Summary
Background
Measuring anti-spike protein antibodies in human plasma or serum is commonly used to determine prior exposure to SARS-CoV-2 infection and to assess the anti-viral protection capacity. According to the mass-action law, a lesser concentration of tightly binding antibody can produce the same quantity of antibody-antigen complexes as higher concentrations of lower affinity antibody. Thus, measurements of antibody levels reflect both affinity and concentration. These two fundamental parameters cannot be disentangled in clinical immunoassays, and so produce a bias which depends on the assay format.
Methods
To determine the apparent affinity of anti-spike protein antibodies, a small number of antigen-coated magnetic microparticles were imaged by fluorescence microscopy after probing antigen-antibody equilibria directly in patient plasma. Direct and indirect anti-SARS-CoV-2 immunoassays were used to measure antibody levels in the blood of infected and immunised individuals.
Findings
We observed affinity maturation of antibodies in convalescent and vaccinated individuals, showing that higher affinities are achieved much faster by vaccination. We demonstrate that direct and indirect immunoassays for measuring anti-spike protein antibodies depend differently on antibody affinity which, in turn, affects accurate interpretation of the results.
Interpretation
Direct immunoassays show substantial antibody affinity dependence. This makes them useful for identifying past SARS-CoV-2 exposure. Indirect immunoassays provide more accurate quantifications of anti-viral antibody levels.
Funding
The authors are all full-time employees of Abbott Laboratories. Abbott Laboratories provided all operating funds. No external funding sources were used in this study.
Keywords: Antibody affinity, Immunoassays, HDIA, SARS-CoV-2 vaccination, Diagnostics
Research in context.
Evidence before this study
Affinity values of anti-SARS-CoV-2 antibody in blood plasma/serum of convalescent patients and vaccinated individuals had not been previously measured. The effect of antibody affinity on the results of diagnostic quantitative SARS-CoV-2 antibody assays was unknown. Affinity maturation patterns in people exposed to the infection had not been documented.
Added value of this study
We present a straightforward method for affinity determination directly in patient plasma/serum samples and use it to characterise the humoral response of SARS-CoV-2 infected patients and vaccinated individuals. We also demonstrate how the affinity affects the results of quantitative diagnostic antibody assays.
Implications of all the available evidence
Antibody affinity affects the results of diagnostic immunoassays for measuring levels of anti-SARS-CoV-2 antibody. As affinity changes with time, interpreting assay results may depend on how long after activation of the immune response the test was performed. Indirect sandwich immunoassays are better suited for quantifying anti-viral antibody levels.
Alt-text: Unlabelled box
Introduction
Antibody affinity and concentration are fundamental characteristics of adaptive immunity against infections. The SARS-CoV-2 pandemic has underscored the need for quick and practical methods for assessing these parameters in COVID-19 patients and vaccinated individuals. In particular, the ability to quickly screen a large number of patient samples is critical, especially for population surveillance,1 which per force must come second to clinical needs in emergency conditions. Initially, SARS-CoV-2 qualitative antibody assays came into their own as an emergency means for determining seroprevalence.2,3 Lately, the need has been increasingly shifting toward quantitative antibody measurements to monitor vaccine-acquired immune protection.4, 5, 6 These quantitative tests, being developed under less urgent conditions, must be scrutinised for their ability to accurately assess the level of immune protection for tested individuals.
Immunometric (or sandwich) immunoassays have proven to be preeminent for measuring concentrations of diagnostically important macromolecules.7 In these assays, the analyte is sandwiched between the capture reagent (to concentrate the target) and the detection reagent (to generate a signal). Sandwich formation is driven by the affinity and concentration of the interacting molecules. For instance, a few antibodies that bind extremely well can create as many immune complexes as a higher concentration of poorly binding antibodies. Therefore, quantifications of anti–SARS-CoV-2 antibody depend on both these parameters. The measured value reflects a convolution of patient antibody concentration and affinity, which both vary widely with time and the strength of the immune response.8, 9, 10
Furthermore, the assay design also plays a role. There are two common immunometric assay formats utilised for SARS-CoV-2 antibody detection on automated diagnostic platforms. Indirect assays capture anti-spike antibodies from patient plasma/serum on microparticles coated with recombinant spike protein and detect them with labelled anti-human IgG or IgM secondary antibody (e.g., mouse anti-human). Direct immunoassays also capture patient antibodies with spike protein-coated microparticles but utilise a different detection approach. Instead of secondary antibody, the same recombinant spike protein, labelled with a signal generating group, is employed. In this format, the sample, microparticles, and labelled conjugate are all combined simultaneously, which allows the conjugate to fill antibody binding sites that remain free from interacting with microparticles. Clearly, both immunoassay formats rely on the patient antibody's ability to bind antigen. Measurements of antibody concentration are inextricably linked to their affinity.
When interpreting measured antibody levels, it is important to understand the degree to which affinity biases the reported values. To address this challenge, we developed a research method to quickly estimate antibody affinity directly in patient samples and used these data for evaluating the affinity dependence of diagnostic immunoassays. As is well-known from literature (see examples11,12), decoupling affinity and concentration requires a multi-point titration experiment. In each individual measurement, the signal is indeed a convolution of affinity and concentration. However, by controlling the reagent concentrations at each titration point (antigen is varied, antibody held fixed, or vice versa) and globally fitting the data to a binding model, it is possible to extract the apparent affinity value independently from the concentration. To avoid stoichiometric binding and other artifacts, it is important to keep the antibody concentration at or below the expected dissociation constant (Supplementary Figure 1).
