Abstract
Nucleic acids do not fold into a single conformation, and dynamic ensembles are needed to describe their propensities to cycle between different conformations when performing cellular functions. We review recent advances in solution state NMR methods and their integration with computational techniques that are improving the ability to probe the dynamic ensembles of DNA and RNA. These include computational approaches for predicting chemical shifts from structure and generating conformational libraries from sequence, measurements of exact NOEs, development of new probes to study chemical exchange using relaxation dispersion, faster and more sensitive real-time NMR techniques, and new NMR approaches to tackle large nucleic acid assemblies. We discuss how these advances are leading to new mechanistic insights into gene expression and regulation.
Introduction
Composed of only four chemically similar building block nucleotides, the 3 dimensional (3D) structures of nucleic acids are nowhere near as diverse or complex as those of proteins with their 20 chemically diverse amino-acids. Yet nucleic acids are specifically recognized by the cellular machinery and participate in highly complex reactions, both as substrate and enzyme. During cellular processes, DNA and RNA cycle through different conformations each satisfying distinct requirements of multi-step biochemical reactions. These structural dynamics provide a means for amplifying specificity and functionality beyond that which can be attained based on a rigid 3D structure [1,2].
To deeply understand how nucleic acids fold and function at the atomic level, we need to reimagine their 3D structures to include a description regarding propensities to undergo conformational changes over timescales spanning twelves orders of magnitude in time, from picoseconds to seconds. The dynamic ensemble provides one such unified description [1,3,4].
In solution, a nucleic acid molecule will sample a vast number of different conformations. Some conformations may be highly populated (>10%) while others may be vanishingly low in abundance (<0.00001%). Some may form on the picosecond timescale while others may form slowly on the second timescale. Some may differ markedly from the native state while others may only differ by the relocation of a single proton. The Boltzmann-weighted distribution of interconverting conformations is referred to as a ‘dynamic ensemble’. The dynamic ensemble provides a statistical mechanical description that can be used to rigorously compute the thermodynamic cost accompanying conformational transitions and also the rates with which they occur [1].
Because a continuous landscape comprising thousands of 3D structures needs to be specified along with the Boltzmann weighted populations and kinetic rates of interconversion, determining dynamic ensembles continues to be an outstanding challenge in structural biology and biophysics. Many biologically important conformations exist in low-abundance and/or are exceptionally short-lived, and are therefore difficult to detect experimentally. Over the past five years, significant progress has been made toward the development of experimental and computational approaches that provide different cross-sectional views in space and time of the dynamic ensembles formed by DNA and RNA. Here, we highlight recent advances in solution state NMR methods for ensemble determination, and when applicable, their integration with computational techniques. We also discuss how the characterization of dynamic ensembles is leading to a deeper and more quantitative understanding regarding biochemical processes mediated by nucleic acids.
Dynamic ensembles from ensemble-averaged NMR data
Approaches for ensemble determination
Many NMR observables including residual dipolar couplings (RDCs), the nuclear Overhauser effect (NOE), and chemical shifts are averaged over many conformations in the dynamic ensemble. In conventional NMR structure determination protocols, a single conformation is identified that satisfies the ensemble-averaged experimental data. A collection of such structures each satisfying the data within experimental uncertainty are often pooled together in a bundle, and such ensembles need to be distinguished from the statistical mechanical definition in terms of a Boltzmann distribution used throughout this review [1,3,4].
Three different approaches have been developed to interpret NMR data in terms of a Boltzmann distribution of nucleic acid conformations. In one approach, the NMR data is used to guide the assignment of Boltzmann weights to a library of conformations generated using computational methods, typically molecular dynamics (MD) simulations [5,6]. In a second approach, the data is applied as a restraint during MD simulations [7]. Finally, the NMR data can also be used in conventional structure refinement programs to explicitly solve for multiple conformations [8]. While the experimental data is typically not used to encode timescale information [9], they are often sensitive to motions occurring over sub-millisecond timescales.
Dynamic ensembles that satisfy different types of NMR data are being reported for a small but steadily growing number of nucleic acid motifs, including apical loops [10-13], bulges [14-16], single-strands [17], duplexes [18], and a G4 quadruplex [19] (Figure 1a). Together, these ensembles are beginning to outline the conformations that dominate the landscape of nucleic acid ensembles.
Figure 1.
Determining dynamic ensembles of nucleic acids using NMR and computational methods. (a) Shown are NMR-derived dynamic ensembles for RNA (UUCG apical loop [13] and HIV-TAR bulge [14]) and DNA (duplexes [18] and a G-quadruplex [19]). The figures were adapted from original publication. (b) Representative fragments of an RNA hairpin structure used to compute chemical shift using the AF-QM/MM approach (top). The figure was adapted from [22]. Also shown is a comparison between measured and AF-QM/MM predicted 13C chemical shifts for a conventional NMR structure and an RDC-derived dynamic ensemble of a DNA duplex (bottom). The data shown in the figure was adapted from [23]. (c) The principle of eNOE. A mobile proton H1 samples positions that are close to both protons H2 and H3. The eNOE data is better explained by a 2-state ensemble shown in blue and red relative to a model which assumes a single-state structure. The figure was adapted from [25]. (d) The Rosetta FARFAR structure prediction algorithm uses an RNA secondary structure to rapidly generate a conformation library which can be used in conjunction with NMR data to determine a dynamic ensemble. The figure was adapted from [14].
Nucleotides in single-strand and loop regions frequently transition between a narrow distribution of stacked conformations and a broader distribution of unstacked extra-helical conformations [14,16,17,19] (Figure 1a). Extra-helical flipping can be accompanied by changes in sugar pucker and coaxial inter-helical stacking [14,17] (Figure 1a). The ensembles can also include partially melted states [13] or non-native secondary structures [14] (Figure 1a). Dynamic ensembles of canonical DNA duplexes reveal sequence-specific variations particularly for backbone torsion angles ε and ζ [18] (Figure 1a).
