Abstract
Aim
The aim of this study was to isolate multi-drug-resistant p. aeruginosa from dental implant, and control the growth and biofilm of isolated p. aeruginosa by silver nanoparticles.
Materials and methods
Thirty specimens from patients with Peri-implantitis were taken for isolation of p. aeruginosa. Bacterial samples were obtained from the infected peri-implant pocket with sterile paper points (size 30–45 mm). Samples were cultured for isolation of Multi-drug resistance P. aeruginosa. Phenotypical identification was done by the VITEK 2 system. DNA was extracted from the isolates and 16S rDNA-based PCR assay was used to confirm the identification. Susceptibility of isolated p. aeruginosa to 16 antibiotics was evaluated using the VITEK 2 system. The growth inhibition of isolated bacteria by AgNPs was tested by disk-diffusion method. The microtiter plate assay was used to estimate the capacity of P. aeruginosa to from biofilms. Antibiofilm activity of AgNPs was determined by microtiter plate assay.
Results
Three P. aeruginosa were successfully isolated from 30 clinical specimens. P. aeruginoas isolates were resistance to most of used antibiotics. Silver nanoparticles exerted an inhibitory effect on all isolated bacteria. All tested concentration of AgNPS exhibited a greatest anti-biofilm activity against multi-drug resistance (MDR) p. aeruginosa.
Conclusion
Current findings highlight the role of AgNPS in growth inhibition of P. aeruginosa and reveal a potential application of AgNPS in eradication of p. aeruginosa biofilms.
Keywords: Pseudomonas aeruginosa, Multi-drug resistant, Peri-implantitis, Nanoparticles, Biofilms
Graphical abstract
1. Introduction
The public health problem of antimicrobial resistance (AMR) has promoted the examination of alternative therapies to control infections caused by multidrug-resistant (MDR) pathogenic bacteria.1 Losing of supporting bone due to the inflammation which affects the tissues around an osseointegrated implant leads to implant failure because of the inadequacy of the host tissue to establish or sustain osseointegration.2 There are many risk factors for peri-implant diseases such as poor personal hygiene, periodontitis, Diabetes and smoking. Pathogenic oral microbiota plays a major role in the increasing of the implant failure.3 Gingivitis and gingival inflammations due to bacterial plaque or bacterial biofilms is considered a main risk factor for periodontitis and implant-failure.4 Bacteria residing in biofilms are difficultly treated with antibacterial agents and not accessible to the immune system, and lead to chronic infections throughout the body.5 Bacteria which lodged in biofilms are involved in the failure of implant treatment. Isolates from oral infections may carry certain antibiotic resistant determinant that has the ability to be transferred to other pathogenic bacteria in biofilm communities.6,7 It is noteworthy that opportunistic microorganisms such as Enterobacteriaceae, Staphylococci and Enteric bacteria are found at infection sites, especially after a peri-implant lesion occurs.8 Several studies have indicated the role of opportunistic pathogens in causing orthopedic device-related infection such as mandibular osteomyelitis after implant surgery, which leads to implant failure.9 Some cases may harbor microorganisms, which not usually found among essential oral flora, such as Enterobacteriaceae, Pseudomonas aeruginosa, Staphylococcus aureus, Enterococci, and Candida albicans.10 P. aeruginosa an opportunistic pathogen which usually carry virulence factors related to adhesions and biofilm formation, which facilitate its colonization of different oral sites and enable it to cause severe infections such as respiratory infections, and septicemia.11 In recent years, P. aeruginosa causes severe nosocomial infections due to its tendency to resist a broad range of antibiotics, and also its capacity to acquire a high resistance to the most effective antibiotics.12
Most of studies focused on anaerobic gram-negative bacteria, so that there are a limited data about the prevalence of opportunistic pathogens in patients with peri-implantitis. In terms of biomedical applications, AgNPs have been widely among nanoparticles biomedical applications.13 Studies have reported that the use of AgNPs in dental applications in order to eliminate dental biofilms and decreasing the probability of serious disease such as: dental caries and periodontal disease.14 However, there is not enough information that has determined the antimicrobial and anti-biofilm capacity of AgNPs against clinical dental biofilms associated with dental implant diseases. The quest for new therapy, to treat the over-growing of pathogenic bacteria and antimicrobial-resistant, is urgent. This study was aimed to evaluate the ability of AgNPS to inhibit the growth of p. aeruginosa, and to destruct the biofilms which formed by P. aeruginosa isolated from patients presenting active peri-implant diseases.
