Abstract
A second-generation hepatitis C virus (HCV) quantitative assay (COBAS AMPLICOR HCV MONITOR Test, version 2.0; COBAS HCM-2) has been developed, with the intention of achieving equivalent quantification of all HCV genotypes and improving assay performance. To evaluate the clinical performance of COBAS HCM-2 and its utility in predicting the response to alpha interferon treatment, sera from 215 chronic hepatitis C patients were analyzed and the results were compared with those obtained by the Quantiplex bDNA HCV RNA, version 2.0, assay (bDNA-2). The COBAS HCM-2 had significantly greater sensitivity than bDNA-2 (94.9 versus 88.4%; P < 0.001) when performed with sera from chronic hepatitis C patients who were viremic by a qualitative PCR test. The standard deviations for the within-run and between-run reproducibilities of COBAS HCM-2 were <0.1 and <0.2, respectively, and it showed an improved linear range between genotypes with the threefold serial dilutions tested (r2 = 0.986 to 0.995). The COBAS HCM-2 results were positively correlated with the bDNA-2 results, but the values for COBAS HCM-2 were on average 0.96 log lower than the values for bDNA-2. The mean difference in quantification values between these two assays did not differ among samples with different genotypes (0.70 to 1.00 log). No genotype-dependent difference in viral load was observed. The pretreatment viral load was significantly lower in complete responders. By using multivariate analysis, the viral load 2 weeks after the initiation of alpha interferon treatment was the strongest predictor of a complete response. In conclusion, COBAS HCM-2 demonstrated good sensitivity, linearity, and reproducibility and efficiency equal to that of bDNA-2 for the quantification of HCV genotypes 1 and 2. Hence, this assay provides a rapid and reliable method for the quantification of HCV RNA in serum and is useful for the planning of interferon treatment.
Hepatitis C virus (HCV) is the major etiologic agent in parenterally transmitted non-A, non-B hepatitis and frequently causes persistent infection, which leads to chronic liver disease and primary hepatocellular carcinoma (2, 3). Since serum HCV RNA levels (viral load) may correlate with clinical manifestations and virologic characteristics (10, 11) and have been reported to be an important factor predictive of the response to alpha interferon (IFN-α) therapy in patients with chronic HCV infection (8, 28), easy, reliable, and standardized tests with good reproducibilities are needed for routine clinical use. Several systems have been developed for quantitation of HCV RNA, including competitive reverse transcription (RT)-PCR (10, 11), the branched DNA (bDNA) assay (Quantiplex HCV RNA assay; Bayer, Emeryville, Calif.) (5, 9, 13, 17, 28), a noncompetitive RT-PCR assay (AMPLICOR HCV MONITOR assay; Roche, Branchburg, N.J.) (4, 17, 28), and a real-time RT-PCR technique (15). Differences in the performances of these systems due to fundamentally different technologies and variations in the efficiency of hybridization of HCV RNA to complementary nucleotide sequences in these assays may lead to method-related discrepancies in absolute quantification values and in the correlation between HCV genotypes and viral load (9, 14, 28).
We have previously undertaken a comparative study to examine the performance characteristics and clinical utility of the current version of the bDNA assay (the Quantiplex HCV RNA, version 2.0, assay, referred to as bDNA-2) and the first version of the AMPLICOR HCV MONITOR (AMPLICOR, version 1.0) assay (28). In agreement with other reports, significant differences in the efficiencies of detection of genotype 1 and non-genotype 1 isolates were observed with the first version of the AMPLICOR assay (9, 14, 17). Recently, a second version of the AMPLICOR HCV MONITOR Test (COBAS AMPLICOR HCV MONITOR Test, version 2.0, referred to as COBAS HCM-2; Roche, Branchburg, N.J.) has been produced. The assay has been placed onto the automated COBAS AMPLICOR system (19) with the intention of achieving more equivalent quantification of all HCV genotypes, improving assay performance, and saving hands-on time. In addition, previous studies with the qualitative AMPLICOR HCV test have demonstrated that the automated COBAS AMPLICOR system, with the incorporation of the uracil-N-glycosylase procedure for the prevention of carryover contamination, provides a reliable and specific RT-PCR method for the detection of HCV RNA (1, 6).
