Abstract
Upon infection, the herpes simplex virus (HSV) transcriptional activator VP16 directs the formation of a multiprotein-DNA complex—the VP16-induced complex—with two cellular proteins, the host cell factor HCF-1 and the POU domain transcription factor Oct-1, on TAATGARAT-containing sequences found in the promoters of HSV immediate-early genes. HSV VP16 contains carboxy-terminal sequences important for transcriptional activation and a central conserved core that is important for VP16-induced complex assembly. On its own, VP16 displays little, if any, sequence-specific DNA-binding activity. We show here that, within the VP16-induced complex, however, the VP16 core has an important role in DNA binding. Mutation of basic residues on the surface of the VP16 core reveals a novel DNA-binding surface with essential residues which are conserved among VP16 orthologs. These results illuminate how, through association with DNA, VP16 is able to interpret cis-regulatory signals in the DNA to direct the assembly of a multiprotein-DNA transcriptional regulatory complex.
In eukaryotes, regulation of gene expression often involves the assembly of multiprotein-DNA complexes consisting of trans-acting proteins, called transcription factors, bound to cis-acting DNA elements in promoters. To assemble a proper transcriptional regulatory complex, transcription factors must recognize and interpret regulatory signals encoded in the DNA. An example of such a multiprotein transcriptional regulatory complex is the herpes simplex virus (HSV) VP16-induced complex, which regulates the transcription of HSV immediate-early (IE) genes.
The VP16-induced complex is composed of three proteins—the viral protein VP16 and two cellular proteins, HCF-1 and Oct-1 (see references 14, 31, and 43 for reviews). Upon HSV infection, VP16 (also known as αTIF and Vmw65), a component of the virus particle, is released into the cell, where it first binds to HCF-1 (17, 20, 37, 50), a protein involved in cell proliferation (9). HCF-1 (also known as HCF, C1, VCAF, and CFF) is a dimeric complex of polypeptides that arises from internal proteolytic cleavage of a single large precursor protein of over 2,000 amino acids (22, 48, 49). Although HCF-1 is large and complex, its amino-terminal 380 residues are sufficient for association with VP16 and VP16-induced complex formation (23, 47).
HCF-1 association with VP16 leads to the subsequent interaction of the VP16–HCF-1 complex with Oct-1 on DNA, creating a multiprotein HCF-1–VP16–Oct-1–DNA complex on VP16 IE gene response elements. Oct-1 is a broadly expressed transcription factor that binds the octamer sequence ATGCAAAT with high affinity through a POU DNA-binding domain (12, 39). The POU domain is a bipartite DNA-binding domain composed of an amino-terminal POU-specific domain and a carboxy-terminal POU homeodomain, joined by a flexible linker region (18, 40). Residues on the solvent-exposed surface of the Oct-1 POU homeodomain direct VP16-induced complex formation (24, 34, 38).
The cis-regulatory target of the VP16-induced complex is referred to as the TAATGARAT element because it often contains the core consensus sequence TAATGARAT (where R is purine). TAATGARAT elements contain three sequence determinants important for VP16-induced complex formation. First, the TAAT segment is involved in Oct-1 POU homeodomain binding; many, but not all, TAATGARAT response elements also contain an adjacent 5′ ATGC POU-specific domain binding site (ATGCTAATGARAT), which also contributes to Oct-1 DNA binding (1, 4, 6). Second, the GARAT sequence is important for VP16-induced complex formation but not necessarily Oct-1 binding—some point mutations within the GARAT sequence disrupt VP16-induced complex formation with little, if any, effect on the affinity of Oct-1 for the TAATGARAT element (8, 19, 32, 37). Third, the D element, a short 3-bp sequence located 3′ of the GARAT element, is also involved in VP16-induced complex formation and can determine the selectivity of VP16 association with TAATGARAT elements by different VP16 orthologs, in particular, the HSV-1 and bovine herpesvirus type 1 (BHV-1) VP16 proteins (16, 29).
Like other transcription factors, HSV-1 VP16, a 490-amino-acid protein, has a modular structure (5, 35, 44). It contains a carboxy-terminal transcriptional activation domain and a central conserved core (residues 49 to 385), which is sufficient for VP16-induced complex formation (10, 27). Recently, the crystal structure of the conserved core (residues 49 to 412) was solved to 2.1 Å resolution (27), revealing a protein with a novel structure containing ordered (residues 49 to 349) and disordered (residues 350 to 394) regions important for VP16-induced complex formation and two small ordered (residues 395 to 402) and disordered (residues 403 to 412) regions which are dispensable for VP16-induced complex formation. The larger disordered region has been analyzed extensively by mutagenesis, by protease sensitivity, and with synthetic peptides (see reference 25 for references); it is involved in interactions with each of the other components of the VP16-induced complex—HCF-1, Oct-1, and DNA—and probably adopts an ordered structure upon VP16-induced complex formation. In contrast, the large ordered region, which has a structure resembling a seat, has been much less well characterized, although it is known to be involved in recognition, either directly or indirectly, of the D element (27).
VP16 has little DNA-binding activity on its own, which has made analysis of its DNA-binding activity difficult (20, 28, 37, 45). Thus, the mechanism by which VP16 recognizes DNA within the VP16-induced complex is unknown, and models for direct (6, 13, 20, 27, 32, 43) and indirect (29, 45) recognition of DNA have been proposed. Here, we describe molecular studies that support the model for direct DNA contact by VP16 in the VP16-induced complex. Our results show that, on a TAATGARAT site, VP16 is positioned over the D element adjacent to the Oct-1 POU domain. VP16 associates with the DNA through a novel DNA-binding surface, indicating that one role for VP16 in assembly of the VP16-induced transcriptional regulatory complex is DNA binding.
MATERIALS AND METHODS
Expression constructs.
Plasmids pNCITE-HCF-1N380 (47), pET11c.G.POU-1 (24), pET11c.ori+(−)/VP1649–412 (27), pET11c.ori+(−)/VP16ΔC, pCGNVP16 (25), pU2/β 6xTAAT-1 (2), and pα4x(A+C) (42) have been described previously. Full-length VP16 for expression in Escherichia coli was generated by transferring the VP16 coding sequence (residues 5 to 490) as an XbaI-BamHI DNA fragment from pCGNVP16 into the 5′ XbaI and 3′ BamHI sites of pET11c.ori+(−); creating the plasmid pET11c.ori+(−)/VP16. For HCF-1 synthesis in E. coli, the XbaI-BamHI DNA fragment from pNCITE-HCF-1N380 (amino acids 2 to 380) was inserted into the 5′ NheI and 3′ BamHI sites of pET28a(+) (Novagen), creating the plasmid pET28a(+)/HCF-1N380. For HCF-1 synthesis in baculovirus-infected Sf9 cells, HCF-1N380 sequences were amplified by PCR and inserted into the 5′ XbaI and 3′ BamHI sites of the modified baculovirus transfer vector pAcUW51 (11) such that the HCF-1 open reading frame continued into 10 in-frame histidine codons 3′ of the BamHI site.
VP16 mutagenesis.
Mutant VP16ΔC expression plasmids were generated by oligonucleotide-mediated site-directed mutagenesis of the plasmid pET11c.ori+(−)/VP16ΔC, which encodes HSV-1 VP16 (residues 5 to 412) fused to the glutathione S-transferase (GST) gene product. The mutations are named according to the identity of the wild-type amino acid, followed by its position in VP16 and the identity of the amino acid substitution. The following sequences were created to generate the mutant proteins: R64A, AAC gcg tTa CTC (MluI); R100A, TAC gcG GAG (BstUI); K103A, TGc gcA TTC (FspI); R129A, ATT gcC GCC (Fnu4HI); R143A, ACa gct GAC (PvuII); R155A, CTC TCa GcT (DdeI); R184A, CTa gcC GCC (BfaI); R208A, ATG CTa gcC (NheI); R214A, GAC gcG TAC (MluI); R217A, TAC gct GAG (DdeI); R221A, GCa gcg CTG (Eco47III); R224, GCG gcg GTT (EaeI); R236A, ACC gcg GAG (SacII); R290A, GCC gcg CGc CTG (BssHII). In this notation scheme, the bases that are altered in wild-type VP16 are shown in lowercase, the new amino acid codon is in boldface, and the engineered restriction site for the nuclease indicated in parentheses is underlined. The mutations E361A/385Ala3, 366Ala3, and G374A have been described previously (25). The deletion mutations were generated by oligonucleotide-mediated loop-out mutagenesis by using oligonucleotides Δ198–201 (CAGGCGCACATGCGCGACCTGGGAGAAATG) and Δ187–206 (TACCTGCGCGCCAGCCTGCGCGCCACGATC). Mutant pCGNVP16 expression plasmids were generated by the QuikChange site-directed mutagenesis protocol (Stratagene). All mutations were verified by DNA sequence analysis.
Protein synthesis and purification.
Wild-type and mutant VP16 proteins and the Oct-1 POU domain were synthesized as GST fusion proteins and purified from E. coli BL21(DE3) as described previously (24). The GST moiety of the GST-VP16 fusion proteins used in the experiments whose results are shown in Fig. 2 to 4 and of the Oct-1 POU domain was removed by thrombin digestion.
