Abstract
A widely distributed strain designated 210 was identified in a study of the diversity of Mycobacterium tuberculosis DNA fingerprints from three geographically separate states in the United States. This strain is characterized by a 21-band fingerprint pattern when probed with IS6110, and the pattern is similar to that displayed by strains designated W. Intracellular growth of strain 210 isolates in human macrophages is significantly faster than that of isolates from other clusters or nonclustered isolates. The purpose of this study was to identify the sites of IS6110 insertions in strain 210 and compare these to IS6110 insertion sites in strain W. Our hypothesis is that an IS6110 insertion site(s) could possibly be responsible for a strain's increased capacity for transmission and/or replication. In this report, the insertion sites in strains 210 and W are described and referenced to their location in the M. tuberculosis H37Rv genome sequence. The W and 210 strains have 17 identical sites of IS6110 insertion and additional sequence not found in H37Rv but present in other clinical isolates. The IS6110 insertion site in the 36-bp direct repeat (DR) region of strains 210 and W has 15 spacers in the left flanking region. The DR region on the right side of IS6110 has been deleted. Five sites of insertion in strain 210 not found in strain W are described, as well as two unique sites in strain W. One copy of IS6110 was found to reside 55 bp in the ctpD gene. This gene is expressed, indicating that IS6110 can provide a promoter sequence for the transcription of genes.
Recent studies have indicated that some strains of Mycobacterium tuberculosis may be more readily transmitted and ultimately more successful in causing infection and/or disease (23). In 1995, we examined the diversity of M. tuberculosis DNA fingerprints from three geographically separate states in the United States. The results revealed a widely distributed strain, designated 210, which was isolated from 57 patients out of a total of 1,324 (28). The strain was characterized by a 21-band fingerprint pattern when probed with IS6110. All of the isolates shared a common polymorphic-GC-rich sequence pattern when probed with pTBN12 (a probe commonly used for secondary genotyping) (6). Spoligotyping (spacer oligotyping) detects DNA polymorphism in the direct repeat (DR) region of the M. tuberculosis genome (14). The isolates shared a spoligotype characteristic of the Beijing strain, the predominant family of strains in China that is found throughout Asia (26, 28). Currently, the 210 strain has been shown to account for 215 cases of tuberculosis in five states (unpublished data from M. D. Cave).
In a study of tuberculosis in central Los Angeles, Calif., the largest cluster (43 of 96 patients) was comprised of the 210 strain (1). Epidemiological analysis indicated that strain 210 caused disease in homeless patients in several shelters, as well as nonhomeless patients who used daytime services at the same shelters. To determine whether the dissemination of this strain correlated with its capacity for replication, the intracellular growth rate of strain 210 in human macrophages was measured. Compared to isolates from other clusters or from nonclustered patients, strain 210 grew significantly faster (29).
Strain W is a multidrug-resistant strain that caused disease in more than 350 patients in New York City, N.Y., during the early 1990s (19). Recently, variants of W (usually pansensitive) have been found in 42 patients in New Jersey (4). Strains 210 and W and the W variants are all members of the Beijing family and share many characteristics (16, 23). These strains all have similar IS6110 fingerprint patterns consisting of 16 to 23 bands. In addition, each isolate has an insertion of IS6110 in the origin of replication and a single copy of IS6110 in the NTF1 region (556-bp intervening sequence between two copies of IS6110) (15, 20). When characterized by secondary typing methods, they share the Beijing family spoligotype, a common polymorphic-GC-rich sequence pattern, and an identical variable number tandem repeat type (4, 25).
The genome of M. tuberculosis is highly conserved among different strains, and mutation does not appear to offer an explanation for differences in the success of one strain over another (22). There is evidence that IS6110 plays an important role in mediating genomic rearrangements and deletions (5). Our hypothesis is that an IS6110 insertion site(s) could be responsible for a strain's increased capacity for transmission and/or replication. The first step in this investigation was to identify the sites of IS6110 insertions in strain 210 and compare them to the IS6110 insertions in strain W. We have described and referenced these sites to their location in the M. tuberculosis H37Rv genome sequence (7). We found that the 210 and W strains share 17 identical insertion sites. Since several copies of IS6110 are integrated in or closely adjoin genes, we used reverse transcription-PCR (RT-PCR) to examine the expression of a gene in which IS6110 resides.