Measurements of ensemble-averaged NMR data and their use in ensemble determination
Because determining a dynamic ensemble is an inherently underdetermined problem, much effort has been directed toward measuring new types of ensemble averaged NMR data so as to improve the accuracy of the determined ensembles. A recent study underscored this point [20], showing that increasing the amount of input RDC data used to determine dynamic ensembles of HIV-1 TAR RNA results in ensembles that better predict the binding of small molecules using ensemble-based virtual screening.
NMR chemical shifts can be measured with great ease and abundance and carry rich information regarding the distribution of torsion angles, sugar puckers, hydrogen-bonding, and ionization/tautomerization states of the nucleobases [21]. Recent advances in predicting chemical shifts from 3D structure are enabling their application in determining and assessing dynamic ensembles of nucleic acids.
Frank et al [16] developed an empirical approach for rapidly predicting 1H and 13C chemical shift based on a 3D RNA structure. This made it possible to apply the chemical shifts as restraints during an MD simulation. The approach was used to determine a dynamic ensemble for the U6 intramolecular stem-loop RNA, which is a key component of the spliceosome.
Empirical approaches are constrained to the available database of nucleic acid chemical shifts with known 3D structures and by difficulties in taking into account motional averaging contributions. Some of these limitations can be overcome using ab initio approaches for computing chemical shifts based on 3D structures. However, these calculations can be prohibitively time-consuming for large biomolecules, and until recently, the agreement between predicted and measured chemical shifts was not sufficiently good for nucleic acids to allow quantitative applications.
To accelerate the ab initio calculations, Swails et al [22] developed automated fragmentation quantum mechanics/molecular mechanics or AF-QM/MM, in which a nucleic acid is divided into small quantum fragments each of which is subjected to quantum mechanical calculation while treating the remaining parts of the molecule using molecular mechanics (Figure 1b). For four DNA duplexes [18] as well as the HIV-1 TAR RNA [14], quantitative agreement was obtained when averaging the predicted 1H, 13C, and 15N chemical shifts over an ensemble of conformations determined using RDCs and MD simulations (Figure 1b) [14,23]. For both the DNA and RNA examples above, the agreement deteriorated considerably for the conventional single NMR structure, underscoring the importance of treating motional averaging when calculating chemical shifts, in addition to validating the accuracy of the calculations.
Ensemble-averaged inter-proton distances derived from quantitative measurements of exact NOEs (eNOE) [24] have been applied to determine nucleic acid ensembles (Figure 1c). Relative to semi-quantitative standard NOEs, eNOE can be more readily interpreted quantitatively in terms of many conformational states in an ensemble. eNOEs have been used to determine ensembles for a 14-mer UUCG tetraloop either through multi-state ensemble refinement [25] or posteriori refinement of an MD trajectory using a maximum entropy/Bayesian approach [26] establishing the robustness of these measurements in RNA ensemble determination.
Dynamic ensembles by combining NMR data with structure prediction
The accuracy of dynamic ensembles obtained with the aid of NMR data can be highly dependent on the accuracy of the force field used to generate the starting conformational library or to run restrained MD simulations. Despite improvements in nucleic acid force fields [27], even for simple motifs, ensembles of nucleic acids generated using MD often poorly predict NMR data [10,14,17,19,23,28]. By examining what aspects of the MD-derived ensembles need to be altered to satisfy experimental NMR data, studies are beginning to pinpoint potential biases in nucleic acid force fields, including a tendency to form intercalated structures [17] and certain biases in sugar pucker [14]. These findings should help guide and accelerate future advances in force field development.
Recent studies have demonstrated the power of combining NMR data, including 1H chemical shifts [29] and imino proton NOEs [30] with structure prediction algorithms when solving a 3D structure of an RNA. Shi et al [14] showed the utility of combining RNA structure prediction algorithms with RDC and chemical shift measurements in determining dynamic ensembles of RNAs. Rather than using the time-consuming step of solving a 3D structure then subjecting the structure to MD simulations, Rosetta’s Fragment Assembly of RNA with Full-Atom Refinement (FARFAR) RNA structure prediction algorithm [31] was used to rapidly and directly generate a conformational library for an RNA secondary structure [14] (Figure 1d). FARFAR [31] generates physically plausible conformations by stitching together fragments from a library of existing structures in the protein data bank (PDB) [32], and it was shown to overcome some of the biases and inaccuracies in MD force fields as supported by comparisons to quantum-mechanical calculations of NMR chemical shifts [14].
Excited conformational states from relaxation dispersion
The dynamic ensembles of nucleic acids also include low abundance excited conformational states (ESs) that have exceptionally low populations (as low as 0.01%) and short lifetimes (as low as a few microseconds) [1,2]. Methods based on NMR relaxation dispersion (RD) are enabling the characterization of excited conformational states with unprecedented detail providing information regarding their structure, population, and kinetic rates of interconversion [33-35].