FIG. 2.
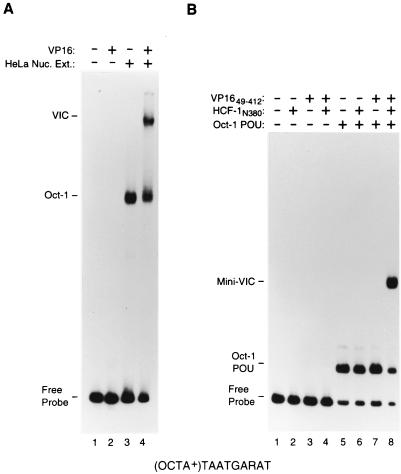
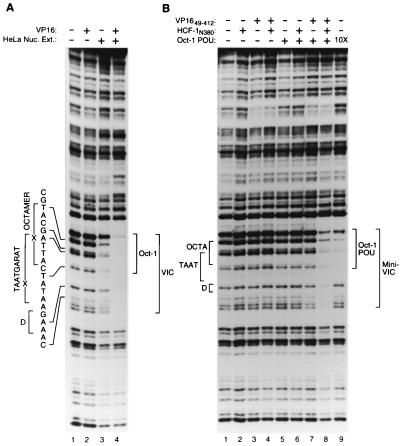
A VP16-induced complex can be assembled with proteins synthesized in E. coli. (A) Electrophoretic mobility retardation assay of a VP16-induced complex assembled from full-length proteins. All samples contain the (OCTA+)TAATGARAT probe either alone (lane 1) or incubated with HeLa cell nuclear extract (Nuc. Ext.; lanes 3 and 4) or full-length VP16 (lanes 2 and 4), as indicated above each lane. (B) Electrophoretic mobility retardation assay of a VP16-induced complex assembled from recombinant proteins synthesized in E. coli representing the minimal domains sufficient for VP16-induced complex formation. All samples contain the (OCTA+)TAATGARAT probe either alone (lane 1) or with HCF-1N380 (lanes 2, 4, 6, and 8). VP1649–412 (lanes 3, 4, 7, and 8), or the Oct-1 POU domain (lanes 5 to 8), as indicated above the lanes. The positions of the free (OCTA+)TAATGARAT probe (Free Probe), the Oct-1–DNA complex (Oct-1), the Oct-1 POU domain–DNA complex (Oct-1 POU), the VP16-induced complex (VIC), and the VP16-induced complex with minimal domains (Mini-VIC) are shown at the left.
FIG. 4.
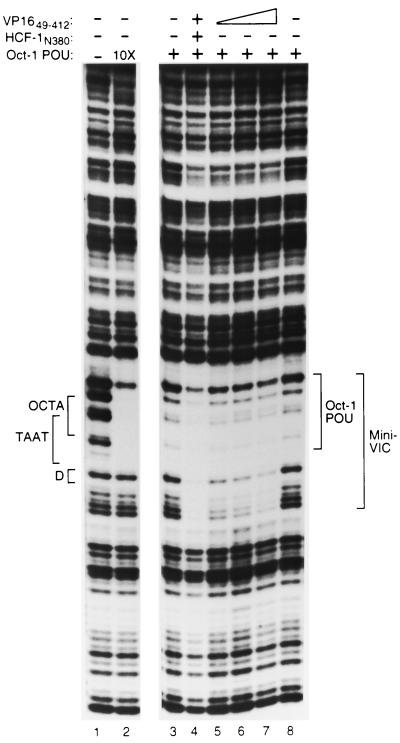
The VP16 core is sufficient to extend the DNase I protection pattern generated by the Oct-1 POU domain. DNase I protection analysis of Oct-1 POU domain–VP16 core complexes on an (OCTA+)TAATGARAT-containing probe. The Oct-1 POU domain was assayed at either a low (lanes 3 to 8) or a high (10X; lane 2) concentration along with VP1649–412 (lanes 4 to 7) either in the absence (lanes 1 to 3 and 5 to 8), or in the presence (lane 4) of HCF-1N380. As indicated, lanes 6 and 7 contain two- or threefold more VP1649–412 protein, respectively, than lanes 4 and 5. The positions of the DNA sites and DNase I protection patterns are as described in the legend to Fig. 3.
HCF-1N380 used in the experiment whose results are shown in Fig. 2 was synthesized and purified from E. coli BL21(DE3). Typically, cells grown continuously at 18°C to an optical density at 600 nm of 0.4 to 0.6 were induced at 18°C for ∼12 h with 0.4 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested by centrifugation and resuspended in 1/10 volume of the culture medium with a buffer containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 20% glycerol, and 10 mM imidazole. Following lysozyme treatment, NP-40 was added to 0.1% and the lysate was sonicated five times with 40-s pulses (20% duty on a Tekmar CV26 sonicator). Insoluble material was removed by centrifugation. Soluble histidine-tagged proteins were bound to Ni-nitrilotriacetic acid agarose (Qiagen) at 4°C for 2 h, washed, and subsequently eluted with 500 mM imidazole in 50 mM NaH2PO4 (pH 8.0)–300 mM NaCl–20% glycerol. Protein integrity, purity, and relative concentration were determined by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
The HCF-1N380 used in subsequent experiments was produced in baculovirus-infected Sf9 cells. Recombinant HCF-1N380-containing viruses were generated from an HCF-1N380-containing transfer vector by using a BaculoGold transfection kit (Pharmigen). Positive viruses were plaque purified twice and amplified. Sf9 cells were infected at a multiplicity of infection of 10, and cells were harvested 36 h postinfection. Cells (5 × 107) were washed and lysed in 1 ml of a buffer containing 10 mM Tris (pH 7.6), 10 mM NaH2PO4 (pH 7.6), 500 mM NaCl, 10 mM NaF, 10 mM Na pyrophosphate, 5 mM 2-mercaptoethanol, 1% Triton X-100, 10% glycerol, 500 μg of Pefablock per ml, and 10 mM imidazole for 1 h on ice with periodic vortexing. Cellular debris was removed by centrifugation, and the remaining lysate was dialyzed for ∼12 h against buffer BAC (10 mM Tris [pH 8.0], 10 mM NaH2PO4 [pH 8.0], 500 mM NaCl, 5 mM 2-mercaptoethanol, 10% glycerol, 500 μg of Pefablock per ml, 10 mM imidazole). Histidine-tagged proteins were bound to Ni-nitrilotriacetic acid agarose (Qiagen) at 4°C for 3 h, washed twice with 10 bead volumes of buffer BAC containing 50 mM imidazole, eluted with buffer BAC containing 1 M imidazole, and collected in ∼0.5-ml fractions. Protein integrity, concentration, and purity were judged by VP16-induced complex formation, Western blotting using the anti-HCF-1N18 antibody (9), and Coomassie blue staining after SDS-PAGE. For proteins used in immunoprecipitations, VP16ΔC and HCF-1N380 were synthesized by transcription and translation in vitro as described previously (3, 47). HeLa cell nuclear extract was a kind gift of Xuemei Zhao, Cold Spring Harbor Laboratory.
DNase I protection analysis.
Protein-DNA binding was performed in 50-μl reaction mixtures containing 2 × 104 cpm of DNA probe, 10 mM Tris-HCl (pH 7.9), 50 mM KCl, 2 mM dithiothreitol, 1 mM EDTA, 0.1% NP-40, 2% glycerol, 2% Ficoll, 3 mg of fetal bovine serum per ml, 50 ng of fish sperm DNA, 8.0 μg of unsonicated poly(dI-dC) (Pharmacia), and 2% polyvinyl alcohol. The purified recombinant Oct-1 POU domain (40 ng), VP16 (80 ng), HCF-1N380 (4 μl), and HeLa cell nuclear extract (35 μg) were used as indicated in Results. Reaction mixtures were processed as described previously (2).
Coimmunoprecipitation assays.
In vitro-translated hemagglutinin (HA)-tagged HCF-1N380 (5 μl) and VP16ΔC (5 μl) were mixed in 40 μl of 50 mM Tris (pH 8.0)–200 mM KCl–1 mM EDTA–0.05% NP-40–1 mM phenylmethylsulfonyl fluoride and incubated at 4°C for 30 min with rotation. Subsequently, 10 μl of precoupled anti-HA antibody (12CA5)–protein G–agarose beads was added and allowed to incubate for a further 2 h at 4°C with rotation. After incubation, the beads were washed four times with 500 μl of 50 mM Tris (pH 8.0)–200 mM KCl–1 mM EDTA–0.05% NP-40–1 mM phenylmethylsulfonyl fluoride and complexes were resolved by SDS–8% PAGE and visualized by fluorography.
Electrophoretic mobility retardation assays.
