Summary
Clinical interpretation of missense variants is challenging because the majority identified by genetic testing are rare and their functional effects are unknown. Consequently, most variants are of uncertain significance and cannot be used for clinical diagnosis or management. Although not much can be done to ameliorate variant rarity, multiplexed assays of variant effect (MAVEs), where thousands of single-nucleotide variant effects are simultaneously measured experimentally, provide functional evidence that can help resolve variants of unknown significance (VUSs). However, a rigorous assessment of the clinical value of multiplexed functional data for variant interpretation is lacking. Thus, we systematically combined previously published BRCA1, TP53, and PTEN multiplexed functional data with phenotype and family history data for 324 VUSs identified by a single diagnostic testing laboratory. We curated 49,281 variant functional scores from MAVEs for these three genes and integrated four different TP53 multiplexed functional datasets into a single functional prediction for each variant by using machine learning. We then determined the strength of evidence provided by each multiplexed functional dataset and reevaluated 324 VUSs. Multiplexed functional data were effective in driving variant reclassification when combined with clinical data, eliminating 49% of VUSs for BRCA1, 69% for TP53, and 15% for PTEN. Thus, multiplexed functional data, which are being generated for numerous genes, are poised to have a major impact on clinical variant interpretation.
Keywords: BRCA1, TP53, PTEN, functional data, variant interpretation, MAVE
Introduction
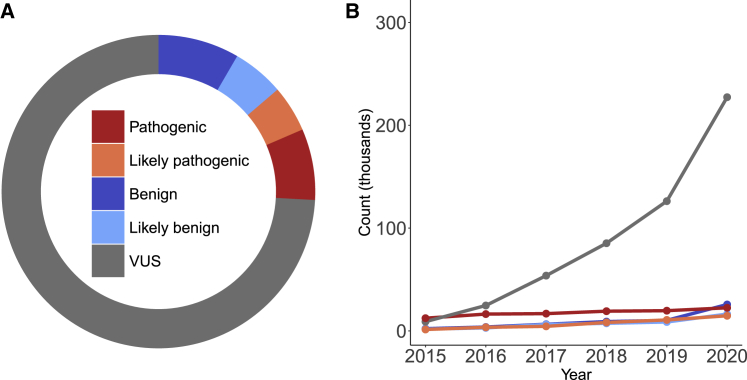
Targeted panel testing for cancer predisposition is now widespread, and as panels increase in size, the likelihood of identifying a rare missense variant of uncertain significance (VUS) also increases.1,2 As a result, these inconclusive VUS results are commonly returned to individuals. Multigene panel testing for hereditary cancer predisposition, for example, identifies one or more VUS in 40% of individuals who are tested for suspicion of cancer predisposition.2 Most VUSs are missense variants, which can be challenging to interpret as pathogenic or benign according to the American College for Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines.3 This challenge is largely due to the fact that missense variants are typically rare, making clinical evidence such as segregation and case-control data scarce. Of the nearly 330,000 missense variants from clinical testing in the ClinVar database, 70% are VUSs4 (Figure 1A). The VUS problem has grown exponentially over time (Figure 1B) and exists even among extremely well-studied cancer predisposition genes such as BRCA1, TP53, and PTEN, where the majority of missense variants reported in ClinVar from clinical genetic testing are VUSs (BRCA1 80%, n = 2,395; TP53 64%, n = 589; and PTEN 72%, n = 411).
Figure 1.
Missense variants of uncertain significance are a large and growing problem
(A) Single-nucleotide missense variants colored by ClinVar classifications (benign = 25,707; likely benign = 16,377; VUSs = 227,365; likely pathogenic = 14,716; pathogenic = 22,489; conflicting interpretations = 20,026). ClinVar data downloaded on 10/27/2020.
(B) Missense variants in ClinVar from 2015 to 2020 shown by clinical significance.
Definitive variant interpretation in BRCA1, TP53, and PTEN is critical because morbidity and mortality can be reduced for individuals known to harbor pathogenic variants through increased cancer surveillance and preventative measures.5, 6, 7 In contrast, medical management is not changed for individuals who carry a VUS, and the uncertainty surrounding these variants can provoke anxiety8 at best or represent a missed opportunity to provide life-saving care at worst. Thus, improved interpretation of VUSs directly impacts the well-being of carriers and their families. Furthermore, the ACMG recommends that pathogenic variants in all three of these genes be returned to individuals regardless of the indication for testing.9,10 However, the recommendations for return of secondary findings are limited to established pathogenic and likely pathogenic variants and secondary VUSs are not typically shared with individuals.9 Thus, individuals may be left in the dark about increased risk if their VUSs are reclassified as pathogenic, further highlighting the need for timely VUS resolution.
While little can be done to change the lack of information arising from a variant’s rarity, it is now possible to generate variant functional data at scale. Models suggest that functional evidence could lead to the reclassification of most VUSs,11 however that has not been tested on a large scale with real-world data. Multiplexed assays of variant effect (MAVEs), where thousands of single-nucleotide variants are assayed simultaneously, have been applied to BRCA1 (MIM: 113705),12 TP53 (MIM: 191170),13,14 and PTEN (MIM: 601728),15,16 producing functional annotations for thousands of variants that can be used as evidence to resolve VUSs.17, 18, 19, 20 Recently, guidelines for both generating and using multiplexed functional data have been developed and create the opportunity to systematically explore the clinical value of multiplexed functional data in the reinterpretation of VUSs.21,22 However, the extent to which multiplexed functional data can result in medically significant variant reinterpretation has not been systematically evaluated.
Thus, we assessed the clinical value of BRCA1, TP53, and PTEN multiplexed functional data by integrating them with existing lines of evidence from clinical variant interpretations for these genes. First, we curated 49,281 variant functional scores for clinical integration from multiplexed assays across the three genes.21 Then, we determined the strength of evidence for the functional evidence component of variant interpretation provided by each multiplexed functional dataset based on its ability to predict established pathogenic and benign variants22 and reevaluated 324 VUS classifications (BRCA1 = 110, TP53 = 166, PTEN = 48) (Figure 2). Multiplexed functional data, when combined with existing lines of evidence, resulted in reclassification of 49% of VUSs for BRCA1, 69% for TP53, and 15% for PTEN. Thus, multiplexed functional data can help to resolve a large percentage of VUSs, highlighting the utility of generating and curating multiplexed functional data. Our analysis revealed two major factors that limited the utility of multiplexed functional data: the modest predictive value of some MAVEs and the scarcity of established pathogenic or benign variants that serve as validation controls for some genes. On the basis of these findings, we discuss how MAVEs should be designed and piloted with clinical utility in mind, prioritizing genes with established pathogenic and benign variants. We conclude by discussing the overall implications of multiplexed functional data for variant reclassification.
