 were titrated to identify multiplicities of infection that would achieve an equivalent level of production of the DNA-binding activities. For one experiment, multiplicities of 600 for E2F1 and 250 for E2F2 were used; a second experiment employed a multiplicity of 600 for E2F1 and 400 for E2F2.
Nuclear extract preparation and E2F DNA-binding assays.
Nuclear extracts from 2r15 cells were prepared as described (13). E2F DNA-binding assays were performed as described (9) using dihydrofolate reductase promoter DNA fragment as the probe.
RNA preparation.
Total RNA was prepared by treating cells with Trizol reagent (Gibco). mRNA was selected from total RNA with the polyATract mRNA isolation system (Promega) according to the manufacturer's instructions.
Northern analysis.
Northern analysis was performed as described (13).
DNA microarray analysis.
The targets for Affymetrix DNA microarray analysis were prepared according to the manufacturer's instructions. Either the Mu6500 or the Mu11K DNA Affymetrix GeneChip microarray was hybridized with the targets at 45°C for 16 h and then washed and stained using the GeneChip Fluidies station according to the manufacturer's instructions. DNA chips were scanned with the GeneChip scanner, and signals obtained by scanning were processed by the GeneChip expression analysis algorithm (version 3.2) (Affymetrix).
Cluster analysis of cell cycle expression patterns.
The data acquired through absolute analysis of the Affymetrix GeneChip expression analysis algorithm (version 3.2) was imported to the GeneSpring analysis program (Silicon Genetics). The average difference value of each gene at each time point during the serum stimulation experiment, as well as the HU release experiment, was used. If the average difference value at a given time point was below the raw Q value, that number was replaced with the raw Q value. If the average difference value of a given gene was below the raw Q value at all time points, that gene was excluded from the clustering. The genes that showed substantial induction after serum stimulation were selected based on the following criteria: the maximum of the average difference value after serum stimulation should be 2.5-fold greater than the average difference value of quiescent state, and the difference of the maximum of the average difference value after serum stimulation and the average difference value of quiescent state should be greater than or equal to 50. A total of 578 of approximately 6,200 clones met both conditions. The expression pattern of each gene was normalized across the experiments by dividing the average difference value at each time point by the median of every average difference value through the serum stimulation and HU experiments with the same gene. Those genes were initially ordered hierarchically by applying the tree-making program (GeneSpring; Silicon Genetics) to the normalized expression patterns. The genes were then clustered into 16 sets by applying the k-mean clustering algorithm (GeneSpring; Silicon Genetics). The average at each time point of each set was calculated to generate the template patterns for the further clustering. Clones that showed an expression pattern similar to these 16 template patterns were then selected among the 578 genes described above. The similarity of the expression pattern to the template pattern was evaluated by calculating the standard correlation coefficient. Genes with a coefficient greater than or equal to 0.88 of the standard correlation coefficient were selected and clustered. If a given clone showed similarity to several template patterns, the pattern that gave the highest standard correlation coefficient was selected for that gene. To select the “growth” gene, the ratio of the standard deviation of the average difference values of HU experiment and the average of those values were calculated. Genes that had a ratio of less than or equal to 0.185 were selected. After this clustering and selection, clusters were grouped by eye. Finally, the expression pattern of each gene was examined, and a few outlying genes were excluded. The G0 group was identified separately by applying a similar clustering approach but focusing on the genes expressed at a higher level in quiescent state than after serum stimulation.
Selection of E2F target genes.
An analysis of the data from the E2F expression samples and the control sample was performed using the comparison analysis of the Affymetrix GeneChip expression analysis algorithm (version 3.2). Genes that fit the following criteria were considered induced genes in a given experiment: the change call was either induced or marginally induced; the induction was greater than or equal to twofold: and the average difference value of E2F-expressing sample was at least 50. In order to determine how many calls were needed for statistical significance, we made the following statistical considerations. Let D denote the total number of genes on the chip, and let F denote the number of false-positive calls. Then q = F/D is the relative frequency of false-positives. For an arbitrary gene, the probability P that there are at least k false-positive calls for this gene out of the six comparisons can be directly derived from a binomial distribution with success parameter q, assuming q is an accurate estimate of the underlying false-positive probability. From this, we would expect, on average, D*p genes with at least k false-positive calls in the entire set of experiments. Since P is typically small, we assume that the number of these genes is roughly distributed according to a Poisson distribution with mean D*p. Hence, we find that the probability of identifying a gene with at least k false-positive calls in the list of D genes is approximately 1 − exp(−D*p). Using this formula, we conclude that four or more calls out of six cannot be explained by chance, with probabilities in the range of 10−4. Although we do not consider a single occurrence of a gene with three of six induced calls significant, it is likely that the majority of genes that are called as induced in three of six experiments are true positives, since we can assume that the number of false-positives with at least that many calls is Poisson distributed.
Supplementary material.
The entire dataset for both the cell cycle analysis and the E2F-induced gene analysis is available at http://cgt.duke.edu.
RESULTS
Analysis of gene expression during the cell cycle.
When cells are cultured in the absence of normal concentrations of growth factors, they enter a quiescent state usually referred to as G0. Upon the addition of serum, the cells reenter a growth state and progress synchronously through G1 into S phase and then G2 and mitosis. Although a large number of studies have employed this experimental strategy to study the molecular events associated with a proliferative response, there are at least two limitations to this approach. First, gene expression changes that can be measured following the stimulation of quiescent cells to enter a proliferative cycle (serum stimulation) do not distinguish between regulation that is strictly related to growth stimulation versus cell cycle control. For instance, genes induced during G1, including at G1/S, may reflect the fact that the cells are reentering a cell cycle as opposed to passing through G1 from a previous cell cycle; genes induced during this time might not be cell cycle regulated but rather growth regulated. Second, it is largely impossible to measure the events associated with continued cycling in serum-stimulated cultures, in particular the changes taking place at the second G1/S transition, due to a loss of synchrony as the population of cells proceed into the cell cycle.
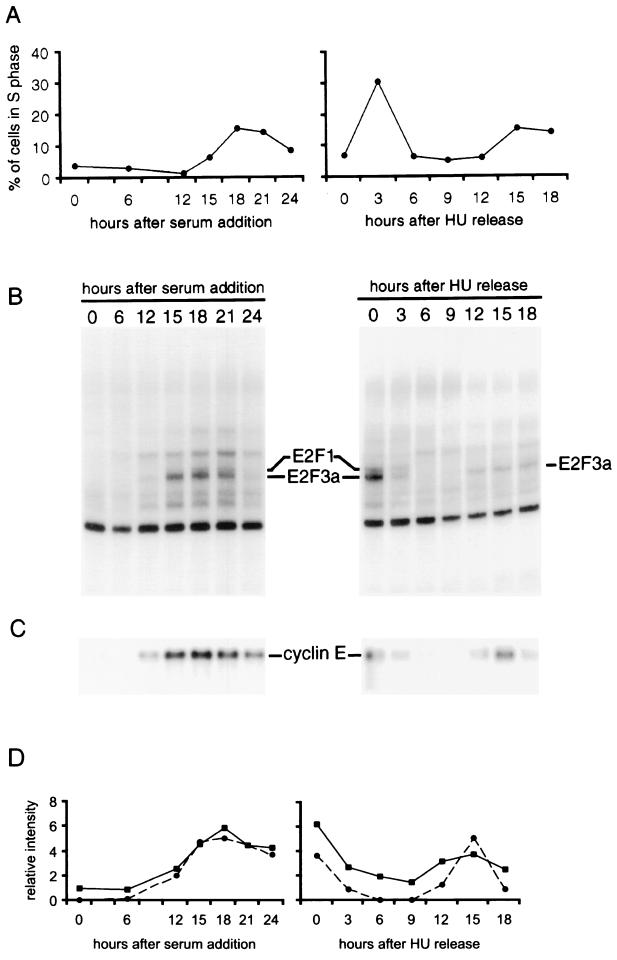
To address these issues, we have combined two forms of analysis to study the events associated with cell cycle reentry and cell cycle progression. In the first instance, MEFs were brought to quiescence by serum starvation and then stimulated to grow by the addition of serum. Samples were taken through 24 h after serum addition and analyzed by flow cytometry. Under the conditions of this experiment, cells began to enter S phase at 15 h following serum addition, as indicated by a determination of DNA content by flow cytometry (Fig. 1A). To analyze events specific to the cell cycle and apart from control related to stimulation out of a quiescent state, a second population of MEFs were synchronized at the beginning of S phase by arresting the cells in the presence of HU. Upon removal of the drug, these cells then progressed through S phase, G2, and mitosis and into the next G1 and second S phase. We have previously described the use of this experimental approach for the analysis of cell cycle regulation of E2F activity as well as certain E2F target genes (13). Flow cytometry analysis demonstrated that the cells completed the initial S phase by 6 h following release from the HU block and then entered the second S phase approximately 15 h following release (Fig. 1A).
