Abstract
The composition of the intestinal microbiome affects health from the prenatal period throughout childhood, and many diseases have been associated with dysbiosis. The gut microbiome is constantly changing, from birth throughout adulthood, and several variables affect its development and content. Features of the intestinal microbiota can affect development of the brain, immune system, and lungs, as well as body growth. We review the development of the gut microbiome, proponents of dysbiosis, and interactions of the microbiota with other organs. The gut microbiome should be thought of as an organ system that has important effects on childhood development. Dysbiosis has been associated with diseases in children and adults, including autism, attention deficit hyperactivity disorder, asthma, and allergies.
Keywords: gut microbiome, childhood development, microbiota, infancy, childhood diseases
The intestinal microbiome plays an important role in childhood development. Childhood development refers to the biological, psychological, and emotional changes that occur between birth and the end of adolescence. There are set milestones to ensure that developmental trajectories are on target and development in each area is closely tracked. In addition, anthropometric markers typically follow along established growth curves. At each pediatrician’s visit children are tracked for growth, gross motor development, fine motor development, social and emotional development, language development, and cognitive development. Childhood development can be divided into four distinct periods: infancy, preschool years, middle childhood years, and adolescence. Deviation from preset milestone markers can be an early sign of disease development, whether for malnutrition or obesity, social developmental delay in Autism Spectrum Disorder (ASD), food allergies, or asthma. And for each of these issues, the gut microbiome factors in significantly. The proposed link between the gut microbiome and childhood development offers a unique opportunity to modify health prevention, a cornerstone of pediatric medicine (1).
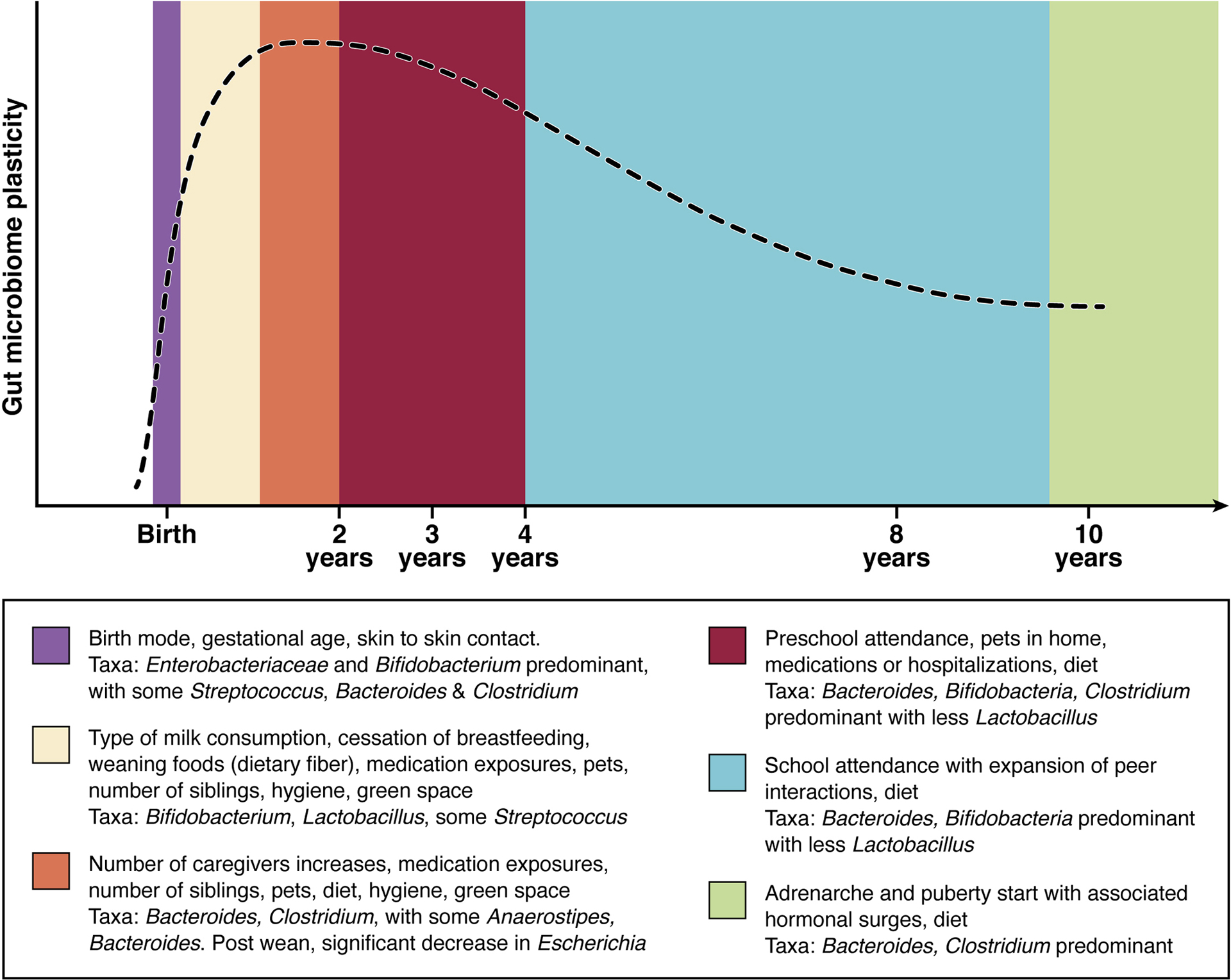
The environment influences microbiome development (Figure 1). While there is conflicting data as to whether or not the microbiome is influenced prior to birth via in utero microbial colonization of the placenta (2,3,4), there is agreement that there is significant increase in microbial colonization immediately after birth, in which initial colonization is by facultative anaerobes, followed by obligate anaerobes (5). Additionally, there is evidence suggesting that exposure to bacteria and bacterial products via cord blood affects risk for childhood wheezing, and primes IL-13 responses.(6,7).
Figure 1. Factors influencing Microbiome development throughout childhood.
For simplicity, this figure represents the most commonly proposed dominant taxa of a healthy full-term breast fed infant along with the events of childhood or exposures which likely contribute to alterations in taxa predominance (112, 113). A comparison of Adult gut microbiome and that of the developing microbiome throughout childhood shows there is a temporal progression. The microbiome in infancy is less diverse and less stable than that of an adult. The microbiome of an infant is dependent on many factors- mode of delivery, milk consumption, medication exposure, environment. Weaning signals a significant change in diet, after which Proteobacteria become far less abundant. During childhood years the diversity increases as does the stability. By adolescence the gut microbiome does not yet resemble that of an adult but does show a shift toward an overall decrease in numbers of aerobes and facultative anaerobes, as well as concurrent increases in anaerobic species (113, 114). While the exact population and shifts may not yet be known, it is clear that environmental and physiologic triggers induce changes.
For full term infants, factors that contribute to early intestinal colonization include mode of delivery and infant diet (5). An infant delivered vaginally has colonization representative of the mother’s vaginal tract, including Lactobacillus, Prevotella, or Sneathia spp (8). Infants born via cesarean section have colonization more consistent with maternal skin and oral microbes, such as Enterobacter hormaechei/E. cancerogenus, Haemophilus parainfluenzae/H. aegyptius/H. influenza/H. haemolyticus, Staphylococcus saprophyticus/S. lugdunensis/S. aureus, Streptococcus australis, and Veillonella dispar/V. parvula (9). An infant delivered by C- section misses out on contact with the microbes in the mother’s vaginal canal, and one study shows that these infants have a significantly lower abundance of Bacteroides over time, regardless of feeding mode (8). Infants born via C-section bypass microbes found in mom’s stool as well, including Escherichia/Shigella, Bifidobacterium longum, Enterococcus faecalis, Bacteroides fragilis, B. thetaiomaomicron, and Bilophila wadsworthia, which have increased prevalence in the intestinal microbiomes of infants born vaginally (9). Moreover, children born via C-section tended to have a higher need for antibiotics due to respiratory infections in the first year of life (10). This was attributed to differences in acquisition of bacteria. In children born via C- section the gut microbiota was less stable with delay to Bifidobacterium emergence and higher pathogen abundance, from the genera Klebsiella and Enterococcus. These taxa are specifically noted to be associated with higher incidence of respiratory infections within the first year of life (10). A review of samples from nearly 600 infants in the UK by Shao et al (11) suggests that this could be due to the fact that these children lack commensal maternal Bacteroides strains of gut flora and are instead dominated with opportunistic pathogens (including Enterococcus, Enterobacter and Klebsiella species) associated with hospital environments. However Chu et al (12) argue this difference disappears by 6 weeks of age.