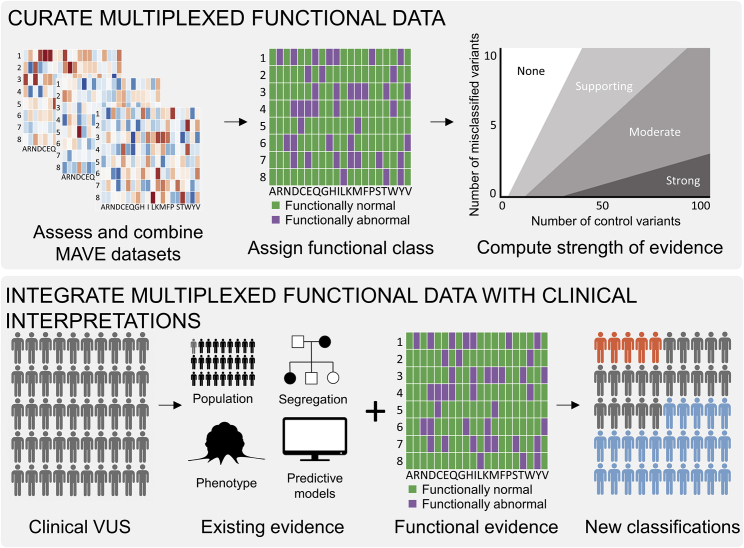
Figure 2.
Schematic for integration of multiplexed functional data into clinical variant interpretation
Top: we first collected variant function scores and determined assay dynamic range and sensitivity and specificity for established pathogenic and benign variants. If a single assay had high sensitivity and specificity, we used the function scores directly to determine which variants were functionally normal and functionally abnormal. Where possible, we combined multiple MAVE datasets to increase predictive value of function scores and determine the functional class of variants. Finally, we computed the odds of pathogenicity for the assigned functional classes to determine the strength of evidence assigned to each dataset. Bottom: existing evidence for 324 VUSs were combined with the MAVE functional evidence to reinterpret variants as either likely pathogenic (orange), likely benign (blue), or VUSs (gray).
Material and methods
Human subjects
This study was approved by the University of Washington (UW) Institutional Review Board STUDY00003598. Data were deidentified for transfer to UW, and additional consent was not required.
Clinical data collection
Data were provided from 9,234 individuals who were found to have a variant in BRCA1, TP53, and/or PTEN by Ambry Genetics in multigene panel testing for cancer predisposition on or before 9/25/2019. Interpretation of sequence variations was performed according to the American College of Medical Genetics and Genomics guidelines.3 Variants were classified as pathogenic, likely pathogenic, variant of unknown significance, likely benign, or benign according to the Ambry 5-tier variant classification protocol.23 Ambry classifications follow a modified version of ACMG/AMP guidelines.
Clinical data filtering
Individual phenotype and variant data from clinical testing were excluded from this analysis when they met certain exclusion criteria: (1) an individual with a VUS in BRCA1, TP53, or PTEN was excluded if that individual was found to have a pathogenic or likely pathogenic variant in another cancer-predisposing gene on the multigene panel, (2) an individual with a VUS in BRCA1, TP53, or PTEN was excluded if they were found to have one or more additional VUSs in other cancer-predisposing genes, and (3) an individual with a pathogenic variant in BRCA1, TP53, or PTEN were excluded if they were found to have a pathogenic variant in any other gene. In total, 2,437 of the 9,234 individuals from the clinical data were excluded in our analysis. We used data from 4,723 individuals with a BRCA1 variant, 1,334 individuals with a TP53 variant, and 740 individuals with a PTEN variant.
Naive Bayes classifier
Naive Bayes classification of TP53 variants was conducted with the Gaussian naive Bayes functionality from the Python SciKit Learn library.24 Prior probabilities were set at 0.5, the default for binary classification, for functionally normal and functionally abnormal classes. Variants used for training were all of the clinically derived ClinVar pathogenic/likely pathogenic and benign/likely benign variants. After training the classifier on function scores from all four TP53 MAVEs, performance was evaluated with leave-one-out cross-validation. Additional naive Bayes classifiers were trained on function scores from single TP53 MAVEs and evaluated with leave-one-out cross-validation. Because the four-feature classifier had the highest overall accuracy, we used this classifier to make predictions of TP53 variant functional effects.
Variant reinterpretation
Variants from multigene cancer panels were reinterpreted with multiplexed functional data following ACMG/AMP rules-based guidelines and a Bayesian adaptation for ACMG/AMP rules.3,25 For TP53 and PTEN, ClinGen variant curation expert panel (VCEP) adaptations of the ACMG rules were used in reinterpretation with the exception of the functional data evidence code where we exclusively used MAVE data.17,26 For BRCA1, BayesDel27 predictions were used as computational predictive model data with thresholds of less than 0.147 for benign evidence (BP4) and greater than 0.425 for pathogenic evidence (PP3).27,28 For BRCA1 splice region variants, SpliceAI29 predictions were used as computational predictive model evidence with a threshold of greater than 0.8 for pathogenic evidence (PP3).29 In addition, absence from gnomAD30 was used as pathogenic population data for BRCA1 and restricted to supporting level of evidence (PM2_P). All other evidence codes were applied as recommended in the original ACMG guidelines or VCEP adaptations. The Bayesian implementation of the ACMG guidelines was performed as previously described, and prior probability of pathogenicity was set at 0.1 for all variants.25
Results
Multiplexed functional data curation
First, we evaluated whether each multiplexed functional dataset was compatible with three key recommendations for clinical integration.21 All assays used in this analysis met the first criterion: the function scores generated for each variant must be a direct measure of variant effect. Next, the dynamic range of the assay must be sufficient to separate variant effects targeted by the assay from synonymous variants. For example, an assay designed to detect loss-of-function variants must have a readout that is able to separate nonsense variants from synonymous variants. When this criterion is met, variants that score like nonsense variants are called “functionally abnormal” and variants that score like synonymous variants are called “functionally normal.” Finally, the assay should have high sensitivity and specificity for clinically ascertained control variants, where benign variants are scored as functionally normal and pathogenic variants are scored as functionally abnormal.
