Abstract
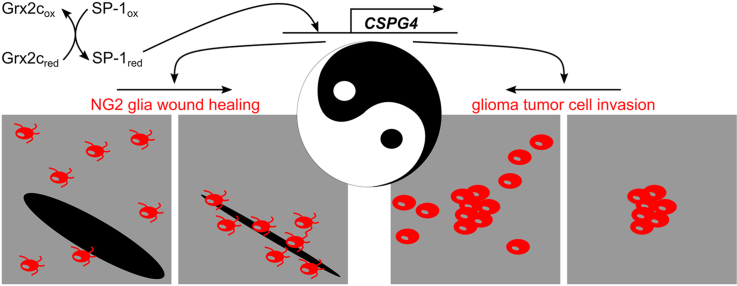
Redox regulation of specific cysteines via oxidoreductases of the thioredoxin family is increasingly being recognized as an important signaling pathway. Here, we demonstrate that the cytosolic isoform of the vertebrate-specific oxidoreductase Glutaredoxin 2 (Grx2c) regulates the redox state of the transcription factor SP-1 and thereby its binding affinity to both the promoter and an enhancer region of the CSPG4 gene encoding chondroitin sulfate proteoglycan nerve/glial antigen 2 (NG2). This leads to an increased number of NG2 glia during in vitro oligodendroglial differentiation and promotes migration of these wound healing cells. On the other hand, we found that the same mechanism also leads to increased invasion of glioma tumor cells. Using in vitro (human cell lines), ex vivo (mouse primary cells), and in vivo models (zebrafish), as well as glioblastoma patient tissue samples we provide experimental data highlighting the Yin and Yang of redox signaling in the central nervous system and the enzymatic Taoism of Grx2c.
Keywords: Redox signaling, Transcription, Oligodendrocytes, Glioblastoma, Cancer invasion, Zebrafish
Abbreviations: CNPase, 2′,3′-cyclic nucleotide 3′-phosphodiesterase; CSPG4, chondroitin sulfate proteoglycan 4; Grx, Glutaredoxin; IDH, isocitrate dehydrogenase; MBP, myelin basic protein; NG2, nerve/glial antigen 2; OPCs, oligodendrocyte progenitor cells; PLP, proteolipid protein; Trx, Thioredoxin
Graphical abstract
Highlights
-
•
CSPG4 promoter binding of the transcription factor SP-1 depends on glutaredoxin 2
-
•
Cytosolic glutaredoxin 2 promotes oligodendrocyte differentiation into NG2 glia
-
•
Migration and wound healing capacity of NG2 glia is increased by glutaredoxin 2
-
•
Glutaredoxin 2 increases invasion of human glioblastoma cells in vitro and in vivo
1. Introduction
1.1. Redox signaling
In all kingdoms of life, the importance of redox signaling has been reported; from sulfate reduction in bacteria [1] to brain development in vertebrates [2]. Redox signaling emerged as an essential signaling event of cellular processes via regulation of posttranslational oxidative thiol modifications. These modifications consist of disulfide or sulfenic acid formation, glutathionylation, nitrosylation, and persulfidation. Oxidoreductases of the thioredoxin family are key enzymes in redox signaling by reducing these modifications. Glutaredoxin 2 (Grx2) is such an oxidoreductase and present in three isoforms in mammalian cells, the mitochondrial isoform Grx2a and the cytosolic/nuclear isoforms Grx2b and c [3,4]. Of note, Grx2c was originally described as cancer-specific isoform [3]. Meanwhile, Grx2c is connected to cytoskeletal rearrangements leading to increased cellular migration [5] underlying processes such as heart development [6]. These Grx2-dependent rearrangements are also of high importance for differentiation processes such as axon and vessel outgrowth [2,7]. In general, the importance of redox regulation for cell fate decision of neural stem and progenitor cells including differentiation of oligodendrocyte progenitor cells (OPCs) (summarized in e.g. Refs. [[8], [9], [10]]), but also for de-differentiation processes leading to the formation of gliomas is established [11].
1.2. Differentiation of oligodendrocyte progenitor/precursor cells
Oligodendrocytes, the myelinating cells of the central nervous system, arise around birth from OPCs that are formed by neuroectodermal stem cells that also generate neurons and astrocytes [12]. Differentiation of oligodendrocytes follows a stepwise morphological transformation from bipolar progenitors to myelinating oligodendrocytes that is accompanied by the sequential expression of molecular markers characteristic for the differentiation state, e.g. A2B5 and nerve/glial antigen 2 (NG2) in progenitors, 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) in mature oligodendrocytes as well as myelin basic protein (MBP) and proteolipid protein (PLP) in further developed myelinating oligodendrocytes [13]. Interestingly, even after the myelination process is completed, the NG2-positive fraction of OPCs named NG2 glia form the largest population of progenitor cells in the brain with up to 10% of the total brain cell population [14,15]. First discovered in the beginning of the 1980s [16,17], NG2 glia are known today to have important functions in the pathological context, e.g. wound closure [18] and remyelination [19] in the adult brain. However, NG2 glia have been suggested as possible cells of orgin of distinct types of gliomas, including glioblastomas and oligodendrogliomas [20,21].
1.3. De-differentiation of glia cells
Glioblastoma is a highly malignant astrocytic glioma without mutation of the isocitrate dehydrogenase (IDH) 1 or 2 genes and belongs to the overall most lethal cancer types with a median survival of less than two years after diagnosis [22]. In contrast, oligodendroglioma carries mutations in IDH1 or IDH2 combined with a co-deletion of chromosomal arms 1p and 19q and is clinically characterized by a less malignant phenotype and higher sensitivity to radio- and chemotherapy [23]. However, both glioma types share highly invasive growth properties in the surrounding brain parenchyma, which presents one of the core challenge for their successful treatment. Molecular networks promoting glioma cell invasion, such as mesenchymal reprogramming of tumor cells, niche factors stimulating a pro-invasive cellular phenotype, or redox sensitive processes have been identified [[24], [25], [26]]. Interestingly, elevated expression of NG2, also known as chondroitin sulfate proteoglycan 4 (CSPG4), has been linked to increased invasion of glioma cells and to clinical outcome [[27], [28], [29]].
In this project, we identified Grx2c as a novel regulator of OPC motility, a function that was also observed in human glioma models, thus demonstrating the enzymatic Taoism of specific redox signaling in normal glial cell function and glioma pathogenesis. Enzymatic Taoism refers to the Chinese philosophy/religion represented by the Yin and Yang (negative-positive) symbol.
2. Materials and methods
Materials – Chemicals were purchased from Sigma-Aldrich unless otherwise stated and were at least of analytical grade. Primary antibodies were anti-A2B5 (Sigma-Aldrich, USA), -actin (Abcam, UK), - acetylated histone 3 (Abcam, UK), -α-tubulin (Acris, Germany), -BrdU (Sigma-Aldrich, USA), -CNPase (Sigma-Aldrich, USA), -zebrafish, mouse and human Grx2 (Abcam, UK or selfmade [30]), -H4 (Millipore, USA), -MBP (Merck Millipore, USA), -NG2 (Merck Millipore, USA), -SP-1 (Santa Cruz, USA). Secondary antibodies used after western transfer or for immunocytochemistry were from LI-COR (USA) or from Millipore (USA), respectively.
Protein expression and purification – Grx2, Grx2C37S, and Grx2C40S were expressed as His-tagged proteins and purified via IMAC as described before [31].
