Abstract
Background & Aims
It is well established that chronic inflammation promotes gastric cancer–associated metaplasia, but little is known regarding the mechanisms by which immune cells and cytokines regulate metaplastic cellular changes. The goals of this study were to identify interleukin 13 (IL13)-producing immune cells, determine the gastric epithelial cell response(s) to IL13, and establish the role(s) of IL13 in metaplasia development.
Methods
Experiments used an established mouse model of autoimmune gastritis (TxA23), TxA23×Il4ra-/- mice, which develop gastritis but do not express the IL4/IL13-receptor subunit IL4Rα, and TxA23×Il13-Yfp mice, which express yellow fluorescent protein in IL13-producing cells. Flow cytometry was used to measure IL13 secretion and identify IL13-producing immune cells. Mouse and human gastric organoids were cultured with IL13 to determine epithelial cell response(s) to IL13. Single-cell RNA sequencing was performed on gastric epithelial cells from healthy and inflamed mouse stomachs. Mice with gastritis were administered IL13-neutralizing antibodies and stomachs were analyzed by histopathology and immunofluorescence.
Results
We identified 6 unique subsets of IL13-producing immune cells in the inflamed stomach. Organoid cultures showed that IL13 acts directly on gastric epithelium to induce a metaplastic phenotype. IL4Rα-deficient mice did not progress to metaplasia. Single-cell RNA sequencing determined that gastric epithelial cells from IL4Rα-deficient mice up-regulated inflammatory genes but failed to up-regulate metaplasia-associated transcripts. Neutralization of IL13 significantly reduced and reversed metaplasia development in mice with gastritis.
Conclusions
IL13 is made by a variety of immune cell subsets during chronic gastritis and promotes gastric cancer–associated metaplastic epithelial cell changes. Neutralization of IL13 reduces metaplasia severity during chronic gastritis.
Keywords: IL13, Gastritis, Metaplasia
Abbreviations used in this paper: BSA, bovine serum albumin; cDNA, complementary DNA; DMEM, Dulbecco’s modified Eagle medium; GEM, gel beads in emulsion; Gkn3, gastrokine3; GSII, Griffonia simplicifolia; IFNγ, interferon γ; IL, interleukin; ILC2, innate lymphoid cell type 2; MHC, major histocompatibility complex; MUC, mucin; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; scRNA-seq, single-cell RNA sequencing; SPEM, spasmolytic polypeptide-expressing metaplasia; TFF2, trefoil factor 2 (or spasmolytic polypeptide); Th, T helper; UEA-I, Ulex europaeus agglutinin-1; YFP, yellow fluorescent protein
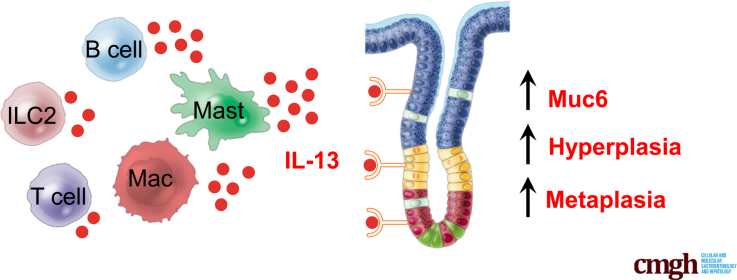
Graphical abstract
Summary.
Interleukin 13 (IL13) is secreted by multiple subsets of immune cells and acts directly on the gastric epithelium to promote neck cell expansion and metaplasia. Mice with autoimmune gastritis lacking the IL13 receptor do not develop neck cell expansion or metaplasia, and neutralization of IL13 inhibits and reverses disease development during chronic gastritis.
Gastric cancer is the fifth most common neoplasm and the fourth leading cause of cancer-associated deaths, resulting in more than 1 million new cases and more than 750,000 deaths yearly worldwide.1 Gastric cancer is most common in males and individuals older than age 50. Although cases have been steadily decreasing over the years, an increase in the incidence of gastric cancer among young adults and women (age, <50 y) was reported recently.2,3 Gastric cancer can be classified into distinct subtypes based on the mechanisms of initiation (sporadic vs inherited) and anatomically (cardia vs noncardia). Sporadic noncardia-type adenocarcinomas are associated most commonly with Helicobacter pylori infection.4,5 H pylori infects 50% of the world’s population, yet only approximately 1%–3% of individuals with chronic H pylori infections develop gastric cancer, showing that infection with H pylori alone is not sufficient to induce gastric cancer development.6 Individuals with sustained H pylori infection develop chronic inflammation. Chronic inflammation in the stomach (gastritis), as a result of H pylori infection or autoimmune gastritis, is the major risk factor for gastric cancer development.5, 6, 7
Chronic gastritis can cause the loss of parietal cells (oxyntic atrophy), expansion of mucous neck cells, and, ultimately, the replacement of normal gastric lineages with metaplastic cell types.8, 9, 10 Metaplasia is the conversion of a terminally differentiated cell type into another not found in a given tissue under homeostatic conditions. Metaplasia develops as a result of external stimuli, such as cytokines and reactive oxygen species, produced by infiltrating immune cells at the site of injury. When injury-inducing stimuli are removed, metaplastic cells may revert back to their originally differentiated state.11, 12, 13 However, when damage persists, as occurs during chronic inflammation, metaplastic cells proliferate and may accumulate mutations that drive progression to dysplasia and eventually adenocarcinoma. Metaplasia plays a key role in the development of multiple diseases including Barrett esophagus, lung cancer, pancreatic cancer, and gastric cancer.12,14, 15, 16
There are 2 main forms of metaplasia found in the stomach: intestinal metaplasia and spasmolytic polypeptide-expressing metaplasia (SPEM). Studies have identified SPEM as a potential precursor to intestinal metaplasia and an important biomarker of gastric cancer development.17, 18, 19 SPEM is characterized by the presence of mucus-expressing and trefoil factor 2 (TFF2)-expressing (also known as spasmolytic polypeptide) chief cells at the base of gastric glands. Gastrokine 3 (Gkn3) expression was identified recently as a reliable identifier of SPEM in both mouse and human stomachs with chronic gastritis and we recently determined that Gkn3 can be used to identify SPEM cells in single-cell RNA sequencing data sets.20,21 Gkn3 expression is virtually undetectable in healthy corpus; however, in the presence of chronic inflammation, neck and chief cells differentiate into a TFF2- and GKN3-expressing metaplastic cell type.19,20,22 These preneoplastic epithelial cell changes occur as a result of exposure to inflammatory mediators and cytokines released by infiltrating immune cells during chronic inflammation. Although genetic studies have associated single-nucleotide polymorphisms of inflammatory cytokines with increased risk of gastric cancer, we are just beginning to understand the mechanism(s) by which immune cells and cytokines contribute to the development of SPEM and progression to gastric cancer. We recently showed that interferon (IFN)γ and interleukin (IL)17A accelerate SPEM induction by causing parietal cell atrophy and increasing CD4+ effector T-cell activities in the gastric mucosa.23, 24, 25
To define the immune cells and cytokines that regulate SPEM development, our laboratory uses a mouse model of autoimmune gastritis referred to as TxA23. TxA23 mice express a transgenic CD4+ T-cell receptor that targets a peptide from the parietal cell–specific H+/K+ adenosine triphosphatase proton pump; the same autoantigen targeted in human beings with autoimmune gastritis. Similar to mice and human beings with autoimmune gastritis and/or chronic H pylori infection, the inflammation present in the mucosa of the TxA23 mouse is driven by IFNγ- and IL17A-producing CD4+ T cells. As a result, TxA23 mice develop autoimmune-mediated chronic gastritis, parietal cell atrophy, mucous neck cell expansion, and SPEM by 4 months of age. These mice have been a useful tool to study immune cells and cytokines that regulate gastritis and gastric metaplasia.26, 27, 28, 29, 30
IL4 and IL13 are cytokines involved in allergic responses and IgE production that signal via an IL4Rα and IL13Rα1 heterodimeric receptor.31, 32, 33, 34 IL4 and IL13 secretion typically is associated with the T helper 2 (Th2) subsets of CD4+ T cells. However, IL13 is produced by cell types other than CD4, including innate lymphoid cells type 2 (ILC2), macrophages, and mast cells. IL13 has been identified as having a role in SPEM development and increased IL13 levels have been found in peripheral blood of patients with gastric cancer.35,36 Still, the molecular mechanisms by which IL13 and IL13-producing immune cells regulate SPEM development and gastric cancer progression remain complex and largely unknown.
In this study, we identified multiple subsets of IL13-producing immune cells in the gastric mucosa of mice with autoimmune-induced chronic gastritis. We used 3-dimensional gastric organoid cultures to show that IL13 acts directly on gastric epithelial cells to increase organoid size, viability, and up-regulation of Muc6, all of which are features of mucous neck cell expansion and SPEM. We also showed that knocking out a receptor required for IL13 signaling in mice with autoimmune gastritis significantly reduced neck cell expansion and SPEM development. In addition, single-cell RNA sequencing showed that murine gastric epithelial cells respond to inflammatory stimuli but do not up-regulate SPEM-associated transcripts in the absence of IL13 signaling. Finally, we showed that administering IL13-neutralizing antibodies significantly reduced and reversed neck cell expansion and SPEM development in mice with autoimmune gastritis. Together, these data show that IL13 plays an important role in promoting the metaplastic epithelial cell changes that develop in response to chronic gastritis.
Results
Identifying IL13-Producing Immune Cell Subsets in the Gastric Mucosa of Mice With Chronic Gastritis
The purpose of these studies was to identify cytokines secreted by immune cell subsets in the gastric mucosa of mice with chronic gastritis. To do this, we isolated immune cells from the chronically inflamed gastric mucosa of TxA23 mice. TxA23 mice develop autoimmune-mediated chronic atrophic gastritis, which shares many of the features observed in the human disease including immune responses, gastric pathology, and SPEM development. By 2 months of age, TxA23 mice have superficial gastritis with patchy areas of atrophy and metaplasia. Gastric disease progresses in severity as the mice age and is present throughout the gastric mucosa by 4 months of age.26, 27, 28 Immune cells were isolated from the stomachs of 2- to 4-month-old TxA23 mice. After isolation, immune cell subsets were identified using flow cytometry and cytokine secretion was quantified after restimulation.
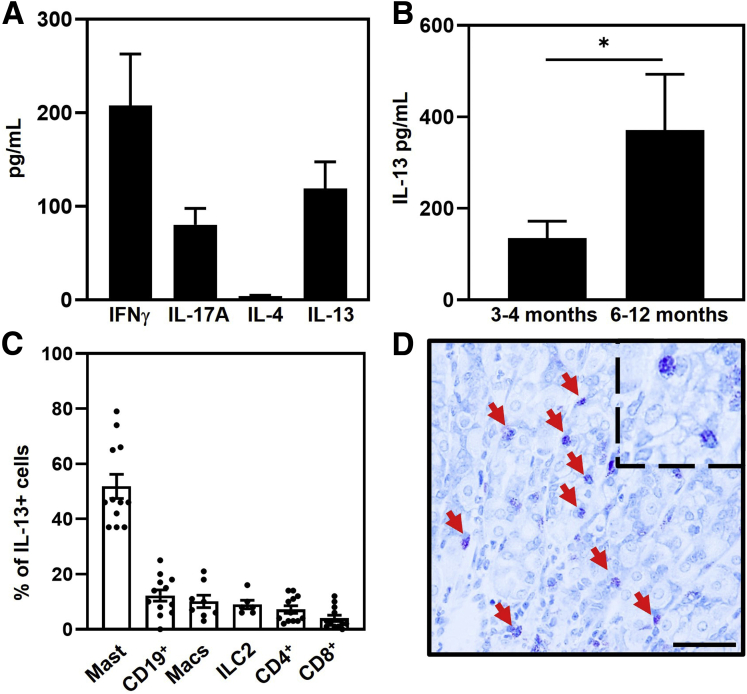
Analyses of supernatants collected 72 hours after restimulation identified relatively high concentrations of IFNγ and IL17A, and undetectable IL4 secretion by mucosal immune cells (IFNγ, 208 ± 55 pg/mL; IL17A, 81 ± 18 pg/mL; and IL4, <10 pg/mL). These data are consistent with previous reports that disease in this and other models is associated with Th1 and Th17 CD4+ T cells while IL4-producing Th2 cells are largely absent.7,27,37, 38, 39, 40, 41, 42, 43 Unexpectedly, relatively high levels of IL13 were secreted by cells from the gastric mucosa (119 ± 29 pg/mL) (Figure 1A). SPEM-containing glands appear in the gastric mucosa of TxA23 mice at 2–4 months of age and increase in frequency as the mice age.26,27 To determine whether IL13 secretion correlated with the frequency of SPEM+ glands, immune cells were isolated from the stomachs of 3- to 4-month-old and 6- to 12-month-old TxA23 mice. Supernatants collected 72 hours after restimulation identified a more than 2.5-fold increase in IL13 secretion in mice 6–12 months of age compared with 3- to 4-month-old mice (135 ± 37 pg/mL vs 371 ± 122 pg/mL; P = .039) (Figure 1B). These data show that an increase in IL13 secretion correlates with more severe disease and SPEM development.