Infant diet also affects microbiome development. Breast-fed infants have a microbiome predominantly consisting of Lactobacillus, Staphylococcus, and Bifidobacterium, whereas infants that are formula-fed have microbiomes consistent with Roseburia, Clostridium, and Anaerostipes (4,9). The cessation of breast-feeding induces marked changes. The microbiome of a one-year-old infant who stops breast-feeding evolves towards a more adult-like one, and consists of microbes that degrade dietary fibers and produce short chain fatty acids (SCFAs) (9). Formula-fed infants may have a more rapid maturation of their microbiome towards that of an adult, and has been shown to have more organisms associated with inflammation (4).
For preterm infants microbiome development is primarily driven by gestational age, with progression from Bacilli to Gammaproteobacteria to Clostridia occurring in all infants, but with differences in pace of progression (13). Preterm infant colonization is dominated by Enterobacter, Staphyloccoccus, and Enterococcus (4), as opposed to Bacteroides, Bifidobacterium, Parabacteroides, and Escherichia in full term infants (9).
An infant’s neonatal microbiome matures into a more complex one within the first year of life. Additional factors that can influence this time period include treatment with antibiotics, whether the child has siblings, or exposure to household pets (14,15). Antibiotics cause the microbiome of infants to seem even less developed than those without exposure, and have been associated with an increased risk of various problems like obesity, asthma, and allergies (16). Antibiotics can also alter the gut virome, and enrich phage-encoded genes that influence antibiotic resistance (17). Having older siblings correlates with increased bacterial diversity (alpha diversity), attributed to more microbe exposure (4,18). The infant microbiome converges to adult patterns by age three (19).
The microbiota found within the human gut have many important functions. Apart from inter-bacterial communication, these taxa interact with the host in many important ways. The gut microbiome functions in providing essential nutrients, metabolizing dietary fiber to SCFAs, educating the host immune system, and producing bioactive neurotransmitters such as gamma-aminobutyric acid (GABA), tryptophan metabolites, and histamine (20).
Below, we will go into further detail regarding these aspects of the gut microbiome, and their role biologically and in childhood diseases.
Microbiome and Body Growth
The gut microbiome plays key roles in regulating energy harvest from nutrients, growth hormone signaling, and prevention of pathogen colonization. It has been proposed that disturbance of gut microbiota development, particularly during the first two years of life, can affect growth trajectories. Some of the proposed mechanisms of how the gut microbiome affects weight include increased dietary energy acquisition, promotion of fat deposition, locomotor activity modification, satiety effects, and systemic inflammation activation (21,22,23,24).
Studies of the role of the microbiome and normal weight gain and length growth parameters have been performed in preterm infants. Lu et al (25) showed that the microbiome was important for growth by demonstrating that the microbiome community from a good growth (by weight) preterm infant could be transfaunated to a germ free mouse dam, resulting in mouse pups with a good growth phenotype. In contrast, the microbiome from a poor growth infant resulted in poor growth mouse pups after transfaunation, and this poor growth was associated with increased inflammation (25). Yee et al (26) showed that the microbiome of preterm infants influences both weight gain and length, and that infants with improved length have increased volatility of beta diversity and less microbiome maturity.
Further studies of the effect of the microbiome on weight and growth can be extrapolated from studies involving overweight and underweight subjects. Studies in mouse models have shown that transplanting fecal matter from obese subjects (human or murine) leads to an increase in total body fat when compared to recipients of fecal matter from lean subjects (27). The microbiota of an obese individual is particularly adept at harvesting energy via fermentation of dietary polysaccharides not usually digestible by the host. The resultant intestinal absorption of monosaccharides and SCFAs leads to increased hepatic conversion of more complex lipids, which in turn stimulates deposition of adipocytes (28). Dysbiosis has been associated with increased SCFAs, obesity, and other metabolic changes, but the connection between SCFAs and obesity is still being explored (29, 30). SCFA’s, the principal metabolites of gut microbiota, are important in controlling intestinal permeability, inflammation, bile acid metabolism, and immunological functions, and it is believed that obese individuals produce more colonic SCFA for increased microbial energy harvest (30). However, SCFAs can also activate certain peptide hormones that stimulate satiety and increase peripheral glucose disposal (30). The gut microbiota of obese individuals is dominated by Firmicutes (29, 31), with also a reduction in Akkermansia muciniohila, an organism thought to be protective against excess adipose tissue and inflammation in adipose tissue (29).
Early exposure to antibiotics has been shown to alter both the composition and metabolic activity of gut microbiota (32) and has been associated with increased adipose tissue, metabolic hormone levels, and SCFA levels (3). This can lead to either an increase or decrease in body mass, resulting in weight gain or stunted growth (33,34). Interestingly, a study of 28,354 mother–child dyads from the Danish National Birth Cohort found that antibiotics in the first 6 months of life led to an increased risk of being overweight only if the mothers were of normal weight. However, antibiotics were actually associated with a decreased risk of being overweight if the mothers were overweight or obese (35). This highlights the complex role of the microbiome in growth and the influence of early colonization. Fecal samples during infancy of normal weight children have a higher number of Bifidobacteria, while samples from overweight children are noted to have higher numbers of Staphylococcus aureus (36).
There have also been studies examining the influence of the gut microbiome on malnutrition and poor growth. Dietary interventions alone have not been shown to effectively correct weight in malnutrition, and more recently the gut microbiome has been investigated as a key component of growth trajectories. In a study of twins in Malawi discordant for kwashiorkor, it was found that the malnourished twin had an abnormal microbiome (3). While no specific taxa were noted to be consistently related to kwashiorkor, to prove that the microbiome was a cause for kwashiorkor, gnotobiotic mice were transplanted with frozen feces from the affected twins, and subsequently developed significant weight loss and changes in metabolism (4). A study involving children in Bangladesh with severe acute malnutrition (SAM) identified 220 bacterial taxa that were significantly different in their proportional representation to those of healthy children (37). The study profiled the development of the gut microbiome in the first 2 years of life and attributed a “maturity index” dependent on taxa proportionally represented. When comparing the SAM group to healthy controls, a persistence in immaturity of gut microbiome taxa was found in children with SAM. Interestingly this immaturity persisted even after the introduction of a nutritional intervention that led to temporary weight gain, suggesting that maturity of gut microbiome is necessary for long-term expected growth (37). The hypothesis that the microbiome of an undernourished host perpetuates a state of under-nutrition is mirrored in murine models, with a similar lag in microbiome changes despite growth catch up. Undernourished mice were noted to have lower abundance of Bacteroidetes and higher proportions of Firmicutes than controls –a difference which persisted despite recovery of growth (38, 39).
The gut virome might also play a role in growth and malnutrition (17). The virome develops in parallel with the infant and its gut bacterial microbiome (17). In a study done by Reyes et al (40) on healthy Malawi twins and those discordant for Kwasiorker, it was found that Anelloviridae and Circoviridae were present in the microbiota of the twins with SAM, although the impact is unknown.