Cell culture – A2B5+ and NG2+ primary cells were isolated from P8-9 mouse brains using the Neural Tissue Dissociation kit (Miltenyi Biotec, Germany) and magnetic associated cell sorting via either anti-A2B5 or anti-NG2 microbeads (Miltenyi Biotec, Germany). All mouse experiments were conducted according to the guidelines and protocols approved by the local animal welfare committee “Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen” (LANUV) under the protocol number O74/08. Isolation of OPCs led to cell cultures with high purity (A2B5: 94 ± 1% [32], NG2: 91 ± 1%, Supplemental Fig. S1A). The isolated cells were cultivated on poly(l)-lysine coated surfaces either in proliferation (DMEM/F-12 medium with 1% HEPES, 1% GlutaMax, 1% Pen/Strep, 2% B27 supplement without Vitamin A, 0.1% FGF, 0.1% PDGFα) or differentiation medium (DMEM/F-12 medium with 1% HEPES, 1% GlutaMax, 1% Pen/Strep, 2% B27 supplement with Vitamin A, 1% T3, 1% T4). Hela cells (wildtype and Grx2 overexpressing cells [33]) were cultivated in DMEM medium with 1 g/L d-glucose, 10% FCS, 1% Pen/Strep, GBM-1 cells in DMEM/F-12 medium with 2% B27 supplement with Vitamin A, 0.1% EGF, 0.1% FGF, 0.1% Heparin, 1% Pen/Strep, and GBM18 as well as U343-MGA in MEM with 10% FCS, 1% Pen/Strep. All cells were kept at 37 °C in a humidified atmosphere containing 5% CO2. Grx2 knockdown in primary cells was performed using Accell siRNA (Horizon Discovery, USA). To knock out or overexpress Grx2 in GBM-18 and U343-MGA cells, the cells were chemically transfected using lipofectamin 2000 (Thermo Scientific, USA) with siRNA [34] and a Grx2c-encoding pExpress plasmid [33]. Grx2 was knocked out in HeLa cells via electroporation as described before [34].
Uptake of Grx2c – Cells were treated with 2 or 5 μM recombinant Grx2c for 24 and/or 2 h. To remove recombinant proteins from the cell surface an acidic wash was performed before western transfer. For acidic wash the medium was aspirated, cells were washed twice with PBS and for 30 s with 200 mM glycine, 150 mM NaCl, pH 3.0. Afterwards, cells were neutralized by adding cell culture medium. These steps were performed two times before lysates were prepared. Cytosolic and nuclear fractions were extracted using the NE-PER Nuclear and Cytosolic Extraction Reagent Kit (Thermo Scientific, USA).
Migration/invasion – 2D-migration was followed in a Boyden chamber assay using ThinCert™ cell culture inserts with 8 μm pores (Greiner, Switzerland). 50,000 cells were treated with or without 2 μM recombinant Grx2 or transfected either with control siRNA or Grx2siRNA before. After 24 h (NG2+-cells) or 48 h (GBM-18 and U343-MAG cells) inserts were removed and the cells were incubated for another 24 h to attach to the bottom of the wells before they were fixed with 4% Roti-Histofix (Carl Roth, Germany) for 15 min. After staining nuclei with Hoechst 33342 (Molecular Probes, USA), cells were counted. To follow 3D-migration, spheroids of glioblastoma cells were generated in 96 wells coated with 50 μl 1.5% agar noble solution. 5,000 cells in 50 μl MEM were seeded per well and spheroids were harvested after 48 h. For the collagen mix 420 μl 10x EMEM, 38 μl l-glutamine (200 nM), 462 μl FCS, 78 μl NaHCO3 (7.5%) and 3.5 ml collagen (2 mg/ml in PBS) were prepared. NaHCO3 was added until the color of the mixture appeared orange. 300 μl of the collagen mixture were added to each well of a 24 well plate. Spheroids were resuspended in 300 μl collagen mix and added on top of the first solid collagen layer and 500 μl MEM were added on top.
Scratch assay – 400,000 NG2+ cells were seeded on poly(l)-lysine coated glasses and treated with or without 2 μM recombinant Grx2 for 2 h. The scratch assay was performed according to Ref. [35]. To mimic a wound, a 200 μl pipet tip was used to scrape the cell monolayer in a straight line. Wound/scratch closure was followed for 48 h.
Cell survival and proliferation – Cell survival was measured in 96 well plates using the Cell Titer Blue assay (Promega, USA) and proliferation using BrdU. 30,000 cells on a glass plate were incubated with or without 2 μM recombinant Grx2c for 24 h before 10 μM BrdU was added for 2 h. After fixation, washing, and blocking (PBS, 0.1% Triton-X 100, 5% normal goat serum), primary antibodies detecting BrdU and histone 4 were applied over night. After applying secondary antibodies, pictures were taken with a BX51 fluorescent microscope (Olympus, Japan) and F-view CCD camera (Olympus, Japan).
Immunocytochemistry – Cells were grown on poly(l)-lysine coated glasses, washed with PBS and fixed for 15 min with 4% Roti-Histofix (Carl Roth, Germany). After washing (PBS, 0.1% Triton-X 100) samples were blocked (PBS, 0.1% Triton-X 100, 5% normal goat serum) and primary antibodies were added over night. After washing secondary antibodies were added, nuclei were stained with Hoechst and samples were covered with ImmuMount (Thermo Fisher, USA). Pictures of dried samples were taken with a BX51 fluorescent microscope (Olympus, Japan) and F-view CCD camera (Olympus, Japan).
Identification of interaction partners - The intermediate trapping was performed by immobilizing Grx2C40S on a HisTrap column (Qiagen, Germany). Following reduction with 10 mM DTT, 10–20 mg HeLa cell lysate were loaded on the column. Following extensive washing, trapped proteins were eluted with 10 mM GSH, precipitated over night at 4 °C with 20% TCA and were centrifuged for 15 min at 13,000 rpm and 4 °C. The pellet was washed with ice cold acetone, centrifuged again and resuspended in 50 μl urea containing lysis buffer. The presence of SP-1 was analyzed by SDS-Page and western transfer or by mass spectrometry. Mass spectrometric analysis was essentially as described before [36]. Briefly, samples were reduced and alkylated, in-gel digested with trypsin and resulting peptides separated over 2 h on an C18 material using an Ultimate 3000 Rapid Separation Liquid Chromatography system (Thermo Fisher Scientific). Peptides were injected via a nano-electrospray interface into a Q Exactive Plus (Thermo Fisher Scientific) mass spectrometer and analyzed in data dependent top ten mode (MS1 resolution 140000, MS2 resolution 17500). Protein identification and quantification was done with MaxQuant version 1.6.17.0 using standard parameters if not stated otherwise. The “match between runs” and LFQ quantification were enabled and sequences from the UniProt KB proteome section (UP000005640, 75777 entries, downloaded on 2021-01-27).
Electrophoresis and western transfer - Cell lysates were thawn on ice and centrifuged at 4 °C for 15 min at 13,000 rpm. The protein content of the supernatants was analyzed according to the BC Assay Protein Quantification Kit (Interchim, France). 20–50 μg of total protein were diluted in sample buffer (0.3 M Tris/HCl, pH 7, 50% glycerol, 5% SDS, 1 mM EDTA, 0.1% bromphenol blue), were reduced with 100 mM DTT for 20 min at RT. Proteins were separated for approximately 30 min at 150 V using any KD Bio-RAD mini-PROTEAN TGX Gels (Bio-Rad, USA) in mini-PROTEAN Tetran System (Bio-Rad, USA) and Tris/Glycine running buffer (25 mM Tris, 192 mM Glycine, 0.1% (w/v) SDS, pH 8.3). For western transfer, we used 0.2 μm Nitrocellulose membrane (Bio-Rad Trans- Blot Turbo Transfer Pack mini format) and the TransBlot Turbo Transfer System (Bio-Rad). Membranes were blocked with PBS, 0.05% Tween 20, 5% bovine serum albumin before applying antibodies. Proteins were visualized with the Odyssey Infrared Imaging System (LI-COR Bioscience, USA). For the electrophoretic mobility shift assay (EMSA), we synthesized IRD700 labelled duplex DNA containing a NG2 promoter (ggctgggacagttcatgcctcctccctgagggggtctcc; ccgaccctgtcaagtacggaggagggactcccccagagg) or enhancer region (tgctgaaccagctgggccgcccccgcgcagatggtctgca; acgacttggtcgacccggcgggggcgcgtctaccagacgt) with several SP-1 binding sites (Metabion, Germany). The DNA was diluted in annealing buffer (60 mM KCl, 6 mM HEPES, 0.2 mM MgCl2) to 0.1 μM. DNA was reannealed by heating for 1 min at 95 °C and cooling down for more than 30 min to RT. 1 μl DNA and 20 μg cell lysate were incubated for 15 min at RT in the dark. Native PAGE was run in the dark for 45 min at 120 V in 25 mM Tris, 192 mM Glycine, pH 8.3. Afterwards, the probe was detected at 700 nm using the Odyssey Infrared Imaging System (LI-COR Bioscience, USA).