Figure 1.
Mast cells are the major source of IL13 in the gastric mucosa. (A) Cytokine secretion from immune cells isolated from the stomachs of 2- to 4-month-old TxA23 mice (N = 19–25). (B) IL13 secretion from immune cells isolated from the stomachs of 3- to 4-month-old TxA23 and 6- to 12-month-old TxA23 mice (N = 9–14). ∗P = 0.039. (C) CD45+IL13+ cells in the stomach of TxA23 mice are composed of multiple cell types. Immune cell types were identified as follows: mast cells (FcεR1+cKit+), B cells (CD19+), macrophages (CD11b+), ILC2 (CD25+CD127+CD3-CD19-CD11b-GR-1-CD49b-), and T cells (CD8+ and CD4+). Each dot represents 1 mouse (N = 6–12). (D) Representative toluidine blue–stained corpus section showing mast cells (red arrow) in 4-month-old TxA23 stomach. Enlarged inlay section shown in top right corner. Scale bar: 50 μm. Data show means ± SEM.
The absence of IL4 secretion and the secretion of large amounts of IL13 prompted us to identify the cell type(s) responsible for IL13 secretion in the gastric mucosa. To identify IL13-secreting immune cell subsets, we used a line of TxA23×Il13-Yfp/Cre mice. Cells in these mice express yellow fluorescent protein (YFP) driven by the IL13 promoter.44 For these studies, immune cells were isolated from the mucosa, restimulated, and analyzed via flow cytometry. After gating on immune cells (CD45+) and IL13/YFP+ cells, we identified immune cell subsets using antibodies to lineage-specific markers (Figure 1B). These analyses showed that IL13 was produced by a variety of immune cell subsets, the majority of which was produced by mast cells (52% ± 4.3%), and smaller subsets of CD19+ B cells (12% ± 2.1%), macrophages (10% ± 2.3%), ILC2 (9.0% ± 1.5%), CD4+ T cells (7.2% ± 1.3%), and CD8+ T cells (4.0% ± 1.2%) (Figure 1C). These data show that a variety of different immune cell types, including mast cells, B cells, macrophages, ILC2s, and CD4+ and CD8+ T cells, secrete IL13 in the gastric mucosa of mice with autoimmune gastritis and inflammation-induced metaplasia.
Mast cells are the largest producers of IL13 in the inflamed gastric mucosa. To determine the location of mast cells in the stomach, corpus tissue sections from healthy BALB/c mice and 4-month-old TxA23 mice were stained with toluidine blue. Toluidine blue is a metachromatic dye that identifies mast cells by reacting with the heparin and histamine in mast cell cytoplasmic granules. Although only a limited number of mast cells were present in the submucosa (but not the mucosa) of healthy BALB/c mice (data not shown), mast cells were found infiltrating into the mucosal gastric glands of the inflamed corpus of TxA23 mice (Figure 1D). These data show that IL13-secreting mast cells infiltrate into the gastric glands during chronic inflammation.
IL13 Acts Directly on Both Mouse and Human Gastric Epithelial Cells to Alter Organoid Cellular Composition and Increase Organoid Size, Viability, and Up-Regulation of Metaplasia-Associated Genes
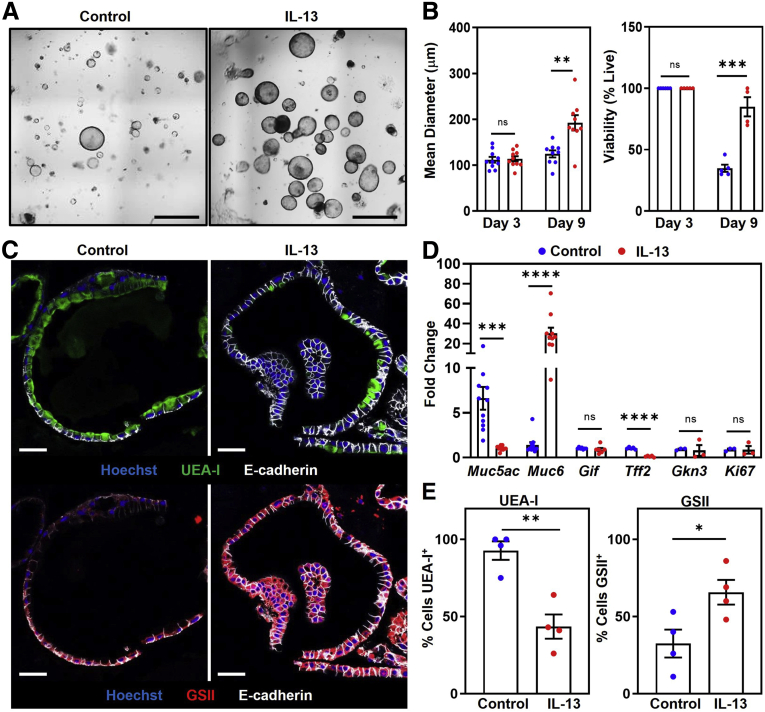
Having identified IL13-producing immune cells in the stomachs of mice with chronic gastritis, we tested whether IL13 can act directly on gastric epithelium. Gastric organoids were cultured with and without recombinant mouse IL13. Photographs of organoids were taken on days 3 and 9 (Figure 2A) to assess the size and number of viable organoids. On day 3, control organoids and IL13-cultured organoids were of similar diameter (112 ± 6.3 μm vs 114 ± 5.7 μm; P = 0.8). However, on day 9, organoids cultured with IL13 had 1.5-fold larger mean diameters compared with control organoids (124 ± 7.7 μm vs 192 ± 17 μm; P = .002). The proportion of collapsed (nonviable) vs viable organoids also was determined. Control and IL13-cultured organoids had 100% viability on day 3. By day 9, organoids cultured with IL13 had a more than 2-fold increase in viability compared with control organoids (85% ± 7.8% vs 35% ± 2.9%; P = .0003) (Figure 2B). Together, these results show that IL13 increases the size and viability of gastric organoids.
Figure 2.
IL13 regulates murine organoid cellular composition, promotes growth and survival, and alters Muc6 and Muc5ac expression. (A) Representative organoid photographs on day 9 cultured without (left) and with (right) 5 ng recombinant mouse IL13. Scale bars: 200 μm. (B) Left: Quantification of the mean diameter of organoids cultured without (blue) and with (red) 5 ng recombinant mouse IL13. Each dot represents the mean diameter from 1 independent experiment (N = 10). ∗∗P = 0.002. Right: Quantification of organoid viability. Each dot represents the proportion of viable organoids from 1 independent experiment (N = 5–6). ∗∗∗P = 0.0003. (C) Representative immunofluorescent staining of organoids cultured without (left) or with (right) 5 ng recombinant mouse IL13 on day 7. Top: MUC5AC expression represented by UEA-I (green), anti–E-cadherin (white), and Hoechst (blue). Bottom: MUC6 expression represented by GSII (red). Scale bars: 50 μm. (D) qPCR analysis of gene expression in organoids cultured without (blue) or with (red) 5 ng recombinant mouse IL13 on day 7. Data show fold change expression. Each dot represents 1 independent experiment (N = 3–11). Muc5ac: ∗∗∗P = 0.0003; Muc6: ∗∗∗∗P <0.0001; Tff2: ∗∗∗∗P < 0.0001. (E) Proportion of UEA-I+ cells (left) and GSII+ cells (right). Each dot represents the percentage of positive cells from 1 independent experiment (N = 4). ∗P = 0.033 ∗∗P = 0.002. Significance was determined using a Student t test. Data show means ± SEM.
During organoid observation under light microscopy, we noted a change in the appearance of IL13-cultured organoids compared with controls. This observation led us to perform additional analyses of the IL13-cultured organoids, including transcriptional and imaging analyses. We cultured organoids under the same conditions described earlier, only instead of imaging, we isolated RNA and performed quantitative polymerase chain reaction (qPCR) to compare the relative expression of different gastric epithelium-associated transcripts. Expression of Muc5ac, a mucin-encoding transcript expressed by foveolar cells, was decreased 7-fold in organoids cultured with IL13 (6.6 ± 1.2 vs 1.0 ± 0.07; P = .0003). In contrast, expression of Muc6, a mucin-encoding transcript expressed by neck cells and induced in metaplastic cells (SPEM in particular), was increased 20-fold in IL13-cultured organoids compared with controls (1.4 ± 0.30 vs 30 ± 5.5; P < .0001). Gastric intrinsic factor, expressed by murine chief cells, was very low in organoids and not significantly different between IL13-cultured and control organoids (1.0 ± 0.04 vs 0.90 ± 0.18; P = .47). Expression of Tff2 was decreased in organoids cultured with IL13 compared with controls (1.00 ± 0.06 vs 0.12 ± 0.04; P < .0001), and Gkn3 expression was similar between the 2 cultures (0.93 ± 0.07 vs 0.80 ± 0.6; P = .84). Expression of the proliferation marker Ki67 was unchanged on day 7 between control and IL13-cultured organoids (0.90 ± 0.10 vs 0.87 ± 0.42; P = 0.94) (Figure 2D). These data showed that in addition to increasing the size and viability of gastric organoids, IL13 also increased Muc6 expression and decreased Muc5ac levels.
The decrease in Muc5ac and increase in Muc6 expression could be caused by changes in mucus production or by alterations in the number of MUC5AC- and/or MUC6-expressing cells in IL13-cultured organoids. To distinguish between these possibilities, conditions were repeated, and organoids were evaluated using immunofluorescence staining. Organoids cultured with and without IL13 were fixed, embedded in paraffin, and stained with anti–E-cadherin (white), Ulex europaeus agglutinin-1 (UEA-I) lectin (green), which binds MUC5AC, Griffonia simplicifolia (GSII) lectin, which binds MUC6, and Hoechst nuclear stain (Figure 2C). Using the immunofluorescent images, the percentage of MUC5AC- or MUC6-positive cells was determined from the total number of E-cadherin–positive cells in control and IL13-cultured organoids. In IL13-cultured organoids, the percentage of UEA-I+ (MUC5AC+) cells was decreased roughly 2-fold compared with control organoids (43% ± 7.8% vs 93% ± 5.9%; P = .002), while the percentage of GSII+ (MUC6+) cells more than doubled compared with control organoids (66% ± 8.0% vs 32% ± 9.0%; P = .033) (Figure 2E). These data show that IL13 acts directly on gastric epithelium in organoid cultures to increase the proportion of MUC6-expressing cells.
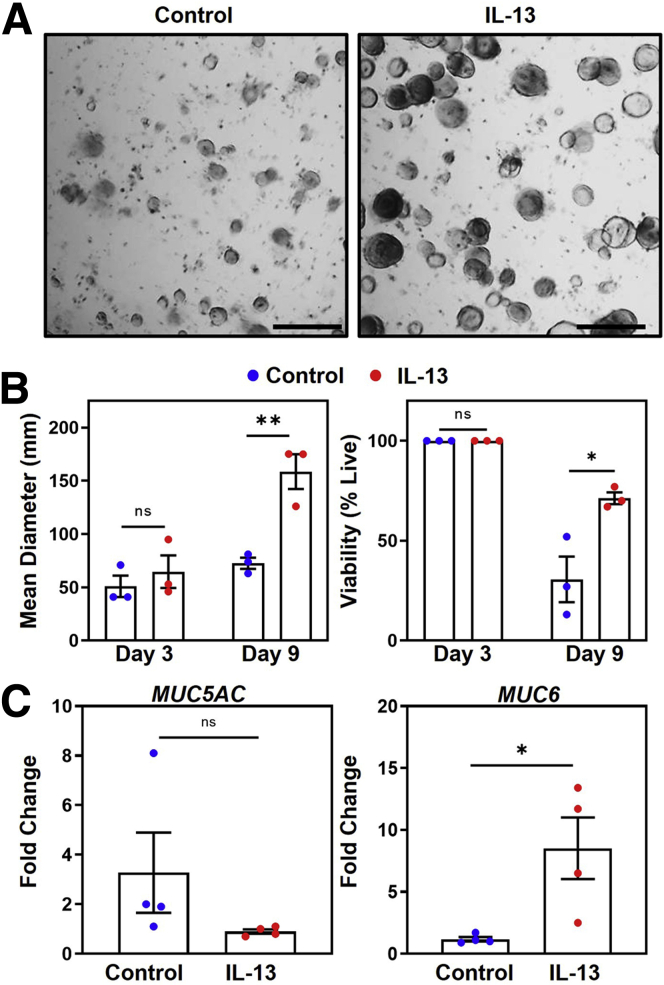
To determine whether IL13 has similar effects on human gastric epithelium, we repeated the previously described experiments using human corpus–derived gastric organoids. Photographs of organoids cultured with and without recombinant human IL13 were taken on days 3 and 9 (Figure 3A) to assess the size and viability of organoids over the culture period. Similar to results observed in mouse-derived organoids, by day 9, IL13-cultured organoids were twice as large in diameter (159 ± 16 μm vs 73 ± 5.2 μm; P = .007) and had increased viability (71% ± 2.9% vs 31% ± 11%; P = .026) compared with control organoids (Figure 3B). The qPCR analyses showed an increase in MUC6 expression (1.2 ± 0.18 vs 8.5 ± 2.5; P = .026) in IL13-cultured human organoids (Figure 3C). Together, these data show that IL13 elicits similar responses of increased organoid size, viability, and increasing MUC6-expressing cells in both mouse and human organoids.
