Summary
The 2-oxoglutarate dehydrogenase-like (OGDHL) protein is a rate-limiting enzyme in the Krebs cycle that plays a pivotal role in mitochondrial metabolism. OGDHL expression is restricted mainly to the brain in humans. Here, we report nine individuals from eight unrelated families carrying bi-allelic variants in OGDHL with a range of neurological and neurodevelopmental phenotypes including epilepsy, hearing loss, visual impairment, gait ataxia, microcephaly, and hypoplastic corpus callosum. The variants include three homozygous missense variants (p.Pro852Ala, p.Arg244Trp, and p.Arg299Gly), three compound heterozygous single-nucleotide variants (p.Arg673Gln/p.Val488Val, p.Phe734Ser/p.Ala327Val, and p.Trp220Cys/p.Asp491Val), one homozygous frameshift variant (p.Cys553Leufs∗16), and one homozygous stop-gain variant (p.Arg440Ter). To support the pathogenicity of the variants, we developed a novel CRISPR-Cas9-mediated tissue-specific knockout with cDNA rescue system for dOgdh, the Drosophila ortholog of human OGDHL. Pan-neuronal knockout of dOgdh led to developmental lethality as well as defects in Krebs cycle metabolism, which was fully rescued by expression of wild-type dOgdh. Studies using the Drosophila system indicate that p.Arg673Gln, p.Phe734Ser, and p.Arg299Gly are severe loss-of-function alleles, leading to developmental lethality, whereas p.Pro852Ala, p.Ala327Val, p.Trp220Cys, p.Asp491Val, and p.Arg244Trp are hypomorphic alleles, causing behavioral defects. Transcript analysis from fibroblasts obtained from the individual carrying the synonymous variant (c.1464T>C [p.Val488Val]) in family 2 showed that the synonymous variant affects splicing of exon 11 in OGDHL. Human neuronal cells with OGDHL knockout exhibited defects in mitochondrial respiration, indicating the essential role of OGDHL in mitochondrial metabolism in humans. Together, our data establish that the bi-allelic variants in OGDHL are pathogenic, leading to a Mendelian neurodevelopmental disease in humans.
Keywords: OGDHL, mitochondria, α-ketoglutarate, bi-allelic, developmental and epileptic encephalopathy, DEE, exome sequencing, Drosophila, CRISPR-Cas9 gene editing, neurodevelopmental disease
Introduction
The Krebs cycle, also known as the tricarboxylic acid (TCA) cycle, plays an essential role in mitochondrial metabolism in all metazoans. Pathogenic variation in genes encoding enzymes or subunits of enzyme complexes of the Krebs cycle causes recessive neurometabolic diseases. These diseases and the genes that underlie their etiology include Leigh syndrome or Leigh-like disease (DLD [MIM: 238331], SUCLG1 [MIM: 611224], SUCLA2 [MIM: 603921], SDHA [MIM: 600857], and FH [MIM: 136850]1, 2, 3, 4, 5), infantile cerebellar-retinal degeneration (ACO2 [MIM: 100850]6), infantile leukoencephalopathy (SDHAF1 [MIM: 612848]7), and a specific subtype of developmental and epileptic encephalopathy (MDH2 [MIM: 154100]8). These diseases are characterized developmental delay and neurological manifestations that include seizures, ataxia, optic atrophy, and encephalopathy. Onset is typically in infancy.
All Krebs cycle enzymes and subunits of enzyme complexes have a single copy of genes in humans but not the genes encoding the enzymes having 2-oxoglutarate dehydrogenase (OGDH) activity for the α-ketoglutarate dehydrogenase complex. OGDH (MIM: 613022) and OGDH-like (OGDHL) (MIM: 617513) appear to have evolved by duplication of a single ancestral gene.9 OGDH and OGDHL play an essential part in a rate-limiting enzymatic reaction for the conversion of α-ketoglutarate (α-KG) to succinyl-CoA. Similar to other Krebs cycle enzymes, OGDH is widely expressed in most tissues.10,11 Two individuals carrying a homozygous missense variant in OGDH (GenBank: NM_002541.3; c.959A>G [p.Asn320Ser]) presented with developmental delay, seizure, and ataxia.12 In contrast to the ubiquitous expression of OGDH, OGDHL expression is mainly restricted to the brain.11,13 Higher expression of OGDHL in human brains suggests the involvement of OGDHL in brain development and function.
Here, we report nine individuals from eight unrelated families with bi-allelic variants in OGDHL. The individuals presented with diverse neurodevelopmental phenotypes including epileptic encephalopathy, gait ataxia, hearing loss, and optic atrophy, combined with microcephaly and hypoplastic corpus callosum. Functional studies in Drosophila melanogaster and fibroblasts showed that all nine single-nucleotide variants (SNVs) identified from individuals carrying bi-allelic OGDHL variants are loss-of-function (LOF) alleles, indicating that loss of OGDHL function underlies a neurodevelopmental disease in humans.
Subjects and methods
Subjects
Clinical ascertainment included physical examination, medical history interviews, and specialized consultation by pediatric neurologists and clinical geneticists. This study was approved by local institutional review board (IRB)/ethical review boards, and written informed consent was obtained prior to genetic testing from the families involved. Clinical details were obtained through medical file review and clinical examination. Family 1 provided consent according to the protocol of Columbia University (IRB-AAAS2343) approved by the IRB at Columbia University. Family 2 provided consent according to the protocol (ethics committee [EC] reference #21, 9/9/15, Project MitMed) approved by the IRB at University of Milan. Family 3 provided consent according to the IRB protocol (A115/02, A116/02) at the University of Kiel. Family 4 was consented for the research in accordance with IRB-approved protocol (15-012226) of the Epilepsy Genetics Research Project (EGRP) at Children’s Hospital of Philadelphia. Families 5, 6, 7, and 8 were consented to the research in accordance with IRB-approved protocol (research ethics committee [REC] reference number: 07/Q0512/26; University College London Hospital [UCLH] project ID number: 07/N018) of University College London.
Exome and Sanger sequencing
DNA was extracted from venous blood according to standard procedures. Exome sequencing (ES) was performed on genomic DNA. We performed ES on a DNA sample from individual 1 from family 1 by using libraries prepared with the NimblegenV2 library preparation kit (Roche, Basel, Switzerland), following the manufacturer’s protocol. Sequencing was performed by 100 bp paired-end sequencing on a HiSeq2500 instrument (Illumina, San Diego, California, USA) with a mean on-target coverage of 50×. Filtered reads were aligned to the human genome (Hg19/GRC37) via the Burrows-Wheeler transform method (BWA-MEM). Reads were sorted, and PCR duplicates were marked with Picard. Base quality recalibration and insertion/deletion (InDel) realignments were performed with the Genome Analysis Toolkit (GATK). SNVs and InDel variants were called jointly with HaplotypeCaller and recalibrated with GATK. Annotation and filtering were performed with ANNOVAR and custom scripts. Homozygous and potentially compound heterozygous variants with a population-specific minor allele frequency (MAF) < 0.005 in the Genome Aggregation Database (gnomAD) were selected and prioritized on the basis of their annotation. For the affected probands of family 2, ES was performed as described before.14 For the affected proband of family 3, ES was performed with Nextera Rapid Capture Expanded Exome Enrichment Kit.15 For family 4, a duo-ES analysis was performed with genomic DNA from the proband and mother. The exonic regions and flanking splice junctions of the genome were captured the Clinical Research Exome kit (Agilent Technologies, Santa Clara, CA). Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants with a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described.16 For families 5, 7, and 8, exome sequencing and data analysis were performed as previously described.17,18 For family 6, exome-coding DNA was captured with the Agilent SureSelect Clinical Research Exome (CRE) kit (v2) and sequenced on an Illumina HiSeq 4000 with 150 bp paired-end reads as described before.19 The candidate variants were confirmed and segregation analysis was performed with traditional Sanger sequencing for families 1, 2, 3, 5, and 8 (Figure S1, supplemental information). For family 6, trio-whole-exome sequencing was performed at diagnostics.
3D modeling of protein structure
Two homology models were generated with the atomic coordinates of alpha-ketoglutarate decarboxylase (PDB: 2XT6, 2JGD) as input into Modeler.20 We combined two models to maximize the predicted ordered region. The final dimer model includes TPP (thiamine pyrophosphate) and magnesium derived from the coordinates of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase (PDB: 2XYT).
Cloning and transgenesis
pUASTattB-dOgdhP867A-FLAG, pUASTattB-dOgdhR688Q-FLAG, pUASTattB-dOgdhF749S-FLAG, pUASTattB-dOgdhA343V-FLAG, pUASTattB-dOgdhW237C-FLAG, pUASTattB-dOgdhD507V-FLAG, pUASTattB-dOgdhR261W-FLAG, and pUASTattB-dOgdhR316G-FLAG were generated by site-directed mutagenesis PCR with pUASTattB-dOgdh (WT)-FLAG DNA construct as a template.21 The primers are listed in Table S3 (supplemental information). A series of pUASTattB-mutant dOgdh constructs were injected into y,w,ΦC31; VK37 embryos,22,23 and transgenic flies were selected.
The plasmid pCFD3.1-w-dU:3gRNA-dOgdh was cloned as follows. pCFD3.1-w-dU6:3gRNA was a gift from Simon Bullock (Addgene plasmid # 123366; RRID: Addgene_123366). We phosphorylated, annealed, and cloned 20 bp oligonucleotides containing guide RNA (gRNA) targeting dOgdh genomic locus with 4 nt overhangs for BbsI site, into the BbsI site of the pCFD3.1-w-dU:3gRNA vector. The oligonucleotide sequences used for dOgdh gRNA are dOgdh gRNA-F: 5′-GTCG AGGGATACTTACGGTGTGCA-3′ and gRNA-R: 5′-AAAC TGCACACCGTAAGTATCCCT-3′. The pCFD3.1-w-dU:3-gRNAdOgdh construct was injected into y,w,ΦC31; P{CaryP}attP40 embryos, and transgenic flies were selected.
Fly strains and maintenance
The following stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University (BDSC): elavC155-Gal4, elavC155-Gal4; UAS-Cas9.P2 (on III). All flies were maintained at room temperature (21°C) and crosses were kept at 25°C.