Zebrafish – Zebrafish (wildtype, Tg(olig2:GFP) [37], Tg(ßact:Grx2)) were maintained under standard conditions (28.5 °C, dark:light cycle of 14:10 h, pH 7.2, 500 μS) [38]. Zebrafish ubiquitously overexpressing zfGrx2 (Tg(ßact:Grx2)) have been established using the Gateway/Tol2 system. In brief, a construct in which zfGrx2 expression is driven by the ubiquitous beta-actin promoter has been cloned into the Tol2 destination vector and injected together with transposase into fertilized zebrafish eggs of the AB strain according to published protocols (http://tol2kit.genetics.utah.edu/index.php/Main_Page). Zebrafish with germ-line transmission of the transgene were selected and incrossed. Line 588 showed strong overexpression of zfGrx2 (Supplemental Fig. S1B) and was used for experiments. Grx2 expression was downregulated by injection of morpholinos (Genetools, USA) as described before [2]. Approximately 300 U343-MGA cells transfected with either control siRNA or Grx2siRNA were injected into the blastula stage to follow tumor growth as described in Ref. [39]. Time-lapse images were acquired on a Zeiss Lightsheet Z.1 using a 10x detection objective and 10x illumination objectives. Samples were illuminated from one or both directions and Z-stacks were acquired every 15 min using a 1x optical zoom, 6.4 μm light-sheet thickness and 1.8 μm Z-interval. Max intensity projections were generated and sample drift was corrected for using a rigid body transformation in the StackReg plugin of ImageJ. All zebrafish experiments were performed in accordance with the German and European animal welfare legislation. According to the EU Directive 2010/63/EU on the protection of animals used for scientific purposes, early life-stages of zebrafish are not protected as animals until the stage of being capable of independent feeding, i.e. 5 days post fertilization. Housing of zebrafish in Stockholm was approved by Norra Stockholms etiska nämnd under the number 1049–2019.
Primary human tumor tissue samples – Patients tissue samples were retrieved from the CNS tumor tissue bank Düsseldorf and investigated in a pseudonymous manner as approved by the institutional review board of the Medical Faculty, Heinrich Heine University Düsseldorf (study number 2019-725). All patients gave their written informed consent to the use of their tissue specimens for research puposes. Deep frozen tissue samples of untreated primary glioblastomas, IDH-wildtype, were retrieved from the CNS tumor tissue bank Düsseldorf. A representative section of each tissue sample used for RNA extraction was histologically investigated to ensure a sufficient tumor cell content of at least 80%. Control mRNA from total human brain tissue was purchased from Takara (Japan).
RNA isolation, qRTPCR – RNA was isolated using Trizol® and chloroform. 1–2 μg of mRNA were reverse transcribed. 5 μl of 1:3 diluted cDNA were analyzed in the AB 7500 Real-Time PCR System (Life Technologies, USA) using SYBR Green or TaqMan Master Mixes (Applied Biosystems, USA). Data were normalized and analyzed using the ΔΔCt method. Primers are listed in Supplemental Table S1. Further minimum information for publication of quantitative real-time PCR experiments are available upon request.
Bioinformatic and statistical analyses – Patient data in Fig. 6D were calculated from IVY GAP project (https://glioblastoma.alleninstitute.org). Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software). All graphs show mean values with standard error of the mean (SEM). Statistical significance was determined only when controls were not set to 100% using the two-tailed Student's t-test or ANOVA with the following p values: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001.
Fig. 6.
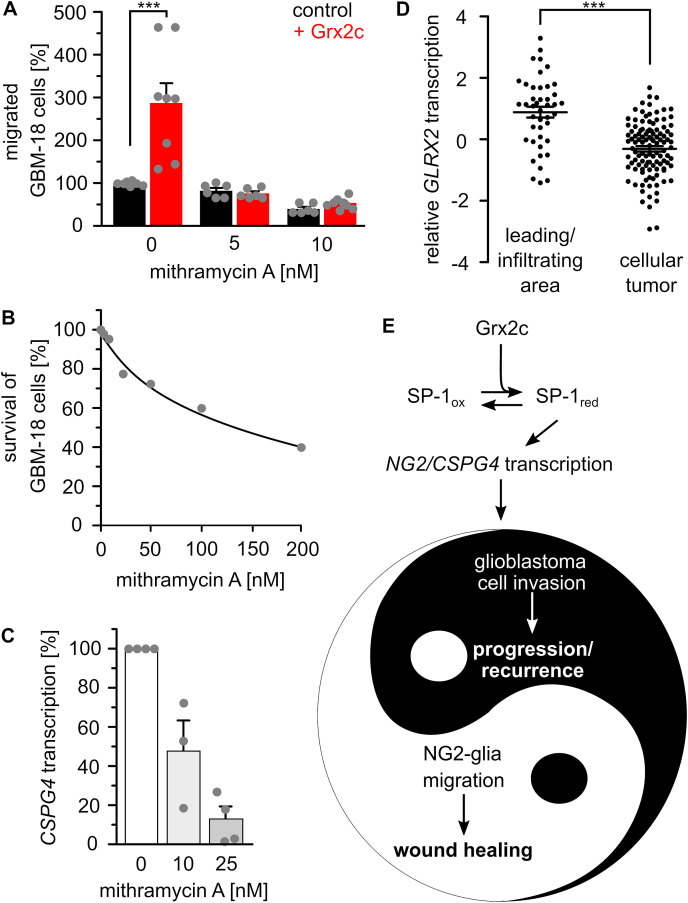
SP-1 is a main target of glutaredoxin 2′s regulation of migration. A) Treatment with 5 and 10 nM of the SP-1 inhibitor mithramycin A abolished the positive effect of Grx2c incubation on GBM-18 cell migration, ***: p < 0.001 (Student's t-test). B) Survival of GBM-18 cells treated with indicated concentrations of mithramycin A was tested using the Cell Titer Blue assay. C) CSPG4 transcripts in GBM-18 cells treated with 10 or 25 nM mithramycin A measured by qRTPCR and normalized to GAPDH transcripts. D) GLRX2 transcripts in the invading areas compared to all other areas of a tumor according to the IVY GAP project. ***: p < 0.001 (ANOVA). E) The Yin and Yang of the (patho-) physiological consequences of Grx2c regulated CSPG4 transcription. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Results