MATERIALS AND METHODS
Mycobacterial strains and culture medium.
M. tuberculosis strain H37Rv was obtained from the American Type Culture Collection (Manassas, Va.). M. tuberculosis clinical isolates were submitted to the laboratory as part of our M. tuberculosis genotyping network. M. tuberculosis strain 210, a clinical isolate from a Texas outbreak, was selected for study. This strain has the same 21-band IS6110 fingerprint pattern as that of other isolates found to be associated with outbreaks in five states. Two additional clinical isolates with the same IS6110 fingerprint pattern as strain 210 were used in the gene expression studies. H1110 is a clinical isolate from a Texas outbreak, and TB545 is a clinical isolate from an outbreak in Los Angeles. Strain W is represented by M. tuberculosis isolate H2255, a clinical isolate from Puerto Rico. This isolate has the same characteristic IS6110 fingerprint reported for W strains and is multidrug resistant. Cells were cultivated in Dubos broth containing 0.1% Tween and albumin enrichment (Difco Laboratories, Detroit, Mich.).
Isolation of IS6110 flanking sequences.
DNA was isolated as described previously (3). The amplification of the right side of IS6110 along with a variable-size flanking sequence was performed by mixed-linker PCR (13).
Some of the IS6110 flanking sequences were obtained by cloning IS6110-containing PvuII fragments representing particular IS6110-hybridizing bands. DNA from strain 210 was digested with PvuII and electrophoresed on a 20-cm 0.8% agarose gel for 18.5 h at 50 V using a 1-kb DNA ladder as a size standard (Gibco Life Technologies, Grand Island, N.Y.). A region of the gel known to contain the hybridizing fragment of interest was excised, and the DNA was isolated (Qiagen, Valencia, Calif.). DNA was cloned into the pSTBlue-1 Perfectly Blunt PCR cloning vector (Novagen, Milwaukee, Wis.), and colonies were checked directly in PCRs for the presence of IS6110 (10). Plasmid DNA was isolated from colonies positive for IS6110 and sequenced with the IS6110 primer IS#3 (5′-CCATCGCCGCCTCTACCAGT-3′) as described below.
Nucleotide sequencing.
Sequencing was performed on an ABI 373 DNA Sequencer with the Taq DyeDeoxy Terminator Cycle sequencing kit (Perkin-Elmer, Applied Biosystems Inc., Foster City, Calif.). Sequencing primers were designed with the Oligo 5.1 Primer Analysis software (National Biosciences, Inc., Plymouth, Minn.). Primers were synthesized by Gibco BRL Life Technologies (Gaithersburg, Md.). Sequence analysis was performed using DNAstar (Madison, Wis.). The primer used for sequencing the mixed-linker PCR products was based on IS6110 (13). The flanking sequences were analyzed for homology in the National Center for Biotechnology Information (NCBI) GenBank and The Institute for Genomic Research (TIGR) databases by gapped Blast analysis.
Verification of insertion sites.
When a match was found for a flanking region in the databases, additional primers were designed to verify the site of insertion on the right side and in some cases on the left side. Most PCR products were in the range of 300 to 600 bp and included approximately 70 to 100 bp of IS6110, depending upon primer design. Each PCR product was sequenced and analyzed for homology with the NCBI and TIGR databases by gapped Blast analysis.
Southern blot hybridization.
Right-side IS6110 DNA fingerprinting analysis was performed as previously described (24). To identify which IS6110 band matched each flanking sequence, the right-side PCR product for each flank was used to probe PvuII-restricted DNA from strains 210 and W. The size of the fragments hybridizing with the right-side PCR product of each flank was compared with the size of the IS6110-hybridizing fragment.
RT-PCR.