Figure 3.
IL13 increases organoid size and viability and alters MUC5AC and MUC6 expression in human organoids. (A) Representative organoid photographs on day 9 cultured without (left) and with (right) 5 ng recombinant human IL13. Scale bars: 200 μm. (B) Left: Quantification of the mean diameter of organoids cultured without (blue) and with (red) 5 ng recombinant human IL13. Each dot represents the mean diameter from 1 independent experiment (N = 3). ∗∗P = 0.007. Right: Quantification of organoid viability. Each dot represents the proportion of viable organoids from 1 independent experiment (N = 3). ∗P = 0.026. (C) qPCR analysis of MUC5AC expression (left) and MUC6 expression (right) in organoids cultured without (blue) or with (red) 5 ng recombinant human IL13 on day 7. Data show fold change expression. Each dot represents 1 organoid culture from 1 independent experiment (N = 4). ∗P = 0.026. Significance was determined using a Student t test. Data show means ± SEM.
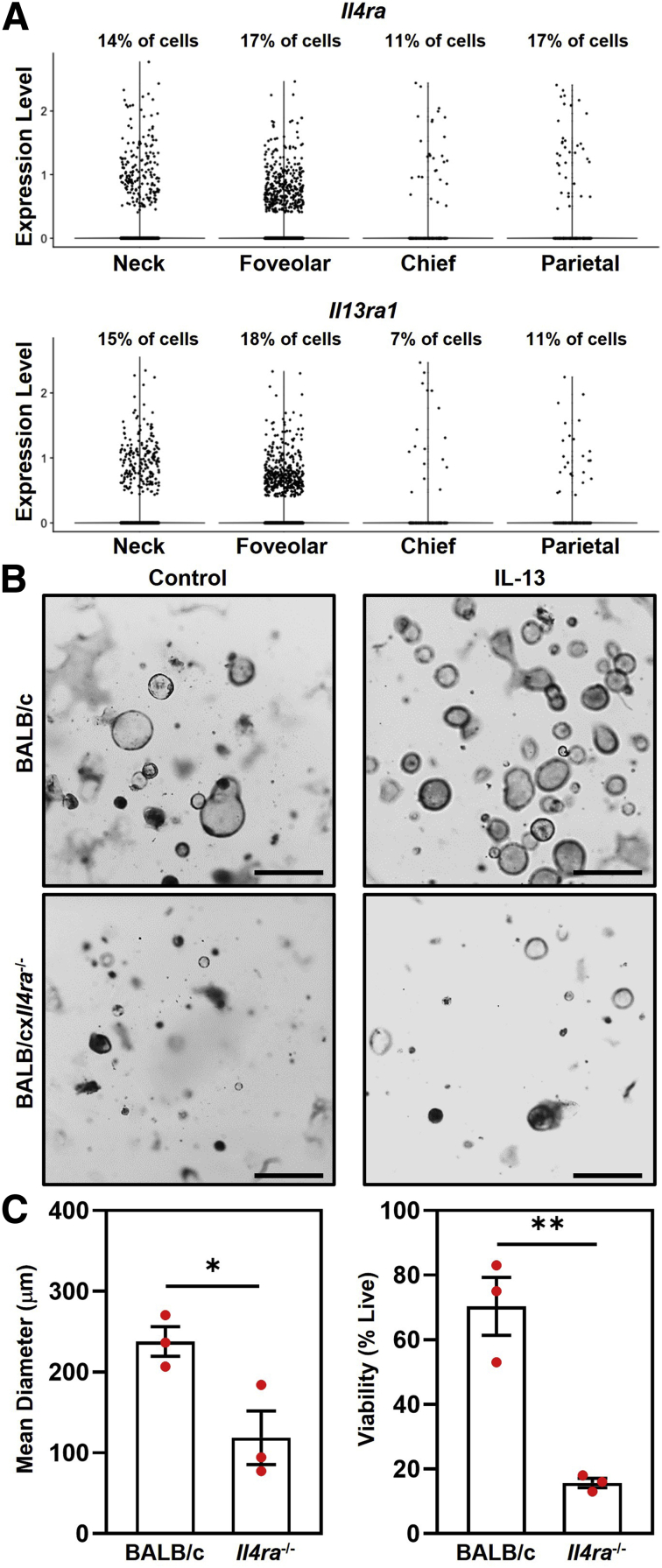
The effects of IL13 in organoid cultures indicated that IL13 affected the number of mucous neck cells (Muc6+) and foveolar cells (Muc5ac+). To determine whether mucous neck and/or foveolar cells express the IL13 receptor (Il4ra and Il13ra1), we analyzed our single-cell RNA sequencing (scRNA-seq) data sets of gastric epithelial cells isolated from stomachs of TxA23 mice. We detected Il4ra and Il13ra1 expression in both neck and foveolar cells, as well as in chief and parietal cells.
Isolation of gastric glands may result in carryover of gastric immune cells into organoid cultures. Although organoid culture conditions are not ideal for the survival of immune cells, we determined whether IL13 may be affecting organoids indirectly by acting on contaminating immune cells in IL13-cultured organoids. Immune cells were isolated from the gastric lymph nodes of TxA23 mice and cultured with organoids derived from glands from BALB and BALB/cxIl4ra-/- mice. Il4ra-/- organoids do not express the IL4Rα subunit and cannot respond to IL13. If IL13 acted on contaminating immune cells to promote larger and more viable organoids, then Il4ra-/- organoids would have a phenotype like that of BALB/c organoids after the addition of IL13. Photographs were taken on day 9 of culture (Figure 4B), and organoid diameter and viability were determined as previously described. The mean organoid diameter was 2-fold higher in BALB/c organoids compared with Il4ra-/- organoids (238 ± 18 μm vs 119 ± 33 μm; P = .03). BALB/c organoids also had more than a 3-fold increase in viability compared with Il4ra-/- organoids cultured with IL13 (70% ± 8.9% vs 16% ± 1.5%; P = .004) (Figure 4C). Culturing Il4ra-/- organoids with wild-type immune cells did not rescue the organoid phenotype. These data show that IL13 acts directly on the gastric epithelium, and not on surrounding immune cells, to promote increased organoid size and viability.
Figure 4.
IL13 acts on gastric epithelial cells to increase organoid size and viability. (A) Summary violin plots showing transcript expression in TxA23 mice of the IL13-receptor subunits Il4ra (top) and Il13ra1 (bottom) on the corpus epithelial cell subsets: neck cells, foveolar cells, chief cells, and parietal cells. Above each plot the cell subset frequency of cells with a transcript expression level >0.1 is labeled. (B) Representative photographs of BALB/c (top) and BALB/cxIl4ra-/- (bottom) organoids cultured with immune cells isolated from TxA23 mice. Photographs are of day 9 organoids cultured without (left) and with (right) 5 ng recombinant human IL13. Scale bars: 200 μm. (C) Left: Quantification of the mean diameter of organoids cultured with (red) 5 ng recombinant human IL13. Each dot represents the mean diameter from 1 independent experiment (N = 3). ∗P = 0.03. Right: Quantification of organoid viability. Each dot represents the proportion of viable organoids from 1 independent experiment (N = 3). ∗∗P = 0.004. Significance was determined using a Student t test. Data show means ± SEM.
IL4Rα Expression Is Critical for Inflammation-Induced Neck Cell Expansion and Metaplasia Development
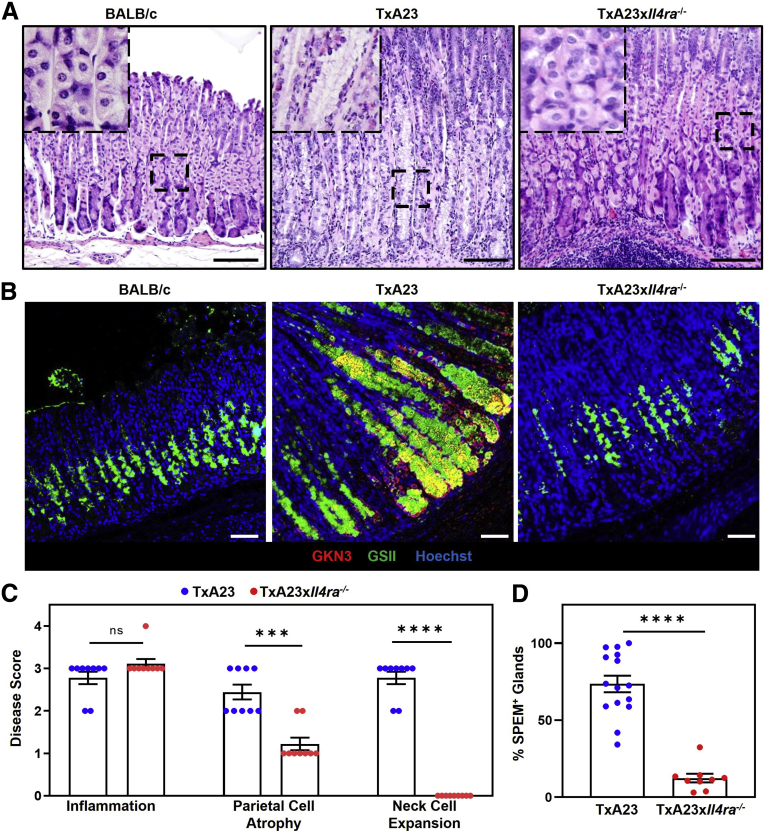
Having identified IL13-producing immune cells in the gastric mucosa and having shown that IL13 acts directly on the gastric epithelium in organoid cultures, we designed experiments to determine the importance of IL13 signaling in gastric pathology induced by chronic gastritis. We crossed TxA23 mice with IL4Rα-deficient mice to generate a new line of TxA23×Il4ra-/- mice. Disease in the TxA23 mouse model has been well established at 4 months of age.26 The degrees of inflammation, atrophy, and mucous neck cell expansion were compared in cohorts of 4-month-old TxA23 and TxA23×Il4ra-/- mice. As expected, TxA23 mice had moderate to severe inflammation, parietal cell atrophy, expansion of mucous neck cells, and loss of chief cells in the gastric mucosa. Although age-matched TxA23×Il4ra-/- mice had similar degrees of inflammation compared with TxA23 mice (2.8 ± 0.15 vs 3.1 ± 0.11; P = .26), and slightly less parietal cell atrophy (2.4 ± 0.18 vs 1.2 ± 0.15; P = .001), they had a complete lack of neck cell expansion (2.8 ± 0.15 vs 0.0 ± 0.0; P <.0001) (Figure 5A and C). Immunofluorescent staining was performed to compare the development of SPEM. SPEM can be identified by the expression of a number of proteins not expressed in healthy epithelium (CD44v9, GKN3).20,21,45 As previously reported, SPEM staining was present throughout the corpus of TxA23 mice.20 In contrast, there was little to no SPEM observed in age-matched TxA23×Il4ra-/- mice (Figure 5B). SPEM was quantitated by calculating the proportion of corpus glands containing GKN3+ cells: 74% of glands in TxA23 contained SPEM, while very few SPEM-containing glands were observed in TxA23×Il4ra-/- mice (74 ± 5.3 vs 12 ± 2.8; P <.0001) (Figure 5D). These results showed that, although signaling through IL4Rα is not required to develop moderate–severe gastritis with atrophy, signals through this receptor are important for the development of neck cell expansion and induction of metaplasia (SPEM) in a chronic inflammatory setting.
Figure 5.
IL4Ra is critical for inflammation-induced neck cell expansion and metaplasia development. (A) H&E corpus sections from healthy 4-month-old BALB/c (left), TxA23 (center), and TxA23×Il4ra-/- (right). Enlarged inlay section shown in top left corner. Scale bars: 200 μm. (B) Immunofluorescent staining with anti-GKN3 (red), GSII (green), and Hoechst (blue) showing SPEM development (GKN3+GSII+, yellow) in BALB/c mice (left) and 4-month-old TxA23 (center) and TxA23×Il4ra-/- mice (right). Scale bars: 50 μm. (C) Disease scores of 4-month-old TxA23 and TxA23×Il4ra-/- mice. Each dot represents 1 mouse (N = 9). Significance was determined using a Mann–Whitney U test. ∗∗∗P = 0.001 ∗∗∗∗P <0.0001. (D) Quantification of SPEM+ (GKN3+) glands in 4-month-old TxA23 and TxA23×Il4ra-/-. Each dot represents 1 mouse (N = 9–15). Significance was determined using a Student t test. ∗∗∗P <0.0001. Data show means ± SEM.