Recent developments in methods
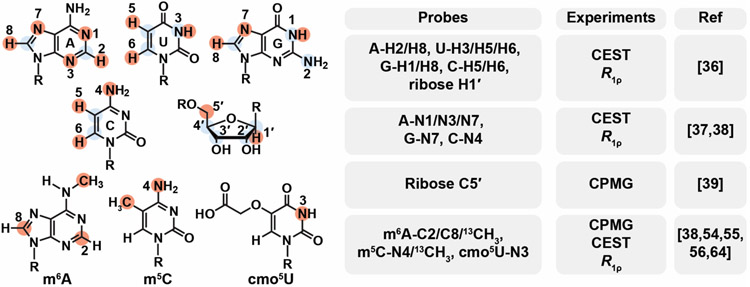
NMR techniques to characterize ESs come in four flavors optimized for different exchange rates (kex= 1 s−1 to 105 s−1) and/or type of nuclei all of which are now routinely applied to study nucleic acids. These include spin relaxation in the rotating frame (R1ρ), Chemical Exchange Saturation Transfer (CEST), Carr-Purcell-Meiboom-Gill (CPMG), and ZZ-exchange experiments [33-35]. While early studies focused on the protonated base (U/C-C5, T/U/C-C6, A/G-C8, A-C2) and sugar (C1', C2', C3' and C4') carbons, and the base imino (G-N1, T/U-N3) as well as amino nitrogen (G-N2 [33]), the list of RD probes now includes base and sugar protons (U-H3, G-H1, A-H2; A/G-H8, U/C-H5/H6; and ribose H1') [36], non-protonated (G-N7 and A-N1/N7) [37] and amino nitrogen (C-N4) [38] as well as the ribose C5' [39] (Figure 2). Leveraging the practical advantages of CEST, a recent study broadened the timescale sensitivity of the experiment to fast exchange through use of high radiofrequency fields [40].
Figure 2.
A growing number of nuclei are targeted for relaxation dispersion measurements (CEST, CPMG and R1ρ) in studies of nucleic acid ESs. The list of probes is growing steadily from an initial focus on protonated carbons, imino and amino (G-N2) nitrogens (highlighted using blue circles) to now include protons, non-protonated and amino (C-N4) nitrogens, ribose C5', and nuclei in modified bases including m6A, m5C and cmo5U (highlighted using red circles). Also shown are the types of RD experiments performed for the various probes.
Approaches to stabilize ESs using mutations or chemical modifications [41] and improvements in predicting chemical shifts based on secondary structures (Y Wang et al., unpublished) are making it possible to determine the structure and dynamic properties of nucleic acid ESs at atomic resolution [23]. Techniques have also been developed that rely on biasing the conformational equilibria through the use of mutations to assess the abundance of RNA ESs within the physiological cellular context [42,43].
Biological activities of ESs
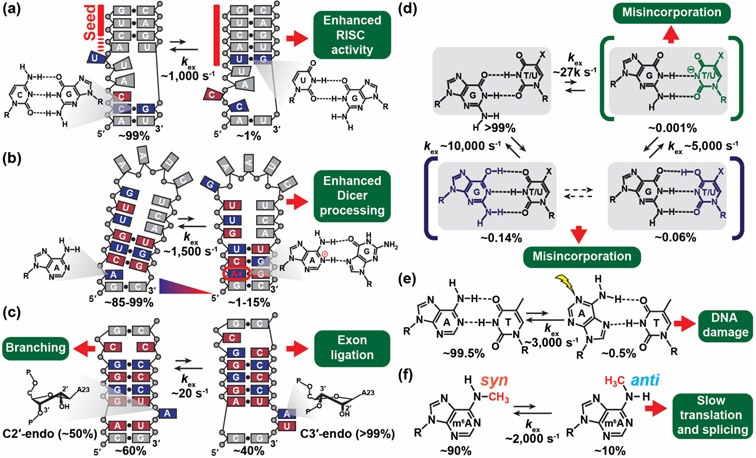
Several recent examples highlight how the cellular machinery recognizes and acts on the ESs of RNAs that form by reshuffling base pairs in and around non-canonical motifs. For example, microRNAs (miRNA) downregulate gene expression by binding to the three prime untranslated regions (3'-UTRs) of mRNAs and activating the RNA-induced silencing complexes (RISCs), which promote mRNA degradation and translation repression [44]. Using R1ρ RD, Baronti et al [43] showed that the catalytically active RISC conformation acts on an ES formed by miR-34a:mRNA duplex. The ES features an elongated 8-mer seed with a coaxially stacked topology that better satisfies the substrate requirements for binding the active conformation of RISC (Figure 3a). Mutations to various mRNA targets that stabilize the ES of the miR-34a:mRNA duplex were shown to robustly increase the miR-34a mediated downregulation of the target genes in human cells.
Figure 3.
NMR reveals RNA and DNA ESs that play important roles in biochemical processes. (a-c) RNA ESs form by reshuffling base pairs. (a) The miR-34a:mRNA duplex undergoes conformational exchange between the GS and the ES, which has an elongated 8-mer seed and altered RNA topology, leading to formation of the active RISC complex. The figure was adapted from [43]. (b) pre-miR-21 forms a pH dependent protonated ES with an A+(anti)·G(syn) base pair (bp) that is more efficiently recognized and processed by Dicer. The figure was adapted from [45]. (c) Domain 6 of type II intron forms an ES with a bulge adenine that has a predominantly (>99%) C3' endo sugar pucker conformation, suitable for exon ligation during splicing and reverse splicing. The figure was adapted from [47]. (d) Wobble G·T and G·U mismatches form tautomer (blue) and anionic (green) Watson-Crick-like ES conformations that can evade fidelity checkpoints during DNA replication and cause spontaneous mutations. cmo5U increases the population of the anionic Watson-Crick ES conformation. The figure was adapted from [50]. (e) Watson-Crick A·T bps form Hoogsteen ES conformations which increase the damage susceptibility of canonical duplex DNA. (f) The methylamino group of m6A forms an anti ES needed for base pairing, thereby slowing down duplex hybridization and conformational transitions.
Using CEST, Baisden et al [45] showed that the Dicer protein more efficiently recognizes and processes a pH dependent ES in pre-miR-21 that features a protonated A+(anti)·G(syn) mismatch, and which also better satisfies its substrate requirements (Figure 3b). Mutants that stabilize the ES were shown to be more readily processed by Dicer relative to mutants that favor the GS. It was proposed that pH-dependent ESs may provide a general mechanism for regulating miRNA biogenesis in response to environmental stimuli.
