Abstract
Background
Autophagosome, the central organelle in autophagy process, can assemble via canonical pathway mediated by LC3-II, the lipidated form of autophagy-related protein LC3/ATG8, or noncanonical pathway mediated by the small GTPase Rab9. Canonical autophagy is essential for exocrine pancreas homeostasis, and its disordering initiates and drives pancreatitis. The involvement of noncanonical autophagy has not been explored. We examine the role of Rab9 in pancreatic autophagy and pancreatitis severity.
Methods
We measured the effect of Rab9 on parameters of autophagy and pancreatitis responses using transgenic mice overexpressing Rab9 (Rab9TG) and adenoviral transduction of acinar cells. Effect of canonical autophagy on Rab9 was assessed in ATG5-deficient acinar cells.
Results
Pancreatic levels of Rab9 and its membrane-bound (active) form decreased in rodent pancreatitis models and in human disease. Rab9 overexpression stimulated noncanonical and inhibited canonical/LC3-mediated autophagosome formation in acinar cells through up-regulation of ATG4B, the cysteine protease that delipidates LC3-II. Conversely, ATG5 deficiency caused Rab9 increase in acinar cells. Inhibition of canonical autophagy in Rab9TG pancreas was associated with accumulation of Rab9-positive vacuoles containing markers of mitochondria, protein aggregates, and trans-Golgi. The shift to the noncanonical pathway caused pancreatitis-like damage in acinar cells and aggravated experimental pancreatitis.
Conclusions
The results show that Rab9 regulates pancreatic autophagy and indicate a mutually antagonistic relationship between the canonical/LC3-mediated and noncanonical/Rab9-mediated autophagy pathways in pancreatitis. Noncanonical autophagy fails to substitute for its canonical counterpart in protecting against pancreatitis. Thus, Rab9 decrease in experimental and human pancreatitis is a protective response to sustain canonical autophagy and alleviate disease severity.
Keywords: Autophagosome, Alternative Autophagy, Rab GTPase, RabGDI
Abbreviations used in this paper: ANOVA, analysis of variance; AP, acute pancreatitis; Arg-AP, L-arginine–induced acute pancreatitis; ATG, autophagy-related (proteins); Cat, cathepsin; CCK, cholecystokinin-8; CER, cerulein (ortholog of CCK); ER, endoplasmic reticulum; GTPase, guanosine triphosphatase; IB, immunoblot; IF, immunofluorescence; LC3, microtubule-associated protein 1 light chain 3; RabGDI, Rab guanosine dissociation inhibitor; Rab9TG, transgenic mice overexpressing Rab9; SEM, standard error of the mean; WT, wild-type
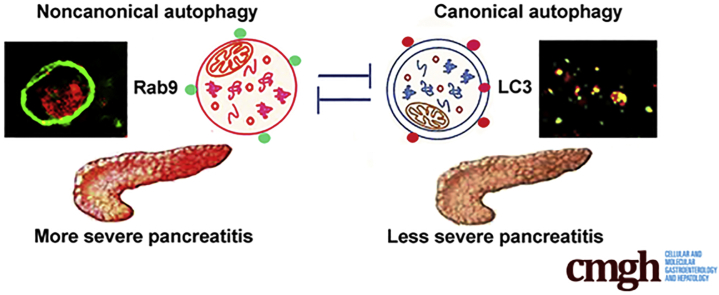
Graphical abstract
Summary.
Autophagy is essential for pancreas homeostasis. Autophagosome, key organelle in this process, forms via canonical or noncanonical pathways. We show that the small GTPase Rab9 inhibits canonical and promotes noncanonical autophagy in pancreas; Rab9 overexpression causes organ damage and worsens pancreatitis.
The pathogenic mechanism of pancreatitis, a common and potentially fatal disease of the exocrine pancreas, is incompletely understood, and no specific/effective treatment is available.1,2 The disease is believed to initiate in injured acinar cells. Because of the general lack of access to human tissue, most studies use animal or ex vivo models (on acinar cells subjected to pancreatitis stressors) to elucidate the mechanism of pancreatitis.3 These models reproduce pathologic responses of human disease and the spectrum of its severity. Major pancreatitis responses include increased serum levels of digestive enzymes (ie, hyperamylasemia), inappropriate/intra-acinar trypsinogen activation (its conversion to trypsin), acinar cell vacuolization and death, and inflammation. Recent studies have revealed that macroautophagy (herein, autophagy) plays a critical role in acinar cell homeostasis, and its disruption mediates the development of pancreatitis.4, 5, 6, 7, 8, 9
Autophagy is a principal degradative, lysosome-driven mechanism that eliminates damaged or unneeded cellular components and transports degradation products, such as amino acids and fatty acids, back to the cytoplasm to re-enter cellular metabolism.4,9, 10, 11, 12, 13 Autophagy mediates key cellular homeostatic functions, acting as an organellar “quality control” system, and underlies adaptive cell response to a variety of stress conditions. Autophagy can remove cytoplasmic components in a nonselective manner, as with starvation; or selectively degrade damaged/dysfunctional mitochondria (mitophagy), endoplasmic reticulum (ER), and other organelles, or ubiquitinated protein aggregates (aggrephagy). Selective autophagy requires adaptor molecules that deliver cargo to the autophagic machinery; for example, the protein p62/SQSTM1 (sequestosome 1) mediates the removal of toxic protein aggregates.10,12,13
Autophagosome is the central organelle in the autophagy process; it sequesters biological material destined for degradation and then fuses with late endosomes and lysosomes, forming autolysosomes in which lysosomal hydrolases degrade cargo.4,10, 11, 12, 13 Two major pathways of autophagosome formation have been discerned.11, 12, 13, 14, 15, 16 In the canonical pathway, autophagosomes are built through sequential recruitment of several complexes involving evolutionary conserved autophagy-related (ATG) proteins, such as the ATG5-ATG12-ATG16 complex that mediates nascent autophagosomal membrane elongation. The final step in canonical autophagosome formation is the conversion of microtubule-associated protein 1 light chain 3 (LC3) (ATG8) from its soluble/cytosolic LC3-I form to the lipidated, membrane-bound LC3-II, which is critical for autophagosome closure.11,12,16 Recently, noncanonical (alternative) autophagy has been discovered that uses a different mechanism of autophagosome formation.14, 15, 16 The “noncanonical” autophagosomes could be built without the hierarchical recruitment of ATG protein complexes; instead, their formation is mediated by the small guanosine triphosphatase (GTPase) Rab9.14 Importantly, in both canonical and noncanonical pathways, cargos are enclosed by autophagosomes and are degraded in autolysosomes by lysosomal lytic enzymes. Rab GTPases are major regulators of membrane transport, particularly post-Golgi trafficking and endocytosis.17,18 Some Rab family members, ie, Rab5, Rab7, Rab11, are involved in the formation of canonical autophagosomes.19, 20, 21, 22 The effects of Rab9 on canonical autophagy have not been examined, whereas its mediatory role in noncanonical autophagy is well-established.14,23,24
LC3-mediated autophagosome formation is stimulated in pancreatitis, but autophagic degradation is impaired in both experimental and human pancreatitis.4, 5, 6,9,25,26 Furthermore, pancreas-specific genetic ablation of ATG5 or ATG7, blocking the canonical autophagy pathway, triggers spontaneous pancreatitis.7,8 These data demonstrate the essential role of canonical autophagy in maintaining exocrine pancreas homeostasis and implicate its impairment in pancreatitis initiation and progression.4,9,27 By contrast, the role of Rab9 in pancreatic autophagy, basal pancreas homeostasis, and the responses and severity of pancreatitis has not been explored. In general, specific physiological or pathophysiological functions of noncanonical, Rab9-mediated autophagy are largely unknown. There is little known on the interrelationship between the 2 autophagy pathways, not only in the pancreas but in general.
Here, we show that Rab9 regulates the pattern of autophagy in pancreas. Rab9 overexpression (using transgenic mice overexpressing Rab9 [Rab9TG]28 or adenoviral transduction of acinar cells) inhibited LC3-mediated and activated Rab9-mediated autophagosome formation, causing autophagy shift from canonical to the noncanonical pathway. The shift to noncanonical pathway in Rab9TG mice aggravated experimental pancreatitis and elicited pancreatitis-like damage (in particular, inflammation) in control pancreas. Both wild-type (WT) experimental and human pancreatitis caused marked decreases in pancreatic Rab9, indicating a protective response to sustain canonical autophagy and alleviate disease severity. The results reveal mutually antagonistic relationship between the canonical and noncanonical pathways of autophagosome formation in pancreas.
Results
Pancreatic Levels of Membrane-Bound Rab9 Decrease in Experimental and Human Pancreatitis
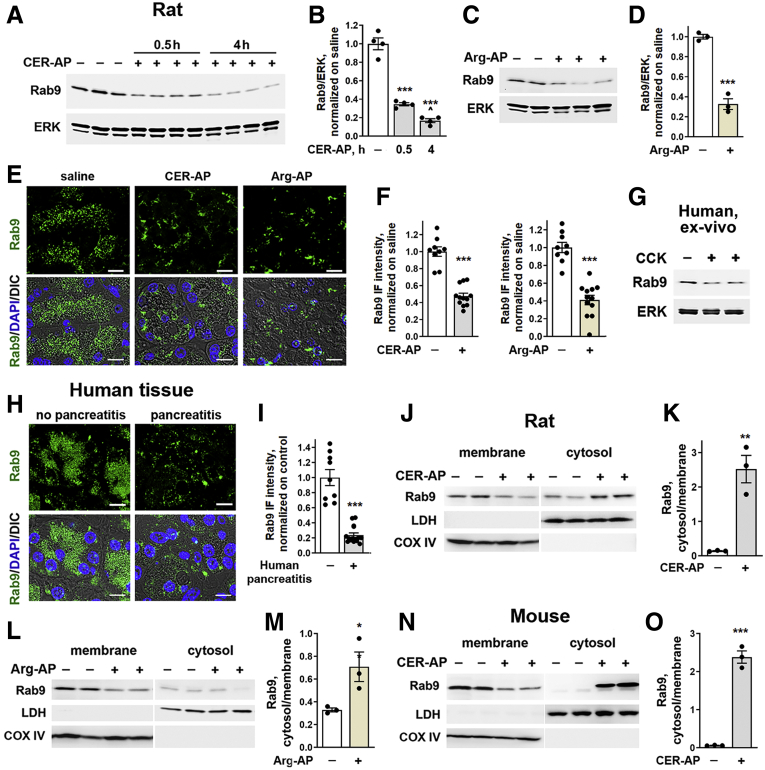
Immunoblot (IB) analysis (Figure 1A–D) of pancreas tissue homogenates showed a pronounced decrease in Rab9 in 2 dissimilar rodent models of acute pancreatitis (AP) induced with cerulein (CER-AP), an ortholog of cholecystokinin-8 (CCK), and L-arginine (Arg-AP). Rab9 reduction was prominent at 30 minutes after the start of CER injections and was sustained in full-blown pancreatitis (Figure 1A and B). Correspondingly, Rab9 levels decreased in the ex vivo pancreatitis models on human (Figure 1G) and mouse (Figure 2E and F) acinar cells at 0.5-to 1-hour incubation with supramaximal (100 nmol/L) CCK. These data indicate that Rab9 reduction is an early pancreatitis event on the time scale of trypsinogen and nuclear factor kappa B activation.9,25,26
Figure 1.
Pancreatic Rab9 levels decrease in rodent models and human pancreatitis. Rab9 levels were measured by IB in the whole tissue (A–D) and pancreatic membrane and cytosolic fractions of (J–M) rats and (N and O) mice subjected to CER-AP (4 hours in rats and 7 hour in mice or for indicated times) and Arg-AP (for 24 hours). In this and other figures, each lane on tissue IB represents an individual animal; ERK1/2, lactate dehydrogenase (LDH), and cyclooxygenase IV (COX IV) serve as loading controls and to validate the quality of subcellular fractionation; a narrow white space (as in J) indicates that the lanes are on the same gel but not contiguous. (B and D) Densitometric band intensities for Rab9 were normalized to ERK in the same sample and the Rab9/ERK ratios further normalized to control (saline-treated) group. (E, F, H, and I) IF analysis of Rab9 in (E and F) pancreas of rats subjected to CER-AP or Arg-AP and (H and I) human pancreas with no pancreatitis (control) and pancreatitis (from a total of 21 patients). In this and other figures, nuclei are stained with DAPI; DIC denotes differential interference contrast microscopy, prominently displaying zymogen granules area in acinar cells; scale bars are 10 μm if not stated otherwise. (F and I) IF intensity of Rab9 in rodent and human pancreatic tissue sections was quantified, normalized to the number of nuclei (DAPI) in the field, and expressed relative to control. (G) Rab9 levels were measured by IB in human pancreatic acinar cells incubated with and without 100 nmol/L CCK for 1 hour (ex vivo pancreatitis). (K, M, and O) Densitometric band intensities for Rab9 in membrane and cytosolic fractions were normalized to loading control in the same sample and presented as cytosol to membrane ratios. Values are mean ± SEM from at least 3 animals or cell preparations for each condition. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs control (saline-treated animals or human pancreas with no pancreatitis). ˆP < .01 vs 0.5-hour CER-AP (B). Significance was determined by 2-tailed Student t test or 1-way ANOVA, followed by Tukey multiple comparisons test (B).
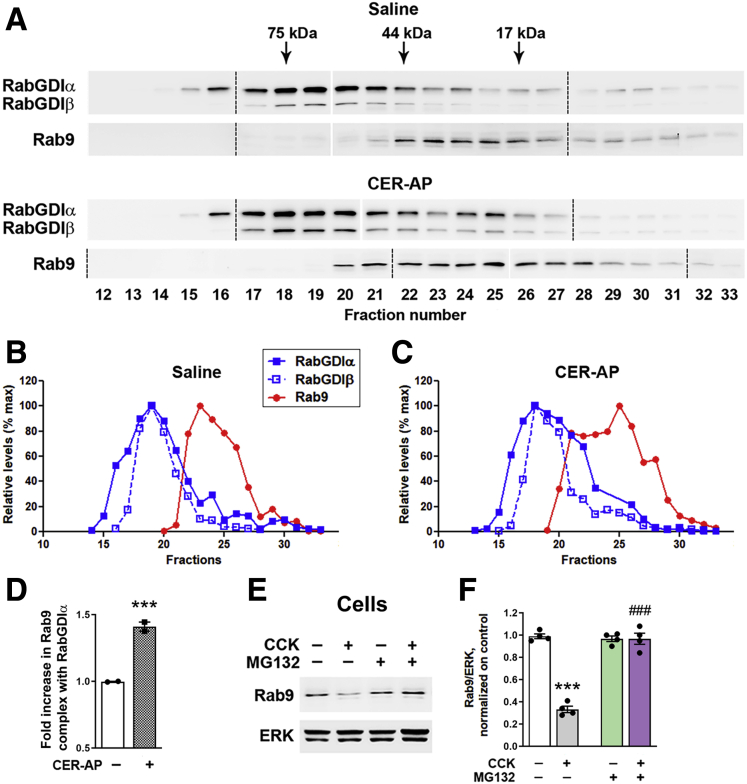
Figure 2.
Pancreatitis increases cytosolic Rab9 complex formation with RabGDI and stimulates Rab9 proteasomal degradation. (A–D) Pancreas cytosolic fractions from control animals and those subjected to CER-AP (4 hours) were resolved on a Superdex S200 gel-exclusion column using SMART system (details in Methods). Data are representative of 2 experiments on different animals, with similar results. (A) Eluted fractions were analyzed by IB using antibodies against Rab9 and RabGDIα/β. Samples were run on 3 gels (see full gels in Figure 3), and all the IBs were developed together. Dashed lines denote the first lane in each gel; narrow white space indicates omitted lanes in which protein size ladder was run (see Figure 3). (B and C) Band intensities of Rab9 and RabGDI in each fraction were densitometrically quantified and presented as % of maximal for each protein. (D) Amount of Rab9 in complex with RabGDIα was quantified as a ratio of sum of Rab9 band intensities co-fractionated with RabGDIα to that of total Rab9 (in all fractions 12–33) and presented relative to saline control. In the control (saline-treated) group, the bulk of Rab9 complex with RabGDIα eluted in fractions 22–24; in CER-AP, in fractions 20–25. (E and F) Acinar cells incubated for 30 minutes with and without the proteasomal inhibitor MG132 (50 μmol/L), followed by 30-minute incubation with and without 100 nmol/L CCK. Rab9 level was measured by IB. Values are mean ± SEM from 3 cell preparations. ∗∗∗P < .001 vs control. ###P < .001 vs CCK alone (no MG132). Significance was determined by 2-tailed Student t test (D) or 1-way ANOVA, followed by Tukey multiple comparisons test (F).
Immunofluorescence (IF) analysis also showed prominent Rab9 decrease in experimental (Figure 1E and F) and, importantly, human (Figure 1H and I) pancreatitis. In particular, IF intensity of punctate Rab9 decreased >4-fold in human pancreatitis tissue.