Immune Cell Subsets and Protein Secretion in the Gastric Mucosa Are Similar Between Control and IL4Rα-Deficient Mice
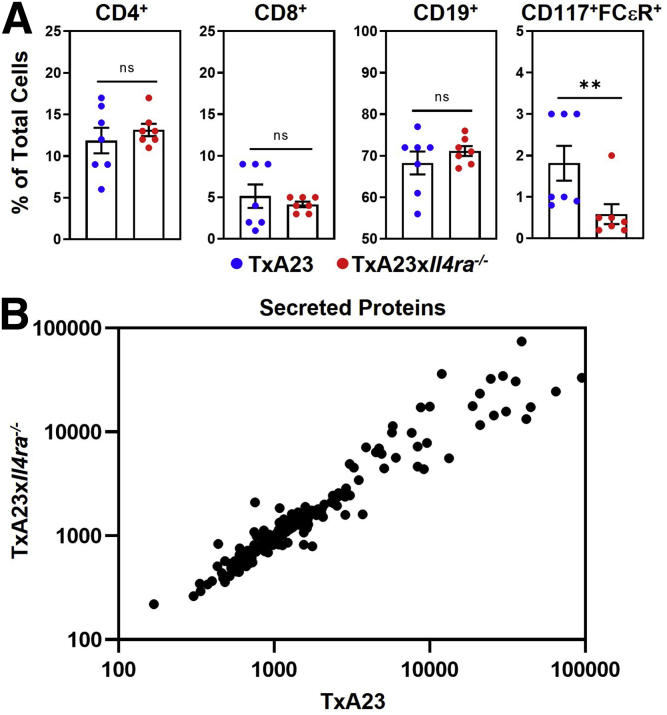
In the TxA23×Il4ra-/- mouse, all cells lack the ability to receive signals through IL4 and IL13. This is relevant because IL4Rα is expressed on both immune and epithelial cells, so it is possible that the phenotype of the TxA23×Il4ra-/- is owing to a lack of signaling into immune cells, epithelial cells, or both. To determine whether the lack of IL4Rα affected inflammation, we first compared immune cell subsets infiltrating the gastric mucosa of TxA23 and TxA23×Il4ra-/- mice. Immune cells were isolated from the gastric mucosa of 4-month-old TxA23 and TxA23×Il4ra-/- mice and identified by flow cytometry. After gating on CD45+ immune cells, additional markers were used to identify immune cell subsets including: CD4+ and CD8+ T cells, CD19+ B cells, and FcεR+CD117+ mast cells (Figure 6A). There were no significant differences in the percentages of CD4+ T cells (12 ± 1.5 vs 13 ± 0.74; P = .5), CD8+ T cells (5.1 ± 1.4 vs 4.1 ± 0.34; P = .5), and CD19+ B cells (68 ± 2.8 vs 71 ± 1.2; P = .4) between control and IL4Rα-deficient mice. A 2-fold increase in the percentage of mast cells in TxA23 mice compared with TxA23×Il4ra-/- mice was observed (1.8 ± 0.42 vs 0.6 ± 0.24; P = .03). Overall, these data show that the major subsets of immune cells infiltrating the gastric mucosa (CD4+ and CD8+ T cells, B cells) are similar in control and IL4Rα-deficient mice.
Figure 6.
Immune cell subsets and protein secretion in the gastric mucosa was similar between control and IL4Ra-deficient mice. (A) CD45+ immune cells isolated from the stomach of TxA23 and TxA23×Il4ra-/- mice. Data show the percentage of total cells that are CD4+, CD8+, CD19+, and mast cells (FceR+CD117+). Each dot represents 1 mouse (N = 7). ∗∗P = 0.03. (B) Secretion of 200 individual proteins from TxA23 and TxA23×Il4ra-/- mucosal immune cells using a mouse cytokine array GS4000 assay (RayBiotech). Each dot represents the detected secretion level of individual proteins. Significance was determined using a Student t test. Data show means ± SEM.
To determine whether the lack of IL4Rα signaling affected cytokine secretion by immune cells infiltrating the gastric mucosa, immune cells were isolated, restimulated, and supernatants were collected at 72 hours. Supernatants were analyzed using a multiplex protein array to identify and compare the levels of 200 proteins secreted by immune cells from control TxA23 and TxA23×Il4ra-/- mice. In Figure 6B, each dot represents an individual protein, with the relative levels in the supernatants from activated immune cells from TxA23 and TxA23×Il4ra-/- quantified on the axes. The expression levels of all 200 proteins were nearly identical. Only 3 proteins (IL6, IL1RA, and vascular endothelial growth factor), were significantly different and all differences were less than 3-fold. Together, these data identify the relative expression levels of 200-secreted proteins by immune cells isolated from the gastric mucosa and show that the immune cell types and secreted proteins are nearly identical between TxA23 and TxA23×Il4ra-/- mice.
IL4Rα-Deficient Neck and Chief Cells Up-Regulate Inflammation-Associated Gene Transcripts and Adopt a Pre-SPEM Phenotype, but Do Not Progress to SPEM During Chronic Inflammation
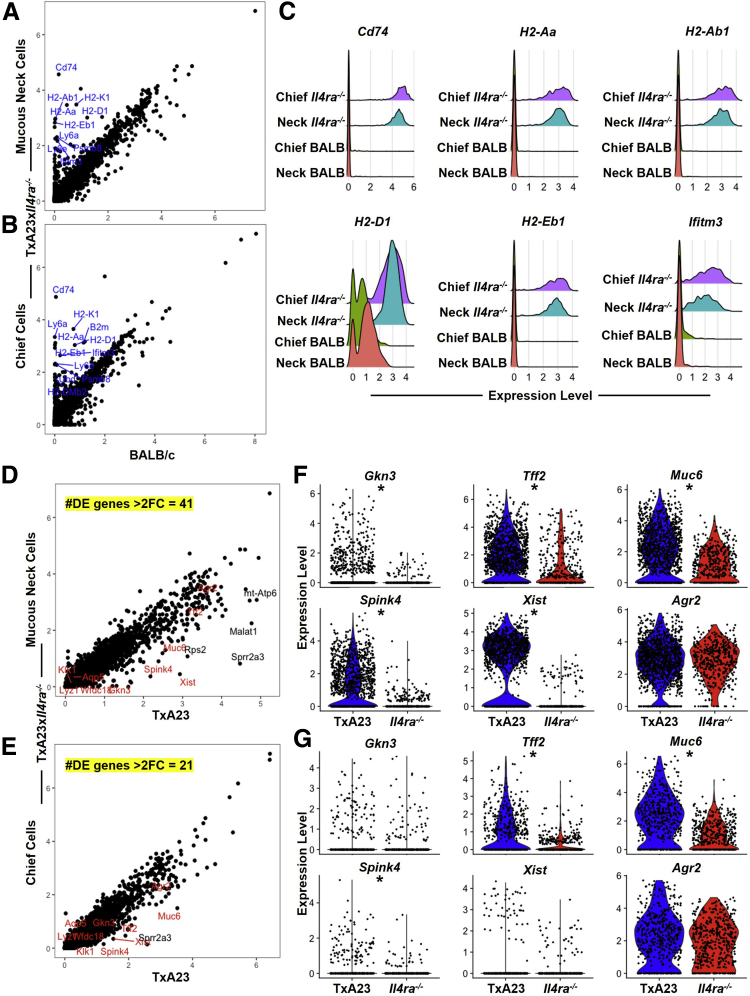
Previous experiments identified cellular changes in organoid cultures induced by IL13. Experiments were designed to determine how signaling through IL4Rα regulates transcriptional changes in mucous neck cells and chief cells within a disease setting. To do this, we performed scRNA-seq on gastric epithelium isolated from the stomach of BALB/c mice (healthy controls), TxA23, and TxA23×Il4ra-/- mice. We previously used scRNA-seq to define the transcriptional responses of gastric epithelium to acute and chronic inflammation, and showed that in a chronic inflammatory setting, both mucous neck cells and chief cells undergo transcriptional changes that result in SPEM.20,29 Although the pathologic evaluation of TxA23×Il4ra-/- mice indicated very little SPEM, as defined by a lack of GKN3 staining (Figure 4), it was unclear how the lack of IL4/IL13 signaling affected the transcriptional responses of mucous neck cells and chief cells to chronic inflammation. To address this, the average per-cell expression level of transcripts detected was determined and compared between various populations of epithelial cells isolated from stomachs of TxA23×Il4ra-/- and BALB/c mice. This analysis identified several inflammation-associated transcripts that were significantly higher in mucous neck cells and chief cells from TxA23×Il4ra-/- stomachs relative to BALB/c stomachs (Figure 7A and B). These inflammation-associated transcripts included invariant chain CD74, major histocompatibility complex (MHC) class II H2-Aa, H2-Ab1, H2-Eb1, MHC class I H2-D1, and interferon-associated Ifitm3 compared with cells from healthy stomachs (Figure 7C). This analysis showed that although TxA23×Il4ra-/- gastric epithelium appeared phenotypically healthy, transcriptionally, mucous neck and chief cells that lack IL4/IL13 signaling up-regulate inflammation response genes, indicating a response to inflammatory cytokines.
Figure 7.
(See previous page). IL4Ra-deficient mucous neck and chief cells up-regulate inflammation-associated transcripts and adopt a pre-SPEM phenotype but do not progress to SPEM during chronic inflammation. (A and B) Dot plots of per-cell average relative expression of all detected transcripts from neck (A) and chief (B) cells identified in BALB/c and TxA23×Il4ra-/- libraries. Statistically significant inflammatory-associated transcripts with a greater than 3-fold expression level increase in TxA23×Il4ra-/- over BALB/c (blue). (C) Ridge plots summarizing the expression level of selected inflammation-associated genes of interest in neck and chief cells from BALB/c and TxA23×Il4ra-/- stomachs. (D and E) Dot plots of per-cell average relative expression of all detected transcripts from (D) neck and (E) chief cells identified in TxA23 and TxA23×Il4ra-/- libraries. Statistically significant SPEM-associated transcripts (red) and other transcripts (black) with a greater than 3-fold expression level increase in TxA23×Il4ra-/- over TxA23. The number of statistically significant and greater than 2-fold differentially expressed genes between TxA23 and TxA23×Il4ra-/- mice (yellow). (F and G) Summary violin plots showing expression level differences of selected SPEM-associated genes of interest in (F) neck and (G) chief cells of TxA23 and TxA23×Il4ra-/- mice. ∗P < .0001 by Wilcoxon rank-sum test. DE, Differentially Expressed; FC, Fold Change.
We next set out to determine whether IL4/IL13 signaling regulated SPEM-associated transcript expression. To do this, we compared single-cell gastric epithelial libraries from TxA23 and TxA23×Il4ra-/-. We previously identified SPEM-associated transcripts expressed in both neck and chief cells during chronic inflammation.20 These transcripts include SPEM-associated transcripts such as Muc6, Agr2, and Tff2; the metaplasia-associated transcript Gkn3; the long noncoding RNA transcript Xist, which has been implicated in gastric cancer cell biology by promoting proliferation and progression; and Spink4, a serine peptidase inhibitor associated with malignant colorectal cancer.46,47 Analysis of the transcriptional differences between TxA23 and TxA23×Il4ra-/- mucous neck cells showed a total of 41 differentially expressed genes with a greater than 2-fold change (Figure 7D). SPEM-associated transcripts Gkn3, Tff2, Muc6, Spink4, and Xist were up-regulated significantly in TxA23 neck cells relative to that of TxA23×Il4ra-/-, while expression of Agr2 was similar between the 2 libraries (Figure 7F). Chief cell comparison of transcript expression identified 21 differentially expressed genes with a greater than 2-fold change (Figure 7E). Tff2, Muc6, and Spink4 were expressed at significantly higher levels in TxA23 chief cells compared with TxA23×Il4ra-/-, while Gkn3, Xist, and Agr2 had similar expression levels (Figure 7G). Together, these data show that neck and chief cells deficient in IL4/IL13 signaling respond to inflammatory stimuli, but fail to up-regulate SPEM-associated transcripts during chronic inflammation.