RNA was extracted from M. tuberculosis strains grown in vitro, from approximately 5 × 107 cells (9). RT mixtures consisted of total RNA, 2 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris [pH 8.3], 0.1% gelatin), 10 pmol of random hexamer oligonucleotides (Amersham Pharmacia Biotech, Piscataway, N.J.), 1 μl of 40 mM deoxynucleoside triphosphates (10 mM [each] dATP, dCTP, dGTP, and dTTP), 0.5 μg of RNase-free bovine serum albumin, 1 μl of Prime RNase inhibitor, and 2.5 U of avian myeloblastosis virus reverse transcriptase (Boehringer-Mannheim, Roche Molecular Biochemicals, Indianapolis, Ind.) in a 20-μl total volume. RT was performed at 42°C for 1 h followed by 95°C for 5 min. RT-PCR controls included the above reaction without avian myeloblastosis virus reverse transcriptase to monitor for the presence of contaminating DNA.
DNA amplification was performed on a GeneAmp PCR system 2400 (Perkin-Elmer, Norwalk, Conn.) in a 50-μl reaction mixture containing 10 pmol of each primer, 5 μl of RT reaction mix or 1 ng of genomic DNA, 200 μM deoxyribonucleoside triphosphates, 1× PCR buffer (Perkin-Elmer, pH 8.3, 1.5 mM MgCl2), and 1 U of Taq polymerase (Perkin-Elmer). The primers for the ctpD gene were designed to amplify a 177-bp fragment within the coding region of the gene. Primer sequences used were as follows: ctpD upper, 5′-GTGGCGCTGCTGCTGTTTCT-3′, and ctpD lower, 5′-CGCGGCAATCATCAGCAGAT-3′. The cycling conditions were initial denaturation at 95°C for 5 min and 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 60°C, and 2 min of extension at 72°C, followed by a single 5-min extension at 72°C. The PCR products were electrophoresed on a 2% agarose gel, with a 1-kb DNA ladder for size determination (Gibco Life Technologies), stained with ethidium bromide, and photographed using the Eagle Eye II gel documentation system (Stratagene, La Jolla, Calif.).
Nucleotide sequence accession numbers.
All sequence data described were submitted to the NCBI GenBank database. The following accession numbers (followed by fragment number/strain) were assigned: AF257113 (1/210), AF227480 (2/210), AF227481 (3/210), AF227482 (4/210), AF227483 (5/210), AF227484 (6/210), AF227485 (7/210), AF227486 (8/210), AF227487 (9/210), AF227488 (10/210), AF227489 (11/210), AF227479 (12/210), AF227490 (13/210), AF230202 (14/210), AF227491 (15/210), AF227492 (16/210), AF227493 (17/210), AF227494 (18/210), AF227495 (19/210), AF227496 (20/210), AF228670 (21/210), AF270485 (22/210), AF227497 (DR region, left side/210), AF257114 (1/W), AF228678 (2/W), AF228671 (3/W), AF228672 (4/W), AF228673 (5/W), AF230204 (6/W), AF283212 (8/W), AF228674 (9/W), AF230205 (10/W), AF230206 (11/W), AF230210 (12/W), AF230203 (14/W), AF230207 (15/W), AF228675 (16/W), AF230208 (17/W), AF228676 (19/W), AF228677 (20/W), AF230209 (21/W), AF270486 (22/W), and AF2274978 (DR region, left side/W).
RESULTS
IS6110 sites of insertion in 210 and W strains.