VP16-induced complex formation assays were performed under conditions described previously (25), with the following modifications. For the experiment whose results are shown in Fig. 2A, ∼2 μg of HeLa cell nuclear extract and 20 ng of full-length VP16 were used, and for Fig. 2B, 10 ng each of Oct-1 POU, VP1649–412, and HCF-1N380 were used as indicated. For Fig. 5, 25 ng of wild-type or mutant GST-VP16ΔC, 5 ng of Oct-1 POU domain, and 0.5 μl of baculovirus expressed HCF-1N380 were used. For VP16–Oct-1–DNA complex formation assays, GST-VP16ΔC (200 ng) and the Oct-1 POU domain (0.1 ng) were incubated with 2 × 104 cpm of DNA probe, as indicated, in 10 mM Tris-HCl (pH 7.9)–50 mM KCl–2 mM dithiothreitol–1 mM EDTA–0.1% NP-40–2% glycerol–2% Ficoll–100 μg of bovine serum albumin–10 ng of fish sperm DNA–10 ng of unsonicated poly(dI-dC) (Pharmacia) for 30 min at 30°C. After incubation, the reaction mixtures were loaded onto a 6% polyacrylamide gel (acrylamide-bisacrylamide ratio, 19:1) in 0.25× TBE (22.5 mM Tris-borate, 0.5 mM EDTA) which had been subjected to prior electrophoresis for 30 min at room temperature. For VP16-DNA complex formation assays, a larger amount of VP16 (∼1 μg) was used and incubated under the same conditions as for the VP16–Oct-1–DNA complex formation assay. After incubation, the reaction mixtures were loaded onto a 3% glycerol–6% polyacrylamide gel (acrylamide-bisacrylamide ratio, 39:1) in 0.25× TBE which had been subjected to prior electrophoresis for 30 min at room temperature.
FIG. 5.
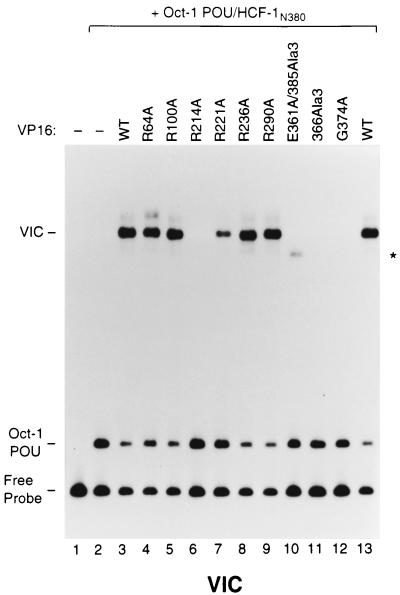
Some basic amino acids in the structured VP16 core are important for VP16-induced complex formation. An electrophoretic mobility retardation assay for VP16-induced complex formation of a representative set of alanine mutations in the VP16 core was performed. An (OCTA+)TAATGARAT-containing DNA probe was incubated in the absence (lane 1) or presence of the Oct-1 POU domain and HCF-1N380 (lanes 2 to 13) along with either wild-type (WT; lanes 3 and 13) or mutant VP16 protein (lanes 4 to 12), whose identity is indicated above each lane. The positions of the free (OCTA+)TAATGARAT probe (Free Probe), the Oct-1 POU domain–DNA complex (Oct-1 POU), and the VP16-induced complex (VIC) are shown at the left. The asterisk indicates a likely HCF-1-independent VP16-induced complex (lane 10; see reference 25).
In vivo transcription assays.
HeLa cells were seeded at 5 × 105/10-cm-diameter dish and transfected after 24 h with 2 μg of reporter plasmid pU2/β6xTAAT-1 150 ng of internal reference plasmid pα4x(A+C), and 1 μg of wild-type or mutant expression plasmid pCGNVP16 by using FUGENE 6 transfection reagent (Roche) in accordance with manufacturer's recommendations. To assay reporter gene expression, cytoplasmic RNA was isolated and analyzed by RNase protection (41) with the α98 and β134 probes (2). Unprotected RNA was digested with RNases A and T1, and protected fragments were visualized by denaturing PAGE. Levels of reporter gene expression were quantified on a Fuji BAS1000 phosphorimager. Expression of epitope-tagged VP16 proteins was measured by immunoblot analysis using the monoclonal antibody 12CA5. All proteins were expressed at similar levels.
RESULTS
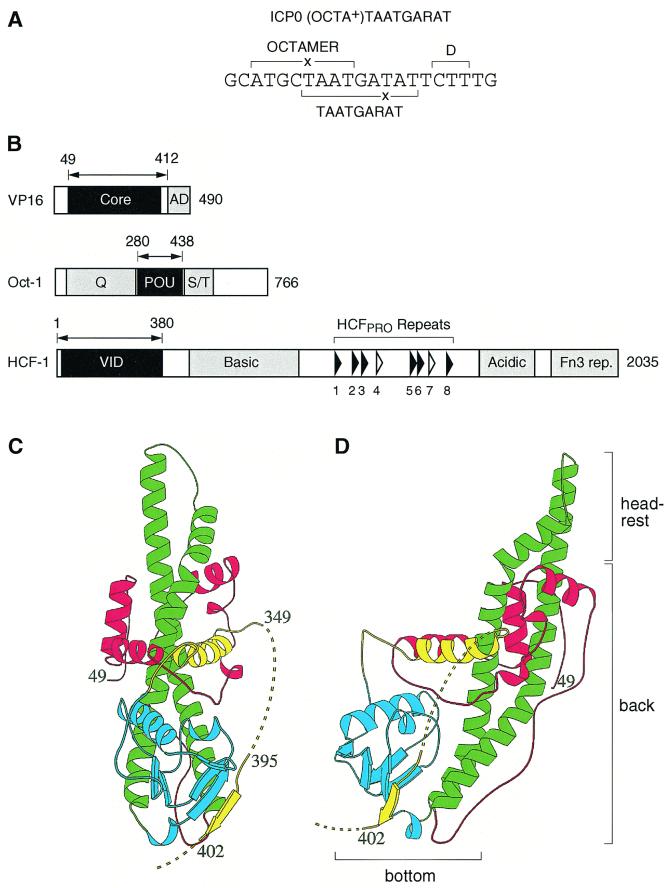
We have analyzed how VP16 recognizes DNA within the VP16-induced complex. Figure 1 illustrates the four components of the VP16-induced complex: the cis-regulatory TAATGARAT element (Fig. 1A) and the three trans-acting proteins VP16, Oct-1, and HCF-1 (Fig. 1B). Figure 1C and D show two views, the front and left, respectively, of the determined VP16 core structure (27). The TAATGARAT element derives from the HSV-1 ICP0 IE gene promoter and contains an imperfect TAATGARAT core sequence (TAATGATAT) and an imperfect overlapping ATGCTAAT octamer sequence (imperfections underlined), for which it is referred to as an (OCTA+)TAATGARAT site. On the 3′ side of the TAATGARAT core sequence is the three-nucleotide D element sequence CTT, which is responsible for differential recognition of TAATGARAT elements by the HSV-1 and BHV-1 VP16 proteins (16). Figure 1B shows a scaled schematic of VP16, Oct-1, and HCF-1. The minimal regions required for VP16-induced complex formation determined previously (10, 19, 23, 27, 38, 47) are highlighted in black.
FIG. 1.
Representation of TAATGARAT and VP16-induced complex components. (A) Nucleotide sequence of an (OCTA+)TAATGARAT element from the HSV-1 ICP0 promoter. The octamer TAATGARAT and D elements are indicated by the brackets. ×, nucleotide substitution from a consensus site. (B) Structural features of the three proteins, VP16, Oct-1, and HCF-1, required for VP16-induced complex formation. The minimal domain within each protein that is required for VP16-induced complex formation is shown in black, and the amino acid boundries are indicated by the double-headed arrow. Regions such as the VP16 transcriptional activation domain (AD), the Oct-1 amino- and carboxy-terminal transcriptional activation domains (Q and S/T, respectively), the HCF-1 Fn3 repeats (rep.); and regions enriched in basic or acidic amino acids are highlighted in gray. The positions of the six nearly perfect (filled arrowheads) and two nonfunctional (open arrowheads) HCF-1PRO repeats are indicated. (C and D) Ribbon diagrams of the front (C) and left-side (D) views of the VP16 core (27). The diagram is colored red, green, blue, and yellow from the amino terminus to the carboxy terminus. Disordered regions are represented by the broken lines (adapted from reference 27). The bottom, back, and headrest of the seat-like structure are identified in panel D.
A VP16-induced complex can be assembled from recombinant proteins synthesized in E. coli.
Figure 2A illustrates the assembly of a VP16-induced complex using a HeLa cell nuclear extract as a source of native Oct-1 and HCF-1 and full-length VP16 synthesized in E. coli. VP16-induced complex formation was measured in an electrophoretic mobility retardation assay using the ICP0 (OCTA+)TAATGARAT site. As expected, VP16 alone failed to bind the (OCTA+)TAATGARAT probe effectively (compare lanes 1 and 2). Addition of HeLa cell nuclear extract generated an Oct-1–DNA complex (lane 3; labeled Oct-1), and further addition of VP16 generated the slower-migrating VP16-induced complex (lane 4; labeled VIC).