2. Material and method
2.1. Patient selection
Thirty patients with peri-implantitis were selected to join this study from the Periodontal clinic at the faculty of Dentistry, Zagazig University, Egypt, after approval the approval of Ethics Committee of the University, and the study procedures were explained to all patients, and a written informed consent was obtained from them. This investigation was designed as a cross-sectional clinical study to assess the presence of multi-drug resistance P. aeruginosa in peri-impalntitis, and also to evaluate the effectiveness of AgNPs in eradication of p. aeruginosa biofilms in peri-implant.
To be included in the study, a minimum age of 25 years were required for all subjects. The peri-implant tissue should be inflamed (probing depth more than 4 mm with bleeding on probing) and there should be evidence of radiographic bone loss beyond bone remodeling. Individuals should be periodontally unhealthy (presence of periodontal pockets ≥4 mm and bleeding) and present full mouth plaque scores and bleeding scores more than 20%.
The exclusion criteria were systemic conditions reported during anamneses that could affect the progression of peri-implant diseases and bone metabolism, the long-term use of anti-inflammatory medications, antibiotic therapies in the previous 6 months, patients who required bone grafts before or alongside the implant surgery and a history of previous regenerative procedures in the area treated with implant therapy. On the radiographs, mandibular bone density was measured by the DIGORA® software (Soredex, Tuusula, Finland).
2.2. Sample collection and processing
Probing depth (PD) measurements of all implants and teeth were done. Measurements were carried out by using plastic Williams's periodontal probes (Hu-Friedy, Chicago, IL, USA). Specimens were taken from the implants of the participants and microbiological analysis was performed.
2.3. Sampling procedures
The bacterial specimens were gathered from the peri-implant pocket by using sterile absorbent paper point (size 30–45 mm) (Paper Points, Dia Dent, Korea), paper points were placed in each pocket for about 20 s. After that, paper points were removed from the affected area, contamination from the surrounding tissues and saliva was avoided, and then the collected samples were transferred into in 1.5-ml micro centrifuge tubes. Then, 200 μL of phosphate buffered were added to paper points and vortexed to separate bacteria from the paper points.
3. Microbiological analysis
3.1. Isolation and identification of MDR P. aeruginosa
Selective media like MacConkey‐agar and Cetrimide agar (Sigma-Aldrich, St Louis, MO) were used for isolation of P. aeruginosa. The isolates were identified by conventional criteria, including morphology of colonies, production of pigments, and positive oxidase test. VITEK method was used for confirmed identification, according to manufacturer's recommendations at the Biotechnology unit, Animal Health Research Institute, Dokki, Giza, Egypt.
3.2. PCR analysis of P.aeruginosa genomic DNAs
Extraction of DNA from samples was done by using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) according to the manufacturer's recommendations.
Briefly, 200 μl of the sample suspension were pipetted into the bottom of a 1.5 ml microcentrifuge tube, 20 μl of the protease and 200 μl of lysis buffer were added, and mixed by pulse vortexing for 15 s. The mixture was then incubated at 56 °C for 10 min. After incubation, 200 μl of ethanol (96%) were added to the sample, and mixed again by pulse vortexing for 15 s. After mixing, the samples were centrifuged to remove drops from the inside of the lid. 100 μl of elution buffer pro-vided in the kit was added. PCR assay that based on 16S rDNA as described before.15 using the forward primer 5′- GGGGGATCTTCGGACCTCA-3′ and reverse primer 5′- TCCTTAGAGTGCCCACCCG-3′ (Midland Certified Reagent Company, USA).