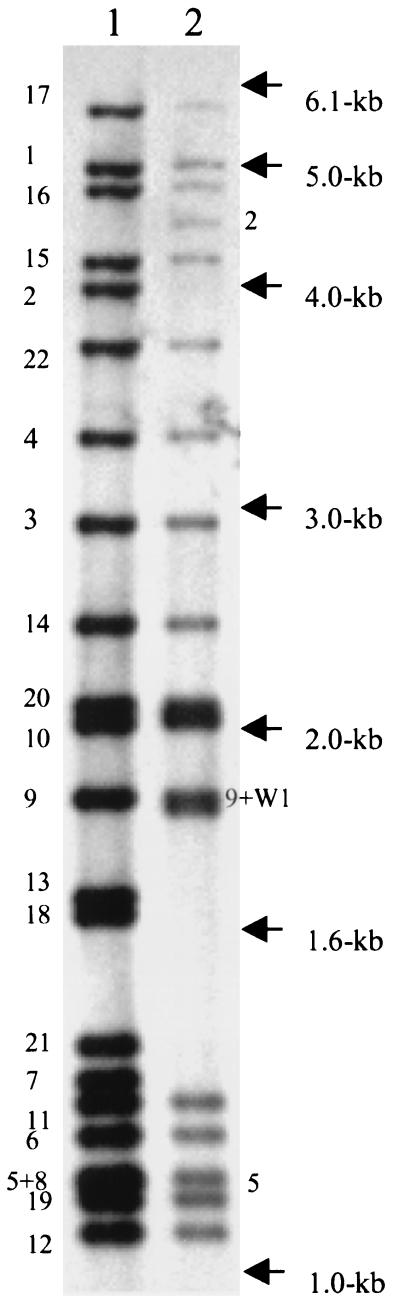
Table 1 lists the sites of insertion of IS6110 in the 210 and W strains, mapped to the genome of M. tuberculosis H37Rv (7). Figure 1 shows the IS6110 fingerprint patterns for strains 210 and W and correlates the band number with the sequence shown in Table 1. There are actually 22 copies of IS6110 in strain 210 and 19 copies in strain W.
TABLE 1.
Insertion sites of IS6110 in 210 and W strains of M. tuberculosisa
| No. | Wb | Rv gene no.c | Cosmid | Gene name(s)d |
|---|---|---|---|---|
| 21-4 | + | Rv0001:Rv0002 | MTV029 | dnaA/dnaN |
| 21-1 | + | Rv1135c | MTCI65 | PPE |
| 21-5 | + | Rv1371 | MTCY2B12 | Unknown |
| 21-20 | + | Rv1469 | MTV007 | ctpD |
| 21-16 | + | Rv1754c | MTCY28 | ATP-GTP binding motif |
| 21-7 | Rv1798:Rv1799 | MTV049 | ATP-GTP binding motif/lppT | |
| 21-18 | Rv1917c | MTCY180 | PPE | |
| 21-2 | + | Rv1917c | MTCY180 | PPE |
| 21-6 | + | Rv2016 | MTV018 | Unknown |
| 21-21 | Rv2104c:Rv2107 | MTCY261 | Unknown/PE | |
| 21-8 | Rv2107:Rv2108 | MTCY261 | PE/PPE | |
| 21-22 | + | Rv2352c | MTCY98 | PPE |
| 21-12 | + | Rv2813:Rv2820c | MTCY16B7 | DR region/unknown |
| 21-14 | + | Rv3018c:Rv3019c | MTV012 | PPE/ESAT6-like gene |
| 21-11 | + | Rv3019c:Rv3020c | MTV012 | ESAT6-like gene/PE |
| 21-19 | + | Rv3128c | MTCY164 | Unknown |
| 21-13 | Rv3178:Rv3179 | MTV014 | Unknown/ATP-GTP binding motif | |
| 21-9 | + | Rv3179:Rv3180c | MTV014 | ATP-GTP binding motif/probable coiled-coil domain |
| 21-10 | + | Rv3326:Rv3327 | MTV016 | ipl5 site |
| 21-3 | + | Rv3383c | MTV004 | idsB |
| 21-15 | + | Rv3427c:Rv3428c | MTCY78 | IS21-like element/IS1532 |
| 21-17 | + | Not identified | Unknown | |
| W-1 | Rv3128c:Rv3129 | MTCY164 | Unknown/unknown | |
| W-2 | Rv2104c:2107 | MTCY261 | Unknown/PE |
Sites are in order of appearance on the M. tuberculosis H37Rv genome map (6).
The plus sign indicates that this site of insertion of IS6110 is identical in the W strain.
One gene is listed when the site of insertion is in that gene. Two genes are listed to indicate the ORFs on either side of the insertion site of IS6110.
Gene names come from the M. tuberculosis H37Rv genome map (6).
FIG. 1.
IS6110 fingerprint patterns of strains 210 (lane 1) and W (lane 2). Size markers are indicated on the right. Numbers 1 to 21 correspond to the flanking sequences listed in Table 1. These were numbered in the order in which they were isolated.
(i) Shared sites of insertion.