Finally, the strength of evidence that could be applied to multiplexed functional data for variant interpretation was determined following recommendations from the ClinGen Sequence Variant Interpretation (SVI) Working Group.22 According to these recommendations, the strength of evidence generated by a functional assay is determined by how well the assay can distinguish between control benign and pathogenic variants and how many control variants are available for this comparison. The resulting odds of pathogenicity (OddsPath) corresponds to strength of evidence codes from the original ACMG/AMP guidelines for variant interpretation: supporting, moderate, or strong.3,25
BRCA1 multiplexed functional data curation
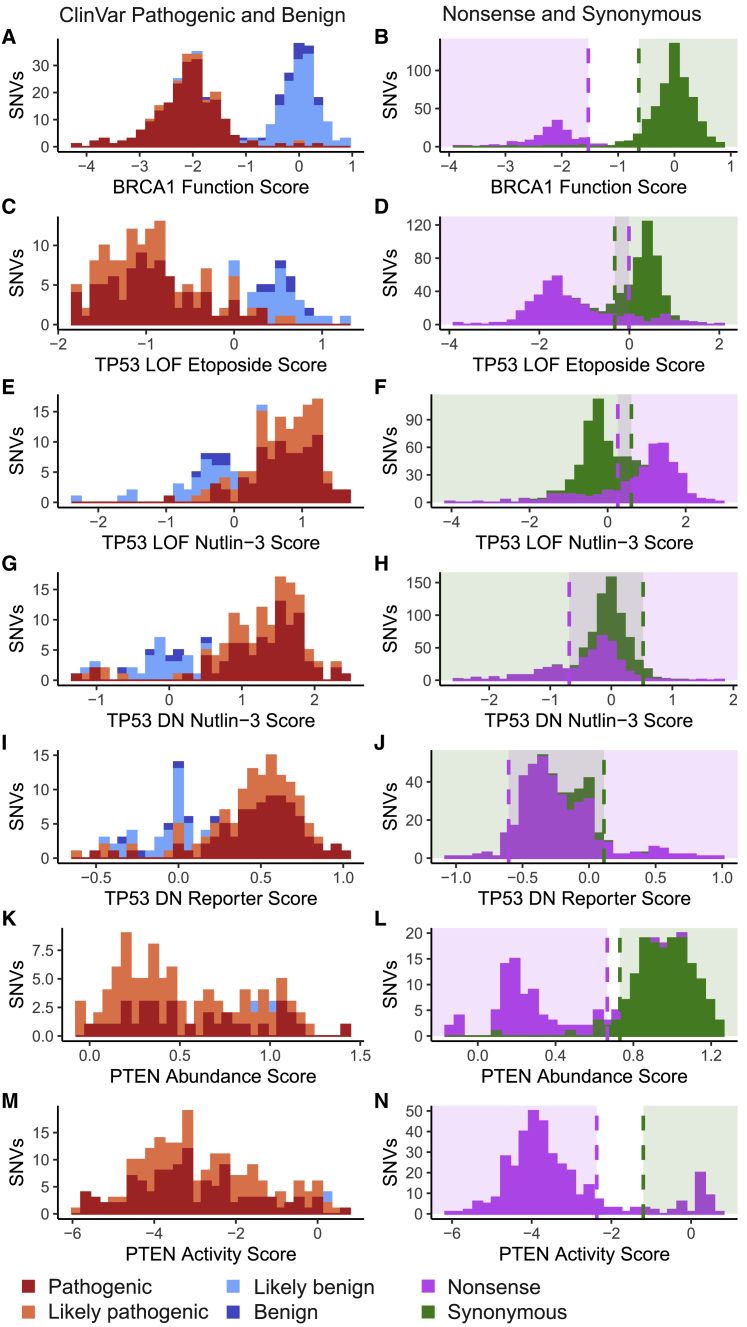
Multiplexed functional data for 3,893 single-nucleotide variants (SNVs) of BRCA1 were generated with saturation genome editing12 (Table 1). We chose this MAVE to assess because it is the largest and most accurate for BRCA1. These functional data were suitable for clinical integration because the assay result was directly linked to variant effect. Here, cells with functionally abnormal BRCA1 variants were depleted relative to wild type after growth selection. Furthermore, the dynamic range of function score distributions of the assay was sufficient to separate functionally abnormal nonsense variants from functionally normal synonymous variants, yielding clear thresholds for functionally normal and functionally abnormal variants (Figures 3A and 3B). With these thresholds, the functional data cleanly separated ClinVar pathogenic and benign variants (area under the precision-recall curve [AUCPR] = 0.97) (Figure S1). Thus, we used the published BRCA1 functional classifications for this study.
Table 1.
Functionally abnormal variant selection
| Gene | Number of variants tested | Cell type | Functionally abnormal variant selection | Selection | Functional relevance | Readout | Reference |
|---|---|---|---|---|---|---|---|
| BRCA1 | 3,893 | HAP1 | negative | cell growth | BRCA1 is essential for survival | sequencing of variants, enrichment after growth | Findlay et al., 2018,12 PMID: 30209399 |
| TP53 | 8,258 | A549 | negative | etoposide toxicity | etoposide is more toxic to cells with TP53 loss-of-function mutations | sequencing of variants, enrichment after growth | Giacomelli et al., 2019,13 PMID: 30224644 |
| TP53 | 8,258 | A549 | positive | nutlin-3 toxicity | nutlin-3 is less toxic to cells with TP53 loss-of-function mutations | sequencing of variants, enrichment after growth | Giacomelli et al., 2019,13 PMID: 30224644 |
| TP53 | 8,258 | A549 | positive | nutlin-3 toxicity | nutlin-3 is less toxic to cells with dominant negative TP53 mutations | sequencing of variants, enrichment after growth | Giacomelli et al., 2019,13 PMID: 30224644 |
| TP53 | 8,258 | MOLM13 | negative | nutlin-3 toxicity | nutlin-3 is less toxic to cells with dominant negative TP53 mutations | sequencing of variants, binned by P21-GFP intensity | Boettcher et al., 2019,14 PMID: 31395785 |
| PTEN | 4,112 | HEK293 | negative | cell growth | PTEN-GFP fusion protein abundance | sequencing of variant barcodes, binned by PTEN-GFP intensity | Matreyek et al., 2018,15 PMID: 29785012 |
| PTEN | 7,244 | YPH-499 | negative | PI3K toxicity | human PI3K is toxic to yeast in absence of functional human PTEN | sequencing of variants, enrichment after growth | Mighell et al., 2018,16 PMID: 29706350 |
Positive indicates cells harboring functionally abnormal variants have prolonged survival upon functional selection compared to wild type. Negative indicates cells harboring functionally abnormal variants are more rapidly depleted than wild type upon functional selection.