PCR was carried out in 25 μL reaction volumes, (12.5 μl of PCR Master Mix (Takara, Japan), 1 μl of forward primer (20 pmol), 1 μl of forward primer (20 pmol), 4.5 μl of water, and 6 μl of extracted DNA, and 4.5 μl of water was added). An Applied Biosystems 2720 thermal cycler (Biometra, Germany) was used to perform the reaction. After electrophoresis grade agarose was prepared, 0.5 μg/ml ethidium bromide was added and mixed thoroughly. Twenty μl of each PCR product samples were loaded to the gel. The fragment sizes were determined by using a Gelpilot 1000 bp Ladder (Qiagen GmbH, Germany). The results were analyzed using associated software.
3.3. Antimicrobial susceptibility test
Antimicrobial susceptibility of P. aeruginosa was estimated to 16 antibiotics; Norfloxacin, Ampicillin/Sulbactam, Chloramphenicol, Streptomycin, Cefobid, Nitrofurantoin, Clindamycin, Amoxicillin/Clavulanic acid, E−moxclav, Amikacin, Kanamycin, Ofloxacin, Oxytetracyclin, Rifampicin, Enrofloxacin and Trimethoprim, by using VITEK 2 system, according to Clinical Laboratory Standard Institute (CLSI) guidelines, 2012.16
3.4. Antibacterial effect of AgNPs
Antimicrobial activity of AgNPs was determined by using disc diffusion method.17 Mueller-Hinton agar (Sigma-Aldrich, St Louis, MO) were inoculated with pseudomonas sp. through dipped sterile swab into the inoculum and then streaked the swab all over the surface of the medium 2 times, rotated the plates through an angle of 60 °C after each application. In order to prepare disks, what man filter papers no.1 were used approximately 6 mm in diameter and placed in a petri dish and then sterilized in autoclave for 40 min. After this, disks impregnated with AgNPs were placed on the surface of the agar using a pair of sterile forceps. The plates were incubated in an incubator at 37 °C. After overnight incubation, the diameter of each inhibition zone (including the diameter of disk) was measured with a ruler and recorded in mm.
3.5. Biofilm formation capacity
Microtiter plate technique was used to evaluate the biofilms formed by P. aeruginosa, as previously described.18 Briefly, overnight cultures of pseudomonas grown at 37 °C in modified LB broth were adjusted to an optical density equal to 0.5 McFarland standards. Bacterial cultures were further diluted in LB broth (1:100) (Oxoid, UK) and aliquots of 200 μL of each culture were loaded into wells of microtiter plate (Costar, Corning Inc., Corning, NY, USA). Sterile uninoculated LB broth (Oxoid, UK) was used a control. In order to allow biofilm formation, the plates were incubated 37 °C for 24 h. After that, the wells were washed gently with sterile PBS to remove planktonic cells. After air drying, the 200 μL of methanol were added to each well in order to fix the formed biofilms. The wells again were washed twice with sterile PBS. After air drying, 200 μL of 0.1% crystal violet (Sigma-Aldrich) were added to each well for 20 min in order to quantify the biofilm biomass. After washing with PBS, 95% ethanol was added to each well and left for 20 min and the optical density of each well was determined using an ELISA plate reader at OD600. Experiments were performed in triplicate & the data were expressed as means ± SD.
3.6. Assessment of silver nanoparticles activity in biofilms
The established P. aeruginosa biofilms were treated with AgNPs, by using the microtiter plate assay as previously described.19 The diluted bacterial culture (1:100) was added to the wells of the plates and incubated for 24 h in order to allow the formation of biofilms. The planktonic cells were removed via washing twice with sterile PBS. After biofilms were established, diluted AgNPs (200 μg/ml, 100 μg/ml and 50 μg/ml) were added to each well, while normal saline solution was considered a positive control in other wells. The plates were further incubated for 24 h at 37 °C, and then washed again with PBS. After that, crystal violet (Sigma) (1% W⁄ V) was added to each well for 20 min, and then ethanol (95%) was added, 200μL/well. The absorbance was measured using an ELIZA plate reader at OD600 at Biotechnology Lab., Faculty of Pharmacy, Zagazig University, Zagazig, Egypt.