In eukaryotic cells Rabs cycle between 2 states, the active (GTP-loaded) in membranes and inactive (guanosine diphosphate–loaded) in the cytosol.17,18,29 Active Rabs localize to specific organelles to facilitate their function (eg, Rab4 in early and Rab11 in recycling endosomes). IB analysis of pancreas membrane and cytosolic fractions (Figure 1J–O) and Rab9 immunostaining (Figure 1E, F, H, and I) both showed a decrease in membrane-bound (active) Rab9 in pancreatitis models and in human disease. In rat and mouse CER-AP the decrease in membrane-bound Rab9 was associated with an increase in its cytosolic counterpart (Figure 1J, K, N, and O). Arg-AP caused reductions in both membrane-bound and cytosolic Rab9 (Figure 1L and M). Importantly, the cytosol/membrane Rab9 ratio increased in all pancreatitis models, indicating a reduction in active Rab9.
Rab cytosol/membrane cycling is tightly regulated.17,18,29, 30, 31 Rabs are synthesized as soluble proteins; their recruitment to membranes requires geranylgeranyl post-translational modification, followed by complex formation of the prenylated Rab with Rab guanosine dissociation inhibitor (RabGDI). The latter protein delivers Rabs to and extracts them from membranes and is thus a key regulator of Rab activity.17,18,31 We analyzed the effect of pancreatitis on Rab9-RabGDI complex formation by using gel filtration of pancreatic cytosolic fraction from rats subjected to CER-AP (Figure 2A–D, Figure 3). Pancreas cytosolic proteins were resolved on a Superdex S200 gel-exclusion column using the SMART system32 as detailed in Methods; the eluted fractions were analyzed by IB with antibodies against Rab9 and the α/β isoforms of RabGDI (Figure 2A). CER-AP did not significantly change the elution profile of the RabGDIα and RabGDIβ isoforms (Figure 2A–C). In contrast, the amount of cytosolic Rab9 and its complex with RabGDI both increased in CER-AP (Figure 2A and D); the latter effect was similar for α and β isoforms. In pancreas of control/saline-treated mice the bulk of Rab9 complex with RabGDI clustered in 3 fractions (#22–24), whereas in CER-AP Rab9-RabGDI complex eluted in 6 fractions (#20–25) (Figure 2A–C). The mechanisms underlying deranged Rab9 cycling in CER-AP, namely the increases in cytosolic Rab9 and Rab9-RabGDI complex formation, remain to be determined. However, the results indicate that pancreatitis does not impair Rab9 prenylation, a prerequisite for Rab complex formation with RabGDI.30, 31, 32
Figure 3.
Full IBs used for panels (A–D) inFigure 2. Data are representative of 2 different experiments, which both gave similar results.
The levels of both membrane and cytosolic Rabs are regulated by ubiquitin-proteasomal system29,33; in particular, proteasomal degradation eliminates protein aggregates formed by Rabs that excessively accumulate in the cytosol (although not shown for Rab9). Notably, CCK-induced Rab9 decrease in ex vivo pancreatitis was entirely prevented by the proteasomal inhibitor MG132 (Figure 2E and F), implicating the above mechanism.
Rab9 Overexpression Causes Pancreas Damage and Aggravates Experimental Pancreatitis
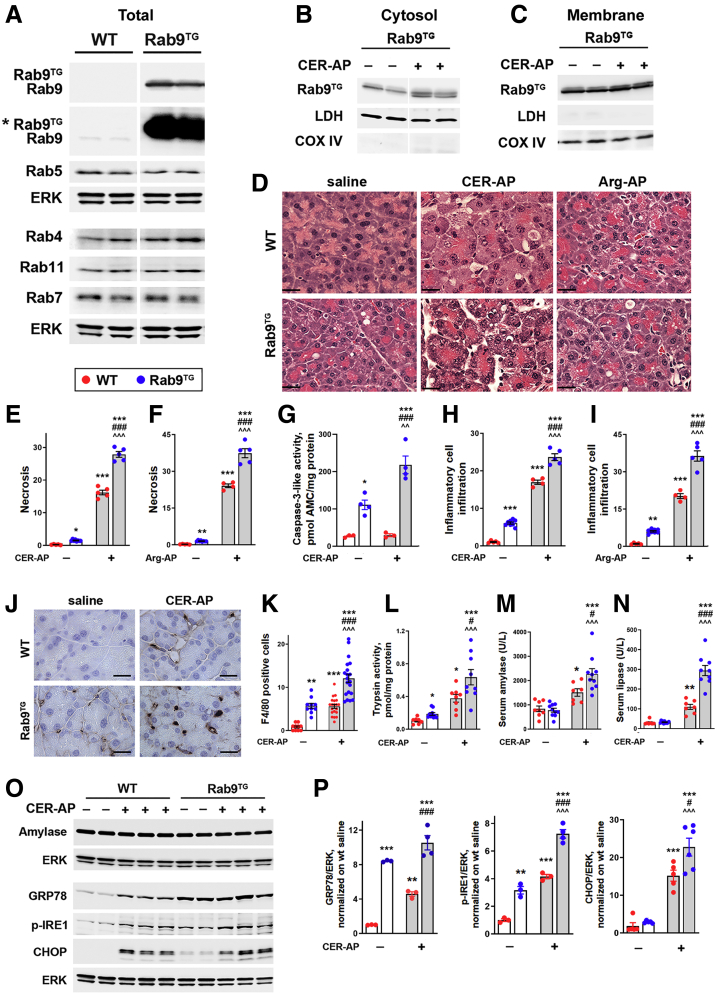
To elucidate the role of Rab9 in pancreatitis we used Rab9TG mice overexpressing Rab9.28 Whereas pancreatic Rab9 greatly increased in Rab9TG mice as compared with WT, the levels of other Rab proteins tested did not change (Figure 4A). Notably, there was no Rab9 decrease with CER-AP in Rab9TG mice (Figure 4B and C). As seen on H&E-stained pancreatic tissue sections, histopathologic alterations caused by CER-AP and Arg-AP were more severe in Rab9TG than in WT mice (Figure 4D). Rab9 overexpression increased the extent of acinar cell necrotic and apoptotic death in AP models (Figure 4E–G) (necrosis was quantified on H&E-stained tissue sections; apoptosis by measuring caspase-3-like [DEVDase] activity). Compared with WT, the inflammatory response was aggravated in Rab9TG mice with pancreatitis (Figure 4H and I), driven predominantly by macrophages as shown by immunostaining for the macrophage marker F4/80 (Figure 4J and K). Inappropriate/intrapancreatic trypsin activity, a hallmark response of pancreatitis, and serum levels of amylase and lipase, a signature diagnostic indicator of acute pancreatitis, were all significantly greater in experimental pancreatitis in Rab9TG mice (Figure 4L–N). Pancreatitis did not change the Rab9TG intrapancreatic amylase level, same as in WT (Figure 4O).
Figure 4.
Rab9 overexpression aggravates experimental pancreatitis. (A–C, O, and P) IB analysis of indicated proteins in the whole tissue (A, O, and P) and pancreatic membrane and cytosolic pancreas fractions (B and C) from WT and Rab9TG mice subjected to CER-AP (7 hours). In (A), the asterisk (∗) indicates longer exposure. (D–P) Parameters of CER-AP (7 hours) and Arg-AP (24 hours) in WT and Rab9TG mice. Histopathologic changes (D), necrosis (E and F), and inflammatory cell infiltration (H and I) were measured on H&E-stained pancreatic tissue sections; macrophage infiltration (J and K), by immunostaining for macrophage marker F4/80; intrapancreatic caspase-3-like (G) and trypsin (L) activities, with enzymatic assays. (P) Densitometric band intensities for indicated proteins were normalized to ERK in the same sample and further to those in WT control (saline-treated) group. Values are mean ± SEM from 3–10 mice per group. In (E–I and L–N), each symbol represents an individual mouse; in (K), the number of positively stained macrophages in a different high-power field (n = 10–20 fields from 3 to 5 mice per group). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs WT control (saline-treated) mice. #P < .05, ###P < .001 vs WT CER-AP. ˆˆP < .01, ˆˆˆP < .001 vs Rab9TG control (saline-treated) mice. Significance was determined by 1-way ANOVA, followed by Tukey multiple comparisons test. Scale bars, 20 μm.
Although some of pancreatitis responses, eg, CER-induced hyperlipasemia, were much stronger in Rab9TG mice (9.0-fold) than in WT (3.6-fold; Figure 4N), others were of similar or even lesser magnitude. For example, the increase in inflammatory cell infiltration induced by CER-AP or Arg-AP in Rab9TG pancreas was much less than in WT (Figure 4H and I). Such non-additive effects suggest the involvement of common/overlapping mechanisms engaged by Rab9 overexpression and experimental pancreatitis.
Of note, Rab9 overexpression itself caused pancreatitis-like damage in pancreas, which was manifested by increased basal level of necrosis and apoptosis, macrophage infiltration, and trypsinogen activation (Figure 4E–L), and acinar cell vacuolization (Figure 5). For some of these parameters, the effect size was similar to or even greater than that of CER-AP in WT mice.
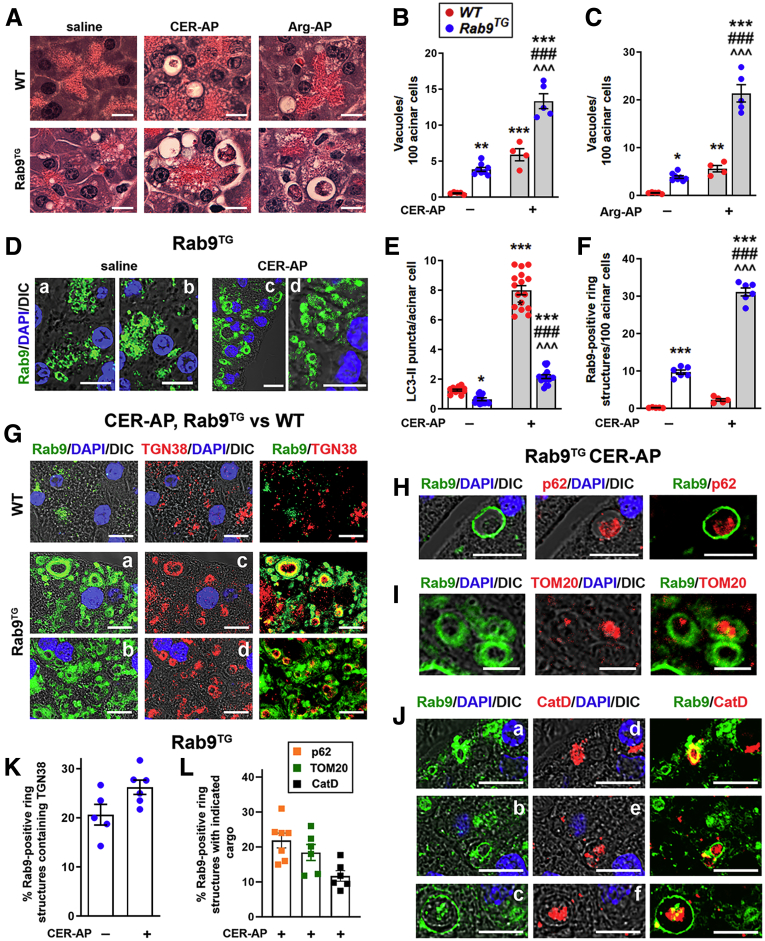
Figure 5.
Rab9 overexpression activates noncanonical autophagy in the exocrine pancreas. WT and Rab9TG mice were subjected to CER-AP (7 hours) and Arg-AP (24 hours). (A–C) Acinar cell vacuolization was quantified on H&E-stained pancreatic tissue sections. Scale bars, 20 μm. (D–L) IF analysis of indicated proteins in control pancreas (saline) and CER-AP. Scale bars, 10 μm. Number of LC3 puncta (E) was quantified from images illustrated in Figure 6F. Panels D (a-d), G (a-d), and J (a-f) show different fields displaying Rab9-positive ring structures on pancreatic tissue sections. The number of these structures (F) and the percentage of them (K and L) containing markers of trans-Golgi network (TGN38), protein aggregates (p62/SQSTM1), mitochondria (TOM20), or lysosomal proteases (CatD) were quantified from images illustrated in panels (D, G–J). Values are mean ± SEM from 3 to 8 mice per group. In (B and C), each symbol represents an individual mouse. In (E, F, K, and L), each symbol corresponds to 20–30 cells in a different field (n = 12–16 fields from at least 3 mice per group). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs WT control (saline) group. ###P < .001 vs WT CER-AP or Arg-AP. ˆˆˆP < .001 vs Rab9TG control (saline) group. Significance was determined by 1-way ANOVA, followed by Tukey or Holm-Sidak multiple comparisons test.
ER stress, a prominent feature of acute pancreatitis, mediates acinar cell damage.9,34 It was manifested in WT CER-AP by up-regulation of ER stress markers GRP78, phosphorylated (p)-IRE1, and especially CHOP, a mediator of apoptosis (Figure 4O and P). Rab9 overexpression triggered ER stress in basal conditions and worsens it in CER-AP (Figure 4O and P). The effects of Rab9TG and CER-AP on ER stress markers were non-additive (Figure 4P), indicating overlapping mechanisms of ER stress (such as the IRE1-mediated pathway) triggered by both Rab9 up-regulation and experimental pancreatitis.
Rab9 Overexpression Perturbs Canonical/LC3-Mediated Autophagy in Experimental Pancreatitis
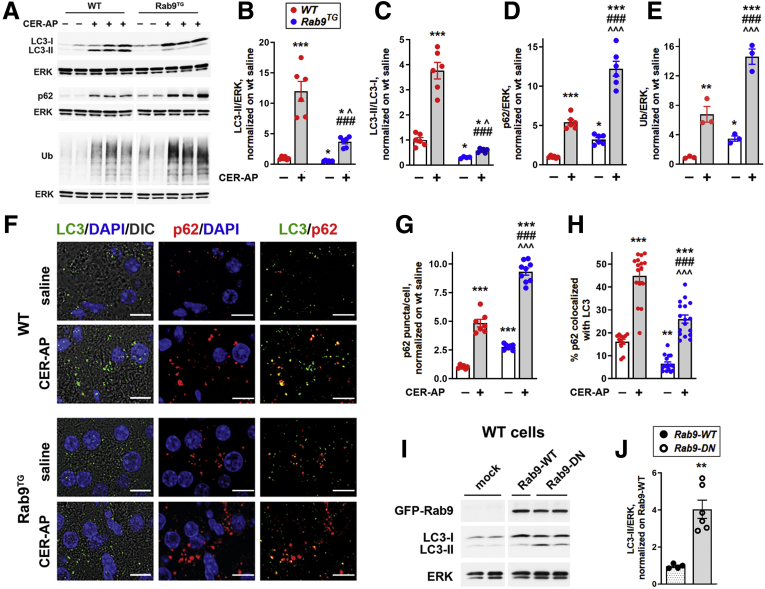
We have previously shown that pancreatitis stimulates the formation of canonical/LC3-positive autophagosomes but inhibits autophagic degradation (because of lysosomal dysfunction), resulting in impaired, retarded autophagic flux.4, 5, 6,9,25, 26, 27 The impaired/inefficient autophagy is recognized as a critical pathologic response of pancreatitis.4,9,27 In WT CER-AP, the impaired autophagy is evidenced by concomitant increases in LC3-II, p62/SQSTM1, and ubiquitinated proteins and accumulation in acinar cells of LC3-positive autophagic vacuoles, all demonstrating retarded autophagic flux (Figure 6A–H). Compared with WT, experimental pancreatitis in Rab9TG mice caused lesser increases in pancreatic LC3-II level and greater increases in p62/SQSTM1 and ubiquitinated proteins (Figure 6A–E). Congruent with the IB data, IF analysis showed reduced number of LC3 puncta, increased staining for p62, and decreased p62 colocalization with LC3 in pancreas of Rab9TG mice with CER-AP (Figure 6F–H; see LC3 puncta quantification in Figure 5).
Figure 6.
Rab9 overexpression perturbs canonical/LC3-mediated autophagy in experimental pancreatitis and in control pancreas. (A–E) IB analysis of markers/mediators of canonical autophagy in pancreas of WT and Rab9TG mice subjected to CER-AP (7 hours). Densitometric band intensities for indicated proteins were normalized to ERK in the same sample and further to those in WT control (saline-treated) group. Ub, ubiquitinated proteins. (F–H) IF analysis of pancreatic LC3 and p62/SQSTM1. Scale bars, 10 μm. IF colocalization (H) was quantified with Volocity image analysis software using Manders-Costes coefficients. (I and J) IB analysis and densitometric quantification of LC3-II in WT acinar cells transduced with adenoviral vectors containing GFP-Rab9 (Rab9-WT), GFP-Rab9 dominant-negative S21N mutant (Rab9-DN), or GFP alone (mock), as described in Methods. Values are mean ± SEM from at least 3 animals or cell preparations per group. In (G and H), at least 10 high-power fields per animal were quantified. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs WT control (saline) group (B–H) or cells transduced with Rab9-WT vector (J). ###P < .001 vs WT CER-AP. ˆP < .05, ˆˆˆP < .001 vs Rab9TG control (saline) group. Significance was determined by 2-tailed Student t test (J) or 1-way ANOVA, followed by Tukey or Holm-Sidak multiple comparisons test.
