Abstract
The pathogenic Neisseria species N. meningitidis and N. gonorrhoeae possess an outer membrane lipoprotein, designated Lip, which is present in all strains tested. The predicted protein sequence of Lip consists of a consensus AAEAP amino acid repeat. The objective of this study was to determine the feasibility of using the Lip repeat number and sequence for subtyping of Neisseria gonorrhoeae. The lip genes of each isolate were amplified by PCR and sequenced to determine the repeat number and sequence. Among the 46 strains we examined, eight different Lip repeat numbers were identified, with lengths of 11 (1 strain), 12 (14 strains), 13 (2 strains), 14 (10 strains), 15 (5 strains), 16 (10 strains), 17 (3 strains), and 20 (1 strain) repeats. Analysis indicated differences in the sequences within the repeats that resulted in amino acid alterations in repeat classes that contained multiple strains. Among the 46 isolates examined, we were able to identify 17 unique Lip subtyping patterns.
The pathogenic Neisseria species Neisseria meningitidis and Neisseria gonorrhoeae possess an outer membrane lipoprotein, designated Lip, which is present in all strains tested. Characterization of purified Lip has revealed that the protein has no A280; glutamic acid, alanine, and proline account for 85% of the amino acids in the protein; no aromatic and sulfur-containing amino acids are present; and Lip copurifies with lipid (2). The apparent molecular mass of Lip demonstrates strain variation, with a size range of 18 to 30 kDa (3, 6). Three gonococcal lip genes have been sequenced and have been shown to be highly homologous (1, 18). The predicted proteins from these sequences consist entirely of a consensus AAEAP amino acid pentamer repeat with the predicted proteins varying between 13 and 19 repeats in length (1, 18). In this study, we have used Lip repeat number variation and sequencing to develop a new gonococcal subtyping method which we can use in combination which the other subtyping methods, such as auxotype-serovar (A/S) classification, plasmid profile, TetM typing, antimicrobial susceptibilities, and GyrA and ParC mutations, that we currently use in our laboratory to increase our ability to differentiate gonococcal strains in outbreak situations.
MATERIALS AND METHODS
Bacterial strains.
A total of 46 gonococcal isolates were analyzed in this study: (i) 22 isolates from Cleveland, Ohio, isolated in 1994; (ii) 12 isolates from Orange County, Calif., collected in 1995; (iii) 10 isolates from Hawaii collected from 1991 to 1994; and (iv) 2 isolates that were an epidemiologically linked contact pair isolated in Hawaii in 1999. Isolates were provided to the Centers for Disease Control and Prevention (CDC [Atlanta, Ga.]) frozen in Trypticase soy broth containing 20% glycerol and were stored at −70°C.
Strain characterization and culture.
Isolates were characterized by A/S as described previously (8, 10, 15). Agar dilution susceptibilities of isolates to penicillin G, tetracycline, spectinomycin, and ciprofloxacin were determined and interpreted by National Committee for Clinical Laboratory Standards-recommended methods, as described previously (11, 12). Isolates were assigned to penicillin-tetracycline resistance phenotypes as described previously (7, 13, 14). The TetM subtype was determined as previously described (16). Isolates were subcultured daily on GC base-IsoVitalex medium at 37°C in a 5% CO2 atmosphere. Harvested cells were frozen in Trypticase soy broth containing 20% glycerol and were stored at −70°C.
PCR analysis.
The lip genes were amplified by PCR after extraction of the chromosomal DNA of each gonococcal strain. PCR primers specific for lip, H8-1 (5′-CAAATTCAGCGATGAATTTCCAACCC 3′) and H8-4 (5′-TATGAAGGTCAGGCATGTTTGTCGG 3′), were synthesized in the National Center for Infectious Diseases Core Facility, CDC. PCR amplification consisted of 35 cycles of denaturation at 94°C for 30 s, annealing at 64°C for 30 s, and extension at 72°C for 60 s in reaction aliquots of 50 μl. Each PCR mixture contained 5 μl of 10× PCR buffer (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), 1 μl of deoxynucleoside triphosphates (dNTP) stock solution (containing 10 mM each dATP, dCTP, dGTP, and dTTP) (Boehringer Mannheim), 0.5 μl of Taq DNA polymerase (5 U/μl) (Boehringer Mannheim), 2.5 μl of each of the appropriate primers (20 mM), 28.5 μl of sterile water, and 10 μl of template DNA. Ten microliters of PCR product from each isolate was visualized after electrophoresis on a 7.5% polyacrylamide gel, with size being determined by comparison to DNA molecular weight marker VIII (Boehringer Mannheim).
Sequencing analysis.