3.1. Grx2c inhibits formation of mature oligodendrocytes
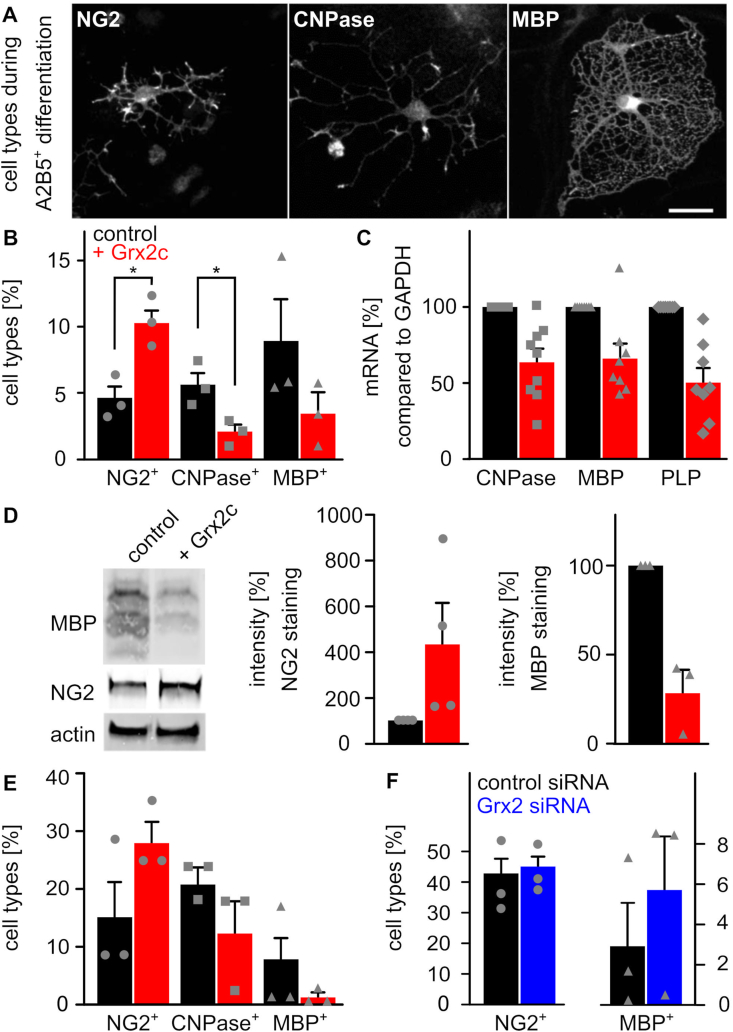
During in vitro differentiation of isolated mouse A2B5+-cells to MBP expressing oligodendrocytes, these cells form intermediate states such as NG2 glia. These different cell types can be distinguished by cell shape (Fig. 1A, Supplemental Fig. S2A), mRNA (Supplemental Fig. S2B) as well as protein levels of markers (Supplemental Figs. S2C and D). During differentiation of OPCs (A2B5+-cells in Supplemental Fig. S2E, NG2 glia in Supplemental Fig. S2F), transcription of GLRX2c is upregulated in an intermediate state and downregulated in later stages. Therefore, we investigated the impact of increased Grx2c on differentiation of oligodendrocytes. Incubation of OPCs with Grx2c lead to an increase in NG2 glia in expense of more mature oligodendrocytes as seen at cellular (Fig. 1B), mRNA (Fig. 1C), and protein levels (Fig. 1D) starting with either isolated A2B5+-cells or NG2 glia (Fig. 1E). Both, A2B5+-cells [32] and NG2 glia (Supplemental Fig. S2G) took up recombinant Grx2c added to the medium. While increased intracellular Grx2c levels lead to a decreased fraction of MBP+-cells during differentiation of NG2 glia (Fig. 1E), lowered intracellular levels after transfection with Grx2-specific siRNA increased the fraction of MBP+-cells (Fig. 1F).
Fig. 1.
Glutaredoxin 2c inhibits differentiation of mouse oligodendrocyte progenitor cells into mature oligodendrocytes. A) Primary mouse A2B5+ cells differentiate into mature MBP+ oligodendrocytes via NG2−glia and CNPase+ cells. Immunocytochemical analyses were performed after 5 days of differentiation, scale bar: 10 μm. B) Differentiation of A2B5+ cells with or without treatment with 2 μM recombinant Grx2c, quantification after immunohistochemistry from 3 independent biological samples. C) qRT-PCR of indicated genes after 5 days of differentiation of A2B5+ cells using GAPDH for normalization. D) Representative western blot analyses and quantification of 3 independent experiments after 5 days of differentiation of A2B5+ cells. E) Differentiation of NG2+ cells with or without treatment with 2 μM recombinant Grx2c, quantification after immunohistochemistry from 3 independent biological samples. F) qRT-PCR of indicated genes after 5 days of differentiation of NG2+ cells treated either with control siRNA or Grx2siRNA. Data were normalized to transcripts of GAPDH. *: p < 0.05 (Student's t-test).
3.2. Grx2c promotes migration of and wound healing by NG2 glia
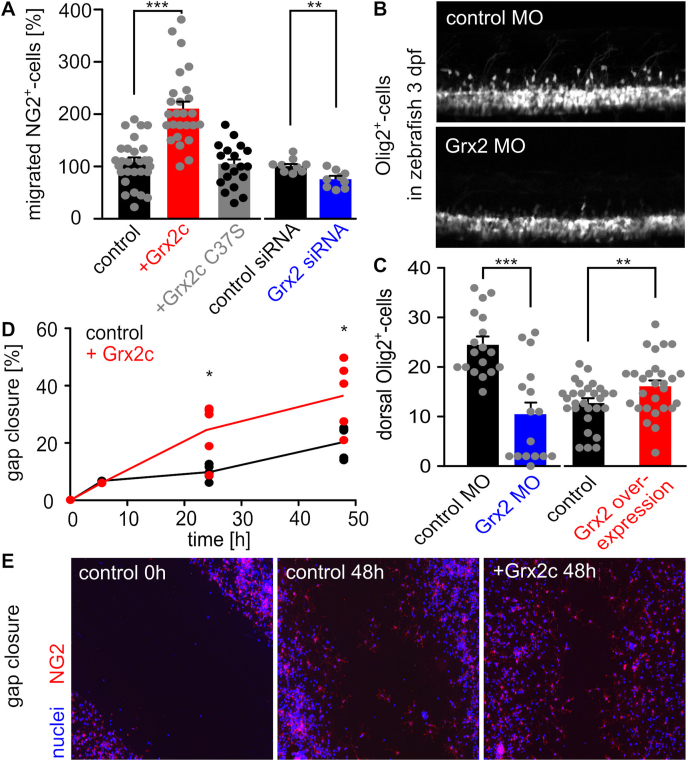
Since NG2 glia are the migrating cells of the oligodendroglial lineage, we tested whether Grx2c affects the migratory capacity of these cells. Using a Boyden chamber assay, we demonstrated the importance of the enzymatic activity of Grx2c for migration of NG2 glia as we found a significant positive correlation between migratory capacity and intracellular levels of Grx2 and no effect after addition of an enzymatically inactive Grx2c mutant (Fig. 2A). To confirm this result in vivo, we modulated Grx2 levels in zebrafish embryos and counted dorsally migrating OPCs. As shown in Fig. 2B and C, we observed the same correlation as described before for NG2 glia in vitro. Next, we investigated whether increased migration is connected to the CNS wound healing capacity of NG2 glia. We found that increased Grx2c levels promoted wound closure in a scratch assay (Fig. 2D and E).
Fig. 2.
Glutaredoxin 2c increases migratory capacity of mouse NG2+ glia. A) NG2+ cells were seeded in a Boyden chamber and incubated with 2 μM Grx2cwt or Grx2cC37S or transfected either with control siRNA or Grx2siRNA. Transmigrated cells were counted. B) Two-photon-microscopy of dorsally migrating oligodendrocyte progenitor cells in Tg(olig2-GFP) zebrafish at 3 dpf with and without morpholino-induced Grx2 knock-down. C) Migrated cells shown in b) were counted in zebrafish 3 dpf comparing Grx2 knock-down or Grx2 overexpression with respective controls. D) Quantified gap closure after scratch assay at indicated time points by NG2+ cells treated with 2 μM recombinant Grx2c or not. E) Immunocytochemistry of control and Grx2c-treated NG2+ cells after scratch assay at indicated time points. *: p < 0.05; **: p < 0.01; ***: p < 0.001 (Student's t-test).