In contrast to VP16-induced complex assembly with native human HCF-1 and Oct-1 proteins as shown in Fig. 2A, attempts to assemble a VP16-induced complex with all three protein components prepared from E. coli have reportedly failed (23, 33). These and other results (21, 33) have led to the suggestion that eukaryotic cell-specific posttranslational modifications may be required for VP16-induced complex assembly. In preliminary experiments, we found the synthesis of the HCF-1 VP16 interaction domain (VID) in E. coli in an active form to be difficult to achieve. By lowering the temperature at which E. coli synthesizes the HCF-1N380 VID-containing protein (residues 1 to 380; Fig. 1B), however, we were able to obtain soluble and active HCF-1N380 protein. By using this HCF-1N380 protein and the Oct-1 POU domain (residues 280 to 438) and VP1649–412 core (residues 49 to 412) proteins (Fig. 1B) also synthesized in E. coli, we examined whether the minimal regions of VP16, Oct-1, and HCF-1 synthesized solely in E. coli can form a VP16-induced complex.
Figure 2B shows the activity of the E. coli-synthesized minimal-region proteins used at approximately equimolar levels (10 ng of each protein). Neither the minimal VP16 (VP1649–412) protein nor the minimal HCF-1 (HCF-1N380) protein, alone or in combination, bound the DNA probe, as expected (compare lanes 1 to 4). In contrast, the Oct-1 POU domain bound the (OCTA+)TAATGARAT probe to form an Oct-1 POU domain–DNA complex with a majority of the probe (lane 5; labeled Oct-1 POU). Further addition of either VP1649–412 or HCF-1N380 alone did not affect the Oct-1 POU domain–DNA complex (lanes 6 and 7). When all three E. coli-derived VP1649–412, HCF-1N380, and Oct-1 POU domain proteins were added together, however, efficient levels of VP16-induced complex formation were achieved (lane 8; labeled mini-VIC). Thus, a VP16-induced complex can be faithfully and efficiently reproduced in vitro by using proteins that have been synthesized in E. coli. We conclude, therefore, that no eukaryotic cell-specific protein modifications are essential for the minimal regions of VP16, HCF-1, and Oct-1 to form a VP16-induced complex. These results validate the use of E. coli-derived proteins for the analysis of VP16-induced complex formation.
A minimal VP16-induced complex produces an extended DNase I protection pattern.
Previous DNA cleavage protection studies have shown that, on (OCTA+)TAATGARAT and related sites, Oct-1 alone protects nucleotides centered over the octamer sequence and addition of VP16 and HCF-1 extends the Oct-1 protection pattern unidirectionally over the entire TAATGARAT element, including the D element (Fig. 1A) (2, 19, 20, 45). The component of the VP16-induced complex responsible for extension of the protection pattern is not known. One explanation for the extension is that it represents nonspecific interactions, resulting simply from the creation of a large multiprotein complex. To determine if the regions of HCF-1, Oct-1, and VP16 that are dispensable for VP16-induced complex formation are responsible for the extended footprint, we compared the DNase I protection patterns of VP16-induced complexes assembled from full-length proteins with those of complexes assembled from recombinant minimal-region proteins.
Figure 3A shows the DNase I protection pattern produced by the native complex of full-length proteins. As expected, addition of VP16 alone did not affect the DNase I cleavage pattern of naked DNA (compare lanes 1 and 2). Addition of HeLa cell nuclear extract resulted in weak protection of the octamer sequence (lane 3), but further addition of VP16 enhanced the footprint over the entire octamer (TAATGARAT) and D element sequences (compare lanes 3 and 4). Figure 3B shows the protection pattern produced by the recombinant minimal-region proteins. Addition of VP1649–412 and HCF-1N380, either alone or in combination, failed to protect the DNA probe (lanes 2 to 4). At the concentrations of protein used in this experiment, the Oct-1 POU domain alone (lane 5) or in the presence of either VP1649–412 or HCF-1N380 (lanes 6 and 7) produced little protection of the TAATGARAT element. A 10-fold higher concentration of the Oct-1 POU domain, however, protected nucleotides over the entire octamer sequence from DNase I digestion but failed to protect nucleotides in the D element (compare lanes 1 and 9). Addition of all three minimal-region proteins together, however, resulted in the same extended protection of the TAATGARAT and D element sequences as elicited by the VP16-induced complex assembled with full-length proteins (compare Fig. 3A, lane 4, with Fig. 3B, lane 8). These results show that regions in HCF-1, Oct-1, and VP16 dispensable for VP16-induced complex formation are not responsible for the extended TAATGARAT-D element VP16-induced complex footprint. There may be an effect of the full-length proteins (e.g., Oct-1) over the octamer sequence, because the ATGC position of the octamer sequence was not as well protected from digestion with the minimal complex as with the native complex (compare Fig. 3A, lane 4, and B, lane 8). We conclude, however, that to a large degree, in a native VP16-induced complex, the regions of HCF-1, Oct-1, and VP16 that are not essential for VP16-induced complex formation are probably not associated with the DNA in any specific and stable manner.
FIG. 3.
A VP16-induced complex assembled from full-length proteins and one assembled from the minimal domains sufficient for VP16-induced complex formation show similar DNase I protection patterns. (A) DNase I protection analysis of a VP16-induced complex assembled from full-length proteins. All samples contain the (OCTA+)TAATGARAT probe incubated either alone (lane 1) or with HeLa cell nuclear extract (Nuc. Ext.; lanes 3 and 4) or full-length VP16 (lanes 2 and 4), as indicated above each lane. The same amount of DNase I was used in each sample, resulting in decreased nuclease activity in the samples containing nuclear extract (lanes 3 and 4). (B) DNase I protection analysis of a VP16-induced complex assembled from recombinant proteins representing the minimal domains sufficient for VP16-induced complex formation. All samples contain the (OCTA+)TAATGARAT probe incubated either alone (lane 1) or with HCF-1N380 (lanes 2, 4, 6, and 8), VP1649–412 (lanes 3, 4, 7, and 8), or the Oct-1 POU domain (lanes 5 to 8), as indicated above each lane. Lane 9 contains 10 times the amount of the Oct-1 POU domain as lane 5. The positions of the octamer (OCTA), TAATGARAT (TAAT), and D (D) elements as determined by chemical sequencing of the same probe, are indicated at the left. The identities of the sites of DNase I cleavage are indicated by the sequence and brackets to the far left. The boundaries of the regions protected from DNase I digestion by Oct-1, Oct-1 POU, the VP16-induced complex (VIC), and the minimal VP16-induced complex (Mini-VIC) are indicated to the right.
VP16 is responsible for the extended VP16-induced complex footprint.
In the aforementioned studies, HCF-1 and VP16, when added together, generated the extension of the TAATGARAT protection pattern. To determine whether it is the HCF-1 or VP16 protein alone or in combination that is responsible for the extended DNase I protection pattern, we investigated whether, in the absence of HCF-1N380, VP1649–412 can extend the Oct-1 POU domain DNase I protection pattern. To accomplish this, we added high concentrations of VP16 to overcome the lack of HCF-1 stabilization. Figure 4 shows the results of this experiment. As expected, high concentrations of the Oct-1 POU domain protected the octamer sequence but not the D element sequence (compare lanes 1 and 2), and with lower concentrations of the Oct-1 POU domain (lanes 3 and 8), the presence of VP1649–412 and HCF-1N380 produced the extended VP16-induced complex footprint over the D element sequence and beyond (lane 4). Consistent with its role in stabilizing the VP16-induced complex, removal of HCF-1 (lane 5) resulted in a general decrease in the overall protection pattern (compare lanes 3 to 5). At these concentrations of the Oct-1 POU domain and VP16, however, an extended VP16-induced complex protection pattern (mini-VIC) is evident (compare lanes 3 and 5), and increasing concentrations of VP16 enhanced the D element protection pattern (compare lanes 7 and 8) without decreasing the protection pattern over the octamer sequence. Thus, in the absence of HCF-1, the Oct-1 POU domain and the VP16 core (lane 7), but not the Oct-POU domain alone (lane 2), are sufficient to protect the entire ICP0 TAATGARAT site. These results suggest that it is VP16 that is directly responsible for the protection of 3′ residues flanking the DNA-bound Oct-1 POU domain, consistent with previous models of the VP16-induced complex (6, 13, 20, 27, 32, 43).
VP16 mutagenesis.
To explore how VP16 coordinates assembly of the VP16-induced complex, we used the structure of the VP16 core as a guide for a mutational analysis of its surface. The structure of the VP16 core, as shown in Fig. 1C and D, has been likened to a seat, possessing a back, a bottom, and a headrest (27; Fig. 1D). The seat back is formed by two long, antiparallel α helices (shown in green) forming a coiled-coil structure that runs the length of the protein. The seat bottom projects out from the two long α helices. The seat back and bottom form a concave surface which has been proposed by molecular modeling to be involved in DNA binding (27). The position of the unstructured regions of the VP16 core is indicated by the dashed lines. In an earlier mutational analysis of VP16 (25), we showed that the major unstructured region of the VP16 core is involved in DNA binding, as well as association with Oct-1 and HCF-1. To identify residues in the ordered region of the VP16 core that are involved in DNA binding, we selected many of the basic amino acids across its surface for replacement with alanine because, within DNA-binding proteins, basic residues are often critical for binding to DNA.