RNA ESs can also feature non-canonical sugar puckers [46]. Using ZZ-exchange, Plangger et al [47] showed that the group II intron ribozyme exists as a dynamic equilibrium between two conformations in which a catalytic adenine residue adopts either ~50:50 C2'-endo:C3'-endo or >99% C3'-endo sugar pucker conformations. The equilibrium was proposed to regulate the switch between branching and exon ligation during splicing and reverse splicing (Figure 3c).
Combining RD with chemical shift perturbations and Bayesian inference, two ligand molecules were shown to recognize and bind an ES of HIV-1 TAR that has an exceptionally low population of <0.001%, highlighting how ligands with apparently weak binding affinity actually recognize exceptionally lowly populated conformations in the dynamic ensemble with high binding affinity [48]. On the other hand, the ESs of HIV-1 TAR and RRE stem IIB[49] were shown to disrupt key elements of structure needed for protein recognition and cellular activity making them interesting HIV-1 drug targets [42].
The cellular machinery can also act on the ESs of DNA. ES conformations of the G·T mismatch that satisfy the Watson-Crick geometry through tautomerization or ionization of the bases were shown to be recognized by high fidelity polymerases leading to copying errors during DNA replication [50] (Figure 3d). A kinetic mechanism based on the NMR derived exchange kinetics was shown to predict copying errors across a range of high-fidelity polymerases, conditions, and modifications [50]. Damaging reagents were also shown to act on ES Hoogsteen base pairs, which expose the Watson-Crick face of nucleotide bases in duplex DNA to solvent [51,52] (Figure 3e).
Impact of epigenetic and post-transcriptional modifications on excited conformational states
Application of NMR RD is helping reveal how epigenetic and post-transcriptional modifications of DNA and RNA reshape the dynamic ensembles of nucleic acids providing new insights into their biological mechanisms [53]. Key to many of these studies has been the organic synthesis of 13C and/or 15N labeled phosphoramidites bearing these different modifications [38,54-56] (Figure 2).
The highly abundant epigenetic and epitranscriptomic modification m6A was shown to slow annealing of DNA and RNA duplexes by ~5-20-fold [55] through isomerization of the methylamino group [56], potentially explaining why the modification slows a variety of biochemical processes (Figure 3f). On the other hand, 5-formylcytosine (5fC), a semipermanent epigenetic modification which also forms as an intermediate along the 5-methylcytosine (m5C) demethylation pathway, was shown to both slow DNA duplex annealing and to also accelerate duplex melting [57].
Uridine 5-oxyacetic acid (cmo5U), a naturally occurring tRNA modification, was shown to increase the population of an anionic Watson-Crick like G·U ES mismatch (Figure 3d), potentially explaining why the modification enhances the decoding of mRNAs containing G·U mismatches at the third wobble position [54]. By stabilizing paired nucleotides, 2'-O-methylation of the sugar moiety was shown to increase the abundance and prolonged the lifetime of the TAR ESs [58]. Conversely, m5C was shown to minimally impact exchange kinetics between Watson-Crick and Hoogsteen conformations in duplex DNA [38].
Real-Time NMR
Within cells, many RNAs function while they fold in a process referred to as ‘co-transcriptional folding’. Many biochemical processes are coupled to co-transcriptional folding and require a kinetic description in terms of dynamic ensembles that evolve with time.
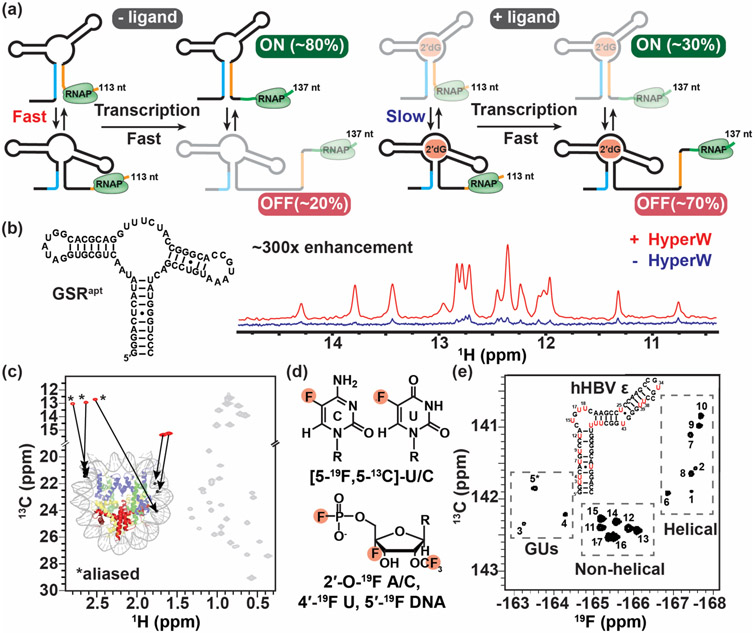
Time-resolved NMR experiments employing photolabile protection groups are providing a window into the time-evolution of biologically important meta-stable intermediates during co-transcriptional folding that play important roles in the mechanisms of riboswitches [59,60]. Helmling et al [60] showed that formation of an antiterminator structure that ensures gene expression in the esoplasma florum 2'-deoxyguanosine (2'dG)-sensing riboswitch is slowed down below the rate of transcription when binding to its cognate ligand, thus mediating the switch into the functional OFF state (Figure 4a).
Figure 4.