These results indicate that Rab9 overexpression reduced the efficiency of canonical, LC3-mediated autophagy in pancreatitis. Rab9 overexpression itself caused up-regulation of p62 and ubiquitinated proteins in pancreas, indicating perturbed basal autophagy (Figure 6D–G).
To further probe the effect of Rab9 on LC3-II level in acinar cells, we transduced WT mouse acinar cells with adenoviruses harboring Rab9 or its dominant-negative mutant. Rab9 dominant-negative increased LC3-II level ∼4-fold (Figure 6I and J), in accord with the negative effect of Rab9 overexpression on LC3-II level in basal condition and CER-AP (Figure 6A and C).
Rab9 Inhibits Canonical Autophagosome Formation in Pancreatic Acinar Cells Through ATG4B-Dependent Mechanism
To quantitatively analyze the effect of Rab9 overexpression on canonical autophagosome formation, we measured changes in LC3-II level in acinar cells isolated from WT and Rab9TG mice and subjected to the ex vivo CCK-induced pancreatitis (Figure 7A–F). Cells were incubated with and without 100 nmol/L CCK in the presence and absence of the lysosomal v-ATPase inhibitor bafilomycin (Baf) A1, which blocks lysosomal acidification and thus lysosomal functions. This type of analysis is widely used to obtain information on 2 key parameters of canonical autophagy, the LC3-mediated autophagosome formation and the efficiency of autophagic flux.12,26,35 The number of autophagic vacuoles in a cell is a net balance between autophagosome formation and their degradation through autophagic flux; thus, an increase in LC3-II level can result from both induction of autophagosome formation and inhibition of flux. BafA1 blocks lysosomal/autophagic degradation but does not affect autophagosome formation; therefore, in the presence of BafA1 any difference in cellular LC3-II levels between 2 conditions (eg, WT cells with and without CCK) is due solely to changes in autophagosome formation.12,26 On the other hand, the difference in LC3-II levels in the absence and presence of BafA1 in a given condition (eg, CCK-treated WT cells) provides a measure of the efficiency of autophagic degradation in this condition; the bigger the difference, the greater was the contribution of autophagic flux abrogated by BafA1.12,26
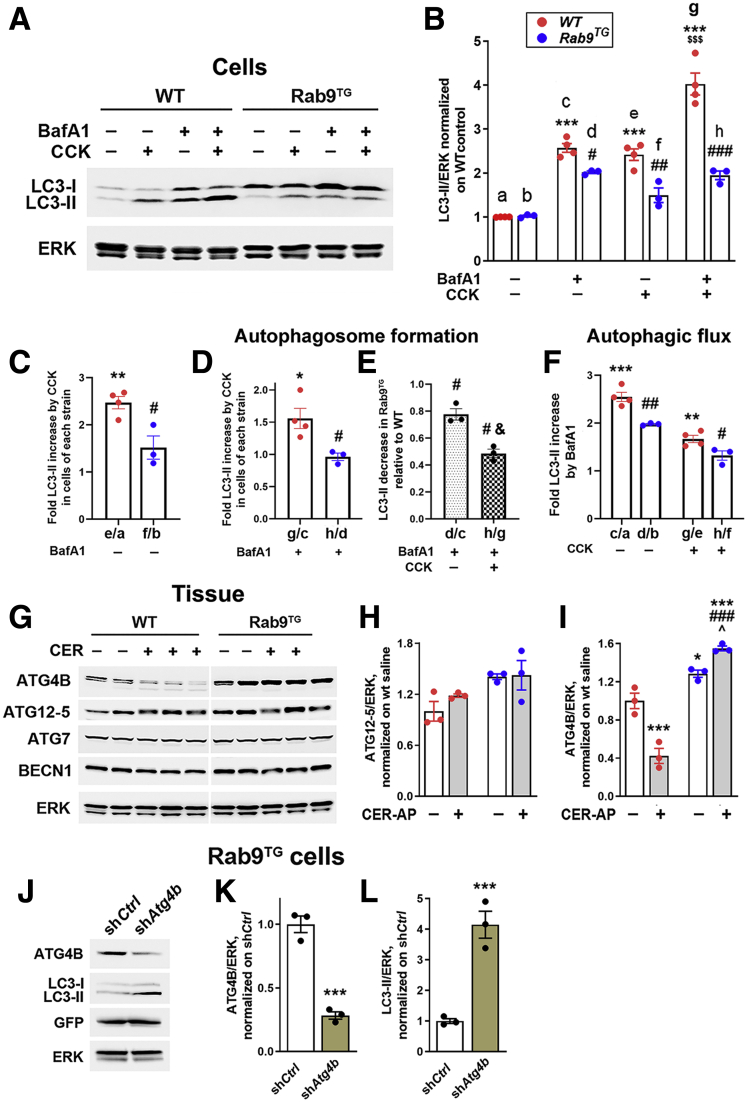
Figure 7.
Rab9 overexpression inhibits canonical autophagosome formation in acinar cells through up-regulation of ATG4B. (A–F) Autophagic flux analysis in WT and Rab9TG acinar cells. Cells were incubated for 30 minutes in presence and absence of the lysosomal inhibitor Bafilomycin A1 (BafA1; 20 nmol/L), followed by 1-hour incubation with or without 100 nmol/L CCK. (A and B) Representative IB and densitometric quantification of LC3-II level used in the autophagic flux analysis. LC3-II band intensities were normalized to those of ERK in the same sample and further normalized to control (untreated) WT cells. Data in (B) were used as ratios between values in indicated columns to quantify overall effect of ex vivo CCK pancreatitis on LC3-II in WT and Rab9TG cells (C; in the absence of BafA1), the effects of CCK (D) and Rab9 overexpression (E) on autophagosome formation (all in the presence of BafA1), and the efficiency of autophagic flux/degradation (F; BafA1 vs no inhibitor). (G–I) Levels of indicated ATG proteins were measured by IB in pancreas of mice subjected to CER-AP (7 hours). (J–L) Levels of indicated proteins were measured by IB in Rab9TG acinar cells transduced with adenoviral vectors containing ATG4B shRNA (shAtg4b) or control/scrambled (shCtrl) shRNA. Values are mean ± SEM from 3 cell preparations (or 3 animals) per group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs control (untreated) WT cells (B–F), vs WT control (saline) group (I), or vs Rab9TG cells transduced with control shCtrl (K and L). #P < .05, ##P < .01, ###P < .001 vs the same parameter in WT cells (B–F) or WT pancreas (I). ˆP < .05 vs control (saline) Rab9TG group (I). $$$P < .001 vs WT cells treated with CCK alone (no BafA1; B). &P < .05 vs d/c (E). Significance was determined by 2-tailed Student t test (C–E, K, and L) or 1-way ANOVA, followed by Tukey or Holm-Sidak multiple comparisons test (B, F, and I).
Figure 7A shows a representative IB used for densitometric quantification (Figure 7B) of the changes in LC3-II level. In accord with previous results,26 we measured a ∼2.6-fold increase in LC3-II induced by the ex vivo CCK pancreatitis in WT cells (Figure 7C, e/a), whereas in Rab9TG cells the LC3-II response to CCK was greatly reduced (Figure 7C, f/b), corroborating the results on tissue (Figure 6). Notably, the data obtained in the presence of BafA1 show that CCK increased LC3-mediated autophagosome formation in WT cells ∼1.5-fold (Figure 7D, g/c) but failed to activate canonical autophagy in Rab9TG cells (Figure 7D, h/d). Thus, canonical autophagosome formation in Rab9TG cells subjected to the ex vivo pancreatitis was ∼50% of that in WT (Figure 7E, h/g). Rab9 overexpression also decreased canonical autophagosome formation in the basal state (Figure 7E, d/c), but the effect was smaller than in CCK-treated cells.
The basal autophagic flux in WT acinar cells was quite robust, because BafA1 treatment increased LC3-II level ∼2.7-fold in this condition (Figure 7F, c/a). In accord with the previous report,26 CCK reduced autophagic flux efficiency in WT cells; the fold increase of LC3-II elicited by BafA1 in the ex vivo pancreatitis was ∼40% lower than in control cells (Figure 7F, g/e vs c/a). The inhibitory effect of CCK on autophagic flux efficiency was even more pronounced in Rab9TG cells (Figure 7F, h/f vs g/e). Rab9 overexpression also decreased autophagic flux efficiency in the basal state (Figure 7F, d/b vs c/a).
The results indicate that Rab9 overexpression inhibited the formation of canonical, LC3-positive autophagosomes and decreased the efficiency of LC3-mediated autophagic flux in acinar cells; the inhibition was in both the ex vivo pancreatitis and (to a lesser extent) the basal state.
In search of mechanisms underlying the effect of Rab9 on LC3-II, we measured changes in the levels of ATG proteins involved in canonical autophagosome formation (Figure 7G–I). Rab9 overexpression had no effect on pancreatic levels of ATG7 and Beclin1/ATG6; the level of ATG5 (more precisely, the ATG12-ATG5 conjugate) increased slightly (Figure 7G and H), which could only enhance the canonical autophagy. However, we found a pronounced effect of Rab9 overexpression on ATG4B (Figure 7G and I), a cysteine protease that regulates the balance between LC3-I and LC3-II, which is critical for canonical autophagy.26,36,37 We previously showed26 that in WT pancreas ATG4B acts as a negative regulator of LC3-mediated autophagosome formation; its overall effect is de-conjugation of LC3-II from autophagic membranes back to the cytosolic LC3-I. WT pancreatitis greatly decreased ATG4B level (Figure 7G and I), thus sustaining the formation of LC3-positive autophagosomes.26 In stark contrast, Rab9 overexpression elevated pancreatic ATG4B (in both the basal state and CER-AP) and, notably, abrogated ATG4B decrease in pancreatitis (Figure 7G and I). The up-regulation of ATG4B could account for the effect of Rab9 on LC3-II; therefore, we examined the effect of down-regulating ATG4B on LC3-II level in Rab9TG acinar cells. Transduction of Rab9TG cells with Atg4B shRNA reduced ATG4B and markedly increased LC3-II (Figure 7J–L). We have reported similar results in WT acinar cells, in which ATG4B knockdown with shRNA increased LC3-II level, whereas adenoviral ATG4B overexpression caused a marked reduction in LC3-II.26
Taken together, our present (Figure 7G–L) and previous26 results indicate that the inhibition of canonical autophagosome formation in Rab9TG pancreas is mediated through Rab9-induced up-regulation of ATG4B.
Rab9 Overexpression Activates Noncanonical Autophagy in Experimental Pancreatitis
Acinar cell vacuolization seen on H&E-stained pancreatic tissue sections (as well as with electron microscopy) is a hallmark of experimental and human pancreatitis. The data4, 5, 6 indicate that it is caused by accumulation of large autolysosomes containing partially degraded material resulting from impaired autophagic flux in pancreatitis. Rab9 overexpression markedly increased acinar cell vacuolization, as seen on H&E-stained pancreatic tissue sections in CER-AP and Arg-AP; there were also more vacuoles in the basal state in Rab9TG compared with WT pancreas (Figure 5A–C). Many vacuoles in Rab9TG acinar cells contained cargo, indicating their autophagic nature. However, our results (Figures 4 and 6) showed that Rab9 overexpression significantly reduced the formation of LC3-positive autophagic vacuoles. Indeed, the number of immunolabeled LC3 puncta decreased >2 times in Rab9TG pancreas vs WT, and the decrease was even greater in CER-AP (Figure 6F and Figure 5D and E). Together, the results indicate that the autophagic vacuoles in Rab9TG pancreas are formed through noncanonical pathway. Supporting this notion, IF analysis of Rab9TG pancreas showed the appearance of Rab9-positive ring-like structures that were evident in the basal state and prominently accumulated in CER-AP (Figure 5D–J). Such structures were not observed in WT pancreas in the basal state or in pancreatitis (Figure 1C, Figure 5F and G).
Next, we examined the identity of the cargo in Rab9-positive vacuoles. trans-Golgi, a major site of Rab9 localization, is a membrane source for noncanonical autophagosome formation.14,38 IF analysis showed that ∼25% of all Rab9-positive vacuoles in Rab9TG pancreas contained TGN38, a marker of trans-Golgi network38 (Figure 5G and K). Although there were many more Rab9-positive ring structures in CER-AP than in control (saline-treated) Rab9TG mice (Figure 5F), the same percentage of them contained TGN38 (Figure 5K). These data indicate trans-Golgi as a source/substrate of Rab9-mediated autophagy. Rab9-positive vacuoles in Rab9TG mice with CER-AP also contained protein aggregates and mitochondria, the “classical” autophagy cargo, as manifested by the presence inside these vacuoles of their respective markers, p62/SQSTM1 and the mitochondrial resident protein TOM20 (Figure 5H, I, and L). Rab9-positive vacuoles also contained the lysosomal protease cathepsin D (CatD), identifying them as autolysosomes (Figure 5J and L).
Inhibition of LC3-mediated autophagy concomitant with the accumulation in acinar cells of Rab9-positive autophagic vacuoles, and especially the appearance of ring-like structures containing autophagic cargo, indicates that Rab9 overexpression causes a shift from canonical to noncanonical autophagy pathway in the exocrine pancreas.
Mutually Antagonistic Relationship Between Canonical and Noncanonical Autophagy in Experimental Pancreatitis
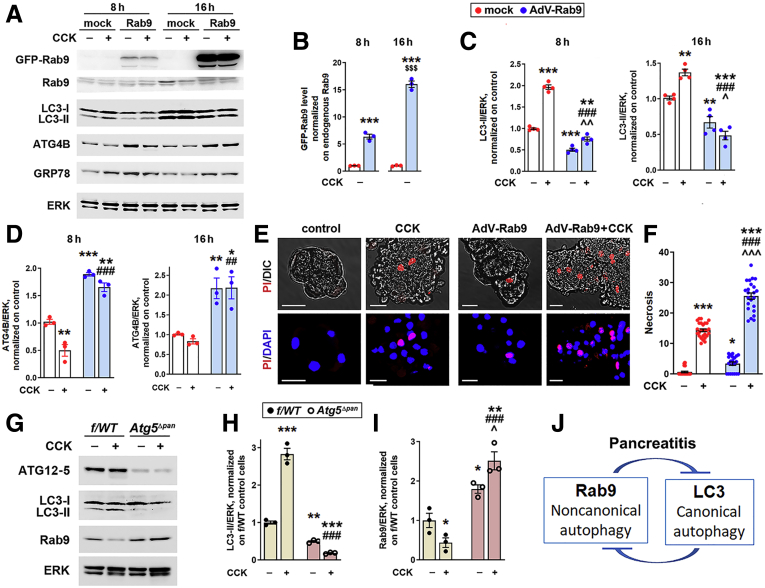
In pancreas of Rab9TG mice, Rab9 protein increased ∼35-fold compared with its endogenous level (Figure 4A), in agreement with data in the study28 that generated these mice. We examined the effects of lesser Rab9 increases in acinar cells transduced with adenovirus bearing WT GFP-Rab9. In transduced cells, ∼6-fold and ∼18-fold increases in Rab9 (over the basal level) both elicited similar effects on LC3-II and ATG4B (Figure 8A–D). Importantly, the magnitudes of these effects were comparable to those observed in vivo in Rab9TG mice (Figure 6B and C and Figure 7I). Furthermore, the effect on necrosis, a key pancreatitis response, of the smaller (∼6-fold) Rab9 overexpression in acinar cells (Figure 8E and F) was similar to that in Rab9TG pancreas (Figure 4E and F). In Rab9-transduced cells, basal necrosis increased 5.2-fold (over WT) and that elicited by ex vivo pancreatitis increased 1.8-fold (Figure 8F). In CER-AP model in Rab9TG mice these increases were 6.3-fold and 1.7-fold, respectively (Figure 4E). Collectively, these data suggest a threshold character of Rab9 effects in acinar cells.
Figure 8.
Mutually antagonistic relationship between canonical and noncanonical autophagy in the exocrine pancreas. (A–F) WT mouse pancreatic acinar cells were transduced with adenovirus bearing mouse GFP-Rab9 (or “mock”) for indicated times and treated (or not) for 1 hour with 100 nmol/L CCK. (G–I) Acinar cells were isolated from Atg5Δpan and f/WT (control littermate) mice and treated (or not) for 1 hour with 100 nmol/L CCK. (A–D, G–I) IB analysis of indicated proteins. Densitometric band intensities were normalized to endogenous Rab9 (B) or ERK in the same sample and further normalized to those in control, mock-transduced (B–D) or f/WT (H and I) cells. (E and F) Necrosis was measured as the percentage of propidium iodide (PI)-permeable acinar cells. Scale bars, 10 μm. (J) Schematic depicting mutually antagonistic relationship between noncanonical and canonical autophagy in acinar cells. Values are mean ± SEM from 3 to 4 cell preparations for each condition. In (F), each symbol corresponds to a different group of acinar cells (n = 21–32 groups of cells analyzed per condition). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 vs the same parameter in control (no CCK) cells, mock-transduced (B–F), or f/WT (H and I). ##P < .01, ###P < .001 vs CCK-treated mock-transduced (B–F) or f/WT (H and I) cells. ˆP < .05, ˆˆP < .01, ˆˆˆP < .001 vs control (no CCK) cells transduced with AdV-Rab9 (C and F) or f/WT cells (I). $$$P < .001 vs the same parameter in 8-hour transduced cells (B). Significance was determined by 1-way ANOVA, followed by Tukey or Holm-Sidak multiple comparisons test.