FIG. 1.
Analysis of cell cycle progression in MEFs. (A) MEF cells were synchronized either by serum starvation or by HU block and brought back to the cell cycle progression either by adding serum (left panel) or by adding the fresh medium containing serum without HU (right panel). Cells were harvested at the indicated time points, stained with propidium iodide, and processed for flow cytometry. Percentage of cells in S phase at each time point is plotted. (B) E2F DNA-binding activity in the samples described in panel A. Nuclear extracts prepared from the indicated samples were assayed for E2F DNA-binding activity by electrophoretic mobility shift assay as described in the text. The identity of the indicated E2F binding activities is based on relative gel mobility and identification with specific antibodies. (C) Cyclin E expression during the cell cycle progression. RNA was prepared from the indicated samples and analyzed by Northern blotting, using a cyclin E cDNA probe. An equal amount of mRNA was loaded in each lane. (D) Comparison of gene expression measurement by the Affymetrix GeneChip cyclin E array to that obtained by densitometric scanning of a Northern blot of the same RNA sample. The average difference values of cyclin E gene calculated by the Affymetrix GeneChip expression analysis algorithm were normalized across the two experiments and plotted (■). The intensity of the cyclin E bands in the Northern blot shown in panel C was measured by densitometric scanning. The values are normalized across each experiment and plotted (●).
Aliquots of these samples from the two experiments were also assayed for E2F DNA-binding activity as a measure of progression through the proliferative response. As shown in Fig. 1B, E2F activities previously shown to accumulate at G1/S, including E2F1 and E2F3a, were first observed at 12 h following serum stimulation and then peaked at 18 h, coinciding with G1/S, defined by DNA synthesis measurements. These activities were also elevated in the HU-arrested cells and declined as the cells entered S phase, and then E2F3a activity reaccumulated at the second G1/S transition. These observations parallel results described previously that demonstrate a cell cycle control of E2F3a activity (13, 14). In addition, an assay for cyclin E RNA accumulation by Northern blot revealed an accumulation at G1/S that parallels the accumulation of E2F activity at G1/S (Fig. 1C). As such, this experimental approach, which combines analysis of cells reentering a cell cycle from a quiescent state with analysis of proliferating cells leaving a G1/S arrest, provides a comprehensive view of cell cycle progression.
We next used the RNA from each of these samples to hybridize to high-density DNA microarrays in order to provide a broader examination of the changes in gene expression as cells enter a proliferative state and also pass through a cell cycle. We made use of Affymetrix GeneChip DNA arrays that contained approximately 6,200 murine gene sequences and ESTs. RNA from each of the samples was converted to target following established procedures and then used to hybridize to the GeneChip arrays. The hybridized chips were then processed and analyzed as described in Materials and Methods. The hybridization quantified by the Affymetrix software is shown in Fig. 1D and compared to a densitometric analysis of the cyclin E Northern blot shown in Fig. 1C. It is evident from this analysis that the microarray analysis closely matches the Northern analysis.
In order to identify groups of genes with a similar pattern of expression within the cell cycle, the Affymetrix average difference values for each gene, as calculated by the GeneChip expression analysis algorithm, were plotted as a function of time following serum stimulation or time after HU release. Preliminary visual inspection of the data indicated the existence of distinct patterns of gene expression. We have clustered genes based on vectors of expression levels consisting of Affymetrix average difference values for all time points in both the growth stimulation and the cell cycle experiments. This was done using k-means clustering as implemented in the GeneSpring software (Silicon Genetics). This approach is a self-organization of the measured gene expression data and is hence not biased by any prior expectations of how genes might be regulated. Criteria were set to eliminate genes that failed to show significant induction in the serum stimulation experiment. Expression patterns of genes that met these criteria were normalized across the experiments and then clustered by a k-mean clustering algorithm. We have tested several values for the total number of clusters in the k-means clustering procedure. The final analysis was based on 16 clusters; with fewer clusters, we could not identify a unique course of up- and downregulation within each individual cluster, while a larger number of clusters led to distinct clusters with a similar course of gene expression. For this setup, we can summarize each cluster of genes by a characteristic sequence of up- and downregulation at specific time points in the experiments.
Delineation of multiple, distinct patterns of expression within the mammalian cell cycle.
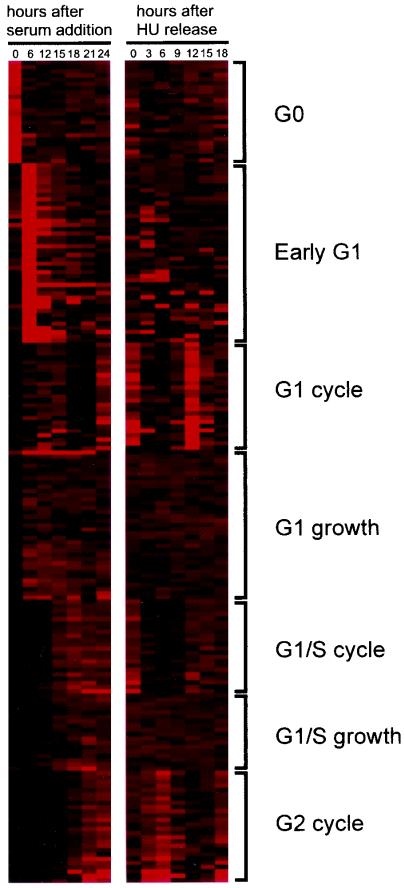
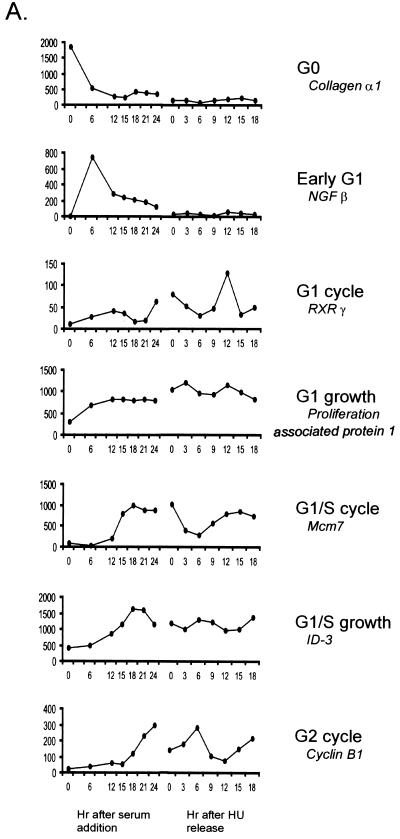
Figure 2 displays the clusters as a function of the time of expression through the two experiments. As indicated in the figure, clusters could be identified that included genes expressed highly in quiescent cells and then turned off once the cells begin to proliferate (G0); genes whose expression increased soon after the stimulation of growth and then fell to basal levels (early G1); genes whose expression increased in G1, declined, and then increased again during the second G1 (G1 cycle); genes whose expression increased in G1 and then remained constant thereafter (G1 growth); genes whose expression increased at the G1/S transition, declined, and then increased again at the second G1/S transition (G1/S cycle); genes whose expression increased at G1/S and then remained constant (G1/S growth); and finally, genes whose expression increased at a time coincident with the end of S phase, declined, and then increased again at the second G2 (G2 cycle). Examples of patterns of expression for specific genes within each cluster are shown in Fig. 3A. The identities of the genes in these clusters, together with information regarding functional properties, were obtained from a search of the UniGene database and are listed in Table 1.
FIG. 2.
Identification of patterns of gene expression following growth stimulation and during the mammalian cell cycle. Expression profile of individual genes in the serum stimulation and HU release experiments, clustered according to the methods described in the text. The expression level of each gene in the two experiments was displayed by a pseudo-color visualization matrix (6). In each experiment, a vertical column represents all of the clustered genes for a given time point. The intensity of expression, as determined from the average difference values calculated by GeneChip expression analysis (Affymetrix), is depicted by the intensity of red color.
FIG. 3.