The objectives of the current study were (i) to investigate the clinical sensitivity, reproducibility, and linearity of COBAS HCM-2 and to examine the correlation of the results of COBAS HCM-2 and bDNA-2 with serum samples from patients with different HCV genotypes and various clinical settings and (ii) to evaluate the clinical utility of COBAS HCM-2 in predicting the response to IFN-α treatment before and during therapy. Because specificity studies were performed by the manufacturers during the test registration procedure, specificity was not a focus of the present study.
MATERIALS AND METHODS
Patients.
Two hundred fifteen Taiwanese patients with chronic hepatitis C (126 men and 89 women; age range, 15 to 73 years; mean age, 46.3 ± 12.0 years) were enrolled in the study. Among the patients included 57 had chronic persistent hepatitis, 115 had chronic active hepatitis, and 43 had liver cirrhosis. Histologic diagnosis of chronic hepatitis was made on the basis of standard criteria (12). Sixty-two patients (28.8%) had a history of blood transfusion.
Seventy-nine patients with chronic hepatitis C were treated with 6 million units (MIU) of recombinant IFN-α (IFN-α 2b) thrice weekly for 24 weeks, followed by 3 MIU thrice weekly for the next 12 weeks. The presence of HCV RNA in serum was assessed by qualitative RT-PCR every 3 months for 21 months. Complete responders were defined as patients showing normal alanine aminotransferase levels and clearance of serum HCV RNA at the end of the therapy and for 12 months after the cessation of therapy, as measured by qualitative COBAS AMPLICOR HCV Test, version 2.0 (Roche). All other patients were classified as nonresponders.
Clinical samples.
Serum samples from 215 chronic hepatitis C patients were analyzed for their HCV RNA levels. Serum was separated from whole blood collected in a serum separation tube at the time that a liver biopsy was performed and was immediately stored at −70°C in several aliquots. All the samples were reactive for antibodies to HCV by using the second-generation HCV antibody enzyme immunoassay (Abbott Laboratories, North Chicago, Ill.) and were positive for HCV RNA by qualitative COBAS AMPLICOR HCV Test, version 2.0.
Samples were available at 2 and 4 weeks for a subset of 30 unselected patients treated with IFN-α. These 60 samples were analyzed for serum HCV RNA levels by COBAS HCM-2. Samples with viral loads under the detection limit of COBAS HCM-2 (<1,000 copies/ml) were further tested for the presence of HCV RNA by using the qualitative COBAS AMPLICOR HCV Test, version 2.0.
Detection, quantification, and genotyping of HCV RNA in serum.
Detection of HCV RNA in serum was performed by a standardized, automated, qualitative RT-PCR assay (COBAS AMPLICOR HCV Test, version 2.0; Roche) (6). The detection limit was 100 copies/ml.
HCV genotypes were determined by amplification of the core region with the genotype-specific primers that distinguish between genotypes 1a, 1b, 2a, 2b, and 3a described by Okamoto et al. (18).
Two commercial assays for the quantification of serum HCV RNA were used: COBAS HCM-2 and bDNA-2. Both assays were performed strictly in accordance with the manufacturers' instructions. HCV RNA was detected directly in bDNA-2 by a series of probe hybridizations that boost the signal coming from each HCV RNA strand. The relative intensity of the signal was compared with that on a curve prepared with an external standard, giving a quantification range from 0.2 million to 120 million equivalents (MEq) of HCV RNA per ml.
COBAS HCM-2 is based on RT and amplification of HCV RNA with primers that target a 244-base region located within the highly conserved 5′ noncoding region of the HCV genome. An internal quantitation standard (HCM QS) was added to every sample, via the lysis buffer, and was coamplified with the HCV RNA target. The HCM QS consists of a noninfectious RNA transcript that has primer binding regions identical to those of the HCV target sequence and that generates a product of the same length and base composition as that generated by the HCV target RNA but a probe binding region different from that of the target amplicon. Both the amplified products of the internal standard and sample RNA were serially diluted and detected by probe hybridization.