3.3. Grx2c promotes invasion of glioblastoma cells
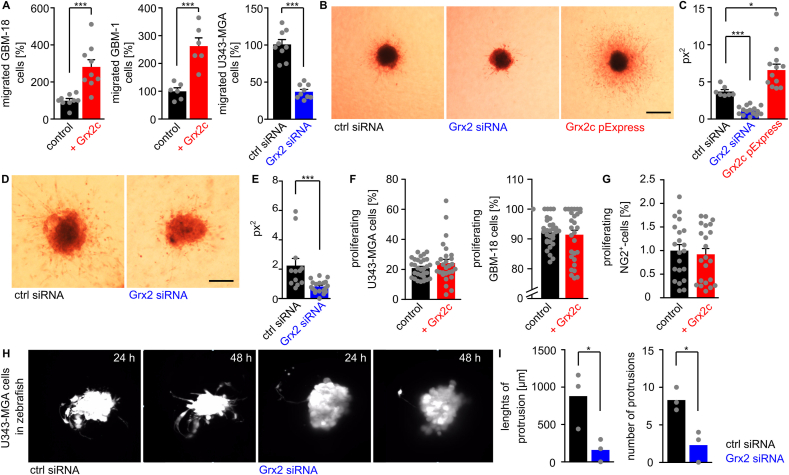
NG2/CSPG4 is not only linked to migration of OPCs but also of cancer cells. Since cell models for IDH-mutant and 1p/19q-co-deleted oligodendroglioma are limited, we investigated the connection between Grx2c and migration of glioma cells derived from the most common type of glioma, namely IDH-wildtype glioblastoma. We used Boyden chambers as 2D migration assay and a collagen matrix as 3D invasion assay. Thereby, we demonstrated a significant positive correlation between Grx2 levels, modulated by siRNA, overexpression, or incubation with recombinant Grx2c, and the migratory and invasive capacity of the human glioblastoma cell lines GBM-1, GBM-18, and U343-MGA (Fig. 3A–E). The increased area covered by cells is not caused by increased proliferation of glioblastoma cells (Fig. 3F). No effect on proliferation, we also observerd in NG2 glia (Fig. 3G). To validate elevated invasion of glioma tumor cells in vivo, we chose again the zebrafish model. As shown before [39], injection of human glioblastoma cells into the 1000-cell stage of zebrafish embryos lead to tumor formation in the developing fish. We injected U343-MGA cells treated wither either control siRNA or Grx2siRNA and followed the formation of protrusions out of the developed tumor between 24 and 48 h post fertilization by light-sheet microscopy. As shown in Fig. 3H and I (and supplemental movies), number and length of protrusions were significantly diminished in fish embryos injected with Grx2-deficient glioblastoma cells.
Fig. 3.
Glutaredoxin 2c promotes invasion of human glioblastoma cells. A) Quantification of transmigrated glioblastoma cells using a Boyden chamber assay, different glioblastoma cell lines, as well as recombinant protein and siRNA to modulate Grx2 levels. B-E) U343-MGA (B) or GBM-18 (D) spheres with modulated Grx2 levels (down-regulation by siRNA, up-regulation by overexpression via a pExpress plasmid) were embedded in a 3D collagen matrix and invasion of cells after 72 h is shown, scale bars: 200 μm. C/E) Quantification of the areas of migrated cells (px2) based on B/D. F,G) U343-MGA, GBM-18 cells (F), and NG2+-cells (G) were treated with or without 2 μM recombinant Grx2c and BrdU+-cells were quantified. H) Fluorescent labelled U343-MGA cells transfected with either control siRNA or Grx2 siRNA were injected into zebrafish at the 1000 cell stage. Pictures show stills from supplemental movies 1 obtained by light sheet microscopy between 24 and 48 hpf. I) Quantification of invasion in 3 zebrafish embryos based on supplemental movies 2. *: p < 0.05; ***: p < 0.001 (Student's t-test).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.redox.2021.102221
The following is/are the supplementary data related to this article:
3.4. Modulation of Grx2 levels regulates NG2/CSPG4 levels
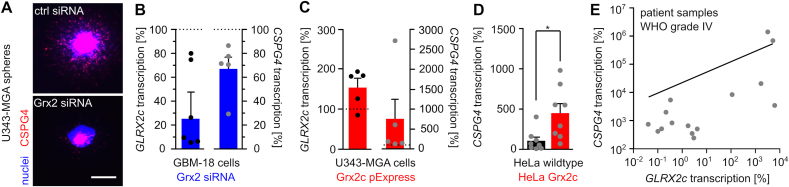
To investigate the underlying mechanism of Grx2c-dependent increase in migration/invasion, we determined a potential link between Grx2c and CSPG4 expression. In tumors formed by U343-MGA cells, Grx2 knock-down via siRNA decreased the number of CSPG4+ cells compared to treatment with control siRNA (Fig. 4A). Decreased transcript levels of Grx2c in siRNA-treated U343-MGA cells were accompanied by decreased transcript levels of CSPG4 (Fig. 4B), whereas Grx2c-overexpressing U343-MGA cells showed increased CSPG4 transcript levels (Fig. 4C). This correlation was also demonstrated in cervix cancer HeLa cells (Fig. 4D). In addition, we were able to validate these findings in primary glioblastoma tissue samples, in which we found that higher GLRX2c transcript levels were associated with higher CSPG4 transcript levels (Fig. 4E, Supplemental Fig. S3A).
Fig. 4.
Glutaredoxin 2c regulates CSPG4 transcription. A) Nuclei and CSPG4+ cells were stained in U343-MGA spheres with or without decreased Grx2 levels, scale bar: 200 μm. B) - E) Determination of GLRX2c and CSPG4 transcripts in GBM-18 (B), U343-MGA cells (C), or HeLa cells (D) with and without modulated Grx2 levels and in mRNA isolated from patient samples (E, R2 = 0.358). Data in B) - E) were normalized to GAPDH transcripts.
3.5. Grx2c increases SP-1-dependent transcription of CSPG4
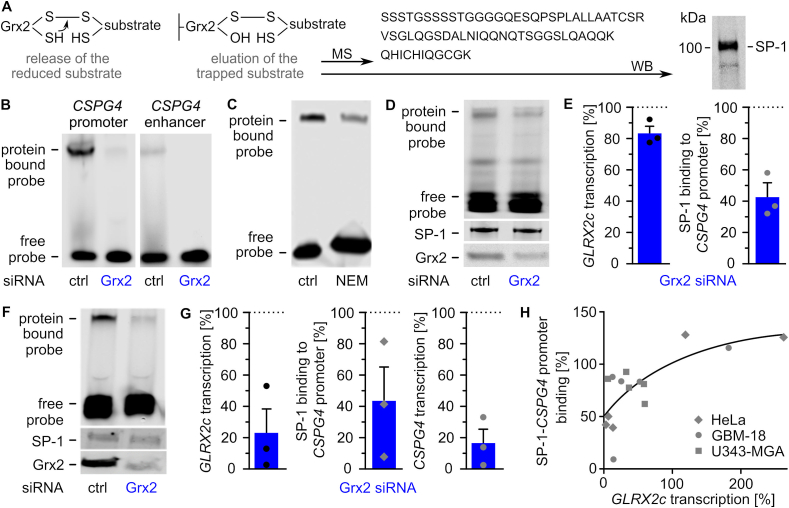
The DNA binding of SP-1, a transcription factor regulating the expression of various genes including CSPG4, has been described as being redox-regulated via thioredoxin 1 [40]. We demonstrated that incorporated Grx2c reaches the nucleus to modify the redox state of SP-1 (Supplemental Fig. S3B) and SP-1 was identified as a Grx2c substrate by the formation of stable intermediate disulfides using the Grx2c C40S mutant by western blot and mass spectrometry (Fig. 5A, supplemental Fig. S3C) and is a potential substrate in cells since recombinant. We generated infrared labelled probes covering areas of the promoter and an enhancer region of the CSPG4 gene containing several SP-1 binding sites. Both probes showed a Grx2-dependent shift in electrophoretic mobility shift assays (EMSAs) (Fig. 5B), however, further experiments were performed only with the promoter probe. We confirmed that SP-1- binding to the probe depends on thiols. Fig. 5C shows inhibited probe binding after thiol blocking using N-ethyl-maleimide. In Hela, GBM-18 and U343-MGA cells, SP-1 binding and subsequent CSPG4 transcription was regulated by the level of available Grx2 (Fig. 5D–H), whereas SP-1 expression was not affected (Fig. 5D, F). The promoting effect of Grx2c on migration of GBM-18 cells was abolished after inhibition of SP-1 by mithramycin A (Fig. 6A) using concentrations not affecting cell survival but inhibiting CSPG4 transcription (Fig. 6B and C). In line with the described increase of Grx2 on glioblastoma cell invasion, GLRX2 transcription is higher in the infiltrating area than in the cellular tumor parts of glioblastomas according to the IVY GAP project (https://glioblastoma.alleninstitute.org) (Fig. 6D).