Specific examples of genes regulated during the cell cycle. (A) Representative example of expression profile among each cluster is shown with its identification. The average difference value at each time point across the cell cycle experiments is plotted for each gene. NGF, nerve growth factor; RXR, retinoid X receptor. (B) Northern analysis for selected G2 cell cycle cluster genes. RNA samples prepared from the indicated times points of the HU release experiment were analyzed by Northern blotting, using probes for cdc2 and importin-α2. The cyclin E profile is shown for comparison. An equal amount of mRNA was loaded in each lane.
TABLE 1.
Identification of genes regulated during the cell cycle
| Accession no. | Gene description | Functional category |
|---|---|---|
| G0 cluster | ||
| W30101 | EST; similar to calcium/calmodulin-dependent protein kinase type 1 (R. norvegicus) | Signal transduction |
| L07924 | Ral guanine nucleotide dissociation stimulator | Signal transduction |
| W09590 | EST; protein-tyrosine phosphatase zeta precursor (R. norvegicus) | Signal transduction |
| U40576 | SIM2 | Transcription control |
| U80036 | Paired-like homeodomain transcription factor 2 | Transcription control |
| M74570 | Aldehyde dehydrogenase | Metabolism |
| W14823 | EST; similar to integrin beta 5 | Cell adhesion |
| W87253 | EST; integrin beta 5 | Cell adhesion |
| U16162 | Prolyl 4-hydroxylase alpha-1 subunit | Collagen synthesis |
| AA000961 | EST; legumain | Cysteine endopeptidase |
| D85414 | NEDD-4 | E3 ubiquitin-protein ligase (by similarity) |
| W62849 | EST; similar to 4F2 cell surface antigen heavy chain | Amino-acid transport |
| AA014996 | EST; similar to apolipoprotein B (R. norvegicus) | Lipoprotein catabolism |
| X66976 | Procollagen, type VIII, alpha 1 | Major constituent of Descemet's membrane |
| M26270 | Acyl-CoA desaturase 2 | |
| X97755 | MSI | |
| M32240 | Peripheral myelin protein, 22 kDa | |
| U00674 | Syndecan 2 | |
| D16262 | Mesoderm-specific transcript | |
| AA109109 | EST; similar to myosin regulatory light chain 2 (H. sapiens) | |
| AA062342 | EST; similar to RAB-28 (R. norvegicus) | |
| AA014656 | EST | |
| AA117286 | EST | |
| AA061086 | EST | |
| Early G1 cluster | ||
| U59876 | mGCM1 | Transcription control |
| M82874 | Helix loop helix 1 | Transcription control |
| M83380 | RelB | Transcription control |
| X75014 | Phox2 | Transcription control |
| M95200 | Vascular endothelial growth factor | Signal transduction |
| X62701 | Urokinase plasminogen activator receptor | Signal transduction |
| D30782 | Epiregulin | Signal transduction |
| Z80941 | Semaphorin H | Signal transduction |
| V00836 | Nerve growth factor beta | Signal transduction |
| X69619 | Inhibin beta-A | Signal transduction |
| U43186 | MAPK/ERK kinase kinase 2 | Signal transduction |
| AA107252 | EST; phosphatidylinositol 4-kinase (R. norvegicus) | Signal transduction |
| L01695 | Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase 1B | Signal transduction |
| M14872 | Gonadoliberin I | Signal transduction |
| K02108 | Keratin complex 2, gene 6a | Filament assembly |
| M88242 | Prostaglandin G/H synthase 2 precursor | Prostaglandin synthesis |
| M91443 | Gap junction beta-5 | Transmembrane channels |
| U66865 | Vacuolar protein sorting-associated protein 45 | Vesicular transportation |
| X73985 | Calretinin | Calcium binding protein |
| X79003 | Integrin alpha 5 | Cell adhesion |
| L42293 | Sterol O-acyltransferase | Cholesterol synthesis |
| M55154 | Transglutaminase 2, C polypeptide | Protein cross link |
| M55669 | Neuroendocrine convertase 2 | Prohormone processing |
| M73696 | Glvr-1 | Inorganic phosphate transporter |
| Z30970 | TIMP-3 | Regulation of matrix metalloproteinase activity |
| D10727 | NPC-derived proline-rich protein | |
| D50263 | DAN | |
| U68267 | Myosin binding protein H | |
| X04123 | DNA nucleotidylexotransferase | |
| AA028547 | EST; hypothetical protein | |
| AA051231 | EST; similar to zinc finger protein | |
| AA097292 | EST; beta-tubulin | |
| AA137667 | EST; similar to gamma-filamin (H. sapiens) | |
| W09925 | EST; gamma-filamin (H. sapiens) | |
| W97775 | EST; similar to gamma-filamin (H. sapiens) | |
| AA002759 | EST | |
| AA023458 | EST | |
| AA062328 | EST | |
| AA106256 | EST | |
| AA106913 | EST | |
| W08057 | EST | |
| W34915 | EST | |
| G1 cycle cluster | ||
| X66225 | Retinoid X receptor γ | Nuclear hormone receptor/transcription control |
| M26053 | T-cell receptor germline beta-chain gene constant region | Signal transduction |
| U70622 | Lysophosphatidic acid receptor | Signal transduction |
| M21532 | Purkinje cell protein 2 | Signal transduction |
| M12836 | T-cell receptor gamma, variable 4 | Signal transduction |
| W83658 | EST; similar to G protein gamma 2 subunit | Signal transduction |
| L36135 | T-cell receptor delta chain | Signal transduction |
| M26417 | T-cell receptor beta chain | Signal transduction |
| D13867 | Integrin alpha 3 | Cell adhesion |
| W50665 | EST; similar to T-cadherin | Cell adhesion |
| AA028501 | EST; cytochrome c oxidase, subunit VIIIb | Mitochondrial electron transport |
| M60429 | Immunoglobulin heavy chain C region | Immunoglobulin |
| K02785 | cDNA induced by PDGF with some homology to c-fos | |
| M96430 | Acrosin | |
| M35667 | Lysozyme-binding Ig κ chain (HyHEL-10) V23-J2 region | |
| M23236 | Proline-rich protein M14 | |
| W62585 | EST; plenty-of-prolines-101 | |
| W65899 | EST; similar to guanine nucleotide-binding protein g(i)/g(s)/g(t) beta subunit (H. sapiens) | |
| W89939 | EST | |
| W91095 | EST | |
| W98265 | EST | |
| W98496 | EST | |
| G1 growth cluster | ||
| U27457 | ORC2 | Replication initiation |
| U20225 | adl | DNA synthesis |
| M11310 | APRT | DNA synthesis |
| W34756 | EST; uridine kinase (R. norvegicus) | DNA synthesis |
| U44426 | D52 | Signal transduction |
| U28151 | p130Cas | Signal transduction |
| U02313 | MAST205 | Signal transduction |
| W15568 | EST; similar to U1 snRNP-specific protein C | Pre-mRNA processing |
| D87691 | eRF1 | Transcription termination |
| U51002 | DLX-2 | Transcription control |
| J04179 | HMG-Y | Transcription control |
| W13191 | EST; similar to thyroid hormone receptor alpha-2 | Nuclear hormone receptor/transcription control |
| AA062397 | EST; similar to RAB-5B | Protein trafficking |
| U07951 | GDP-dissociation inhibitor | Protein trafficking |
| X01237 | P53 | Cell cycle checkpoint |
| D63784 | Id-associated protein 1 | Cell growth |
| W85270 | EST; similar to inorganic pyrophosphatase (H. sapiens) | Lipoprotein catabolism |
| D85904 | APG-2 | Heat shock protein |
| U21673 | T complex testis-specific protein 2 | |
| U77415 | Bop1 | |
| AA111149 | EST; similar to tubulin alpha 2 | |
| U43918 | Proliferation-associated protein 1 | |
| U49720 | Blue-sensitive opsin | |
| W18778 | EST; similar to tubulin Mbeta 1 | |
| W41072 | EST; similar to ubiquitin conjugating enzyme (H. sapiens) | |
| AA138226 | EST; clathryn light chain (R. norvegicus) | |
| AA051583 | EST | |
| AA062365 | EST | |
| AA073743 | EST | |
| AA088121 | EST | |
| AA110046 | EST | |
| W35742 | EST | |
| W49178 | EST | |
| W56929 | EST | |
| G1/S cycle cluster | ||
| X15666 | Ribonucleotide reductase M2 | DNA replication |
| U63337 | Cdk2 | DNA replication |
| X75888 | Cyclin E | DNA replication |
| X62154 | MCM3 | DNA replication |
| D26089 | MCM4 | DNA replication |
| D26090 | MCM5 | DNA replication |
| D86726 | MCM6 | DNA replication |
| D26091 | MCM7 | DNA replication |
| X57800 | PCNA | DNA replication |
| X72711 | Replication factor C, 140 kDa | DNA replication |
| W20901 | EST; similar to replication factor C 38-kDa subunit (H. sapiens) | DNA replication |
| D17384 | DNA polymerase alpha 1, 180 kDa | DNA replication |
| U19604 | DNA ligase I | DNA replication |
| L26320 | Flap endonuclease-1 | DNA replication |
| D13473 | Rad51 | DNA repair |
| U42190 | EST; MSH6 | DNA repair |
| X70472 | B-myb | Transcription control |