IL13 Is Critical for Inflammation-Induced Mucous Cell Expansion and Metaplasia Development
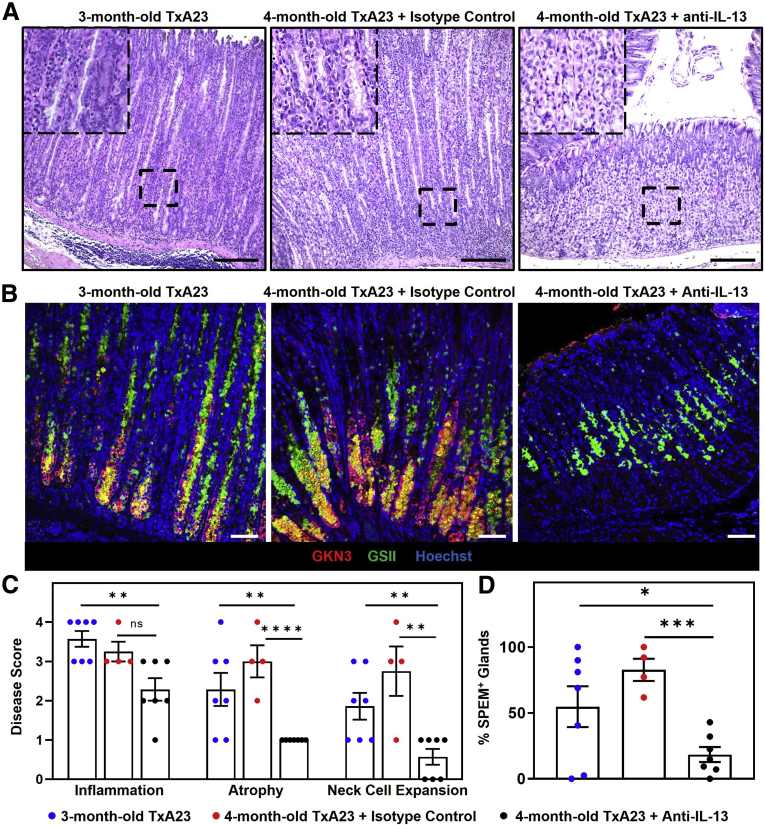
Both IL13 and IL4 signal through the IL4Rα subunit, however, undetectable levels of IL4 and relatively high levels of IL13 secreted by immune cells led us to focus on IL13 (Figure 1A). To determine if IL13 signaling contributes to disease pathology during chronic inflammation, we treated TxA23 mice with an IL13-neutralizing antibody and determined the impact of treatment on disease development. Three-month-old TxA23 mice were treated twice a week with 10 mg/kg of IL13-neutralizing antibody or IgG isotype control. At the time of treatment (age, 3 mo), TxA23 mice had moderate degrees of inflammation, atrophy, mucous cell expansion, and SPEM. After 4 weeks of treatment, gastric pathology was assessed in isotype control and anti–IL13-treated mice. As expected, 3-month-old and isotype control-treated 4-month-old TxA23 mice had severe inflammation, moderate–severe atrophy, and expansion of neck cells throughout the glands. After 4 weeks of treatment, TxA23 mice treated with anti-IL13 had a similar degree of inflammation compared with 3-month-old TxA23 (3.6 ± 0.2 vs 2.3 ± 0.29; P = .003) and isotype control-treated TxA23 (3.3 ± 0.25 vs 2.3 ± 0.29; P = .05). Atrophy scores were lower in anti–IL13-treated mice compared with 3-month-old TxA23 (2.3 ± 0.41 vs 1.0 ± 0.0; P = .01) and control-treated TxA23 (3.0 ± 0.41 vs 1.0 ± 0.0; P <.0001). The striking result, however, was that mucous neck cell expansion was nearly absent in anti–IL13-treated mice compared with isotype control-treated (2.8 ± 0.63 vs 0.57 ± 0.2; P = .002) and 3-month-old TxA23 (1.9 ± 0.34 vs 0.57 ± 0.2; P = .007) (Figure 8A and C).
Figure 8.
IL13 is critical for inflammation-induced mucous cell expansion and metaplasia development. (A) H&E corpus sections showing the degree of inflammation, parietal cell atrophy, and mucous cell expansion in 3-month-old TxA23 (left) and 4-month-old TxA23 treated with isotype control (center) or anti-IL13 (right). Enlarged inlay section shown in top left corner. Scale bars: 200 μm. (B) Immunofluorescent staining with anti-GKN3 (red), GSII (green), and Hoechst (blue) showing SPEM development (GKN3+GSII+, yellow). Scale bars: 50 μm. (C) Disease scores of TxA23 mice at 3 months of age before treatment (blue), and 4 months of age after treatment with isotype control (red) or anti-IL13 (black). Each dot represents 1 mouse (N = 4–7). Significance was determined using the Mann–Whitney U test. Inflammation: ∗∗P = 0.003; Atrophy: ∗∗P = 0.01 ∗∗∗∗P < 0.0001; Neck Cell Expansion: ∗∗P = 0.002 ∗∗P = 0.007. (D) Quantification of SPEM+ (GKN3+) glands in 3-month-old TxA23 (blue) and 4-month-old TxA23 treated with isotype control (red) or anti-IL13 (black). Each dot represents 1 mouse (N = 4–7). Significance was determined using a Student t test. ∗P = 0.04 ∗∗∗P = 0.0001. Data show means ± SEM.
Next, we evaluated the stomachs for the presence of SPEM using immunofluorescent staining. Three-month-old and isotype-treated TxA23 mice had extensive SPEM (GKN3+GSII+) throughout the gastric glands; however, mice treated with anti-IL13 had little to no SPEM development (Figure 8B). More than 50% of glands in 3-month-old (55 ± 16 vs 18 ± 5.7; P = .04) and isotype-treated (83 ± 8.5 vs 18 ± 5.7; P = .0001) TxA23 mice were SPEM+, whereas mice treated with anti-IL13 developed very few SPEM-containing glands (18 ± 5.7) (Figure 8D). Together, these results show an important role for IL13 in promoting neck cell expansion and SPEM development during chronic inflammation. In addition, treatment with anti-IL13 prevented neck cell expansion and the development of SPEM, showing that IL13 inhibition may be a potential therapeutic strategy for preventing progression from atrophic gastritis to metaplasia.
Discussion
Th1 and Th17 are the primary immune responses in both mice and human beings during chronic H pylori infections and autoimmune gastritis.27 We previously determined that both IFNγ and IL17 act directly on gastric epithelial cells to promote parietal cell atrophy, and recent studies have begun to investigate the role of type 2 cytokines in SPEM development.24,25,36,48 IL4 and IL13 are type 2 cytokines often thought to have similar effector functions, however, studies conducted using knockout mice and neutralizing antibodies have discovered that IL13 display several unique effector functions that distinguish it from IL4.31 A study using cytotoxic T-lymphocyte-associated protein 4 (CTLA4)-knockdown mouse models of chronic inflammation highlighted the importance of IL4 and IL13 in the development of gastric atrophy and neck cell hyperplasia and that Th2 CD4+ T cells may be the driving force behind the progression from gastritis to tumorigenesis.48 In this study, we characterized cytokine production from mucosal immune cells isolated from the stomachs of mice with autoimmune gastritis. High levels of IFNγ, IL17A, and IL13 were secreted from the isolated immune cells, while IL4 secretion was nearly undetectable. This finding showed that IL13 was not being secreted by canonical CD4+ Th2 cells. The lack of IL4, but the presence of IL13, led us to investigate and identify IL13-producing immune cells in the gastric mucosa of mice with chronic gastritis. Other studies have focused primarily on IL13 secretion from M2 macrophages, ILC2s, and eosinophils, and highlight the important role of IL13 and the IL33/ST2 axis.36,49, 50, 51, 52 In this study, we identified 6 IL13-secreting immune cell types in the inflamed gastric mucosa. Of these newly identified IL13-secreting immune cells, we discovered that mast cells were the largest producers of IL13. Furthermore, in healthy BALB/c stomachs, mast cells were confined to the submucosa (data not shown), but infiltrated into the gastric glands in the chronically inflamed stomach of TxA23 mice. Petersen et al49 identified an important role for IL33 and M2 macrophages in SPEM development, and Eissmann et al53 stated in a recent publication that IL33-mediated mast cell activation promotes macrophage recruitment to the site of gastric tumors. In addition, mast cell density is increased in well-differentiated gastric tumors.54 However, the role of mast cells and whether mast cells or mast cell–derived IL13 is critical for SPEM development and gastric tumor progression remains unknown. The novel finding that IL13 is produced by multiple immune cell subsets that may contribute to SPEM development during chronic inflammation adds to the complexity of the immune response during chronic inflammation.
Overexpression of IL13 in the lung increases mucous cell hyperplasia, a key pathologic feature of inflammation-induced metaplasia; however, whether or not IL13 can act directly on the gastric epithelium has yet to be determined.55 In this study, we discovered that multiple gastric epithelial cell types express IL13 receptors and may be capable of responding to IL13 signals in different ways. Our study also determined that IL13 acts directly on mucous neck cells in organoid cultures to increase size and survival. The addition of IL13 to organoids decreased expression of the mucin gene Muc5ac and the number of UEAI+ cells. This important finding shows that IL13 acts directly on the gastric epithelium and may be contributing to mucous neck cell hyperplasia and/or SPEM development by affecting epithelial cell growth, survival, and/or differentiation.
IL4 and IL13 signal via an IL4Rα-containing heterodimeric receptor to initiate phosphorylation of signal transducer and activator of transcription 6 (STAT6). Recent observations have found that phospho-STAT6 regulates prometastatic behaviors such as proliferation, migration, and tissue invasion.33 This study identified an important role for IL4Rα in promoting inflammation-induced mucous neck cell expansion and SPEM development. Mice with autoimmune gastritis deficient in IL4Rα had no neck cell expansion and very little SPEM development despite the presence of gastritis. In addition, IL4Rα deficiency did not significantly alter the infiltrating immune cell subsets or secreted proteins and is not a likely cause for the decrease in disease pathology severity. These results show that IL4/IL13 signaling via IL4Rα is not required for gastritis, but that signals through this receptor regulate neck cell expansion and metaplasia (SPEM), establishing an important role for IL4/IL13 in SPEM development.
In autoimmune gastritis mouse models of inflammation-induced metaplasia, both neck and chief cells begin to express inflammation-associated genes (ie, MHC class I, MHC class II), followed by up-regulation of metaplasia-associated genes (Tff2, Muc6, Gkn3, and Xist).20 SPEM is associated with 90% of resected gastric cancers, yet the mechanisms by which inflammation regulates the epithelial cell changes that promote SPEM development are poorly understood.56 In mouse models of CTLA4 knockdown-induced gastritis, mice deficient in IL4/IL13 or IL4Rα had decreased parietal cell atrophy and mucous cell hyperplasia, but the molecular characterization of SPEM was not determined.48 The current study confirmed that SPEM and SPEM-associated transcripts are decreased in IL4Rα-deficient mice with autoimmune gastritis. Another key takeaway from this study is that although neck and chief cells in autoimmune gastritis mice lacking the IL4/IL13 receptor appeared healthy by H&E staining, we identified numerous inflammation-associated transcripts that were up-regulated compared with healthy cells. Although these cells were responding to inflammation; they did not transition to SPEM in the absence of the IL4Rα receptor. Finally, this study showed that a therapeutic antibody developed to neutralize IL13 is effective at inhibiting neck cell expansion and preventing SPEM development.
SPEM can develop as a result of H pylori– or autoimmune-induced chronic inflammation and there are multiple immune pathways that contribute to SPEM development. Previous studies have found that T cells are necessary for parietal cell atrophy during H pylori infection, identifying a role for the adaptive immune system in the progression to SPEM.57 T-cell–derived IFNγ promotes parietal cell atrophy and macrophage activation, and infiltrating M2 macrophages have been found to contribute to a metaplastic phenotype in models of drug-induced parietal cell atrophy and SPEM.25,49,58 The IL33/IL13 signaling pathway has been identified as an important immune component in the polarization of M2 macrophages and progression of metaplasia after drug-induced parietal cell atrophy.36 Our study identified mast cells as the largest producers of IL13 in a model of autoimmune gastritis and identified mast cells infiltrating into the gastric mucosa during chronic inflammation. ILC2s also are found in the inflamed gastric tissue and are another source of IL13. Furthermore, depletion of ILC2s inhibits the development of metaplasia after gastric injury.52 The findings in this study further underscore the critical role of IL13 in the progression of gastritis to metaplasia development and show it can serve as a therapeutic target. IL13- and IL4Rα-neutralizing antibodies (lebrikizumab, dupilumab) are currently in use in clinical settings and this study identifies IL13 and its receptor as potential therapeutic targets for halting and/or reversing progression from atrophic gastritis to metaplasia.