4. Results
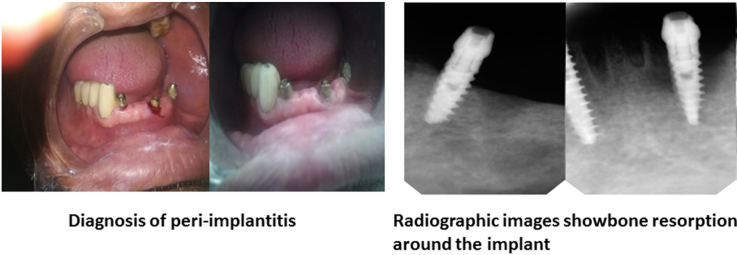
In the present study, only 3 P. aeruginosa isolates (10%) were obtained from 30 samples from patients with peri-implantitis (Fig. 1), with gingival inflammations around implant. Radiographic images showed bone resorption (data not shown).
Fig. 1.
Shows bleeding with probing which indicates inflammation. Also, reveals the presence of plaques around the implant.
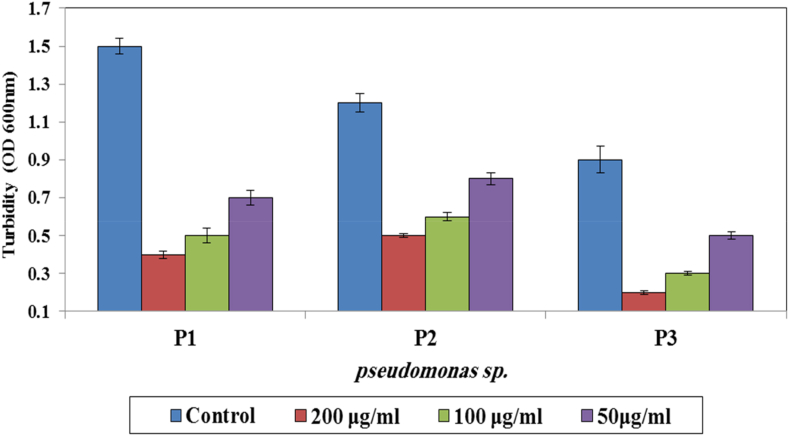
Isolates were identified via pigment production, positive oxidase test. The identification was confirmed by VITEK 2 system (Table 1). The isolated P. aeruginosa were further confirmed by 16s rDNA‐based PCR assay (data not shown). The three isolates showed band representing to special band of P. aeruginosa (956 bp). P. aeruginosa (accession number: LC514698) was used as positive control. The susceptibility or resistance pattern of isolates to different classes of antibiotics was determined by using VITEK 2 system. As shown in Table 2, P. aeruginosa was susceptible only to Nitrofloxacin, Streptomycin and Enrofloxacin but resistant to Amoxicillin/Clavulanic acid, E−moxclav, Kanamycin, Ofloxacin, Oxytetracyclin, Ampicillin/Sulbactam, Chloramphenicol, Cefobid, Streptomycin, Amikacin, Rifampicin, Clindamycin and Trimethoprim/sulfamethoxazole . The results show the highly resistance of bacteria to conventional antibiotics. Estimation of the biological potential of AgNPs against pseudomonas strains was carried out by disk diffusion method. The results revealed that the AgNPs have noticeable bactericidal activity and inhibit bacterial growth. AgNPs showed an inhibition zone of 25 ± 5.9 mm for isolates no.1, 17.5 ± 4.3 mm for isolates no.2, and 30 ± 6.5 mm for isolates no.3. Biofilm formation was investigated in vitro by crystal violet staining technique. The optical density value was measured to observe the ability of isolates to form biofilms. The results showed the formation of biofilm in three strains of P. aeruginosa; strain no. 1 recorded the highest biofilm (O.D = 1.5), followed by strain no.2 (O.D = 1.2) and strain no.3 (O.D = 0.9). The ability of AgNPs against isolated bacteria biofilms was evaluated by using the microtiter plate assay. Results in (Fig. 2) show the effectiveness of AgNPS in reduction of bacterial biofilms. AgNPs showed increasingly reduction of P. aeruginosa biofilms. Treatment of bacterial biofilms with AgNPs revealed the efficacy of AgNPS in eradication of their biofilm, and the results were shown by estimation of OD measurements.