Briefly, a known concentration of HCM QS and 100 μl of patient serum were coincubated with a lysis buffer; and the RNA was precipitated with isopropanol, pelleted by centrifugation, washed once with 70% ethanol, and resuspended in 1 ml of specimen diluent. The isolated RNA was then added to a master mix solution that contained rTth DNA polymerase and dimethyl sulfoxide, which decreases the inter- and intrastrand reanealing (25), resulting in equal access and amplification of the target sequences (24). This was then loaded onto the COBAS AMPLICOR system, which performed the remainder of the procedure. After RT and amplification of the specimen or the control in the thermal cycler section, the COBAS AMPLICOR system denatured and serially diluted the double-stranded, biotinylated amplified products (four HCV amplicon dilutions and two HCM QS amplicon dilutions) and used a suspension of magnetic particles coated with multiple copies of an oligonucleotide probe specific for HCV (or HCM QS) to capture the amplified amplicons. Unbound material was removed by washing, and the biotinylated amplicon was detected with an avidin-horseradish peroxidase conjugate–tetramethylbenzidine–hydrogen peroxide colorimetric reaction. The absorbance (660 nm) at each dilution was recorded, and the COBAS AMPLICOR system automatically selected the optical density and dilution, according to the manufacturer's criteria, to be used for the calculation of the copy number for each specimen and control. The manufacturer's stated quantification range was 103 to 106 HCV RNA copies/ml.
Samples with titers outside the linear range by either of the two quantitative assays were retested following dilution 1/10 and 1/100.
To investigate the reproducibility of COBAS HCM-2, three samples (low-, medium-, and high-titer sera) were tested 10 times in one test run and then one time in each of five test runs. A separate aliquot of each sample was processed for each test; thus, the variation between results includes variation due to sample processing, amplification, and detection. To assess the linearity of HCV RNA quantification for different genotypes, four dilution panels were made by seven threefold serial dilutions of high-titer serum samples from patients infected with genotype 1b, 2a, 2b, or both 1b and 2a, which are the common genotypes in Taiwan.
Statistical analysis.
Data were expressed as means ± standard deviations after logarithmic transformation of the original values. The chi-square test with Yates' correction, the chi-square test with linear correlation, Fisher's exact test, Student's t test, analysis of variance, Spearman's rank correlation coefficient, simple linear regression, stepwise multiple linear regression, and multiple logistic regression were used. For the purpose of analyzing the data with suitable statistical methods, we assigned a nominal value of 0.1 MEq/ml to samples that were negative for HCV RNA by bDNA-2 but positive for HCV RNA by qualitative RT-PCR and a nominal value of 500 copies/ml to samples which were COBAS HCM-2 negative but positive by qualitative RT-PCR. For the 60 serum samples collected during IFN-α treatment, a nominal value of 50 copies/ml was assigned to those negative for HCV RNA by both COBAS HCM-2 and qualitative RT-PCR.
RESULTS
Performance characteristics of COBAS HCM-2.
The performance characteristics of COBAS HCM-2 and bDNA-2 were analyzed with samples collected at the time that liver biopsy was performed but before the initiation of therapy. Of the 215 patients, HCV RNA was quantifiable in 204 patients (94.9%) by COBAS HCM-2 and in 190 patients (88.4%) by bDNA-2. The difference was significant (P < 0.001; chi-square test). One hundred eighty-six (86.5%) samples were quantifiable by both of the assays. Seven (3.3%) had HCV RNA levels below the detection limits of both assays but were positive by the qualitative COBAS AMPLICOR HCV Test, version 2.0. The quantitative range observed with clinical samples was 1 × 103 to 3.88 × 106 copies/ml for COBAS HCM-2 and 0.2 to 62.93 MEq/ml for bDNA-2. Six (5.9%) of the 101 genotype 1b-infected samples, 2 (3.1%) of the 65 genotype 2a-infected samples, 1 (3.7%) of the 27 genotype 2b-infected samples, 1 (9.1%) of the 11 samples infected with mixed genotypes, and 1 (10%) of the 10 samples infected with unclassified genotypes had viral loads below the detection limit of COBAS HCM-2. Fifteen (14.9%) genotype 1b-infected samples, six (9.2%) genotype 2a-infected samples, one (9.1%) sample infected with mixed genotypes, and three (30%) samples infected with unclassified genotypes had viral loads below the detection limit of bDNA-2. The clinical sensitivities of COBAS HCM-2 and bDNA-2 for the detection of HCV RNA did not differ for the samples infected with different genotypes.