Development of the Central Nervous System
The existence of the gut brain axis is something long proposed in literature. The model of communication proposed is bi-directional. Top down signaling is from the brain influencing motor, sensory, and secretory functions of the GI tract via afferent fibers of the vagus nerve. Bottom up communication affects function of the brain, especially the amygdala and hypothalamus, via efferent vagal fibers. Bioactive metabolites are produced by microbiota in the gut and can function as neurotransmitters. Serotonin, dopamine, noradrenaline, acetylcholine and GABA are produced by gut microbiota (41, 42, 43). Microglia are integral to neurodevelopment both prenatally and continuing after birth into adolescence. Microglia are the resident central nervous system macrophages and have important roles in innate immunity and neuroprotection, phagocytosis of cellular debris, and synaptic pruning. They drive neuroinflammation coordinated with systemic inflammation (44).
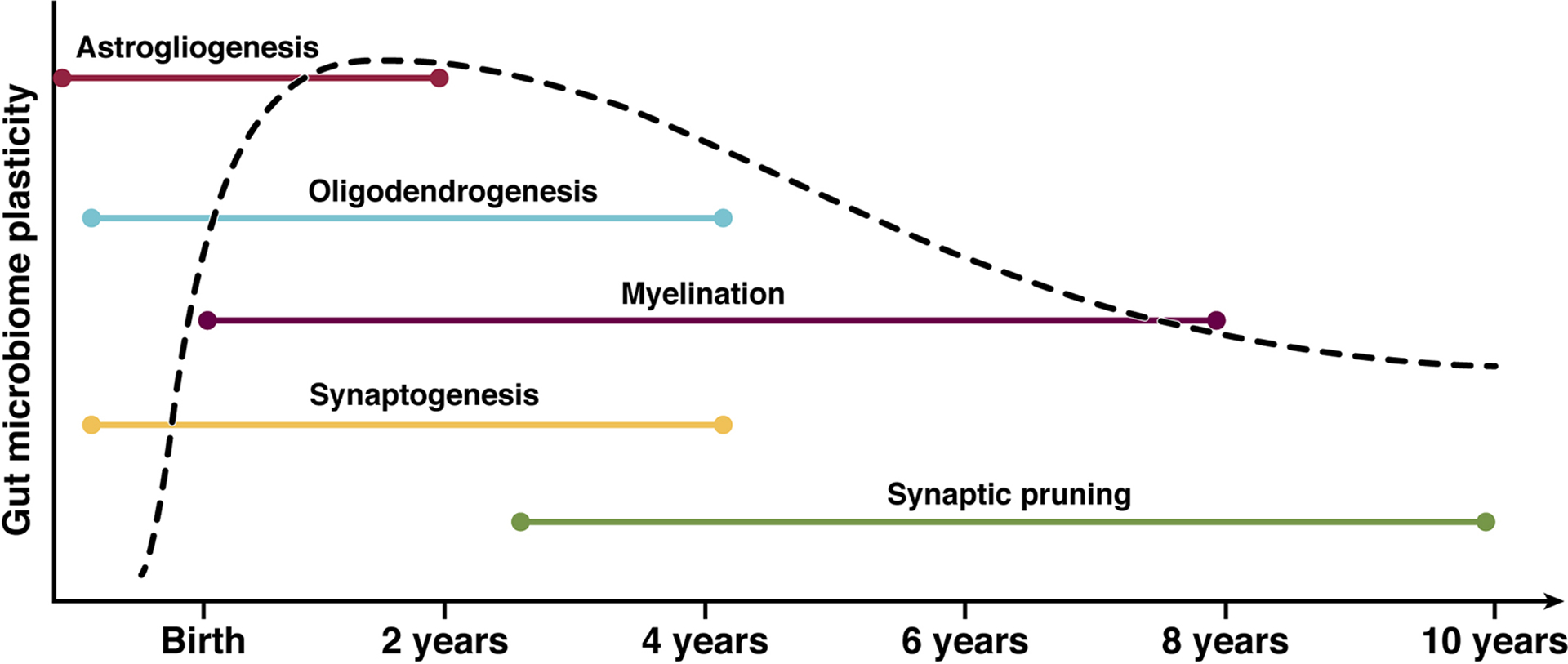
Neurodevelopment progresses over time in parallel with microbiome development (Figure 2). While neuronal migration and neurogenesis occur during fetal development, gliogenesis, synaptogenesis, myelination and synaptic pruning continue throughout childhood and into young adulthood (45). This ongoing neuronal development throughout childhood allows for various factors to impact the developmental trajectory, one such factor being gut microbiota. Dysbiosis can lead to an alteration of metabolite profiles, thus influencing the enteric nerve CNS communication, brain immune function, CNS inflammation, and function and integrity of the blood-brain barrier.
Figure 2. Parallel neurodevelopment and microbiome development during childhood.
Astrogliogenesis begins in utero and continues until approximately 2 years. Astrocytes function to shape neural circuits in the developing brain by coordinating synapse formation and function, neuronal survival, and axon guidance. Oligodendrogenesis continues from the prenatal period through 4 years and is responsible for myelin sheath production. Myelination occurs from birth through 8 years, with 80% of adult myelination being achieved between ages 2–3 years. Synaptogenesis begins prenatally and continues through 4 years, with synaptic pruning occurring from 3 years through 10 years. Here we can see the microbiotal changes that are occurring concurrently with neurodevelopment. The periods of highest growth and plasticity represent crucial windows in which the gut-brain axis can be impacted (115, 116, 117, 118).
Lu et al (46) determined that germfree mice, when inoculated with fecal matter from preterm babies with poor NICU growth, demonstrated delayed brain development in neuronal differentiation, oligodendrocyte differentiation, and myelination when compared to recipients of donors with appropriate growth. The microbiota from infants with low growth also resulted in alteration of neurotransmitter pathways, increases in neuroinflammation and decreased levels of IGF-1 (46).
Further studies on germfree mice have shown an increase in motor dysfunction and lack of appropriate anxiety-like behavior when compared to their normally colonized counterparts (47). Germfree mice were noted to have an increase in neurotransmitters associated with a decreased situation appropriate anxiety and an increase in dopamine receptors via immunohistochemical staining. These motor function and behavioral abnormalities were prevented by early exposure to gut microbiota, further implying a critical window in brain development (47). Animal models, both germfree and those with early antibiotic exposure, have shown that dysbiosis leads to an impairment of social development preserved across species. In murine models dysbiosis results in impairments that last into adulthood, including in fear extinction, object recognition memory and working memory (48,49,50).
There are suggestions that dysbiosis may play a role in the etiology and development of neurodevelopment disorders and neuropsychiatric diseases as well (51). In human studies, dysbiosis has been associated with attention deficit hyperactivity disorder (ADHD), lower cognitive performance in receptive and expressive language domains, and an increase in fear reactivity and oppositional behavior (52,53). A case control study comparing Chinese children with ADHD to healthy controls found those with ADHD have lower Faecalibacterium and Veillonellaceae, while Enterococcus and Odoribacter were significantly increased (54). Two large human studies have noted associations between gut microbiome and temperament in infants, where dysbiosis increased the traits that may precede psychopathology, such as in depression or anxiety (55,56). In a study conducted by Carlson et al (18), the microbiomes of 89 one-year-old infants were analyzed along with Mullen Scales testing for neurodevelopment. Alpha diversity at age one was associated with poorer scores on an Early Learning Composite score, visual reception, and expressive language scales at age two. A recent study of Serbian children comparing patients with neurodevelopmental disorders to typically developing counterparts found those with neurodevelopmental disorders to have less butyrate producing taxa and an increase in clostridium like species (57).