Real-time NMR and methods to study large nucleic acid assemblies. (a) Schematic representation of the interconverting transcriptional intermediates of 2′dG-sensing riboswitch at transcript lengths 113 and 137 nt with and without ligand. Ligand binding slows the conformational exchange between the OFF and ON state below the transcription rate, triggering the OFF state function. The figure was adapted from [60]. (b) 1D imino spectra of the guanine-sensing riboswitch (GSR) aptamer domain (GSRapt) of the Bacillus subtilis xpt-pbuX operon with (red) and without (blue) injection of hyperpolarized water (HyperW). The figure was adapted from [62]. (c) The 2D 13C-1H HMQC spectrum of a nucleosome core particle (NCP) (shown is a cartoon representation of NCP, PDB ID 6ESF), in which the 153-bp Widom DNA is highly deuterated with 13CH3-methyl labeled at five sites indicated by black circles. The methyl group resonances belonging to the DNA and protein components are shown in red and gray, respectively, with the positions of the m6A methyls aliased (*). The figure was adapted from [65]. (d) Recently developed 19F labeling schemes for nucleic acids. (e) 2D 19F-13C TROSY spectra of 5-fluorouracil (red) modified human hepatitis B virus encapsidation signal epsilon (hHBV ε) element. The figure was adapted from [72].
The scope of time-resolved NMR experiments to study RNA folding was recently dramatically improved through the application of the hyperpolarized water technique [61] to boost NMR sensitivity by 300-fold [62] (Figure 4b). With this approach, real-time NMR was successfully used to monitor RNA refolding with subsecond time-resolution [62]. Time-resolved NMR experiments have also provided fresh insights into the folding landscape of DNA G-quadruplexes, reveling multiple folding pathways [63,64].
Applications to large molecular weight nucleic acids
While most studies of nucleic acid dynamics have focused on relatively small motifs, recent advances push application to much larger molecular weight assemblies. Methyl-TROSY–based NMR techniques, which have been used with great success to probe the dynamics of large protein assembles [2], have recently been successfully deployed to probe the dynamics of high-molecular-weight (as large as 200 kDa) DNA and DNA-protein complexes such as the nucleosome core particle [65]. The approach employs enzymatic methods to introduce m6A and m5C into DNA and then uses the methyl groups to probe dynamics using methyl-TROSY–based NMR experiments (Figure 4c). Developments in isotopic labelling strategies [66-69] as well as experiments for measuring RDCs in large molecular weight systems (up to 78 kDa [70]) are also making it possible to quantitatively measure NMR data in nucleic acid systems as large as 232 kDa [71].
Other exciting developments to probe the structural dynamics of nucleic acid include methods that introduce fluorine 19F into the bases [72,73], sugars [74-76] and the phosphate backbone [77] of nucleic acids (Figure 4d). These approaches take advantage of the high sensitivity and broad chemical shift range of 19F nuclei as well as the favorable relaxation properties of the aromatic 19F-13C spin pair in 2D transverse relaxation-optimized spectroscopy experiments (Figure 4e) [72,73,78]. Coupled with advances in the synthesis of site-specifically labeled and/or modified nucleotides and phosphoramidites, and methods to incorporate them into large nucleic acids, these approaches should streamline dynamics studies of large nucleic acid assemblies.
Future Perspective
Owing to continued advances in NMR methods, studies are beginning to view the vast landscape of conformations in the dynamic ensembles of nucleic acids. Using the dynamic ensemble description, a deeper and more quantitative understanding of nucleic acid mechanisms is beginning to emerge. The continued development of computational methods to model nucleic acid structure and dynamics and their integration with data from NMR and other biophysical approaches not reviewed here will be necessary to increase the accuracy, throughput, and robustness of current ensemble determination methods. We can anticipate new insights into how nature amplifies the specificity and functional complexity of nucleic acids by evolving machinery capable of ‘reading out’ features of their dynamic ensembles.
Highlights:
Advances in NMR and computational methods improve the accuracy of nucleic acid ensemble determination.
New NMR methods help uncover ‘invisible’ nucleic acid states that play important biological roles.
Fast and sensitive NMR techniques facilitate studies of large nucleic acids assemblies.
Acknowledgements
We thank Prof David A. Case (Rutgers University), Prof. Daniel Herschlag (Stanford University), Prof Christoph Kreutz (University of Innsbruck), and Prof. Joseph D. Yesselman (University of Nebraska-Lincoln) for their collaboration on determining nucleic acid ensembles. We thank Atul Rangadurai (Duke University) for his input. We thank Prof. Kwaku Dayie (University of Maryland, College Park), Prof. Lucio Frydman (Weizmann Institute of Science), Prof. Lewis Kay (University of Toronto), Prof. Kresten Lindorff-Larsen (University of Copenhagen), Prof. Harald Schwalbe (Johann Wolfgang Goethe-Universität) and Prof. Michele Vendruscolo (University of Cambridge) for providing editable figures. We acknowledge support from US National Institute for General Medical Sciences (R01GM132899) and US National Institute of Health (U54 AI150470, R01GM089846, R21AI143460, R21AI149112).
Footnotes
Conflict of interest statement
H.M.A. is an adviser to and holds an ownership interest in Nymirum Inc., an RNA-based drug discovery company. The other authors declare no competing interests.