The W and 210 strains have 17 identical sites of IS6110 insertion (Table 1 and Fig. 1). The flanking regions on each side of IS6110 were obtained for 12 of the 22 sites, and in each case a 3- or 4-bp DR was present at the junction of IS6110 and the flanking sequence. The band representing site 21-2 in strain 210 is smaller than the corresponding band in the W strain (Fig. 1). The sites of insertion are identical in these two fragments, and so the size difference may be due to an alteration in the flanking PvuII site. The sequence flanking the right side of IS6110 (in both strains) does not completely match the H37Rv sequence and appears to have undergone rearrangement.
The 210 and W strains have an insertion of IS6110 in the area of the chromosome containing oriC, as opposed to H37Rv, which does not (21–24). Both strains contain a copy of IS6110 between the dnaA and dnaN genes, with an insertion in the previously described A1 site (15).
Strains 210 and W have a sequence that is also present in the clinical isolate CSU#93 and three other clinical isolates (TIGR, personal communication; Z. Fang, direct submission to NCBI GenBank, accession no. MTY16254, MTY17220, and MBY17219). This sequence is not found in strain H37Rv. The sequence has been mapped as a hypothetical gene in CSU#93 and appears to reside near genes present in the H37Rv cosmid MTV016. In strains 210 and W, a copy of IS6110 is inserted in the middle of this open reading frame (ORF) (site 21-17). Therefore, these strains share genes that are not present in the H37Rv genome.
(ii) The DR region.
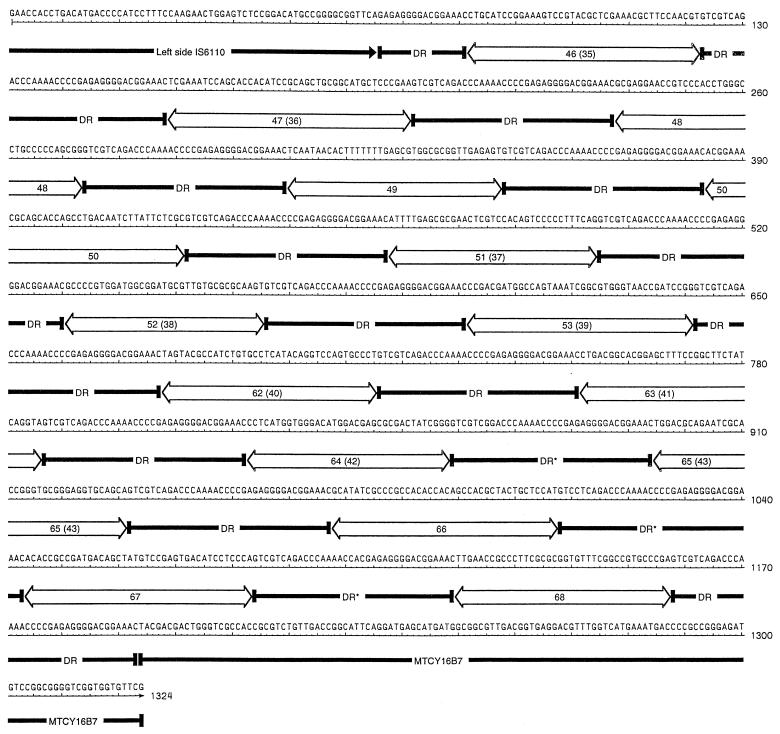
As in most M. tuberculosis strains, the 210 and W strains contain a copy of IS6110 in the highly conserved 36-bp DR region (14). The DR region flanking the left side of IS6110 has 15 spacer sequences interrupting 16 DRs (Fig. 2). In contrast to H37Rv, Mycobacterium bovis BCG, and M. bovis 401, this copy of IS6110 has been inserted in a different site within a copy of the DR (2, 7, 12, 17). The DR sequence interrupted by IS6110 in the above strains is flanked by three C's. These nucleotides are absent in the DR sequence directly adjacent to the left side of IS6110 in the 210 and W strains. The first five spacers between the DRs on the left side of IS6110 are not present in H37Rv. Four are identical to those described for M. bovis strain 401, and one is a unique spacer recently reported (2, 7, 12, 17, 25). The sequence from spacer 51 to the region of MTCY16B7 outside the last DR is identical to that in H37Rv. This includes three DRs that are not identical to the consensus 36-bp sequence (Fig. 2).