Serum HCV RNA levels, tested 10 times in the same round for within-run reproducibility of COBAS HCM-2, showed that the standard deviations were 0.04, 0.06, and 0.09 for low-, medium- and high-titer sera, respectively (Table 1). Those tested five times in five different rounds to determine the between-run reproducibility of COBAS HCM-2 showed that the standard deviations were 0.03, 0.09, and 0.12 for low-, medium-, and high-titer sera, respectively. Because a separate sample aliquot was processed for each test performed, these variations represent the total variability of the assay.
TABLE 1.
Reproducibility of COBAS HCM-2
| Reproducibility and RNA titer | HCV RNA load (log10 no. of copies/ml) in sample:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | Mean ± SD | |
| Within run | |||||||||||
| Low | 3.65 | 3.67 | 3.62 | 3.69 | 3.71 | 3.69 | 3.67 | 3.72 | 3.62 | 3.74 | 3.68 ± 0.04 |
| Medium | 4.92 | 4.75 | 4.83 | 4.94 | 4.85 | 4.90 | 4.76 | 4.86 | 4.84 | 4.85 | 4.85 ± 0.06 |
| High | 5.98 | 5.84 | 5.80 | 5.82 | 5.94 | 5.89 | 5.87 | 5.94 | 5.69 | 5.81 | 5.86 ± 0.09 |
| Between run | |||||||||||
| Low | 3.67 | 3.74 | 3.69 | 3.68 | 3.59 | 3.67 ± 0.03 | |||||
| Medium | 4.98 | 4.77 | 4.95 | 4.80 | 4.88 | 4.88 ± 0.09 | |||||
| High | 5.98 | 5.68 | 5.84 | 5.72 | 5.79 | 5.80 ± 0.12 | |||||
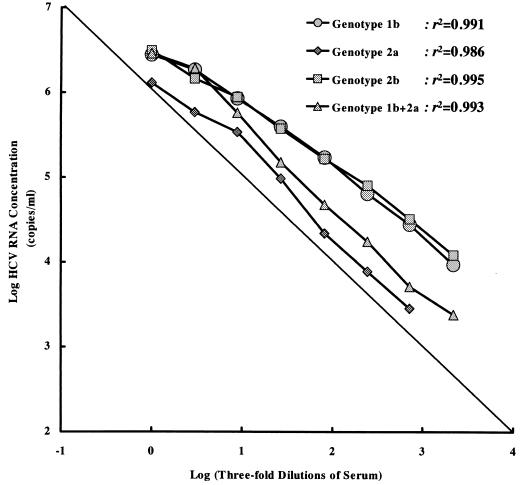
The linearity of HCV RNA quantification was assessed with dilution panels of high-titer sera infected with different HCV genotypes. The undiluted high-titer samples contained HCV genotypes 1b, 2a, 2b, and both 1b and 2a and had viral loads of 6.44, 6.11, 6.49, and 6.46 logs, respectively, as determined by COBAS HCM-2 and viral loads of 7.80, 6.83, 7.74, and 6.95 logs, respectively, as determined by bDNA-2. As shown in Fig. 1, COBAS HCM-2 showed a good linear response (r2 = 0.986 to 0.995) in the threefold serial dilutions, although the undiluted high-titer serum samples did demonstrate some underquantification (especially in the sample with genotype 2a infection). However, this is to be expected, as the viral loads were above the upper detection limit of the assay.
FIG. 1.
Analysis of HCV RNA in four dilution panels of high-titer sera infected with different HCV genotypes by COBAS HCM-2.
For 186 specimens positive by both COBAS HCM-2 and bDNA-2, HCV RNA quantification values by COBAS HCM-2 were positively correlated with those measured by bDNA-2 [log(bDNA-2) = 0.47 × log(COBAS HCM-2) + 3.86; n = 186; Spearman's rank correlation coefficient r = 0.693; P < 0.0001]. The values obtained by COBAS HCM-2 were on average 0.96 log lower than the values obtained by bDNA-2 (Table 2). Nonparametric tests of correlation gave a high correlation coefficient upon pairwise comparison of the results for samples with different genotypes (for genotypes 1b, 2a, and 2b, P < 0.0001; for unclassified genotypes and mixed genotypes, P < 0.001). The mean difference in quantification values between these two assays ranged from 0.70 to 1.00 log. The mean difference in the values between the two assays did not differ for samples infected with different genotypes.