In other systems, noncanonical autophagy was activated in conditions in which canonical autophagy was blocked.14,15 To examine whether this occurs in pancreatitis, we isolated acinar cells from Atg5Δpan mice with pancreas-specific ablation of ATG5, an essential mediator of canonical autophagy, and subjected them to ex vivo pancreatitis (Figure 8G–I). The reduction in LC3-II in ATG5-deficient acinar cells, caused by blockage of canonical autophagosome formation, was associated with marked up-regulation of Rab9 (Figure 8H and I). The effect was particularly prominent in the ex vivo pancreatitis; loss of canonical autophagy completely prevented CCK-induced decrease in Rab9 observed in WT; as a result, Rab9 level in CCK-treated ATG5-deficient acinar cells was 5.5-fold greater than that in WT cells (Figure 8I).
The results demonstrate a mutually antagonistic relationship between canonical and noncanonical autophagy in pancreatitis (Figure 8J), specifically the suppression of noncanonical/Rab9-mediated pathway in acinar cells by the canonical/LC3-mediated autophagy. This negative regulation provides an explanation for several-fold Rab9 decrease in both in vivo (Figure 1B, D, and I) and ex vivo (Figure 2F, Figure 8I) WT experimental pancreatitis, in which canonical autophagosome formation is stimulated. By contrast, CER/CCK-induced pancreatitis did not decrease Rab9 level in Rab9TG pancreas (Figure 4A) or Rab9-overexpressing acinar cells (Figure 8A), in which canonical autophagy is inhibited, and it even increased Rab9 in ATG5-deficient acinar cells in which canonical autophagosome formation is blocked (Figure 8I).
Discussion
The principal function of Rab-GTPases is to coordinate all aspects of the endolysosomal system, including lysosomal function and autophagy.17, 18, 19 The roles of Rab5, Rab7, and Rab11 in the endolysosomal system are well-established, and they have been shown to promote canonical autophagy.19, 20, 21, 22 In contrast, the role of Rab9 in endosomal trafficking is still being debated; it has been implicated in the endosome-to-trans-Golgi transport and in the transition from early to late endosomes.38,39 Although the role of Rab9 in alternative, LC3-independent autophagy is well-established,14,15,23,24 its effect on canonical autophagy has not been explored.
Our results show that Rab9 inhibits canonical/LC3-mediated autophagosome formation in both control pancreas and experimental pancreatitis. The underlying mechanism involves ATG4B, a cysteine protease that regulates the balance between the cytosolic LC3-I and lipidated LC3-II.36,37 In the canonical pathway, LC3-II translocation to nascent autophagosomes is necessary for their closure.11,16 ATG4B regulates LC3-II level both positively and negatively, and the outcome is cell and context dependent.26,36,37 ATG4B can increase LC3-II level by cleaving LC3 pro-protein to create a pool of LC3-I available for subsequent conversion to LC3-II; on the other hand, it de-conjugates (delipidates) LC3-II from autophagic membranes, converting it back to LC3-I.36 We have shown26 that in WT acinar cells the second effect, ie, LC3-II delipidation, predominates; ATG4B suppresses canonical autophagosome formation in these cells. The results in the present study indicate that Rab9 reduces LC3-II through up-regulating the pancreatic level (and thereby, activity26,36,37) of ATG4B. Rab9 overexpression decreased both LC3-II level and the number of LC3-positive autophagic vacuoles (LC3 puncta), which was especially pronounced in experimental pancreatitis. LC3-II level in Rab9TG acinar cells was restored by shRNA knockdown of ATG4B.
Concurrent with reducing LC3-positive autophagosomes, Rab9 overexpression stimulated the formation of Rab9-positive vacuoles in acinar cells, thus shifting autophagy from the canonical/LC3-mediated to noncanonical pathway. IF analysis revealed ring-like Rab9-positive vacuoles, especially abundant in pancreatitis, which sequestered protein aggregates and mitochondria, classical autophagy substrates.10,13 A large portion of these vacuoles also contained the trans-Golgi marker TGN38. It is yet to be determined whether, in general, noncanonical autophagy can be selective, for example, in eliminating damaged/dysfunctional mitochondria as mediated by mitophagy. In canonical autophagy, the receptors mediating selective cargo sequestration typically have LC3-interacting region, a common structural motif that enables their recognition by LC3.40,41 In particular, the interaction between LC3-II and LC3-interacting region domains in p62/SQSTM1 and the mitophagy receptor Parkin enables the sequestration of p62-decorated protein aggregates or Parkin-decorated mitochondria.40, 41, 42, 43, A recent study23 showed that noncanonical autophagy in cardiomyocytes can selectively eliminate damaged mitochondria through a Rab9-mediated, LC3-interacting region–independent mechanism. A similar mechanism may operate in Rab9TG pancreas. Interestingly, that study reported ring-like Rab9-positive vacuoles containing mitochondrial cargo.23 How exactly Rab9 recognizes p62/SQSTM1 is not known.
Collectively, our results reveal mutually antagonistic relationship between canonical and noncanonical pathways in pancreatitis, a finding of general interest for autophagy research. Indeed, blockage of canonical autophagy in ATG5-deficient acinar cells caused several-fold increase in Rab9 in ex vivo CCK pancreatitis. Conversely, adenoviral expression of Rab9-DN in WT acinar cells greatly increased LC3-II level.
The inhibition of LC3-positive autophagosome formation by Rab9 contrasts the stimulatory effect of Rab5, Rab7, and Rab11 (shown for Rab7 in pancreatic acinar cells22). It remains to be determined whether the inhibition of canonical autophagosome formation by Rab9, which we found in pancreas, also operates in other organs and cell types, because most studies investigating the effects of Rab9 on autophagy were performed on cells in which canonical autophagy was already blocked by genetic or molecular means.14,15
Experimental pancreatitis in rodents caused marked reduction in membrane-bound (active) Rab9. Similarly, human pancreatitis tissue displayed several-fold decrease in Rab9-positive puncta. One possible mechanism mediating such a decrease could be reduced complex formation of Rab9 with RabGDI, a key process regulating Rabs’ translocation to and from membranes.29, 30, 31 However, our gel-filtration experiments showed that, to the contrary, Rab9-RabGDI complex formation increased in CER-AP. Of note, this indicates that Rab9 prenylation, a prerequisite for its interaction with RabGDI,32 is not blocked in pancreatitis. The data suggest that CCK-induced Rab9 decrease in acinar cells involves proteasomal degradation because it was prevented with the proteasomal inhibitor MG132. Excessive Rab accumulation in the cytosol is known to result in Rab aggregates, which are targeted by ubiquitin and removed by proteasomal degradation.33 The observed increase in cytosolic Rab9 in CER-AP may promote the formation of such aggregates, stimulating Rab9 proteasomal degradation.
As shown by our group and others, canonical autophagosome formation is stimulated in pancreatitis, both experimental, in WT rodent in vivo and ex vivo models, and in human disease.4, 5, 6,9,25, 26, 27,44 Activation of this pathway is a protective mechanism, because its blockage in mice with pancreas-specific ablation of the key canonical autophagy mediators ATG5 or ATG7 causes accumulation of damaged mitochondria, ER and oxidative stress, and the development of spontaneous pancreatitis with all classical disease responses: hyperamylasemia, trypsinogen activation, inflammation, fibrosis, and acinar cell death.7, 8, 9,27 These findings reveal an essential role of canonical autophagy in maintaining pancreatic acinar cell homeostasis and protection against pancreatitis. By contrast, our data indicate that the involvement of noncanonical, Rab9-mediated autophagy in WT pancreatitis is minimal. Indeed, we could not detect any significant number of Rab9-positive vacuoles (particularly ring structures) in either basal state or WT experimental pancreatitis or in human pancreas. Furthermore, we observed pronounced decreases in pancreatic Rab9 level in several WT pancreatitis models, as well as in human disease. The results indicate that Rab9 decrease in pancreatitis is a protective response, whereas activation of noncanonical autophagy is damaging to pancreas. Rab9 overexpression, which stimulated the noncanonical pathway and inhibited canonical autophagosome formation, markedly worsened pancreatitis responses such as inflammation, acinar cell death (particularly necrosis), and trypsinogen activation. Rab9 overexpression also caused pancreatitis-like damage in control pancreas or acinar cells, namely ER stress, caspase activation, necrosis, and inflammation. Noncanonical autophagy in pancreas cannot substitute for its canonical counterpart, because Rab9TG pancreas displayed greater levels of p62 and ubiquitinated proteins (compared with WT).
In conclusion, our study shows a major role of Rab9 in determining the pattern of autophagy in pancreas. The data reveal mutually antagonistic interrelationship between canonical and noncanonical autophagy in pancreatitis. Rab9 overexpression inhibited LC3-mediated and promoted Rab9-mediated autophagosome formation; on the other hand, genetic blockage of canonical/LC3-mediated autophagosome formation caused up-regulation of Rab9. Autophagy switch from canonical to the noncanonical pathway aggravated experimental pancreatitis and induced pancreatitis-like damage in control pancreas. Both experimental and human pancreatitis displayed marked decreases in pancreatic Rab9, indicating a protective response to sustain canonical autophagy and alleviate disease severity. The results further underscore a critical homeostatic role of canonical autophagy in the exocrine pancreas and show that the noncanonical pathway fails to substitute for its canonical counterpart in protecting against pancreatitis. They suggest Rab9 down-regulation as a therapeutic approach to mitigate the severity of pancreatitis, because high levels of Rab9 may portend a more severe disease.
Materials and Methods
Experimental Models of Pancreatitis
Rats (Sprague-Dawley; Envigo, Indianapolis, IN; 002), WT mice (C57BL/6J; Jackson Laboratory, Bar Harbor, ME; 000664), and transgenic Rab9TG mice28 expressing HA-tagged human Rab9 (Jackson Laboratory, 009678) on the same background were subjected to CER-AP and Arg-AP, as detailed previously.5,6,25,26,44,45 CER-AP was induced by hourly intraperitoneal injections of 50 μg/kg CER. If not stated otherwise, rats were euthanized 4 hours and mice 7 hours after the first injection. Arg-AP was induced in rats by 2 hourly intraperitoneal injections of 3 g/kg L-arginine and in mice by 3 hourly intraperitoneal injections of 3.3 g/kg L-arginine. The animals were euthanized 24 hours after the first injection. Control mice received similar injections of physiological saline. We also applied ex vivo CCK-AP model on acinar cells isolated from mice using a standard collagenase digestion procedure and incubated for 0.5–1 hour with supramaximal concentration (100 nmol/L) of CCK.6,25,26,44, 45, 46 The ex vivo model was performed on human acinar cells and acinar cells from WT, Rab9TG mice, and from Atg5Δpan mice with pancreas-specific ATG5 ablation. To generate the latter, we crossed Atg5flox/- mice (generated by M.O.47) with Pdx1-Cre driver mice to generate Atg5flox/-;Pdx1-Cre mice (termed Atg5Δpan) in which the Atg5 gene is specifically ablated in pancreatic epithelial cells. Control mice were littermates with WT alleles. All animal experiments were approved by the Institutional Animal Care and Use Committee of VA Greater Los Angeles Healthcare System.
Human acinar cells were analyzed on pancreatic slices48 from portions of normal human pancreata obtained from pre-terminal donors that were not used for transplantation and diverted to research or from normal portions of pancreatic cancer operations. Pancreas was cut into 3 × 3 × 3 mm pieces and embedded with 37°C low melting 3.8% agarose gel in extracellular solution containing (mmol/L) 125 NaCl, 2.5 KCl, 1 MgCl2, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, 2 sodium pyruvate, 0.25 ascorbic acid, 3 myo-inositol, 6 lactic acid, and 7 glucose, and placed on ice to solidify the gel. The gelled tissue blocks were sliced with Vibratome into 120-μm-thick slices; slices were washed with the extracllular solution, placed in growth factor- and serum-free Waymouth’s MB 752/1 medium containing 5.5 mmol/L glucose and 0.1 mg/mL soybean trypsin inhibitor, and treated with supramaximal (100 nmol/L) CCK for ex vivo pancreatitis. All the experiments with human pancreas slices were approved by Institutional Review Boards of the University of Toronto and Toronto General Hospital (H.Y.G.).
Formalin Fixed Human Pancreas Specimens
Formalin fixed paraffin-embedded samples of human normal pancreas and pancreatitis from non-cancer patients were provided de-identified by the Pancreas Tissue Bank of the UCLA Department of Pathology (D.D.), according to a protocol approved by the UCLA Institutional Review Board. Normal pancreas and pancreatitis samples were procured from surgical resection specimens where malignant disease was absent or unlikely to be a confounding variable (ie, Whipple for ampullary or duodenal adenoma, well-circumscribed neuroendocrine mass, benign cystic lesions, or other non-malignant presentations). All analyzed samples of normal pancreas and pancreatitis were taken >1 cm away from any associated lesion. Because of the inhomogeneous nature of pancreas damage due to necrosis and fibrosis seen at various stages of disease, only samples with large acinar component (evaluated by D.D.) were used in the study.
Adenoviral Transductions
Mouse pancreatic acinar cells were transduced with adenoviruses (2–5 × 109 pfu/mL) bearing GFP-Rab9 WT and GFP-Rab9 DN (dominant negative mutant S21N). The advantage of using S21N mutation is that it only inactivates Rab9, whereas Rab9 shRNA may affect the expression of other Rabs. To produce recombinant adenovirus overexpressing Rab9 WT or Rab9 DN, the plasmids encoding Rab9 WT (Addgene, Watertown, ME; 12663) and Rab9 DN (Addgene, 12664) were subcloned into the pShuttle-CMV vector (Agilent Technologies, Santa Clara, CA; 240007) and transferred into the pAdEasy adenoviral vector by using the AdEasy XL adenoviral vector system (Agilent Technologies, 240010). For transduction with recombinant shRNA adenoviral vectors (3–4 × 1010 pfu/mL) encoding GFP-MmAtg4B shRNA (Vector Biolabs, Malvern, PA; shADV-253222) or “scrambled” (control) GFP-shRNA (Vector Biolabs, 1122), acinar cells were incubated for 38–40 hours at 37°C. Purification and concentration of adenoviruses were performed at the UCLA Integrated Molecular Technologies Core. In these “prolonged-culture” experiments,5 acinar cells were cultured on collagen IV coated plates (BD Biosciences, Franklin Lakes, NJ; 354428) in Dulbecco modified Eagle medium (Thermo Fisher Scientific, Waltham, MA; 11995) containing 10% fetal bovine serum (Thermo Fisher Scientific, 10437028), 5 ng/mL EGF (Sigma-Aldrich, St Louis, MO; SRP3196), 200 μg/mL soybean trypsin inhibitor (Worthington Biochemicals, Lakewood, NJ; LS003571), 100 U/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific, 10378016).
Preparation of Tissue and Cell Lysates and Membrane and Cytosolic Fractions
Tissue or cell samples were homogenized on ice in RIPA buffer supplemented with 1 mmol/L phenylmethylsulfonyl fluoride and protease inhibitors’ cocktail (Roche, Basel, Switzerland). Protein concentration in the supernatants was determined by Bradford assay (Bio-Rad Laboratories, Hercules, CA). To obtain cytosolic and membrane fractions, pancreatic tissue was homogenized in a buffer containing 25 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L MgCl2, and protease inhibitor cocktail. Postnuclear supernatant was centrifuged for 2 hours at 150,000g, and both the pellet (membrane fraction) and supernatant (cytosolic fraction) were collected. The quality of the fractions was evaluated by IB analysis for specific protein markers, for example, the absence of the mitochondrial protein COX IV in cytosolic fraction.
IB Analysis
Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose membranes. Nonspecific binding was blocked by 1-hour incubation of the membranes in 5% (w/v) nonfat dry milk or 5% bovine serum albumin in Tris-buffered saline (pH 7.5). The blots were then incubated for 2 hours or overnight with primary antibodies in the antibody buffer containing 1% (w/v) nonfat dry milk in 0.05% (v/v) Tween 20 in Tris-buffered saline, washed, and finally incubated for 1 hour with a peroxidase-labeled secondary antibody. The blots were developed for visualization using an enhanced chemiluminescence detection kit (Pierce). Band intensities in the IBs were quantified by densitometry using the FluorChem HD2 imaging system (Alpha Innotech/ProteinSimple, San Leandro, CA).
Gel-Filtration Chromatography
The assay was performed as previously.32 Pancreatic tissue cytosolic fractions were loaded onto a Superdex S200 column using the SMART system (Amersham Biosciences, Amersham, UK). The column was equilibrated in a buffer containing (mmol/L) 50 Tris/HCl (pH 7.5), 100 NaCl, 8 MgCl2, 2 EDTA, and 1 dithiothreitol, supplemented with 10 μmol/L GDP, at a flow rate of 50 μL/min. The samples were injected, and the material eluting between 1.2 mL and 3.3 mL was collected in 42 50-μL fractions. Ten-μL aliquots of the fractions were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (on 4%–20% gel), transferred to nitrocellulose membranes, and Rab9 and RabGDIα/β proteins identified by IB. All the IBs (Figure 3 for the full, uncropped IBs) were developed together. Fractions 1–5 and 39–42 had no proteins recognized by the antibodies; therefore, these fractions are not shown in the figures.