While the design of these titration experiments is constrained by the laws of chemistry, the choice of detection strategy is more flexible. There are a host of biophysical methods commonly used to perform affinity studies with purified reagents.13, 14, 15, 16, 17, 18, 19 However, measuring antibody affinity directly in blood plasma and other complex media presents a challenge. Here, we introduce a new approach to probe binding equilibria with minimal perturbation. We show that the affinity of unpurified antibody can be reliably estimated by performing solution-phase binding, probing the reactions with microparticles, and detecting the complexes by imaging.20 The technique offers high sensitivity, simple protocols, and minimal reagent consumption. We validate this method over an affinity range of 0·05–50 nM using previously well-characterised monoclonal antibodies.21,22 We present this approach as a tool to measure the apparent affinity of patient antibody directly in blood plasma or serum without the need for purification or additional labelling steps.20
First, we applied the technique to monitoring antibody maturation in SARS-CoV-2 seroconversion samples and found that high antibody affinities can be reached much faster by vaccination than by overcoming infection. Then, the method was used to determine antibody affinities in a patient panel. By correlating these data with the results of the indirect and direct antibody assays run on the same panel, we found that the indirect immunoassay format is less dependant on antibody affinity and is thus better suited for measuring antibody concentrations.
Methods
Fluorescence imaging
Fluorescently labelled magnetic microparticles were placed in an optical-bottom 96-well plates (Nunc, Thermo Fisher Scientific, Waltham, MA), and placed on a 96-well neodymium magnet array for 3 s to pull microparticles down to the glass surface. An IX83 inverted microscope (Olympus, Tokyo, Japan), equipped with ZeroDrift IX3-ZDC2 continuous focus, was used to measure samples with bright-field illumination from a pE-100 LED (CoolLED, Andover, UK) and epifluorescence excitation light produced by an X-Cite LED illumination system (Excelitas, Wheeling, IL) passing through a Cy3 Olympus dichroic filter cube set (Edmond Optics, Barrington, NJ). Images were acquired through a 20x air objective (UPlanXApo, NA=0·80, Olympus) and recorded on a scientific CMOS PCO.panda camera (PCO, Kelheim, Germany). MetaMorph software (Molecular Devices, San Jose, CA) coordinated imaging measurements, directing a ProScan III xy-stage (Prior Scientific Instruments, Cambridge, UK) to acquire one bright-field image (100 ms, 1·3% LED power) and one (or two) fluorescence image(s) (10 and 100 ms, 25% LED power) at 9 positions within each 96-well. Images were analysed as described previously.20
SARS-CoV-2 patient samples
Seroconversion patient samples were purchased from New York Biologics, Inc. (Southampton, NY), in which patients having a positive PCR SARS-CoV-2 test had blood drawn every few days for ∼ 2–6 weeks under IRB approved protocols. Moderna vaccinated patient samples were purchased from Access Biologicals, LLC (Vista, CA), in which blood was drawn just prior to the first dose of vaccine, ∼3–4 weeks after dose 1, and ∼2–3 weeks after dose 2. The convalescent patient plasma samples were purchased from New York Blood centre (New York, NY). The samples were collected under IRB approved protocols.
Apparent affinity titration experiments
Patient samples were diluted in Architect wash buffer (phosphate buffer containing detergent, Abbott Laboratories, Abbott Park, IL) to achieve a maximum fluorescence intensity signal of ∼2000 counts per particle (CPP), an antibody concentration estimated to be approximately 60 pM and well below the characteristic antibody affinity range. The diluted patient antibody was aliquoted into 23 wells of two 96-well plates. The CHO-cell expressed recombinant receptor binding domain (RBD) of the S1 subunit of spike protein was generated in-house (Abbott Laboratories, Abbott Park, IL) and titrated into these aliquots in two-fold dilutions from 10 µM down to 5 pM. The 23rd aliquot, left free of RBD, was the zero-titration point, and a final well, containing pooled negative human plasma, served as a background control. Each of these samples was incubated at 37 °C for 30 min to equilibrate the antibody-RBD binding reaction. Magnetic microparticles (4·7 µm) were coated with the same recombinant RBD protein (coating conditions: 50 µg/mL, 1% solids) using standard carbodiimide (EDAC) coupling. Then, 2 µL of RBD-coated magnetic microparticles (0·1% solids) were incubated for 5 min to probe these equilibria by capturing unbound patient antibody. Following this short probe step, the microparticles were washed and reacted for 10 min with Cy3-labelled donkey anti-human-IgG antibody (Jackson Immunoresearch, West Grove, PA) to detect the captured patient antibody, and then washed 3 more times in wash buffer. All assay steps were carried out on a KingFisher magnetic microparticle plate processor (Thermo Fisher Scientific), maintained at 37 °C, and then the microparticles were imaged as described in our high-definition immunoassay (HDIA) approach20 on the fluorescence microscope. The resulting binding data (fluorescence signal vs. antigen concentration) were fit to a standard four-parameter logistic model.