Autism spectrum disorder (ASD) is a specific childhood condition characterized by social and communication abnormalities that has been connected to the microbiome. The microbiome of children with autism has higher levels of Bacteroidetes and lesser levels of Firmicutes (4). There are also increased amounts of Clostridial species found in fecal matter of children with late-onset autism (4). Previous studies have reported mechanisms of dysregulation of dopamine, serotonin, and norepinephrine (58,59,60). Sharon et al (61) demonstrated that germfree mice that underwent fecal matter transplant from human donors with ASD displayed ASD-like behavior, while the mice that received transplants from typically developing individuals did not exhibit any change. The brains of mice with ASD-like behavior were notable for several ASD relevant genes with extensive alternative splicing. This study furthered the hypothesis that the gut microbiota regulates behaviors via production of neuroactive metabolites and specifically that gut brain connections contribute to the pathophysiology of ASD (61). Furthermore, although the exact cause is still unclear, it is believed that an altered gut microbiome causes many ASD individuals to have associated gastrointestinal issues like irritable bowel syndrome (IBS) (62). The gut microbiota may contribute to central perception of pain via modulating receptor expression. Studies on IBS and pain have noted that certain Lactobacillus strains alter mu-opioid and cannabinoid receptor expression of intestinal epithelial cells, effects mimicking those of morphine (64). Findings in human studies support that the gut brain connection is important in the etiology and treatment of ASD. In an open label trial, Kang et al (64) established that children with autism who received fecal transplantation showed improvements in social skills and adaptive behaviors.
Development of the Immune System
The immune system is responsible for recognizing and responding to innumerable self versus non-self molecules. The gut microbiome, as the largest surface area of exposure to non-self, is crucial for this function. Infancy and early childhood is a key time period for education of the immune system and establishing tolerance to commensal organisms, yet developing the ability to defend the host from pathogens.
Infants are initially primarily protected by the innate immune system, which governs interaction of the host and the microbiome in a non-specific manner. Pattern recognition receptors (PRRs) are expressed by innate immune cells like dendritic cells (DCs), macrophages, and natural killer (NK) cells in the host, and recognize microbe associated molecular patterns (MAMPS) (65). This symbiotic relationship of gut bacteria and humans maintains homeostasis. MAMPs initially stimulate innate cells and program them to handle non-specific responses to subsequent pathogenic exposures (65). Other important components of the innate immune system include Paneth cells that produce microbicidal peptides such as α-defensins, goblet cells which produce mucins to limit contact between microbes and enterocytes, and tight junctions between intestinal epithelial cell (IEC) to limit translocation of bacteria across the intestinal epithelial barrier. Cytokines also provide protection via inflammasomes, IL-22 (maintains mucosal barrier integrity), IL-17 (limits invasion), and IL-10 (limits inflammation and prevent host damage) (66).
Post-natal development of the immune system then relies on both the development of secondary immune organs (lymph nodes, Peyers patches, thymus), and the education of immune cells to differentiate “self” from “non-self” and thus appropriately contain “non-self.”
Gut microbiota are crucial to structuring gut-associated lymphoid tissues (GALTs), which are the front-liners in gut mucosal defense (67). The innate immune cells in GALTs non-specifically recognize pathogens, initiate a response, and activate a downstream response; they are also vital in sustainment of immune tolerance to commensal flora (67). The intestinal microbiome is also necessary for stimulation of IgA production from B cells and memory B cell formation within GALT (68,69). Experimental studies in germfree mice have demonstrated under-developed lymphatic organs (spleen, thymus, lymph nodes and GALT tissue), confirming the importance of the microbiome in their development (70).
Studies in mice have further shown that there is a communication between the gut microbiome and the developing thymus. This enterothymic-communication-axis is mediated by plasmacytoid dendritic cells (pDCs). These antigen-presenting cells from the intestinal mucosa are thought to carry bacteria and bacterial-related products to secondary lymphoid tissue, altering the development of precursor T cells. In murine models, when this axis was interrupted by antibiotics in early life, the mice were more prone to colitis in adulthood compared to mice with normal colonization in early life (71).
Mother’s breast milk offers important initial immune protection to the infant through passive transmission of immunoglobulins, microbes, and prebiotics (72). Oligosaccharides function as prebiotics to maintain the growth of specific bacteria such as Bifidobacteria (73). As the infant starts eating solid foods, the microbiome evolves toward an adult one. Changes in the gut microbiome specifically at weaning are critical in development of the immune system, and initiating immune tolerance to commensal bacteria is perhaps more powerful at that time (74). In a study done on mice by Al Nabhani et al (75), it was found that weaning induces a rapid expansion of gut microbiota with an associated systemic inflammatory activation termed the “weaning reaction.” Disruptions of the developmentally appropriate inflammation response to weaning results in what has been termed a “pathological imprinting.” This imprinting increases the risk of inflammatory disorders at a later point as a result of subsequent immune challenges. Induction of protective regulatory T cell lymphocytes (Treg) cells is time sensitive and reliant on gut microbiota. Exposure to gut microbiota after the time of typical weaning does not protect from increased susceptibility to allergic inflammation later, confirming the presence of a critical window (75). This window for weaning begins to open as a response to growth factors in maternal milk leading to increased permeability of antigen passages. The subsequent increase in exposure of the host immune system to a variety of bacteria and bacterial products results in maintenance of long-term tolerance to similar bacteria. The closure of this critical window is not dependent on microbiota but was observed to be time dependent and is likely due to some unknown encoded timed response (75).
Allergies and asthma are related to changes in the gut microbiome. The first suggestion of early microbial exposure shaping immune development was the “hygiene hypothesis.” This hypothesis originally claimed that if infections early in life were limited, the natural immune system does not develop fully, leading to allergic disease (76). The modified version, the “microflora hypothesis,” asserts that a Western lifestyle that is overly hygienic limits the general microbial exposure and hence alters the infant gut microbiome, subsequently disrupting the development of the immune system, and ultimately leading to allergic disease (76). The observation that a decrease in microbial exposure led to increased susceptibility to allergic conditions later in life has been supported by many observational studies noting children raised near farms or those given non-sterile pacifiers developing fewer allergies (77,78,79,80). The existence of a critical window for modulation of microbiome-immune system development prior to weaning has been supported in preclinical studies. Germfree mice exposed to microbiota before weaning, but not after, are protected from increased susceptibility to allergic inflammation (81,82).
In germfree animals there is a skew in T-helper cell population from T-helper 1 cells (Th-1), responsible for cell-mediated immunity and host defenses against intracellular viral and bacterial pathogens, to that of T-helper 2 cells (Th-2), responsible for host defense against helminthes, tissue repair; this then contributes to development of allergy and asthma (83). Additionally, it has been shown in animal models that germfree mice develop high levels of IgE, which has also been associated with allergic responses (84).
While germ free animal models demonstrate the consequences of limited microbial colonization at key time points, a potential clinical correlate is that in human infants, early exposure to antibiotics also limits microbial colonization at a key time point. Lactobacilli and Bifidobacteria, key organisms in an infant gut important for immune system development are reduced in infants exposed to antibiotics in utero (85). Antibiotic exposure makes an infant susceptible to Clostridium difficile infections, and infants colonized with C. difficile are at an increased risk for atopic consequences like eczema, wheezing, allergic sensitization, and asthma (4). Antibiotic treated mice have also been observed to have an increased susceptibility to peanut allergy, with increased peanut IgE and anaphylactic symptoms to peanut exposure (86). Colonization of mice previously exposed to antibiotics with Clostridia rich microbiota resulted in induction of Treg cells, which led to a food allergy protective phenotype (87). Based on work in murine models, it has been hypothesized that commensal bacteria impact development of food allergy through activation of toll-like receptors (TLR) on intestinal epithelial cells. The resultant Th2 skewed response leads to increased IgE production and increased susceptibility to food allergy (87). Additionally a specific microbiota signature has been linked to a gain of function mutation in mice, resulting in increased susceptibility to oral allergic sensitization and anaphylaxis (88).
As another example of the connection between the early microbiome and later allergy, in a study conducted by Feehley et al (89), germ free mice were colonized with feces from either healthy infants or infants with cow’s milk allergy (CAM). The mice colonized with bacteria from the healthy infants were protected against anaphylactic reactions to cow’s milk allergen, due to the differences in bacterial composition of the healthy human donors and the CMA donors. The healthy colonized mice were enriched with Lachnospiraceae, a microbe that protects against allergic sensitization to food.