Measurement of Protease Activities
Caspase-3-like and trypsin activities were measured in pancreatic tissue homogenates using fluorogenic substrates specific for caspase-3 (Z-VAD-fmk; Cayman, Ann Arbor, MI) and trypsin (Boc-Gln-Ala-Arg- AMC; Bachem, Bubendorf, Switzerland), as described.5, 6, 7,25,26,44,45
IF and immunohistochemical analyses were performed as described.5, 6, 7,25,26,44 Pancreatic tissue sections incubated with primary antibodies overnight at 4°C, followed by incubation with secondary antibodies conjugated with FITC or Texas Red. Nonspecific binding was blocked with 5% rabbit or goat serum. IF images were acquired with a Zeiss (Oberkochen, Germany) LSM 710 confocal microscope using ×63 objective.
For immunohistochemical detection of macrophages, endogenous peroxidase was blocked with 3% hydrogen peroxide, followed by incubation of pancreatic tissue sections with primary antibody against F4/80 and visualization with streptavidin-biotin immunoenzymatic antigen detection system (Vector Laboratories).
Analysis of necrosis, vacuolization, and inflammatory cell infiltration in the pancreas was performed on H&E-stained pancreatic tissue on the basis of morphologic criteria as described.5, 6, 7,25,26,44, 45, 46 Serum amylase and lipase were measured in a Hitachi 707 analyzer.
Acinar cell necrosis in ex vivo CCK-AP was quantified by propidium iodide uptake, as described.44 Briefly, cells were loaded with propidium iodide (2 mg/mL medium) for 10 minutes, washed with phosphate-buffered saline to remove excess propidium iodide, and visualized with confocal microscopy, and the percentage of propidium iodide–positive cells was quantified.
Antibodies and Reagents
Antibodies against the following proteins were purchased from Cell Signaling Technology, Danvers, MA: LC3B (catalog #2775), SQSTM1/p62 (5114), ATG4B (5299), ATG12-5 (4180), Beclin1 (3495), ATG7 (2631), Rab9 (5118), CHOP (5554), p44/42 MAPK (ERK1/2; 9102), GRP78 (BiP; 3177), and F4/80 (70076). Antibodies against the following proteins were purchased from Abcam, Cambridge, UK: Rab5 (catalog #ab18211), Rab9 (ab2810), TGN38 (ab16059). Antibodies against the following proteins were purchased from Santa Cruz Biotechnology, Dallas, TX: ubiquitin (catalog sc-8017), LDH (sc-33781), TOM20 (sc-11415), CatD (sc-6487), trypsinogen (sc-67388), and GFP (sc-9996). Antibodies against other proteins were purchased from the following companies: α-amylase (Sigma-Aldrich; A8273), Rab7 (Sigma-Aldrich; R8779), Rab4 (BD Biosciences; 610888), Rab11 (Proteintech, Rosemont, IL; 20229-1-AP), phospho-IRE1 (Novus Biologicals, Littleton, CO; NB100-2323), COX IV (Thermo Fisher Scientific; A21348).
CCK-8 was from Research Plus (Farmingdale, NJ), and CER was from Bachem. All other reagents were from Sigma-Aldrich.
Statistical analysis of the results was performed with Prizm8 (GraphPad Software, San Diego, CA) using 2-tailed Student t test for comparison between 2 groups and 1-way analysis of variance (ANOVA) with Tukey or Holm-Sidak post hoc test for comparison between multiple groups. Values are expressed as mean ± standard error of the mean (SEM); P < .05 was considered significant.
All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors (H.Y.G.) thank the Trillium Gift of Life Network (Toronto, Ontario; 2020) and the Toronto General Hospital/University Health Network Program in Biospecimen Sciences for providing human pancreatic tissue samples. The authors thank Drs Janet Treger and Emmanuelle Faure-Kumar for generating the adenoviral vectors (UCLA Integrated Molecular Technologies Core supported by NIH grant P30DK041301 to CURE: Digestive Diseases Research Center).
Current affiliation: Iskandar Yakubov, National University of Uzbekistan, Tashkent, Uzbekistan.
CRediT Authorship Contributions
Olga Mareninova (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Supporting; Funding acquisition: Supporting; Methodology: Lead; Supervision: Supporting; Validation: Equal; Writing – review & editing: Supporting)
Dustin Dillon (Data curation: Supporting; Formal analysis: Supporting)
Carli Wightman (Data curation: Supporting; Formal analysis: Supporting)
Iskandar Yakubov (Data curation: Supporting; Formal analysis: Supporting)
Toshimasa Takahashi (Data curation: Supporting; Methodology: Supporting; Resources: Supporting)
Herbert Gaisano (Methodology: Supporting; Resources: Supporting)
Keith Munson (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting)
Masaki Ohmuraya (Resources: Supporting)
David Dawson (Resources: Supporting)
Ilya Gukovsky (Conceptualization: Supporting; Formal analysis: Lead; Funding acquisition: Supporting; Validation: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Anna S. Gukovskaya, PhD (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported, fully or in part, by NIH/NIDDK grant P01DK098108 and the US Veterans Administration Merit Review award BX004306 (both to A.S.G.), and the NIH/NIAAA funded (P50AA1199) Southern California Research Center for Alcoholic Liver and Pancreatic Diseases and Cirrhosis (A.S.G., I.G., and O.A.M.)
References
- 1.Pandol S.J., Saluja A.K., Imrie C.W., Banks P.A. Acute pancreatitis: bench to the bedside. Gastroenterology. 2007;132:1127–1151. doi: 10.1053/j.gastro.2007.01.055. Erratum: ibid, 133:1056. [DOI] [PubMed] [Google Scholar]
- 2.Peery A.F., Crockett S.D., Murphy C.C., Lund J.L., Dellon E.S., Williams J.L., Jensen E.T., Shaheen N.J., Barritt A.S., Lieber S.R., Kochar B., Barnes E.L., Fan Y.C., Pate V., Galanko J., Baron T.H., Sandler R.S. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2018. Gastroenterology. 2019;156:254–272 e11. doi: 10.1053/j.gastro.2018.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerch M.M., Gorelick F.S. Models of acute and chronic pancreatitis. Gastroenterology. 2013;144:1180–1193. doi: 10.1053/j.gastro.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 4.Gukovskaya A.S., Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G993–G1003. doi: 10.1152/ajpgi.00122.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mareninova O.A., Hermann K., French S.W., O’Konski M.S., Pandol S.J., Webster P., Erickson A.H., Katunuma N., Gorelick F.S., Gukovsky I., Gukovskaya A.S. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119:3340–3355. doi: 10.1172/JCI38674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mareninova O.A., Sendler M., Malla S.R., Yakubov I., French S.W., Tokhtaeva E., Vagin O., Oorschot V., Lüllmann-Rauch R., Blanz J., Dawson D., Klumperman J., Lerch M.M., Mayerle J., Gukovsky I., Gukovskaya A.S. Lysosome associated membrane proteins maintain pancreatic acinar cell homeostasis: LAMP-2 deficient mice develop pancreatitis. Cell Mol Gastroenterol Hepatol. 2015;1:678–694. doi: 10.1016/j.jcmgh.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonucci L., Fagman J.B., Kim J.Y., Todoric J., Gukovsky I., Mackey M., Ellisman M.H., Karin M. Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress. Proc Natl Acad Sci U S A. 2015;112:E6166–E6174. doi: 10.1073/pnas.1519384112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diakopoulos K.N., Lesina M., Wormann S., Song L., Aichler M., Schild L., Artati A., Römisch-Margl W., Wartmann T., Fischer R., Kabiri Y., Zischka H., Halangk W., Demir I.E., Pilsak C., Walch A., Mantzoros C.S., Steiner J.M., Erkan M., Schmid R.M., Witt H., Adamski J., Algül H. Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology. 2015;148:626–638 e17. doi: 10.1053/j.gastro.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Gukovskaya A.S., Gorelick F., Groblewski G.E., Mareninova O., Lugea A., Antonucci L., Waldron R.T., Habtezion A., Karin M., Pandol S.J., Gukovsky I. Recent insights into the pathogenic mechanism of pancreatitis: role of acinar cell organelle disorders. Pancreas. 2019;48:459–470. doi: 10.1097/MPA.0000000000001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morishita H., Mizushima N. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol. 2019;35:453–475. doi: 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y.G., Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol. 2019;218:757–770. doi: 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., Adamopoulos I.E., Adeli K., Adolph T.E., Adornetto A., Aflaki E., Agam G., Agarwal A., Aggarwal B.B., Agnello M., Agostinis P., Agrewala J.N., Agrotis A., Aguilar P.V., Ahmad S.T., Ahmed Z.M., Ahumada-Castro U., Aits S., Aizawa S., Akkoc Y., Akoumianaki T., Akpinar H.A., Al-Abd A.M., Al-Akra L., Al-Gharaibeh A., Alaoui-Jamali M.A., Alberti S., Alcocer-Gómez E., Alessandri C., Ali M., Alim Al-Bari M.A., Aliwaini S., Alizadeh J., Almacellas E., Almasan A., Alonso A., Alonso G.D., Altan-Bonnet N., Altieri D.C., Álvarez É.M.C., Alves S., Alves da Costa C., Alzaharna M.M., Amadio M., Amantini C., Amaral C., Ambrosio S., Amer A.O., Ammanathan V., An Z., Andersen S.U., Andrabi S.A., Andrade-Silva M., Andres A.M., Angelini S., Ann D., Anozie U.C., Ansari M.Y., Antas P., Antebi A., Antón Z., Anwar T., Apetoh L., Apostolova N., Araki T., Araki Y., Arasaki K., Araújo W.L., Araya J., Arden C., Arévalo M.A., Arguelles S., Arias E., Arikkath J., Arimoto H., Ariosa A.R., Armstrong-James D., Arnauné-Pelloquin L., Aroca A., Arroyo D.S., Arsov I., Artero R., Asaro D.M.L., Aschner M., Ashrafizadeh M., Ashur-Fabian O., Atanasov A.G., Au A.K., Auberger P., Auner H.W., Aurelian L., Autelli R., Avagliano L., Ávalos Y., Aveic S., Aveleira C.A., Avin-Wittenberg T., Aydin Y., Ayton S., Ayyadevara S., Azzopardi M., Baba M., Backer J.M., Backues S.K., Bae D.H., Bae O.N., Bae S.H., Baehrecke E.H., Baek A., Baek S.H., Baek S.H., Bagetta G., Bagniewska-Zadworna A., Bai H., Bai J., Bai X., Bai Y., Bairagi N., Baksi S., Balbi T., Baldari C.T., Balduini W., Ballabio A., Ballester M., Balazadeh S., Balzan R., Bandopadhyay R., Banerjee S., Banerjee S., Á Bánréti, Bao Y., Baptista M.S., Baracca A., Barbati C., Bargiela A., Barilà D., Barlow P.G., Barmada S.J., Barreiro E., Barreto G.E., Bartek J., Bartel B., Bartolome A., Barve G.R., Basagoudanavar S.H., Bassham D.C., Bast R.C., Jr., Basu A., Batoko H., Batten I., Baulieu E.E., Baumgarner B.L., Bayry J., Beale R., Beau I., Beaumatin F., Bechara L.R.G., Beck G.R., Jr., Beers M.F., Begun J., Behrends C., Behrens G.M.N., Bei R., Bejarano E., Bel S., Behl C., Belaid A., Belgareh-Touzé N., Bellarosa C., Belleudi F., Belló Pérez M., Bello-Morales R., Beltran J.S.O., Beltran S., Benbrook D.M., Bendorius M., Benitez B.A., Benito-Cuesta I., Bensalem J., Berchtold M.W., Berezowska S., Bergamaschi D., Bergami M., Bergmann A., Berliocchi L., Berlioz-Torrent C., Bernard A., Berthoux L., Besirli C.G., Besteiro S., Betin V.M., Beyaert R., Bezbradica J.S., Bhaskar K., Bhatia-Kissova I., Bhattacharya R., Bhattacharya S., Bhattacharyya S., Bhuiyan M.S., Bhutia S.K., Bi L., Bi X., Biden T.J., Bijian K., Billes V.A., Binart N., Bincoletto C., Birgisdottir A.B., Bjorkoy G., Blanco G., Blas-Garcia A., Blasiak J., Blomgran R., Blomgren K., Blum J.S., Boada-Romero E., Boban M., Boesze-Battaglia K., Boeuf P., Boland B., Bomont P., Bonaldo P., Bonam S.R., Bonfili L., Bonifacino J.S., Boone B.A., Bootman M.D., Bordi M., Borner C., Bornhauser B.C., Borthakur G., Bosch J., Bose S., Botana L.M., Botas J., Boulanger C.M., Boulton M.E., Bourdenx M., Bourgeois B., Bourke N.M., Bousquet G., Boya P., Bozhkov P.V., Bozi L.H.M., Bozkurt T.O., Brackney D.E., Brandts C.H., Braun R.J., Braus G.H., Bravo-Sagua R., Bravo-San Pedro J.M., Brest P., Bringer M.A., Briones-Herrera A., Broaddus V.C., Brodersen P., Brodsky J.L., Brody S.L., Bronson P.G., Bronstein J.M., Brown C.N., Brown R.E., Brum P.C., Brumell J.H., Brunetti-Pierri N., Bruno D., Bryson-Richardson R.J., Bucci C., Buchrieser C., Bueno M., Buitrago-Molina L.E., Buraschi S., Buch S., Buchan J.R., Buckingham E.M., Budak H., Budini M., Bultynck G., Burada F., Burgoyne J.R., Burón M.I., Bustos V., Büttner S., Butturini E., Byrd A., Cabas I., Cabrera-Benitez S., Cadwell K., Cai J., Cai L., Cai Q., Cairó M., Calbet J.A., Caldwell G.A., Caldwell K.A., Call J.A., Calvani R., Calvo A.C., Calvo-Rubio Barrera M., Camara N.O., Camonis J.H., Camougrand N., Campanella M., Campbell E.M., Campbell-Valois F.X., Campello S., Campesi I., Campos J.C., Camuzard O., Cancino J., Candido de Almeida D., Canesi L., Caniggia I., Canonico B., Cantí C., Cao B., Caraglia M., Caramés B., Carchman E.H., Cardenal-Muñoz E., Cardenas C., Cardenas L., Cardoso S.M., Carew J.S., Carle G.F., Carleton G., Carloni S., Carmona-Gutierrez D., Carneiro L.A., Carnevali O., Carosi J.M., Carra S., Carrier A., Carrier L., Carroll B., Carter A.B., Carvalho A.N., Casanova M., Casas C., Casas J., Cassioli C., Castillo E.F., Castillo K., Castillo-Lluva S., Castoldi F., Castori M., Castro A.F., Castro-Caldas M., Castro-Hernandez J., Castro-Obregon S., Catz S.D., Cavadas C., Cavaliere F., Cavallini G., Cavinato M., Cayuela M.L., Cebollada Rica P., Cecarini V., Cecconi F., Cechowska-Pasko M., Cenci S., Ceperuelo-Mallafré V., Cerqueira