| U75680 | Histone stem-loop binding protein | Pre-mRNA processing |
| U27177 | p107 | Cell cycle regulation |
| X14805 | DNA methyltransferase (cytosine-5) | DNA synthesis |
| AA096762 | EST; similar to NASP | |
| AA154451 | EST | |
| G1/S growth cluster | ||
| K02927 | Ribonucleotide reductase M1 | DNA replication |
| U05823 | Pericentrin | Mitois/meiosis |
| D12646 | KIF4 | Mitosis |
| X72310 | DP-1 | Transcription control |
| M60523 | ID-3 | Transcription control |
| AA023287 | EST; similar to TFIIB chain A (H. sapiens) | Transcription initiation |
| U60530 | MAD homolog 2 | Signal transduction |
| X56045 | RAN binding protein 1 | RNA export/protein import |
| X63100 | Gap junction alpha-6 | Transmembrane channels |
| X13752 | Delta-aminolevulinate dehydratase | Porphyrin and heme biosynthesis |
| U70494 | Histone H2A.Z | |
| Y09419 | Spermine synthase | |
| K03235 | Proliferin 2 | |
| X16151 | T-lymphocyte activation protein | |
| M72394 | Phospholipase A2 | |
| AA138105 | Similar to NOVEL ANTIGEN 2 (H. sapiens) | |
| L15429 | L6 antigen | |
| AA118151 | EST | |
| G2 cycle cluster | ||
| U58633 | Cdc2 | Mitosis |
| L16926 | Cdc25C | Mitosis |
| AA000468 | EST; similar to p55CDC (cdc20) | Mitosis |
| AA008043 | EST; similar to p55CDC (cdc20) (R. norvegicus) | Mitosis |
| Z26580 | Cyclin A2 | Mitosis |
| X64713 | Cyclin B1 | Mitosis |
| U20636 | Cyclin F | Mitosis |
| D55720 | Importin alpha 2 | Mitosis |
| X82786 | Ki-67 | Mitosis |
| W70733 | EST; similar to KIFC1 | Mitosis |
| W98100 | EST; similar to KIF3B | Mitosis |
| L06144 | Polo-like kinase | Mitosis |
| AA050055 | EST; Ayk1 | Mitosis/melosis |
| Z46757 | HMG2 | Transcription control |
| L11316 | Ect2 | Signal transduction |
| M86377 | TTK | Signal transduction |
| D21099 | STK-1 | Signal transduction |
| X64550 | Intracellular hyaluronic acid binding protein | Signal transduction |
| AA023099 | EST; similar to dUTPase (R. norvegicus) | DNA synthesis |
| U20497 | p19-INK4D | Cell cycle regulation |
| AA032836 | Similar to cyclin-dependent kinases regulatory subunit 1 (H. sapiens) | Cell cycle regulation |
| Y09632 | Rab6 | Protein trafficking |
| W99981 | EST; similar to peptidyl-protyl cis-trans isomerase a | Protein folding |
| AA067813 | Glutamate cysteine ligase | Glutathione biosynthesis |
| AA028398 | EST; tubulin beta-3 | |
| X58069 | Histone H2A.X |
Although there were clusters identified in the k-means clustering analysis whose biological relevance was not immediately apparent, other clusters clearly related to known functional properties. For instance, the G1/S and G2 clusters included a number of genes encoding replication and mitotic activities, respectively. The relationship between the time of RNA accumulation and the time when the gene product functions, at least for replication activities controlled at G1/S, has not always been seen in past work studying yeast cell cycle control. In addition, past experiments have not clearly detailed a role for transcriptional regulation during G2 in mammalian systems. In large part this is a reflection of the experimental strategy, which generally examines gene expression following serum stimulation of quiescent cells. Simply examining the pattern of gene expression following stimulation of cell growth does not reveal a clear pattern of gene control at G2, a situation most likely due to loss of cell synchrony. That is, such genes are induced by serum addition, but whether they are activated at G1/S, in S phase, or later is difficult to discern (for instance, compare the G1/S and G2 clusters in Fig. 3A).
Previous work has suggested that some of the genes in the G2 cluster are induced at either G1/S of the cell cycle or late in G1 (8). In order to confirm that the microarray analysis did indeed reflect the true behavior of these genes, we assayed the samples from the HU release experiment by Northern analysis, using probes for several genes categorized as G2 regulated. As shown in Fig. 3B, it is apparent that both the cdc2 gene and the importin-α2 gene are indeed activated at G2, consistent with the microarray assays. These patterns are in sharp contrast to the pattern for cyclin E expression, which is regulated at G1/S. We believe that the discrepancy between these data and previous studies very likely reflects the method of cell synchronization and the ambiguity of cell cycle position when only a serum stimulation experiment is employed.
The importance of combining the HU-synchronized samples with the serum-induced samples is clearly illustrated by the last three clusters identified in Fig. 2. An analysis of only the serum-induced samples would not distinguish these genes. Rather, they would be grouped together as genes induced late in G1. But by combining these data with the HU-synchronized analysis, it becomes readily apparent that there are in fact three distinct clusters—genes induced late in G1 that remain constant, genes induced late in G1 that cycle, and genes induced in G2 that cycle.
Finally, a particularly revealing relationship can be seen in those genes that are activated at G1/S. One group includes genes activated during G1 whose expression levels remain high as cells continue to proliferate (G1/S growth cluster). This group includes genes encoding a variety of proteins that function in transcription, signal transduction, and RNA metabolism (Table 1). In contrast, a second group is also activated at G1/S, but expression of this group oscillates as the cells continue to cycle in the presence of growth factors (G1/S cycle cluster). This group includes genes whose function is distinct from the other G1/S-induced group of genes in that these genes encode proteins that are almost exclusively involved in DNA replication. We do note that there is some discrepancy between these results and past experiments that identified several of these DNA replication genes as showing constant expression following G1/S (13). In particular, the previous work suggested that the expression of a subset of the Mcm genes was constant following the initial G1/S, whereas the analyses performed here with DNA microarrays revealed an oscillation in the expression of each of the Mcm genes, as shown for mcm7 in Fig. 3A. Although we cannot identify a clear distinction in the two analyses that would explain this difference other than a cell type difference, the fact that a substantial number of additional genes encoding replication proteins are coordinately regulated in this manner leads us to believe that the G1/S oscillating pattern of expression may be a common aspect of control of replication activities.
Identification of genes induced by expression of E2F activities.
We have previously described the use of recombinant adenovirus vectors as a means to efficiently produce proteins in otherwise quiescent cells (4). The strategy takes advantage of the ability of adenoviruses to infect quiescent cell populations and do so with an efficiency that allows a biochemical analysis of the entire population of cells. Given the fact that the E2F1, E2F2, and E2F3a activities normally only accumulate at G1/S of the cell cycle, as demonstrated previously and as shown by the data here in Fig. 1, overproduction of these proteins in a quiescent cell allows an analysis of the induction of potential target genes by these E2F proteins in the absence of other growth regulation activities. Indeed, we have made use of this approach in past experiments to study the induction of various E2F target genes (3, 4, 12). We have now extended this work through the use of DNA microarrays to facilitate the assay of large numbers of genes in order to gain a more comprehensive view of the pathway of gene control involving E2F activities. Moreover, by performing these analyses in conjunction with the cell cycle determinations, they provide an opportunity to establish a context for understanding previously characterized as well as uncharacterized E2F-regulated genes.