We tested these substitution mutant proteins in four separate in vitro assays: (i) VP16-induced complex formation; (ii) VP16 binding to HCF-1; (iii) VP16 binding to Oct-1 and DNA in the absence of HCF-1; and (iv) VP16 binding to DNA on its own. Table 1 lists all of the point mutations that were tested and summarizes their effects in each of the four VP16 activity assays. Two mutations of the surface of the VP16 core, R331A and R341A, have been described and characterized in similar assays previously (25); the results for these two mutant proteins from that study are included in Table 1 for comparison. In the activity assays performed here, we included three previously characterized mutations of the disordered VP16 core region that have been shown to selectively prevent VP16 binding to HCF-1 (E361A/385Ala3), DNA (366Ala3), and an Oct-1–DNA complex (G374A) (25). Described below are the activities of a representative set of six mutant proteins (R64A, R100A, R214A, R221A, R236A, and R290A) in each of the four activity assays. All of the mutant proteins lacked the carboxy-terminal transcriptional activation domain (VP16ΔC) and were synthesized in E. coli as fusions to GST.
TABLE 1.
Activities associated with VP16 proteins containing amino acid substitutions in the core domaina
| VP16 protein | VP16-induced complex formation | Interaction with:
|
Relative in vivo activityd | Colore | ||
|---|---|---|---|---|---|---|
| HCF-1 | DNA | Oct-1–DNA | ||||
| Wild type | ++ | ++ | ++ | ++ | 1 | Blue |
| Substitutions | ||||||
| R64A | ++ | ++ | − | − | 0.64 | Yellow |
| R100A | ++ | ++ | + | ++ | 0.87 | Blue |
| K103A | ++ | ++ | + | ++ | ND | Blue |
| R129A | ++ | ++ | + | ++ | ND | Blue |
| R143A | ++ | ++ | ++ | ++ | ND | Blue |
| R155A | ++ | ++ | ++ | ++ | ND | Blue |
| R184A | + | ++ | − | − | ND | Yellow |
| R208A | ++ | ++ | ± | + | ND | Blue |
| R214A | − | ++ | − | − | 0.01 | Red |
| R217A | − | ++ | − | − | ND | Red |
| R221A | + | ++ | − | − | 0.05 | Yellow |
| R224A | − | ++ | − | − | ND | Red |
| R236A | ++ | ++ | ± | + | 1.07 | Blue |
| R290A | ++ | ++ | ++ | ++ | 1.04 | Blue |
| R331Ab | − | ++ | − | − | ND | Red |
| R341Ab | + | ++ | − | + | ND | Yellow |
| E361A/385Ala3c | − | − | +++ | +++ | 0.02 | |
| 366Ala3c | − | ++ | − | − | <0.01 | |
| G374A | − | ++ | ++ | − | 0.01 | |
| Deletions | ||||||
| Δ198-201 | − | ++ | − | − | ND | |
| Δ187-206 | − | + | − | − | ND | |
Wild-type activity is set at ++ for each assay. +++, more active than wild-type VP16; +, less active than wild-type VP16; ±, barely detectable activity; − undetectable activity.
Results from Lai and Herr (25).
E361/385Ala3 contains alanine substitutions at residues E361, D385, D386, and D387, and 366Ala3 contains alanine substitutions at residues R366, R368, and K370.
The results shown are averages of two independent experiments and are within ±20%. ND; not done.
Color of residue in Fig. 10.
Some, but not all, basic residues on the surface of the VP16 core are important for VP16-induced complex formation.
Figure 5 shows the abilities of the selected mutant proteins to support VP16-induced complex formation. As expected, addition of the Oct-1 POU domain and HCF-1N380 generated an Oct-1 POU domain–DNA complex alone (lane 2; labeled Oct-1 POU) and further addition of wild-type VP16 protein produced a slower-migrating VP16-induced complex (lane 3; labeled VIC). As expected (25), the three previously characterized mutant proteins (E361A/385Ala3, 366Ala3, and G374A) failed to support VP16-induced complex formation (lanes 10 to 12). Owing to the increased ability of the HCF-1 association-defective E361A/385Ala3 mutant protein to associate with DNA in the absence of HCF-1, as observed previously (25), it formed a weak, faster-migrating VP16-induced complex that probably lacks HCF-1 (asterisk in Fig. 5). Of the representative set of mutations, only one prevented VP16-induced complex formation entirely (R214A; lane 6); another had an intermediate effect (R221A; lane 7). The remaining selected proteins behaved similarly to wild-type VP16. As shown in Table 1, alanine substitutions for residues 217, 224, and 331 (see reference 25) also prevented VP16-induced complex assembly whereas replacement of residues 184 and 341 (see reference 25) had partial effects on complex assembly. These results suggest that some, but not all, basic amino acids on the surface of the VP16 core are involved in VP16-induced complex formation.
The basic-residue substitutions do not affect HCF-1 association.
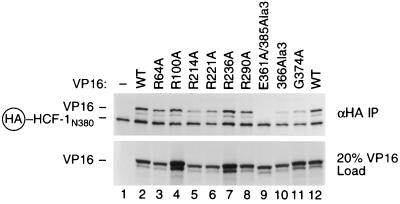
To examine how VP16 association with HCF-1 might be affected by the VP16 core surface mutations, we used a coimmunoprecipitation assay based on the ability of VP16 to bind HCF-1 in the absence of DNA (47). HCF-1N380 was tagged at its amino terminus with an HA epitope (HA–HCF-1N380) and both HA–HCF-1N380 and VP16 were produced by in vitro translation. Figure 6 shows the effects of the selected set of mutations on HCF-1N380 association. The lower panel shows 20% of the input VP16 protein, and the upper panel shows the complexes that were recovered after immunoprecipitation of HA–HCF-1. As expected, the HCF-1 association-defective E361A/385Ala3 VP16 mutant protein was not recovered in this assay (lane 9). All of the remaining mutant proteins were capable of associating with HCF-1N380, most at or near the wild-type level (lanes 3 to 8 and 10 and 11; Table 1). These results show that replacement of many basic amino acids in the structured portion of the VP16 core with alanine does not prevent VP16 association with HCF-1.
FIG. 6.
Basic amino acid substitutions in the structured VP16 core do not prevent association with HCF-1. Coimmunoprecipitation of VP16 core mutants protein with HA epitope-tagged HCF-1. HCF-1N380 and the wild-type (WT) and mutant VP16 proteins were translated in vitro and labeled with [35S]methionine as described in Materials and Methods. HCF-1N380 was incubated with either unprogrammed lysate (lane 1) or wild-type (lanes 2 and 12) or mutant (lanes 3 to 11) VP16 protein, as indicated above each lane. Immune complexes were recovered by immunoprecipitation with an anti-HA monoclonal antibody (top; αHA IP), resolved by SDS–8% PAGE, and detected by fluorography. The lower blot shows 20% of the VP16 starting material (20% VP16 Load). The mobility of VP16 and HA epitope-tagged HCF-1N380 is indicated at the left.
Some basic-residue substitutions resulting in competence for VP16-induced complex formation cause a defect in DNA binding by VP16 on its own or with the Oct-1 POU domain.
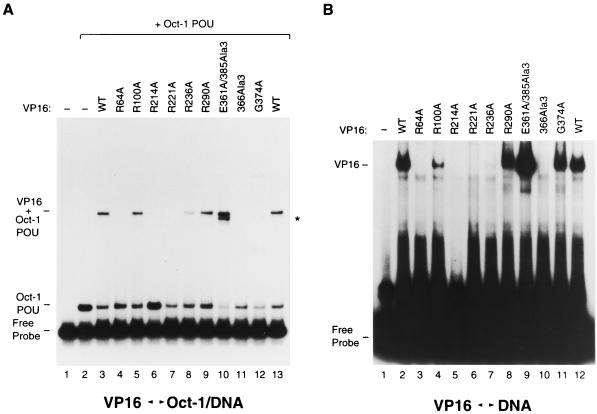
We next tested the effects of the mutations on VP16 binding to DNA either alone or in the presence of Oct-1. As shown in Fig. 7A, addition of an amount of VP16 eightfold greater than that required for VP16-induced complex formation (i.e., 200 ng), in the presence of the Oct-1 POU domain, stimulates the formation of an HCF-1-independent VP16–Oct-1 POU domain–DNA complex (37; compare lanes 2 and 3, labeled VP16 + Oct-1 POU). As expected (25), the mutation 366Ala3 and G374A, which disrupt interaction with the DNA and the Oct-1 POU domain, respectively, prevented the formation of the VP16–Oct-1 POU complex (lanes 11 and 12), whereas the E361A/385Ala3 mutation (lane 10) resulted in cooperative binding with the Oct-1 POU domain and DNA with increased affinity (a slightly faster-migrating complex [asterisk] is probably owing to E361A/385Ala3 VP16 bound to DNA alone). From the selected set of VP16 core mutations, R214A, which prevented the formation of a VP16-induced complex, also prevented binding to the Oct-1 POU domain–DNA complex (lane 6). The substitution mutation R217A, R224A, and R331A (see reference 25) behaved similarly (Table 1). Two additional selected mutations, however, R64A and R221A, which were able to support VP16-induced complex formation, albeit to various degrees, also prevented binding to the Oct-1 POU domain–DNA complex (lanes 4 and 7). The R184A substitution mutation produced a similar phenotype (Table 1). The remainder of the substitution mutant proteins could still cooperate for binding of the Oct-1 POU domain–DNA complex, although on occasion with reduced affinity (e.g., R236A, lanes 5, 8, and 9 and Table 1).