Figure 3.
Function scores for BRCA1, TP53, and PTEN variants of known effect
Histograms of function scores for variants colored by their ClinVar interpretations for each multiplexed functional assay in the left column and nonsense and synonymous variant distributions in the right column.
(A and B) Function scores for BRCA1 derived from saturation genome editing in a BRCA1-deficient HAP1 cell line.
(C–J) Function scores for TP53 derived from four different assays. From top to bottom: TP53-null A549 cell line with positive selection for loss-of-function variants with etoposide. TP53-null A549 cell line with negative selection for loss-of-function variants with nutlin-3. TP53-wild-type A549 cell line with positive selection for dominant negative variants with nutlin-3. TP53-wild-type AML reporter cell line with positive selection for dominant negative variants with nutlin-3.
(K–N) Function scores for PTEN derived from two different assays. From top to bottom: PTEN variant abundance assayed in a HEK293 cell line and PTEN variant phosphatase activity in a humanized yeast system. Histogram color indicates known clinical effect as reported in the ClinVar database (dark blue, benign; light blue, likely benign; light red, likely pathogenic; dark red, pathogenic).
To determine the strength of evidence applied to BRCA1 multiplexed functional data for variant interpretation, we calculated the OddsPath by using all clinically derived pathogenic/likely pathogenic (n = 209) and benign/likely benign (n = 163) variants from ClinVar as BRCA1 control variants. The multiplexed functional data correctly assigned 198 of the 209 pathogenic/likely pathogenic controls to the functionally abnormal class and 159 of the 163 benign/likely benign controls to the functionally normal class, resulting in an OddsPath of 52.4 and 0.02, respectively (Tables S1 and S2). These OddsPath values correspond to strong evidence for pathogenic assessment (PS3) of functionally abnormal variants and strong evidence for benign assessment (BS3) of functionally normal variants.22 Thus, we applied PS3 and BS3 evidence codes to BRCA1 variants with functionally abnormal and normal scores, respectively.
TP53 multiplexed functional data curation
We chose to explore four existing MAVEs generated to interrogate TP53’s multiple functions. Here, multiplexed functional data for 8,258 SNVs of TP53 was generated with two assays that queried loss-of-function variants and two that queried dominant negative variants.13,14 Although these TP53 functional datasets enabled powerful dissections of the molecular mechanisms of TP53 variant effect, they do not individually meet recommendations for multiplexed functional data clinical integration. In particular, while each assay was conducted in relevant human cell lines and was designed with readouts directly linked to multiple molecular consequences of TP53 variant pathogenesis, none cleanly separated nonsense and synonymous variants nor known pathogenic from known benign variants (Figures 3C–3J). Because of the complex landscape of TP53 functional effects, we could not make accurate predictions of variant functional effect for clinical variant interpretation based on any single assay. Therefore, we trained a classifier to predict variant function. Because the function score generated for each variant in each assay represents an independent data point, we used a Gaussian naive Bayes classifier to make predictions of functional effect for each variant by using their scores from all four assays without transformation. We trained the classifier on the 161 TP53 missense variants from ClinVar (129 pathogenic/likely pathogenic and 32 benign/likely benign) (Table S3) and assessed accuracy of predictions with leave-one-out cross-validation (accuracy = 96%, AUCPR = 0.92). When compared with classifiers trained with function scores from any single assay, the classifier using scores from all four assays performed with greater accuracy (Figure S2). Thus, we used the naive Bayes classifier trained on all four assays to generate predictions of variant functional effect for the 7,893 TP53 variants with function scores in each of the four assays (Table S4).
We determined the strength of evidence yielded by our TP53 classifier’s functional predictions based on cross-validation performance. The overall accuracy of the predictions was 96%, and all seven misclassified variants were likely pathogenic variants that were classified as functionally normal (Tables S4 and S5). This corresponds to an OddsPath of 30.3 for variants predicted to be functionally abnormal and 0.054 for variants predicted to be functionally normal (Table S1). Thus, we applied strong functional evidence (PS3) to the variants predicted to be functionally abnormal and moderate functional evidence (BS3_M) to the variants predicted to be functionally normal.22 Although the classifier performs with high overall accuracy, some clinically interpreted pathogenic/likely pathogenic variants cannot be detected by the multiplexed assays by which it was trained. Four of these incorrectly classified variants retain partial function in other functional assays, which may lead to the functionally normal output in the human cell line overexpression systems used in the TP53 MAVEs (GenBank: NM_000546.5 [TP53], c.1000G>T [p.Gly334Trp]; GenBank: NM_000546.6 [TP53], c.579T>A [p.His193Gln]; GenBank: NM_000546.6 [TP53], c.542G>A [p.Arg181His]; GenBank: NM_000546.6 [TP53], c.1010G>A [p.Arg337His]).31, 32, 33
PTEN multiplexed functional data curation
Finally, we explored the existing MAVEs for PTEN. Effects of 8,088 SNVs were measured with two assays: one for variant effects on protein abundance and the another for variant effects on PTEN phosphatase activity levels.15,16 Both protein abundance and phosphatase activity scores for PTEN variants were direct measures of variant effect. The dynamic range of both assays are sufficient to separate nonsense variants from synonymous variants. However, the sensitivity of both assays is only 0.69 for abundance and 0.71 for activity, respectively, when validated against established pathogenic variants in ClinVar (Figures 3K–3N). Due to the low sensitivity for pathogenic variants of each assay and the dearth of benign variants to calculate OddsPath to determine strength of evidence, the multiplexed functional data do not provide any evidence toward pathogenicity and are capped at BS3 moderate for the phosphatase activity assay and BS3 supporting for the abundance assay (Tables S1 and S6). Although combining the two datasets with a probabilistic classifier might improve the sensitivity, specificity, and strength of evidence, there are too few benign missense PTEN variants (n = 2) available for training.
Variant reinterpretation
To understand the impact of adding evidence acquired from multiplexed functional data to the reinterpretation of VUSs, we gathered all available evidence for the 324 VUSs identified in BRCA1, TP53, and PTEN by a single diagnostic testing laboratory that overlapped these datasets. We then reinterpreted these VUSs by using multiple established variant interpretation guidelines to ensure that our conclusions were independent of the approach taken. First, we reinterpreted variants by using guidelines either from the ACMG/AMP3 or from ClinGen VCEPs.17,26 We also reinterpreted variants by using a Bayesian adaptation of the ACMG/AMP guidelines.25 This strategy differs from original, rules-based ACMG/AMP guidelines because the final interpretation is made on the basis of a posterior probability that a variant is pathogenic after combining all evidence in a quantitative framework instead of using evidence codes.