References
- 1.Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM: The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Biol 2019, 20:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderson TR, Kay LE: NMR spectroscopy captures the essential role of dynamics in regulating biomolecular function. Cell 2021, 184:577–595. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD, Nussinov R, Wright PE: The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 2009, 5:789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frauenfelder H, Sligar SG, Wolynes PG: The energy landscapes and motions of proteins. Science 1991, 254:1598–1603. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM: Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 2007, 450:1263–1267. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Campbell SL, Dokholyan NV: Deciphering protein dynamics from NMR data using explicit structure sampling and selection. Biophys J 2007, 93:2300–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M: Simultaneous determination of protein structure and dynamics. Nature 2005, 433:128–132. [DOI] [PubMed] [Google Scholar]
- 8.Schwieters CD, Clore GM: A physical picture of atomic motions within the Dickerson DNA dodecamer in solution derived from joint ensemble refinement against NMR and large-angle X-ray scattering data. Biochemistry 2007, 46:1152–1166. [DOI] [PubMed] [Google Scholar]
- 9.Smith CA, Mazur A, Rout AK, Becker S, Lee D, de Groot BL, Griesinger C: Enhancing NMR derived ensembles with kinetics on multiple timescales. J Biomol NMR 2020, 74:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borkar AN, Vallurupalli P, Camilloni C, Kay LE, Vendruscolo M: Simultaneous NMR characterisation of multiple minima in the free energy landscape of an RNA UUCG tetraloop. Phys Chem Chem Phys 2017, 19:2797–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesari A, Bottaro S, Lindorff-Larsen K, Banas P, Sponer J, Bussi G: Fitting Corrections to an RNA Force Field Using Experimental Data. J Chem Theory Comput 2019, 15:3425–3431. [DOI] [PubMed] [Google Scholar]
- 12.Reisser S, Zucchelli S, Gustincich S, Bussi G: Conformational ensembles of an RNA hairpin using molecular dynamics and sparse NMR data. Nucleic Acids Res 2020, 48:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bottaro S, Nichols PJ, Vogeli B, Parrinello M, Lindorff-Larsen K: Integrating NMR and simulations reveals motions in the UUCG tetraloop. Nucleic Acids Res 2020, 48:5839–5848. • Combining molecular dynamic simulations, NMR eNOE data, and machine learning, atomistic ensembles were generated for the UUCG RNA tetraloop.
- 14. Shi H, Rangadurai A, Abou Assi H, Roy R, Case DA, Herschlag D, Yesselman JD, Al-Hashimi HM: Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction. Nat Commun 2020, 11:5531. •• Combining NMR RDC and cheimcal shift data with Rosetta based RNA structure prediction yields atomistic RNA ensembles with imporved speed and accuracy relative to conventional approaches.
- 15.Fonseca R, Pachov DV, Bernauer J, van den Bedem H: Characterizing RNA ensembles from NMR data with kinematic models. Nucleic Acids Res 2014, 42:9562–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank AT, Law SM, Brooks CL 3rd: A simple and fast approach for predicting (1)H and (13)C chemical shifts: toward chemical shift-guided simulations of RNA. J Phys Chem B 2014, 118:12168–12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottaro S, Bussi G, Kennedy SD, Turner DH, Lindorff-Larsen K: Conformational ensembles of RNA oligonucleotides from integrating NMR and molecular simulations. Sci Adv 2018, 4:eaar8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sathyamoorthy B, Shi H, Zhou H, Xue Y, Rangadurai A, Merriman DK, Al-Hashimi HM: Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res 2017, 45:5586–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portella G, Orozco M, Vendruscolo M: Determination of a Structural Ensemble Representing the Dynamics of a G-Quadruplex DNA. Biochemistry 2020, 59:379–388. [DOI] [PubMed] [Google Scholar]
- 20.Ganser LR, Lee J, Rangadurai A, Merriman DK, Kelly ML, Kansal AD, Sathyamoorthy B, Al-Hashimi HM: High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble. Nat Struct Mol Biol 2018, 25:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case DA: Using quantum chemistry to estimate chemical shifts in biomolecules. Biophys Chem 2020, 267:106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swails J, Zhu T, He X, Case DA: AFNMR: automated fragmentation quantum mechanical calculation of NMR chemical shifts for biomolecules. J Biomol NMR 2015, 63:125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H, Clay MC, Rangadurai A, Sathyamoorthy B, Case DA, Al-Hashimi HM: Atomic structures of excited state A-T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations. J Biomol NMR 2018, 70:229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogeli B, Segawa TF, Leitz D, Sobol A, Choutko A, Trzesniak D, van Gunsteren W, Riek R: Exact distances and internal dynamics of perdeuterated ubiquitin from NOE buildups. J Am Chem Soc 2009, 131:17215–17225. [DOI] [PubMed] [Google Scholar]
- 25.Nichols PJ, Henen MA, Born A, Strotz D, Guntert P, Vogeli B: High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints. Commun Biol 2018, 1:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bottaro S, Bengtsen T, Lindorff-Larsen K: Integrating Molecular Simulation and Experimental Data: A Bayesian/Maximum Entropy Reweighting Approach. Methods Mol Biol 2020, 2112:219–240. [DOI] [PubMed] [Google Scholar]