Immunoblot
Fly heads were homogenized in 1× Laemmli sample buffer containing 2.5% β-mercaptoethanol with pellet pestles (Sigma-Aldrich). After boiling for 10 min, samples were briefly centrifuged. Supernatants loaded into 4%–20% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad), separated by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad). The primary antibodies were used for overnight shaking at 4°C by the following dilution: mouse anti-FLAG M2 (Sigma Cat# F1804 RRID: AB_262044) 1:1,000; rabbit anti-OGDHL (Proteintech Cat# 17110-1-AP) 1:1,000; rabbit anti-OGDH (Abcam Cat# ab137773) 1:1,000; mouse anti-Actin (MP Biomedicals Cat# 8691002). HRP-conjugated goat anti-mouse (Thermo Fisher Scientific Cat# A-28177 RRID: AB_2536163) and goat anti-rabbit antibodies (Thermo Fisher Scientific Cat# A10547 RRID: AB_2534046) were used at 1:7,000 and visualized with enhanced chemiluminescence (ECL) (Bio-Rad).
Drosophila bang sensitivity assay
Methods were adapted from Howlett et al., 2013.24 Briefly, 20 flies were anesthetized with CO2 and were allowed to rest in fresh food vials for 24 h at 25°C before the assay. The male and female flies used were maintained at 1:1 ratio so there would be less chance of gender differences. On the day of the assay, flies were transferred into empty polystyrene vials without the use of CO2 and left to acclimatize to the surrounding for 15 min. They were then vortexed (Vortex-Genie 2 Scientific Industries) for 10 s at maximum strength.25 For data analysis, we recorded a 40 s video to measure the number of flies able to recover and climb at each time point. Five trials were conducted (n ≈ 100 for each genotype).
cDNA analysis
Fibroblasts from individual 3 and two age-matched controls (without any rare variant in OGDHL) were grown in DMEM supplemented with 1 mM pyruvate, 4.5 g/L glucose, 10% fetal calf serum, and 1% penicillin/streptomycin at 37°C in 5% CO2. For RNA purification from fibroblasts and cDNA retrotranscription, we used RNeasy mini kit (QIAGEN) and GoTaq 2-Step RT-qPCR System (Promega), respectively, according to the manufacturer’s protocols. For OGDHL cDNA amplification (GenBank: NM_018245.3), we used the following primer pair: 589F/2413R (589F: 5′-TGGAGAACACCTACTGCCAG-3′; 2413R: 5′-CCTCGAAGTCCTTGGTGAATG-3′). For sequencing of OGDHL transcript, PCR products were processed with Nextera XT DNA sample preparation kit (Illumina). Next-generation sequencing (NGS) was performed on an Illumina MiSeq instrument and reads were then aligned with BWA and visualized with Integrating Genomics Viewer (IGV).
Metabolite analysis with liquid chromatography/mass spectrometry (LC/MS)
Metabolites in brain tissues were extracted by 45 μL of ice-cold acetonitrile:methanol:water (40:40:20) solution containing 0.5% formic acid. We added 4 μL of 15% NH4HCO3 (w/v) in acetonitrile:methanol:water (40:40:20) solution to neutralize pH. Following vortexing and centrifugation at 16,000 g for 10 min at 4°C, extract was loaded to individual vials. Metabolites were analyzed by quadrupole-orbitrap mass spectrometers (Q-Exactive Plus Hybrid Quadrupole-Orbitrap, Thermo Fisher Scientific) coupled to hydrophilic interaction chromatography (HILIC) via electrospray ionization. Liquid chromatography (LC) separation was performed on an Xbridge BEH amide column (2.1 mm × 150 mm, 2.5 μm particle size, 130 Å pore size; waters) at 25°C with a gradient of solvent A (5% acetonitrile in water with 20 mM ammonium acetate and 20 mM ammonium hydroxide) and solvent B (100% acetonitrile). Flow rate was 350 μL/min. The LC gradient was as follows: 0 min, 75% B; 3 min, 75% B; 4 min, 50% B; 5 min, 10% B; 7 min, 10% B; 7.5 min, 75% B; 11 min, 75% B. Autosampler temperature was set at 4°C and the injection volume of the sample was 5 μL. Mass spectrometry (MS) analyses were acquired in negative ion mode with MS full-scan mode from m/z 70 to 830 and 140,000 resolution. Data analysis was performed with MAVEN software.
OGDHL knockout human neuronal cells
Creation of OGDHL knockout SH-SY5Y cell was performed as previously described.26,27 Briefly, we selected two guide RNAs targeting the exon 2 genomic region of human OGDHL with high activity and minimum off-target by using Synthego prediction tools. Then, we cloned double-strand DNA fragments for gRNA1OGDHL (5′-GCCTGTACCCCAAGACGGGA-3′), gRNA2OGDHL (5′-TCGCAGCCTGTACCCCAAGA-3′), and scramble (control) (5′-GCACTACCAGAGCTAACTCA-3′) into LentiCRISPRv2 vector. By transfecting these vectors into HEK293T, we generated lentiviruses, which we used for transduction of SH-SY5Y cells with puromycin selection. OGDHL knockout were confirmed by immunoblot and ICE (inference of CRISPR edits) analysis (Synthego Performance Analysis, ICE Analysis. 2019. v2.0. Synthego; [6.11.2021]).
Cellular respiration assay
We used XFe24 analyzer from Seahorse Biosciences to measure the rate of oxygen consumption, according to the manufacture’s protocol. Briefly, control and OGDHL knockout cells were seeded at a density of 8 × 104 cells per each well of a XFe24 cell culture plate (Agilent 100777-004). The next day, the cells were pre-incubated for 1 h with complete XF DMEM (Agilent 103575-100) containing 10 mM glucose, 1 mM sodium pyruvate, and 2mM L-glutamine. We used the complete XF DMEM to prepare cellular stress reagents to provide the following final concentration: 1 μM oligomycin, 1 μM FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone), and 1 uM antimycin A. All the reagents were loaded in the ports in cartridge unit. Oxygen consumption rates were measured for 3 min for mixing and 2 min of waiting period. Basal respiration was calculated by subtracting non-mitochondrial respiration rate from last rate measurement before oligomycin injection. Maximal respiration was calculated by subtracting non-mitochondrial respiration rate from maximum rate measurement after FCCP injection. We calculated ATP production by subtracting minimum rate measurement after oligomycin injection from last rate measurement before oligomycin injection.
Results
Clinical findings
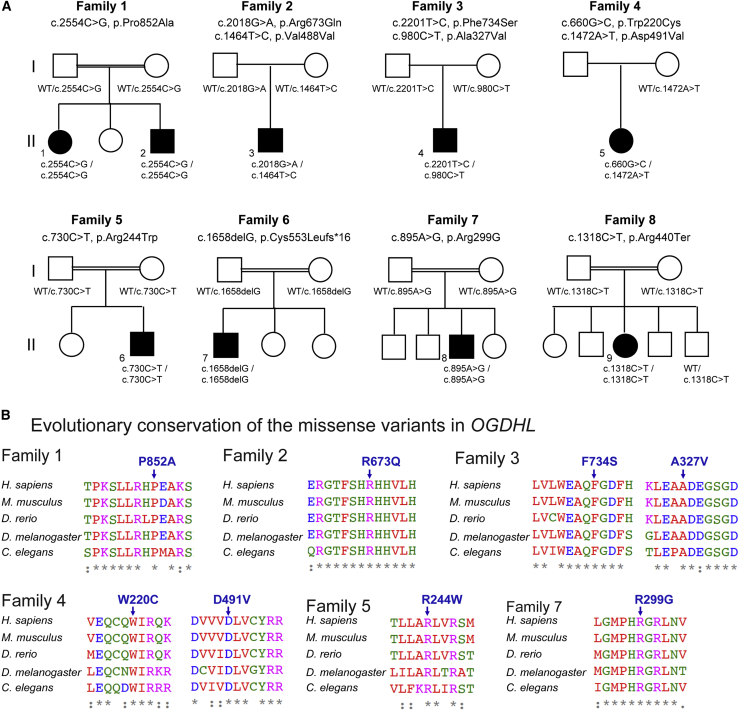
We identified nine individuals from eight families carrying bi-allelic variants in the OGDHL gene through whole-exome sequencing (WES) (Figure 1A). All groups were connected to study the OGDHL gene through GeneMatcher.28,29 For recruiting individual 7, we have contacted close collaborators from genomic centers in Europe and the USA. Parental consanguinity was reported in five of the eight families (families 1, 5, 6, 7, and 8), all of whom had homozygous variants in OGDHL (Figure 1A). The remaining three individuals showed compound heterozygous variants (families 2, 3, and 4). The clinical features of the affected individuals are summarized in Tables 1 and S1 and are described further in the supplemental notes.
Figure 1.
Identification of individuals with neurodevelopmental phenotypes with SNVs in OGDHL
(A) Eight pedigrees drawings, indicating bi-allelic variants in OGDHL identified in nine individuals from eight families. Homozygous missense variants were identified in families 1, 5, and 7, a homozygous frameshift variant in family 6, a homozygous stop-gain variant in family 8, and compound heterozygous variants in families 2, 3, and 4.
(B) Multiple-sequence alignment confirms evolutionary conservation of the eight missense variants in the animal kingdom.
Table 1.