J.J., Cerutti J.M., Cervia D., Cetintas V.B., Cetrullo S., Chae H.J., Chagin A.S., Chai C.Y., Chakrabarti G., Chakrabarti O., Chakraborty T., Chakraborty T., Chami M., Chamilos G., Chan D.W., Chan E.Y.W., Chan E.D., Chan H.Y.E., Chan H.H., Chan H., Chan M.T.V., Chan Y.S., Chandra P.K., Chang C.P., Chang C., Chang H.C., Chang K., Chao J., Chapman T., Charlet-Berguerand N., Chatterjee S., Chaube S.K., Chaudhary A., Chauhan S., Chaum E., Checler F., Cheetham M.E., Chen C.S., Chen G.C., Chen J.F., Chen L.L., Chen L., Chen L., Chen M., Chen M.K., Chen N., Chen Q., Chen R.H., Chen S., Chen W., Chen W., Chen X.M., Chen X.W., Chen X., Chen Y., Chen Y.G., Chen Y., Chen Y., Chen Y.J., Chen Y.Q., Chen Z.S., Chen Z., Chen Z.H., Chen Z.J., Chen Z., Cheng H., Cheng J., Cheng S.Y., Cheng W., Cheng X., Cheng X.T., Cheng Y., Cheng Z., Chen Z., Cheong H., Cheong J.K., Chernyak B.V., Cherry S., Cheung C.F.R., Cheung C.H.A., Cheung K.H., Chevet E., Chi R.J., Chiang A.K.S., Chiaradonna F., Chiarelli R., Chiariello M., Chica N., Chiocca S., Chiong M., Chiou S.H., Chiramel A.I., Chiurchiù V., Cho D.H., Choe S.K., Choi A.M.K., Choi M.E., Choudhury K.R., Chow N.S., Chu C.T., Chua J.P., Chua J.J.E., Chung H., Chung K.P., Chung S., Chung S.H., Chung Y.L., Cianfanelli V., Ciechomska I.A., Cifuentes M., Cinque L., Cirak S., Cirone M., Clague M.J., Clarke R., Clementi E., Coccia E.M., Codogno P., Cohen E., Cohen M.M., Colasanti T., Colasuonno F., Colbert R.A., Colell A., Čolić M., Coll N.S., Collins M.O., Colombo M.I., Colón-Ramos D.A., Combaret L., Comincini S., Cominetti M.R., Consiglio A., Conte A., Conti F., Contu V.R., Cookson M.R., Coombs K.M., Coppens I., Corasaniti M.T., Corkery D.P., Cordes N., Cortese K., Costa M.D.C., Costantino S., Costelli P., Coto-Montes A., Crack P.J., Crespo J.L., Criollo A., Crippa V., Cristofani R., Csizmadia T., Cuadrado A., Cui B., Cui J., Cui Y., Cui Y., Culetto E., Cumino A.C., Cybulsky A.V., Czaja M.J., Czuczwar S.J., D’Adamo S., D’Amelio M., D’Arcangelo D., D’Lugos A.C., D'Orazi G., da Silva J.A., Dafsari H.S., Dagda R.K., Dagdas Y., Daglia M., Dai X., Dai Y., Dai Y., Dal Col J., Dalhaimer P., Dalla Valle L., Dallenga T., Dalmasso G., Damme M., Dando I., Dantuma N.P., Darling A.L., Das H., Dasarathy S., Dasari S.K., Dash S., Daumke O., Dauphinee A.N., Davies J.S., Dávila V.A., Davis R.J., Davis T., Dayalan Naidu S., De Amicis F., De Bosscher K., De Felice F., De Franceschi L., De Leonibus C., de Mattos Barbosa M.G., De Meyer G.R.Y., De Milito A., De Nunzio C., De Palma C., De Santi M., De Virgilio C., De Zio D., Debnath J., DeBosch B.J., Decuypere J.P., Deehan M.A., Deflorian G., DeGregori J., Dehay B., Del Rio G., Delaney J.R., Delbridge L.M.D., Delorme-Axford E., Delpino M.V., Demarchi F., Dembitz V., Demers N.D., Deng H., Deng Z., Dengjel J., Dent P., Denton D., DePamphilis M.L., Der C.J., Deretic V., Descoteaux A., Devis L., Devkota S., Devuyst O., Dewson G., Dharmasivam M., Dhiman R., di Bernardo D., Di Cristina M., Di Domenico F., Di Fazio P., Di Fonzo A., Di Guardo G., Di Guglielmo G.M., Di Leo L., Di Malta C., Di Nardo A., Di Rienzo M., Di Sano F., Diallinas G., Diao J., Diaz-Araya G., Díaz-Laviada I., Dickinson J.M., Diederich M., Dieudé M., Dikic I., Ding S., Ding W.X., Dini L., Dinić J., Dinic M., Dinkova-Kostova A.T., Dionne M.S., Distler J.H.W., Diwan A., Dixon I.M.C., Djavaheri-Mergny M., Dobrinski I., Dobrovinskaya O., Dobrowolski R., Dobson R.C.J., Đokić J., Dokmeci Emre S., Donadelli M., Dong B., Dong X., Dong Z., Dorn Ii G.W., Dotsch V., Dou H., Dou J., Dowaidar M., Dridi S., Drucker L., Du A., Du C., Du G., Du H.N., Du L.L., du Toit A., Duan S.B., Duan X., Duarte S.P., Dubrovska A., Dunlop E.A., Dupont N., Durán R.V., Dwarakanath B.S., Dyshlovoy S.A., Ebrahimi-Fakhari D., Eckhart L., Edelstein C.L., Efferth T., Eftekharpour E., Eichinger L., Eid N., Eisenberg T., Eissa N.T., Eissa S., Ejarque M., El Andaloussi A., El-Hage N., El-Naggar S., Eleuteri A.M., El-Shafey E.S., Elgendy M., Eliopoulos A.G., Elizalde M.M., Elks P.M., Elsasser H.P., Elsherbiny E.S., Emerling B.M., Emre N.C.T., Eng C.H., Engedal N., Engelbrecht A.M., Engelsen A.S.T., Enserink J.M., Escalante R., Esclatine A., Escobar-Henriques M., Eskelinen E.L., Espert L., Eusebio M.O., Fabrias G., Fabrizi C., Facchiano A., Facchiano F., Fadeel B., Fader C., Faesen A.C., Fairlie W.D., Falcó A., Falkenburger B.H., Fan D., Fan J., Fan Y., Fang E.F., Fang Y., Fang Y., Fanto M., Farfel-Becker T., Faure M., Fazeli G., Fedele A.O., Feldman A.M., Feng D., Feng J., Feng L., Feng Y., Feng Y., Feng W., Fenz Araujo T., Ferguson T.A., Fernández Á.F., Fernandez-Checa J.C., Fernández-Veledo S., Fernie A.R., Ferrante A.W., Jr., Ferraresi A., Ferrari M.F., Ferreira J.C.B., Ferro-Novick S., Figueras A., Filadi R., Filigheddu N., Filippi-Chiela E., Filomeni G., Fimia G.M., Fineschi V., Finetti F., Finkbeiner S., Fisher E.A., Fisher P.B., Flamigni F., Fliesler S.J., Flo T.H., Florance I., Florey O., Florio T., Fodor E., Follo C., Fon E.A., Forlino A., Fornai F., Fortini P., Fracassi A., Fraldi A., Franco B., Franco R., Franconi F., Frankel L.B., Friedman S.L., Fröhlich L.F., Frühbeck G., Fuentes J.M., Fujiki Y., Fujita N., Fujiwara Y., Fukuda M., Fulda S., Furic L., Furuya N., Fusco C., Gack M.U., Gaffke L., Galadari S., Galasso A., Galindo M.F., Gallolu Kankanamalage S., Galluzzi L., Galy V., Gammoh N., Gan B., Ganley I.G., Gao F., Gao H., Gao M., Gao P., Gao S.J., Gao W., Gao X., Garcera A., Garcia M.N., Garcia V.E., García-Del Portillo F., Garcia-Escudero V., Garcia-Garcia A., Garcia-Macia M., García-Moreno D., Garcia-Ruiz C., García-Sanz P., Garg A.D., Gargini R., Garofalo T., Garry R.F., Gassen N.C., Gatica D., Ge L., Ge W., Geiss-Friedlander R., Gelfi C., Genschik P., Gentle I.E., Gerbino V., Gerhardt C., Germain K., Germain M., Gewirtz D.A., Ghasemipour Afshar E., Ghavami S., Ghigo A., Ghosh M., Giamas G., Giampietri C., Giatromanolaki A., Gibson G.E., Gibson S.B., Ginet V., Giniger E., Giorgi C., Girao H., Girardin S.E., Giridharan M., Giuliano S., Giulivi C., Giuriato S., Giustiniani J., Gluschko A., Goder V., Goginashvili A., Golab J., Goldstone D.C., Golebiewska A., Gomes L.R., Gomez R., Gómez-Sánchez R., Gomez-Puerto M.C., Gomez-Sintes R., Gong Q., Goni F.M., González-Gallego J., Gonzalez-Hernandez T., Gonzalez-Polo R.A., Gonzalez-Reyes J.A., González-Rodríguez P., Goping I.S., Gorbatyuk M.S., Gorbunov N.V., Görgülü K., Gorojod R.M., Gorski S.M., Goruppi S., Gotor C., Gottlieb R.A., Gozes I., Gozuacik D., Graef M., Gräler M.H., Granatiero V., Grasso D., Gray J.P., Green D.R., Greenhough A., Gregory S.L., Griffin E.F., Grinstaff M.W., Gros F., Grose C., Gross A.S., Gruber F., Grumati P., Grune T., Gu X., Guan J.L., Guardia C.M., Guda K., Guerra F., Guerri C., Guha P., Guillén C., Gujar S., Gukovskaya A., Gukovsky I., Gunst J., Günther A., Guntur A.R., Guo C., Guo C., Guo H., Guo L.W., Guo M., Gupta P., Gupta S.K., Gupta S., Gupta V.B., Gupta V., Gustafsson A.B., Gutterman D.D., H B R, Haapasalo A., Haber J.E., Hać A., Hadano S., Hafrén A.J., Haidar M., Hall B.S., Halldén G., Hamacher-Brady A., Hamann A., Hamasaki M., Han W., Hansen M., Hanson P.I., Hao Z., Harada M., Harhaji-Trajkovic L., Hariharan N., Haroon N., Harris J., Hasegawa T., Hasima Nagoor N., Haspel J.A., Haucke V., Hawkins W.D., Hay B.A., Haynes C.M., Hayrabedyan S.B., Hays T.S., He C., He Q., He R.R., He Y.W., He Y.Y., Heakal Y., Heberle A.M., Hejtmancik J.F., Helgason G.V., Henkel V., Herb M., Hergovich A., Herman-Antosiewicz A., Hernández A., Hernandez C., Hernandez-Diaz S., Hernandez-Gea V., Herpin A., Herreros J., Hervás J.H., Hesselson D., Hetz C., Heussler V.T., Higuchi Y., Hilfiker S., Hill J.A., Hlavacek W.S., Ho E.A., Ho I.H.T., Ho P.W., Ho S.L., Ho W.Y., Hobbs G.A., Hochstrasser M., Hoet P.H.M., Hofius D., Hofman P., Höhn A., Holmberg C.I., Hombrebueno J.R., Yi-Ren Hong C.H., Hooper L.V., Hoppe T., Horos R., Hoshida Y., Hsin I.L., Hsu H.Y., Hu B., Hu D., Hu L.F., Hu M.C., Hu R., Hu W., Hu Y.C., Hu Z.W., Hua F., Hua J., Hua Y., Huan C., Huang C., Huang C., Huang C., Huang C., Huang H., Huang K., Huang M.L.H., Huang R., Huang S., Huang T., Huang X., Huang Y.J., Huber T.B., Hubert V., Hubner C.A., Hughes S.M., Hughes W.E., Humbert M., Hummer G., Hurley J.H., Hussain S., Hussain S., Hussey P.J., Hutabarat M., Hwang H.Y., Hwang S., Ieni A., Ikeda F., Imagawa Y., Imai Y., Imbriano C., Imoto M., Inman D.M., Inoki K., Iovanna J., Iozzo R.V., Ippolito G., Irazoqui J.E., Iribarren P., Ishaq M., Ishikawa M., Ishimwe N., Isidoro C., Ismail N., Issazadeh-Navikas S., Itakura E., Ito D., Ivankovic D., Ivanova S., Iyer A.K.V., Izquierdo J.M., Izumi M., Jäättelä M., Jabir M.S., Jackson W.T., Jacobo-Herrera N., Jacomin A.C., Jacquin E., Jadiya P., Jaeschke H., Jagannath C., Jakobi A.J., Jakobsson J., Janji B., Jansen-Dürr P., Jansson P.J., Jantsch J., Januszewski S., Jassey A., Jean S., Jeltsch-David H., Jendelova P., Jenny A., Jensen T.E., Jessen N., Jewell J.L., Ji J., Jia L., Jia R., Jiang L., Jiang Q., Jiang R., Jiang T., Jiang X., Jiang Y., Jimenez-Sanchez M., Jin E.J., Jin F., Jin H., Jin L., Jin L., Jin M., Jin S., Jo E.K., Joffre C., Johansen T., Johnson G.V.W., Johnston S.A., Jokitalo E., Jolly M.K., Joosten L.A.B., Jordan J., Joseph B., Ju D., Ju J.S., Ju J., Juárez E., Judith D., Juhász G., Jun Y., Jung C.H., Jung S.C., Jung Y.K., Jungbluth H., Jungverdorben J., Just S., Kaarniranta K., Kaasik A., Kabuta T., Kaganovich D., Kahana A., Kain R., Kajimura S., Kalamvoki M., Kalia M., Kalinowski D.S., Kaludercic N., Kalvari I., Kaminska J., Kaminskyy V.O., Kanamori H., Kanasaki K., Kang C., Kang R., Kang S.S., Kaniyappan S., Kanki T., Kanneganti T.D., Kanthasamy A.G., Kanthasamy A., Kantorow M., Kapuy O., Karamouzis M.V., Karim M.R., Karmakar P., Katare R.G., Kato M., Kaufmann S.H.E., Kauppinen A., Kaushal G.P., Kaushik S., Kawasaki K., Kazan K., Ke P.Y., Keating D.J., Keber U., Kehrl J.H., Keller K.E., Keller C.W., Kemper J.K., Kenific C.M., Kepp O., Kermorgant S., Kern A., Ketteler R., Keulers T.G., Khalfin B., Khalil H., Khambu B., Khan S.Y., Khandelwal V.K.M., Khandia R., Kho W., Khobrekar N.V., Khuansuwan S., Khundadze M., Killackey S.A., Kim D., Kim D.R., Kim D.H., Kim D.E., Kim E.Y., Kim E.K., Kim H.R., Kim H.S., Kim Hyung-Ryong, Kim J.H., Kim J.K., Kim J.H., Kim J., Kim J.H., Kim K.I., Kim P.K., Kim S.J., Kimball S.R., Kimchi A., Kimmelman A.C., Kimura T., King M.A., Kinghorn K.J., Kinsey C.G., Kirkin V., Kirshenbaum L.A., Kiselev S.L., Kishi S., Kitamoto K., Kitaoka Y., Kitazato K., Kitsis R.N., Kittler J.T., Kjaerulff O., Klein P.S., Klopstock T., Klucken J., Knævelsrud H., Knorr R.L., Ko B.C.B., Ko F., Ko J.L., Kobayashi H., Kobayashi S., Koch I., Koch J.C., Koenig U., Kögel D., Koh Y.H., Koike M., Kohlwein S.D., Kocaturk N.M., Komatsu M., König J., Kono T., Kopp B.T., Korcsmaros T., Korkmaz G., Korolchuk V.I., Korsnes M.S., Koskela A., Kota J., Kotake Y., Kotler M.L., Kou Y., Koukourakis M.I., Koustas E., Kovacs A.L., Kovács T., Koya D., Kozako T., Kraft C., Krainc D., Krämer H., Krasnodembskaya A.D., Kretz-Remy C., Kroemer G., Ktistakis N.T., Kuchitsu K., Kuenen S., Kuerschner L., Kukar T., Kumar A., Kumar A., Kumar D., Kumar D., Kumar S., Kume S., Kumsta C., Kundu C.N., Kundu M., Kunnumakkara A.B., Kurgan L., Kutateladze T.G., Kutlu O., Kwak S., Kwon H.J., Kwon T.K., Kwon Y.T., Kyrmizi I., La Spada A., Labonté P., Ladoire S., Laface I., Lafont F., Lagace D.C., Lahiri V., Lai Z., Laird A.S., Lakkaraju A., Lamark T., Lan S.H., Landajuela A., Lane D.J.R., Lane J.D., Lang C.H., Lange C., Langel Ü., Langer R., Lapaquette P., Laporte J., LaRusso N.F., Lastres-Becker I., Lau W.C.Y., Laurie G.W., Lavandero S., Law B.Y.K., Law H.K., Layfield R., Le W., Le Stunff H., Leary A.Y., Lebrun J.J., Leck L.Y.W., Leduc-Gaudet J.P., Lee C., Lee C.P., Lee D.H., Lee E.B., Lee E.F., Lee G.M., Lee H.J., Lee H.K., Lee J.M., Lee J.S., Lee J.A., Lee J.Y., Lee J.H., Lee M., Lee M.G., Lee M.J., Lee M.S., Lee S.Y., Lee S.J., Lee S.Y., Lee S.B., Lee W.H., Lee Y.R., Lee Y.H., Lee Y., Lefebvre C., Legouis R., Lei Y.L., Lei Y., Leikin S., Leitinger G., Lemus L., Leng S., Lenoir O., Lenz G., Lenz H.J., Lenzi P., León Y., Leopoldino A.M., Leschczyk C., Leskelä S., Letellier E., Leung C.T., Leung P.S., Leventhal J.S., Levine B., Lewis P.A., Ley K., Li B., Li D.Q., Li J., Li J., Li J., Li K., Li L., Li M., Li M., Li M., Li M., Li M., Li P.L., Li M.Q., Li Q., Li S., Li T., Li W., Li W., Li X., Li Y.P., Li Y., Li Z., Li Z., Li Z., Lian J., Liang C., Liang Q., Liang W., Liang Y., Liang Y., Liao G., Liao L., Liao M., Liao Y.F., Librizzi M., Lie P.P.Y., Lilly M.A., Lim H.J., Lima T.R.R., Limana F., Lin C., Lin C.W., Lin D.S., Lin F.C., Lin J.D., Lin K.M., Lin K.H., Lin L.T., Lin P.H., Lin Q., Lin S., Lin S.J., Lin W., Lin X., Lin Y.X., Lin Y.S., Linden R., Lindner P., Ling S.C., Lingor P., Linnemann A.K., Liou Y.C., Lipinski M.M., Lipovšek S., Lira V.A., Lisiak N., Liton P.B., Liu C., Liu C.H., Liu C.F., Liu C.H., Liu F., Liu