- 27.Tan D, Piana S, Dirks RM, Shaw DE: RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc Natl Acad Sci U S A 2018, 115:E1346–E1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sponer J, Bussi G, Krepl M, Banas P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurecka P, et al. : RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview. Chem Rev 2018, 118:4177–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sripakdeevong P, Cevec M, Chang AT, Erat MC, Ziegeler M, Zhao Q, Fox GE, Gao X, Kennedy SD, Kierzek R, et al. : Structure determination of noncanonical RNA motifs guided by (1)H NMR chemical shifts. Nat Methods 2014, 11:413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams B 2nd, Zhao B, Tandon A, Ding F, Weeks KM, Zhang Q, Dokholyan NV: Structure modeling of RNA using sparse NMR constraints. Nucleic Acids Res 2017, 45:12638–12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins AM, Rangan R, Das R: FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure 2020, 28:963–976 e966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res 2000, 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangadurai A, Szymaski ES, Kimsey IJ, Shi H, Al-Hashimi HM: Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1rho relaxation dispersion. Prog Nucl Magn Reson Spectrosc 2019, 112-113:55–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B, Zhang Q: Characterizing excited conformational states of RNA by NMR spectroscopy. Curr Opin Struct Biol 2015, 30:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marusic M, Schlagnitweit J, Petzold K: RNA Dynamics by NMR Spectroscopy. Chembiochem 2019, 20:2685–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlagnitweit J, Steiner E, Karlsson H, Petzold K: Efficient Detection of Structure and Dynamics in Unlabeled RNAs: The SELOPE Approach. Chemistry 2018, 24:6067–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, Baisden JT, Zhang Q: Probing excited conformational states of nucleic acids by nitrogen CEST NMR spectroscopy. J Magn Reson 2020, 310:106642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangadurai A, Kremser J, Shi H, Kreutz C, Al-Hashimi HM: Direct evidence for (G)O6…H2-N4(C)(+) hydrogen bonding in transient G(syn)-C(+) and G(syn)-m(5)C(+) Hoogsteen base pairs in duplex DNA from cytosine amino nitrogen off-resonance R1rho relaxation dispersion measurements. J Magn Reson 2019, 308:106589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeBlanc RM, Longhini AP, Tugarinov V, Dayie TK: NMR probing of invisible excited states using selectively labeled RNAs. J Biomol NMR 2018, 71:165–172. [DOI] [PubMed] [Google Scholar]
- 40.Rangadurai A, Shi H, Al-Hashimi HM: Extending the Sensitivity of CEST NMR Spectroscopy to Micro-to-Millisecond Dynamics in Nucleic Acids Using High-Power Radio-Frequency Fields. Angew Chem Int Ed Engl 2020, 59:11262–11266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM: Visualizing transient low-populated structures of RNA. Nature 2012, 491:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganser LR, Chu CC, Bogerd HP, Kelly ML, Cullen BR, Al-Hashimi HM: Probing RNA Conformational Equilibria within the Functional Cellular Context. Cell Rep 2020, 30:2472–2480 e2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baronti L, Guzzetti I, Ebrahimi P, Friebe Sandoz S, Steiner E, Schlagnitweit J, Fromm B, Silva L, Fontana C, Chen AA, et al. : Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA. Nature 2020, 583:139–144. •• Combining NMR RD, molecular simulations and cell-based assay, the miRNA-mRNA duplex is shown to form an ES featuring an elongated 8-mer seed and coaxially stacked topology that results in activation of the RNA-induced silencing complex and mRNA downregulation.
- 44.Gebert LFR, MacRae IJ: Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019, 20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baisden JT, Boyer JA, Zhao B, Hammond SM, Zhang Q: Visualizing a protonated RNA state that modulates microRNA-21 maturation. Nat Chem Biol 2021, 17:80–88. •• NMR RD shows that pre-miR-21 forms a pH-dependent ES, featuring a noncanonical protonated A+·G mismatch that is more efficiently processed relative to the ground state by Dicer.
- 46.White NA, Sumita M, Marquez VE, Hoogstraten CG: Coupling between conformational dynamics and catalytic function at the active site of the lead-dependent ribozyme. RNA 2018, 24:1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Plangger R, Juen MA, Hoernes TP, Nussbaumer F, Kremser J, Strebitzer E, Klingler D, Erharter K, Tollinger M, Erlacher MD, et al. : Branch site bulge conformations in domain 6 determine functional sugar puckers in group II intron splicing. Nucleic Acids Res 2019, 47:11430–11440. • Using ZZ-exchange, it is shown that group II intron domain 6 RNA exists as a dynamic equilibrium of two conformations that differ in bulge length and sugar pucker that modulates transesterification activity in splicing and reverse-splicing.
- 48.Orlovsky NI, Al-Hashimi HM, Oas TG: Exposing Hidden High-Affinity RNA Conformational States. J Am Chem Soc 2020, 142:907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu CC, Plangger R, Kreutz C, Al-Hashimi HM: Dynamic ensemble of HIV-1 RRE stem IIB reveals non-native conformations that disrupt the Rev-binding site. Nucleic Acids Res 2019, 47:7105–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimsey IJ, Szymanski ES, Zahurancik WJ, Shakya A, Xue Y, Chu CC, Sathyamoorthy B, Suo Z, Al-Hashimi HM: Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature 2018, 554:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y, Manghrani A, Liu B, Shi H, Pham U, Liu A, Al-Hashimi HM: Hoogsteen base pairs increase the susceptibility of double-stranded DNA to cytotoxic damage. J Biol Chem 2020, 295:15933–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben Imeddourene A, Zargarian L, Buckle M, Hartmann B, Mauffret O: Slow motions in A.T rich DNA sequence. Sci Rep 2020, 10:19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michalak EM, Burr ML, Bannister AJ, Dawson MA: The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol 2019, 20:573–589. [DOI] [PubMed] [Google Scholar]
- 54.Strebitzer E, Rangadurai A, Plangger R, Kremser J, Juen MA, Tollinger M, Al-Hashimi HM, Kreutz C: 5-Oxyacetic Acid Modification Destabilizes Double Helical Stem Structures and Favors Anionic Watson-Crick like cmo(5) U-G Base Pairs. Chemistry 2018, 24:18903–18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi H, Liu B, Nussbaumer F, Rangadurai A, Kreutz C, Al-Hashimi HM: NMR Chemical Exchange Measurements Reveal That N(6)-Methyladenosine Slows RNA Annealing. J Am Chem Soc 2019, 141:19988–19993. • NMR RD was used to measure nucleic acid hybridization kinetics and to show that the epitranscriptomic modification m6A slows duplex annealing, potentially explaining why the modification slows a variety of biochemical processes.