Table 1.
Identification of isolated bacteria by VITEK 2 system (GN card).
| Biochemical Tests | Isolated bacteria |
|---|---|
| P. aeruginosa | |
| APPA | – |
| H2S | – |
| BGLU | – |
| ProA | + |
| SAC | – |
| ILATK | + |
| GLYA | – |
| O129R | + |
| ADO | – |
| BNAG | – |
| dMAL | – |
| LIP | – |
| dTAG | – |
| AGLU | – |
| ODC | – |
| GGAA | – |
| PyrA | – |
| AGLTP | – |
| dMAN | – |
| PLE | – |
| dTRE | – |
| SUCT | + |
| LDC | – |
| IMLTa | + |
| IARL | – |
| dGLU | + |
| dMNE | + |
| TyrA | + |
| CIT | + |
| NAGA | – |
| IHISa | – |
| ELLM | – |
| dCEL | – |
| GGT | + |
| BXYL | – |
| URE | – |
| MNT | + |
| AGAL | – |
| CMT | + |
| ILATa | + |
| BGAL | – |
| OFF | – |
| BAlap | + |
| dSOR | - |
| 5 KG | - |
| PHOS | - |
| BGUR | - |
| Probability (%) | 89% |
- = Negative, + = positive.
Abbreviations as follow: APPA: Ala-Phe-Pro Arylamidase, H2S: Hydrogen sulfide production, BGLU:BETA-Glucosidase,ProA:l-ProlineArylamidase,SAC: Saccharose-Sucrose, ILATK: l-Lactate Alkalization, GLYA: Glycine Arylamidase, O129R: O̸ 129 Resistance(Comp. Vibrio), ADO: Adonitol, BNAG: BETA-N-Acetyl-Glucosaminidase.
dMAL: d-Maltose, LIP: Lipase, dTAG: d-Tagatose, AGLU: ALPHA-Glucosidase,ODC: Ornithine Decarboxylase.
GGAA: Glu-Gly-Arg-Arylamidas, PyrA: L-Pyrrolydonyl-Arylamidase, AGLTP: Glutamyl Arylamidase pNA, dMAN: D-Mannitol.
PLE: Palatinose, dTRE: d-Trehalose,SUCT: SUCCINATE alkalinization, LDC: Lysine Decarboxylase. IMLTa: l-MALATE assimilation, IARL: L-Arabitol, dGLU: d-Glucose, dMNE: d-MANNOSE, TyrA:Tyrosine Arylamidase.
CIT:Citrate (sodium),NAGA: Beta-N-Acetyl-Galactosaminidase, IHISa: L-Histidineassimilation, ELLM:Ellman, dCEL: d-cellobiose, GGT: Gamma-Glutamyl-Transferase, BXYL: Beta-Xylosidase, URE: Urease, MNT: Malonate, AGAL: ALPHA-GALACTOSIDASE, CMT:Courmarate, ILATa: l-LACTATE assimilation, BGAL: Beta- Galactosidase, OFF: Fermentation/Glucose, BAlap: BETA-Alanine arylamidase, dSOR: D-sorbitol, 5 KG: 5-Keto-d-Gluconate, PHOS: Phosphatase, BGUR: B-Glucoronidase.
Table 2.