FIG. 7.
Multiple basic amino acids in the structured VP16 core are important for DNA binding. (A) Cooperative VP16 binding to the Oct-1 POU domain–DNA complex. The (OCTA+)TAATGARAT probe was incubated either in the absence (lane 1) or in the presence (lanes 2 to 13) of the Oct-1 POU domain and wild-type (WT; lanes 3 and 13) or mutant (lanes 4 to 12) VP16, whose identity is indicated above each lane. (B) VP16 binding to DNA alone at high concentrations of VP16. The (OCTA+)TAATGARAT probe was incubated either in the absence (lane 1) or in the presence of wild-type (lanes 2 and 12) or mutant (lanes 3 to 11) VP16, whose identity is indicated above each lane. Complexes were resolved on a 6% gel as described in Materials and Methods. The positions of the free (OCTA+)TAATGARAT probe (Free Probe) and the Oct-1 POU domain–DNA (Oct-1 POU), VP16-DNA (VP16), and VP16–Oct-1 POU domain–DNA (VP16 + Oct-1 POU) complexes are shown at the left. The asterisk indicates a likely complex of VP16-DNA alone (lane 10; see reference 25).
Figure 7B shows the results of assaying the ability of the VP16 mutant proteins to bind DNA on their own. An amount of VP16 fivefold greater than that used in the Oct-1 POU domain–DNA complex-binding assay (i.e., 1 μg) bound to the probe nonspecifically under these conditions (37; lanes 2 and 12, labeled VP16). As previously reported, the G374A mutant protein (lane 11) bound DNA similarly to wild-type VP16, whereas the E361A/385Ala3 mutant protein bound DNA with increased affinity (compare lanes 9 and 2) and the 366Ala3 mutant protein failed to bind DNA on its own (lane 10). Of the representative set of mutant proteins, R64A, R214A, and R221A (lanes 3, 5, and 6) failed to bind DNA on their own and mutant proteins R184A, R217A, R224A, R331A, and R341A (see reference 25) behaved similarly (Table 1). The remaining mutant proteins, R100A, R236A, and R290A (lanes 4, 7, and 8; Table 1), bound with various reduced affinities (weak but detectable binding by R236A is evident on longer exposure of the same gel; data not shown). These results suggest that the inability of mutant proteins R64A and R221A to interact cooperatively with the Oct-1 POU domain and DNA (Fig. 7A) is due to their reduced ability to bind DNA on their own. The ability of these VP16 mutant proteins to form a complete VP16-induced complex but not subcomplexes lacking HCF-1 suggests that HCF-1 is able to suppress the defects of some of the VP16 point mutant proteins for interaction with DNA or Oct-1. These results contrast with those of mutations in the disordered region of the VP16 core, where DNA- protein or Oct-1-binding defects were not overcome by HCF-1 (25). Four of the substitution mutant proteins, however, did display loss of both DNA binding alone and VP16-induced complex formation, as in the case of the disordered region mutations (25). We suggest below that the residues affected in these mutant proteins, R214, R217, R224, and R331, form the TAATGARAT-binding surface on the concave portion of the VP16 seat.
Deletions within the VP16 headrest prevent VP16 binding to DNA.
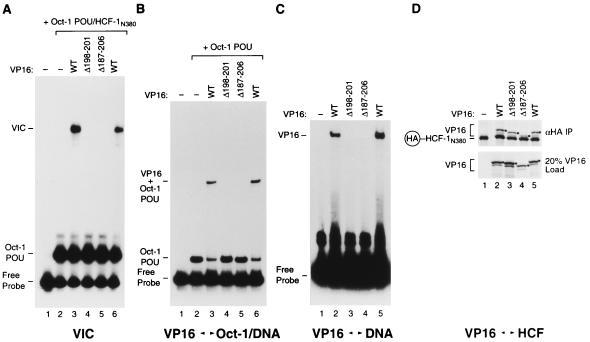
Amino acid sequence alignment of VP16 core domains from HSV-1, BHV-1, varicella-zoster virus, gallus herpesvirus type 1, and equine herpesvirus type 1 VP16 orthologs shows sequence similarity scattered throughout the VP16 core (27). One interesting feature, however, of the HSV-1 VP16 structure, the headrest (Fig. 1D), displays little, if any, amino acid sequence similarity. The HSV-1 VP16 headrest, at 27 amino acids (residues 187 to 213), is the longest nonconserved region of the VP16 core. Indeed, the equine herpesvirus type 1 and BHV-1 VP16 proteins even contain small three- and four-amino-acid deletions, respectively, in this region (see reference 27). Although the headrest is not conserved, it is flanked by basic residues important for VP16-induced complex formation and DNA binding (e.g., R184, R214, and R217; Table 1). Therefore, we examined, by deletion mutagenesis, whether this variable region is important for VP16-induced complex formation, particularly DNA binding.
We created two deletions: a small deletion, called Δ198–201, which mimics the four-amino-acid BHV-1 VP16 ortholog deletion, and a more radical deletion, called Δ187–206, which removes most of the headrest region. Both of the resulting mutant proteins were defective for VP16-induced complex formation (Fig. 8A), Oct-1 POU domain–DNA complex binding (Fig. 8B), and DNA binding on their own (Fig. 8C) but were active for HCF-1 association (Fig. 8D), although the Δ187–206 mutant, which is difficult to see owing to its faster electrophoretic migration, is partially defective for HCF-1 association (lane 4 and data not shown). These results show that even headrest sequences which have no apparent counterpart in a VP16 ortholog—the BHV-1 VP16 protein—are important either directly or indirectly (e.g., for structural stability) for VP16 binding to DNA.
FIG. 8.
The VP16 core headrest is important for DNA binding. Wild-type (WT) and mutant (Δ198–201 and Δ187–206) VP16 molecules were tested for VP16-induced complex formation (A) and for cooperative binding to the Oct-1 POU domain–DNA complex (B), DNA alone (C), and HCF-1 (D). (A) VP16-induced complex formation was assayed as described in the legend to Fig. 5. The presence or absence of wild-type or mutant VP16 along with the Oct-1 POU domain and HCF-1N380 is indicated above each lane. (B) The ability of VP16 to interact cooperatively with the Oct-1 POU domain was assayed as described in the legend to Fig. 7A. The presence or absence of wild-type or mutant VP16 and the Oct-1 POU domain is indicated above each lane. (C) The binding of VP16 alone to DNA was assayed as described in the legend to Fig. 7B. The presence or absence of wild-type or mutant VP16 is indicated above each lane. (D) VP16 binding to HCF-1 was assayed as described in the legend to Fig. 6. The presence or absence of wild-type or mutant VP16 and HA epitope-tagged HCF-1N380 is indicated above each lane. The black dots indicate the position of the recovered VP16 proteins. The positions of the different electrophoretic species are as described in the Fig. 5 to 7 legends.
VP16 mutant proteins defective for VP16-induced complex formation in vitro are defective for transcriptional activation in vivo.
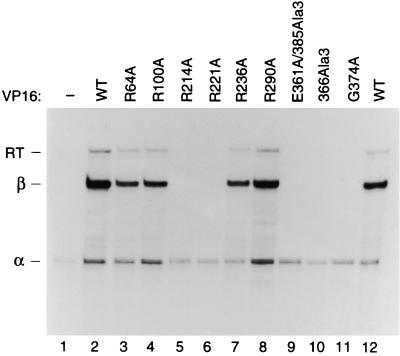
To determine the effects of the selected set of mutant VP16 proteins on transcriptional activation in vivo, we assayed their abilities to activate transcription in human cells. Figure 9 shows the results of VP16-dependent transcriptional activation of a β-globin promoter containing multiple tandem TAATGARAT sites after transient expression of wild-type and mutant VP16 proteins in HeLa cells. As expected, expression of wild-type VP16 strongly activated the reporter (compare lanes 1 and 2) and the E361A/385Ala3, 366Ala3, and G374A VP16 mutant proteins (lanes 9 to 11), which failed to form a VP16-induced complex, failed to stimulate transcriptional activity from the reporter (25). Similar to wild-type VP16, the R64A, R100A, R236A, and R290A VP16 mutant proteins stimulated transcription of the reporter to nearly wild-type levels (lanes 3, 4, 7, and 8; Table 1). In contrast, R214A, which failed to form a VP16-induced complex in vitro, failed to stimulate reporter gene expression in vivo (lanes 5; Table 1) and the R221A mutant, which showed reduced levels of VP16-induced complex formation in vitro, displayed weak but detectable levels of transcriptional activity in vivo (lane 6; Table 1). Therefore, the abilities of the selected set of mutant VP16 proteins to stimulate transcription in vivo closely parallel their abilities to form a VP16-induced complex in vitro.