Fig. 5.
Glutaredoxin 2 acts on CSPG4 transcription and migration via SP-1. A) Scheme of the trapping experiment revealing presence of SP-1 in the eluted substrate after western blot (WB) and mass spectrometry (MS). B) Electrophoretic mobility shift assays (EMSAs) using cell lysates of HeLa cells transfected with either control siRNA or Grx2 siRNA and probes consisting of the promoter or an enhancer region of the CSPG4 gene. C) EMSAs comparing SP-1 probe binding in HeLa lysates treated with or without 100 mM N-ethyl-maleimide. D) EMSAs of GBM-18 cell lysates transfected with either control siRNA or Grx2 siRNA. Shown is the binding of a CSPG4 promoter probe containing SP-1 binding sites during the shift assay and staining of SP-1 as well as Grx2 after western transfer of the same lysate. E) Quantification of D) and qRTPCR was performed to quantify GLRX2c transcription in the cells. F) EMSAs using extracts of HeLa cells transfected with either control siRNA or Grx2siRNA and the probe described in D). The levels of SP-1 and Grx2 were determined after western transfer using the same lysates. G) Quantification of GLRX2c and CSPG4 transcripts, as well as binding between SP-1 and probe in HeLa cells with decreased Grx2 levels compared to control cells. H) SP1-binding to the probe was plotted against GLRX2c transcripts based on all shift assays performed with HeLa, GBM-18, and U343-MGA cell lysates, R2 = 0.543.
4. Discussion
The data reported here identify the cytosolic/nuclear Grx2 isoform Grx2c as a regulator of DNA binding activity of the transcription factor SP-1. We demonstrate that the redox-regulated binding of SP-1 to the promoter and an enhancer region of the CSPG4 gene leads to increased levels of the encoded protein and subsequent enhanced migratory ability of glial and glioma cells. The enzymatic Taoism of this Grx2c-dependent redox signaling pathway is based on the finding that the same signaling event can lead to beneficial or detrimental effects depending on the context and the state of the glia cell (Fig. 6E).
DNA binding of SP-1 is regulated by several posttranslational modifications, such as acetylation, sumoylation, ubiquitylation, glycosylation, and redox regulation [[40], [41], [42]]. It was shown that thioredoxin 1 increases SP-1 activity [40]. SP-1 belongs to the specificity protein/Krüppel-like factor (Sp/KLF) family, is ubiquitously expressed in mammalian tissues, and regulates the transcription of numerous genes involved in a variety of cellular processes including early embryonic development [43,44]. In glioma patients, SP-1 levels are upregulated and associated with poor prognosis [45]. In addition, one study reported that SP-1 may mediate glioblastoma resistance against temozolomide independent from the O6-methylguanine-DNA methyltransferase (MGMT) status [46]. This could suggest that treatments leading to decreased SP-1 activity might overcome this therapy resistance. Modulation of SP-1 levels in glioma cell lines revealed higher or lower migration capacity after overexpression or knock out of SP-1, respectively, which was linked to transcription of matrix metalloproteinase 2 [45]. Our study links SP-1-dependent migration to transcription of CSPG4. The encoded protein is known as being relevant for increased invasion of glioma cells and has been associated with poor prognosis of patients [[27], [28], [29]]. High expression of Grx2 as activator of the SP-1/CSPG4 axis was also linked to shorter patient survival in two independent cohorts of glioblastoma patients (Supplemental Fig. S4A) [47,48], as well as another cohort of oligodendroglioma patients [47]. In general, Grx2 was reported to be down-regulated in glioma tumor tissue [47,49], but its transcription appears to increase in parallel to the WHO grade of the tumors (Supplemental Fig. S4B) [47]. Moreover, Grx2 is increased in the infiltrating area (Fig. 6D). Although these data suggest Grx2 as a marker of aggressiveness in gliomas, they do not distinguish between the different Grx2 isoforms. Only the cytosolic isoform Grx2c affects the cytoskeleton and increases migration of cells, whereas the ubiquitous mitochondrial isoform Grx2a does not [2,5]. Our study links Grx2c to SP-1 activity and increased CSPG4 levels, making Grx2c a promising marker for invasion and therapy resistance of malignant gliomas, and suggests interference with specific redox signaling as a promising therapeutic target. Grx2 and other oxidoreductases of the thioredoxin family were described to be upregulated in a variety of carcinomas leading to increased chemotherapy resistance and decreased patient survival [50,51]. Cells with stem cell properties are considered to be a main driver for the development and progression of gliomas [52]. Therefore, with the GBM-1 and GBM-18 we used glioblastoma stem cell models known to possess functional and molecular features of stemness such as high in vivo tumorigenicity, aberrant activation of phylogenetically conserved developmental stem cell pathways or elevated resistance to standard of care therapies as well as highly invasive behavior [53,54]. Stem and progenitor cells, especially NG2 glia, might be one potential cell of origin for the formation of glioma [20,21]. Blockage/dysregulation of oligodendroglial differentiation by Grx2 might contribute to glioma initiation, however, this was not part of our study and needs further investigation. Here, we strengthen the important role of Grx2c during developmental processes and of redox signaling during glial differentiation and tumorigenesis. Signaling events initiated by oxidoreductases like Grx2c or enzymatically formed oxidizing molecules such as hydrogen peroxide, hydrogen sulfide, and nitric oxide, as well as redox-dependent activities of proteins like sirtuin 1 or collapsin response mediator protein 2 control the differentiation and fate of neural stem and progenitor cells including OPCs [8,10,[55], [56], [57], [58], [59]]. NG2 glia are present in all regions of the brain between postnatal development and death, and provide beneficial support during different damages and diseases of the central nervous system [15,60]. After traumatic injury, for example, these cells react very fast, migrate to the damaged area and contribute to wound closure [18,61,62]. This function was accelerated by increased levels of Grx2c (Fig. 2D and E).
In summary, we report that Grx2c increases migration of glioma cells and OPCs, namely A2B5+-cells and NG2 glia, via activation of a common signaling pathway and thereby either may support a detrimental phenotype, i.e., glioma cell migration and invasion, or have benefical functions, such as wound healing, a Yin and Yang of redox regulation (Fig. 6E).