We want to underscore that the potentially multivalent nature of the RBD antigen and the polyclonal antibody response considerably complicate affinity determinations. The system does not follow the “one antibody binding site per one antigen molecule” requirements and therefore violates the simple bi-molecular binding model. As a protein of considerable size, RBD likely contains several antigenic epitopes. When interacting with polyclonal antibodies, it may form multi-molecular immune complexes with variable stoichiometry. As a result, we had to apply a heuristic, four parameter logistic model to fit the binding data.
SARS-CoV-2 antibody diagnostic immunoassay data
Aliquots from all patient samples presented in this work underwent SARS-CoV-2 diagnostic testing on an Architect instrument (Abbott). The SARS-CoV-2 IgM, and the indirect format SARS-CoV-2 IgG II Quant Architect assays were performed using commercially available assay kits, according to the package instructions (link to the package insert: https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease). The direct format SARS-CoV-2 Total Ig (S) research assay was developed in-house (Abbott) to detect antibodies against RBD of the S1 subunit of the spike protein of SARS-CoV-2 virus in patient plasma. In this one-step assay, patient sample (35 µL), SARS-CoV-2 spike protein-coated magnetic microparticles (50 µL, 0·075% solids) and acridinium-labelled spike protein conjugate (50 µL, 80 ng/mL) were combined, incubated, washed, and measured on an Abbott Architect analyser.
Ethics
Patient plasma/serum samples used for this study were purchased from New York Biologics (Southampton, NY), Access Biologicals, LLC (Vista, CA) and New York Blood centre (New York, NY). The samples purchased from Access Biologicals LLC were collected under an IRB protocol approved by Ballad Health System IRB. The samples purchased from New York Biologics were collected under an IRB approved by Ethical and Independent Review Services (E&I IRB #2-IRB00007807). The patient plasma samples purchased from New York Blood centre (New York, NY), were collected from volunteer blood donors who consented to the use of their samples for research purposes at the time of collection.
Statistics
Fluorescence signals and apparent affinity values are reported as the mean ± Fluorescence signal uncertainties were obtained from multiple images per sample (n = 9), as described above. We used Pearson's correlation analysis to determine the relationship between the direct/indirect assay ratio and the affinity of antibody in patient samples drawn less than 20 days after the onset of SARS-CoV-2 infection and in samples drawn more than 20 days after onset.
Role of the funders
The authors are all full-time employees of Abbott Laboratories. Abbott Laboratories provided all operating funds. No one other than the authors had any role in study design, data interpretation or reporting.
Results
Solution-phase reaction solid-phase detection (SRSD)
Blood plasma, or serum, is the natural environment for antibody-antigen interactions. However, characterizing antibody affinity directly in blood plasma presents a serious challenge. Many other plasma constituents (e.g., albumin, lipids, bilirubin) are present at much higher concentrations than immunoglobulins. This creates a significant background which complicates detection of specific antibodies. Purification is the typical solution, however, purifying polyclonal antibodies from patient samples is always prone to substantial losses and may systematically perturb the distribution of antibody subpopulations. Additionally, when working with small-volume patient samples, antibody purification is not an option.
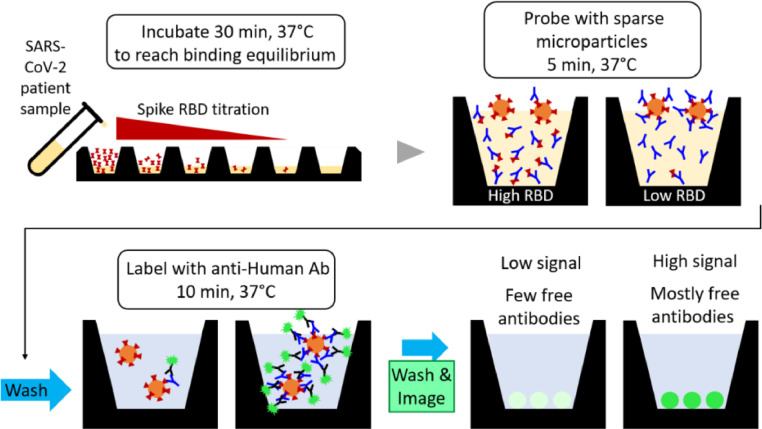
Here, we present a method, termed solution-phase reaction solid-phase detection (SRSD), to overcome these obstacles. Figure 1 outlines the SRSD methodology. After achieving equilibrium during the initial, solution-phase binding, SRSD employs antigen-coated magnetic microparticles to quantify unbound patient antibody. Unlike kinetic exclusion assays,23 we employ an extremely small number of microparticles to minimise perturbation of the equilibrium. Briefly, the microparticles are coated with a large amount of spike protein antigen and so have very high binding capacities (Supplementary Figure 2). Each microparticle acts independently to capture and hold all the free antibody which it encounters. The minimal sampling approach results in a relatively small amount of captured patient antibody on the microparticle surface. Therefore, following a second incubation with fluorescently labelled, anti-human-IgG conjugate, we employ highly sensitive imaging-based detection.20 By measuring a series of patient sample aliquots with antigen concentrations ranging from micromolar to picomolar, as well as zero antigen concentration control, a binding curve can be obtained and fit to determine the apparent affinity.
Figure 1.