FIG. 9.
In vivo transcriptional activation by selected amino acid substitutions in the VP16 core. A β-globin (OCTA+)TAATGARAT-containing reporter plasmid and an α-globin internal reference plasmid were transfected into HeLa cells in the absence (lane 1) or presence of wild-type (WT; lanes 2 and 12) or mutant (lanes 3 to 11) VP16 expression plasmids. RNA was collected 48 h after transfection and analyzed by RNase protection assay as described in Materials and Methods. The positions of correctly initiated α-globin (α), β-globin (β), and β-globin readthrough (RT) transcripts are indicated at the left.
DNA-binding mutations in VP16 map to the concave surface formed by the seat bottom and back of the VP16 core structure.
Figure 10 summarizes the activities of the structured VP16 core region alanine substitution mutant proteins with three representations of the free form of the structured VP16 core. Included are the two previously characterized mutant proteins, R331A and R341A, which prevent VP16 DNA binding (Table 1) (25). The structured VP16 core is shown as a molecular surface representation (30) in white, with residues whose replacement with alanine prevents both VP16-induced complex formation and VP16 DNA binding shown in red those whose replacement with alanine prevents DNA binding but whose inactivity can be suppressed by HCF-1 shown in yellow, and those whose replacement with alanine has no evident effect shown in blue. Figure 10B shows the same projection of the VP16 core as in Fig. 1C.
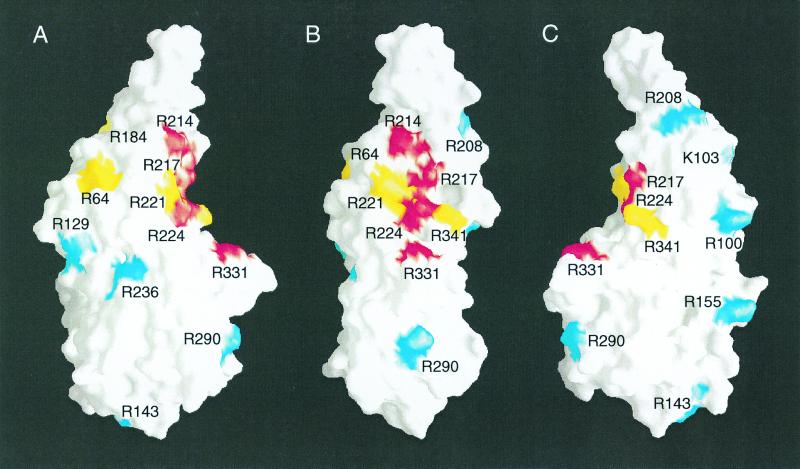
FIG. 10.
Mutations that disrupt DNA binding map to the concave surface of the structured VP16 core. A molecular surface representation (30) was used to generate three projections of the VP16 core. The surface of the VP16 core is shown in white. The surfaces of amino acids whose replacement with alanine prevents DNA binding and VP16-induced complex formation are shown in red, those whose replacement prevents DNA binding but not full VP16-induced complex formation are shown in yellow, and those whose replacement results in wild-type (WT) or nearly wild-type VP16 activity are shown in blue. A, right-side view; B, front view; C, left-side view.
The basic-residue substitutions whose DNA-binding defects are suppressed by HCF-1 (yellow residues) form a ring around the VP16 protein. We do not know the reason for the HCF-1-suppressible phenotype of these mutant proteins, although it could reflect destabilization of a critical and sensitive region of the VP16 structure by the mutations. The basic residues that when replaced with alanine affect DNA binding by VP16 in the presence or absence of HCF-1 and Oct-1 all lie in close proximity on the concave part of the seat-like structure. This putative DNA-binding surface is most clearly visible when viewed from either side (Fig. 10A and C). We know of no known DNA-binding structure that resembles that identified here in VP16. Comparison of the VP16 core structure with other known protein structures by using the DALI server (15; http://www.ebi.ac.uk/dali/) did not reveal extensive similarity to any known DNA-binding protein structures. This result is consistent with the hypothesis that VP16 binds DNA through a novel DNA-binding structure.
DISCUSSION
We have described the role of VP16 in DNA recognition of its cis-regulatory site, the TAATGARAT element. We have shown that a VP16-induced complex can be assembled from recombinant proteins synthesized in E. coli that represent minimal regions necessary for VP16-induced complex formation. By using these minimal regions, we have shown that VP16 is sufficient to extend the DNase I protection pattern of the Oct-1 POU domain alone, positioning VP16 over the D element. In addition, we have defined a surface on the concave side of the VP16 core that is involved in DNA binding. These results suggest that VP16 directly recognizes its cis-regulatory site, the TAATGARAT element, by using a novel DNA-binding structure.
VP16-induced complex formation in the absence of eukaryotic cell-specific modification.
The ability to assemble efficiently a VP16-induced complex with recombinant proteins synthesized in E. coli suggests that eukaryotic cell-specific posttranslational modifications, such as protein phosphorylation, are not required for VP16-induced complex assembly. These results do not support previous suggestions that either VP16 (33) or HCF-1 (21) needs to be modified to support VP16-induced complex formation. In the studies described here, we used the minimal region of each protein synthesized in E. coli to assemble an unmodified complex. It is therefore possible that full-length proteins possess inhibitory regions that need to be inactivated by posttranslational modification to permit assembly of the VP16-induced complex. Elsewhere, however, we have shown that dephosphorylation of native full-length human HCF-1 does not affect its ability to stabilize a VP16-induced complex (49a). We conclude that posttranslational modifications are most likely not required for VP16-induced complex formation.
Previous attempts to form a VP16-induced complex with proteins synthesized in E. coli (see references 23 and 33) may have failed owing to the difficulty of producing active HCF-1 in E. coli. The HCF-1 VP16-interaction domain is predicted to form a six-bladed β propeller structure (47) similar to those formed by WD40 repeat-containing proteins such as the β subunit of heterotrimeric G proteins (26, 36, 46). Studies by Garcia-Higuera and colleagues (7) have shown that it is often difficult to synthesize properly folded WD40 repeat-containing proteins. For example, after synthesis in E. coli, the G protein β subunit fails to fold properly; instead, it requires synthesis in a eukaryotic system in the presence of the γ subunit for proper folding. Therefore, β propeller structures may be inherently difficult to fold into active proteins. By synthesizing the His-tagged HCF-1N380 protein described here in E. coli grown at a low temperature (i.e., 18°C), we may have promoted proper folding of the HCF-1N380 protein by slowing its rate of synthesis.
VP16 is responsible for the extended VP16-induced complex footprint.
Results from the DNase I protection experiments showed that the Oct-1 POU domain with high concentrations of the VP16 core protein in the absence of HCF-1 can generate a protection pattern similar to that obtained with a complete VP16-induced complex assembled with full-length proteins (Fig. 3 and 4). This result shows that VP16 is sufficient to extend the Oct-1 footprint. Walker and colleagues (45) failed to see such an extended footprint in the presence of VP16 and the Oct-1 POU domain alone and speculated that HCF-1 is responsible for the footprint extension. One possible explanation for the failure of Walker and colleagues to observe an effect of VP16 on the Oct-1 protection pattern in the absence of HCF-1 (45) is that, in those studies, the DNA cleavage reactions were performed not in solution at equilibrium as described here but, instead, on protein-DNA complexes in polyacrylamide gels after electrophoretic separation. Perhaps the fractionated complexes dissociate prior to or during postelectrophoresis chemical treatment. A second difference is the use of the VP16 core in this study, as opposed to the use of full-length VP16 with its acidic transcriptional activation domain by Walker et al. (45), which may destabilize the VP16–Oct-1 complex (see reference 23). Whichever the case, the results presented here clearly demonstrate that, in the absence of HCF-1 but in combination with the Oct-1 POU domain, VP16 can reproduce the complete DNase I protection pattern of a complete VP16-induced complex, indicating that VP16 does, indeed, contact the DNA.
The conclusion that VP16 contacts DNA is consistent with other findings; including those of the abilities (i) of VP16 to cross-link to DNA (20), (ii) of VP16 to bind DNA on its own (20, 37), and (iii) of VP16 orthologs to discriminate among different TAATGARAT elements (16, 29). Although VP16 can bind DNA on its own at high concentration, this binding displays little, if any, sequence specificity (37, 45). Thus, association with Oct-1 apparently enhances the sequence-specific DNA binding of VP16, perhaps by stabilizing a structure of VP16 that has increased sequence-specific DNA recognition properties and/or by recruiting VP16 to a particular site on the DNA, thus limiting its freedom to bind to other, nonspecific DNA sequences.
Amino acids in the VP16 core responsible for DNA binding are conserved among VP16 orthologs.