Methods
Mice
TxA23 mice are a transgenic mouse model with T-cell receptors specific for a peptide from the H+/K+ adenosine triphosphatase α chain on a BALB/c background and have been described previously.26, 27, 28,59 Additional mice include BALB/c (000651; Jackson Laboratories, Bar Harbor, ME), BALB/c-Il4ratm1SzIJ (003514; Jackson Laboratories), and C.129S4(B6)-Il13tm1(YFP/cre)LkyIJ (017353; Jackson Laboratories). BALB/c-Il4ratm1SzIJ was crossed with TxA23 mice to generate the TxA23×Il4ra-/- mouse line. C.129S4(B6)-Il13tm1(YFP/cre)LkyIJ was crossed with TxA23 mice to generate the TxA23×Il13-Yfp/Cre mice.44 All TxA23 mice develop CD4-mediated autoimmune gastritis. All mice were maintained in the Saint Louis University Medical School animal facility and cared for in accordance with institutional guidelines. Mixed groups of age-matched male and female co-housed littermates were used for all experiments. All strains were housed under specific pathogen-free conditions.
Histopathology
Stomachs were removed from mice, cut along the lesser curvature, thoroughly washed in a phosphate-buffered saline (PBS) bath pH 7.2–7.4, and then fixed in 10% neutral-buffered formalin for 24 hours. After 24 hours, the forestomach and antrum were removed, leaving only the corpus region for paraffin embedding, sectioning, and staining with H&E or toluidine blue. H&E- and toluidine blue–stained corpus tissue section photographs were taken using a Leica (Wetzlar, Germany) SP6 Epifluorescence microscope. A 10× objection was used for all photographs. For disease scoring, slides were blinded and sections from individual mice were assigned scores between 0 (absent) and 4 (severe) to indicate the severity of inflammation, oxyntic atrophy, and mucous cell hyperplasia.60
Immunofluorescence
Stomachs were prepared, stained, and imaged using methods modified from Ramsey et al.61 Briefly, paraffin-embedded corpus stomach tissue sections were deparaffinized through a series of isopropanol and xylene washes, and 10 μmol/L sodium citrate pH 6.0 was used for antigen retrieval. Slides were washed with PBS and blocked for 1 hour with blocking buffer (PBS, 1% bovine serum albumin [BSA]; 810033; Millipore, Burlington, MA; 0.1% Triton X-100; VW3929-2; VWR International; Radnor, PA). The primary antibodies and lectins used for immunofluorescence were as follows: GSII (1:500, L21415; Life Technologies, Eugene, OR), Hoechst 33342, trihydrochloride trihydrate (1:20,000, H1399; Life Technologies), rabbit anti-GKN3 (1:400, a kind contribution from Jason C. Mills lab, Washington University, St Louis, MO), UEA-I (1:500, DY488-2201-1; EY Laboratories, Inc, San Mateo, CA), and purified mouse anti–E-cadherin (1:100, 610181; BD Biosciences, San Jose, CA). Secondary antibodies were as follows: donkey anti-rat IgG (1:500, A21209; Life Technologies), rabbit anti-goat IgG (1:500, A11080; Life Technologies), donkey anti-mouse IgG (1:500, A31571; Invitrogen, Carlsbad, CA), and chicken anti-rabbit IgG (1:500, A21443; Life Technologies). Immunofluorescent corpus stomach tissue section photographs were taken using a Leica TCS SP8 confocal microscope. A 20× objective was used for all photographs.
Isolation of Immune Cells
Isolating immune cells from the stomach tissue has been described previously.62,63 Briefly, stomachs were harvested from mice, the gastric lymph node was removed, and the stomach was dissected along the lesser curvature. After rinsing in a PBS bath, the stomach tissue was repeatedly injected with a total of 10 mL cold MACS buffer (PBS, 0.5% BSA; 810033; Millipore; 2 mmol/L EDTA; 15575020; Invitrogen) using a 27-gauge needle. The mucosa was injected in such a way as to inflate and rupture the tissue to free the infiltrating immune cells. After repeated injections, the stomach was cut into small sections and collected with the 10 mL of MACS buffer, transferred to a 50-mL conical tube, and then vortexed for 30 seconds to create a single-cell suspension. The cells then were passed through a 70-μm pore size nylon filter and washed with buffer and used for analysis.
To isolate immune cells from the gastric lymph node, stomachs were harvested from TxA23 mice and the gastric lymph node was removed. Using the plunger end of a 3-mL syringe, the lymph node then was gently smashed against a pre-wet, 70-μm pore size nylon filter to free the immune cells. Advanced Dulbecco’s modified Eagle medium (DMEM) (10 mL, 12492013; ThermoFisher, Waltham, MA) then was used to wash the cells through the filter and collect them in a 50-mL conical tube. Cells were centrifuged at 1300 rpm and resuspended at 1 × 106 cells/mL and used in subsequent experiments.
Flow Cytometry
Immune cells were isolated from the gastric mucosa following the protocol previously described and cell surface staining was performed according to standard procedures.62,63 Immune cell subsets were identified using the following antibodies: CD45 (564225; BD Horizon, San Diego, CA), CD45.2 (552950; BD Pharmingen, San Diego, CA), CD4 (562891; BD Horizon), CD4 (553730; BD Pharmingen), CD8 (563046; BD Horizon), CD19 (551001; BD Pharmingen), CD19 (48-0193-82; Invitrogen), CD19 (563148; BD Horizon), FceR1 (47-5898-80; eBioscience, San Diego, CA), CD117 (553869; BD Pharmingen), Fc Block (553142; BD Pharmingen), live/dead (L34966; Invitrogen), CD11b (48-0112-82; Invitrogen), CD11c (550261; BD Pharmingen), I-A/I-E (557000; BD Pharmingen), F4/80 (123147; BioLegend, San Diego, CA), CD90.1 (740917; BD OptiBuild, San Diego, CA), CD 90.1 (563772; BD Horizon), CD3e (48-0031-82; eBioscience), GR-1 (48-9891-82; eBioscience), CD127 (15-1271-82; Invitrogen), CD49b (17-5971-82; eBioscience), and CD25 (551071; BD Pharmingen). Intracellular cytokine staining was performed using antibodies against IFNγ (560660; BD Pharmingen), IL17A (564170; BD Horizon), IL4 (554436; BD Pharmingen), and IL13 (12-7133-81; eBioscience). All flow cytometry was performed on a BD LSRII (BD Biosciences) or BD FACSCanto (BD Biosciences) and analyzed using FlowJo (FlowJo, Ashland, OR). For intracellular staining, cells were stimulated with 50 ng/mL phorbol myristate acetate (p8139; Sigma, St. Louis, MO) and 1 μg/mL ionomycin (10634; Sigma) at 1 × 106 cells/mL for 4 hours at 37°C. Golgi-stop (554724; BD Biosciences) was added after 1 hour of incubation. Cells then were washed with PBS containing 0.5% BSA (810033; Millipore) and fixed and permeabilized using eBioscience forkhead box P3 (FOXP3)/Transcription Factor Staining Buffer Set (00-5523-00; Invitrogen). Cells were incubated overnight with the anticytokine antibodies, and then washed and analyzed by flow cytometry.
For cytokine analysis, cells were cultured at 1 × 106 cells/mL in vitro in 15-mL conical tubes and stimulated with 25 ng/mL phorbol myristate acetate and 500 ng/mL ionomycin for 72 hours at 37°C. Supernatants were collected and cytokine secretion was measured using a Th1, Th2, Th17 (560485; BD Biosciences) and IL13 flex set (558349; BD Biosciences) cytometric bead array kit and analyzed by flow cytometry or sent to RayBiotech (Peachtree Corners, GA) for analysis using a 200-mouse cytokine array (GS4000; RayBiotech; Peachtree Corners, GA).
Gland/Organoid Isolation and Culture
Murine gastric gland isolation has been described previously.64,65 Briefly, stomachs were isolated from healthy BALB/c mice, the forestomach and antrum were removed, and the stomach remnants were washed with PBS. The corpus then was placed in a 50-mL conical tube containing gland digest media (Advanced DMEM, 12492013; ThermoFisher), 20 mmol/L HEPES (15630080; ThermoFisher), 0.2% BSA (810033; Millipore), 1% penicillin/streptomycin (P7081; Sigma Aldrich, St. Louis, MO), 50 μg/mL gentamycin (G-1914; Sigma Aldrich), and 1 mg/mL collagenase 1A (C9891; Sigma Aldrich) and vigorously shaken at 37°C for 30 minutes. Stomach tissue was removed and the remaining gastric glands were washed with gland rinse buffer (Advanced DMEM/F12 Ham, D6421; Sigma Aldrich), 50 μg/mL gentamycin, 1% penicillin/streptomycin, and 0.5 mmol/L dithiothreitol (GE17-1318-01; Sigma Aldrich). Whole gastric glands were cultured in Matrigel (356234; Corning, Tewksbury, MA) with Advanced DMEM/F12 Ham supplemented with 25% conditioned Wnt media, 12% conditioned noggin/R-spondin media, 100 U/mL penicillin/streptomycin, 10 nmol/L HEPES (15630080; ThermoFisher), 1% N-2 supplement (17502048; Fisher, Hampton, NH), 2% B-27 supplement (17504044; Fisher), 10 nmol/L gastrin (4011661; Bachem, Bubendorf, CH), 100 ng/mL fibroblast growth factor (100-26; Peprotech, Rocky Hill, NJ), 50 ng/mL epidermal growth factor (E9644; Sigma Aldrich), 1.25 mmol/L N-acetylcysteine (A7250; Sigma Aldrich), 2 mmol/L GlutaMAX (35050-061; Gibco, Waltham, MA). Glands were plated in a 24-well plate and incubated at 37°C in 5% CO2 for the duration of the experiment.
For human gastric organoid experiments, de-identified human stomach samples were supplied through an existing material transfer agreement with Mid-America Transplant (St. Louis, MO). Gastric biopsy specimens were supplied at the time of organ donation and used for research purposes. For human gastric gland isolation, human corpus tissue was washed with cold PBS, cut into 5-cm2 sections, and glands were isolated following the murine gland isolation protocol previously described. Whole gastric glands were cultured in Growth Factor Reduced Matrigel (356231; Corning) with Advanced DMEM/F12 Ham supplemented with 25% conditioned WNT media, 12% conditioned noggin/R-spondin media, 100 U/mL penicillin/streptomycin (P7081; Sigma Aldrich), 10 nmol/L HEPES (15630080; ThermoFisher), 1% N-2 supplement (17502048; Fisher), 2% B-27 supplement (17504044; Fisher), 10 nmol/L gastrin (4011661; Bachem), 100 ng/mL fibroblast growth factor (100-26; Peprotech), 50 ng/mL epidermal growth factor (E9644; Sigma), 1.25 mmol/L N-acetylcysteine (A7250; Sigma), 2 mmol/L GlutaMAX (35050-061; Gibco), 10 mmol/L ROCK inhibitor Y-27632 (Y0503; Sigma) (added only upon initial plating), 100 mg/mL Primocin (ant-pm-1; InvivoGen; San Diego, CA), 75 mg/mL Fungin (ant-fn-1; InvivoGen; San Diego, CA), 100 mg/mL Normocin (anr-nr-1; InvivoGen; San Diego, CA), and 10 mmol/L nicotinamide (N0636; Sigma). Glands were plated in a 24-well plate and incubated at 37°C and 5% CO2 for the duration of the experiment.
For co-culturing of immune cells and organoids, immune cells were isolated from TxA23 mice as previously described. Gastric glands were isolated from BALB/c and BALB/cxIl4ra-/- following the murine gland isolation protocol previously described. Whole gastric glands were cultured in Matrigel (356234; Corning) and 25,000 immune cells were added to the Matrigel. Organoids were cultured with previously described murine organoid conditioned media, plated in a 24-well plate, and incubated at 37°C in 5% CO2 for the duration of the experiment.
To study the effect of IL13 on epithelial cells, glands were isolated and cultured as described with the addition of 5 ng/mL recombinant mouse IL13 (413-ML; R&D Systems, Minneapolis, MN) or recombinant human IL13 (213-ILL-005; R&D Systems) upon initial plating.64 Organoids formed over a 48- to 72-hour period. Photographs were taken using Olympus Live Cell Imager microscope (Olympus, Tokyo, Japan) or Cytation 5 Cell Imaging Multi-Mode Reader (BioTek; Winooski, VT), and media was replaced every 3 days for 9 days. Organoid diameter was determined using cellSens Imaging Software (Olympus). For paraffin-embedded organoids, glands were isolated according to standard protocol and cultured for 7 days. To harvest organoids, media was removed, the well was washed with ice-cold PBS, and then gently pipetted to dissociate Matrigel. Contents of the well then were transferred to a 1.7-mL tube and placed on ice to allow organoids to settle. PBS was removed and organoids were fixed with 10% formalin and washed with 70% ethanol. Organoids were stained with eosin, washed with 70% ethanol, and embedded in paraffin for sectioning. For organoid gene expression analysis, glands were isolated and cultured according to the protocol described previously. Glands were cultured in 200 uL Matrigel in a 15-mL conical tube for 7 days. On day 7, conical tubes were placed on ice and ice-cold PBS was added and gently pipetted to dissociate Matrigel. Tubes then were centrifuged at 1300 rpm and supernatant was discarded. RNA was extracted from organoids using the PureLink RNA Mini Kit (12183025; Ambion).