SRSD protocol for determining apparent affinity of antibodies in patient samples. SARS-CoV-2 spike RBD antigen is titrated into aliquots of diluted patient serum/plasma and incubated to equilibrate antibody-antigen binding. These equilibria are probed with RBD-coated microparticles to sample the unbound patient antibody that remains in solution (using sparse microparticles and a short incubation time to minimise perturbation of the equilibrium). Subsequently, the microparticle-collected antibody is quantified with Cy3-labelled anti-human-IgG conjugate and fluorescence imaging. Finally, we calculate the fraction of RBD-bound antibody that remains in solution and plot it as a function of total RBD concentration.
As mentioned in the introduction, for accurate measurements it is important to keep the patient sample antibody concentration at or below the expected dissociation constant of the binding reaction. This can be achieved by diluting the patient sample (antibody) as much as possible while still maintaining a measurable signal, and then confirming the necessary conditions (no dependence on antibody concentration) by showing identical titration curves at different patient antibody dilutions (Supplementary Figure 1). Additionally, the fluorescence intensity signals in the titration experiments are proportional to the patient antibody concentration. While we cannot assign an antibody concentration value based on these experiments, we can establish a valid signal range, and thus ensure that all subsequent patient samples are diluted into the correct experimental regime.
All antibody-antigen interactions follow universal principles and the laws of physical chemistry. Thus, the best way to evaluate a new method for measuring affinity is to compare it with an established technique using a well characterised antibody-antigen system. Measuring binding parameters in solution is standard biophysical practice as it avoids the inhomogeneous binding and surface effect artifacts often observed in ELISA and SPR measurements.24, 25, 26 Therefore, the widely accepted, solution-phase spectroscopic approach12 along with previously characterised monoclonal antibodies21,22 were used to validate the SRSD method. Validation experiments were designed to cover a broad range of dissociation constants (0·05–50 nM) and yielded nearly identical results from both techniques (Supplementary Figure 3).
Measuring anti-SARS-CoV-2 antibody affinities in patient samples
Given the impact of the SARS-CoV-2 pandemic and the ongoing interest in understanding the level of immunity conveyed by having recovered from a SARS-CoV-2 infection, and/or from receiving doses of vaccine, we employed SRSD to probe the affinity of patient antibody against the receptor binding domain (RBD) of the spike protein. The SARS-CoV-2 virus gains entry to humans cells via the spike protein's interaction with cell surface ACE2 receptors,27,28 and thus spike protein is the primary target in current vaccines.29,30 The time course for affinity maturation of anti-RBD antibodies provides information about the humoral immune response of convalescent patients and vaccinated individuals. Moreover, as we will demonstrate later, affinity data can be used to clarify the apparently contrasting results from different formats of SARS-CoV-2 antibody diagnostics assays.
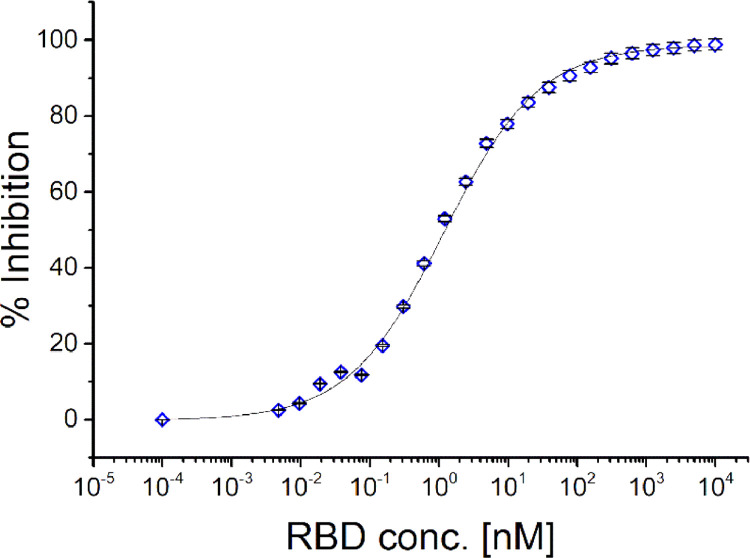
SRSD is simple and straightforward. Figure 2 shows the SRSD results from measuring equilibrated solutions of RBD titrated into patient plasma. SRSD linearity and validation experiments supporting these results can be found in the Supplementary Information (Supplementary Figs. 2 and 3). Recombinant RBD protein was titrated into twenty-three aliquots of a SARS-CoV-2 patient sample, and each reaction was incubated for 30 min at 37 °C to achieve equilibrium. Next, 2 µL of magnetic microparticles (0·1% solids) coated with the same recombinant RBD were added and incubated for 5 min to probe these equilibria by capturing unbound patient antibody. We optimised the number of microparticles and incubation time to limit perturbation effects to a few percent, thereby falling into the measurement error (Supplementary Figure 4). Following this short probe step, the microparticles were reacted with Cy3-labelled anti-human antibody, washed, and imaged. Taking the zero RBD control sample to establish the fully unbound—or maximum— patient antibody signal, one can plot the percent of signal inhibited (by formation of antibody-RBD complex) as a function of total RBD concentration. The result can be fit with a standard logistic function to determine the 50% binding point, which in this case suggests the patient has an apparent affinity constant of 0·91 × 109 M−1 (SD/mean: ± 13%, n = 9).
Figure 2.