TABLE 2.
Correlation between HCV genotypes and quantification values by COBAS HCM-2 and bDNA-2 with 186 specimens positive by both COBAS HCM-2 and bDNA-2
| Genotype | No. of specimens | HCV RNA concna
|
Spearman's rank correlation coefficient | P value | ||
|---|---|---|---|---|---|---|
| bDNA-2 | COBAS HCM-2 | Differenceb | ||||
| All types | 186 | 6.39 ± 0.68 | 5.44 ± 0.84 | 0.96 ± 0.69 | 0.693 | <0.0001 |
| 1b | 84 | 6.44 ± 0.70 | 5.46 ± 0.81 | 1.00 ± 0.69 | 0.714 | <0.0001 |
| 2a | 58 | 6.24 ± 0.63 | 5.34 ± 0.89 | 0.92 ± 0.75 | 0.608 | <0.0001 |
| 2b | 27 | 6.57 ± 0.67 | 5.61 ± 0.90 | 0.96 ± 0.75 | 0.701 | <0.0001 |
| Mixed | 10 | 6.22 ± 0.67 | 5.62 ± 0.42 | 0.70 ± 0.34 | 0.852 | <0.001 |
| Unclassified | 7 | 6.34 ± 0.77 | 5.13 ± 0.87 | 1.21 ± 0.43 | 0.860 | <0.001 |
Expressed as mean ± standard deviation of logarithmic transformation (log number of copies or equivalent per milliliter).
Difference = log(bDNA-2) − log(COBAS HCM-2).
Viral load and clinical manifestations.
To evaluate the relationship between the HCV load and the clinical manifestations of HCV infection, patient age, sex, history of transfusion, liver biochemistry, liver histology, and viral genotype were analyzed (Table 3). By univariate and multivariate analyses, none of the factors analyzed was found to be correlated with viral load, as measured by both COBAS HCM-2 and bDNA-2.
TABLE 3.
Backgrounds of HCV-infected patients and their serum HCV RNA levels measured by COBAS HCM-2 and bDNA-2
| Characteristic | No. of patients | HCV RNA levela
|
|
|---|---|---|---|
| COBAS HCM-2 | bDNA-2 | ||
| All patients | 215 | 5.21 ± 1.03 | 6.22 ± 0.78 |
| Age (yr) | |||
| Below 40 | 70 | 5.27 ± 0.98 | 6.26 ± 0.80 |
| 40 to 60 | 119 | 5.25 ± 1.00 | 6.20 ± 0.77 |
| Above 60 | 26 | 4.87 ± 1.26 | 6.18 ± 0.81 |
| Sex | |||
| Male | 126 | 5.29 ± 0.98 | 6.30 ± 0.79 |
| Female | 89 | 5.09 ± 1.10 | 6.11 ± 0.75 |
| History of transfusion | |||
| Yes | 62 | 5.25 ± 1.11 | 6.28 ± 0.77 |
| No | 153 | 5.20 ± 1.00 | 6.20 ± 0.78 |
| Biochemistry | |||
| ASTb | |||
| Normal (≤25 IU/liter) | 25 | 5.22 ± 1.30 | 6.26 ± 0.91 |
| Abnormal (>25 IU/liter) | 190 | 5.21 ± 1.00 | 6.22 ± 0.76 |
| ALTc | |||
| Normal (≤25 IU/liter) | 23 | 5.01 ± 1.28 | 6.03 ± 1.00 |
| Abnormal (>25 IU/liter) | 192 | 5.24 ± 1.00 | 6.25 ± 0.74 |
| Liver histology | |||
| Chronic persistent hepatitis | 57 | 5.30 ± 0.99 | 6.21 ± 0.76 |
| Chronic active hepatitis | 115 | 5.25 ± 1.06 | 6.28 ± 0.79 |
| Liver cirrhosis | 43 | 5.00 ± 1.02 | 6.07 ± 0.75 |
Mean ± standard deviation of logarithmic transformation of log number of copies or equivalents per milliliter.
AST, aspartate aminotransferase.