Quantitative Reverse-Transcription PCR
Complementary DNA (cDNA) was prepared from 1 μg RNA using TaqMan Reverse Transcription Reagents (N808-0234; Applied Biosystems, Branchburg, NJ) according to the manufacturer’s protocol. Reverse-transcription qPCR was performed using 10 ng cDNA in technical duplicates with TaqMan Fast Advanced Master Mix (4444964; Applied Biosystems). TaqMan primer probes used were as follows: Gapdh (Mm99999915_g1; Thermo Fisher Scientific, Waltham, MA), Muc5ac (Mm01276718_m1; Thermo), Muc6 (Mm01276718_m1; Thermo), Tff2 (Mm01348265_m1; Thermo), Gkn3 (Mm01183934_m1; Thermo), Ki67 (Mm01278617_m1; Thermo), GAPDH (Hs02786624; Thermo), MUC5AC (Hs01365616; Thermo), and MUC6 (Hs01674026; Thermo).
Neutralization of IL13
For neutralization studies, mice were treated with an intraperitoneal injection of 10 mg/kg anti-mouse IL13 or IgG isotype control (both gifts from Janssen Research & Development, LLC, Spring House, PA) twice a week for 4 weeks beginning at 3 months of age. At 4 months, the mice were killed, and the stomachs were removed and prepared for disease analysis following previously mentioned histopathology methods.
Isolation of Gastric Epithelial Single-Cell Suspensions
Stomachs were harvested from 3 mice per group and gastric lymph nodes were removed. The forestomach and antrum were removed, and the corpus was cut along the inner curvature. The corpus then was washed in a bath of PBS and immediately washed in a secondary bath of advanced DMEM (12492013; Thermo Fisher Scientific, Waltham, MA). Dissected corpuses then were placed in a 50-mL conical tube with 10 mL prewarmed gland digestion media (Advanced DMEM, 12492013; ThermoFisher), 20 mmol/L HEPES (15630080; ThermoFisher), 0.2% BSA (810033; Millipore), 1× penicillin/streptomycin (P7081; Sigma Aldrich), 50 μg/mL gentamycin (G-1914; Sigma Aldrich), and 1 mg/mL collagenase 1A (C9891; Sigma Aldrich). Stomachs were agitated at 37°C for 30 minutes. Connective tissue was removed, leaving behind gastric glands, which then were transferred to a 15-mL conical tube using gland rinse media (DMEM/F12 Ham, D6421; Sigma Aldrich), 1% penicillin/streptomycin, 50 μg/mL gentamicin, and 0.5 mmol/L dithiothreitol (GE17-1318-01; Sigma Aldrich). Glands were resuspended in 1 mL prewarmed Trypsin-EDTA (T3924; Millipore-Sigma) and incubated for 20 minutes at 37°C until only single cells were present. Every 10 minutes, glands were agitated using a 1-mL pipette to aid in digestion. After agitation, aliquots were taken, and the digestion progress was measured using a hemocytometer and trypan blue staining. Digestion was stopped when trypan blue staining showed complete digestion of glands into single cells. Cells then were washed twice in advanced DMEM 0.1% fetal bovine serum to quench enzyme activity. Single cells then were resuspended in DMEM + 10% fetal bovine serum to a final cell concentration of 1000 cells/mL.
Chromium Single-Cell 5’ Library Construction
The Chromium Single Cell Controller instrument was used in these studies according to recommended manufacturer protocols and has been described previously.20,29 Briefly, gastric corpus epithelial single-cell suspensions were loaded into a Chromium Single Cell Controller instrument (10x Genomics, Pleasanton, CA) to generate single-cell gel beads in emulsion (GEMs). scRNA-seq libraries were prepared using the Chromium Single Cell 5’ Library & Gel Bead Kit v1 (PN-1000006; 10x Genomics). GEM reverse transcription (GEM-RT) was performed in a Veriti 96-Well Thermal cycler (4375786; Applied Biosystems): 53°C for 45 minutes, 85°C for 5 minutes, held at 4°C, and stored at −20°C. The GEMs then were broken, and the single-strand cDNA was cleaned up with DynaBeads MyOne Silane Beads (37002D; Thermo Fisher Scientific). Barcoded full-length cDNA was amplified using the Veriti 96-Well Thermal Cycler: 98°C for 45 seconds, cycled 13 times: 98°C for 20 seconds, 67°C for 30 seconds, and 72°C for 1 minute; 72°C for 1 minute; held at 4°C. Amplified cDNA product was cleaned up with the SPRIselect Reagent Kit (0.6 × SPRI, P/N B23318; Beckman Coulter). The 5′ gene expression libraries were constructed using the reagents in the Chromium Single Cell 3′/5′ Library Construction kit (PN-1000020; 10x Genomics; Pleasanton, CA). For 5′ gene expression library construction, these steps were followed: (1) fragmentation, end repair, and A-tailing; (2) postfragmentation, end repair, and A-tailing double-sided size selection with SPRIselect; (3) adaptor ligation; (4) postligation clean-up with SPRIselect; and (5) sample index PCR and double-sided cleanup with SPRIselect. Final quality control and Illumina (San Diego, CA) sequencing of the prepared libraries was performed by the Washington University in St. Louis Genome Technology Access Center.
scRNA-seq Data Processing and Analysis
Raw data were processed through the CellRanger 3.0 pipeline (10x Genomics), and secondary clustering and differential expression analysis were conducted in Seurat/R.66 Before clustering, all libraries and subsets were processed to ensure quality. Genes relating to mitochondria-localized proteins are markers for broken or low-quality cells.67 Consequently, low-quality cells expressing high levels of mitochondrial markers above a majority threshold unique to each library/subset were filtered out before downstream analysis. Each library then was globally scaled and normalized by a scale factor of 1 × 104 and log transformation. In addition, unwanted sources of variation attributed to biological noise and batch effect were identified and regressed out to improve downstream analysis and dimensionality reduction.68 Multiple data sets were integrated together through dimension reduction identification of anchors between data sets, followed by filtering, scoring, and weighting of anchors.69 Components for clustering were generated by canonical-correlation analysis. High signal canonical correlates explaining the most variance in comparison identity classes were aligned by dynamic time warping, and their dimensions were used for subsequent shared nearest neighbor clustering and visualization by uniform manifold approximation and projection for dimension reduction.70,71 Globally distinguishing genes for each cluster and comparison identity class were identified by calculating the normalized gene expression for the average single cell. Significant genes with at least a 2-fold change and corrected P value less than .01 were identified via Wilcoxon rank-sum test with Bonferroni correction for multiple comparisons. Data sets presented in this study are available at https://ncbi.nlm.nih.gov/bioproject under the BioSample/accession numbers SAMN13152839 (BALB/c), SAMN13152840 (TxA23), SAMN19814216 (Il4ra-/-).
Statistical Analysis
Data are expressed as means of individual determination ± SEM. Statistical analysis was performed using an unpaired Student t test using GraphPad Prism 8 (GraphPad Software, San Diego, CA). Histopathology score significance was performed using the Mann–Whitney U test. For reverse-transcription qPCR analysis, crossing thresholds were normalized to housekeeping gene (Gapdh) expression and fold change was determined by comparison of delta cycle threshold (ΔCT) values of untreated and treated samples. All authors had access to the study data and have reviewed and approved the final manuscript.
Acknowledgments
The authors thank Sherri Koehm and Joy Eslick for assistance with flow cytometry; Grant Kolar, MD, PhD, and Caroline Murphy from the Saint Louis University Research Microscopy and Histology Core for generation of tissue sections and assistance with confocal microscopy; and the Saint Louis University Comparative Medicine Department for assistance in maintaining mouse colonies. The authors also thank the Washington University in St. Louis Genome Technology Access Center/McDonnell Genome Institute for assistance with 10x Genomics Chromium Single Cell RNA isolation and Illumina sequencing; Jason Mills’ lab at Washington University for supplying conditioned organoid media and GKN3 serum for immunofluorescence studies; Mid-America Transplant (St. Louis, MO) for supplying human corpus sections; Janssen Research & Development, LLC (Spring House, PA), for supplying the IL13-neutralizing antibody and isotype control; and Jose B. Saenz’s lab at Washington University for assistance with organoid imaging. The authors also thank Joel Eissenberg, PhD, for critical reading of the manuscript.
CRediT Authorship Contributions
Christine Noto, BS (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead)
Stella Hoft, BA (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Kevin Bockerstett, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Nicholas Jackson, BS (Formal analysis: Supporting; Investigation: Supporting)
Eric Ford, N/A (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Luke Vest, BS (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Richard DiPaolo, PhD (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Previously supported by American Cancer Society grant RSG-12-171-01-LIB and the National Institutes of Health / National Institute of Diabetes and Digestive and Kidney Diseases grant R01 DK110406; a grant from the Digestive Diseases Research Core Center of the Washington University School of Medicine National Institute of Diabetes and Digestive and Kidney Diseases P30DK52574, by the American Gastroenterological Association Funderburg Research Award (R.J.D.).