Representative antibody-RBD binding titration curve (single blood draw). Using the protocol described in Figure 1 and the maximum intensity from the zero RBD concentration control, we calculate the fraction of unbound patient antibody, blocked from binding to the microparticles due to antibody-RBD complex formation. Plotting the percent inhibition as a function of added RBD protein generates a binding curve. A heuristic, standard four-parameter logistic fit is used to obtain the 50%-inhibition concentration, the inverse of which is a measure of the apparent affinity of this patient's anti-spike antibody, 0·91 × 109 M−1. error bars: ±.
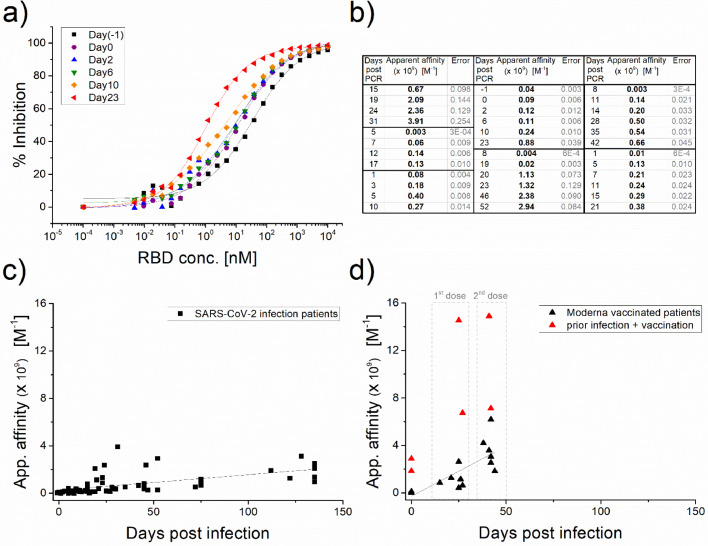
We next applied SRSD to SARS-CoV-2 seroconversion patient samples. Figure 3a displays RBD titration curves for multiple blood draws from a single patient over the three weeks following a positive PCR SARS-CoV-2 test. The curves clearly shift to the left with time, showing increasing affinity of the antibody. As seen in Figure 3b, this trend was observed in all sequential blood draws for a given patient, and the final apparent affinities achieved are similar, with estimated constants on the close order of 109 M−1. It is also worth noting that the data indicate that affinities typically change by a few orders of magnitude over the course of the infection and convalescence. The apparent affinity results of Figure 3c, along with additional, single point, patient plasma samples, are plotted as a function of days following the onset of infection. A positive PCR test was used to define infection onset, which is rather imprecise as it depends on when the patient was tested. Nevertheless, the data clearly show that the apparent affinities of the patients’ antibodies are increasing with time.
Figure 3.
Anti-SARS-CoV-2 antibody affinities in convalescent and vaccinated patient plasma/serum. (a) Binding curves measured using a SARS-CoV-2 seroconversion panel from a single patient, consisting of blood samples drawn at intervals following a positive PCR test. The shift of the curves to the left indicates that as time progresses, there is a pronounced decrease in apparent anti-SARS-CoV-2 antibody dissociation constants (i.e., strong increase in affinities). (b) The table shows data from additional seroconversion patient samples supporting this observed trend (36 samples, 7 patients). (c) Apparent affinity data consolidated from both seroconversion panels and standalone blood draw samples (squares, 63 samples, 21 patients) are plotted as a function of time. The plot demonstrates that anti-SARS-CoV-2 antibody maturation is observed over several months following infection onset. (d) Similar apparent affinity data from blood draws following the first and second doses of Moderna vaccine (triangles, 18 samples, 8 patients) reveal that high anti-SARS-CoV-2 affinities were reached in a much shorter time. Two patients who appear to have experienced a SARS-CoV-2 infection prior to vaccination (red triangles) demonstrated the highest affinities. The Pearson's correlation coefficient is 0.575 (t-test p-Value = <0.0001, 95% CI: [0.381, 0.720]) for the infected patients, and 0.761(t-test p-Value<0.001, 95% CI: [0.408, 0.916]) for the vaccinated individuals (not including red triangles).
Having demonstrated that we can obtain anti-spike antibody affinity values that follow the expected maturation pattern, we next apply SRSD to study vaccinated individuals. Each individual provided three blood samples: pre-vaccination, three weeks following dose 1 of the Moderna vaccine, and two weeks post dose 2. The same SRSD protocol as described above was utilised. The pre-vaccine plasma sample was used as the background control in place of negative human plasma, except in the cases where significant anti-spike IgG levels were already present in the pre-vaccination sample, suggesting the patient had previously been infected with SARS-CoV-2. In each of the measured patients, the apparent affinity following the second dose was higher than after the first dose. The vaccinated individual affinity datapoints vs. days post vaccination are plotted in Figure 3d for comparison with infected patient affinities (Figure 3c). It is clear, that vaccinated people (triangles) are achieving affinities equivalent to, or tighter than, those of the infected patients (squares) and doing so in a much shorter time frame. Most vaccinated individuals developed high affinity antibodies by the third week following the first dose. Convalescent patients do not achieve comparable affinities until 2–4 months after infection onset.