BRCA1 variant reinterpretation
We obtained data for 286 BRCA1 single-nucleotide variants identified in clinical multigene cancer panels from 6,490 individuals that have multiplexed functional data. These variants were classified by a single diagnostic laboratory (Ambry Genetics) as pathogenic (n = 56), likely pathogenic (n = 44), VUS (n = 110), likely benign (n = 16), and benign (n = 60). Of these, 93% were scored as functionally normal or abnormal in the multiplexed functional assay (functionally normal n = 156, functionally abnormal n = 109). The remaining 7% scored in the intermediate range (n = 21) of the assay between the thresholds defining functionally normally and abnormal variants.12 The clinical interpretations were highly concordant with the multiplexed functional data: 54 of the 56 pathogenic variants scored as functionally abnormal and 57 of the 60 benign variants scored as functionally normal. All five of the discordant pathogenic and benign variants had intermediate functional scores.
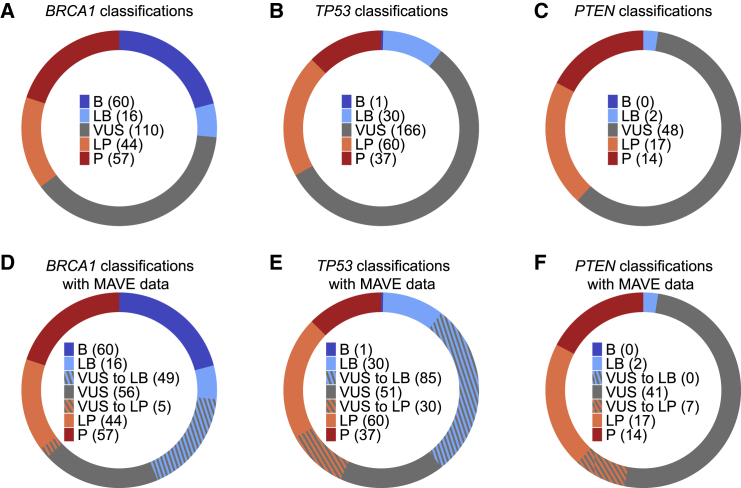
Reinterpretation of the BRCA1 VUSs from the laboratory dataset with the multiplexed functional data following the ACMG/AMP guidelines3 resulted in the reclassification of 49% (54/110) VUSs as likely pathogenic (n = 5) or likely benign (n = 49) (Figure 4A; Table S7). Of the 110 VUSs, 15 scored as functionally abnormal, 82 scored as functionally normal, and 13 scored as intermediate. In addition to the multiplexed functional data, other existing lines of evidence used to reclassify VUSs to likely pathogenic included the following: missense variant at a position where another missense variant is classified as pathogenic (PM5, n = 3), variant absent in population databases (PM2_P, n = 4), and agreement between computational predictive models (PP3, n = 3). For VUSs reclassified as likely benign, the additional evidence was agreement between computational predictive models (BP4, n = 49). In cases where variants had enough evidence to be classified as likely benign and absence from population databases was the only conflicting evidence in favor of pathogenicity, we ignored the conflicting evidence and classified these variants as likely benign (n = 26).26 We note that our reinterpretation of variants with strict adherence to the ACMG/AMP guidelines limits the allowable evidence for variant interpretation that many clinical laboratories routinely use in their interpretations including data siloed to protect private health information and other conflicting lines of evidence. Thus, we expect the application of this ACMG/AMP framework to yield a different proportion of VUSs as compared to reinterpretation of the same variants by a clinical laboratory.
Figure 4.
Reinterpretation of BRCA1, TP53, and PTEN VUSs with multiplexed functional data
(A–C) Original variant classifications from Ambry Genetics.
(D–F) Variant classifications after reinterpretation with existing evidence and multiplexed functional data. Dashed sections represent the proportion of VUSs reclassified to either likely pathogenic or likely benign.
We also reinterpreted variants following the Bayesian implementation of the ACMG/AMP guidelines and resolved 80% of the (91/110) BRCA1 VUSs with this method (likely pathogenic, n = 5; likely benign, n = 84, Table S2). The additional VUSs classified as likely benign with the Bayesian method were variants that scored as functionally normal in the multiplexed functional data but had no other evidence in favor of benign interpretation. Because the Bayesian method allows for likely benign interpretation for any variant with a posterior probability of pathogenicity < 0.1, variants with only functional evidence from an assay achieving strong functional evidence can be classified as likely benign even in the absence of other evidence types. Thus, with either the original or Bayesian adaptation of the ACMG guidelines, it is clear that the multiplexed functional data has a high impact in the clinical significance interpretation for BRCA1 variants.
TP53 variant reinterpretation
We obtained data for 294 TP53 SNVs identified in clinical multigene cancer panels from 1,828 individuals that have multiplexed functional data. These variants were classified by Ambry Genetics as pathogenic (n = 37), likely pathogenic (n = 60), VUS (n = 166), likely benign (n = 30), and benign (n = 1). Our classifier predictions were largely concordant with these clinical interpretations. Of the 49 variants in this clinical dataset that are absent from the classifier training set, all of the likely benign variants were functionally normal (n = 18), 21 of the 26 likely pathogenic variants were functionally abnormal, and all pathogenic variants were functionally abnormal (n = 5). All five of the discordant variants have conflicting interpretations in ClinVar, and one is a ClinGen expert-panel-reviewed VUS (Table S8). Two of these discordant variants (GenBank: NM_000546.5 [TP53], c.1000G>C [p.Gly334Arg]; GenBank: NM_000546.6 [TP53], c.542G>A [p.Arg181His]) have been described as reduced penetrance variants.34, 35, 36 The remaining 166 TP53 variants from the diagnostic laboratory dataset were VUSs, and our classifier predicted 120 to be functionally normal and 46 to be functionally abnormal. To determine the value of TP53 multiplexed functional data in variant interpretation, we reevaluated the VUSs with functional evidence from the classifier.