Summary of the clinical and genetic information of the analyzed cohort
| Individual 1 (family 1) | Individual 2 (family 1) | Individual 3 (family 2) | Individual 4 (family 3) | Individual 5 (family 4) | Individual 6 (family 5) | Individual 7 (family 6) | Individual 8 (family 7) | Individual 9 (family 8) | Individual 10 (Yoon et al., 201721) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Variant GenBank: NM_018245 | c.2554C>G (p.Pro852Ala) | c.2554C>G (p.Pro852Ala) | c.2018G>A (p.Arg673Gln); c.1464T>C (p.Val488Val) | c.2201T>C (p.Phe734Ser); c.980C>T (p.Ala327Val) | c.660G>C (p.Trp220Cys); c.1472A>T (p.Asp491Val) | c.730C>T (p.Arg244Trp) | c.1658delG (p.Cys553Leufs∗16) | c.895A>G (p.Arg299Gly) | c.1318C>T (p.Arg440Ter) | c.2333C>T (p.Ser778Leu) |
| Inheritance | hom. | hom. | com. het. | com. het. | com. het. | hom. | hom. | hom. | hom. | hom. |
| AoO | birth | 1 year | 8 years | 6 years | 6 months | 5 months | 3 months | 3–4 months | 2 months | 1 year |
| DD/ID | mild DD | mild DD | mild DD | no | global DD | global DD and ID | mild ID | moderate to severe ID | global DD and ID | severe DD and ID |
| Seizures (onset) | 1X TC | no | no | abs., MA, TC, SE (N/A) | IS, TC (6 months) | focal-motor, TC (2 years) | no | no | yes (12 months) | no |
| Neurologic examination | mild GA | mild GA | GA, axonal neuropathy, spasticity | normal | hyp., no walking | spastic, quadriplegic, no walking | normal | hyp., walk with help | hyp., GA, walk with help | hyp., hypertonicity, spasticity, GA |
| EEG features | N/A | N/A | normal | N/A | hypsarrhytmia, multifocal/gen. spikes | diffuse spikes, slowed background activity | N/A | mild slowed background | T-R spikes | N/A |
| Hearing involvement | profound bi. | profound bi. | profound | no | no | no | no | no | bi. hearing deficit | no |
| Ophthalmological involvement | VI | no | retinopathy | no | VI, nystagmus | bi. optic atrophy | no | mild nystagmus | poor vision | no |
| Dys. features | no | no | hypomimia, scoliosis, pes cavus | no | bi. hip dysplasia, clonus with extension of right foot | high arched palate, scoliosis, microcephaly | scaphocephaly | no | bi. ptosis, highly arched palate, down slanting of eyes | some synophrys, large mouth, microcephaly |
| MRI | N/A | N/A | bi. unspecific T2 lesions | normal | global atrophy, diffuse T2 hyperintens., focus frontal | periv. leukomalacia, corpus callosum hypoplasia | R-F glioma | normal | normal | hypo plastic corpus callosum, global atrophy |
| Main phenotype | hearing deficit | hearing deficit | dysmorphic features | DEE | DEE | DEE | ID | ID | hearing and vision deficit, DEE | dysmorphic features, ID |
abs., absence; AoO, age of onset; bi., bilateral; com. het., compound heterozygous; DD, developmental delay; GA, gait ataxia; hom., homozygous, hyp., hypotonia; hyperintense., hyperintensity; ID, intellectual disability; IS, infantile spasms; MA, myoclonic-atonic; DEE, developmental and epileptic encephalopathy; N/A not available; periv., periventricular; R-F, right frontal; SE, status epilepticus; TC, tonic-clonic; VI, visual impairment.
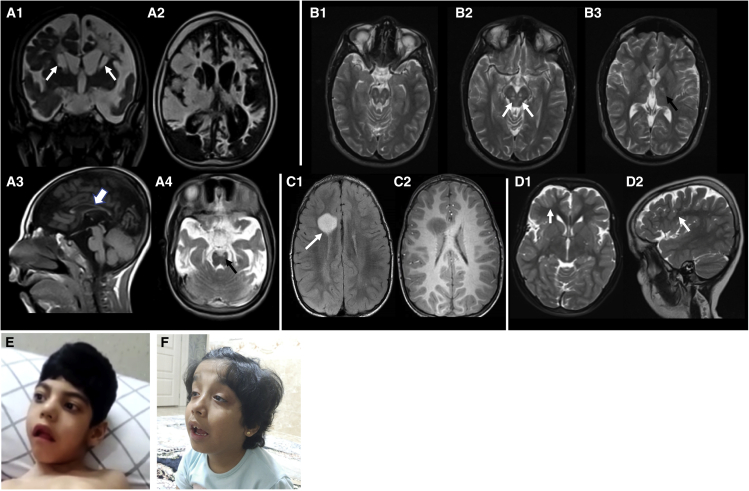
The individuals presented between 2 months and 8 years with a median onset of 1.9 years. Eight of nine individuals presented with mild to severe developmental delay. Feeding difficulties in four individuals (3, 5, 6, and 8) were noted, and excessive laughing/crying was observed in individual 8. Six individuals (individuals 1, 3, 5, 6, 8, and 9) showed an ophthalmological involvement that included visual impairment, nystagmus, and optic atrophy. Profound bilateral sensorineural hearing loss was found in four individuals (individuals 1, 2, 3, and 9). Dysmorphic features were reported in five of nine individuals (individuals 3, 4, 5, 6, 7, and 9) mainly consisting of distinctive craniofacial features including highly arched palate, bilateral ptosis, and downward slanting of eyes (Figures 2E and 2F). Neurological examination showed gait ataxia, spasticity, neuropathy, and hypotonia in seven individuals (individuals 1, 2, 3, 5, 6, 8, and 9). Five individuals (1, 4, 5, 6, and 9) experienced seizures at least once in their life with variable presentations (absences-, tonic-, tonic-clonic-, myoclonic-atonic-, atonic-, focal motor seizures, and infantile spasms). In addition, one person (individual 4) showed a severe obtundation status. Electroencephalogram (EEG) for individuals 5, 6, and 9 were available, and their EEGs were variable with generalized multifocal slowing intermixed with multi-focal sharp discharge (Figures S2–S4). This could be suggestive of a severe bilateral cerebral dysfunction with multi-focal and generalized epileptiform potential, consistent with a transition from infantile spasms to Lennox-Gastaut Syndrome. Brain MRI was available in four individuals (individuals 3, 6, 7, and 9) (Figures 2A–2D). A diverse spectrum of abnormalities was observed on neuroimaging. Individual 6 in family 5 presented with microcephaly (<3rd percentile of head circumference from age 6.5 to 8.5 years). MRI scan on individual 6 in family 5 showed a slender corpus callosum (Figure 2A3), which is similar to those in the previously identified person carrying a homozygous p.Ser778Leu variant.21,30 Frank basal ganglia cavitating damage and cystic leukomalacia was present in individual 6 (Figure 2A). Signal abnormality was noted in the small brain stem nuclei in both individual 6 (family 5) and individual 3 (family 2) (Figures 2A and 2B). The MRI scan on individual 7 in family 6 revealed a low-grade neoplasm with no additional findings (Figure 2C), and the imaging on individual 9 in family 8 showed subtle cortical dysgyria (Figure 2D).
Figure 2.
Clinical findings of affected individuals
(A) MRI of family 5 (individual 6). Coronal T2 (A1) and axial FLAIR (A2) scans show extensive scarring involving the white and gray matter structures of the brain with frank cavitation involving the caudate nuclei and the adjacent deep white matter of the cerebral hemispheres (arrows-A1). Sagittal T1 (A3) shows a slender corpus callosum (arrow) and axial T2 (A4) image shows symmetric involvement of brain stem nuclei (black arrow).
(B) MRI of family 2 (individual 3). Axial T2 sections showing symmetric involvement of the brain stem nuclei (arrows-B2) and the thalamo-capsular regions bilaterally (black arrow-B3).
(C) MRI of family 6 (individual 7). Axial FLAIR (C1) and post-contrast T1 (C2) images show a low-grade astrocytoma adjacent to the lateral ventricle on the right (arrow-C1).
(D) MRI of family 8 (individual 9). Axial T2 (D1) and sagittal T2 (D2) images show cortical dysgyria (arrows).
(E) Individual 6 in family 5 presents with microcephaly. He has a highly arched palate. He has no visual interaction but makes roving eye movements.
(F) Individual 9 in family 8 presents with macrocephaly and mild dysmorphic features, including bilateral ptosis, highly arched palate, and eye downslanting.
In summary, our individuals with bi-allelic OGDHL variants display a broad spectrum of neurological features including epilepsy, gait ataxia, hearing loss, visual impairment, malformations of cortical development, microcephaly, hypoplastic corpus callosum, and dysmorphic signs.
Genetic findings
WES in the affected probands revealed 11 novel bi-allelic variants in OGDHL (Figure 1, Table S2, supplemental information). Five of the identified variants were homozygous: three missense variants (c.2554C>G [p.Pro852Ala] in individuals 1 and 2 [family 1]; c.730C>T [p.Arg244Trp] in individual 6 [family 5]; c.895A>G [p.Arg299Gly] in individual 8 [family 7]), a frameshift variant (c.1658delG [p.Cys553Leufs∗16] in individual 7 [family 6]), and a stop-gain variant (c.1318C>T [p.Arg440Ter] in individual 9 [family 8]). Three individuals from families 2, 3, and 4 carry compound heterozygous single-nucleotide variants (SNVs): c.2018G>A (p.Arg673Gln)/c.1464T>C (p.Val488Val) in individual 3, c.2201T>C (p.Phe734Ser)/c. 980C>T (p.Ala327Val) in individual 4, and c.660G>C (p.Trp220Cys)/c.1472A>T (p.Asp491Val) in individual 5 (Figure 1A). All of the altered residues are evolutionarily conserved across species (Figure 1B). The heterozygous Phe734Ser variant has been previously implicated in eosinophilic esophagitis.31 The other variants have not been reported previously as pathogenic variants. Variants identified in families 1, 2, 3, and 6 (Pro852Ala, Arg673Gln, Ala327Val, and p.Cys553Leufs∗16) were absent in publicly available population databases such as gnomAD, Exome Sequencing Project (ESP), GME Variome, and Iranome, as well as in over 16,000 in-house exomes at Queen Square Genomics, UCL. On the basis of in silico predictions, all variants had high CADD scores (Table S2), with an average CADD score of 31.2, and are thus most likely damaging and deleterious variants.