FIG. 2.
Map of the DR region of strains 210 and W on the left side of IS6110. The sequence of IS6110 is underlined with a black arrow. Each 36-bp DR is underlined with a black bar. The three DR sequences that do not have the consensus sequence are marked with an asterisk. The spacer sequences are underlined with an open bar. The spacers are labeled with the new spoligotyping designation system with the old number in parentheses.
The DR region on the right side of IS6110 has been deleted in both the 210 and W strains. This is a deletion of 4,355 bp and includes the genes Rv2816c-Rv2819c and part of Rv2820c. This same deletion was observed by van Embden et al. for two other Beijing strains (25). This suggests that a genomic deletion or rearrangement has occurred due to the presence of IS6110, the 36-bp DRs, or both. Genomic deletions and rearrangements have been found in different members of the M. tuberculosis complex and have been correlated with attenuation and loss of virulence (11, 16). Spoligotyping of strains 210 and W demonstrates the common Beijing strain pattern, with the presence of nine characteristic spacer sequences. These nine sequences represent the spoligotyping spacers 35 to 43 (25).
(iii) Differences in sites of insertion.
The W-1 insertion site resides in the MTCY164 cosmid and is 526 bases downstream from the 21-19 insertion that is present in both strains. This IS6110 DR in strain W is the basis for the W-strain-specific multiplex PCR assay (20). As expected, strain 210 and other clinical isolates with the 21-band fingerprint pattern give a negative result in this assay (data not shown).
The insertion site W-2 is near the 21-21 site in strain 210 and a site of insertion in H37Rv. This places IS6110 93 bp upstream from Rv2107, which is a member of the PE gene family. The members of this family of genes have a highly conserved N-terminal domain with the motif Pro-Glu and a variable C-terminal domain and may be involved in antigenic variation of the organism (7). It is possible that the insertion of IS6110 in front of this PE gene is affecting its transcription.
The 210 strains carry an additional five insertions of IS6110 which have been identified as 21-7, 21-8, 21-13, 21-18, and 21-21 (Fig. 1 and Table 1). In 21-7, IS6110 is inserted at base 26346 in the cosmid MTV049 between two ORFs. The upstream ORF is Rv1798, which contains the ATP-GTP binding motif and may produce a protein that is part of the CBXX-CFQX family. The lppT gene lies 261 bases downstream from IS6110 and is characterized as a probable lipoprotein. The lppT protein sequence indicates that the protein contains the prokaryotic membrane lipoprotein lipid attachment site, and a possible signal peptide may be present.
In 21-13, IS6110 is inserted at base 25119 between the ORFs designated Rv3178 and Rv3179 in the cosmid MTV014. This places IS6110 259 bases upstream from Rv3179, which has the PS00017 ATP-GTP-binding site motif A (P loop). This copy of IS6110 resides 1,857 bp downstream from the 21-9 site of insertion.
21-18 is an insertion into Rv1917c, a very large gene and a member of the PPE protein family. In strain 210, there are two copies of IS6110 in this gene, separated by 2,272 bp (the other insertion is represented by 21-2). The W strain has only one insertion in Rv1917c. Unlike the 21-2 site, the sequence flanking 21-18 is identical to that of H37Rv for at least 300 bp.
The site of insertion of 21-21 is in cosmid MTCY261 at base 1698 between Rv2104 and Rv2107. H37Rv and the W strain also have a copy of IS6110 in this site but at different locations. This places two copies of IS6110 123 bp apart in the same orientation in the 210 strain (sites 21-8 and 21-21). This insertion of IS6110 places it 467 bp in front of Rv2107, which is a member of the PE family of genes as described above (7).
The 21-8 site of insertion is very near the 21-21 and W-2 sites described above, with IS6110 residing on the opposite side of the PE gene Rv2107.
The occurrence of three pairs of IS6110 insertions in close proximity in strain 210 suggests that it would be possible to distinguish 210 from other strains as described previously for strain W (20). We are developing a multiplex PCR assay for the rapid detection of strain 210. Such an assay would be beneficial in settings where the strain is prevalent, as it would focus contact investigations and subsequent preventative therapy.