Reevaluation of TP53 VUSs following an adapted version of the ClinGen VCEP recommendations26 resulted in the reinterpretation of 69% (115/166) of VUSs to likely pathogenic (n = 30) or likely benign (n = 85) (Figure 4B, Table S9). Of the 166 TP53 VUSs, 120 were predicted to be functionally normal and 46 were predicted to be functionally abnormal by our classifier. The adapted reinterpretation strategy followed the VCEP guidelines for TP53 variant interpretation except for the functional evidence component, where we used the functional evidence corresponding only to our classifier predictions. We made this distinction to assess the impact of data from only multiplexed functional assays. In addition to the multiplexed functional data, existing lines of evidence used for reclassification of VUSs to likely pathogenic included the following: missense variant at a position where another missense variant is classified as pathogenic (PM5_P, n = 3), variant absent in population databases (PM2_P, n = 28), missense variant in a mutational hotspot (PM1, n = 9), and agreement between computational predictive models (PP3/PP3_M, n = 28). For VUSs reclassified as likely benign, additional evidence used was agreement between computational predictive models (BP4, n = 85) and variant observed in adults unaffected with cancer in population datasets (BS2_P, n = 4). We also reinterpreted variants with strict adherence to the TP53 VCEP functional evidence recommendation, which includes two of the multiplexed datasets used in our analysis, and reclassified 60% of VUSs to likely pathogenic (n = 19) or likely benign (n = 81) (Table S10).
Finally, reinterpretation of TP53 variants following the Bayesian adaptation increased the proportion of resolved VUSs to 85% (likely pathogenic [n = 30], likely benign [n = 111]). Similar to BRCA1, the majority (25 of 26) of additional VUSs reclassified as likely benign with the Bayesian approach were variants where no additional evidence could be applied. Because the posterior probability of pathogenicity was below 0.1, these variants were classified as likely benign. The lone variant with additional benign evidence (GenBank: NM_000546.5 [TP53], c.328C>A [p.Arg110Ser]) was predicted to be benign by computational predictive models but was also absent from population databases and occurs at the amino acid position of another pathogenic missense variant. These conflicting pieces of evidence result in a VUS interpretation following rules-based methods, but with the Bayesian method, this variant has a posterior probability of 0.05 and is interpreted as likely benign.
PTEN variant reinterpretation
We obtained data for 74 PTEN missense variants identified in multigene cancer panels from 1,061 individuals that had multiplexed functional data from at least one of the PTEN assays. These variants were classified by Ambry Genetics as pathogenic (n = 7), likely pathogenic (n = 17), VUS (n = 48), and likely benign (n = 2). Both likely benign variants had functionally normal scores from the activity and abundance assays. The seven pathogenic variants were assessed in the phosphatase activity assay. Four scored as low activity and three as intermediate activity. Six of the pathogenic variants were assessed in the abundance assay. Five had low abundance and one had normal abundance. 16 of the likely pathogenic variants were scored in the phosphatase activity assay, and 11 were scored as low activity, two as intermediate, and three as normal activity. Of the 13 likely pathogenic variants that were scored in the abundance assay, seven had low abundance and 6 had normal abundance. This low sensitivity of each assay for pathogenic/likely pathogenic variants from the diagnostic laboratory dataset is consistent with the assessment with variant annotations from ClinVar.
We attempted to reinterpret PTEN VUSs with MAVE data following the ClinGen SVI recommendations and were unable to reclassify any of them because of limited strength of evidence (Tables S1 and S11). For this reason, we employed guidelines developed by the PTEN VCEP for clinical integration of these datasets.17 Here, variants deemed functionally abnormal in the phosphatase activity assay receive PS3 strong evidence. Variants deemed functionally abnormal in the multiplexed protein abundance assay receive PS3 supporting evidence. Variants deemed functionally normal in both assays receive BS3 strong evidence. Following the VCEP recommendations17 with functional evidence restricted to multiplexed functional data resulted in reclassification of 15% of VUSs as likely pathogenic (n = 7) (Figure 4C, Table S12). From the multiplexed functional data, we assigned strong functional evidence for pathogenicity (PS3) to eight of the VUSs, supporting functional evidence for pathogenicity (PS3_P) to five VUSs, and strong functional evidence for benign effect (BS3) to 22 VUSs. In addition to the multiplexed functional data, existing lines of evidence applied to VUSs that were reclassified to likely pathogenic include missense variant in a gene with low rate of benign missense variation (PP2, n = 7), absence in population databases (PM2, n = 7), and missense variant at a position where another missense variant is classified as pathogenic (PM5, n = 3). None of the VUSs with BS3 evidence could be reclassified to likely benign following PTEN VCEP guidelines for two main reasons: (1) there is no consensus for use of in silico predictive models for PTEN missense variants because of the lack of benign control missense variants and (2) these VUSs are present at too low population frequency for BS1 strong evidence. Finally, we note that the PTEN VCEP guidelines predate the updated ClinGen SVI recommendations for the use of functional data, and thus, may not meet current standards for strength of evidence attributable to functional data.
Discussion
Multiplexed functional data is appearing rapidly, but has, so far, not been rigorously evaluated for clinical use. Here, we demonstrated that multiplexed functional data has high utility in clinical variant interpretation by using functional evidence for 49,281 variants derived from MAVEs to reevaluate 324 VUSs in three important cancer predisposition genes, BRCA1, TP53, and PTEN, from 774 individuals. Overall, the multiplexed functional data enabled reclassification of 176 (54%) of the 324 VUSs and had considerable differences in effectiveness across the genes.
Reinterpretation of BRCA1 VUSs with multiplexed functional data was straightforward because a single functional dataset closely adhered to guidelines for multiplexed functional assay design, the data had high sensitivity and specificity for control variants, and a large number of control variants were available for computing the strength of evidence. These factors allowed PS3 strong and BS3 strong levels of evidence to be used for reinterpretation, resulting in the reclassification of 49% of VUSs.
Reinterpretation of TP53 VUSs was complicated by the existence of four distinct multiplexed functional datasets, and no single dataset had a broad enough dynamic range or sufficient sensitivity and specificity to be used alone. We combined these datasets with a naive Bayes classifier, which was possible because TP53 has a large number of established benign and pathogenic variants to use for classifier training. Thus, multiple MAVE datasets can be integrated to produce a single, accurate functional prediction that can be used for variant reinterpretation. This approach is particularly important when each functional assay lacks the requisite dynamic range and sensitivity/specificity alone or when variants in a gene, such as TP53, have multiple mechanisms of pathogenicity that cannot be probed in a single assay. Ultimately, our machine-learning approach, in combination with the large number of available control variants for TP53, allowed strong pathogenic (PS3) and moderate benign (BS3_M) levels of functional evidence to be used for reclassification of 69% of VUSs.