In silico homology modeling and structural analysis of missense variants identified in affected individuals
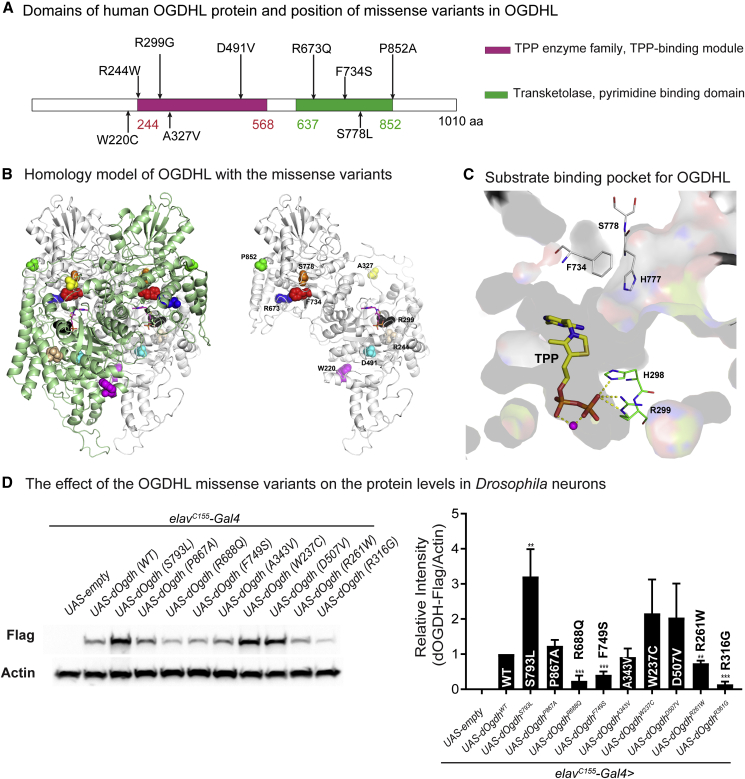
To generate a homology model for human OGDHL, we used the atomic coordinates for alpha-ketoglutarate decarboxylase as input into Modeler.20 In silico structure model suggested that each variant would alter the predicted protein structure (Figures 3B and 3C). The model showed that OGDHL forms a homodimer in which each subunit binds with a TPP (thiamine pyrophosphate) cofactor to form a substrate-binding pocket for OGDHL (Figure 3C). Importantly, the model shows that the mutated residues from two variants Phe734Ser and Arg299Gly are located in the substrate-binding pocket (Figure 3C). The side chain of Phe734 forms a part of the binding site for the pyrimidine ring and thiazole ring of TPP and contributes to the aromatic side-chain relay including the pyrimidine ring of TPP. Serine substitution for Phe734 (F734S) would disrupt this interaction. Arg299 forms ionic interaction with the β-phosphate of the bound TPP as shown in Figure 3C. This interaction is critical for TPP binding, thereby Gly substitution for Arg299 (R299G) is predicted to impair the interaction. Hence, the analysis suggests that Phe734Ser and Arg299Gly may lead to substantial defects in OGDHL function.
Figure 3.
Protein modeling of OGDHL dimer and the effects of eight missense variants on protein levels
(A) Schematic representation of protein domains of human OGDHL and positions of the eight missense variants together with the previously identified p.Ser778Leu variant. Pink indicates a TPP-binding domain. Green indicates a transketolase domain.
(B) In silico protein structure prediction of human OGDHL dimer and position of mutated residues. Dimer of OGDHL is shown in ribbon (left). Each monomer is colored with green and white. Bound TPP is shown in stick model and mutation sites are in CPK model. One monomer is shown for clarity (right).
(C) Only thin section of substrate binding pocket was shown with surface rendering for clarity. Amino acid residues involved in missense variants in the pocket are shown in stick model together with TPP. Residues from cis subunit are shown in green and residues from trans in gray. Bound magnesium is shown in magenta ball.
(D) Immunoblots for Drosophila heads expressing wild-type dOgdh-FLAG or dOgdh-FLAG carrying the homologous missense variants identified from individuals with bi-allelic OGDHL variants. Three biological replicates were quantified. Error bars represent SEM. p values were obtained by ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001.
We examined the other six missense variants, Pro852Ala, Arg673Gln, Asp491Val, Trp220Cys, Arg244Trp, and Ala327Val (Figure 3B). The side chain of Pro852 forms a local aromatic proline interaction. Alanine substitution for proline 852 (Pro852Ala) would abolish this interaction and negatively affect the structural stability. Arg673 forms two ionic interactions with the side chains of Glu644 and Glu731 as well as a hydrogen bond with the main chain carbonyl of Val665. Glutamine substitution for Arg673 (Arg673Gln) is predicted to destabilize the interaction because of the shorter side chain length and lack of a positive charge. The side chain of Asp491 forms hydrogen bonds with side chains of Asn430 and His461 of neighboring β strands, which can contribute to the stability of the β sheet. Thus, aspatate to valine substitution (Asp491Val) would destabilize the β sheet, which also includes the next β strand connected to the helix containing Trp220. Trp220 forms aromatic cluster together with Trp480 and Phe484. Cysteine mutation on Trp220 (Trp220Cys) will abolish the T shape pi network of this aromatic cluster. Arg244Trp substitution would cause losing flexible charged surface residue and adding a rigid bulky tryptophan residue, which may reduce entropy and protein solubility in addition to causing steric clashes with residues in the neighboring helix. Ala327 is surface exposed and the alanine to valine mutation may reduce protein solubility by exposing more hydrophobic side chain to the surface. We also examined the previously identified pathogenic variant Ser778Leu that underlies severe neurodevelopmental defects in humans.21 We found that Ser778Leu would disturb the surrounding local structure because there is not enough space to accommodate the bulky side chain of leucine, which will affect Pro980 and Thr982, destabilizing the dimer interface. The structural change caused by Ser778Leu mutation would affect the conformation of His777, resulting in disturbance of the aromatic cluster formed by side chains of Phe734 and Phe737, which would negatively affect the binding of Phe737 with the pyrimidine ring of TPP, and dimer formation. Collectively, in silico analyses suggest that all these missense variants identified from individuals carrying bi-allelic OGDHL variants could impair OGDHL structure and function.
The effects of the missense variants on protein levels
To determine the effects of a series of missense variants identified from individuals with bi-allelic OGDHL variants on in vivo protein levels, we generated transgenic flies harboring cDNA of dOgdh, the D. melanogaster (Dm) ortholog of human OGDHL, that carry homologous mutations to the human missense variants under the control of upstream activating sequence (UAS) (UAS-dOgdhP867A, UAS-dOgdhR688Q, UAS-dOgdhF749S, UAS-dOgdhA343V, UAS-dOgdhW237C, UAS-dOgdhD507V, UAS-dOgdhR261W, and dOgdhR316G) (Figure 4B). Using pan-neuronal Gal4 driver (elavC155-Gal4), we expressed each mutant transgene, wild-type dOgdh (UAS-dOgdhWT), as well as Dm mutation for the pathogenic S778L variant (UAS-dOgdhS793L)21 (Figure 3D). All transgenes have C-terminal FLAG for protein analysis. Immunoblot for adult fly heads showed that the variants that are predicted to impair TPP-binding, including Dm Arg316Gly (human Arg299Gly) and Dm Phe749Ser (human Phe734Ser), caused a substantial decrease in the protein levels (Figure 3D). In addition, we found that the protein levels of Dm Arg688Gln (human Arg673Gln) are significantly lower than those in wild-type control, and Dm Arg261Trp (human Arg244Trp) caused decreased protein levels in lesser extent (Figure 3D). On the contrary, Dm Ser793Leu (human Ser778Leu) led to a significant increase in the protein levels. This suggest that Ser793Leu may lead to misfolded protein aggregates that cannot be degraded by the protein quality control system. The protein levels for Dm Pro867Ala (human Pro852Ala), Dm Ala343Val (human Ala327Val), Dm Trp237Cys (human Trp220Cys), and Dm Asp507Val (human Asp491Val) were not significantly different from protein levels in the wild-type control (dOgdh WT) (Figure 3D). Hence, these results suggest that Arg673Gln, Arg299Gly, and Phe734Ser may cause loss of OGDHL function.
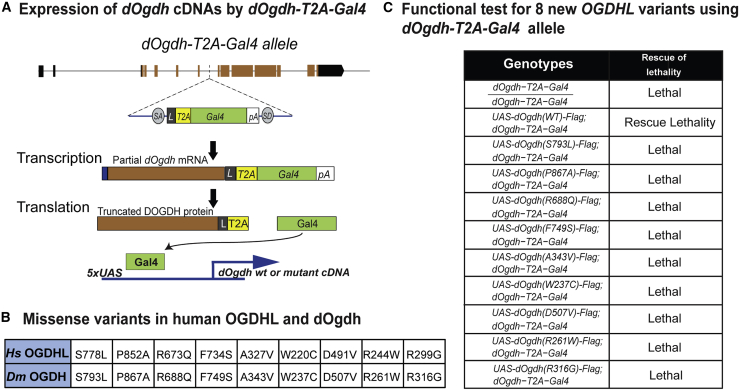
Figure 4.
Functional studies for OGDHL missense variants with dOgdh-null mutant
(A) Schematic of dOgdh-T2A-Gal4 allele and the Gal4/UAS system.
(B) The table shows missense variants in OGDHL and the Drosophila Ogdh variants homologous to the human variants.
(C) The lethality caused by loss of dOgdh was rescued by expression of wild-type dOgdh but not by those carrying any missense variants homologous to those identified from individuals with bi-allelic OGDHL variants.
In vivo functional studies for the variants in OGDHL with dOgdh-null mutant flies
We sought to determine the functional effects of the missense variants in OGDHL. To this end, in our previous work, we created a loss-of-function mutant for dOgdh by insertion of a T2A-Gal4 cassette into the coding intron of dOgdh by using recombination-mediated cassette exchange.21,32,33 The dOgdh mutant allele (dOgdh-T2A-Gal4) expresses the GAL4 transgene under control of the endogenous cis-regulatory elements of dOgdh. The GAL4 protein in turn activates expression of genes downstream of UAS (Figure 4A). We previously showed that loss of dOgdh (homozygous for dOgdh-T2A-Gal4) caused lethality, which was rescued by expression of wild-type dOgdh (dOgdhWT) but not by expression of dOgdh carrying a homologous mutation to human OGDHL (Ser778Leu) (dOgdhS793L).21 Thus, these results demonstrated that dOgdh-T2A-Gal4 mutant is a suitable model for functional interpretation of disease candidate variants in human OGDHL. Using the dOgdh-T2A-Gal4 mutants and transgenic flies harboring UAS-dOgdh cDNA carrying homologous mutations to new OGDHL variants (Figure 4B), we performed the lethality rescue assay. Flies carrying UAS-dOgdhWT and UAS-dOgdhS793L served as controls for the assay. We found that expression of none of eight new missense variants rescued the lethality caused by dOgdh loss (Figure 4C). Thus, the results indicate that all eight missense variants are loss-of-function alleles.