Susceptibility of isolated P. aeruginosa to antibiotics.
| Antimicrobial | MIC | Interpretation | Antimicrobial | MIC | Interpretation |
|---|---|---|---|---|---|
| Norfloxacin | ≥32 | S | E-moxclav | ≥32 | R |
| Ampicillin/Sulbactam | ≤64 | R | Amikacin | ≥32 | R |
| Chloramphenicol | ≥32 | R | Kanamycin | ≤1 | R |
| Streptomycin | ≤1 | S | Ofloxacin | ≤1 | R |
| Cefobid | ≤1 | R | Oxytetracyclin | 128 | R |
| Nitrofurantoin | 8 | R | Rifampicin | ≥32 | R |
| Clindamycin | 0.5 | R | Enrofloxacin | ≥32 | S |
| Amoxicillin/Clavulanic acid | 2 | R | Trimethoprim | 0.5 | R |
(R): Resistant (S): sensitive.
Fig. 2.
AgNPs mediated reduction of P. aeruginosa biofilms. Bacterial biofilms were treated either with AgNPs, and then the crystal violet stainable material was solubilized with ethanol, and the absorbance measured at OD 600 nm (P < 0.02).
The highest reduction of P. aeruginosa biofilm was observed when they treated with 200 μg/ml of AgNPs, followed by 100 μg/ml and 50 μg/ml in comparison with the control. This study represents that all AgNPs concentrations were effective in prevention of P. aeruginosa growth, and in destruction of their biofilms.
5. Discussion
In this study, we evaluated the antibacterial and antibiofilm activity of AgNPs against P. aeruginosa which isolated from dental implant. Previous studies revealed the presence of opportunistic pathogens such as P. aeruginosa, S. aureus and yeast, and showed the high affinity of these pathogens for titanium surfaces.9 According to previous studies, patients have a greater pocket depth, bone loss, more bacterial load, and a high prevalence of the disease.20 In this study, we isolated 3 p. aeruginosa isolates out of 30 samples recovered from dental-implant infections from patient with different ages at the clinics of faculty of Dentistry, Zagazig University, Zagazig, Egypt. P. aeruginosa characterized by its ability to grow in cetrimide agar, and produce oxidase enzyme. Identification was confirmed using VITEK 2 system. Previous studies indicated that, after implant placement, the periodontal pathogens are early transmitted from periodontal to implant sites.21 Present study revealed the presence of p. aeruginosa in dental implantitis, and this finding have been agreed with other studies, which showed a high prevalence of Pseudomonas sp, enteric rods such as Enterobacter sp. or Klebsiella sp.22
The resistance of bacteria to antibiotics represents a major challenge. In our study; resistance of isolated bacteria to 16 antibiotics was estimated. All isolates were resistant to 13 (81%) out of 16 antibiotics. Biofilm formation by these bacteria is difficult to eradicate due to the high level of antimicrobial resistance. It plays a major role in the pathogenesis of infections due to the acquisition of resistance genes. In this study, the isolated bacteria showed exhibited a higher capacity for biofilm formation. This result agreed with results obtained by Elhabibi and Ramzy,23 in which all isolates were recorded as a highly biofilm production. Novel therapeutic strategies are being studied as an alternative treatment to antibiotics to overcome bacterial biofilms in oral cavity in order to treat patients, and avoid the emergence of resistant bacterial populations in oral infections. Current study showed that the AgNPs can increasingly inhibit the growth of p. aeruginosa involved in dental plaque biofilms. Several studies have reported that AgNPs have a great antimicrobial activity against several pathogenic bacteria, including oral bacteria.13 This finding suggests the potential application of AgNPs to prevent the pathogenesis of infections caused by MDR. Studies have revealed that biofilm formation on the surface of the dentine and implants can significantly inhibited by the action of AgNPs.24 In present study, formation of biofilms by p. aeruginosa isolated from implant infections was challenged by AgNPs. The results showed that AgNPs have diminished the biofilm formation by all 3 isolates. There are a variety of studies which reported the efficacy of AgNPs against bacterial biofilms. Our results similar to other studies carried out by El-Shennawy et al.,24 in which AgNPs at different concentrations exhibited a highly anti-biofilm activity against MDR P. aeruginosa. Similarly, Singh et al.25 reported that 100 μg/ml of AgNPs was sufficient to eradicate 90% of bacterial biofilms. Our results suggest that AgNPs have a high potential for applications and might be efficacious to prevent infections of dental implant by MDR. In conclusion, the needy for an additional tool for the clinical management of bacterial infections, is urgent. Nanoparticles offer a promising alternative therapy to antibiotics, and the research on it has been promoted since the emergence of AMR.