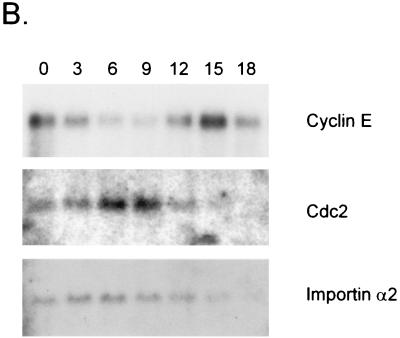
MEFs were brought to quiescence by serum starvation and then infected with either a control adenovirus that expresses green fluorescent protein (GFP) or with viruses that express the E2F1, E2F2, or E2F3 gene products. As shown in Fig. 4A, these conditions allowed an accumulation of E2F1 or E2F2 activity that, at least for E2F1, was at a level similar to that observed when cells normally pass through G1/S. Thus, the experimental approach does not represent a gross overproduction of the proteins but rather an accumulation to near physiological levels in the absence of the other events normally associated with a proliferative response. In contrast to the accumulation of E2F1 and E2F2 activity, the production of E2F3 activity was markedly reduced compared to the others despite the use of a substantial multiplicity of infection (data not shown). Indeed, an increase in E2F3 activity was only clearly evident upon treatment of extracts with deoxycholate, suggesting that the majority of the ectopically expressed protein was bound to Rb. Given this reduced level of E2F3 activity, we have chosen to focus primarily on the analysis of gene induction by E2F1 and E2F2. A virus titration was used to determine the multiplicities of infection needed to achieve an equivalent level of E2F1 and E2F2 activity.
FIG. 4.
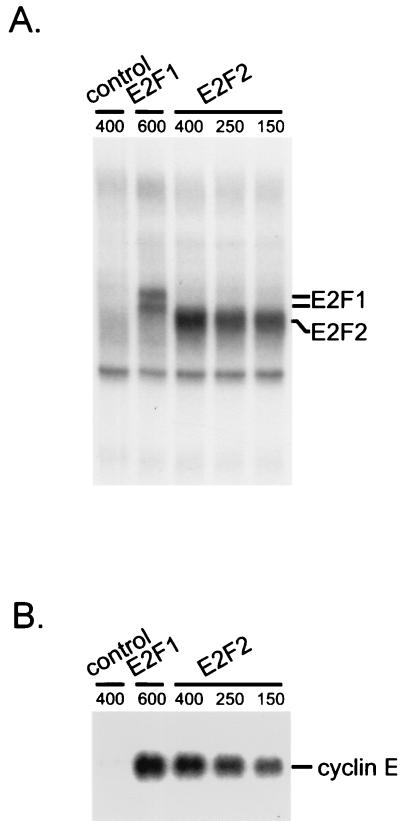
Expression of E2F activities in quiescent fibroblasts. (A) Production of E2F binding activity in cells infected with recombinant adenoviruses containing either the E2F1 or E2F2 gene. Quiescent MEF cells were infected with either E2F1- or E2F2-expressing recombinant adenovirus or with GFP-expressing recombinant adenovirus as a control. E2F2 virus was infected at three different multiplicities to obtain the same E2F DNA-binding activity as with E2F1. E2F binding activities were analyzed by E2F DNA-binding assay with the nuclear extracts prepared from the infected cells. (B) Induction of cyclin E RNA accumulation in cells expressing E2F activities. Expression of cyclin E mRNA was analyzed by Northern analysis in E2F1- and E2F2-expressing cells. mRNA was prepared from the same cells analyzed for E2F DNA-binding activities in panel A and subjected to Northern analysis using a cyclin E probe.
Measurement of the expression of cyclin E, a previously demonstrated E2F target, demonstrated that the production of the E2F1 and E2F2 activities did lead to an induction of cyclin E expression (Fig. 4B). We then used the RNA from these infections to generate target for GeneChip analysis. Targets prepared using the RNA from Ad-E2F-infected cells were hybridized to sets of the Affymetrix murine 11K GeneChips and compared to the hybridization pattern obtained with a control (target prepared from RNA from control-virus-infected cells).
We set the following criteria based on the Affymetrix GeneChip expression analysis software as the basis for identifying genes induced by E2F activities: an intensity of expression (average difference value) that was greater than or equal to 50 in the E2F-expressing cells; the gene was considered increased or marginally increased by comparison analysis using the Affymetrix GeneChip expression analysis algorithm; the fold change, as reported by the Affymetrix comparison analysis, was greater than or equal to 2.0. Of the approximately 11,000 sequences scored in the hybridization assays, a small fraction in any given experiment met these criteria. For instance, in one experiment in which the 11,000 sequences were scored for expression using RNA from E2F1- or E2F2-expressing cells, a total of 255 genes exhibited an induction of at least twofold.
It was also clear from an inspection of the data that there was variation from experiment to experiment in the genes scored as induced in the E2F-expressing cells. Such variation could represent differences in the actual experimental manipulations; alternatively, variations in the hybridization analysis could contribute to the variation. To address the basis for the variation, RNA expression was analyzed from two independent experiments. In addition, the RNA samples from one of these experiments were assayed twice independently. Samples obtained from each of these experiments were used to prepare targets and then used for hybridization to the 11,000 murine gene DNA microarray. Reproducibility was assessed by comparing the duplicate hybridization of a given sample. A comparison of the expression profiles of any given gene sequence in the duplicate hybridizations should, in principle, yield the same value. However, we observed 83 genes scoring as induced in the second hybridization over the first, using the criteria described above for the case of two E2F1-expressing samples, and 69 genes scored as induced in the second hybridization over the first for the E2F2 sample. These false-positives constitute different genes for the E2F1 and E2F2 comparisons, and they do not cluster into any known functional group. In contrast, they appear to represent a random sample from a uniform distribution of the set of genes on the chip.
Clearly, the variation described above leads to statistical significance problems for “calls” of induced genes if they are based on a single comparison. To address this issue, we examined all six analyses of gene expression comparing E2F1 or E2F2 against the control. While we would expect a substantial number of false-positive calls for each individual comparison caused by chance variation in measurement, we do not expect these false-positive calls to refer to the same genes in several comparisons. For instance, cyclin E met the criteria in all six possible comparisons, and there were many more genes that met the criteria in more than one comparison. To ensure maximal confidence in the identification of genes as truly induced by E2F activity, we have combined the data for the E2F1 and E2F2 expression analysis and used a criterion of induction being called in four of the six assays to identify genes as induced by E2F activities (see Materials and Methods for a description of the statistical analysis).
It is evident from the list detailed in Table 2 that many previously identified E2F target genes, including cyclin E, cdk2, and thymidylate synthase, were found in this group. But additional genes were evident as well, including other activities known to function in conjunction with DNA replication, such as DNA primase, DNA ligase, flap endonuclease, and topoisomerase. In addition to these, we also identified a number of E2F-induced genes that encode activities not involved in DNA replication, such as several transcriptional regulatory proteins (HMG proteins, enhancer of zeste), DNA repair (RAD51), and cell cycle control (p18). The largest group of E2F-induced genes apart from those encoding replication activities was, however, a collection of genes that encode proteins that function in mitosis. These include kifC1, cdc2, cyclin B, and cdc20.
TABLE 2.