The gut microbiome also alters immune response to vaccines. Infants with higher relative species within the Actinobacteria phylum have higher humoral and cellular responses to both oral and parenteral vaccines. In contrast, a higher relative abundance of Proteobacteria is associated with lower humoral and cellular response to vaccines. With respect to oral vaccines, a higher relative abundance of Firmicutes is associated with a more favorable response and Bacteroidetes is associated with a less favorable humoral response (90).
Lung Development and Function
The existence of a gut-lung axis has been demonstrated in both murine and human models with the observation that lung diseases can be influenced by gut microbiota changes and vice versa. Stimulation of mouse lungs with lipopolysaccharide leads to a significant increase in number of bacteria within the gut. Additionally, pneumonia induces intestinal injury and decreases gut epithelial proliferation (91). SCFA derived from gut microbiota may have inhibitory effects on lung inflammation, in a mechanism mediated by the liver (92).
Functionally inducible bronchus- associated lymphoid tissue (iBALT) and GALT are similar in structure and function. The main functions of both are to produce and secrete IgA at mucosal surfaces, influence Th and cytotoxic T cell responses, and affect B cell transformation to plasma cells (93). The mechanism by which the communication occurs is not yet clear. It has been proposed that epithelial cells and immune cells absorb signals from the endothelium leading to formation of a local cytokine microenvironment (93). This microenvironment effects changes in the immune response at distal sites. Naïve immune cells activated in the gut, travel via lymph or blood to the lung where they have effector functions. Dendritic cells exposed to microbiota prior to weaning induce a transient increase in a programmed death ligand necessary for the generation of Treg cells. The early generation of Treg cells is associated with protection from allergy long term (94).
The most studied disease state in relation to the gut lung axis is that of asthma, the most common childhood disease (95). In a study reviewing the lifestyle of two U.S. agricultural populations, the Amish and Hutterites, it was revealed that the prevalence of asthma was lower in the Amish, where the children had exposure to more microbes through contact with farm animals (77). Stein et al (77) suggested that the innate immune system was the primary target of protection in the Amish children, demonstrating that in their studies of both humans and mice, protection from asthma was associated with lower levels of eosinophils and higher levels of neutrophils.
In a study looking at early life home environment and the risk of developing asthma among inner city children, it was found that higher house dust concentrations of cockroach, mice, and cat allergens in the first three years of life were associated with a lower risk of asthma (95, 96). The dust in the homes of children that did not develop asthma were enriched in Kocuria, Alloicoccus, Bifidobacterium, and Acinetobacter. The dust in the homes of children that did develop asthma were enriched in Staphylococcus, Haemophilus, Corynebacterium, and Shingomonas (95). Other studies have also demonstrated that exposure to pets like dogs and cats were associated with a lower risk of developing asthma or allergic rhinitis (76). Studies of stool samples of Canadian infants age 3 months showed a reduction of lipopolysaccharide biosynthesis pathways in the microbiota of those identified as high risk for development of asthma (97). Moreover, the genera Lachnospira, Veillonella, Faecalibacterium and Rothia were decreased in relative abundance in children high risk for asthma compared to their counterparts (97).
In studies examining the relationship between SCFA and human airway inflammation, it was noted that children with high amounts of butyrate and propionate at 1 year of age had significantly decreased atopy, and were overall less likely to develop asthma (98). In murine models, SCFA alters gene expression leading to expansion of the Treg population and production of IL-10. A resultant reduction in airway inflammation was observed in mouse models of allergic asthma (99). Human gut microbiota have been shown to produce other metabolites that either promote or suppress inflammation, such as biogenic amines or oxylipins (93,100). Fecal samples of patients with asthma were noted to have a higher number of histamine-secreting bacteria compared with that of healthy volunteers (101).
While bacteria are the most commonly studied component of the microbiome, there have been an increasing number of studies looking into fungal dysbiosis and its impact on development of disease as well. It is unclear whether fungi are permanent colonizers or represent transient changes due to inoculation from food or oral sources, however they still have a capacity to alter the gut ecosystem and subsequent host response. In experimental studies, mice treated with antibiotics were prone to fungal overgrowth with Candida albicans. The mice were subsequently noted to have increase in Th2 cell mediated airway inflammation following airway challenge, via a mechanism thought to involve C. albicans production of prostaglandins (102,103). Similarly, a mouse model treated with antibiotics was noted to have overgrowth of Candida parapsilosis resulting in the prostaglandin mediated divergence of lung macrophages to a M2 phenotype and exacerbating allergic inflammation (104). Two studies recently, in United States and Ecuador, noted fungal dysbiosis as a feature of infant gut microbiota associated with the development of asthma in childhood (105,106). More studies are warranted to further elucidate that mechanism underlying these associations.
Discussion
The gut microbiome plays a crucial role in human development and continued homeostasis, being a foundational part of many different functional axes (Figure 3). We propose that the gut microbiome is in fact an organ system with multiple functions crucial to human development. The definition of an organ system is a group of organs working together as a biological system to perform one or more functions, and the gut microbiome meets that criteria.
Figure 3. Proposed mechanisms of bi- and uni-directional communication from gut microbiome to various organ systems impacting child health and disease.
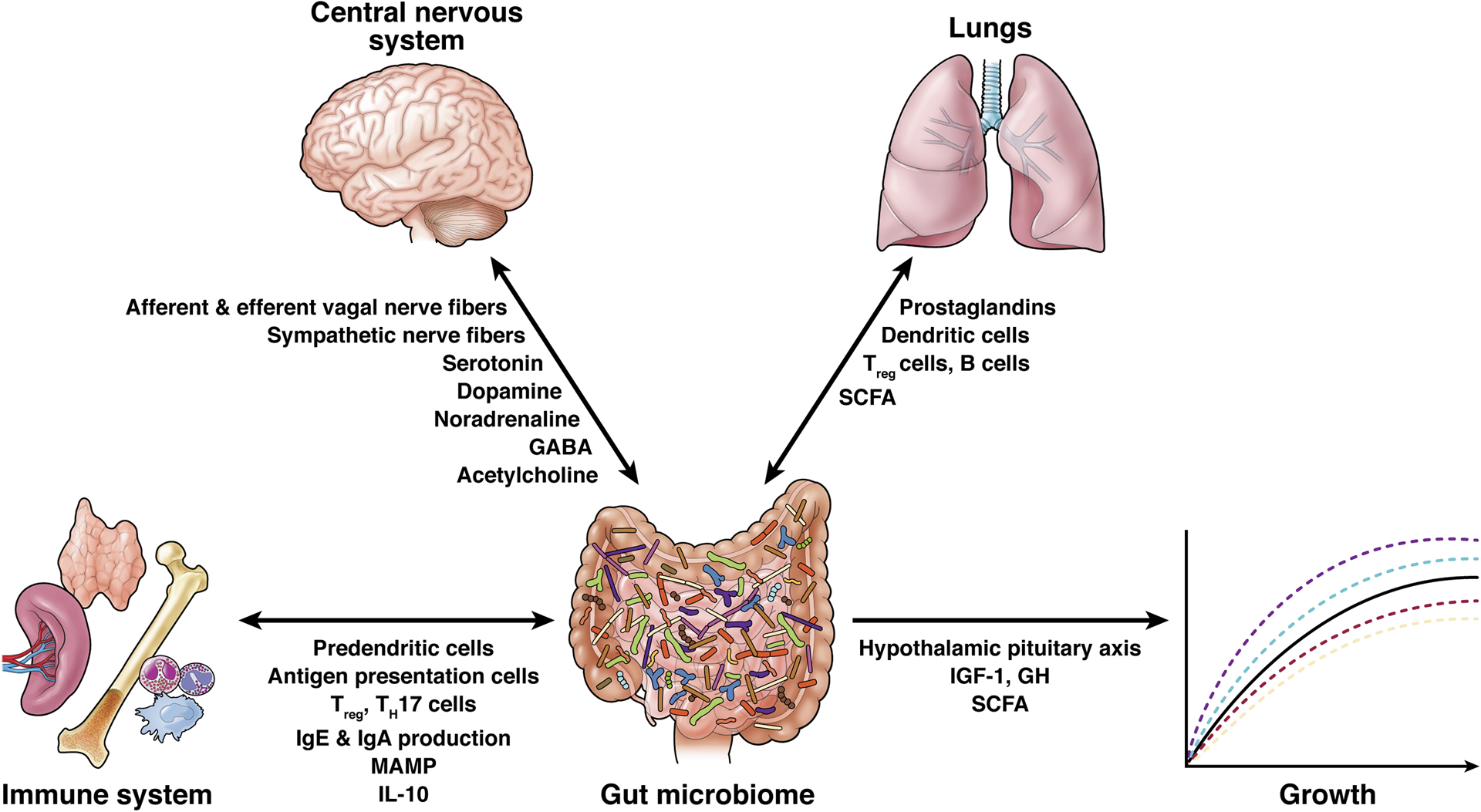
While the exact mechanisms of communication are yet to be understood, these are the most commonly hypothesized pathways. Communication can occur along nerve fibers, hormonal axes and via mediation of many different cytokines, bioactive amines, microbe-associated molecular patterns (MAMP) and short chain fatty acids.