Author contributions
CW, KL, FH, SE, JF, LP, TTL, GP, and KV acquired data; CW, KL, JF, GP, NQ, LB, BO, and CB analyzed data; CB designed the research concept; CW, KL, UDK, MR, TP, OA, GR, LB, BO, and CB designed experiments; CB wrote the manuscript with help of CW, KV, UDK, and GR; SGM and OA reviewed the manuscript and all authors approved its final version.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We thank Monica Nister (Karolinska Institute, Stockholm, Sweden) for providing the glioblasstoma cell lines GBM-18 and U343-MGA. We thank Mary Bayer, Kathrin Brücksken, Thomas Hildebrandt, and Leonie Thewes for experimental and technical assistance and the zebrafish core facilities at the Medical Faculty, University Bonn, Germany, and at the Karolinska Institute, Stockholm, Sweden. Leda Dimou (University Ulm, Germany) and Michael Wegener (Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany) are acknowledged for helpful discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to CB: BE 3259/5-1, BE 3259/5-2, 417677437/GRK2578), the Gemeinnützige Hertie-Stiftung (to CB and OA), and the Karolinska Institute (to LB: KI core-facility grants Dnr 2-5864/2018 and Dnr 2021-00574).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102221.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Russel M., Model P., Holmgren A. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J. Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bräutigam L., Schütte L.D., Godoy J.R., Prozorovski T., Gellert M., Hauptmann G., Holmgren A., Lillig C.H., Berndt C. Vertebrate-specific glutaredoxin is essential for brain development. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20532–20537. doi: 10.1073/pnas.1110085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lönn M.E., Hudemann C., Berndt C., Cherkasov V., Capani F., Holmgren A., Lillig C.H. Expression pattern of human glutaredoxin 2 isoforms: identification and characterization of two testis/cancer cell-specific isoforms. Antioxidants Redox Signal. 2008;10:547–557. doi: 10.1089/ars.2007.1821. [DOI] [PubMed] [Google Scholar]
- 4.Hudemann C., Lönn M.E., Godoy J.R., Zahedi Avval F., Capani F., Holmgren A., Lillig C.H. Identification, expression pattern, and characterization of mouse glutaredoxin 2 isoforms. Antioxidants Redox Signal. 2009;11:1–14. doi: 10.1089/ars.2008.2068. [DOI] [PubMed] [Google Scholar]
- 5.Gellert M., Richter E., Mostertz J., Kantz L., Masur K., Hanschmann E.-M., Ribback S., Kroeger N., Schaeffeler E., Winter S., Hochgräfe F., Schwab M., Lillig C.H. The cytosolic isoform of glutaredoxin 2 promotes cell migration and invasion. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129599. doi: 10.1016/j.bbagen.2020.129599. [DOI] [PubMed] [Google Scholar]
- 6.Berndt C., Poschmann G., Stühler K., Holmgren A., Bräutigam L. Zebrafish heart development is regulated via glutaredoxin 2 dependent migration and survival of neural crest cells. Redox Biol. 2014;2:673–678. doi: 10.1016/j.redox.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bräutigam L., Jensen L.D.E., Poschmann G., Nyström S., Bannenberg S., Dreij K., Lepka K., Prozorovski T., Montano S.J., Aktas O., Uhlén P., Stühler K., Cao Y., Holmgren A., Berndt C. Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20057–20062. doi: 10.1073/pnas.1313753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noble M., Mayer-Pröschel M., Pröschel C. Redox regulation of precursor cell function: insights and paradoxes. Antioxidants Redox Signal. 2005;7:1456–1467. doi: 10.1089/ars.2005.7.1456. [DOI] [PubMed] [Google Scholar]
- 9.Prozorovski T., Schneider R., Berndt C., Hartung H.-P., Aktas O. Redox-regulated fate of neural stem progenitor cells. Biochim. Biophys. Acta. 2015;1850:1543–1554. doi: 10.1016/j.bbagen.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Olguín-Albuerne M., Morán J. Redox signaling mechanisms in nervous system development. Antioxidants Redox Signal. 2018;28:1603–1625. doi: 10.1089/ars.2017.7284. [DOI] [PubMed] [Google Scholar]
- 11.Salazar-Ramiro A., Ramírez-Ortega D., La Pérez de Cruz V., Hérnandez-Pedro N.Y., González-Esquivel D.F., Sotelo J., Pineda B. Role of redox status in development of glioblastoma. Front. Immunol. 2016;7:156. doi: 10.3389/fimmu.2016.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradl M., Lassmann H. Oligodendrocytes: biology and pathology. Acta Neuropathol. 2010;119:37–53. doi: 10.1007/s00401-009-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S.C. Defining glial cells during CNS development. Nat. Rev. Neurosci. 2001;2:840–843. doi: 10.1038/35097593. [DOI] [PubMed] [Google Scholar]
- 14.Dimou L., Simon C., Kirchhoff F., Takebayashi H., Götz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J. Neurosci. 2008;28:10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiyama A., Komitova M., Suzuki R., Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 16.Raff M.C., Miller R.H., Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 17.Ffrench-Constant C., Raff M.C. Proliferating bipotential glial progenitor cells in adult rat optic nerve. Nature. 1986;319:499–502. doi: 10.1038/319499a0. [DOI] [PubMed] [Google Scholar]
- 18.von Streitberg A., Jäkel S., Eugenin von Bernhardi J., Straube C., Buggenthin F., Marr C., Dimou L. NG2-Glia transiently overcome their homeostatic network and contribute to wound closure after brain injury. Front. Cell Dev. Biol. 2021;9:662056. doi: 10.3389/fcell.2021.662056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine J.M., Reynolds R. Activation and proliferation of endogenous oligodendrocyte precursor cells during ethidium bromide-induced demyelination. Exp. Neurol. 1999;160:333–347. doi: 10.1006/exnr.1999.7224. [DOI] [PubMed] [Google Scholar]
- 20.Shoshan Y., Nishiyama A., Chang A., Mörk S., Barnett G.H., Cowell J.K., Trapp B.D., Staugaitis S.M. Expression of oligodendrocyte progenitor cell antigens by gliomas: implications for the histogenesis of brain tumors. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10361–10366. doi: 10.1073/pnas.96.18.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laug D., Glasgow S.M., Deneen B. A glial blueprint for gliomagenesis. Nat. Rev. Neurosci. 2018;19:393–403. doi: 10.1038/s41583-018-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldape K., Zadeh G., Mansouri S., Reifenberger G., von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 23.Wesseling P., van den Bent M., Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:809–827. doi: 10.1007/s00401-015-1424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahlert U.D., Nikkhah G., Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Vollmann-Zwerenz A., Leidgens V., Feliciello G., Klein C.A., Hau P. Tumor cell invasion in glioblastoma. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21061932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescher N., Hänsch S., Knobbe-Thomsen C.B., Stühler K., Poschmann G. The migration behavior of human glioblastoma cells is influenced by the redox-sensitive human macrophage capping protein CAPG. Free Radic. Biol. Med. 2021;167:81–93. doi: 10.1016/j.freeradbiomed.2021.02.038. [DOI] [PubMed] [Google Scholar]
- 27.Svendsen A., Verhoeff J.J.C., Immervoll H., Brøgger J.C., Kmiecik J., Poli A., Netland I.A., Prestegarden L., Planagumà J., Torsvik A., Kjersem A.B., Sakariassen P.Ø., Heggdal J.I., van Furth W.R., Bjerkvig R., Lund-Johansen M., Enger P.Ø., Felsberg J., Brons N.H.C., Tronstad K.J., Waha A., Chekenya M. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 2011;122:495–510. doi: 10.1007/s00401-011-0867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsidulko A.Y., Kazanskaya G.M., Kostromskaya D.V., Aidagulova S.V., Kiselev R.S., Volkov A.M., Kobozev V.V., Gaitan A.S., Krivoshapkin A.L., Grigorieva E.V. Prognostic relevance of NG2/CSPG4, CD44 and Ki-67 in patients with glioblastoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317724282. 1010428317724282. [DOI] [PubMed] [Google Scholar]