Functional studies for the variants in OGDHL in neurons
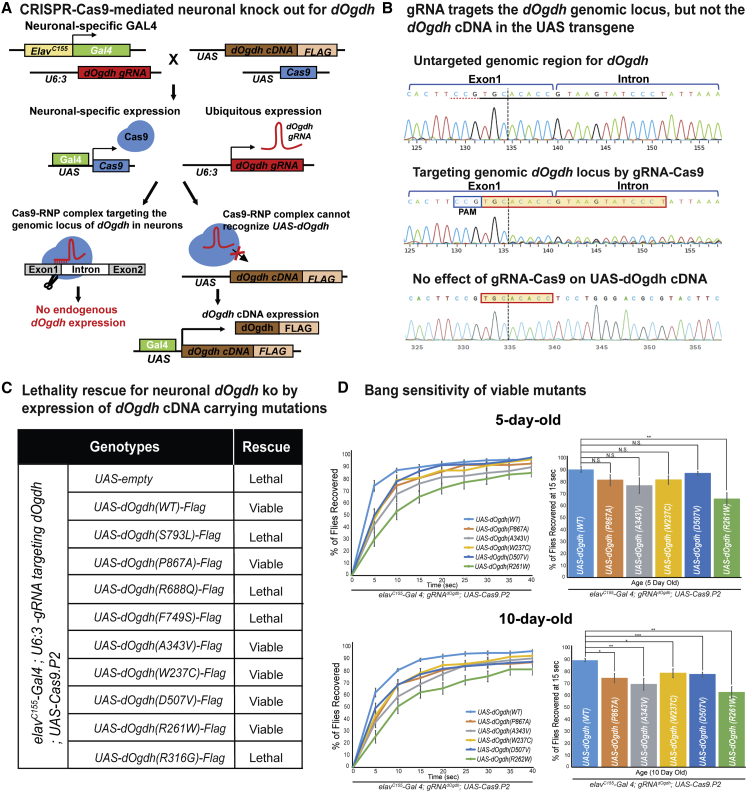
In humans, OGDHL is mainly expressed in the brain, whereas OGDH is expressed ubiquitously.10,13 The single Dm ortholog, dOgdh for human OGDHL and OGDH, is ubiquitously expressed in tissues including nervous and muscular systems.12 Hence, the study from the dOgdh-null mutant (dOgdh-T2A-Gal4) (Figure 4) would not determine the functional effects of the variants specifically on the nervous system. To determine the effects of the identified variants specifically in the nervous system, we developed a novel CRISPR-Cas9-mediated method for tissue-specific knockout (ko) with cDNA rescue (Figure 5A). The current CRISPR-Cas9 method in the Drosophila field permits tissue specificity, but most guide RNAs (gRNAs) target exons and thereby target both genomic loci and cDNA transgenes.34 As such, the current system prevents rescue of ko phenotypes by cDNA transgene expression. We overcame this technical obstacle by developing a strategy that selectively targets genomic loci but not UAS transgenes by using gRNAs that are complementary to the exon-intron junctions of target genes. Figures 5A and 5B illustrate how we applied this new method to study the functional effects of the OGDHL variants on the nervous system. To determine the effect of neuronal ko of dOgdh, we generated transgenic flies harboring the gRNA for dOgdh (gRNAdOgdh) that is ubiquitously expressed by U6:3 promotor and UAS-Cas9.P2 that is expressed by a pan-neuronal Gal4 (elavC155-Gal4). Neuronal ko for dOgdh (elavC155-Gal4; U6:3-gRNAdOgdh/UAS-empty; UAS-Cas9.P2) resulted in lethality, which was fully rescued by expression of wild-type dOgdh cDNA (Figure 5C). By performing Sanger sequencing for fly heads, we demonstrated that gRNAdOgdh and Cas9.P2 efficiently targeted the genomic locus of dOgdh but not dOgdh cDNA in the UAS transgene (Figure 5B). The expression of the known pathogenic dOgdhS793L mutant failed to rescue the lethality caused by neuronal dOgdh ko (Figure 5C). Hence, the results demonstrated that our new system enables determining the functional effects of the missense variants in OGDHL identified from our cohort in neuronal context.
Figure 5.
CRISPR-Cas9-mediated neuronal dOgdh knockout model shows various strength of OGDHL missense variants
(A) Schematic of CRISPR-Cas9-mediated neuron-specific dOgdh knockout and cDNA rescue system.
(B) Sanger sequencing from fly heads carrying elavC155-Gal4, U6:3-gRNAdOgdh, UAS-Cas9.P2, together with UAS-dOgdhWT showed that the gRNA targets the genomic locus of dOgdh but not for the UAS-dOgdh transgene.
(C) The lethality caused by neuronal knockout of dOgdh was rescued by wild-type dOgdh and dOgdh carrying Pro867Ala, Ala343Val, Trp237Cys, Asp507Val, and Arg261Trp but not by those carrying Arg688Gln, Phe749Ser, and Arg316Gly.
(D) Flies with neuronal knockout for dOgdh expressing Pro867Ala, Ala343Val, Trp237Cys, Asp507Val, or Arg261Trp exhibited progressive defects in recovery from bang stress compared to flies expressing WT dOgdh. Six to seven biological replicates (20 flies per genotype) were quantified. Error bars indicate SEM. p values were obtained by the Student’s t test. ∗p < 0.05, ∗∗p < 0.01,∗∗∗p < 0.001. N.S. indicates not statistically significant.
Unlike the dOgdh-T2A-Gal4 assay, the CRISPR-Cas9-mediated neuronal ko with cDNA rescue system revealed various allelic strength for eight missense variants. We first found that three missense variants dOgdhR688Q, dOgdhF749S, and dOgdhR316G failed to rescue the developmental lethality caused by neuronal ko of dOgdh. The results indicate that these three variants are severe loss-of-function alleles. This result is consistent with our 3D homology model that predicted the important role of the residues in TPP-binding (human Phe734Ser and Arg299Gly) (Figure 3C), as well as reduced protein levels (Dm Arg688Gln, Dm Phe749Ser, and Dm Arg316Gly) (Figure 3D). On the contrary, the other five missense variants, dOgdhP867A, dOgdhA343V, dOgdhW237C, dOgdhD507V, and dOgdhR261W, rescued the lethality (Figure 5C). We confirmed that the dOgdh proteins with these five missense variants were expressed in adult brains by performing immunoblot (Figure S5). To determine the effects of the five variants in adult stage, we performed bang-sensitivity assay. We found that flies carrying dOgdhR261W exhibited a recovery defect from mechanical stress at both day 5 and day 10, whereas flies having the other four variants, dOgdhP867A, dOgdhA343V, dOgdhW237C, or dOgdhD507V, did show comparable recovery to wild-type controls at day 5, but exhibited delayed recovery at day 10 (Figure 5D). Hence, the data indicate that Arg261Trp has more defective gene function compared to the other four variants (Pro867Ala, Ala343Val, Trp237Cys, and Asp507Val). Collectively, the results from our new CRISPR-Cas9-mediated neuronal ko system indicate that three missense variants (Arg688Gln, Phe749Ser, and Arg316Gly) are severe loss-of-function alleles, and the other five missense variants (Pro867Ala, Ala343Val, Trp237Cys, Asp507Val, and Arg261Trp) are hypomorphic alleles.
The splicing effect of missense variant p.Val488Val
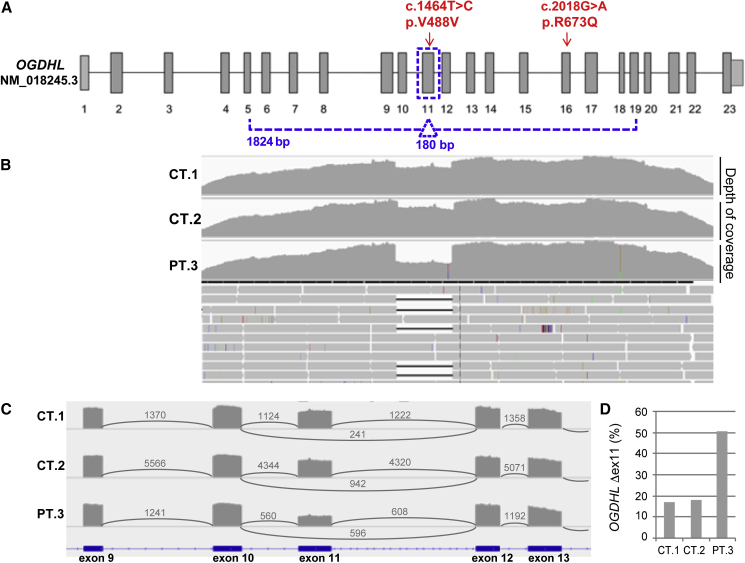
Individual 3 in family 2 carries compound heterozygous variants (c.2018G>A [p.Arg673Gln]/c.1464T>C [p.Val488Val]). Since the synonymous c.1464T>C [p.Val488Val] variant was predicted to cause splice site changes, we assessed its effect on OGDHL transcript. RNA was extracted from fibroblasts of individual 3 and retrotranscribed into cDNA. Despite very low expression of OGDHL in this specimen, we were able to amplify a large portion of the OGDHL transcript (exons from 5 to 19), which showed a single band in controls and a faint double band in individual 3. By performing NGS, besides the full-length transcript we detected an alternative transcript corresponding to exon 11 skipping, Δ exon 11 (Figures 6B and 6C). Interestingly, the Δ exon 11 transcript is present also in controls, but at low percentages (≈15%) (Figure 6D). Contrariwise, in individual 3, the Δ exon 11 transcript is ≈50%, suggesting that this overrepresentation of the Δ exon 11 transcript is favored by the presence of the synonymous variant.