Additionally, we support the proposed hypothesis that there exists a critical window for microbiota modulation of development after which minimal impact is had (107). Buffington et al (108) noted germ-free rodents recolonized with “normal” microbiota at different ages had varying responses to germ free deficits. In fact, social deficits could be reversed with exposure to “normal” microbiota at weaning but not 4 weeks post wean. This sensitive period hypothesis is further supported by the work of Slykerman et al (109, 110) who demonstrated antibiotic exposure in the first year of life but not at later time points has been shown to have a negative impact on cognitive development.
Future directions:
Microbiome studies are relatively new and have primarily focused on bacterial taxa, however as discussed above, the microbiome also includes virome and fungome. Continuing research to elucidate the contribution of both will be crucial moving forward as development of new technology and increased genetic databases emerge.
Though the focus of this review was to discuss existing data on the interplay of the gut microbiome and childhood development, it cannot be ignored that the field is rapidly shifting toward an integrated multi ‘omics and precision medicine model. In one such consideration, Higdon et al (111) describe the stratification of autism spectrum disorder, highlighting the capacity for ‘omics strategies to personalize healthcare for this important population. Beyond this augmented utilization of microbiome data, there is also growing interest in the interaction between microbe and eukaryotic host. As the field continues to expand, each frontier adds a new layer of complexity but also offers hope that we may ultimately come to understand the discrete biologic, genetic, and environmental factors shaping childhood development. There remains a gap in understanding how these organisms exert their effect, and one mechanism by which we can try to bridge this gap is through metabolomics. While we briefly discussed above the various bioactive metabolites produced by gut microbes and the effects they have within the CNS, the immune system, developing respiratory system and on growth, there is much left to understand. The crossover from microbiome to metabolomics is one area in which there may exist potential for optimization particularly during the critical window for modulation of development. Potential modulations include dietary supplements such as probiotics, limiting exposure to antibiotics, and using the microbiome as a biomarker for precision medicine. These mechanisms necessitate further study, and while many studies exist in adult literature in the field of integrated multi omics, there is a paucity of pediatric data.
Clinical studies of the composition and effects of changes in the intestinal microbiome are needed, ideally in large cohorts of healthy children and children with different diseases. We do not know much about the function of the microbiome and interactions between gut dysbiosis and development of disease during childhood. Additionally, further understanding is required regarding the link between the various microbiomes and its constituents – bacterial, viral, and fungal. Identification of deviations that impair development could lead to important development of microbe-based therapies.
Footnotes
Conflict of Interest Statement;
Victoria Ronan, Rummanu Yeasin, and Erika C. Claud declare no conflicts of interest.
References:
- 1.Goulet O, Hojsak I, Kolacek S, et al. Paediatricians play a key role in preventing early harmful events that could permanently influence the development of the gut microbiota in childhood. Acta Paediatr. 2019;108(11):1942–1954. doi: 10.1111/apa.14900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leiby JS, McCormick K, Sherrill-Mix S, et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018;6(1):196. Published 2018 October 30. doi: 10.1186/s40168-018-0575-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ihekweazu FD, Versalovic J. Development of the Pediatric Gut Microbiome: Impact on Health and Disease. Am J Med Sci. 2018;356(5):413–423. doi: 10.1016/j.amjms.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guittar J, Shade A, Litchman E. Trait-based community assembly and succession of the infant gut microbiome. Nat Commun. 2019;10(1):512. Published 2019 February 1. doi: 10.1038/s41467-019-08377-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turturice BA, Gold DR, Litonjua AA, et al. Lower perinatal exposure to Proteobacteria is an independent predictor of early childhood wheezing. J Allergy Clin Immunol. 2019;143(1):419–421.e5. doi: 10.1016/j.jaci.2018.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turturice BA, Ranjan R, Nguyen B, et al. Perinatal Bacterial Exposure Contributes to IL-13 Aeroallergen Response. Am J Respir Cell Mol Biol. 2017;57(4):419–427. doi: 10.1165/rcmb.2017-0027OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17(6):852. doi: 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 10.Reyman M, van Houten M, van Baarle D et al. Author Correction: Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. 2019;10(1). doi: 10.1038/s41467-019-13373-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y, Forster S, Tsaliki E et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574(7776):117–121. doi: 10.1038/s41586-019-1560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu D, Ma J, Prince A, Antony K, Seferovic M, Aagaard K. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. doi: 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Rosa PS, Warner BB, Zhou Y, et al. Patterned progression of bacterial populations in the premature infant gut [published correction appears in Proc Natl Acad Sci U S A. 2014 Dec 2;111(48):17336]. Proc Natl Acad Sci U S A 2014;111(34):12522–12527. doi: 10.1073/pnas.1409497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani C, Duranti S, Bottacini F, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017;81(4):e00036–17. Published 2017 November 8. doi: 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien M, Alvarez AS, de Vos WM. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019;27(12):997–1010. doi: 10.1016/j.tim.2019.08.001 [DOI] [PubMed] [Google Scholar]
- 16.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhya I, Segal JP, Carding SR, Hart AL, Hold GL. The gut virome: the ‘missing link’ between gut bacteria and host immunity?. Therap Adv Gastroenterol. 2019;12:1756284819836620. Published 2019 March 25. doi: 10.1177/1756284819836620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson AL, Xia K, Azcarate-Peril MA, et al. Infant Gut Microbiome Associated With Cognitive Development. Biol Psychiatry. 2018;83(2):148–159. doi: 10.1016/j.biopsych.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. Published 2012 May 9. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan W, Engevik MA, Spinler JK, Versalovic J. Healthy Human Gastrointestinal Microbiome: Composition and Function After a Decade of Exploration. Dig Dis Sci. 2020;65(3):695–705. doi: 10.1007/s10620-020-06118-4 [DOI] [PubMed] [Google Scholar]
- 21.Kimura I, Ozawa K, Inoue D, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun 2013;4:1829. doi: 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Guryn K, Hubert N, Frazier K, et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe. 2018;23(4):458–469.e5. doi: 10.1016/j.chom.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357(6354):912–916. doi: 10.1126/science.aan0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agustí A, García-Pardo MP, López-Almela I, et al. Interplay Between the Gut-Brain Axis, Obesity and Cognitive Function. Front Neurosci. 2018;12:155. Published 2018 March 16. doi: 10.3389/fnins.2018.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu L, Yu Y, Guo Y, Wang Y, Chang EB, Claud EC. Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model. PLoS One. 2015;10(4):e0124504. Published 2015 April 30. doi: 10.1371/journal.pone.0124504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yee AL, Miller E, Dishaw LJ, et al. Longitudinal Microbiome Composition and Stability Correlate with Increased Weight and Length of Very-Low-Birth-Weight Infants. mSystems. 2019;4(1):e00229–18. Published 2019 February 26. doi: 10.1128/mSystems.00229-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006. December 21; 444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 28.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004. November 2; 101(44):15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Shimizu Y, Kimura I. Gut microbial metabolite short-chain fatty acids and obesity. Biosci Microbiota Food Health. 2017;36(4):135–140. doi: 10.12938/bmfh.17-010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murugesan S, Nirmalkar K, Hoyo-Vadillo C, García-Espitia M, Ramírez-Sánchez D, García-Mena J. Gut microbiome production of short-chain fatty acids and obesity in children. Eur J Clin Microbiol Infect Dis. 2018;37(4):621–625. doi: 10.1007/s10096-017-3143-0 [DOI] [PubMed] [Google Scholar]