6. Conclusion and future prospective
The AgNPs used in this study demonstrated to have a significant antimicrobial and anti-biofilm properties to inhibit the P. aeruginosa growth which isolated from subjects with active peri-implantitis disease. Based on our understanding, there is a few microbiological studies that exist on the isolation of these rare bacteria as the causative agent of peri-mplantitis. Although it is still needed to more scientific works in order to investigate microbiological and molecular characterizations of the clinical biofilm samples isolated from patients according to the presence and distribution of the different species included in the clinical samples. Furthermore, more scientific studies wanted to understand the variations and the antimicrobial behavior of AgNPs, this study could suggest the use of AgNPs as a potential antimicrobial agent in the biomedical fields for the prevention and control of peri-implantitis disease.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, ornot-for-profit sectors.
Declaration of competing interest
No potential conflict of interest relevant to this article was reported.
Acknowledgment
We thank Ahmed Tantawy, a Scientific Researcher at American Association for Cancer Research, for assistance with the preparation of the research.
Contributor Information
Mohamed El-Telbany, Email: meltelbany@zu.edu.eg, mohamedsamir9349@yahoo.com.
Ahmed El-Sharaki, Email: dentistshraki@yahoo.com.
References
- 1.Aslam B., Wang W., Arshad M.I., et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang N.P., Berglundh T. Working group 4 of the seventh European workshop on periodontology. Periimplant diseases: where are we now?–consensus of the seventh European workshop on periodontology. J Clin Periodontol. 2011 Mar;38:178–181. doi: 10.1111/j.1600-051X.2010.01674.x. [DOI] [PubMed] [Google Scholar]
- 3.Renvert S., Persson G.R., Pirih F.Q., Camargo P.M. Peri‐implant health, peri‐implant mucositis, and peri‐implantitis: case definitions and diagnostic considerations. J Clin Periodontol. 2018 Jun;45:S278–S285. doi: 10.1111/jcpe.12956. [DOI] [PubMed] [Google Scholar]
- 4.Murakami S., Mealey B.L., Mariotti A., Chapple I.L. Dental plaque–induced gingival conditions. J Clin Periodontol. 2018 Jun;45:S17–S27. doi: 10.1111/jcpe.12937. [DOI] [PubMed] [Google Scholar]
- 5.Davey M.E., O'toole G.A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000 Dec;64(4):847. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlen G., Blomqvist S., Almståhl A., Carlén A. Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J Oral Microbiol. 2012 Jan 1;4(1):10855. doi: 10.3402/jom.v4i0.10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rams T.E., Feik D., Mortensen J.E., Degener J.E., van Winkelhoff A.J. Antibiotic susceptibility of periodontal Enterococcus faecalis. J Periodontol. 2013 Jul;84(7):1026–1033. doi: 10.1902/jop.2012.120050. [DOI] [PubMed] [Google Scholar]
- 8.Hultin M., Gustafsson A., Hallström H., Johansson L.Å., Ekfeldt A., Klinge B. Microbiological findings and host response in patients with peri‐implantitis. Clin Oral Implants Res. 2002 Aug;13(4):349–358. doi: 10.1034/j.1600-0501.2002.130402.x. [DOI] [PubMed] [Google Scholar]
- 9.Renvert S., Lindahl C., Renvert H., Persson G.R. Clinical and microbiological analysis of subjects treated with Brånemark or AstraTech implants: a 7‐year follow‐up study. Clin Oral Implants Res. 2008 Apr;19(4):342–347. doi: 10.1111/j.1600-0501.2007.01476.x. [DOI] [PubMed] [Google Scholar]