Reinterpretation of PTEN VUSs highlighted other limitations of multiplexed functional data. Here, lack of benign control variants constrained assay validation, preventing the use of machine learning for combination of functional data from multiple assays and also meaningful assessment of the strength of evidence. In addition, this lack of benign control variants inhibited the utility of in silico predictors, an important contributor to variant interpretations. Nonetheless, despite these challenges, functional evidence enabled reclassification of 15% of VUSs with application of VCEP recommendations for PTEN variant interpretation. This analysis highlights the important role of ClinGen VCEPs in developing guidelines for functional data integration for genes where control variants are limited, as we could not reclassify any PTEN VUSs following the generalized ClinGen SVI recommendations for use of functional data.22
By analyzing multiple genes and functional datasets, we reveal the key characteristics of each assay and gene that dictate the utility of multiplexed functional data for variant interpretation. Key assay characteristics are the dynamic range of function scores separating functionally normal from abnormal variants, and the predictive value of the assay for correctly identifying known pathogenic and benign variants used as controls. The key gene characteristic is the availability of pathogenic and benign control variants for assay validation. As for PTEN, lack of control variants can severely constrain the strength of evidence arising from functional assays regardless of assay performance.
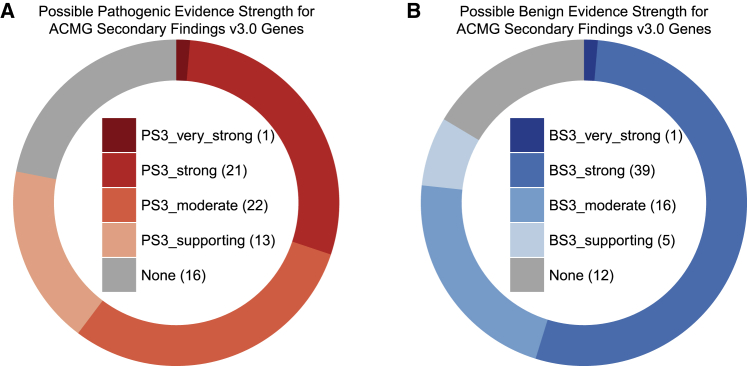
The MAVE community is making progress in developing assays with high dynamic range and high predictive value.37, 38, 39, 40 However, addressing the lack of benign and pathogenic control variants for validation is more challenging. A perfect assay must have at least 19 control benign and pathogenic variants for validation to achieve strong benign and pathogenic functional evidence, whereas at least 11 benign and pathogenic variants are required for an assay to achieve moderate evidence.22 Of the 73 clinically actionable genes on the ACMG Secondary Findings v3.0 list,10 only 23 genes have a sufficient number of control missense variants for a hypothetical MAVE that perfectly distinguishes the pathogenic and benign variants to achieve strong functional evidence (PS3) for interpretation of functionally abnormal variants (Figure 5B, Table S13). The 50 genes that do not reach this threshold are limited by the number of control benign variants available for assay validation or lack established pathogenic missense variants. In addition, just 40 of these genes have sufficient control missense variants for a hypothetical MAVE that perfectly distinguishes pathogenic and benign variants to achieve strong evidence for benign interpretation (BS3) of functionally normal variants (Figure 5A). Thus, most genes lack the control variants required to deploy variant functional data as strong evidence, and closing this control variant gap will likely require active efforts to generate control variants and changes to variant interpretation practices. For genes such as PTEN where multiplexed functional data is available but benign control variants are lacking, expert panels could coordinate review of VUSs and variants discovered in population sequencing studies. Data generators and clinicians should work together to design, pilot, and validate MAVEs as well as to integrate the resulting functional data into interpretation workflows in order to maximize the potential of these powerful data.21,40 Ultimately, a confluence of high-quality multiplexed functional datasets, large numbers of control variants, and reassessment of interpretation guidelines could transform our ability to definitively interpret VUSs.
Figure 5.
Strength of evidence that could be assigned to variants of ACMG Secondary Findings v3.0 genes with hypothetical MAVEs that perfectly distinguish between pathogenic and benign controls
Acknowledgments
We thank Moez Dawood for critical reading of this manuscript and Martha Horike-Pyne for assistance with human subjects. This work was supported by 5T32HG000035 fellowship from the NIH NHGRI administered by UW Genome Sciences to S.F., a Catalytic Collaboration award from the Brotman Baty Institute to D.M.F., 5U01CA242954 from the NIH NCI to L.M.S., and 5RM1HG010461 from the NHGRI to D.M.F. and L.M.S. This research benefitted by support from the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support.
Declaration of interests
J.N.D. is an employee of Adaptive. M.E.R., K.M., F.H., T.P., and R.K. are employees of Ambry Genetics. The remaining authors declare no competing interests.
Published: November 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.11.001.
Contributor Information
Douglas M. Fowler, Email: dfowler@uw.edu.
Lea M. Starita, Email: lstarita@uw.edu.
Data and code availability
Code to reproduce the analysis and regenerate all figures is available on GitHub at https://github.com/bbi-lab/VUS-reinterpretation-with-MAVE.