Figure 6.
NGS analysis of OGDHL transcript for individual 3
(A) Schematic structure of the OGHDL transcript (GenBank: NM_152416.3). The arrows indicate the variants identified in individual 3. The dotted line corresponds to the analyzed PCR amplicon (exons from 5 to 19) and highlights the exon 11.
(B) Profiles of depth of coverage obtained through NGS on OGDHL transcript PCR products encompassing exons from 5 to 19. RNA was extracted from fibroblasts of individual 3 (PT3) and two control samples (CT1 and CT2). The drop in coverage depth corresponds to the alternative transcript with exon 11 skipping (Δex11).
(C) OGDHL Sashimi plot of exon skipping in individual 3’s and control fibroblasts. The number of split reads spanning the given intron is indicated on the exon-connecting lines. The involved exons are depicted at the bottom.
(D) Quantification of exon 11 skipping (Δex11). Graphs of the coverage ratio (in percentage) between the OGDHL Δex11 isoform and the full-length isoform, calculated for all the samples analyzed by NGS (individual 3, PT3, and two controls, CT1 and CT2).
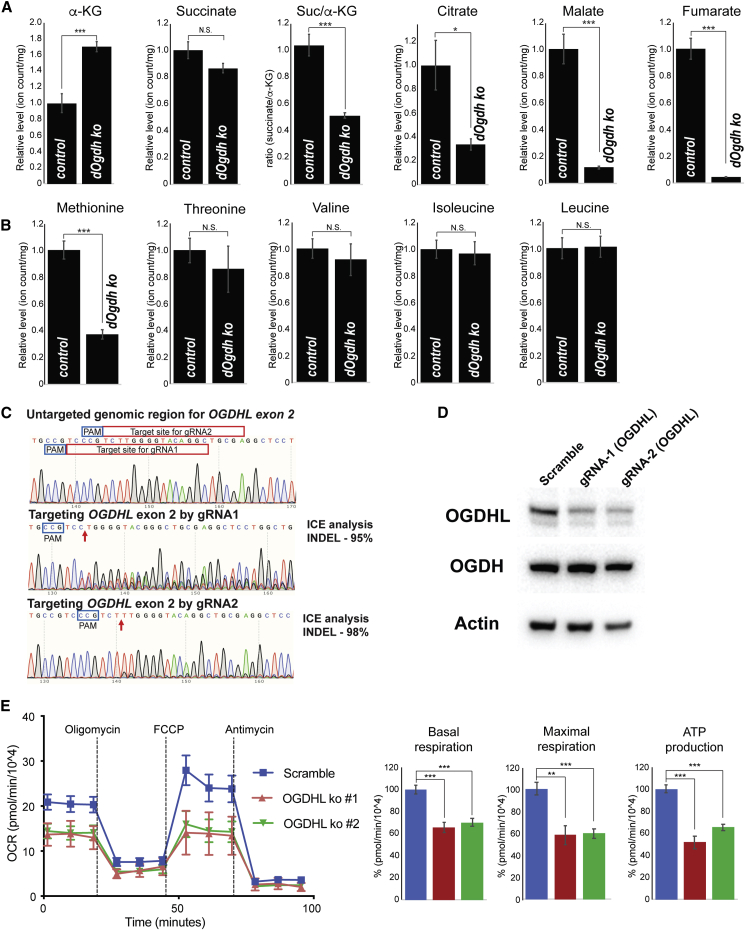
Defective mitochondrial metabolism in neurons lacking OGDHL
To determine whether loss of dOgdh in neurons affect Krebs cycle metabolism, we performed metabolite profiling for Drosophila larval brains with dOgdh ko and control brains by LC/MS. We found that α-KG, the substrate of DOGDH, is significantly elevated in the dOgdh ko brains (Figure 7A). In contrast, the levels of other Krebs cycle metabolites including citrate, malate, and fumarate are decreased in the dOgdh mutant brains compared to those from controls (Figure 7A), indicating that dOgdh is required for normal Krebs cycle metabolism in the brains.
Figure 7.
Neurons lacking functional OGDHL leads to defects in mitochondrial metabolism
(A) Relative levels of the indicated Krebs cycle metabolites in larval brains of neuron-specific dOgdh knockout (elavC155-Gal4, U6:3-gRNAdOgdh, UAS-Cas9.P2) and controls (elavC155-Gal4, U6:3-gRNAdOgdh).
(B) Relative levels of the indicated amino acids in larval brains of neuron-specific dOgdh knockout (elavC155-Gal4, U6:3-gRNAdOgdh, UAS-Cas9.P2) and controls (elavC155-Gal4, U6:3-gRNAdOgdh).
(C) Sanger sequencing results for genomic region for exon 2 of OGDHL of SH-SY5Y cells with or without gene editing. Target sites of gRNA1 and gRNA2 were highlighted with red box. PAM sites were labeled with blue box. Arrows indicate Cas9 cleavage sites.
(D) Immunoblots for protein levels of OGDHL, OGDH, and Actin in SH-SY5Y cells with or without Cas9-directed gene editing.
(E) Oxygen consumption rate (OCR) of SH-SY5Y cells with or without CRISPR-Cas9-directed editing for OGDHL. The cells were treated sequentially (vertical lines) with 1 μM oligomycin, 1 μM FCCP, and 1 μM antimycin A. Quantification of basal respiration, maximal respiration, and ATP production were shown in bar graphs. Error bars indicate SEM. p values were obtained by the Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. N.S. indicates not statistically significant.
We anticipate that dOgdh loss in the brain leads to a decrease in the levels of succinate, the product of DOGDH reaction. The levels of succinate in the dOgdh mutant brains, however, are comparable to those in controls, suggesting that succinate could be replenished with amino acids, including methionine, isoleucine, valine, and threonine, by anaplerotic reactions.35 Interestingly, while the levels of threonine, valine, and isoleucine are similar in the mutants and controls, methionine are markedly decreased in the dOgdh ko brains (Figure 7B). This suggests that methionine catabolism most likely replenishes succinate pools as a compensatory mechanism.
Given that Krebs cycle metabolism is required for mitochondrial respiration, we hypothesize that OGDHL loss in human neurons leads to defects in mitochondrial respiration. To test this, we generated SH-SY5Y, human neuroblastoma cell line, with mutations in OGDHL by using CRISPR-Cas9-mediated gene editing.26,27 Lentivirus-mediated transduction of two gRNAs that target OGDHL exon 2 caused significant fraction of cells to carry InDel mutations (95% InDels by gRNA1; 98% InDels by gRNA2) (Figure 7C). In contrast, scramble gRNA did not lead to gene editing in OGDHL exon 2 (Figure 7C). We confirmed that SH-SY5Y cells carrying mutations in OGDHL exhibited a decrease in the protein levels of OGDHL but not OGDH (Figure 7D). We measured oxygen consumption and found that OGDHL ko cells had significantly lower oxygen consumption rates (OCRs) as well as ATP production compared to the control cells (Figure 7E), indicating that OGDHL is required for normal mitochondrial respiration and energy production in human neurons. Hence, the critical role of OGDHL in mitochondrial metabolism in neurons suggests that defects in the Krebs cycle and mitochondrial respiration contribute to the etiology of persons carrying bi-allelic variants in OGDHL.
Discussion
We previously identified a single individual carrying a recessive variant (p.Ser778Leu) in OGDHL with neurodevelopmental phenotypes.21,30 However, with only a single individual, the causality of this OGDHL variant to human disease was not firmly established and the definition of the associated phenotypes was incomplete. Here, we reported nine individuals from eight unrelated families with bi-allelic variants at the OGDHL locus primarily presenting with developmental delay accompanied with variable neurological and neurodevelopmental phenotypes. The following evidence supports the causality of the bi-allelic variants in OGDHL to the human neurological disease. First, nine individuals in eight unrelated families have been identified in our manuscript with the same consistent genetic defect, derived by independent filtration of WES variants in different neurological centers. Second, all the identified variants are likely to have damaging effects. Indeed, two nonsense variants predict the formation of truncated proteins and a synonymous variant affects splicing. The Drosophila models showed that three missense variants are LOF, whereas five missense variants are hypomorphic alleles. Moreover, all eight missense variants hit highly conserved residues of OGDHL. In addition, we would like to stress the fact that all individuals belong to pedigrees compatible with a recessive trait and harbor bi-allelic variants in OGDHL, and the deleterious effects of the identified variants have been experimentally proven.