- 10.Truong V.K., Lapovok R., Estrin Y.S., et al. The influence of nano-scale surface roughness on bacterial adhesion to ultrafine-grained titanium. Biomaterials. 2010 May 1;31(13):3674–3683. doi: 10.1016/j.biomaterials.2010.01.071. [DOI] [PubMed] [Google Scholar]
- 11.Yang L., Jelsbak L., Marvig R.L., et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108:74816. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nseir S., Ader F., Lubret R., Marquette C.H. Pathophysiology of airway colonization in critically ill COPD patient. Curr Drug Targets. 2011;12 doi: 10.2174/138945011794751537. 514‐20. [DOI] [PubMed] [Google Scholar]
- 13.Şuhani M.F., Băciuţ G., Băciuţ M., Şuhani R., Bran S. Current perspectives regarding the application and incorporation of silver nanoparticles into dental biomaterials. Clujul Med. 2018 Jul;91(3):274. doi: 10.15386/cjmed-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa-Cristóbal L.F., López-Ruiz N., Cabada-Tarín D., et al. Antiadherence and antimicrobial properties of silver nanoparticles against Streptococcus mutans on brackets and wires used for orthodontic treatments. J Nanomater. 2018 Jan 1:2018. [Google Scholar]
- 15.Spilker T., Coenye T., Vandamme P., LiPuma J.J. PCR‐based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42 doi: 10.1128/JCM.42.5.2074-2079.2004. 2074‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI . vol. 32. Clinical and Laboratory Standards Institute; Wayne: 2012. (Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Eleventh Edition). CLSI Document M02-A11. (1) [Google Scholar]
- 17.Wayne P.A. 2010. Clinical and Laboratory Standards Institute: performance Standards for Antimicrobial Susceptibility Testing: 20th Informational Supplement. CLSI Document M100-S20. [Google Scholar]
- 18.Stepanović S., Vuković D., Hola V., et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 19.Palanisamy N.K., Ferina N., Amirulhusni A.N., et al. Antibiofilm properties of chemically synthesized silver nanoparticles found against Pseudomonas aeruginosa. J Nanobiotechnol. 2014;12:2. doi: 10.1186/1477-3155-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zambon J.J., Grossi S.G., Machtei E.E., Ho A.W., Dunford R.R.J.G., Genco R.J. Cigarette smoking increases the risk for subgingival infection with periodontal pathogens. J Periodontol. 1996;67:1050–1054. doi: 10.1902/jop.1996.67.10s.1050. [DOI] [PubMed] [Google Scholar]
- 21.De Boever A.L., De Boever J.A. Early colonization of non‐submerged dental implants in patients with a history of advanced aggressive periodontitis. Clin Oral Implants Res. 2006 Feb;17(1):8–17. doi: 10.1111/j.1600-0501.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 22.Å Leonhardt, Renvert S., Dahlén G. Microbial findings at failing implants. Clin Oral Implants Res. 1999 Oct;10(5):339–345. doi: 10.1034/j.1600-0501.1999.100501.x. [DOI] [PubMed] [Google Scholar]
- 23.Elhabibi T., Ramzy S. Biofilm production by multi drug resistant bacterial pathogens isolated from patients in intensive care units in Egyptian hospitals. J Microb Biochem Technol. 2017;9(4):151–158. [Google Scholar]
- 24.El-Shennawy G., Abd Ellatif R., Badran S., El-Sokkary R. Silver nanoparticles: a potential antibacterial and antibiofilm agent against biofilm forming multidrug resistant bacteria. Microb. Infect Dis. 2020 Aug 1;1(2):77–85. [Google Scholar]
- 25.Singh P., Pandit S., Beshay M., et al. Antibiofilm effects of gold and silver nanoparticles synthesized by the Rhodiola rosea rhizome extracts. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S886–S899. doi: 10.1080/21691401.2018.1518909. [DOI] [PubMed] [Google Scholar]