To identify residues on the surface of VP16 involved in DNA binding, we chose to replace solvent-exposed basic amino acids because basic residues are frequently involved in DNA binding and sequence recognition. Several amino acids involved in DNA binding were identified, and these and a previously identified residue (25) all map on the concave surface of the seat-like VP16 structure (red residues in Fig. 10). Interestingly, the amino acids likely to be directly involved in DNA binding (i.e., R214, R217, R224, and R331) are either universally conserved or, as in one case (R224), always basic residues (i.e., arginine or lysine) among VP16 orthologs, suggesting that these residues define a conserved DNA-binding surface on VP16. Indeed, two of these conserved residues (R214 and R217) are located in the most highly conserved segment of the VP16 core (residues 214 to 218).
Several VP16 mutant proteins (R64A, R184A, R221A, and R341A; the positions colored yellow in Fig. 10) were unable to bind DNA on their own but could still support wild-type or nearly wild-type levels of VP16-induced complex formation. Thus, HCF-1 is able to suppress the defects of some mutant proteins. Examination of the positions of the mutations on the surface of VP16 suggests that they are not located at random but, instead, are clustered near and around the putative DNA-binding surface (Fig. 10). One explanation for the ability of HCF-1 to stabilize these mutations is that they affect the conformation of VP16, and HCF-1 is able to stabilize the active conformation of VP16. Whatever the case, the ability of HCF-1 to suppress defects shown by this class of mutant proteins is not particular to the in vitro assay conditions because mutant proteins of this class could activate transcription in vivo, albeit to differing extents (Fig. 9 and Table 1).
Mutations disrupting the VP16 headrest showed that it also plays a role in VP16-induced complex formation and DNA binding. However, in contrast to other DNA-binding residues, the headrest shows little sequence conservation among VP16 orthologs. An attractive possibility is that the headrest recognizes the D element and is responsible for determining the DNA-binding specificity of VP16 orthologs. The VP16 headrest neighbors amino acids (e.g., R214) that are conserved among VP16 orthologs and are important for DNA binding. Perhaps the sequence variability of the headrest allows it to provide the specificity needed to recognize differentially the D element. In support of this possibility, we have observed that exchanging the HSV-1 headrest (amino acids 186 to 213) with the corresponding amino acids from BHV-1 VP16 resulted in a chimeric HSV-1–BHV-1 protein that could recognize a BHV-1 TAATGARAT element better than an HSV-1 TAATGARAT element, although the overall DNA-binding activity was very low (R. Babb and W. Herr, unpublished results).
Does VP16 possess a bipartite DNA-binding domain?
The previous study of Lai and Herr (25) showed that residues within the unstructured region of the VP16 core are involved in DNA binding, and the results reported here identify a surface of the structured region of VP16 that is involved in DNA binding. These findings suggest a possible bipartite VP16 DNA-binding structure, in which both the structured and unstructured regions of VP16 may contact separate sequences in the TAATGARAT element. For example, the structured region may contact the D element, consistent with its role in discriminating the D element sequence (27), and the unstructured region may adopt a structure upon its association with the Oct-1 POU homeodomain and bind the GARAT sequence abutting the TAAT POU homeodomain binding site. If this is true, then both of the DNA-binding components of the VP16-induced complex, the Oct-1 POU domain and VP16, contain bipartite DNA-binding structures. Such structures may be advantageous because they permit flexibility in protein-DNA interaction and greater combinatorial complexity.
A novel DNA-binding structure.
Cognizant that VP16 can recognize the TAATGARAT D element sequence (16, 27, 29) and that at least one residue on the concave surface of VP16 is involved in DNA binding (R331; 25), Liu et al. (27) proposed a model of VP16 bound to a TAATGARAT element with the Oct-1 POU domain. In that model, the TAATGARAT D element rests on the bottom and back of the VP16 seat, with the disordered VP16 segment positioned for association with the Oct-1 POU homeodomain. The results presented here are consistent with this model because the basic residues that play a role in DNA binding (R214, R217, R224, and R331) are all in close apposition to the DNA in the model. Three of these residues—R214, R217, and R224—are all on one face of an α helix, the second of the two long α helices that make up the back of the seat, that, in the model, crosses the major groove of the DNA. Thus, by using a mechanism common to many DNA-binding proteins, VP16 may recognize the DNA by positioning an α helix within the major groove of the DNA. A more precise model for VP16 binding to DNA, however, requires a high-resolution structure of VP16 bound to DNA and the Oct-1 POU domain. Nevertheless, the studies presented here reveal mechanisms whereby a transcription factor that displays no inherent DNA-binding specificity on its own can interpret signals in the DNA to assemble a transcriptional regulatory complex—in this instance, the VP16-induced complex.
ACKNOWLEDGMENTS
We thank X. Cheng for discussions; A. Bubulya for help with the in vivo transcriptional activation studies; C. Sanders for advice on chemical DNA sequencing; L. Joshua-Tor and M. Wang for help with the VP16 structural similarity analysis; X. Zhao for HeLa cell nuclear extract; N. Hernandez, L. Joshua-Tor, A. Stenlund, and J. Wysocka for comments on the manuscript; J. Duffy and P. Renna for artwork; and J. Reader for help with manuscript preparation.
These studies were funded by U.S. Public Health Service grant CA-13106 from the National Cancer Institute.
REFERENCES
- 1.apRhys C M J, Ciufo D M, O'Neill E A, Kelly T J, Hayward G S. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for cellular nuclear factor III. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 3.Cleary M A, Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol Cell Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleary M A, Pendergrast P S, Herr W. Structural flexibility in transcription complex formation revealed by protein-DNA photocrosslinking. Proc Natl Acad Sci USA. 1997;94:8450–8455. doi: 10.1073/pnas.94.16.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousens D J, Greaves R, Goding C R, O'Hare P. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 1989;8:2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douville P, Hagmann M, Georgiev O, Schaffner W. Positive and negative regulation at the herpes simplex virus ICP4 and ICP0 TAATGARAT motifs. Virology. 1995;207:107–116. doi: 10.1006/viro.1995.1056. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Higuera I, Fenogolia J, Li Y, Lewis C, Panchenko M P, Reiner O, Smith T F, Neer E J. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein β subunit. Biochemistry. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- 8.Gerster T, Roeder R G. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6451. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 10.Greaves R F, O'Hare P. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J Virol. 1990;64:2716–2724. doi: 10.1128/jvi.64.6.2716-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry R W, Mittal V, Ma B, Kobayashi R, Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2 and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 13.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 14.Herr W. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 16.Huang C C, Herr W. Differential control of transcription by homologous homeodomain coregulators. Mol Cell Biol. 1996;16:2967–2976. doi: 10.1128/mcb.16.6.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katan M, Haigh A, Verrijzer C P, van der Vliet P C, O'Hare P. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multiprotein transcription complex. Nucleic Acids Res. 1990;18:6871–6880. doi: 10.1093/nar/18.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 19.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with the HSV αTIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristie T M, Sharp P A. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV α-trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 21.Kristie T M, Sharp P A. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus α-trans-induction factor (VP16) J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 22.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 23.LaBoissière S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai J-S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 25.Lai J-S, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. The 2.0 A crystal structure of a heterotrimeric G protein. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Gong W, Huang C C, Herr W, Cheng X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999;13:1692–1703. doi: 10.1101/gad.13.13.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsden H S, Campbell M E M, Haarr L, Frame M C, Parris D S, Murphy M, Hope R G, Muller M T, Preston C M. The 65,000-Mr, DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misra V, Walker S, Yang P, Hayes S, O'Hare P. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct Funct Genet. 1991;11:281–293. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 32.O'Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly D, Hanscombe O, O'Hare P. A single serine residue at position 375 of VP16 is critical for complex assembly with Oct-1 and HCF and is a target of phosphorylation by casein kinase II. EMBO J. 1997;16:2420–2430. doi: 10.1093/emboj/16.9.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomerantz J L, Kristie T M, Sharp P A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;6:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- 35.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 36.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a GA protein βγ dimer at 2.1A resolution. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 37.Stern S, Herr W. The herpes simplex virus trans-activator VP16 recognizes the Oct-1 homeo domain: evidence for a homeo domain recognition subdomain. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 38.Stern S, Tanaka M, Herr W. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature (London) 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 39.Sturm R A, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 40.Sturm R A, Herr W. The POU domain is a bipartite DNA-binding domain. Nature (London) 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka M, Grossniklaus U, Herr W, Hernandez N. Activation of the U2 snRNA promoter by the octamer motif defines a new class of RNA polymerase II enhancer elements. Genes Dev. 1988;2:1764–1778. doi: 10.1101/gad.2.12b.1764. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 43.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 44.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 45.Walker S, Hayes S, O'Hare P. Site-specific conformational alteration of the Oct-1 POU domain-DNA complex as the basis for differential recognition by Vmw65(VP16) Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 46.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 47.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 49.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 49a.Wysocka J, Liu Y, Kobayashi R, Herr W. Developmental cell-cycle regulation of Caenorhabditis elegans HCF phosphorylation. Biochemistry. 2001;40:5786–5794. doi: 10.1021/bi010086o. [DOI] [PubMed] [Google Scholar]
- 50.Xiao P, Capone J P. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol Cell Biol. 1990;10:4974–4977. doi: 10.1128/mcb.10.9.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]