Promotion of gene expression from IS6110.
The integration of an insertion element can result in the up or down regulation of genes by which it resides. In some cases, activation of an adjacent gene is due to the contribution of a sequence with promoter activity by the insertion element. For example, the insertion element IS1186 has been shown previously to increase the activity of the normally silent cfiA gene (carapenin resistance) in Bacteroides fragilis (21). Transcription of the gene occurs when the insertion sequence element is inserted upstream from the coding region and provides the promoter sequence (21).
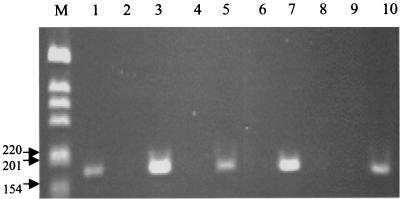
To determine if IS6110 can function in this manner, RT-PCR was performed to assay for the expression of the ctpD gene in three strains of 210 and H37Rv. In the 210 strains, IS6110 has been inserted 55 bp into the ctpD gene (Rv1469). H37Rv does not contain a copy of IS6110 in this region. The ctpD gene is believed to be a cation-transporting ATPase and contains the PS00154 E1-E2 ATPase phosphorylation site (7). Amplification of cDNA with primers ctpD upper and ctpD lower resulted in positive signals for all strains tested (Fig. 3). The negative RT-PCRs gave no signal, indicating that the samples were not contaminated with DNA. The RT-PCR products were sequenced, and the Blast results verified that the product was produced from the ctpD gene. Therefore, IS6110 appears to be providing a promoter sequence for transcription of the ctpD gene.
FIG. 3.
RT-PCRs of the ctpD gene for strains 210 and H37Rv and two 21-band clinical isolates. Lane M, 1-kb ladder; sizes listed are in base pairs. Lane 1, H37Rv positive RT; lane 2, H37Rv negative RT; lane 3, 210 positive RT; lane 4, 210 negative RT; lane 5, H1110 positive RT; lane 6, H1110 negative RT; lane 7, TB545 positive RT; lane 8, TB545 negative RT; lane 9, negative PCR control; lane 10, positive PCR control, 10 pg of 210 DNA.
DISCUSSION
The 210, W, and W variants are important strains of M. tuberculosis. The data presented here and elsewhere indicate that the 210 strain should also be considered a W variant. Because of the large number of shared sites of insertion, the 210 variant probably arose due to transpositional events.
It is interesting that there are numerous insertions in or near PPE genes. The PPE gene Rv1917c appears to have been altered in both strains in the 21-2 region of insertion. It has been hypothesized previously that the PE and PPE genes may be involved in antigenic variation of the organism (7). The presence of IS6110 in these genes could mediate recombination events that would lead to alterations in these genes and a change in the phenotype of the organism. Lack of alteration of sequence adjacent to the second copy of IS6110 in this gene in the 210 strain may be due to its recent acquisition.
Since the 210 strain has been demonstrated to grow more rapidly than other clinical isolates, it would be interesting to examine genes with IS6110 insertions (29). For example, the W and 210 strains share four sites of insertion in unknown genes. Provided that these genes are expressed, the insertion of IS6110 would be expected to alter the phenotype of the organism. The loss of the DR region on the right side of IS6110 in strains 210 and W is probably a feature of all Beijing strains. This is supported by the characteristic spoligotyping pattern of nine spacers (25). van Embden has hypothesized that the DR region may be involved in the regulation of replication similar to the repeats with the same function in other replicons (8, 18, 25). Therefore, the precise alteration in this region of the genome in the 210 strains may provide insight concerning their enhanced replication.
Recently, it was reported that the location of the site of insertion of IS6110 in Mycobacterium smegmatis influences the ability of IS6110 to be transposed (27). Evidence was provided that transposition was due to promotion of IS6110 gene expression by sequences flanking the insertion element. This information combined with the knowledge that IS6110 can mediate large-scale rearrangements and deletions in the chromosome reinforces the need to understand how each copy of this element affects the phenotype of an organism (5). The hypothesis is that an IS6110 insertion site(s) could possibly be responsible for a strain's increased capacity for transmission and/or replication. Our finding suggests that IS6110 may play a role in regulating gene expression and provides additional evidence for the importance of establishing the location of each site of insertion in the chromosome and the effect of this insertion on the phenotype of the organism.