ALT, alanine aminotransferase.
Pretreatment HCV RNA levels and response to IFN-α treatment.
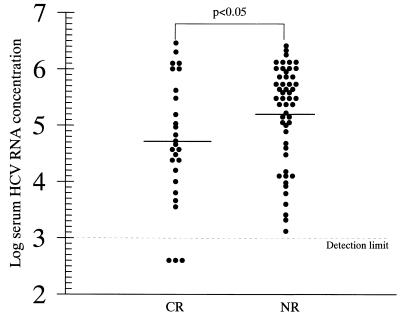
Of the 79 patients who received IFN-α therapy at 6 MIU thrice weekly for 24 weeks, followed by 3 MIU thrice weekly for the next 12 weeks, 27 (34.2%) were complete biochemical and virological responders. The mean pretreatment concentration of HCV RNA was significantly higher in the 52 nonresponders than in the 27 complete responders (5.21 ± 0.87 versus 4.71 ± 1.08 logs for the COBAS HCM-2 results; P < 0.05) (Fig. 2). After the patients were placed into three groups with low (≤3 logs), medium (3 to 5 logs) and high (>5 logs) viral loads, the percentage of complete responders was observed to decrease with higher pretreatment serum HCV RNA concentrations (3 of 3 [100%], 14 of 30 [46.7%], and 10 of 46 [21.7%] for those with low, medium, and high viral loads, respectively; P < 0.01 by the chi-square test with linear correlation).
FIG. 2.
Pretreatment levels of HCV RNA in sera of responders (CR) and nonresponders (NR) to IFN-α therapy at 6 MIU thrice weekly for 24 weeks plus 3 MIU thrice weekly by COBAS HCM-2. Of 79 patients, 27 were responders. The solid lines represent the mean pretreatment serum HCV RNA level in each group.
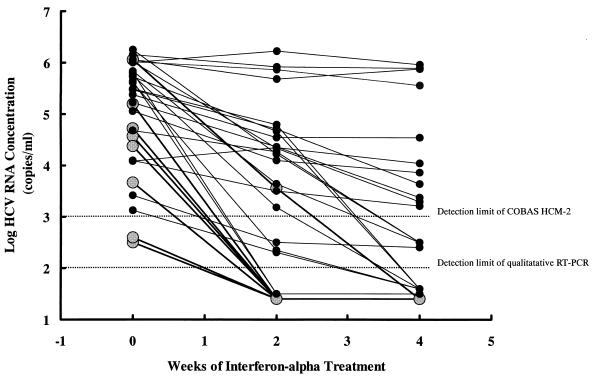
The dynamic changes in serum HCV RNA levels before and at 2 and 4 weeks after IFN-α treatment of 30 unselected patients (8 complete responders and 22 nonresponders) are shown in Fig. 3. Twelve (40%) of 30 serum samples collected 2 weeks after the initiation of IFN-α treatment had loads below the detection limit of COBAS HCM-2. Of these, three were positive for HCV RNA by qualitative RT-PCR. The complete response rate was significantly higher among patients with negative COBAS HCM-2 results at 2 weeks after the initiation of treatment than among those with positive results (7 of 12 [58.3%] versus 1/18 [5.6%]; P < 0.01). Nineteen (63.3%) of 30 serum samples collected 4 weeks after the initiation of IFN-α treatment were COBAS HCM-2 negative, and 4 of these were positive by qualitative RT-PCR. None of the 11 patients with quantifiable viral loads at 4 weeks after the initiation of treatment became complete responders. In contrast, 8 of the 19 (42.1%) patients with negative COBAS HCM-2 results 4 weeks after the initiation of treatment became complete responders (P < 0.05). By univariate analysis, sex, history of transfusion, levels of HCV RNA in serum before and at 2 and 4 weeks after the initiation of treatment, and the ratio of the reduction of the viral load 2 weeks after the initiation of treatment were significant factors associated with the HCV response to IFN-α treatment (Table 4). By further analysis by using multiple logistic regression, the serum HCV RNA level 2 weeks after the initiation of treatment was the only significant factor associated with the HCV response to IFN-α treatment, with an odds ratio and a 95% confidence interval of 0.14 and 0.03 to 0.67, respectively.