Identification of E2F-induced genes
| Accession no. | Gene description | Functional category | Mean induction (fold) ± SD (n = 3)
|
|||
|---|---|---|---|---|---|---|
| E2F1 | E2F2 | Northern | Reference | |||
| M13352 | Thymidylate synthase | DNA replication | 2.5 ± 0.3 | 3.3 ± 0.1 | Yes | 3 |
| M19438 | Thymidine kinase 1 | DNA replication | 5.3 ± 0.8 | 5.9 ± 0.9 | Yes | 4 |
| AA023099 | EST; similar to dUTPase (R. norvegicus) | DNA replication | 15.2 ± 2.8 | 13.7 ± 5.3 | ||
| K02927 | Ribonucleotide reductase M1 | DNA replication | 2.5 ± 0.3 | 3.9 ± 0.4 | ||
| M14223 | Ribonucleotide reductase M2 | DNA replication | 4.3 ± 0.5 | 4.4 ± 0.3 | Yes | 4 |
| X15666 | Ribonucleotide reductase M2 | DNA replication | 19.6 ± 5.4 | 20.7 ± 3.3 | Yes | 4 |
| U63337 | CDK2 | DNA replication | 1.9 ± 0.5 | 2.3 ± 0.3 | Yes | 4 |
| X75888 | Cyclin E | DNA replication | 14.4 ± 3.6 | 22.3 ± 6.2 | Yes | 3; this study |
| X62154 | MCM3 | DNA replication | 2.2 ± 0.2 | 3.1 ± 0.8 | Yes | 13 |
| D26091 | MCM7 | DNA replication | 1.9 ± 0.1 | 2.4 ± 0.1 | Yes | 13 |
| AA690055 | EST; similar to DBF4-related protein | DNA replication | 2.0 ± 0.1 | 2.0 ± 0.1 | ||
| AA692716 | EST; similar to DBF4-related protein | DNA replication | 5.3 ± 2.9 | 4.8 ± 1.2 | ||
| X53068 | PCNA | DNA replication | 2.2 ± 0.4 | 2.6 ± 0.1 | Yes | 2 |
| W20901 | EST; similar to replication factor C 38-kDa subunit | DNA replication | 3.0 ± 0.5 | 3.6 ± 0.8 | ||
| D13544 | DNA primase small subunit | DNA replication | 2.1 ± 0.3 | 2.3 ± 0.2 | ||
| D12513 | DNA topoisomerase II alpha | DNA replication | 3.1 ± 1.1 | 3.9 ± 1.3 | ||
| D12513 | DNA topoisomerase II alpha | DNA replication | 3.5 ± 1.8 | 6.5 ± 4.0 | ||
| U04674 | DNA ligase I | DNA replication | 2.2 ± 0.3 | 3.1 ± 0.5 | ||
| L26320 | Flap endonuclease-1 | DNA replication | 2.8 ± 1.1 | 3.0 ± 1.1 | ||
| D13473 | Rad51 | DNA repair | 2.6 ± 0.9 | 3.6 ± 0.5 | ||
| U58633 | cdc2 | Mitosis | 1.9 ± 0.4 | 2.2 ± 0.4 | Yes | 3; this study |
| AA008043 | EST; similar to p55CDC (cdc20) (R. norvegicus) | Mitosis | 3.3 ± 0.4 | 4.5 ± 0.5 | Yes | This study |
| Z26580 | Cyclin A2 | Mitosis | 2.2 ± 0.1 | 2.7 ± 0.2 | Yes | 3 |
| X58708 | Cyclin B1 | Mitosis | 3.9 ± 0.6 | 4.5 ± 1.7 | Yes | This study |
| X66032 | Cyclin B2 | Mitosis | 1.9 ± 0.2 | 2.5 ± 0.5 | ||
| X82786 | Ki-67 | Mitosis | 2.2 ± 0.1 | 2.7 ± 0.3 | ||
| D49544 | KIFC1 | Mitosis | 5.3 ± 1.1 | 10.7 ± 9.9 | ||
| U42385 | FIN16 | Mitosis | 2.9 ± 0.9 | 4.5 ± 1.4 | ||
| Z31235 | EST; stathmin | Mitosis | 2.1 ± 0.2 | 2.9 ± 0.4 | ||
| AA267955 | EST; similar to HEC | Mitosis | 1.8 ± 0.2 | 2.3 ± 0.0 | ||
| AF002823 | BUB1 | Mitosis | 2.2 ± 1.0 | 3.3 ± 0.3 | ||
| D55720 | Importin alpha-2 subunit | Mitosis | 3.0 ± 1.5 | 3.7 ± 2.7 | Yes | This study |
| Z46757 | HMG2 | Transcription control | 6.2 ± 1.8 | 7.7 ± 1.5 | ||
| U52951 | Enhancer of zeste | Transcription control | 2.7 ± 0.0 | 3.1 ± 0.6 | ||
| D21099 | STK-1 | Signal transduction | 6.1 ± 0.9 | 8.1 ± 2.2 | ||
| AA711028 | EST; similar to pituitary tumor-transforming 1 | Signal transduction | 2.2 ± 0.4 | 2.6 ± 0.5 | ||
| U75680 | Histone stem-loop binding protein | Pre-mRNA processing | 2.7 ± 0.2 | 2.4 ± 0.2 | ||
| M26391 | RB | Cell cycle regulation | 2.2 ± 0.6 | 2.7 ± 0.6 | Yes | 3 |
| U19596 | p18-INK4C | Cell cycle regulation | 2.2 ± 0.3 | 2.0 ± 0.1 | Yes | 4 |
| AA059527 | EST; similar to brain neuron cytoplasmic protein 2 | 2.5 ± 0.3 | 2.9 ± 0.3 | |||
| U69488 | Viral envelope like protein | 3.5 ± 1.0 | 2.8 ± 0.5 | |||
| AJ002390 | Annexin VIII | 6.8 ± 3.4 | 9.9 ± 1.8 | |||
| AA002747 | EST | 2.9 ± 0.1 | 3.3 ± 0.4 | |||
| AA002925 | EST | 5.3 ± 2.4 | 7.2 ± 4.3 | |||
| AA051276 | EST | 3.7 ± 1.0 | 6.0 ± 2.7 | |||
| AA117100 | EST | 1.9 ± 0.2 | 2.2 ± 0.1 | |||
| AA154451 | EST | 2.7 ± 0.9 | 3.5 ± 0.4 | |||
| AA184798 | EST | 2.2 ± 0.6 | 2.9 ± 0.1 | |||
| AA189300 | EST | 3.2 ± 1.1 | 3.4 ± 1.0 | |||
| AA189313 | EST | 3.0 ± 0.6 | 2.8 ± 0.2 | |||
| AA200970 | EST | 2.2 ± 0.3 | 2.4 ± 0.0 | |||
| AA266783 | EST | 2.4 ± 0.3 | 3.1 ± 0.7 | |||
| AA285553 | EST | 3.9 ± 1.4 | 6.3 ± 3.3 | |||
| AA407737 | EST | 7.6 ± 2.6 | 4.6 ± 1.3 | |||
| AA426917 | EST | 2.6 ± 0.3 | 2.7 ± 0.6 | |||
| AA516966 | EST | 11.5 ± 3.5 | 11.2 ± 6.0 | |||
| AA561108 | EST | 2.1 ± 0.2 | 2.7 ± 0.2 | |||
| AA590750 | EST | 2.6 ± 0.2 | 3.1 ± 0.3 | |||
| AA590750 | EST | 2.4 ± 0.5 | 2.6 ± 0.2 | |||
| AA592163 | EST | 8.5 ± 1.0 | 9.3 ± 0.9 | |||
| AA673176 | EST | 2.3 ± 0.2 | 2.4 ± 0.2 | |||
| AA673431 | EST | 2.0 ± 0.1 | 2.6 ± 0.6 | |||
| C76791 | EST | 1.8 ± 0.3 | 2.7 ± 0.1 | |||
| C77497 | EST | 2.8 ± 0.5 | 3.3 ± 1.5 | |||
| C81593 | EST | 3.2 ± 0.7 | 3.3 ± 0.5 | |||
Evidence for induction based on Northern analysis of RNA from Ad-E2F-infected cells.
Relationship of E2F-induced genes to cell cycle control—role for E2F in control of expression of G2 genes.
The finding that many of the genes induced by either E2F1- or E2F2-encoded proteins known to function during mitosis was surprising given the fact that E2F activity, particularly E2F1-3, normally accumulates at G1/S of the cell cycle. As such, it raised the possibility either that the effect of E2F activation on these genes was indirect or that these genes are normally regulated at G1/S even though the products function in mitosis. The latter scenario has precedence, since a number of yeast DNA replication genes are induced in mitosis, well before S phase (1, 23). To address this question, we have examined the relationship between the control of transcription by E2F proteins and the control during the cell cycle.