ACKNOWLEDGMENTS
We thank Laurie Lacer Defoor and Suzhannah Mayo for excellent technical assistance.
REFERENCES
- 1.Barnes P F, Yang Z, Preston-Martin S, Pogoda J M, Jones B E, Otaya M, Eisenach K D, Knowles L, Harvey S, Cave M D. Patterns of tuberculosis transmission in Central Los Angeles. JAMA. 1997;278:1159–1163. [PubMed] [Google Scholar]
- 2.Beggs M L, Cave M D, Marlowe C, Cloney L, Duck P, Eisenach K D. Characterization of the Mycobacterium tuberculosis complex direct repeat sequence for use in the cycling probe reaction. J Clin Microbiol. 1996;34:2985–2989. doi: 10.1128/jcm.34.12.2985-2989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs M L, Stevanova R, Eisenach K D. Speciation and differentiation of Mycobacterium avium complex isolates with a variety of molecular techniques. J Clin Microbiol. 2000;38:508–512. doi: 10.1128/jcm.38.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bifani P J, Mathema B, Liu Z, Moghazeh S, Shosin B, Tempalski B, Driscoll J, Frothingham R, Musser J M, Alcabes P, Kreiswirth B N. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 5.Brosch R, Philipp W J, Stavropoulos E, Colston M J, Cole S T, Gordon S V. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect Immun. 1999;67:5768–5774. doi: 10.1128/iai.67.11.5768-5774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaves F, Yang Z, el Hajj H, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave M D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.del Solar G, Giraldo R, Ruiz-Echevarria M J, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DesJardin L E, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–2439. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenach K D, Cave M D, Bates J H, Crawford J T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1989;161:977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole S T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 12.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 13.Haas W H, Butler W R, Woodley C L, Crawford J T. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1993;31:1293–1298. doi: 10.1128/jcm.31.5.1293-1298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermans P W, van Soolingen D, Bik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurepina N E, Srevatsan S, Plikaytis B B, Bifani P J, Connell N D, Donnelly R J, van Soolingen D, Musser J M, Kreiswirth B N. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 16.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendiola M V, Martin C, Otal I, Gicquel B. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res Microbiol. 1992;143:767–772. doi: 10.1016/0923-2508(92)90104-v. [DOI] [PubMed] [Google Scholar]
- 18.Moijica F J M, Ferrer G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the archea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 19.Moss A R, Alland D, Telzak E, Hewlett D, Jr, Sharp V, Chiliade P, LaBombardi V, Kabus D, Hanna B, Palumbo L, Brudney K, Weltman A, Stoeckle K, Chirgwin K, Simberkoff M, Moghazeh S, Eisner W, Lutfey M, Kreiswirth B. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuberc Lung Dis. 1997;1:115–121. [PubMed] [Google Scholar]
- 20.Plikaytis B, Marden J L, Crawford J T, Woodley C L, Butler W R, Shinnick T M. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:1542–1546. doi: 10.1128/jcm.32.6.1542-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podglajen I, Breuli J, Coliatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1994;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 22.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valway S E, Sanchez M P C, Shinnick T F, Orme I, Agerton T, Hoy D, Jones J S, Westmoreland H, Onorato I M. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- 24.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Embden J D A, van Gorkom T, Kremer K, Jansen R, van der Zeijst B A M, Schouls L M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Enkhasaikan D, van Embden J D. Predominance of a single clone of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wall S, Ghanekar K, McFadden J, Dale J W. Context-sensitive transposition of IS6110 in mycobacteria. Microbiology. 1999;145:3169–3176. doi: 10.1099/00221287-145-11-3169. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Gong J, Yang Z, Samten B, Cave M D, Barnes P F. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J Infect Dis. 1999;179:1213–1217. doi: 10.1086/314738. [DOI] [PubMed] [Google Scholar]