- 29.Schiffer D., Mellai M., Boldorini R., Bisogno I., Grifoni S., Corona C., Bertero L., Cassoni P., Casalone C., Annovazzi L. The significance of chondroitin sulfate proteoglycan 4 (CSPG4) in human gliomas. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19092724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godoy J.R., Funke M., Ackermann W., Haunhorst P., Oesteritz S., Capani F., Elsässer H.-P., Lillig C.H. Redox atlas of the mouse. Immunohistochemical detection of glutaredoxin-, peroxiredoxin-, and thioredoxin-family proteins in various tissues of the laboratory mouse. Biochim. Biophys. Acta. 2011;1810:2–92. doi: 10.1016/j.bbagen.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Lillig C.H., Berndt C., Vergnolle O., Lönn M.E., Hudemann C., Bill E., Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepka K., Volbracht K., Bill E., Schneider R., Rios N., Hildebrandt T., Ingwersen J., Prozorovski T., Lillig C.H., van Horssen J., Steinman L., Hartung H.-P., Radi R., Holmgren A., Aktas O., Berndt C. Iron-sulfur glutaredoxin 2 protects oligodendrocytes against damage induced by nitric oxide release from activated microglia. Glia. 2017;65:1521–1534. doi: 10.1002/glia.23178. [DOI] [PubMed] [Google Scholar]
- 33.Enoksson M., Fernandes A.P., Prast S., Lillig C.H., Holmgren A., Orrenius S. Overexpression of glutaredoxin 2 attenuates apoptosis by preventing cytochrome c release. Biochem. Biophys. Res. Commun. 2005;327:774–779. doi: 10.1016/j.bbrc.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 34.Lillig C.H., Lönn M.E., Enoksson M., Fernandes A.P., Holmgren A. Short interfering RNA-mediated silencing of glutaredoxin 2 increases the sensitivity of HeLa cells toward doxorubicin and phenylarsine oxide. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13227–13232. doi: 10.1073/pnas.0401896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C.-C., Park A.Y., Guan J.-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 36.Brenig K., Grube L., Schwarzländer M., Köhrer K., Stühler K., Poschmann G. The proteomic landscape of cysteine oxidation that underpins retinoic acid-induced neuronal differentiation. J. Proteome Res. 2020;19:1923–1940. doi: 10.1021/acs.jproteome.9b00752. [DOI] [PubMed] [Google Scholar]
- 37.Shin J., Park H.-C., Topczewska J.M., Mawdsley D.J., Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- 38.Westerfield M. fourthth ed. University of Oregon Press; Eugene, USA: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 39.Pudelko L., Edwards S., Balan M., Nyqvist D., Al-Saadi J., Dittmer J., Almlöf I., Helleday T., Bräutigam L. An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 2018;20:1475–1484. doi: 10.1093/neuonc/noy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloomfield K.L., Osborne S.A., Kennedy D.D., Clarke F.M., Tonissen K.F. Thioredoxin-mediated redox control of the transcription factor Sp1 and regulation of the thioredoxin gene promoter. Gene. 2003;319:107–116. doi: 10.1016/s0378-1119(03)00799-6. [DOI] [PubMed] [Google Scholar]
- 41.Tan N.Y., Khachigian L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X., Bishopric N.H., Discher D.J., Murphy B.J., Webster K.A. Physical and functional sensitivity of zinc finger transcription factors to redox change. Mol. Cell Biol. 1996;16:1035–1046. doi: 10.1128/MCB.16.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marin M., Karis A., Visser P., Grosveld F., Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor L., Gilmour J., Bonifer C. The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 2016;89:513–525. [PMC free article] [PubMed] [Google Scholar]
- 45.Guan H., Cai J., Zhang N., Wu J., Yuan J., Li J., Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int. J. Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 46.Chang K.-Y., Hsu T.-I., Hsu C.-C., Tsai S.-Y., Liu J.-J., Chou S.-W., Liu M.-S., Liou J.-P., Ko C.-Y., Chen K.-Y., Hung J.-J., Chang W.-C., Chuang C.-K., Kao T.-J., Chuang J.-Y. Specificity protein 1-modulated superoxide dismutase 2 enhances temozolomide resistance in glioblastoma, which is independent of O6-methylguanine-DNA methyltransferase. Redox Biol. 2017;13:655–664. doi: 10.1016/j.redox.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gravendeel L.A.M., Kouwenhoven M.C.M., Gevaert O., de Rooi J.J., Stubbs A.P., Duijm J.E., Daemen A., Bleeker F.E., Bralten L.B.C., Kloosterhof N.K., de Moor B., Eilers P.H.C., van der Spek P.J., Kros J.M., Sillevis Smitt P.A.E., van den Bent M.J., French P.J. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi A., Yajima N., Tsuchiya N., Homma J., Sano M., Natsumeda M., Takahashi H., Fujii Y., Kakuma T., Yamanaka R. Gene expression signature-based prognostic risk score in patients with glioblastoma. Cancer Sci. 2013;104:1205–1210. doi: 10.1111/cas.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L., Hui A.-M., Su Q., Vortmeyer A., Kotliarov Y., Pastorino S., Passaniti A., Menon J., Walling J., Bailey R., Rosenblum M., Mikkelsen T., Fine H.A. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Fernandes A.P., Capitanio A., Selenius M., Brodin O., Rundlöf A.-K., Björnstedt M. Expression profiles of thioredoxin family proteins in human lung cancer tissue: correlation with proliferation and differentiation. Histopathology. 2009;55:313–320. doi: 10.1111/j.1365-2559.2009.03381.x. [DOI] [PubMed] [Google Scholar]
- 51.Mollbrink A., Jawad R., Vlamis-Gardikas A., Edenvik P., Isaksson B., Danielsson O., Stål P., Fernandes A.P. Expression of thioredoxins and glutaredoxins in human hepatocellular carcinoma: correlation to cell proliferation, tumor size and metabolic syndrome. Int. J. Immunopathol. Pharmacol. 2014;27:169–183. doi: 10.1177/039463201402700204. [DOI] [PubMed] [Google Scholar]
- 52.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vargas-Toscano A., Nickel A.-C., Li G., Kamp M.A., Muhammad S., Leprivier G., Fritsche E., Barker R.A., Sabel M., Steiger H.-J., Zhang W., Hänggi D., Kahlert U.D. Rapalink-1 targets glioblastoma stem cells and acts synergistically with tumor treating fields to reduce resistance against temozolomide. Cancers. 2020;12 doi: 10.3390/cancers12123859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pudelko L., Rouhi P., Sanjiv K., Gad H., Kalderén C., Höglund A., Squatrito M., Schuhmacher A.J., Edwards S., Hägerstrand D., Berglund U.W., Helleday T., Bräutigam L. Glioblastoma and glioblastoma stem cells are dependent on functional MTH1. Oncotarget. 2017;8:84671–84684. doi: 10.18632/oncotarget.19404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R., Zipp F., Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 56.Okamoto S.-i., Lipton S.A. S-Nitrosylation in neurogenesis and neuronal development. Biochim. Biophys. Acta. 2015;1850:1588–1593. doi: 10.1016/j.bbagen.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson C., Muñoz-Palma E., González-Billault C. From birth to death: a role for reactive oxygen species in neuronal development. Semin. Cell Dev. Biol. 2018;80:43–49. doi: 10.1016/j.semcdb.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 58.Kimura H. Signaling by hydrogen sulfide (H2S) and polysulfides (H2Sn) in the central nervous system. Neurochem. Int. 2019;126:118–125. doi: 10.1016/j.neuint.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 59.Berndt C., Wilms C., Thauvin M., Vriz S. Oxidative Stress. Elsevier; 2020. Redox-regulated brain development; pp. 565–582. [Google Scholar]
- 60.Dimou L., Gallo V. NG2-glia and their functions in the central nervous system. Glia. 2015;63:1429–1451. doi: 10.1002/glia.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levine J.M. Increased expression of the NG2 chondroitin-sulfate proteoglycan after brain injury. J. Neurosci. 1994;14:4716–4730. doi: 10.1523/JNEUROSCI.14-08-04716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hughes E.G., Kang S.H., Fukaya M., Bergles D.E. Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 2013;16:668–676. doi: 10.1038/nn.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.