Previous studies have shown that patient antibody levels following SARS-CoV-2 exposure and a single vaccine dose are similar to the levels following two vaccine doses in infection-naïve individuals.31,32 It is interesting to observe that the highest antibodies affinities measured in our study were found in the two patients who appeared to have had a SARS-CoV-2 infection prior to being vaccinated (red triangles). These two patients showed significant levels of high affinity anti-spike IgG in their baseline blood draw. Following the first dose of the vaccine, their antibodies showed an extremely tight apparent affinity (equivalent to dissociation constants of ∼2 and 4 pM). Vaccination appears to further increase the infection-triggered immune response. In these cases, the second vaccine dose only provided a marginal increase in affinity.
Anti- SARS-CoV-2 antibody affinity affects outcomes of diagnostic antibody assays. The ongoing spread of SARS-CoV-2, as well as the existence of current and future variants means that vaccine titre monitoring and antibody assays continue to have an important role in global health. The current generations of commercial COVID-19 antibody immunoassays were developed extremely rapidly under emergency conditions. As the clinical and diagnostic research communities now perform rigorous evaluations of these assays, they are observing discrepancies in the dynamics of the anti-SARS-CoV-2 antibody response, as reported by immunoassays employing two common assay formats.33
Anti-viral antibody levels in patients with an infection are known to have a certain dynamic pattern.8, 9, 10 IgM antibody levels rise shortly after the onset of infection and then fall away as the more specific IgG antibodies go into production. IgG levels in the blood rise higher and last longer to provide ongoing immunity, but how long they persist has been an important point in understanding anti-SARS-CoV-2 immunity and the defence against re-infection.
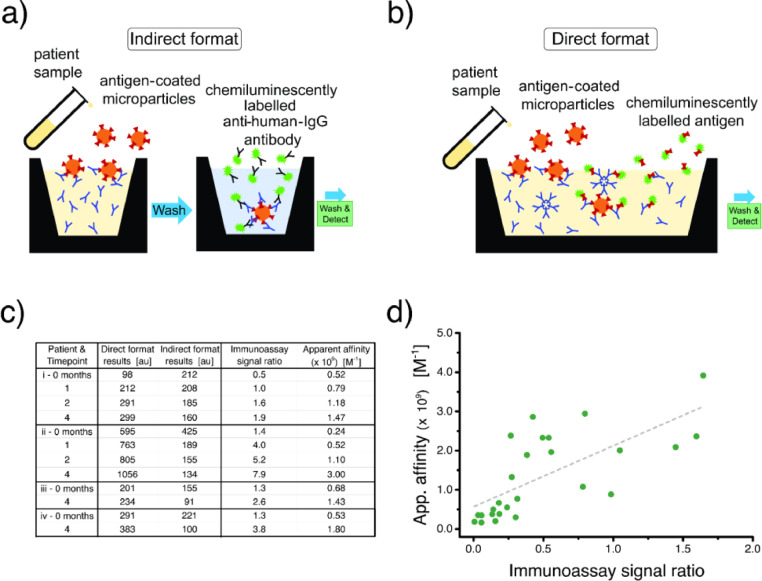
Therefore, it is necessary to monitor not only the presence of anti-SARS-CoV-2 IgG antibody—indicating virus exposure—but also the quantity of IgG, to determine protection levels following infection and/or vaccination. As mentioned in the Introduction, there are two common immunometric assay formats which can be used to detect anti-viral antibodies in patient samples. Both formats employ analyte-specific, RBD-coated microparticles for the capture step but utilise different detection conjugates. Indirect assays use signal generating anti-human IgG secondary antibody (Figure 4a), while direct immunoassays use labelled RBD to fill the remaining binding sites of captured antibodies (Figure 4b). Indirect antibody assays are performed as two-step sandwich immunoassays and are intended to measure a certain immunoglobulin class. Direct immunoassays are also sandwich immunoassays, typically performed in one step, and are designed to measure antibodies of all classes. Many assay developers favour direct assays as using this format often helps to minimise nonspecific background.
Figure 4.
Two formats of SARS-CoV-2 antibody diagnostic assays and their relative dependence on patient antibody affinity. (a) The cartoon depicts an indirect, two-step sandwich assay format to detect patient antibody. A sample containing anti-RBD antibody is incubated with antigen-coated microparticles. After washing, the microparticle-captured antibody is incubated with labelled anti-human IgG conjugate, washed again, and measured. (b) The direct format sandwich assay, typically performed in one step (all reagents added simultaneously) is an alternative means for quantifying anti-RBD antibodies. It uses the same antigen-coated microparticles to capture antibodies, while, in this case, labelled antigen serves as the detection conjugate. (c) Data from four patients (i, ii, iii, iv) who provided blood samples at different times (up to 4 months after infection onset). For each patient, the assay signals (antibody levels) show opposite trends over time when measured by a direct vs. indirect assay. The table also includes the calculated ratio of direct/indirect signals and the measured affinities. (d) Correlating the direct/indirect signal ratio with affinity demonstrates that the direct format assay has a pronounced dependence on patient antibody affinity (26 samples, 16 patients). The Pearson's correlation coefficient is 0.702 (t-test p-Value<0.0001, 95% CI: [0.432, 0.856]).