The clinical findings included developmental and epileptic encephalopathy (DEE) defined by early onset seizures and developmental delay, hearing loss, visual impairment, intellectual disability, gait ataxia, and dysmorphic features (Tables 1 and S1). Individual 6 in family 5 presented with hypoplastic corpus callosum and microcephaly (Table S1, Figures 2A and 2E), which are similar to clinical manifestations found in a previously reported single individual carrying a homozygous variant in OGDHL (p.Ser778Leu).21 Several of the clinical features found in our cohort overlap with phenotypes associated with a pathogenic variant in OGDH (c.959A>G [p.Asn320Ser]), including gait ataxia, tonic-clonic seizures, and brain atrophy.12 Pathogenic variants in Krebs cycle genes and other mitochondrial genes, including AIFM1 (MIM: 300169), also lead to overlapping features found in individuals carrying bi-allelic OGDHL.36, 37, 38 Mutations in MDH2 resulted in DEE (MIM: 154100),8 while mutations in DLD, SUCLG1, SUCLA2, SDHA, and FH lead to Leigh syndrome or Leigh-like disease.1, 2, 3, 4, 5
OGDHL variants p.Arg299Gly (individual 8, family 7), p.Phe734Ser (individual 4, family 3), and p.Arg673Gln (individual 3, family 2) had more deleterious in silico prediction scores (CADD > 30, predicted to be the 0.1% most deleterious substitutions in the human genome), which were consistent with the Drosophila functional studies demonstrating these three variants as severe LOF alleles (Figure 5C). Importantly, these results were also predicted by the 3D protein structure modeling (Figures 3B and 3C) and the protein level test in Drosophila neurons (Figure 3D). Indeed, individual 8, homozygous for p.Arg299Gly, exhibited severe limb and facial muscle weakness and hypotonia as well as severe intellectual disability (Table 1, supplemental notes). On the contrary, the other five missense variants, p.Pro852Ala, p.Ala327Val, p.Trp220Cys, p.Asp491Val, and p.Arg244Trp, had lower CADD prediction scores > 20 (predicted to be the 1% most deleterious substitutions in the human genome) compared to the three severe LOF alleles (CADD > 30) (Table S2). Consistent with the in silico prediction, Drosophila functional studies demonstrated that these five alleles are hypomorphic (Figures 5C and 5D). Similarly, the synonymous variant p.Val488Val, which had a partial impact on the splicing process, may be considered an hypomorphic allele. Individuals 3 and 4, who carry a combination of severe LOF alleles and mild alleles (individual 3, p.Arg673Gln/p.Val488Val; individual 4, p.Phe734Ser/p.Ala327Val), were the only individuals in our cohort presented with a later disease onset (after 8 and 6 years of life in individuals 3 and 4, respectively) compared to others who presented the disease either at birth or within 6 months of life. A milder phenotype without seizures in individual 3 or drug-controlled epileptic encephalopathy in individual 4 has been reported. The latter has been seizure free and his cognitive functions have recovered in the last 7 years (Table 1, supplemental notes). This suggests that alleles in individuals 3 and 4 are still associated with a residual production of functional protein. The frameshift variant in individual 7 (p.Cys553Leufs∗16) and nonsense variant in individual 9 (p.Arg440Ter) lead to early stop codons, which could underlie the cause of disease in early infancy (2–3 months of age), the severe phenotype comprised of visual impairment, bilateral hearing deficit, and seizures in individual 9 as well as the growth retardation and presence of glioma on the brain imaging of individual 7. In families 1 and 5 with functional data suggestive of hypomorphic alleles, we hypothesize that different levels consanguinity in these pedigrees might add difficulty to deduce single disease-causing genes. Overall, the studies on Drosophila strongly supported the pathogenicity of the variants, which correlates well with the severity of clinical manifestations in humans.
While Drosophila has a single dOgdh gene, humans have two paralogs carrying OGDH activity, OGDHL and OGDH. These two genes appear to have evolved by gene duplication of a single ancestral gene during early vertebrate evolution.9 Interestingly, it was inferred from a large-scale Drosophila genetic screen combined with Mendelian disease study that essential fly genes with two or more human homologs have a higher likelihood of being associated with Mendelian diseases than those that only have a single human homolog because redundant function of duplicated genes may cause genes to be more susceptible to pathogenic mutations.39 In contrast to ubiquitous OGDH expression, brain-specific expression of OGDHL suggests that deleterious genetic variants in OGDHL result in human disease phenotypes more frequently than those in OGDH. Neuronal expression of OGDHL also suggests the pivotal role of OGDHL in neuronal development and function, which is in agreement with diverse neurodevelopmental phenotypes observed in our cohort of patients. Besides, our analysis of single-cell RNA sequencing results of the cochlear epithelium in several stages of early postnatal mouse development showed that OGDHL is expressed in the hair cells, inner pillar cells, and the inner border cells/inner phalangeal cells (Figure S6).40,41 These data are consistent with hearing loss found in four individuals of our cohort.
OGDHL is involved in the decarboxylation of α-KG to succinyl-CoA during the Krebs cycle, and its impairment is expected to cause a mitochondrial metabolic defect. Indeed, we found that α-KG is significantly elevated while other Krebs cycle metabolites, including citrate, malate, and fumarate, are decreased in the dOgdh ko brains compared to those from controls (Figure 7A). Notably, succinate levels were comparable to controls despite the fact that dOgdh loss in the brain should lead to a decrease in the levels of succinate, the product of OGDH and OGDHL reaction. This finding suggests that succinate could be replenished with some amino acids by anaplerotic reactions.35 While threonine, valine, and isoleucine levels are similar between mutants and controls, methionine levels are significantly decreased in the dOgdh ko brains, suggesting succinate is mainly replenished with methionine.
The recessive variant in OGDH (GenBank: NM_002541.3; c.959A>G [p.Asn320Ser]) was previously reported to lead to a neurodevelopmental phenotypes12 similar to those carrying bi-allelic variants in OGDHL. The findings suggest that OGDHL in the brain cannot compensate defects of OGDH in spite of their common role in α-KG decarboxylation.11,13 Why does the brain need both OGDHL and OGDH for its normal development and function? The brain needs sustained energy production obtained from mitochondrial respiration to maintain electrochemical gradients and synaptic transmission, which is essential for development and function of the brain. Hence, it is possible to speculate that both isozymes, OGDHL and OGDH, are required for sustained energy production in neurons in response to a wide-range of α-KG concentration. This idea is supported by the following evidence. First, OGDHL and OGDH in rat brain extract exhibited biphasic substrate saturation kinetics with higher α-KG concentration for full saturation (Km1 = 0.4 mM) and lower α-KG concentration (Km2 = 0.07 mM).13 In contrast, OGDH from heart exhibited a standard Michaelis-Menten kinetics with Km (0.2 mM).42 Hence, having two isozymes (OGDHL and OGDH) with the two distinct binding affinities (Km1 = 0.4 mM and Km2 = 0.07 mM) for the substrate enables the brain to utilize a wide range of α-KG levels for running the Krebs cycle for energy production. Importantly, we showed that OGDHL ko human neuronal cells had significantly lower oxygen consumption rates as well as ATP production compared to the control cells (Figure 7E), indicating that both OGDHL and OGDH are required for mitochondrial metabolism in human neurons.
One individual (individual 7 in family 6) exhibited low-grade astrocytoma. In our previous study, we showed that reduction in OGDH activity leads to hyperactive mTORC1 activity in Drosophila and mouse embryonic fibroblasts.21 OGDHL silencing was also shown to induce mTORC1 signaling in hepatocellular carcinoma.43 Interestingly, hyperactivation of mTOR signaling has been associated with other human diseases, including tuberous sclerosis complex-associated subependymal giant cell astrocytomas.44 mTOR inhibitors have been shown to improve the clinical status of subependymal giant cell astrocytomas and tuberous sclerosis patients.45 Hence, it is tempting to speculate that treatment with mTOR inhibitors may be a therapeutic avenue for individual 7 with low-grade astrocytoma.
In summary, we have identified nine individuals in eight unrelated families with bi-allelic variants in OGDHL showing diverse neurological phenotypes including DEE, visual impairment, hearing loss, intellectual disability, and hypoplastic corpus callosum. Functional study with a novel CRISPR-Cas9-mediated tissue knockout with cDNA rescue system revealed that eight missense OGDHL variants are loss-of-function alleles. Further, the analysis of the transcripts from fibroblasts showed that the synonymous variant in family 2 impairs splicing in OGDHL. Taken together, we establish OGDHL as a disease gene whose impairment leads to a distinct Mendelian mitochondrial disease with mainly neurological phenotypes.
Acknowledgments
We thank all individuals and relatives for consent to be part of the study. Families 5–8 were collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA), and research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. We thank former and present Yoon lab members for their input during investigations, particularly Madison Chilian, Yohan Park, David Seo, and Jae Sun Kang. We thank Holly Van Remmen for her support of Seahorse assays. We thank Scott Plafker for his helpful advices during investigation. We thank Hugo Bellen for his help to initiate this study. We thank the “Cell line and DNA Bank of Genetic Movement Disorders and Mitochondrial Diseases” of the Telethon Network of Genetic Biobanks (grant GTB12001J) and Eurobiobank Network, which supplied biological specimens for family 2. C.L. and D.G. are members of the European Reference Network for Rare Neuromuscular Diseases (ERN EURO-NMD). W.H.Y. is supported by the National Institute of General Medical Sciences (5 P20 GM103636-08) and the National Institute of Neurological Disorders and Stroke (1R01 NS121298-01) of the National Institutes of Health. W.H.Y. was also supported by Presbyterian Health Foundation (PHF 4431-04-04-0 and PHF 4411-05-07-0). H.H. is funded by the MRC (MR/S01165X/1, MR/S005021/1, G0601943), the National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), and Muscular Dystrophy Association (MDA USA). Y.W. was funded by the German Research Foundation (DFG; WE4896/3-1) by the DFG/FNR INTER Research Unit FOR2715 (WE4896/4-1) Treat-ION grant (01GM1907) (continued in supplemental acknowledgments).
Declaration of interests
I.H. serves on the Scientific Advisory Board of Biogen. A.R. is an employee of GeneDx. The remaining authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.11.003.
Data and code availability
The variant alleles identified in all families have been deposited on the Leiden Open Variation Database (LOVD) with the following accession numbers: p.Pro852Ala in individual 1 in family 1 (individual ID: 385786; variant ID: 814865); p.Pro852Ala in individual 2 in family 1 (individual ID: 385787; variant ID: 814866); p.Arg673Gln in individual 3 in family 2 (individual ID: 385788; variant ID: 814867); p.Val488Val in individual 3 in family 2 (individual ID: 385788; variant ID: 814868); p.Phe734Ser in individual 4 in family 3 (individual ID: 385790; variant ID: 814869); p.Ala327Val in individual 4 in family 3 (individual ID: 385790; variant ID: 814870); p.Trp220Cys in individual 5 in family 4 (individual ID: 385821; variant ID: 814902); p.Asp491Val in individual 5 in family 4 (individual ID: 385821; variant ID: 814903); p.Arg244Trp in individual 6 in family 5 (individual ID: 385822; variant ID: 814904); p.Cys553Leufs∗16 in individual 7 in family 6 (individual ID: 385823; variant ID: 814905); p.Arg299Gly in individual 8 in family 7 (individual ID: 385824; variant ID: 814906); p.Arg440∗ in individual 9 in family 8 (individual ID: 385825; variant ID: 814907).