Web resources
OMIM, http://www.omim.org
Supplemental information
References
- 1.Maxwell K.N., Hart S.N., Vijai J., Schrader K.A., Slavin T.P., Thomas T., Wubbenhorst B., Ravichandran V., Moore R.M., Hu C., et al. Evaluation of ACMG-Guideline-Based Variant Classification of Cancer Susceptibility and Non-Cancer-Associated Genes in Families Affected by Breast Cancer. Am. J. Hum. Genet. 2016;98:801–817. doi: 10.1016/j.ajhg.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaDuca H., Polley E.C., Yussuf A., Hoang L., Gutierrez S., Hart S.N., Yadav S., Hu C., Na J., Goldgar D.E., et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 2020;22:407–415. doi: 10.1038/s41436-019-0633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurian A.W., Sigal B.M., Plevritis S.K. Survival analysis of cancer risk reduction strategies for BRCA1/2 mutation carriers. J. Clin. Oncol. 2010;28:222–231. doi: 10.1200/JCO.2009.22.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villani A., Tabori U., Schiffman J., Shlien A., Beyene J., Druker H., Novokmet A., Finlay J., Malkin D. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol. 2011;12:559–567. doi: 10.1016/S1470-2045(11)70119-X. [DOI] [PubMed] [Google Scholar]
- 7.Tan M.-H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makhnoon S., Shirts B.H., Bowen D.J. Patients’ perspectives of variants of uncertain significance and strategies for uncertainty management. J. Genet. Couns. 2019;28:313–325. doi: 10.1002/jgc4.1075. [DOI] [PubMed] [Google Scholar]
- 9.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller D.T., Lee K., Chung W.K., Gordon A.S., Herman G.E., Klein T.E., Stewart D.R., Amendola L.M., Adelman K., Bale S.J., et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:1381–1390. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 11.Brnich S.E., Rivera-Muñoz E.A., Berg J.S. Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks. Hum. Mutat. 2018;39:1531–1541. doi: 10.1002/humu.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacomelli A.O., Yang X., Lintner R.E., McFarland J.M., Duby M., Kim J., Howard T.P., Takeda D.Y., Ly S.H., Kim E., et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat. Genet. 2018;50:1381–1387. doi: 10.1038/s41588-018-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boettcher S., Miller P.G., Sharma R., McConkey M., Leventhal M., Krivtsov A.V., Giacomelli A.O., Wong W., Kim J., Chao S., et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365:599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J., et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mighell T.L., Evans-Dutson S., O’Roak B.J. A Saturation Mutagenesis Approach to Understanding PTEN Lipid Phosphatase Activity and Genotype-Phenotype Relationships. Am. J. Hum. Genet. 2018;102:943–955. doi: 10.1016/j.ajhg.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mester J.L., Ghosh R., Pesaran T., Huether R., Karam R., Hruska K.S., Costa H.A., Lachlan K., Ngeow J., Barnholtz-Sloan J., et al. Gene-specific criteria for PTEN variant curation: Recommendations from the ClinGen PTEN Expert Panel. Hum. Mutat. 2018;39:1581–1592. doi: 10.1002/humu.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mighell T.L., Thacker S., Fombonne E., Eng C., O’Roak B.J. An Integrated Deep-Mutational-Scanning Approach Provides Clinical Insights on PTEN Genotype-Phenotype Relationships. Am. J. Hum. Genet. 2020;106:818–829. doi: 10.1016/j.ajhg.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasperini M., Starita L., Shendure J. The power of multiplexed functional analysis of genetic variants. Nat. Protoc. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant Interpretation: Functional Assays to the Rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelman H., Dines J.N., Berg J., Berger A.H., Brnich S., Hisama F.M., James R.G., Rubin A.F., Shendure J., Shirts B., et al. Recommendations for the collection and use of multiplexed functional data for clinical variant interpretation. Genome Med. 2019;11:85. doi: 10.1186/s13073-019-0698-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brnich S.E., Abou Tayoun A.N., Couch F.J., Cutting G.R., Greenblatt M.S., Heinen C.D., Kanavy D.M., Luo X., McNulty S.M., Starita L.M., et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pesaran T., Karam R., Huether R., Li S., Farber-Katz S., Chamberlin A., Chong H., LaDuca H., Elliott A. Beyond DNA: An Integrated and Functional Approach for Classifying Germline Variants in Breast Cancer Genes. Int. J. Breast Cancer. 2016;2016:2469523. doi: 10.1155/2016/2469523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 25.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018;20:1054–1060. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortuno C., Lee K., Olivier M., Pesaran T., Mai P.L., de Andrade K.C., Attardi L.D., Crowley S., Evans D.G., Feng B.J., et al. Specifications of the ACMG/AMP variant interpretation guidelines for germline TP53 variants. Hum. Mutat. 2021;42:223–236. doi: 10.1002/humu.24152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng B.-J. PERCH: A Unified Framework for Disease Gene Prioritization. Hum. Mutat. 2017;38:243–251. doi: 10.1002/humu.23158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y., Pesaran T., Chamberlin A., Fenwick R.B., Li S., Gau C.-L., Chao E.C., Lu H.-M., Black M.H., Qian D. REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci. Rep. 2019;9:12752. doi: 10.1038/s41598-019-49224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiGiammarino E.L., Lee A.S., Cadwell C., Zhang W., Bothner B., Ribeiro R.C., Zambetti G., Kriwacki R.W. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat. Struct. Biol. 2002;9:12–16. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 32.Kato S., Han S.-Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malcikova J., Tichy B., Damborsky J., Kabathova J., Trbusek M., Mayer J., Pospisilova S. Analysis of the DNA-binding activity of p53 mutants using functional protein microarrays and its relationship to transcriptional activation. Biol. Chem. 2010;391:197–205. doi: 10.1515/bc.2010.027. [DOI] [PubMed] [Google Scholar]
- 34.Powers J., Pinto E.M., Barnoud T., Leung J.C., Martynyuk T., Kossenkov A.V., Philips A.H., Desai H., Hausler R., Kelly G., et al. A Rare TP53 Mutation Predominant in Ashkenazi Jews Confers Risk of Multiple Cancers. Cancer Res. 2020;80:3732–3744. doi: 10.1158/0008-5472.CAN-20-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zick A., Kadouri L., Cohen S., Frohlinger M., Hamburger T., Zvi N., Plaser M., Avital E., Breuier S., Elian F., et al. Recurrent TP53 missense mutation in cancer patients of Arab descent. Fam. Cancer. 2017;16:295–301. doi: 10.1007/s10689-016-9951-z. [DOI] [PubMed] [Google Scholar]
- 36.Lolas Hamameh S., Renbaum P., Kamal L., Dweik D., Salahat M., Jaraysa T., Abu Rayyan A., Casadei S., Mandell J.B., Gulsuner S., et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int. J. Cancer. 2017;141:750–756. doi: 10.1002/ijc.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Da Kuang W., Weile J., Kishore N., Rubin A.F., Fields S., Fowler D.M., Roth F.P. MaveRegistry: a collaboration platform for multiplexed assays of variant effect. Bioinformatics. 2021:btab215. doi: 10.1093/bioinformatics/btab215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuang D., Weile J., Li R., Ouellette T.W., Barber J.A., Roth F.P. MaveQuest: a web resource for planning experimental tests of human variant effects. Bioinformatics. 2020;36:3938–3940. doi: 10.1093/bioinformatics/btaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esposito D., Weile J., Shendure J., Starita L.M., Papenfuss A.T., Roth F.P., Fowler D.M., Rubin A.F. MaveDB: an open-source platform to distribute and interpret data from multiplexed assays of variant effect. Genome Biol. 2019;20:223. doi: 10.1186/s13059-019-1845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.AVE Alliance Founding Members . 2021. The Atlas of Variant Effects (AVE) Alliance: understanding genetic variation at nucleotide resolution. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code to reproduce the analysis and regenerate all figures is available on GitHub at https://github.com/bbi-lab/VUS-reinterpretation-with-MAVE.