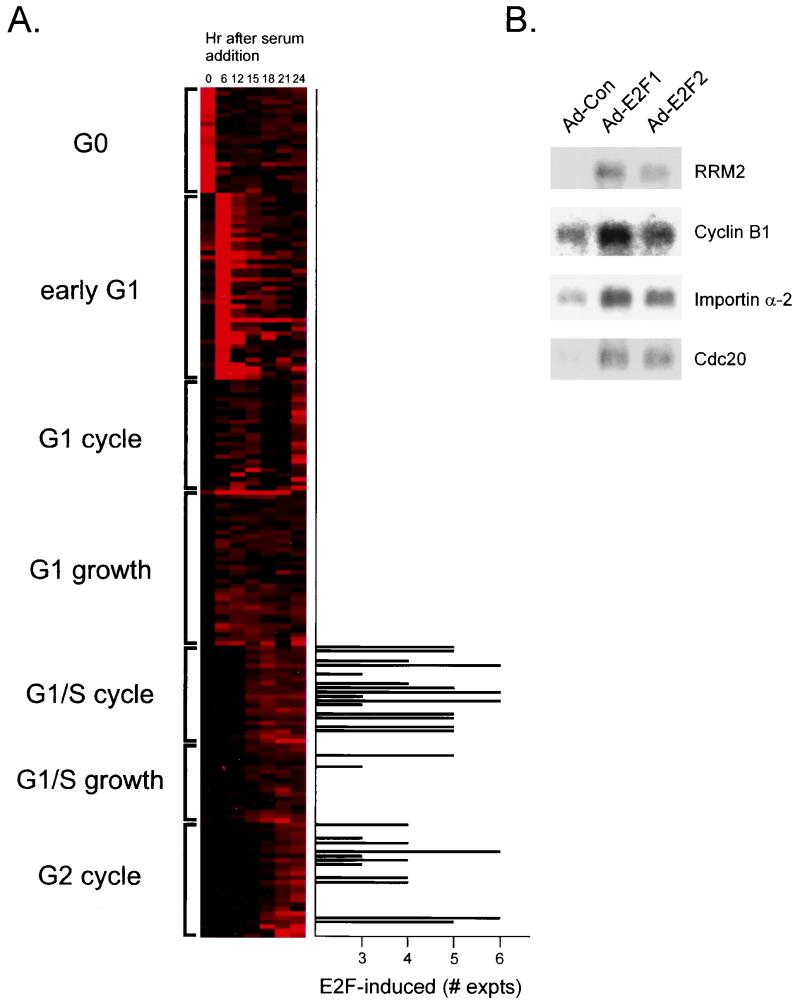
As shown by the data in Fig. 5A, the E2F-induced genes did not distribute uniformly over all clusters derived from the cell cycle analysis. Rather, the majority accumulated in only two of these cell cycle clusters. Most of the E2F-induced genes fell into either the G1/S cell cycle cluster, genes whose expression peaks at the initial G1/S transition upon stimulation of cell proliferation and whose expression then continues to oscillate during the cell cycle with a peak at G1/S, or the G2 cell cycle cluster. The clustering of E2F-induced genes within the G1/S group of cell cycle-regulated genes is consistent with previous work that demonstrates an accumulation of E2F activities at this time of the cell cycle. In contrast, the finding that a number of genes induced by E2F proteins are normally regulated at G2 is surprising in light of the fact that these E2F activities are essentially undetectable at this time of the cell cycle. Although it is possible that there is an accumulation of E2F activity in G2 that has gone undetected in previous work or that there is a role for other E2F activities, such as E2F4 or E2F5, which are present at this time, in transcription activation, it is also possible that the activation of these genes that are normally regulated at G2 during the cell cycle is a secondary effect of E2F accumulation at G1/S.
FIG. 5.
Relationship of E2F-regulated genes to control in the cell cycle. (A) Genes identified as E2F-induced, plotted as a function of the number of experiments in which the gene was induced, are compared to the cell cycle-regulated clusters shown in Fig. 3. For this comparison, we have included those genes that were scored as induced by either E2F1 or E2F2 in at least three of the six assays that were performed. Although there is some probability that a gene induced in only three of six assays could represent a false-positive, the probability is very small. (B) Northern analysis of E2F gene induction. RNA samples prepared from cells infected with either Ad-Con, Ad-E2F1, or Ad-E2F2 virus were analyzed by Northern blotting using probes for RRM2, Cdc20, cyclin B1, and importin-α2. An equal amount of mRNA was loaded in each lane.
To provide further verification of the induction of genes by E2F, particularly those in the G2 category, we analyzed RNA samples by Northern blot assays. As shown by the data in Fig. 5B, the E2F-mediated induction of one of the G1/S-regulated genes (RRM2) was clearly evident, similar to the induction of cyclin E, as seen in the analysis shown in Fig. 4B and consistent with the induction of many others in this category (Table 2). In addition, we also assayed several of the genes identified in the G2 cluster, including cdc20, cyclin B1, and importin-α2. It is evident from these assays that each of these genes was indeed induced by E2F, either E2F1 or E2F2, similar to the induction of the G1/S genes, thus confirming the results of the DNA microarray analysis.
The fact that cells expressing E2F1, E2F2, or E2F3 do not complete S phase or enter mitosis (data not shown) argues that the induction of these G2-specific genes is not the simple consequence of induced cell cycle progression. But whether the G2-specific genes such as cyclin B are directly or indirectly activated by E2Fs is not clear and must await a determination of the promoter elements that are critical for the induction of these genes in G2.
DISCUSSION
A considerable body of work has detailed the transcriptional control properties of the E2F proteins, including the fact that E2F activities are critically important for the activation of genes that encode proteins important for DNA replication. Nevertheless, progress to this point has been incremental and driven largely by prior knowledge. The approach that we describe here represents an unbiased examination of the genes that are subject to E2F control, particularly as they relate to the normal control of the cell cycle. We believe that two important observations derive from these data. First, the logic of gene control during the mammalian cell cycle largely reflects an activation of genes at the time the gene products are required to function. Second, although E2F activity primarily accumulates at G1/S, genes that are normally activated at G2 of the cell cycle are also subject to E2F control.
Gene control during the cell cycle.
Although cell cycle control of gene expression has been studied in detail in yeast, studies in mammalian systems have generally been limited to the initial events following the stimulation of cell proliferation, including recent studies that have employed DNA microarrays to measure the expression of large numbers of genes (7, 10). In general, the experimental approach employed in these studies uses cells synchronized in a quiescent state as a result of growth factor deprivation. When growth factors are then added to such cultures, the cells reenter the cell cycle and maintain a reasonable degree of synchrony through the initial cell cycle. Studies of such cell populations for changes in gene expression have revealed waves of gene expression as the cells move from the quiescent state through G1 and into S phase. This includes genes transcribed in the quiescent cell that are shut off when proliferation is stimulated, genes that are activated early in the proliferation process, and genes that are activated later in G1. The genome-scale analyses recently performed have characterized the regulation of genes involved in fibroblast-specific processes such as wound healing but also a variety of genes involved in events such as cytoskeletal remodeling.
Our analysis of cell cycle control of gene expression extends these studies by combining the assay of gene expression in cells stimulated to reenter a cell cycle by addition of growth factors together with the assay of cells synchronized at G1/S by HU block that are then released and allowed to pass through another cell cycle. This has allowed us to distinguish genes activated following the stimulation of cell growth that either remain constant in their expression as the cells continue to proliferate or oscillate in expression as cells begin to cycle. Two examples of the cell cycle clusters that derive from these analyses are particularly informative. First, for the genes activated at G1/S, two distinct subgroups can be identified—those whose expression remains constant and those that oscillate, with peak expression occurring at the following G1/S transition. Strikingly, this distinction in expression pattern of genes activated at G1/S reflects a distinct grouping of functional activities, at least for the genes that oscillate as cells continue to cycle, since this group largely encode the DNA replication activities as well as DNA repair genes.
The second clear example is the group of genes activated at G2, which then oscillate in expression as cells continue to grow. Once again, these genes, which are clustered according to expression pattern, constitute a functional group. As is evident from our work and consistent with a recently published study that also examined cell cycle-specific gene control (2), genes activated at G2 encode proteins involved in mitotic functions. Cho and colleagues also noted the regulation of genes involved in cell motility and remodeling of the extracellular matrix (2), suggesting a balance between cell proliferation and cell invasion. Taken together, it would appear that the expression of activities during the mammalian cell cycle coordinates synthesis with the time at which the activities are required to function.
Relationships of cell cycle control in yeast and mammalian cells.
The most extensive analyses of cell cycle-regulated gene expression, particularly through the use of DNA microarrays that include the entire set of open reading frames, has been carried out in S. cerevisiae. Two previous studies have detailed the gene expression changes during the S. cerevisiae cell cycle (1, 23). When comparing the results described here for the analysis of mammalian cell growth to these previous studies, it is apparent that there are many similarities in the program of cell cycle regulation in the two systems. For instance, many of the genes that encode the activities directly or indirectly involved in DNA replication are regulated near the G1/S transition in both systems (1, 23). In addition, several DNA repair activities, including Rad51 and Msh6, are similarly controlled at G1/S in yeast and mouse cells. Nevertheless, it is also evident that there are differences. The sharpest contrast between control in yeast and mouse cells is seen for the genes encoding DNA replication initiation proteins. Although each of the genes encoding proteins involved in replication initiation, such as Cdc6, Orc1, and the Mcm proteins, is regulated at G1/S in mammalian cells, the majority of these are regulated either at mitosis or early in G1 in yeast cells. Presumably, this difference in timing of expression of genes encoding the initiation complex proteins reflects a distinction in the mechanisms of prereplication complex assembly in the two systems.
E2F gene control and the cell cycle—role for E2F in control of mitotic activities.