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Anderson W.F., Rabkin C.S., Turner N., Fraumeni J.F., Jr., Rosenberg P.S., Camargo M.C. The changing face of noncardia gastric cancer incidence among US Non-Hispanic whites. J Natl Cancer Inst. 2018;110:608–615. doi: 10.1093/jnci/djx262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heer E.V., Harper A.S., Sung H., Jemal A., Fidler-Benaoudia M.M. Emerging cancer incidence trends in Canada: the growing burden of young adult cancers. Cancer. 2020;126:4553–4562. doi: 10.1002/cncr.33050. [DOI] [PubMed] [Google Scholar]
- 4.Sepulveda A.R. Helicobacter, inflammation, and gastric cancer. Curr Pathobiol Rep. 2013;1:9–18. doi: 10.1007/s40139-013-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox J.G., Wang T.C. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wroblewski L.E., Peek R.M., Jr., Wilson K.T. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peek R.M., Jr., Crabtree J.E. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233–248. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 8.Fox J.G., Sheppard B.J., Dangler C.A., Whary M.T., Ihrig M., Wang T.C. Germ-line p53-targeted disruption inhibits helicobacter-induced premalignant lesions and invasive gastric carcinoma through down-regulation of Th1 proinflammatory responses. Cancer Res. 2002;62:696–702. [PubMed] [Google Scholar]
- 9.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554–3560. [PubMed] [Google Scholar]
- 10.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 11.Slack J.M. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- 12.Giroux V., Rustgi A.K. Metaplasia: tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat Rev Cancer. 2017;17:594–604. doi: 10.1038/nrc.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teal E., Dua-Awereh M., Hirshorn S.T., Zavros Y. Role of metaplasia during gastric regeneration. Am J Physiol Cell Physiol. 2020;319:C947–C954. doi: 10.1152/ajpcell.00415.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigden H.M., Alias A., Havelock T., O'Donnell R., Djukanovic R., Davies D.E., Wilson S.J. Squamous metaplasia is increased in the bronchial epithelium of smokers with chronic obstructive pulmonary disease. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burclaff J., Mills J.C. Plasticity of differentiated cells in wound repair and tumorigenesis, part I: stomach and pancreas. Dis Model Mech. 2018;11 doi: 10.1242/dmm.033373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer A.R., Goldenring J.R. Injury, repair, inflammation and metaplasia in the stomach. J Physiol. 2018;596:3861–3867. doi: 10.1113/JP275512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldenring J.R., Nam K.T., Mills J.C. The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the mist. Exp Cell Res. 2011;317:2759–2764. doi: 10.1016/j.yexcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halldorsdottir A.M., Sigurdardottrir M., Jonasson J.G., Oddsdottir M., Magnusson J., Lee J.R., Goldenring J.R. Spasmolytic polypeptide-expressing metaplasia (SPEM) associated with gastric cancer in Iceland. Dig Dis Sci. 2003;48:431–441. doi: 10.1023/a:1022564027468. [DOI] [PubMed] [Google Scholar]
- 19.Goldenring J.R., Nam K.T., Wang T.C., Mills J.C., Wright N.A. Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology. 2010;138:2207–2210.e1. doi: 10.1053/j.gastro.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bockerstett K.A., Lewis S.A., Noto C.N., Ford E.L., Saenz J.B., Jackson N.M., Ahn T.H., Mills J.C., DiPaolo R.J. Single cell transcriptional analyses identify lineage-specific epithelial responses to inflammation and metaplastic development in the gastric corpus. Gastroenterology. 2020;159:2116–2129.e4. doi: 10.1053/j.gastro.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geahlen J.H., Lapid C., Thorell K., Nikolskiy I., Huh W.J., Oates E.L., Lennerz J.K., Tian X., Weis V.G., Khurana S.S., Lundin S.B., Templeton A.R., Mills J.C. Evolution of the human gastrokine locus and confounding factors regarding the pseudogenicity of GKN3. Physiol Genomics. 2013;45:667–683. doi: 10.1152/physiolgenomics.00169.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam K.T., Lee H.J., Sousa J.F., Weis V.G., O'Neal R.L., Finke P.E., Romero-Gallo J., Shi G., Mills J.C., Peek R.M., Jr., Konieczny S.F., Goldenring J.R. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139:2028–2037.e9. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bockerstett K.A., DiPaolo R.J. Regulation of gastric carcinogenesis by inflammatory cytokines. Cell Mol Gastroenterol Hepatol. 2017;4:47–53. doi: 10.1016/j.jcmgh.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bockerstett K.A., Osaki L.H., Petersen C.P., Cai C.W., Wong C.F., Nguyen T.M., Ford E.L., Hoft D.F., Mills J.C., Goldenring J.R., DiPaolo R.J. Interleukin-17A promotes parietal cell atrophy by inducing apoptosis. Cell Mol Gastroenterol Hepatol. 2018;5:678–690.e1. doi: 10.1016/j.jcmgh.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osaki L.H., Bockerstett K.A., Wong C.F., Ford E.L., Madison B.B., DiPaolo R.J., Mills J.C. Interferon-gamma directly induces gastric epithelial cell death and is required for progression to metaplasia. J Pathol. 2019;247:513–523. doi: 10.1002/path.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T.L., Dipaolo R.J. A new mouse model of inflammation and gastric cancer. Oncoimmunology. 2013;2 doi: 10.4161/onci.25911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen T.L., Khurana S.S., Bellone C.J., Capoccia B.J., Sagartz J.E., Kesman R.A., Jr., Mills J.C., DiPaolo R.J. Autoimmune gastritis mediated by CD4+ T cells promotes the development of gastric cancer. Cancer Res. 2013;73:2117–2126. doi: 10.1158/0008-5472.CAN-12-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHugh R.S., Shevach E.M., Margulies D.H., Natarajan K. A T cell receptor transgenic model of severe, spontaneous organ-specific autoimmunity. Eur J Immunol. 2001;31:2094–2103. doi: 10.1002/1521-4141(200107)31:7<2094::aid-immu2094>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 29.Bockerstett K.A., Lewis S.A., Wolf K.J., Noto C.N., Jackson N.M., Ford E.L., Ahn T.H., DiPaolo R.J. Single-cell transcriptional analyses of spasmolytic polypeptide-expressing metaplasia arising from acute drug injury and chronic inflammation in the stomach. Gut. 2020;69:1027–1038. doi: 10.1136/gutjnl-2019-318930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulnigg-Dabsch S. Autoimmune gastritis. Wien Med Wochenschr. 2016;166:424–430. doi: 10.1007/s10354-016-0515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynn T.A. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 32.Wills-Karp M., Luyimbazi J., Xu X., Schofield B., Neben T.Y., Karp C.L., Donaldson D.D. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 33.Gour N., Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 2015;75:68–78. doi: 10.1016/j.cyto.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick S.M., Heller N.M. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine. 2015;75:38–50. doi: 10.1016/j.cyto.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallett M.A., Venmar K.T., Fingleton B. Cytokine stimulation of epithelial cancer cells: the similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012;72:6338–6343. doi: 10.1158/0008-5472.CAN-12-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen C.P., Meyer A.R., De Salvo C., Choi E., Schlegel C., Petersen A., Engevik A.C., Prasad N., Levy S.E., Peebles R.S., Pizarro T.T., Goldenring J.R. A signalling cascade of IL-33 to IL-13 regulates metaplasia in the mouse stomach. Gut. 2018;67:805–817. doi: 10.1136/gutjnl-2016-312779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge Z., Sheh A., Feng Y., Muthupalani S., Ge L., Wang C., Kurnick S., Mannion A., Whary M.T., Fox J.G. Helicobacter pylori-infected C57BL/6 mice with different gastrointestinal microbiota have contrasting gastric pathology, microbial and host immune responses. Sci Rep. 2018;8:8014. doi: 10.1038/s41598-018-25927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayi A., Kohler E., Hitzler I., Arnold I., Schwendener R., Rehrauer H., Müller A. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J Immunol. 2009;182:7085–7101. doi: 10.4049/jimmunol.0803293. [DOI] [PubMed] [Google Scholar]
- 39.Mohammadi M., Czinn S., Redline R., Nedrud J. Helicobacter-specific cell-mediated immune responses display a predominant Th1 phenotype and promote a delayed-type hypersensitivity response in the stomachs of mice. J Immunol. 1996;156:4729–4738. [PubMed] [Google Scholar]
- 40.DeLyria E.S., Redline R.W., Blanchard T.G. Vaccination of mice against H pylori induces a strong Th-17 response and immunity that is neutrophil dependent. Gastroenterology. 2009;136:247–256. doi: 10.1053/j.gastro.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stummvoll G.H., DiPaolo R.J., Huter E.N., Davidson T.S., Glass D., Ward J.M., Shevach E.M. Th1, Th2, and Th17 effector T cell-induced autoimmune gastritis differs in pathological pattern and in susceptibility to suppression by regulatory T cells. J Immunol. 2008;181:1908–1916. doi: 10.4049/jimmunol.181.3.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caruso R., Pallone F., Monteleone G. Emerging role of IL-23/IL-17 axis in H pylori-associated pathology. World J Gastroenterol. 2007;13:5547–5551. doi: 10.3748/wjg.v13.i42.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawai N., Kita M., Kodama T., Tanahashi T., Yamaoka Y., Tagawa Y., Iwakura Y., Imanishi J. Role of gamma interferon in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Infect Immun. 1999;67:279–285. doi: 10.1128/iai.67.1.279-285.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price A.E., Liang H.E., Sullivan B.M., Reinhardt R.L., Eisley C.J., Erle D.J., Locksley R.M. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertaux-Skeirik N., Wunderlich M., Teal E., Chakrabarti J., Biesiada J., Mahe M., Sundaram N., Gabre J., Hawkins J., Jian G., Engevik A.C., Yang L., Wang J., Goldenring J.R., Qualls J.E., Medvedovic M., Helmrath M.A., Diwan T., Mulloy J.C., Zavros Y. CD44 variant isoform 9 emerges in response to injury and contributes to the regeneration of the gastric epithelium. J Pathol. 2017;242:463–475. doi: 10.1002/path.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D.L., Ju H.Q., Lu Y.X., Chen L.Z., Zeng Z.L., Zhang D.S., Luo H.Y., Wang F., Qiu M.Z., Wang D.S., Xu D.Z., Zhou Z.W., Pelicano H., Huang P., Xie D., Wang F.H., Li Y.H., Xu R.H. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie M., Li K., Li J., Lu D., Hu B. Association and diagnostic value of serum SPINK4 in colorectal cancer. PeerJ. 2019;7:e6679. doi: 10.7717/peerj.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miska J., Lui J.B., Toomer K.H., Devarajan P., Cai X., Houghton J., Lopez D.M., Abreu M.T., Wang G., Chen Z. Initiation of inflammatory tumorigenesis by CTLA4 insufficiency due to type 2 cytokines. J Exp Med. 2018;215:841. doi: 10.1084/jem.20171971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen C.P., Weis V.G., Nam K.T., Sousa J.F., Fingleton B., Goldenring J.R. Macrophages promote progression of spasmolytic polypeptide-expressing metaplasia after acute loss of parietal cells. Gastroenterology. 2014;146:1727–1738.e8. doi: 10.1053/j.gastro.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Salvo C., Buela K.A., Creyns B., Corridoni D., Rana N., Wargo H.L., Cominelli C.L., Delaney P.G., Rodriguez-Palacios A., Cominelli F., Vermeire S., Pizarro T.T. NOD2 drives early IL-33-dependent expansion of group 2 innate lymphoid cells during Crohn's disease-like ileitis. J Clin Invest. 2021;131 doi: 10.1172/JCI140624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Salvo C., Pastorelli L., Petersen C.P., Buttò L.F., Buela K.A., Omenetti S., Locovei S.A., Ray S., Friedman H.R., Duijser J., Xin W., Osme A., Cominelli F., Mahabeleshwar G.H., Mills J.C., Goldenring J.R., Pizarro T.T. Interleukin 33 triggers early eosinophil-dependent events leading to metaplasia in a chronic model of gastritis-prone mice. Gastroenterology. 2021;160:302–316.e7. doi: 10.1053/j.gastro.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer A.R., Engevik A.C., Madorsky T., Belmont E., Stier M.T., Norlander A.E., Pilkinton M.A., McDonnell W.J., Weis J.A., Jang B., Mallal S.A., Peebles R.S., Jr., Goldenring J.R. Group 2 innate lymphoid cells coordinate damage response in the stomach. Gastroenterology. 2020;159:2077–2091.e8. doi: 10.1053/j.gastro.2020.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eissmann M.F., Dijkstra C., Jarnicki A., Phesse T., Brunnberg J., Poh A.R., Etemadi N., Tsantikos E., Thiem S., Huntington N.D., Hibbs M.L., Boussioutas A., Grimbaldeston M.A., Buchert M., O’Donoghue R.J.J., Masson F., Ernst M. IL-33-mediated mast cell activation promotes gastric cancer through macrophage mobilization. Nat Commun. 2019;10:2735. doi: 10.1038/s41467-019-10676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherjee S., Bandyopadhyay G., Dutta C., Bhattacharya A., Karmakar R., Barui G. Evaluation of endoscopic biopsy in gastric lesions with a special reference to the significance of mast cell density. Indian J Pathol Microbiol. 2009;52:20–24. doi: 10.4103/0377-4929.44956. [DOI] [PubMed] [Google Scholar]
- 55.Zhu Z., Homer R.J., Wang Z., Chen Q., Geba G.P., Wang J., Zhang Y., Elias J.A. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi H., Goldenring J.R., Kaminishi M., Lee J.R. Identification of spasmolytic polypeptide expressing metaplasia (SPEM) in remnant gastric cancer and surveillance postgastrectomy biopsies. Dig Dis Sci. 2002;47:573–578. doi: 10.1023/a:1017920220149. [DOI] [PubMed] [Google Scholar]
- 57.Roth K.A., Kapadia S.B., Martin S.M., Lorenz R.G. Cellular immune responses are essential for the development of Helicobacter felis-associated gastric pathology. J Immunol. 1999;163:1490–1497. [PubMed] [Google Scholar]
- 58.Paulnock D.M., Demick K.P., Coller S.P. Analysis of interferon-gamma-dependent and -independent pathways of macrophage activation. J Leukoc Biol. 2000;67:677–682. doi: 10.1002/jlb.67.5.677. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen T.L., Makhlouf N.T., Anthony B.A., Teague R.M., DiPaolo R.J. In vitro induced regulatory T cells are unique from endogenous regulatory T cells and effective at suppressing late stages of ongoing autoimmunity. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rogers A.B. Histologic scoring of gastritis and gastric cancer in mouse models. Methods Mol Biol. 2012;921:189–203. doi: 10.1007/978-1-62703-005-2_22. [DOI] [PubMed] [Google Scholar]
- 61.Ramsey V.G., Doherty J.M., Chen C.C., Stappenbeck T.S., Konieczny S.F., Mills J.C. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 62.DiPaolo R.J., Glass D.D., Bijwaard K.E., Shevach E.M. CD4+CD25+ T cells prevent the development of organ-specific autoimmune disease by inhibiting the differentiation of autoreactive effector T cells. J Immunol. 2005;175:7135–7142. doi: 10.4049/jimmunol.175.11.7135. [DOI] [PubMed] [Google Scholar]
- 63.Alderuccio F., Toh B.H., Gleeson P.A., van Driel I.R. A novel method for isolating mononuclear cells from the stomachs of mice with experimental autoimmune gastritis. Autoimmunity. 1995;21:215–221. doi: 10.3109/08916939509008018. [DOI] [PubMed] [Google Scholar]
- 64.Mahé M.M., Aihara E., Schumacher M.A., Zavros Y., Montrose M.H., Helmrath M.A., Sato T., Shroyer N.F. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol. 2013;3:217–240. doi: 10.1002/9780470942390.mo130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dedhia P.H., Bertaux-Skeirik N., Zavros Y., Spence J.R. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ilicic T., Kim J.K., Kolodziejczyk A.A., Bagger F.O., McCarthy D.J., Marioni J.C., Teichmann S.A. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 69.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waltman L., van Eck N.J. A smart local moving algorithm for large-scale modularity-based community detection. Eur Physical J B. 2013;86:471. [Google Scholar]
- 71.Becht E., McInnes L., Healy J., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]