- 56.Liu B, Shi H, Rangadurai A, Nussbaumer F, Chu CC, Erharter K, Case DA, Kreutz C, Al-Hashimi HM: A quantitative model predicts how m6A reshapes the kinetic landscape of nucleic acid hybridization and conformational transitions. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubini RCA, Schon A, Muller M, Carell T, Rovo P: Impact of 5-formylcytosine on the melting kinetics of DNA by 1H NMR chemical exchange. Nucleic Acids Res 2020, 48:8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abou Assi H, Rangadurai AK, Shi H, Liu B, Clay MC, Erharter K, Kreutz C, Holley CL, Al-Hashimi HM: 2'-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res 2020, 10.1093/nar/gkaa928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binas O, Schamber T, Schwalbe H: The conformational landscape of transcription intermediates involved in the regulation of the ZMP-sensing riboswitch from Thermosinus carboxydivorans. Nucleic Acids Res 2020, 48:6970–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helmling C, Klotzner DP, Sochor F, Mooney RA, Wacker A, Landick R, Furtig B, Heckel A, Schwalbe H: Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat Commun 2018, 9:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K: Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A 2003, 100:10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Novakovic M, Olsen GL, Pinter G, Hymon D, Furtig B, Schwalbe H, Frydman L: A 300-fold enhancement of imino nucleic acid resonances by hyperpolarized water provides a new window for probing RNA refolding by 1D and 2D NMR. Proc Natl Acad Sci U S A 2020, 117:2449–2455. •• Hyperpolarized water (HyperW) NMR technique was used to boost NMR signal sensitivity by 300-fold allowing real-time monitoring of riboswitch folding at subsecond timescales.
- 63.Grun JT, Hennecker C, Klotzner DP, Harkness RW, Bessi I, Heckel A, Mittermaier AK, Schwalbe H: Conformational Dynamics of Strand Register Shifts in DNA G-Quadruplexes. J Am Chem Soc 2020, 142:264–273. [DOI] [PubMed] [Google Scholar]
- 64.Harkness RW, Hennecker C, Grun JT, Blumler A, Heckel A, Schwalbe H, Mittermaier AK: Parallel reaction pathways accelerate folding of a guanine quadruplex. Nucleic Acids Res 2021, 10.1093/nar/gkaa1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abramov G, Velyvis A, Rennella E, Wong LE, Kay LE: A methyl-TROSY approach for NMR studies of high-molecular-weight DNA with application to the nucleosome core particle. Proc Natl Acad Sci U S A 2020, 117:12836–12846. •• Combining a new method to site-specifically introduce methyl groups into DNA with methyl-TROSY, high-quality quantitative NMR dynamics data was obtained for high-molecular weight assemblies comprising the 200kDa nucleosome core particle.
- 66.Lu K, Miyazaki Y, Summers MF: Isotope labeling strategies for NMR studies of RNA. J Biomol NMR 2010, 46:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchanka A, Kreutz C, Carlomagno T: Isotope labeling for studying RNA by solid-state NMR spectroscopy. J Biomol NMR 2018, 71:151–164. [DOI] [PubMed] [Google Scholar]
- 68.Kotar A, Foley HN, Baughman KM, Keane SC: Advanced approaches for elucidating structures of large RNAs using NMR spectroscopy and complementary methods. Methods 2020, 183:93–107. [DOI] [PubMed] [Google Scholar]
- 69.Becette O, Olenginski LT, Dayie TK: Solid-Phase Chemical Synthesis of Stable Isotope-Labeled RNA to Aid Structure and Dynamics Studies by NMR Spectroscopy. Molecules 2019, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marchant J, Bax A, Summers MF: Accurate Measurement of Residual Dipolar Couplings in Large RNAs by Variable Flip Angle NMR. J Am Chem Soc 2018, 140:6978–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown JD, Kharytonchyk S, Chaudry I, Iyer AS, Carter H, Becker G, Desai Y, Glang L, Choi SH, Singh K, et al. : Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 2020, 368:413–417. • Using 2H edited NMR approaches enables the structural characterization of a conformational switch in the 232 kDa HIV-1 RNA that involves changes in one guanosine at the 5′ terminus which directs dimerization of the ~9kb genome to regulate the switch between viral translation and packaging.
- 72. Becette OB, Zong G, Chen B, Taiwo KM, Case DA, Dayie TK: Solution NMR readily reveals distinct structural folds and interactions in doubly (13)C- and (19)F-labeled RNAs. Sci Adv 2020, 6. • 19F-13C uridine labels were introduced into RNA, enabling studies of large RNAs with enhanced sensitivity using 19F─13C TROSY experiment.
- 73. Nussbaumer F, Plangger R, Roeck M, Kreutz C: Aromatic (19) F-(13) C TROSY-[(19) F, (13) C]-Pyrimidine Labeling for NMR Spectroscopy of RNA. Angew Chem Int Ed Engl 2020, 59:17062–17069. • 19F-13C pyrimidine labels were introduced into RNA, enabling studies of large RNAs with enhanced sensitivity using 19F─13C TROSY experiment.
- 74.Maximilian Himmelstoß KE, Renard Eva, Ennifar Eric, Kreutz Christoph and Micura Ronald: 2’-O-Trifluoromethylated RNA – a powerful modification for RNA chemistry and NMR spectroscopy. Chemical Science 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Q, Chen J, Trajkovski M, Zhou Y, Fan C, Lu K, Tang P, Su X, Plavec J, Xi Z, et al. : 4'-Fluorinated RNA: Synthesis, Structure, and Applications as a Sensitive (19)F NMR Probe of RNA Structure and Function. J Am Chem Soc 2020, 142:4739–4748. [DOI] [PubMed] [Google Scholar]
- 76.Dow BJ, Malik SS, Drohat AC: Defining the Role of Nucleotide Flipping in Enzyme Specificity Using (19)F NMR. J Am Chem Soc 2019, 141:4952–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baranowski MR, Warminski M, Jemielity J, Kowalska J: 5'-fluoro(di)phosphate-labeled oligonucleotides are versatile molecular probes for studying nucleic acid secondary structure and interactions by 19F NMR. Nucleic Acids Res 2020, 48:8209–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boeszoermenyi A, Chhabra S, Dubey A, Radeva DL, Burdzhiev NT, Chanev CD, Petrov OI, Gelev VM, Zhang M, Anklin C, et al. : Aromatic (19)F-(13)C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics. Nat Methods 2019, 16:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]