Effect of antibody affinity on diagnostic anti- SARS-CoV-2 antibody assays
When monitoring seroconversion panels with assays in both formats, we observed an apparent discrepancy. In some patients, at a few months post onset of a SARS-CoV-2 infection, the indirect assay showed a decreasing signal, indicating reduced concentration of anti-spike IgG, while the direct assay of the same sample showed a steady, or even increasing, signal (Figure 4c). We hypothesised that the observed discrepancy stemmed from the effect of different dependencies on patient antibody affinity. Since the direct assay format relies on patient antibody binding sites for both capture and detection, it is likely to be more influenced by this important parameter. A similar hypothesis is also indicated in a recent publication by Di Germanio et al.33 To test this hypothesis, we returned to the patient samples that had demonstrated this discrepancy (Figure 4c), and used SRSD to acquire the apparent affinities. Then, when running a given sample with both direct and indirect assays, we calculated the ratio of the output signals. Since both assays are measuring the same sample, the antibody concentration component of that ratio should be a constant. This means that any ratio variations across samples should reflect differences in the affinity component.
Furthermore, using the ratio of the assay signals also allowed us to combine all data obtained from numerous patient samples in a single dataset without bias from the varying antibody concentrations, and to plot them as a function of the disease time course. A positive correlation of the direct/indirect signal ratio with measured affinity indicates that the numerator (direct format signal) has greater affinity dependence. In fact, referring to Figure 4c, we observed that the ratio of the direct/indirect signals is tracking with the affinities. To explore this result further, we collected affinities, as well as direct and indirect assay results, from additional seroconversion and stand-alone patient sample panels and then plotted affinity as a function of signal ratio (Figure 4d). The plot shows a good, positive correlation with apparent affinity, indicating a stronger affinity dependence for the direct format. Note that this plot (Figure 4d) is constructed from patient samples drawn more than 20 days after the onset of SARS-CoV-2 infection. Signal ratios obtained for samples collected during the early stage of the infection (Supplementary Figure 5) are uncorrelated with affinity. This is likely influenced by the increased presence of IgM and low-affinity IgG antibodies, which contribute to the direct but not the indirect assay signal. We should also keep in mind the high variability in patient immune responses, which can be amplified by lower— and thus noisier—IgG signal levels. Nevertheless, these results clearly indicate that the direct immunoassay signal has a strong correlation, and thus dependence, on the affinity of the patient antibody. Therefore, as affinity maturation takes place and affinity improves, the direct format signal will tend to increase despite a drop in actual IgG concentration in the blood plasma. This sensitivity and preference for higher affinity anti-spike antibodies make direct format immunoassays useful for extending the time window in which a previous SARS-CoV-2 infection can be detected. However, for quantifying the level of anti-spike IgG antibody, indirect formats provide more accurate results.
Discussion
We demonstrated a practical research approach for measuring the apparent affinity of antibodies directly in human blood plasma/serum without purification or labelling. It can be used to follow natural antibody maturation, which is a key aspect of the immune response, in patients who suffered and recovered from a SARS-CoV-2 infection and/or upon vaccination. We also found that antibody affinity can impact diagnostic SARS-CoV-2 antibody measurements. We showed that direct immunoassays, which include two antibody-antigen binding reactions, are correlated with the antibody affinity. This improves the ability of direct assays to identify previous SARS-CoV-2 exposure. On the other hand, the indirect immunoassays are better suited for quantifying anti-viral antibody levels.
In general, it is important for the medical community to be aware that antibody affinity affects diagnostic immunoassay outputs. As affinity changes with time, interpreting assay results may depend on how long after activation of the immune response the test was performed.
CRediT authorship contribution statement
Patrick J. Macdonald: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Qiaoqiao Ruan: Conceptualization, Methodology, Formal analysis. Jessica L. Grieshaber: Methodology, Writing – review & editing. Kerry M. Swift: Methodology, Writing – review & editing. Russell E. Taylor: Methodology, Writing – review & editing. John C. Prostko: Conceptualization, Writing – review & editing, Formal analysis. Sergey Y. Tetin: Conceptualization, Writing – original draft.
CRediT authorship contribution statement
Patrick J. : Conceptualization, Methodology, Formal analysis, Writing – review & editing. Qiaoqiao : Conceptualization, Methodology, Formal analysis. Jessica L. : Methodology, Writing – review & editing. Kerry M. : Methodology, Writing – review & editing. Russell E. : Methodology, Writing – review & editing. John C. : Conceptualization, Writing – review & editing, Formal analysis. Sergey Y. Tetin: Conceptualization, Writing – original draft.
Declaration of interests
All authors are employees of Abbott Laboratories.
Acknowledgments
Acknowledgements
We thank all our Abbott colleagues without whom this work could not have been completed. Our gratitude goes to Mary A. Rodgers for providing patient samples, to Yazmin San Miguel and Li Zhou for help with statistics, and to Claudio Galli and Audrey L. Bartnicki for critical reading of the manuscript. Many reagents used in this work were made in our R&D and Operations labs. All funding was provided by Abbott Laboratories.
Data Sharing Statement
Data generated or used in this study are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103796.