- 31.Dugas LR, Lie L, Plange-Rhule J, et al. Gut microbiota, short chain fatty acids, and obesity across the epidemiologic transition: the METS-Microbiome study protocol. BMC Public Health. 2018;18(1):978. Published 2018 August 6. doi: 10.1186/s12889-018-5879-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott FI, Horton DB, Mamtani R, Haynes K, Goldberg DS, Lee DY, Lewis JD. Administration of Antibiotics to Children Before Age 2 Years Increases Risk for Childhood Obesity. Gastroenterology. 2016. July; 151(1):120–129.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zárate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences.Cell. 2014. August 14; 158(4):705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621–626. doi: 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ajslev TA, Andersen CS, Gamborg M, Sørensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond). 2011;35(4):522–529. doi: 10.1038/ijo.2011.27 [DOI] [PubMed] [Google Scholar]
- 36.Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 2008;87:534–8. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian S, Huq S, Yatsunenko T et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Composition and function of the undernourished neonatal mouse intestinal microbiome. J Nutr Biochem. 2015;26(10):1050–1057. doi: 10.1016/j.jnutbio.2015.04.010 [DOI] [PubMed] [Google Scholar]
- 39.Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Microbial-Derived Metabolites Reflect an Altered Intestinal Microbiota during Catch-Up Growth in Undernourished Neonatal Mice. J Nutr. 2016;146(5):940–948. doi: 10.3945/jn.115.229179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes A, Blanton LV, Cao S, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A. 2015;112(38):11941–11946. doi: 10.1073/pnas.1514285112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strandwitz P Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693(Pt B):128–133. doi: 10.1016/j.brainres.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galland L The gut microbiome and the brain. J Med Food. 2014;17(12):1261–1272. doi: 10.1089/jmf.2014.7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Colle A, Israelyan N, Gross Margolis K. Novel aspects of enteric serotonergic signaling in health and brain-gut disease. Am J Physiol Gastrointest Liver Physiol. 2020;318(1):G130–G143. doi: 10.1152/ajpgi.00173.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menassa D, Gomez-Nicola D. Microglial Dynamics During Human Brain Development. Front Immunol. 2018;9. doi: 10.3389/fimmu.2018.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron. 2016;89(2):248–268. doi: 10.1016/j.neuron.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J, Lu L, Yu Y, Cluette-Brown J, Martin C, Claud E. Effects of Intestinal Microbiota on Brain Development in Humanized Gnotobiotic Mice. Sci Rep. 2018;8(1). doi: 10.1038/s41598-018-23692-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108(7):3047–3052. doi: 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu C, Murdock M, Jing D et al. The microbiota regulate neuronal function and fear extinction learning. Nature. 2019;574(7779):543–548. doi: 10.1038/s41586-019-1644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gareau M, Wine E, Rodrigues D et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2010;60(3):307–317. doi: 10.1136/gut.2009.202515 [DOI] [PubMed] [Google Scholar]
- 50.Luk B, Veeraragavan S, Engevik M, et al. Postnatal colonization with human “infant-type” Bifidobacterium species alters behavior of adult gnotobiotic mice. PLoS One. 2018;13(5):e0196510. Published 2018 May 15. doi: 10.1371/journal.pone.0196510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva YP, Bernardi A, & Frozza RL (2020). The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Frontiers in endocrinology, 11, 25. 10.3389/fendo.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aarts E, Ederveen THA, Naaijen J, et al. Gut microbiome in ADHD and its relation to neural reward anticipation. PLoS One. 2017;12(9):e0183509. Published 2017 September 1. doi: 10.1371/journal.pone.0183509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prehn-Kristensen A, Zimmermann A, Tittmann L, et al. Reduced microbiome alpha diversity in young patients with ADHD. PLoS One. 2018;13(7):e0200728. Published 2018 July 12. doi: 10.1371/journal.pone.0200728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan L, Ge WR, Zhang S, Sun YL, Wang B, Yang G. Case-Control Study of the Effects of Gut Microbiota Composition on Neurotransmitter Metabolic Pathways in Children With Attention Deficit Hyperactivity Disorder. Front Neurosci. 2020;14:127. Published 2020 February 18. doi: 10.3389/fnins.2020.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aatsinki AK, Lahti L, Uusitupa HM, et al. Gut microbiota composition is associated with temperament traits in infants. Brain Behav Immun. 2019;80:849–858. doi: 10.1016/j.bbi.2019.05.035 [DOI] [PubMed] [Google Scholar]
- 56.Christian H, Zubrick SR, Foster S, et al. The influence of the neighborhood physical environment on early child health and development: A review and call for research. Health Place. 2015;33:25–36. doi: 10.1016/j.healthplace.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 57.Bojović K, Ignjatović ÐI, Soković Bajić S, et al. Gut Microbiota Dysbiosis Associated With Altered Production of Short Chain Fatty Acids in Children With Neurodevelopmental Disorders. Front Cell Infect Microbiol. 2020;10:223. Published 2020 May 19. doi: 10.3389/fcimb.2020.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magula L, Moxley K, and Lachman A (2019). Iron deficiency in South African children and adolescents with attention deficit hyperactivity disorder. J. Child Adolesc. Ment. Health 31, 85–92. doi: 10.2989/17280583.2019.1637345 [DOI] [PubMed] [Google Scholar]
- 59.Stewart A, Davis GL, Gresch PJ, Katamish RM, Peart R, Rabil MJ, et al. (2019). Serotonin transporter inhibition and 5-HT2C receptor activation drive loss of cocaine-induced locomotor activation in DAT Val559 mice. Neuropsychopharmacology 44, 994–1006. doi: 10.1038/s41386-018-0301-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki C, Ikeda Y, Tateno A, Okubo Y, Fukayama H, and Suzuki H (2019). Acute atomoxetine selectively modulates encoding of reward value in ventral medial prefrontal cortex. J. Nippon Med. Sch 86, 98–107. doi: 10.1272/jnms.JNMS.2019_86-205 [DOI] [PubMed] [Google Scholar]
- 61.Sharon G, Cruz NJ, Kang DW, et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell. 2019;177(6):1600–1618.e17. doi: 10.1016/j.cell.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rousseaux C, Thuru X, Gelot A, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med. 2007;13(1):35–37. doi: 10.1038/nm1521 [DOI] [PubMed] [Google Scholar]