H., Liu H.S., Liu H.F., Liu H., Liu J., Liu J., Liu J., Liu L., Liu L., Liu M., Liu Q., Liu W., Liu W., Liu X.H., Liu X., Liu X., Liu X., Liu X., Liu Y., Liu Y., Liu Y., Liu Y., Liu Y., Livingston J.A., Lizard G., Lizcano J.M., Ljubojevic-Holzer S., LLeonart M.E., Llobet-Navàs D., Llorente A., Lo C.H., Lobato-Márquez D., Long Q., Long Y.C., Loos B., Loos J.A., López M.G., López-Doménech G., López-Guerrero J.A., López-Jiménez A.T., López-Pérez Ó., López-Valero I., Lorenowicz M.J., Lorente M., Lorincz P., Lossi L., Lotersztajn S., Lovat P.E., Lovell J.F., Lovy A., Lőw P., Lu G., Lu H., Lu J.H., Lu J.J., Lu M., Lu S., Luciani A., Lucocq J.M., Ludovico P., Luftig M.A., Luhr M., Luis-Ravelo D., Lum J.J., Luna-Dulcey L., Lund A.H., Lund V.K., Lünemann J.D., Lüningschrör P., Luo H., Luo R., Luo S., Luo Z., Luparello C., Lüscher B., Luu L., Lyakhovich A., Lyamzaev K.G., Lystad A.H., Lytvynchuk L., Ma A.C., Ma C., Ma M., Ma N.F., Ma Q.H., Ma X., Ma Y., Ma Z., MacDougald O.A., Macian F., MacIntosh G.C., MacKeigan J.P., Macleod K.F., Maday S., Madeo F., Madesh M., Madl T., Madrigal-Matute J., Maeda A., Maejima Y., Magarinos M., Mahavadi P., Maiani E., Maiese K., Maiti P., Maiuri M.C., Majello B., Major M.B., Makareeva E., Malik F., Mallilankaraman K., Malorni W., Maloyan A., Mammadova N., Man G.C.W., Manai F., Mancias J.D., Mandelkow E.M., Mandell M.A., Manfredi A.A., Manjili M.H., Manjithaya R., Manque P., Manshian B.B., Manzano R., Manzoni C., Mao K., Marchese C., Marchetti S., Marconi A.M., Marcucci F., Mardente S., Mareninova O.A., Margeta M., Mari M., Marinelli S., Marinelli O., Mariño G., Mariotto S., Marshall R.S., Marten M.R., Martens S., Martin A.P.J., Martin K.R., Martin S., Martin S., Martín-Segura A., Martín-Acebes M.A., Martin-Burriel I., Martin-Rincon M., Martin-Sanz P., Martina J.A., Martinet W., Martinez A., Martinez A., Martinez J., Martinez Velazquez M., Martinez-Lopez N., Martinez-Vicente M., Martins D.O., Martins J.O., Martins W.K., Martins-Marques T., Marzetti E., Masaldan S., Masclaux-Daubresse C., Mashek D.G., Massa V., Massieu L., Masson G.R., Masuelli L., Masyuk A.I., Masyuk T.V., Matarrese P., Matheu A., Matoba S., Matsuzaki S., Mattar P., Matte A., Mattoscio D., Mauriz J.L., Mauthe M., Mauvezin C., Maverakis E., Maycotte P., Mayer J., Mazzoccoli G., Mazzoni C., Mazzulli J.R., McCarty N., McDonald C., McGill M.R., McKenna S.L., McLaughlin B., McLoughlin F., McNiven M.A., McWilliams T.G., Mechta-Grigoriou F., Medeiros T.C., Medina D.L., Megeney L.A., Megyeri K., Mehrpour M., Mehta J.L., Meijer A.J., Meijer A.H., Mejlvang J., Meléndez A., Melk A., Memisoglu G., Mendes A.F., Meng D., Meng F., Meng T., Menna-Barreto R., Menon M.B., Mercer C., Mercier A.E., Mergny J.L., Merighi A., Merkley S.D., Merla G., Meske V., Mestre A.C., Metur S.P., Meyer C., Meyer H., Mi W., Mialet-Perez J., Miao J., Micale L., Miki Y., Milan E., Milczarek M., Miller D.L., Miller S.I., Miller S., Millward S.W., Milosevic I., Minina E.A., Mirzaei H., Mirzaei H.R., Mirzaei M., Mishra A., Mishra N., Mishra P.K., Misirkic Marjanovic M., Misasi R., Misra A., Misso G., Mitchell C., Mitou G., Miura T., Miyamoto S., Miyazaki M., Miyazaki M., Miyazaki T., Miyazawa K., Mizushima N., Mogensen T.H., Mograbi B., Mohammadinejad R., Mohamud Y., Mohanty A., Mohapatra S., Möhlmann T., Mohmmed A., Moles A., Moley K.H., Molinari M., Mollace V., Møller A.B., Mollereau B., Mollinedo F., Montagna C., Monteiro M.J., Montella A., Montes L.R., Montico B., Mony V.K., Monzio Compagnoni G., Moore M.N., Moosavi M.A., Mora A.L., Mora M., Morales-Alamo D., Moratalla R., Moreira P.I., Morelli E., Moreno S., Moreno-Blas D., Moresi V., Morga B., Morgan A.H., Morin F., Morishita H., Moritz O.L., Moriyama M., Moriyasu Y., Morleo M., Morselli E., Moruno-Manchon J.F., Moscat J., Mostowy S., Motori E., Moura A.F., Moustaid-Moussa N., Mrakovcic M., Muciño-Hernández G., Mukherjee A., Mukhopadhyay S., Mulcahy Levy J.M., Mulero V., Muller S., Münch C., Munjal A., Munoz-Canoves P., Muñoz-Galdeano T., Münz C., Murakawa T., Muratori C., Murphy B.M., Murphy J.P., Murthy A., Myöhänen T.T., Mysorekar I.U., Mytych J., Nabavi S.M., Nabissi M., Nagy P., Nah J., Nahimana A., Nakagawa I., Nakamura K., Nakatogawa H., Nandi S.S., Nanjundan M., Nanni M., Napolitano G., Nardacci R., Narita M., Nassif M., Nathan I., Natsumeda M., Naude R.J., Naumann C., Naveiras O., Navid F., Nawrocki S.T., Nazarko T.Y., Nazio F., Negoita F., Neill T., Neisch A.L., Neri L.M., Netea M.G., Neubert P., Neufeld T.P., Neumann D., Neutzner A., Newton P.T., Ney P.A., Nezis I.P., Ng C.C.W., Ng T.B., Nguyen H.T.T., Nguyen L.T., Ni H.M., Ní Cheallaigh C., Ni Z., Nicolao M.C., Nicoli F., Nieto-Diaz M., Nilsson P., Ning S., Niranjan R., Nishimune H., Niso-Santano M., Nixon R.A., Nobili A., Nobrega C., Noda T., Nogueira-Recalde U., Nolan T.M., Nombela I., Novak I., Novoa B., Nozawa T., Nukina N., Nussbaum-Krammer C., Nylandsted J., O’Donovan T.R., O’Leary S.M., O’Rourke E.J., O’Sullivan M.P., O’Sullivan T.E., Oddo S., Oehme I., Ogawa M., Ogier-Denis E., Ogmundsdottir M.H., Ogretmen B., Oh G.T., Oh S.H., Oh Y.J., Ohama T., Ohashi Y., Ohmuraya M., Oikonomou V., Ojha R., Okamoto K., Okazawa H., Oku M., Oliván S., Oliveira J.M.A., Ollmann M., Olzmann J.A., Omari S., Omary M.B., Önal G., Ondrej M., Ong S.B., Ong S.G., Onnis A., Orellana J.A., Orellana-Muñoz S., Ortega-Villaizan M.D.M., Ortiz-Gonzalez X.R., Ortona E., Osiewacz H.D., Osman A.K., Osta R., Otegui M.S., Otsu K., Ott C., Ottobrini L., Ou J.J., Outeiro T.F., Oynebraten I., Ozturk M., Pagès G., Pahari S., Pajares M., Pajvani U.B., Pal R., Paladino S., Pallet N., Palmieri M., Palmisano G., Palumbo C., Pampaloni F., Pan L., Pan Q., Pan W., Pan X., Panasyuk G., Pandey R., Pandey U.B., Pandya V., Paneni F., Pang S.Y., Panzarini E., Papademetrio D.L., Papaleo E., Papinski D., Papp D., Park E.C., Park H.T., Park J.M., Park J.I., Park J.T., Park J., Park S.C., Park S.Y., Parola A.H., Parys J.B., Pasquier A., Pasquier B., Passos J.F., Pastore N., Patel H.H., Patschan D., Pattingre S., Pedraza-Alva G., Pedraza-Chaverri J., Pedrozo Z., Pei G., Pei J., Peled-Zehavi H., Pellegrini J.M., Pelletier J., Peñalva M.A., Peng D., Peng Y., Penna F., Pennuto M., Pentimalli F., Pereira C.M., Pereira G.J.S., Pereira L.C., Pereira de Almeida L., Perera N.D., Pérez-Lara Á, Perez-Oliva A.B., Pérez-Pérez M.E., Periyasamy P., Perl A., Perrotta C., Perrotta I., Pestell R.G., Petersen M., Petrache I., Petrovski G., Pfirrmann T., Pfister A.S., Philips J.A., Pi H., Picca A., Pickrell A.M., Picot S., Pierantoni G.M., Pierdominici M., Pierre P., Pierrefite-Carle V., Pierzynowska K., Pietrocola F., Pietruczuk M., Pignata C., Pimentel-Muiños F.X., Pinar M., Pinheiro R.O., Pinkas-Kramarski R., Pinton P., Pircs K., Piya S., Pizzo P., Plantinga T.S., Platta H.W., Plaza-Zabala A., Plomann M., Plotnikov E.Y., Plun-Favreau H., Pluta R., Pocock R., Pöggeler S., Pohl C., Poirot M., Poletti A., Ponpuak M., Popelka H., Popova B., Porta H., Porte Alcon S., Portilla-Fernandez E., Post M., Potts M.B., Poulton J., Powers T., Prahlad V., Prajsnar T.K., Praticò D., Prencipe R., Priault M., Proikas-Cezanne T., Promponas V.J., Proud C.G., Puertollano R., Puglielli L., Pulinilkunnil T., Puri D., Puri R., Puyal J., Qi X., Qi Y., Qian W., Qiang L., Qiu Y., Quadrilatero J., Quarleri J., Raben N., Rabinowich H., Ragona D., Ragusa M.J., Rahimi N., Rahmati M., Raia V., Raimundo N., Rajasekaran N.S., Ramachandra Rao S., Rami A., Ramírez-Pardo I., Ramsden D.B., Randow F., Rangarajan P.N., Ranieri D., Rao H., Rao L., Rao R., Rathore S., Ratnayaka J.A., Ratovitski E.A., Ravanan P., Ravegnini G., Ray S.K., Razani B., Rebecca V., Reggiori F., Régnier-Vigouroux A., Reichert A.S., Reigada D., Reiling J.H., Rein T., Reipert S., Rekha R.S., Ren H., Ren J., Ren W., Renault T., Renga G., Reue K., Rewitz K., Ribeiro de Andrade Ramos B., Riazuddin S.A., Ribeiro-Rodrigues T.M., Ricci J.E., Ricci R., Riccio V., Richardson D.R., Rikihisa Y., Risbud M.V., Risueño R.M., Ritis K., Rizza S., Rizzuto R., Roberts H.C., Roberts L.D., Robinson K.J., Roccheri M.C., Rocchi S., Rodney G.G., Rodrigues T., Rodrigues Silva V.R., Rodriguez A., Rodriguez-Barrueco R., Rodriguez-Henche N., Rodriguez-Rocha H., Roelofs J., Rogers R.S., Rogov V.V., Rojo A.I., Rolka K., Romanello V., Romani L., Romano A., Romano P.S., Romeo-Guitart D., Romero L.C., Romero M., Roney J.C., Rongo C., Roperto S., Rosenfeldt M.T., Rosenstiel P., Rosenwald A.G., Roth K.A., Roth L., Roth S., Rouschop K.M.A., Roussel B.D., Roux S., Rovere-Querini P., Roy A., Rozieres A., Ruano D., Rubinsztein D.C., Rubtsova M.P., Ruckdeschel K., Ruckenstuhl C., Rudolf E., Rudolf R., Ruggieri A., Ruparelia A.A., Rusmini P., Russell R.R., Russo G.L., Russo M., Russo R., Ryabaya O.O., Ryan K.M., Ryu K.Y., Sabater-Arcis M., Sachdev U., Sacher M., Sachse C., Sadhu A., Sadoshima J., Safren N., Saftig P., Sagona A.P., Sahay G., Sahebkar A., Sahin M., Sahin O., Sahni S., Saito N., Saito S., Saito T., Sakai R., Sakai Y., Sakamaki J.I., Saksela K., Salazar G., Salazar-Degracia A., Salekdeh G.H., Saluja A.K., Sampaio-Marques B., Sanchez M.C., Sanchez-Alcazar J.A., Sanchez-Vera V., Sancho-Shimizu V., Sanderson J.T., Sandri M., Santaguida S., Santambrogio L., Santana M.M., Santoni G., Sanz A., Sanz P., Saran S., Sardiello M., Sargeant T.J., Sarin A., Sarkar C., Sarkar S., Sarrias M.R., Sarkar S., Sarmah D.T., Sarparanta J., Sathyanarayan A., Sathyanarayanan R., Scaglione K.M., Scatozza F., Schaefer L., Schafer Z.T., Schaible U.E., Schapira A.H.V., Scharl M., Schatzl H.M., Schein C.H., Scheper W., Scheuring D., Schiaffino M.V., Schiappacassi M., Schindl R., Schlattner U., Schmidt O., Schmitt R., Schmidt S.D., Schmitz I., Schmukler E., Schneider A., Schneider B.E., Schober R., Schoijet A.C., Schott M.B., Schramm M., Schröder B., Schuh K., Schüller C., Schulze R.J., Schürmanns L., Schwamborn J.C., Schwarten M., Scialo F., Sciarretta S., Scott M.J., Scotto K.W., Scovassi A.I., Scrima A., Scrivo A., Sebastian D., Sebti S., Sedej S., Segatori L., Segev N., Seglen P.O., Seiliez I., Seki E., Selleck S.B., Sellke F.W., Selsby J.T., Sendtner M., Senturk S., Seranova E., Sergi C., Serra-Moreno R., Sesaki H., Settembre C., Setty S.R.G., Sgarbi G., Sha O., Shacka J.J., Shah J.A., Shang D., Shao C., Shao F., Sharbati S., Sharkey L.M., Sharma D., Sharma G., Sharma K., Sharma P., Sharma S., Shen H.M., Shen H., Shen J., Shen M., Shen W., Shen Z., Sheng R., Sheng Z., Sheng Z.H., Shi J., Shi X., Shi Y.H., Shiba-Fukushima K., Shieh J.J., Shimada Y., Shimizu S., Shimozawa M., Shintani T., Shoemaker C.J., Shojaei S., Shoji I., Shravage B.V., Shridhar V., Shu C.W., Shu H.B., Shui K., Shukla A.K., Shutt T.E., Sica V., Siddiqui A., Sierra A., Sierra-Torre V., Signorelli S., Sil P., Silva B.J.A., Silva J.D., Silva-Pavez E., Silvente-Poirot S., Simmonds R.E., Simon A.K., Simon H.U., Simons M., Singh A., Singh L.P., Singh R., Singh S.V., Singh S.K., Singh S.B., Singh S., Singh S.P., Sinha D., Sinha R.A., Sinha S., Sirko A., Sirohi K., Sivridis E.L., Skendros P., Skirycz A., Slaninová I., Smaili S.S., Smertenko A., Smith M.D., Soenen S.J., Sohn E.J., Sok S.P.M., Solaini G., Soldati T., Soleimanpour S.A., Soler R.M., Solovchenko A., Somarelli J.A., Sonawane A., Song F., Song H.K., Song J.X., Song K., Song Z., Soria L.R., Sorice M., Soukas A.A., Soukup S.F., Sousa D., Sousa N., Spagnuolo P.A., Spector S.A., Srinivas Bharath M.M., St Clair D., Stagni V., Staiano L., Stalnecker C.A., Stankov M.V., Stathopulos P.B., Stefan K., Stefan S.M., Stefanis L., Steffan J.S., Steinkasserer A., Stenmark H., Sterneckert J., Stevens C., Stoka V., Storch S., Stork B., Strappazzon F., Strohecker A.M., Stupack D.G., Su H., Su L.Y., Su L., Suarez-Fontes A.M., Subauste C.S., Subbian S., Subirada P.V., Sudhandiran G., Sue C.M., Sui X., Summers C., Sun G., Sun J., Sun K., Sun M.X., Sun Q., Sun Y., Sun Z., Sunahara K.K.S., Sundberg E., Susztak K., Sutovsky P., Suzuki H., Sweeney G., Symons J.D., Sze S.C.W., Szewczyk N.J., Tabęcka-Łonczynska A., Tabolacci C., Tacke F., Taegtmeyer H., Tafani M., Tagaya M., Tai H., Tait S.W.G., Takahashi Y., Takats S., Talwar P., Tam C., Tam S.Y., Tampellini D., Tamura A., Tan C.T., Tan E.K., Tan Y.Q., Tanaka M., Tanaka M., Tang D., Tang J., Tang T.S., Tanida I., Tao Z., Taouis M., Tatenhorst L., Tavernarakis N., Taylor A., Taylor G.A., Taylor J.M., Tchetina E., Tee A.R., Tegeder I., Teis D., Teixeira N., Teixeira-Clerc F., Tekirdag K.A., Tencomnao T., Tenreiro S., Tepikin A.V., Testillano P.S., Tettamanti G., Tharaux P.L., Thedieck K., Thekkinghat A.A., Thellung S., Thinwa J.W., Thirumalaikumar V.P., Thomas S.M., Thomes P.G., Thorburn A., Thukral L., Thum T., Thumm M., Tian L., Tichy A., Till A., Timmerman V., Titorenko V.I., Todi S.V., Todorova K., Toivonen J.M., Tomaipitinca L., Tomar D., Tomas-Zapico C., Tomić S., Tong B.C., Tong C., Tong X., Tooze S.A., Torgersen M.L., Torii S., Torres-López L., Torriglia A., Towers C.G., Towns R., Toyokuni S., Trajkovic