PCR products were purified for sequencing with the High Pure PCR Product Purification kit (Boehringer Mannheim). Sequencing reactions were performed in 20-μl volumes with ABI Prism BigDYE Terminator Cycle Sequencing Ready Reaction kit reagents (Applied Biosystems Division of Perkin-Elmer Corp., Foster City, Calif.) containing 5.0 μl of Terminator Ready Reaction mix, 5 μl of PCR product template (approximately 100 ng), 3.2 μl of 20 μM primer, and 6.8 μl of sterile, deionized water. Reaction mixtures were cycle sequenced on a GeneAmp PCR system 2400 (Perkin-Elmer) in 25 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min. Spin column purification with Centrisep Columns (Princeton Separations, Adelphia, N.J.) was used to remove excess dye terminators and purify reaction mixtures before electrophoresis. Electrophoresis of extension reaction mixtures was performed on the ABI Prism 377 DNA Sequencer (Applied Biosystems) with a 4% polyacrylamide gel (19:1 acrylamide-bisacrylamide; Bio-Rad Laboratories, Hercules, Calif.).
RESULTS AND DISCUSSION
Repeat variation and stability.
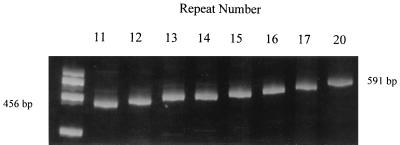
PCR analysis, with sequencing used for confirmation, was used to determine the number of repeats present in the Lip of gonococcal strains. Among the 46 strains examined, eight different Lip repeat patterns were identified, with lengths of 11 (1 strain), 12 (14 strains), 13 (2 strains), 14 (10 strains), 15 (5 strains), 16 (10 strains), 17 (3 strains), and 20 (1 strain) repeats (Fig. 1). To determine the in vitro stability of the repeat lengths, two strains (one with 12 repeats and one with 20 repeats) were subcultured daily for 30 days, with a sample taken and frozen each fifth day. Following the 30-day passage, all samples were subjected to lip PCR and analyzed on 7.5% acrylamide gels. Neither of the strains showed a change in the length of the PCR product during the passage period; the Lip sequences from day 1 and day 30 passages were identical for each strain (data not shown). The in vivo stability of the Lip pattern was examined by subtyping two epidemiologically linked gonococcal isolates which had been collected in Hawaii during an outbreak investigation. Both isolates possessed the 16b Lip pattern as well as having identical A/S class, plasmid profile, antimicrobial susceptibilities, and GyrA and ParC mutations.
FIG. 1.
Polyacrylamide gel showing the size of Lip PCR amplicons versus the number of pentamer repeats present. The 465-bp and 591-bp markers represent the sizes of PCR amplicons for 11 and 20 repeats, respectively.
Sequence analysis of similar repeat lengths.
DNA sequencing data were examined to determine if the amino acid sequences were identical for all Lips with the same number of repeats. Analysis indicated differences in the sequences within the repeats that resulted in amino acid alterations in each of the Lip repeat classes that contained multiple strains. An example of the variation of the amino acid sequence within a repeat number class is shown in Fig. 2 for the sequences observed in the 12-repeat class. In this class sequence, 12a differs from 12b by two amino acids and 12a differs from 12c by eight amino acids. The other repeat classes exhibited similar sequence variation (data not shown). The repeat class-sequence patterns, including the number of strains with each pattern, are summarized in Table 1.
FIG. 2.
Comparison of amino acid sequences of three variations seen among strains with 12 pentamer repeats. Amino acid alterations compared to sequence 12a are shown in lowercase italics for sequences 12b and 12c.
TABLE 1.
Lip repeat-sequence subtyping patterns and number of strains that exhibited each pattern among 46 isolates tested
| No. of repeats | Sequence | No. of strains with pattern |
|---|---|---|
| 11 | a | 1 |
| 12 | a | 4 |
| b | 2 | |
| c | 8 | |
| 13 | a | 1 |
| b | 1 | |
| 14 | a | 8 |
| b | 1 | |
| c | 1 | |
| 15 | a | 3 |
| b | 1 | |
| c | 1 | |
| 16 | a | 5 |
| b | 5 | |
| 17 | a | 2 |
| b | 1 | |
| 20 | a | 1 |
Use of Lip in subtyping.
To determine the feasibility of using the Lip repeat-sequence pattern to subtype apparently related strains of gonococci, we evaluated two subsets of strains which were indistinguishable by A/S classification. One subset of nine strains isolated in Orange County, Calif., all belonged to the Proto IB-3 A/S class. Determination of antimicrobial resistance profiles divided the strains into three groups: three strains were penicillinase producing and tetracycline resistant (PP/TR), five strains were tetracycline-resistant N. gonorrhoeae (TRNG), and one strain was TRNG with chromosomal penicillin resistance. Subtyping of the TetM determinant did not further differentiate the strains, in that all strains contained the “Dutch-type” TetM gene (data not shown). Lip repeat number-sequence analysis was able to identify the three PP/TR strains as individual isolates with repeat-sequence patterns of 11a, 12c, and 15c (Table 2). Additionally, Lip subtyping further divided the five TRNG strains, with three strains having the 14a pattern, one strain having the 14c pattern, and one having the 12c pattern (Table 2).