- 64.Kang DW, Adams JB, Gregory AC, et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5(1):10. Published 2017 January 23. doi: 10.1186/s40168-016-0225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Negi S, Das DK, Pahari S, Nadeem S, Agrewala JN. Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Front Immunol. 2019;10:2441. Published 2019 October 25. doi: 10.3389/fimmu.2019.02441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng HY, Ning MX, Chen DK, Ma WT. Interactions Between the Gut Microbiota and the Host Innate Immune Response Against Pathogens. Front Immunol. 2019;10:607. Published 2019 March 29. doi: 10.3389/fimmu.2019.00607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk Between Gut Microbiota and Innate Immunity and Its Implication in Autoimmune Diseases. Front Immunol. 2020;11:282. Published 2020 February 21. doi: 10.3389/fimmu.2020.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev. 2014;260(1):76–85. doi: 10.1111/imr.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease [published correction appears in Nat Rev Immunol. 2009 Aug;9(8):600]. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Li J, Zheng W, et al. Peripheral Lymphoid Volume Expansion and Maintenance Are Controlled by Gut Microbiota via RALDH+ Dendritic Cells. Immunity. 2016;44(2):330–342. doi: 10.1016/j.immuni.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ennamorati M, Vasudevan C, Clerkin K, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci U S A. 2020;117(5):2570–2578. doi: 10.1073/pnas.1915047117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front Immunol. 2018;9:361. Published 2018 February 28. doi: 10.3389/fimmu.2018.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickramasinghe S, Pacheco AR, Lemay DG, Mills DA. Bifidobacteria grown on human milk oligosaccharides downregulate the expression of inflammation-related genes in Caco-2 cells. BMC Microbiol. 2015;15:172. Published 2015 August 25. doi: 10.1186/s12866-015-0508-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hornef MW, Torow N. ‘Layered immunity’ and the ‘neonatal window of opportunity’ - timed succession of non-redundant phases to establish mucosal host-microbial homeostasis after birth. Immunology. 2020;159(1):15–25. doi: 10.1111/imm.13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al Nabhani Z, Dulauroy S, Marques R, et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 2019;50(5):1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014 [DOI] [PubMed] [Google Scholar]
- 76.Azad MB, Konya T, Maughan H, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9(1):15. Published 2013 April 22. doi: 10.1186/1710-1492-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein MM, Hrusch CL, Gozdz J, et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N Engl J Med. 2016;375(5):411–421. doi: 10.1056/NEJMoa1508749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hesselmar B, Sjöberg F, Saalman R, Aberg N, Adlerberth I, Wold AE. Pacifier cleaning practices and risk of allergy development. Pediatrics. 2013;131(6):e1829–e1837. doi: 10.1542/peds.2012-3345 [DOI] [PubMed] [Google Scholar]
- 79.Riedler J, Braun-Fahrländer C, Eder W, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358(9288):1129–1133. doi: 10.1016/S0140-6736(01)06252-3 [DOI] [PubMed] [Google Scholar]
- 80.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy [published correction appears in Nat Rev Immunol. 2019 Sep;19(Cox et al, 2014):594]. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871 [DOI] [PubMed] [Google Scholar]
- 81.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. Published 2012 May 1. doi: 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee N, Kim WU. Microbiota in T-cell homeostasis and inflammatory diseases. Exp Mol Med. 2017;49(5):e340. Published 2017 May 26. doi: 10.1038/emm.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14(5):559–570. doi: 10.1016/j.chom.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lamont RF, Møller Luef B, Stener Jørgensen J. Childhood inflammatory and metabolic disease following exposure to antibiotics in pregnancy, antenatally, intrapartum and neonatally. F1000Res. 2020;9:F1000 Faculty Rev-144. Published 2020 February 25. doi: 10.12688/f1000research.19954.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 87.de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol. 2014;5:60. Published 2014 February 18. doi: 10.3389/fimmu.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noval Rivas M, Burton OT, Wise P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131(1):201–212. doi: 10.1016/j.jaci.2012.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feehley T, Plunkett CH, Bao R, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25(3):448–453. doi: 10.1038/s41591-018-0324-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zimmermann P, Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine. 2018;36(30):4433–4439. doi: 10.1016/j.vaccine.2018.04.066 [DOI] [PubMed] [Google Scholar]
- 91.Perrone EE, Jung E, Breed E, et al. Mechanisms of methicillin-resistant Staphylococcus aureus pneumonia-induced intestinal epithelial apoptosis. Shock. 2012;38(1):68–75. doi: 10.1097/SHK.0b013e318259abdb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young RP, Hopkins RJ, Marsland B. The Gut-Liver-Lung Axis. Modulation of the Innate Immune Response and Its Possible Role in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2016;54(2):161–169. doi: 10.1165/rcmb.2015-0250PS [DOI] [PubMed] [Google Scholar]
- 93.Pugin B, Barcik W, Westermann P, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health Dis. 2017;28(1):1353881. Published 2017 January 1. doi: 10.1080/16512235.2017.1353881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20(6):642–647. doi: 10.1038/nm.3568 [DOI] [PubMed] [Google Scholar]
- 95.O’Connor GT, Lynch SV, Bloomberg GR, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. 2018;141(4):1468–1475. doi: 10.1016/j.jaci.2017.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson CC, Ownby DR. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res. 2017;179:60–70. doi: 10.1016/j.trsl.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- 98.Roduit C, Frei R, Ferstl R, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74(4):799–809. doi: 10.1111/all.13660 [DOI] [PubMed] [Google Scholar]
- 99.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity. 2020;52(2):241–255. doi: 10.1016/j.immuni.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barcik W, Pugin B, Brescó MS, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74(5):899–909. doi: 10.1111/all.13709 [DOI] [PubMed] [Google Scholar]
- 102.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72(Cox et al, 2014):4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Erb-Downward JR, Noverr MC. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun. 2007;75(7):3498–3505. doi: 10.1128/IAI.00232-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim RY, Pinkerton JW, Essilfie AT, et al. Role for NLRP3 Inflammasome-mediated, IL-1β-Dependent Responses in Severe, Steroid-Resistant Asthma. Am J Respir Crit Care Med. 2017;196(3):283–297. doi: 10.1164/rccm.201609-1830OC [DOI] [PubMed] [Google Scholar]
- 105.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187–1191. doi: 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arrieta MC, Arévalo A, Stiemsma L, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 2018;142(2):424–434.e10. doi: 10.1016/j.jaci.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turroni F, Milani C, Duranti S, et al. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr. 2020;46(1):16. Published 2020 February 5. doi: 10.1186/s13052-020-0781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, & Costa-Mattioli M (2016). Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell, 165, 1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Slykerman RF, Coomarasamy C, Wickens K, Thompson JMD, Stanley TV, Barthow C, … & Mitchell EA (2019). Exposure to antibiotics in the first 24 months of life and neurocognitive outcomes at 11 years of age. Psychopharmacology (Berl), 236, 1573–1582. [DOI] [PubMed] [Google Scholar]
- 110.K. E, Murphy R, Wall C, & Mitchell EA (2017). Antibiotics in the first year of life and subsequent neurocognitive outcomes. Acta Pae- diatrica, 106, 87–94. [DOI] [PubMed] [Google Scholar]
- 111.Higdon R, Earl R, Stanberry L et al. The Promise of Multi-Omics and Clinical Data Integration to Identify and Target Personalized Healthcare Approaches in Autism Spectrum Disorders. OMICS: A Journal of Integrative Biology. 2015;19(4):197–208. doi: 10.1089/omi.2015.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34 Suppl 2:S12–S18. doi: 10.1016/s1590-8658(02)80157-8 [DOI] [PubMed] [Google Scholar]
- 114.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77(2):404–412. doi: 10.1111/j.1574-6941.2011.01120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Menassa DA, Gomez-Nicola D. Microglial Dynamics During Human Brain Development. Front Immunol. 2018;9:1014. Published 2018 May 24. doi: 10.3389/fimmu.2018.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marín O Developmental timing and critical windows for the treatment of psychiatric disorders. Nat Med. 2016;22(11):1229–1238. doi: 10.1038/nm.4225 [DOI] [PubMed] [Google Scholar]
- 117.Hoffmann A, Ziller M, Spengler D. Childhood-Onset Schizophrenia: Insights from Induced Pluripotent Stem Cells. Int J Mol Sci. 2018;19(12):3829. Published 2018 November 30. doi: 10.3390/ijms19123829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deoni SCL, O’Muircheartaigh J, Elison JT et al. White matter maturation profiles through early childhood predict general cognitive ability. Brain Struct Funct 221, 1189–1203 (2016). 10.1007/s00429-014-0947-x) [DOI] [PMC free article] [PubMed] [Google Scholar]