V., Tramontano D., Tran Q.G., Travassos L.H., Trelford C.B., Tremel S., Trougakos I.P., Tsao B.P., Tschan M.P., Tse H.F., Tse T.F., Tsugawa H., Tsvetkov A.S., Tumbarello D.A., Tumtas Y., Tuñón M.J., Turcotte S., Turk B., Turk V., Turner B.J., Tuxworth R.I., Tyler J.K., Tyutereva E.V., Uchiyama Y., Ugun-Klusek A., Uhlig H.H., Ułamek-Kozioł M., Ulasov I.V., Umekawa M., Ungermann C., Unno R., Urbe S., Uribe-Carretero E., Üstün S., Uversky V.N., Vaccari T., Vaccaro M.I., Vahsen B.F., Vakifahmetoglu-Norberg H., Valdor R., Valente M.J., Valko A., Vallee R.B., Valverde A.M., Van den Berghe G., van der Veen S., Van Kaer L., van Loosdregt J., van Wijk S.J.L., Vandenberghe W., Vanhorebeek I., Vannier-Santos M.A., Vannini N., Vanrell M.C., Vantaggiato C., Varano G., Varela-Nieto I., Varga M., Vasconcelos M.H., Vats S., Vavvas D.G., Vega-Naredo I., Vega-Rubin-de-Celis S., Velasco G., Velázquez A.P., Vellai T., Vellenga E., Velotti F., Verdier M., Verginis P., Vergne I., Verkade P., Verma M., Verstreken P., Vervliet T., Vervoorts J., Vessoni A.T., Victor V.M., Vidal M., Vidoni C., Vieira O.V., Vierstra R.D., Viganó S., Vihinen H., Vijayan V., Vila M., Vilar M., Villalba J.M., Villalobo A., Villarejo-Zori B., Villarroya F., Villarroya J., Vincent O., Vindis C., Viret C., Viscomi M.T., Visnjic D., Vitale I., Vocadlo D.J., Voitsekhovskaja O.V., Volonté C., Volta M., Vomero M., Von Haefen C., Vooijs M.A., Voos W., Vucicevic L., Wade-Martins R., Waguri S., Waite K.A., Wakatsuki S., Walker D.W., Walker M.J., Walker S.A., Walter J., Wandosell F.G., Wang B., Wang C.Y., Wang C., Wang C., Wang C., Wang C.Y., Wang D., Wang F., Wang F., Wang F., Wang G., Wang H., Wang H., Wang H., Wang H.G., Wang J., Wang J., Wang J., Wang J., Wang K., Wang L., Wang L., Wang M.H., Wang M., Wang N., Wang P., Wang P., Wang P., Wang P., Wang Q.J., Wang Q., Wang Q.K., Wang Q.A., Wang W.T., Wang W., Wang X., Wang X., Wang Y., Wang Y., Wang Y., Wang Y.Y., Wang Y., Wang Y., Wang Y., Wang Y., Wang Z., Wang Z., Wang Z., Warnes G., Warnsmann V., Watada H., Watanabe E., Watchon M., Wawrzyńska A., Weaver T.E., Wegrzyn G., Wehman A.M., Wei H., Wei L., Wei T., Wei Y., Weiergräber O.H., Weihl C.C., Weindl G., Weiskirchen R., Wells A., Wen R.H., Wen X., Werner A., Weykopf B., Wheatley S.P., Whitton J.L., Whitworth A.J., Wiktorska K., Wildenberg M.E., Wileman T., Wilkinson S., Willbold D., Williams B., Williams R.S.B., Williams R.L., Williamson P.R., Wilson R.A., Winner B., Winsor N.J., Witkin S.S., Wodrich H., Woehlbier U., Wollert T., Wong E., Wong J.H., Wong R.W., Wong V.K.W., Wong W.W., Wu A.G., Wu C., Wu J., Wu J., Wu K.K., Wu M., Wu S.Y., Wu S., Wu S.Y., Wu S., Wu W.K.K., Wu X., Wu X., Wu Y.W., Wu Y., Xavier R.J., Xia H., Xia L., Xia Z., Xiang G., Xiang J., Xiang M., Xiang W., Xiao B., Xiao G., Xiao H., Xiao H.T., Xiao J., Xiao L., Xiao S., Xiao Y., Xie B., Xie C.M., Xie M., Xie Y., Xie Z., Xie Z., Xilouri M., Xu C., Xu E., Xu H., Xu J., Xu J., Xu L., Xu W.W., Xu X., Xue Y., Yakhine-Diop S.M.S., Yamaguchi M., Yamaguchi O., Yamamoto A., Yamashina S., Yan S., Yan S.J., Yan Z., Yanagi Y., Yang C., Yang D.S., Yang H., Yang H.T., Yang H., Yang J.M., Yang J., Yang J., Yang L., Yang L., Yang M., Yang P.M., Yang Q., Yang S., Yang S., Yang S.F., Yang W., Yang W.Y., Yang X., Yang X., Yang Y., Yang Y., Yao H., Yao S., Yao X., Yao Y.G., Yao Y.M., Yasui T., Yazdankhah M., Yen P.M., Yi C., Yin X.M., Yin Y., Yin Z., Yin Z., Ying M., Ying Z., Yip C.K., Yiu S.P.T., Yoo Y.H., Yoshida K., Yoshii S.R., Yoshimori T., Yousefi B., Yu B., Yu H., Yu J., Yu J., Yu L., Yu M.L., Yu S.W., Yu V.C., Yu W.H., Yu Z., Yu Z., Yuan J., Yuan L.Q., Yuan S., Yuan S.F., Yuan Y., Yuan Z., Yue J., Yue Z., Yun J., Yung R.L., Zacks D.N., Zaffagnini G., Zambelli V.O., Zanella I., Zang Q.S., Zanivan S., Zappavigna S., Zaragoza P., Zarbalis K.S., Zarebkohan A., Zarrouk A., Zeitlin S.O., Zeng J., Zeng J.D., Žerovnik E., Zhan L., Zhang B., Zhang D.D., Zhang H., Zhang H., Zhang H., Zhang H., Zhang H., Zhang H., Zhang H., Zhang H.L., Zhang J., Zhang J., Zhang J.P., Zhang K.Y.B., Zhang L.W., Zhang L., Zhang L., Zhang L., Zhang L., Zhang M., Zhang P., Zhang S., Zhang W., Zhang X., Zhang X.W., Zhang X., Zhang X., Zhang X., Zhang X., Zhang X.D., Zhang Y., Zhang Y., Zhang Y., Zhang Y.D., Zhang Y., Zhang Y.Y., Zhang Y., Zhang Z., Zhang Z., Zhang Z., Zhang Z., Zhang Z., Zhang Z., Zhao H., Zhao L., Zhao S., Zhao T., Zhao X.F., Zhao Y., Zhao Y., Zhao Y., Zhao Y., Zheng G., Zheng K., Zheng L., Zheng S., Zheng X.L., Zheng Y., Zheng Z.G., Zhivotovsky B., Zhong Q., Zhou A., Zhou B., Zhou C., Zhou G., Zhou H., Zhou H., Zhou H., Zhou J., Zhou J., Zhou J., Zhou J., Zhou K., Zhou R., Zhou X.J., Zhou Y., Zhou Y., Zhou Y., Zhou Z.Y., Zhou Z., Zhu B., Zhu C., Zhu G.Q., Zhu H., Zhu H., Zhu H., Zhu W.G., Zhu Y., Zhu Y., Zhuang H., Zhuang X., Zientara-Rytter K., Zimmermann C.M., Ziviani E., Zoladek T., Zong W.X., Zorov D.B., Zorzano A., Zou W., Zou Z., Zou Z., Zuryn S., Zwerschke W., Brand-Saberi B., Dong X.C., Kenchappa C.S., Li Z., Lin Y., Oshima S., Rong Y., Sluimer J.C., Stallings C.L., Tong C.K. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolz A., Ernst A., Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 14.Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 15.Codogno P., Mehrpour M., Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer S.R. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell. 2017;28:712–715. doi: 10.1091/mbc.E16-10-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 19.Szatmári Z., Sass M. The autophagic roles of Rab small GTPases and their upstream regulators. Autophagy. 2014;10:1154–1166. doi: 10.4161/auto.29395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou F., Wu Z., Zhao M., Murtazina R., Cai J., Zhang A., Li R., Sun D., Li W., Zhao L., Li Q., Zhu J., Cong X., Zhou Y., Xie Z., Gyurkovska V., Li L., Huang X., Xue Y., Chen L., Xu H., Xu H., Liang Y., Segev N. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol. 2019;218:1908–1927. doi: 10.1083/jcb.201811173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puri C., Vicinanza M., Ashkenazi A., Gratian M.J., Zhang Q., Bento C.F., Renna M., Menzies F.M., Rubinsztein D.C. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev Cell. 2018;45:114–131 e8. doi: 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K., Mashima H., Miura K., Maeda D., Goto A., Goto T., Sun-Wada G.H., Wada Y., Ohnishi H. Disruption of small GTPase Rab7 exacerbates the severity of acute pancreatitis in experimental mouse models. Sci Rep. 2017;7:2817. doi: 10.1038/s41598-017-02988-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito T., Nah J., Oka S.I., Mukai R., Monden Y., Maejima Y., Ikeda Y., Sciarretta S., Liu T., Li H., Baljinnyam E., Fraidenraich D., Fritzky L., Zhai P., Ichinose S., Isobe M., Hsu C.P., Kundu M., Sadoshima J. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest. 2019;129:802–819. doi: 10.1172/JCI122035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C.Y., Kuo W.W., Ho T.J., Chiang S.F., Pai P.Y., Lin J.Y., Lin D.Y., Kuo C.H. Rab9-dependent autophagy is required for the IGF-IIR triggering mitophagy to eliminate damaged mitochondria. J Cell Physiol. 2018;233:7080–7091. doi: 10.1002/jcp.26346. [DOI] [PubMed] [Google Scholar]
- 25.Biczo G., Vegh E.T., Shalbueva N., Mareninova O.A., Elperin J., Lotshaw E., Gretler S., Lugea A., Malla S.R., Dawson D., Ruchala P., Whitelegge J., French S.W., Wen L., Husain S.Z., Gorelick F.S., Hegyi P., Rakonczay Z., Jr., Gukovsky I., Gukovskaya A.S. Mitochondrial dysfunction, through impaired autophagy, leads to endoplasmic reticulum stress, deregulated lipid metabolism, and pancreatitis in animal models. Gastroenterology. 2018;154:689–703. doi: 10.1053/j.gastro.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mareninova O.A., Jia W., Gretler S.R., Holthaus C.L., Thomas D.D.H., Pimienta M., Dillon D.L., Gukovskaya A.S., Gukovsky I., Groblewski G.E. Transgenic expression of GFP-LC3 perturbs autophagy in exocrine pancreas and acute pancreatitis responses in mice. Autophagy. 2020;16:2084–2097. doi: 10.1080/15548627.2020.1715047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gukovskaya A.S., Pandol S.J., Gukovsky I. New insights into the pathways initiating and driving pancreatitis. Curr Opin Gastroenterol. 2016;32:429–435. doi: 10.1097/MOG.0000000000000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaptzan T., West S.A., Holicky E.L., Wheatley C.L., Marks D.L., Wang T., Peake K.B., Vance J., Walkley S.U., Pagano R.E. Development of a Rab9 transgenic mouse and its ability to increase the lifespan of a murine model of Niemann-Pick type C disease. Am J Pathol. 2009;174:14–20. doi: 10.2353/ajpath.2009.080660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Homma Y., Hiragi S., Fukuda M. Rab family of small GTPases: an updated view on their regulation and functions. FEBS J. 2021;288:36–55. doi: 10.1111/febs.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung K.F., Baron R., Seabra M.C. Thematic review series: lipid posttranslational modifications—geranylgeranylation of Rab GTPases. J Lipid Res. 2006;47:467–475. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.DerMardirossian C., Bokoch G.M. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Baron R.A., Seabra M.C. Rab geranylgeranylation occurs preferentially via the pre-formed REP-RGGT complex and is regulated by geranylgeranyl pyrophosphate. Biochem J. 2008;415:67–75. doi: 10.1042/BJ20080662. [DOI] [PubMed] [Google Scholar]
- 33.de la Vega M., Burrows J.F., Johnston J.A. Ubiquitination: added complexity in Ras and Rho family GTPase function. Small GTPases. 2011;2:192–201. doi: 10.4161/sgtp.2.4.16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukas J., Pospech J., Oppermann C., Hund C., Iwanov K., Pantoom S., Petters J., Frech M., Seemann S., Thiel F.G., Modenbach J.M., Bolsmann R., de Freitas Chama L., Kraatz F., El-Hage F., Gronbach M., Klein A., Müller R., Salloch S., Weiss F.U., Simon P., Wagh P., Klemenz A., Krüger E., Mayerle J., Delcea M., Kragl U., Beller M., Rolfs A., Lerch M.M., Sendler M. Role of endoplasmic reticulum stress and protein misfolding in disorders of the liver and pancreas. Adv Med Sci. 2019;64:315–323. doi: 10.1016/j.advms.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 36.Yu Z.Q., Ni T., Hong B., Wang H.Y., Jiang F.J., Zou S., Chen Y., Zheng X.L., Klionsky D.J., Liang Y., Xie Z. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8:883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez A.F., Lopez-Otin C. The functional and pathologic relevance of autophagy proteases. J Clin Invest. 2015;125:33–41. doi: 10.1172/JCI73940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbero P., Bittova L., Pfeffer S.R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kucera A., Bakke O., Progida C. The multiple roles of Rab9 in the endolysosomal system. Commun Integr Biol. 2016;9 doi: 10.1080/19420889.2016.1204498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogov V., Dotsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Kirkin V., Rogov V.V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Sengupta D., Graham M., Liu X., Cresswell P. Proteasomal degradation within endocytic organelles mediates antigen cross-presentation. EMBO J. 2019;38 doi: 10.15252/embj.201899266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knaevelsrud H., Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–2645. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 44.Lugea A., Waldron R.T., Mareninova O.A., Shalbueva N., Deng N., Su H.Y., Thomas D.D., Jones E.K., Messenger S.W., Yang J., Hu C., Gukovsky I., Liu Z., Groblewski G.E., Gukovskaya A.S., Gorelick F.S., Pandol S.J. Human pancreatic acinar cells: proteomic characterization, physiologic responses, and organellar disorders in ex vivo pancreatitis. Am J Pathol. 2017;187:2726–2743. doi: 10.1016/j.ajpath.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mareninova O.A., Sung K.F., Hong P., Lugea A., Pandol S.J., Gukovsky I., Gukovskaya A.S. Cell death in pancreatitis: caspases protect from necrotizing pancreatitis. J Biol Chem. 2006;281:3370–3381. doi: 10.1074/jbc.M511276200. [DOI] [PubMed] [Google Scholar]
- 46.Mareninova O., Orabi A.I., Husain S., Sohail Z. In: Pancreatitis. Mountain View. Williams J.A., editor. Michigan Publishing; , CA: 2016. Experimental acute pancreatitis: in vitro models; pp. 3–14. [Google Scholar]
- 47.Hashimoto D., Ohmuraya M., Hirota M., Yamamoto A., Suyama K., Ida S., Okumura Y., Takahashi E., Kido H., Araki K., Baba H., Mizushima N., Yamamura K. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang T., Dolai S., Xie L., Winter E., Orabi A.I., Karimian N., Cosen-Binker L.I., Huang Y.C., Thorn P., Cattral M.S., Gaisano H.Y. Ex vivo human pancreatic slice preparations offer a valuable model for studying pancreatic exocrine biology. J Biol Chem. 2017;292:5957–5969. doi: 10.1074/jbc.M117.777433. [DOI] [PMC free article] [PubMed] [Google Scholar]