TABLE 2.
A/S classification, MICs of penicillin and tetracycline, and Lip subtypes of nine gonococcal strains isolated in Orange County, Calif., between July and December 1995
| Strain | Mo | A/S classa | MIC (μg/ml)
|
Lip subtype | |
|---|---|---|---|---|---|
| Penicillin | Tetracycline | ||||
| 1663 | October | Proto IB-3 | 32 | 16 | 12c |
| 1667 | October | Proto IB-3 | 32 | 16 | 15c |
| 1676 | November | Proto IB-3 | 32 | 16 | 11a |
| 1662 | October | Proto IB-3 | 2 | 32 | 12c |
| 1665 | October | Proto IB-3 | 0.125 | 16 | 14a |
| 1669 | October | Proto IB-3 | 0.125 | 16 | 14a |
| 1677 | December | Proto IB-3 | 0.125 | 16 | 12c |
| 1652 | July | Proto IB-3 | 0.125 | 32 | 14c |
| 1653 | August | Proto IB-3 | 0.125 | 32 | 14a |
Proto, no requirements.
The second subset of 10 strains were isolated in Cleveland, Ohio, and all belonged to the Pro IA-6 A/S class. Two strains were TRNG, and eight were classified as susceptible. As shown in Table 3, both of the TRNG strains had a 15a Lip pattern, while the susceptible strains could be further subtyped with Lip patterns of 12a (four strains), 13a (one strain), and 14a (three strains).
TABLE 3.
A/S classification, penicillin and tetracycline MICs, and Lip subtypes of 10 gonococcal strains isolated in Cleveland, Ohio, in 1994
| Strain | A/S classa | Penicillin/tetracycline MIC typeb | Lip subtype |
|---|---|---|---|
| 65 | Pro IA-6 | Susceptible | 12a |
| 68 | Pro IA-6 | Susceptible | 12a |
| 70 | Pro IA-6 | Susceptible | 12a |
| 74 | Pro IA-6 | Susceptible | 12a |
| 82 | Pro IA-6 | Susceptible | 13a |
| 85 | Pro IA-6 | Susceptible | 14a |
| 131 | Pro IA-6 | Susceptible | 14a |
| 183 | Pro IA-6 | Susceptible | 14a |
| 157 | Pro IA-6 | TRNG | 15a |
| 141 | Pro IA-6 | TRNG | 15a |
Pro, proline requiring.
Susceptible, no antimicrobial resistance.
In this study, we have evaluated the feasibility of using Lip repeat number and amino acid sequence in the subtyping of N. gonorrhoeae isolates. Of 46 isolates examined, we were able to identify 17 unique Lip subtyping patterns which gave a discriminatory index (5) of 0.923 for this set of strains. The Lip patterns, both repeat number and sequence, of the two isolates tested were stable after 30 days of passage, and the same Lip pattern was observed in an epidemiologically linked contact pair. Additionally, by determining the Lip pattern, we were able to differentiate between strains which had the same A/S class and antimicrobial profile.
It should be noted, however, that Lip subtyping is not sufficiently discriminatory to be used without additional gonococcal subtyping methods, such as A/S classification, TetM subtyping, β-lactamase plasmid profiles, or GyrA-ParC mutation analysis in fluoroquinolone-resistant isolates. An example of the need for combination subtyping is the fact that seven isolates with the 14a Lip pattern belonged to four different A/S classifications. The need to use multiple methods when subtyping gonococci exists, even when one of the methods is genomically based, such as pulsed-field gel electrophoresis (PFGE), discriminatory index of 0.997 (17), in that a report by Marquez et al. (9) identified multiple A/S classes in a single PFGE pattern.
One significant advantage of Lip subtyping over TetM, plasmid, or GyrA-ParC subtyping is that the Lip gene is present in all gonococcal strains and therefore does not require the presence of antimicrobial resistance. Presently, only A/S classification and much more complex methods such as PFGE can be used to subtype susceptible gonococcal isolates.
At this point, we envision Lip subtyping as being extremely useful in situations such as those in Orange County and Cleveland in which a cluster of isolates belong to the same A/S class and therefore may be interpreted as an outbreak caused by a single strain without the use of an additional subtyping system. The PCR stage of Lip subtyping will quickly demonstrate if all isolates contain the same number of repeats. If a number of isolates have the same repeat number, then the second stage of sequencing can be performed in an attempt to further differentiate isolates.