Web resources
GeneDx ClinVar submission page, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
Synthego prediction tools, https://tools.synthego.com/#/
Supplemental information
References
- 1.Grafakou O., Oexle K., van den Heuvel L., Smeets R., Trijbels F., Goebel H.H., Bosshard N., Superti-Furga A., Steinmann B., Smeitink J. Leigh syndrome due to compound heterozygosity of dihydrolipoamide dehydrogenase gene mutations. Description of the first E3 splice site mutation. Eur. J. Pediatr. 2003;162:714–718. doi: 10.1007/s00431-003-1282-z. [DOI] [PubMed] [Google Scholar]
- 2.Ostergaard E., Schwartz M., Batbayli M., Christensen E., Hjalmarson O., Kollberg G., Holme E. A novel missense mutation in SUCLG1 associated with mitochondrial DNA depletion, encephalomyopathic form, with methylmalonic aciduria. Eur. J. Pediatr. 2010;169:201–205. doi: 10.1007/s00431-009-1007-z. [DOI] [PubMed] [Google Scholar]
- 3.Carrozzo R., Dionisi-Vici C., Steuerwald U., Lucioli S., Deodato F., Di Giandomenico S., Bertini E., Franke B., Kluijtmans L.A., Meschini M.C., et al. SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain. 2007;130:862–874. doi: 10.1093/brain/awl389. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., Rötig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeron T., Chretien D., Poggi-Bach J., Doonan S., Rabier D., Letouzé P., Munnich A., Rötig A., Landrieu P., Rustin P. Mutation of the fumarase gene in two siblings with progressive encephalopathy and fumarase deficiency. J. Clin. Invest. 1994;93:2514–2518. doi: 10.1172/JCI117261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiegel R., Pines O., Ta-Shma A., Burak E., Shaag A., Halvardson J., Edvardson S., Mahajna M., Zenvirt S., Saada A., et al. Infantile cerebellar-retinal degeneration associated with a mutation in mitochondrial aconitase, ACO2. Am. J. Hum. Genet. 2012;90:518–523. doi: 10.1016/j.ajhg.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghezzi D., Goffrini P., Uziel G., Horvath R., Klopstock T., Lochmüller H., D’Adamo P., Gasparini P., Strom T.M., Prokisch H., et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 8.Ait-El-Mkadem S., Dayem-Quere M., Gusic M., Chaussenot A., Bannwarth S., François B., Genin E.C., Fragaki K., Volker-Touw C.L.M., Vasnier C., et al. Mutations in MDH2, Encoding a Krebs Cycle Enzyme, Cause Early-Onset Severe Encephalopathy. Am. J. Hum. Genet. 2017;100:151–159. doi: 10.1016/j.ajhg.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsson T.A., Olsson F., Sundstrom G., Lundin L.G., Brenner S., Venkatesh B., Larhammar D. Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol. Biol. 2008;8:184. doi: 10.1186/1471-2148-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap Z.Y., Strucinska K., Matsuzaki S., Lee S., Si Y., Humphries K., Tarnopolsky M.A., Yoon W.H. A biallelic pathogenic variant in the OGDH gene results in a neurological disorder with features of a mitochondrial disease. J. Inherit. Metab. Dis. 2021;44:388–400. doi: 10.1002/jimd.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunik V., Kaehne T., Degtyarev D., Shcherbakova T., Reiser G. Novel isoenzyme of 2-oxoglutarate dehydrogenase is identified in brain, but not in heart. FEBS J. 2008;275:4990–5006. doi: 10.1111/j.1742-4658.2008.06632.x. [DOI] [PubMed] [Google Scholar]
- 14.Legati A., Reyes A., Nasca A., Invernizzi F., Lamantea E., Tiranti V., Garavaglia B., Lamperti C., Ardissone A., Moroni I., et al. New genes and pathomechanisms in mitochondrial disorders unraveled by NGS technologies. Biochim. Biophys. Acta. 2016;1857:1326–1335. doi: 10.1016/j.bbabio.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Helbig I., Lopez-Hernandez T., Shor O., Galer P., Ganesan S., Pendziwiat M., Rademacher A., Ellis C.A., Hümpfer N., Schwarz N., et al. A Recurrent Missense Variant in AP2M1 Impairs Clathrin-Mediated Endocytosis and Causes Developmental and Epileptic Encephalopathy. Am. J. Hum. Genet. 2019;104:1060–1072. doi: 10.1016/j.ajhg.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G., et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 17.Makrythanasis P., Maroofian R., Stray-Pedersen A., Musaev D., Zaki M.S., Mahmoud I.G., Selim L., Elbadawy A., Jhangiani S.N., Coban Akdemir Z.H., et al. Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur. J. Hum. Genet. 2018;26:330–339. doi: 10.1038/s41431-017-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mencacci N.E., Kamsteeg E.J., Nakashima K., R’Bibo L., Lynch D.S., Balint B., Willemsen M.A., Adams M.E., Wiethoff S., Suzuki K., et al. De Novo Mutations in PDE10A Cause Childhood-Onset Chorea with Bilateral Striatal Lesions. Am. J. Hum. Genet. 2016;98:763–771. doi: 10.1016/j.ajhg.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perenthaler E., Nikoncuk A., Yousefi S., Berdowski W.M., Alsagob M., Capo I., van der Linde H.C., van den Berg P., Jacobs E.H., Putar D., et al. Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol. 2020;139:415–442. doi: 10.1007/s00401-019-02109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 21.Yoon W.H., Sandoval H., Nagarkar-Jaiswal S., Jaiswal M., Yamamoto S., Haelterman N.A., Putluri N., Putluri V., Sreekumar A., Tos T., et al. Loss of Nardilysin, a Mitochondrial Co-chaperone for α-Ketoglutarate Dehydrogenase, Promotes mTORC1 Activation and Neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venken K.J., He Y., Hoskins R.A., Bellen H.J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 23.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howlett I.C., Tanouye M.A. Seizure-sensitivity in Drosophila is ameliorated by dorsal vessel injection of the antiepileptic drug valproate. J. Neurogenet. 2013;27:143–150. doi: 10.3109/01677063.2013.817574. [DOI] [PubMed] [Google Scholar]
- 25.Ganetzky B., Wu C.F. Indirect Suppression Involving Behavioral Mutants with Altered Nerve Excitability in DROSOPHILA MELANOGASTER. Genetics. 1982;100:597–614. doi: 10.1093/genetics/100.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M., et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherrill J.D., Kc K., Wang X., Wen T., Chamberlin A., Stucke E.M., Collins M.H., Abonia J.P., Peng Y., Wu Q., et al. Whole-exome sequencing uncovers oxidoreductases DHTKD1 and OGDHL as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight. 2018;3:e99922. doi: 10.1172/jci.insight.99922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venken K.J., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., Bellen H.J. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diao F., Ironfield H., Luan H., Diao F., Shropshire W.C., Ewer J., Marr E., Potter C.J., Landgraf M., White B.H. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Port F., Chen H.M., Lee T., Bullock S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 36.Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D’Adamo P., Novara F., Zuffardi O., Uziel G., Zeviani M. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 2010;86:639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinaldi C., Grunseich C., Sevrioukova I.F., Schindler A., Horkayne-Szakaly I., Lamperti C., Landouré G., Kennerson M.L., Burnett B.G., Bönnemann C., et al. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 2012;91:1095–1102. doi: 10.1016/j.ajhg.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zong L., Guan J., Ealy M., Zhang Q., Wang D., Wang H., Zhao Y., Shen Z., Campbell C.A., Wang F., et al. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. J. Med. Genet. 2015;52:523–531. doi: 10.1136/jmedgenet-2014-102961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto S., Jaiswal M., Charng W.L., Gambin T., Karaca E., Mirzaa G., Wiszniewski W., Sandoval H., Haelterman N.A., Xiong B., et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolla L., Kelly M.C., Mann Z.F., Anaya-Rocha A., Ellis K., Lemons A., Palermo A.T., So K.S., Mays J.C., Orvis J., et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 2020;11:2389. doi: 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubota M., Scheibinger M., Jan T.A., Heller S. Greater epithelial ridge cells are the principal organoid-forming progenitors of the mouse cochlea. Cell Rep. 2021;34:108646. doi: 10.1016/j.celrep.2020.108646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMinn C.L., Ottaway J.H. Studies on the mechanism and kinetics of the 2-oxoglutarate dehydrogenase system from pig heart. Biochem. J. 1977;161:569–581. doi: 10.1042/bj1610569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai W., Xu L., Yu X., Zhang G., Guo H., Liu H., Song G., Weng S., Dong L., Zhu J., et al. OGDHL silencing promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J. Hepatol. 2020;72:909–923. doi: 10.1016/j.jhep.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Franz D.N., Belousova E., Sparagana S., Bebin E.M., Frost M., Kuperman R., Witt O., Kohrman M.H., Flamini J.R., Wu J.Y., et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381:125–132. doi: 10.1016/S0140-6736(12)61134-9. [DOI] [PubMed] [Google Scholar]
- 45.Ebrahimi-Fakhari D., Franz D.N. Pharmacological treatment strategies for subependymal giant cell astrocytoma (SEGA) Expert Opin. Pharmacother. 2020;21:1329–1336. doi: 10.1080/14656566.2020.1751124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant alleles identified in all families have been deposited on the Leiden Open Variation Database (LOVD) with the following accession numbers: p.Pro852Ala in individual 1 in family 1 (individual ID: 385786; variant ID: 814865); p.Pro852Ala in individual 2 in family 1 (individual ID: 385787; variant ID: 814866); p.Arg673Gln in individual 3 in family 2 (individual ID: 385788; variant ID: 814867); p.Val488Val in individual 3 in family 2 (individual ID: 385788; variant ID: 814868); p.Phe734Ser in individual 4 in family 3 (individual ID: 385790; variant ID: 814869); p.Ala327Val in individual 4 in family 3 (individual ID: 385790; variant ID: 814870); p.Trp220Cys in individual 5 in family 4 (individual ID: 385821; variant ID: 814902); p.Asp491Val in individual 5 in family 4 (individual ID: 385821; variant ID: 814903); p.Arg244Trp in individual 6 in family 5 (individual ID: 385822; variant ID: 814904); p.Cys553Leufs∗16 in individual 7 in family 6 (individual ID: 385823; variant ID: 814905); p.Arg299Gly in individual 8 in family 7 (individual ID: 385824; variant ID: 814906); p.Arg440∗ in individual 9 in family 8 (individual ID: 385825; variant ID: 814907).