Consistent with previous work, many of the genes newly identified as induced by the E2F proteins include those encoding DNA replication activities such as replication protein C, DNA ligase, DNA primase, topoisomerase, and flap endonuclease (Fig. 6). In addition, other E2F targets include genes encoding proteins that function in DNA metabolism, such as DNA repair enzymes. As such, it seems possible that the majority of the DNA synthetic machinery, including the apparatus that assembles at origins of replication, is regulated at G1/S by E2F activities. Another recent study using DNA microarrays to analyze E2F-induced gene expression also identified DNA replication and cell cycle genes as induced by E2F proteins (15), but this study also identified a large number of additional genes with roles in apoptosis, differentiation, and development, the majority of which were not scored in our assays. Several reasons could explain the differences, but possibly they reflect differences in the cell type used for the expression of E2Fs as well as the use of actively growing cells instead of quiescent cells in our study.
FIG. 6.
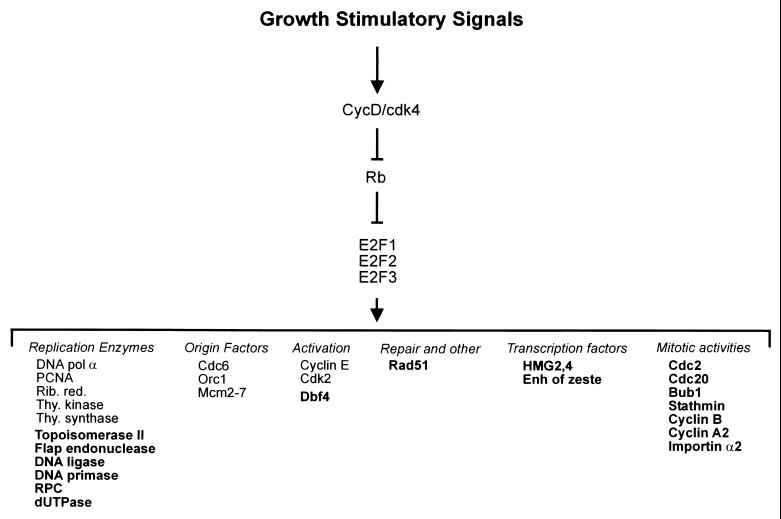
E2F gene regulatory pathway. Schematic representation of the pathway initiated upon stimulation of cell growth that leads to an induction of cyclin D/cdk4 activity. A primary target for D/cdk4 is the Rb protein, inactivating its ability to regulate the accumulation of E2F proteins. Previously identified E2F targets include replication enzymes, proteins that form the initiation complex, and activities that regulate the function of the origin complex. The data presented here now identify additional genes (indicated in bold) that fall within these previously defined groups as well as new groups as indicated. pol, polymerase; Enh, enhancer; Rib. red., ribonucleotide reductase; thy., thymidine.
Perhaps of most interest in the analysis reported here is the finding that many of the E2F-induced genes are normally regulated at G2 in the cell cycle and encode proteins that function in mitosis. Past work has documented changes in E2F activity as cells leave G0 and then as cells pass through G1/S, but there is no evidence for alterations in E2F activity as cells pass through the G2 phase of the cell cycle. In several cases, the E2F-mediated control of these genes has already been recognized, since past work has shown that cyclin A, cyclin B, and cdc2 are regulated by E2F. With the exception of cyclin B, previous work has characterized the cell cycle control of these genes as occurring at G1/S, not G2. We believe this is largely the result of the method of cell synchronization and analysis, making it difficult to discern a peak of induction either in late G1 or in G2.
Although the vast majority of work has focused on the role of E2F in controlling expression of genes at G1/S, it is true that previous work has provided evidence of a connection between E2F activity and the control of mitotic activities, at least in Drosophila melanogaster. In particular, the work of Edgar and colleagues has shown that the cdc25 string product, a rate-limiting activity for progression through mitosis, is a target for E2F in Drosophila cells (16). Moreover, overexpression of E2F was shown to accelerate both G1/S and G2/M, consistent with the ability of E2F to induce both cyclin E and string, rate-limiting activities for transition through these two cell cycle transitions. However, whereas the mammalian cdc25 gene is transcribed at G2/M, the Drosophila cdc25 gene (string) appears to be expressed in G1 (11).
Although it remains possible that there is a particular E2F activity or modified form of an E2F activity that is specifically operational at G2, it is also possible that the induction of these genes normally regulated at G2 is a secondary effect of the E2F activities. A trivial explanation would be that activation of these genes reflects an E2F-induced cell cycle progression. As such, the induction of the mitotic genes would simply reflect the stimulation of cell cycle progression. We believe this possibility is unlikely, since under the conditions of this experiment, there is little evidence for cells progressing through S phase. There is an induction of DNA synthesis, and this does appear to reflect true DNA replication, but the extent of this replication is quite limited. This is perhaps best seen by a cell sorting analysis that measures the DNA content of the cell population following expression of the E2F activities; these assays reveal an increase in the DNA content of the cell population but no evidence for progression to a G2 DNA content. In addition, there is no indication for the appearance of any mitotic cells in the population.
Given these observations, we can envision at least two alternative explanations for the E2F-mediated induction of genes such as cyclin B, cdc2, and Bub1. One possibility is that these genes are activated by transcription factors whose expression is controlled at G1/S by E2F activities. In this scenario, E2F gene control would establish a cascade of events, initially activating the genes encoding DNA replication activities and then secondarily activating genes encoding mitotic activities. Simple kinetic experiments to measure the timing of activation of genes following E2F induction, to determine if the induction of genes such as cyclin E precedes the induction of cdc2, have been inconclusive (data not shown). A second possibility could relate to the recent studies of Dean and colleagues, which provide evidence for two forms of E2F/Rb-mediated transcription repression (25). One repressor complex, which is inactivated by cyclin D/cdk4, appears to control genes normally expressed at G1/S, including cyclin E. A second repressor, which is not affected by cyclin D/cdk4 but is inactivated by cyclin E/cdk2, persists longer in the cell cycle and appears to control genes such as cyclin A. This is thus consistent with the G2 regulation seen in the experiments reported here. Thus, the induction of both groups of genes by E2F overexpression in our experiments could reflect a relief of two distinct types of repression that are normally temporally regulated in the cell cycle. Ultimately, the answer to this question will require a determination of the factors normally responsible for the G2-specific control of genes such as cdc2.
Finally, although the complexity of the E2F family would suggest the potential of specificity in the activation or repression of transcription by the individual E2F family members, there are only hints of such from previous work and from the data generated in the present studies. For instance, previous work employing recombinant adenoviruses to express each of the E2F proteins demonstrated differences in gene induction (4), suggesting the potential for gene-specific activation events. Nevertheless, it is also true that the differences in gene induction by any one member of the E2F family are minimal. Moreover, the loss of function of individual E2F family members also has minimal consequences for gene regulation, with the disruption of E2F3 function appearing to have the most dramatic effect (8). Thus, either there is substantial overlap in gene induction by the individual E2F proteins or the specific targets have not yet been clearly identified.
ACKNOWLEDGMENTS
We thank Kaye Culler for help with preparation of the manuscript and Helena Abushamma for performing the Affymetrix GeneChip analyses.
J.R.N. is an Investigator in the Howard Hughes Medical Institute.
REFERENCES
- 1.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 2.Cho R J, Huang M, Campbell M J, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge S J, Davis R W, Lockhart D J. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 3.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 6.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fambrough D, McClure K, Kazlauskas A, Lander E S. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 8.Humbert P O, Verona R, Trimarchi J M, Rogers C, Dandapani S, Lees J A. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda M-A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 11.Johnston L A, Edgar B A. Wingless and notch regulate cell-cycle arrest in the developing Drosophila wing. Nature. 1998;394:82–84. doi: 10.1038/27925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leone G, DeGregori J, Sears R, Jakoi L, Nevins J R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 13.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins J R. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller H, Bracken A P, Vernell R, Moroni M C, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner J D, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2000;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neufeld T P, de la Cruz A F A, Johnston L A, Edgar B A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 17.Nevins J R. Cell cycle targets of the DNA tumor viruses. Curr Opin Gen Dev. 1994;4:130–134. doi: 10.1016/0959-437x(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 18.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 19.Nevins J R, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F. Methods Enzymol. 1997;283:205–219. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- 20.Robertson E J. Embryo-derived stem cells. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL Press; 1987. pp. 104–108. [Google Scholar]
- 21.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 22.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spellman P T, Sherlock G, Zhnag M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H S, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]