Sequences of all Lip types or sequences will be provided through the internet (4) to allow other investigators to compare the Lip patterns they observe with patterns previously reported. We will continue to add new patterns obtained during our studies to the site and will be willing to add any new patterns identified by other investigators.
ACKNOWLEDGMENTS
This study was supported in part by an appointment (A.J.S.) to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education.
REFERENCES
- 1.Baehr W, Gotschlich E C, Hitchcock P J. The virulence-associated gonococcal H.8 gene encodes 14 tandemly repeated pentapeptides. Mol Microbiol. 1989;3:49–55. doi: 10.1111/j.1365-2958.1989.tb00103.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee A K, Moran E E, Ray J S, Zollinger W D. Purification and characterization of H.8 antigen from group B Neisseria meningitidis. Infect Immun. 1988;56:773–778. doi: 10.1128/iai.56.4.773-778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon J G, Black W J, Nachamkin I, Stewart P W. Monoclonal antibody that recognizes an outer membrane antigen common to the pathogenic Neisseria species but not to most nonpathogenic species. Infect Immun. 1984;43:994–999. doi: 10.1128/iai.43.3.994-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Subtyping of gonococcal isolates by Lip repeat number and sequence. [Online.] Atlanta, Ga: Centers for Disease Control and Prevention; 2000. http://www.cdc.gov/ncidod/dastlr/gcdir /liptyping.html. [Google Scholar]
- 5.Dillon J-A R, Rahman M, Yeung K-H. Discriminatory power of typing schemes based on Simpson's index of diversity for Neisseria gonorrhoeae. J Clin Microbiol. 1993;31:2831–2833. doi: 10.1128/jcm.31.10.2831-2833.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitchcock P, Hayes S, Mayer L, Shafer W, Tessier S. Analyses of gonococcal H.8 antigen: surface location, inter- and intrastrain electrophoresis heterogeneity, and unusual two-dimensional electrophoretic characteristics. J Exp Med. 1985;162:2017–2034. doi: 10.1084/jem.162.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knapp J S, Washington J A, Doyle L J, Neal S W, Parekh M C, Rice R J. Persistence of Neisseria gonorrhoeae strains with decreased susceptibilities to ciprofloxacin and ofloxacin in Cleveland, Ohio, from 1992 through 1993. Antimicrob Agents Chemother. 1994;38:2194–2196. doi: 10.1128/aac.38.9.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knapp J S, Tam M R, Nowinski R C, Holmes K K, Sandström E G. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J Infect Dis. 1984;150:44–48. doi: 10.1093/infdis/150.1.44. [DOI] [PubMed] [Google Scholar]
- 9.Marquez C, Xia M, Borthagaray G, Roberts M C. Conjugal transfer of the 3.05 b-lactamase plasmid by the 25.2 Mda plasmid in Neisseria gonorrhoeae. Sex Transm Dis. 1999;26:157–159. doi: 10.1097/00007435-199903000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Meyers J A, Sanchez D, Elwell L P, Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976;127:1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Approved standard M7-A3. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 13.Rice R J, Knapp J S. Antimicrobial susceptibilities of Neisseria gonorrhoeae strains representing five distinct resistance phenotypes. Antimicrob Agents Chemother. 1994;38:155–158. doi: 10.1128/aac.38.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice R J, Knapp J S. Susceptibility of Neisseria gonorrhoeae associated with pelvic inflammatory disease to cefoxitin, ceftriaxone, clindamycin, gentamicin, doxycycline, azithromycin, and other antimicrobial agents. Antimicrob Agents Chemother. 1994;38:1688–1691. doi: 10.1128/aac.38.7.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Short H B, Ploscowe V B, Weiss J A, Young F E. Rapid method for auxotyping multiple strains of Neisseria gonorrhoeae. J Clin Microbiol. 1977;6:244–248. doi: 10.1128/jcm.6.3.244-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trees D L, Fakile Y, Neal S W, Knapp J S. Prevalence and tetM subtype of tetracycline resistant Neisseria gonorrhoeae in Ohio, 1994. Sex Transm Dis. 2000;27:46–48. doi: 10.1097/00007435-200001000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Van Looveren M, Ison C A, Ieven M, Vandamme P, Martin I M, Vermeulen K, Renton A, Goossens H. Evaluation of the discriminatory power of typing methods for Neisseria gonorrhoeae. J Clin Microbiol. 1999;37:2183–2188. doi: 10.1128/jcm.37.7.2183-2188.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods J P, Spinola S M, Strobel S M, Cannon J G. Conserved lipoprotein H.8 of pathogenic Neisseria consists entirely of pentapeptide repeats. Mol Microbiol. 1989;3:43–48. doi: 10.1111/j.1365-2958.1989.tb00102.x. [DOI] [PubMed] [Google Scholar]