Appendix. Supplementary materials
References
- 1.Berkelman R.L., Bryan R.T., Osterholm M.T., LeDuc J.W., Hughes J.M. Infectious disease surveillance: a crumbling foundation. Science. 1994;264(5157):368–370. doi: 10.1126/science.8153621. [DOI] [PubMed] [Google Scholar]
- 2.Reifer J., Hayum N., Heszkel B., Klagsbald I., Streva V.A. SARS-CoV-2 IgG antibody responses in New York City. Diagn Microbiol Infect Dis. 2020;98(3) doi: 10.1016/j.diagmicrobio.2020.115128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippi G., Plebani M. SARS-CoV-2 antibodies titration: a reappraisal. Ann Transl Med. 2020;8(16):1032. doi: 10.21037/atm-20-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Vu S., Jones G., Anna F., et al. Prevalence of SARS-CoV-2 antibodies in France: results from nationwide serological surveillance. Nat Commun. 2021;12(1):3025. doi: 10.1038/s41467-021-23233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogl T., Leviatan S., Segal E. SARS-CoV-2 antibody testing for estimating COVID-19 prevalence in the population. Cell Rep Med. 2021;2(2) doi: 10.1016/j.xcrm.2021.100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Addison G.M., Hales C.N., Kirkham K.E., Hunter W.M. Radioimmunoassay Methods. Edinburgh. Churchill-Livingstone; 1970. The immunoradiometric assay; pp. 447–461. [Google Scholar]
- 8.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101(8):791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen H.N., Siskind G.W. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3(7):996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 10.Eisen H.N. Affinity enhancement of antibodies: how low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol Res. 2014;2(5):381–392. doi: 10.1158/2326-6066.CIR-14-0029. [DOI] [PubMed] [Google Scholar]
- 11.Weber G., Pullman B.W. Molecular Biophysics. Academic Press; New York: 1965. The binding of small molecules to proteins; pp. 369–397. M. [Google Scholar]
- 12.Tetin S.Y., Hazlett T.L. Optical spectroscopy in studies of antibody–hapten Interactions. Methods. 2000;20(3):341–361. doi: 10.1006/meth.1999.0927. [DOI] [PubMed] [Google Scholar]
- 13.Eisen H.N. Equilibrium dialysis for measurement of antibody-hapten affinities. Methods Med Res. 1964;10:106–114. [PubMed] [Google Scholar]
- 14.Steward M., Steensgaard J. CRC Press, Inc.; Boca Raton, FL: 1983. Antibody Affinity: Thermodynamic Aspects and Biological Significance. [Google Scholar]
- 15.Lebowitz J., Lewis M.S., Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11(9):2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phizicky E.M., Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59(1):94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen H.N. Determination of antibody affinity for haptens and antigens by means of fluorescence quenching. Methods Med Res. 1964;10:115–121. [PubMed] [Google Scholar]
- 18.Dandliker W.B., Kelly R.J., Dandliker J., Farquhar J., Levin J. Fluorescence polarization immunoassay. Theory and experimental method. Immunochemistry. 1973;10(4):219–227. doi: 10.1016/0019-2791(73)90198-5. [DOI] [PubMed] [Google Scholar]
- 19.Tetin S.Y., Swift K.M., Matayoshi E.D. Measuring antibody affinity and performing immunoassay at the single molecule level. Anal Biochem. 2002;307(1):84–91. doi: 10.1016/s0003-2697(02)00011-8. [DOI] [PubMed] [Google Scholar]
- 20.Ruan Q., Macdonald P.J., Swift K.M., Tetin S.Y. Direct single-molecule imaging for diagnostic and blood screening assays. Proc Natl Acad Sci USA. 2021;118(14) doi: 10.1073/pnas.2025033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetin S.Y., Ruan Q., Saldana S.C., et al. Interactions of two monoclonal antibodies with BNP: high resolution epitope mapping using fluorescence correlation spectroscopy. Biochemistry. 2006;45(47):14155–14165. doi: 10.1021/bi0607047. [DOI] [PubMed] [Google Scholar]
- 22.Longenecker K.L., Ruan Q., Fry E.H., et al. Crystal structure and thermodynamic analysis of diagnostic mAb 106.3 complexed with BNP 5-13 (C10A) Proteins. 2009;76(3):536–547. doi: 10.1002/prot.22366. [DOI] [PubMed] [Google Scholar]
- 23.Darling R.J., Brault P.A. Kinetic exclusion assay technology: characterization of molecular interactions. Assay Drug Dev Technol. 2004;2(6):647–657. doi: 10.1089/adt.2004.2.647. [DOI] [PubMed] [Google Scholar]
- 24.Nieba L., Krebber A., Plückthun A. Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal Biochem. 1996;234(2):155–165. doi: 10.1006/abio.1996.0067. [DOI] [PubMed] [Google Scholar]
- 25.Underwood P.A. Problems and pitfalls with measurement of antibody affinity using solid phase binding in the ELISA. J Immunol Methods. 1993;164(1):119–130. doi: 10.1016/0022-1759(93)90282-c. [DOI] [PubMed] [Google Scholar]
- 26.Myszka D.G. Improving biosensor analysis. J Mol Recognit. 1999;12(5):279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Wang W., Chen Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 30.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 31.Anderson M., Stec M., Rewane A., Landay A., Cloherty G., Moy J. SARS-CoV-2 antibody responses in infection-naive or previously infected individuals after 1 and 2 doses of the BNT162b2 vaccine. JAMA Netw Open. 2021;4(8) doi: 10.1001/jamanetworkopen.2021.19741. -e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebinger J.E., Fert-Bober J., Printsev I., et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med. 2021;27(6):981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Germanio C., Simmons G., Kelly K., et al. SARS-CoV-2 antibody persistence in COVID-19 convalescent plasma donors: dependency on assay format and applicability to serosurveillance. Transfusion. 2021;61(9):2677–2687. doi: 10.1111/trf.16555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.