FIG. 3.
Dynamic change in serum HCV RNA levels determined by COBAS HCM-2 before and at 2 and 4 weeks after initiation of IFN-α treatment for 8 complete responders and 22 nonresponders. Black circles linked by bold lines represent complete responders. Solid dots linked by regular lines represent nonresponders.
TABLE 4.
Factors associated with response to IFN-α treatment in 30 chronic hepatitis C patients
| Characteristic | No. of patients | Complete responders (n = 8; 26.7%) | Nonresponders (n = 22; 73.3%) | P value |
|---|---|---|---|---|
| Age (yr) | 30 | 47.9 ± 16.3 | 44.2 ± 11.8 | >0.05 |
| Sex (no. [%] of patients) | <0.05 | |||
| Male | 16 | 7 (43.8) | 9 (56.2) | |
| Female | 14 | 1 (7.1) | 13 (92.9) | |
| History of transfusion (no. [%] of patients) | 0.05 | |||
| Yes | 8 | 0 | 8 (100%) | |
| No | 22 | 8 (36.4) | 14 (63.6) | |
| Biochemistry (IU/liter) | >0.05 | |||
| ASTa | 30 | 50.3 ± 30.2 | 48.6 ± 21.7 | |
| ALTb | 30 | 66.8 ± 40.8 | 51.8 ± 23.1 | |
| Liver histology (no. [%] of patients) | >0.05 | |||
| Chronic persistent hepatitis | 9 | 3 (33.3) | 6 (66.7) | |
| Chronic active hepatitis | 13 | 3 (23.1) | 10 (76.9) | |
| Liver cirrhosis | 8 | 2 (25.0) | 6 (75.0) | |
| HCV genotype (no. [%] of patients) | >0.05 | |||
| 1b | 15 | 3 (20.0) | 12 (80.0) | |
| Non-1b | 15 | 5 (33.3) | 10 (66.7) | |
| HCV viral loadc (log) | ||||
| Pretreatment | 30 | 4.25 ± 1.17 | 5.31 ± 0.89 | 0.01 |
| 2 wk after initiation of treatment | 30 | 1.93 ± 0.66 | 4.08 ± 1.27 | <0.0001 |
| 4 wk after initiation of treatment | 30 | 1.70 ± 0.00 | 3.27 ± 1.50 | <0.01 |
| Ratio of reduction of viral load | ||||
| 2 wk after initiation of treatment | 30 | 0.46 ± 0.14 | 0.22 ± 0.20 | <0.01 |
| 4 wk after initiation of treatment | 30 | 0.49 ± 0.16 | 0.37 ± 0.24 | >0.05 |
AST, aspartate aminotransferase.
ALT, alanine aminotransferase.
As measured by COBAS HCM-2.
DISCUSSION
In the present study, the automated COBAS HCM-2 was shown to be an easy, reliable, standardized test with good performance for routine clinical use. Compared to the manual AMPLICOR microwell plate system, version 1.0, the integrated AMPLICOR HCV assay based on the COBAS AMPLICOR system allowed a significant reduction of hands-on time (1, 19, 28). The turnaround time for the complete COBAS HCM-2 including specimen preparation and the amplification and detection procedures was 8 h for a batch of 21 specimens plus 3 controls. The hands-on time required to perform the manual extraction was about 2 h, so that the overall workload time was 6 min/specimen, whereas it was 21 min/specimen for the manual test.
In comparison to bDNA-2 and the first version of the AMPLICOR HCV MONITOR assay (23, 28), COBAS HCM-2 was found to have greater clinical sensitivity (95%) and was shown to be able to detect genotypes 1b, 2a, and 2b and mixtures of genotypes, which are the common genotypes in Taiwan, with equal sensitivities. In this study, repeat testing of low-, medium-, and high-titer sera in the same run and in different runs revealed standard deviations of less than 0.1 and 0.2 for within-run and between-run tests of COBAS HCM-2, respectively, in contrast to the larger variability found in the manual first version of the AMPLICOR assay observed in previous studies (9, 14). The standard deviations of 0.1 to 0.2 imply that the test can reliably detect 0.5 log10 differences in viral loads. The good reproducibility of this integrated quantitative PCR system for the detection of HCV, regardless of viral load, will enable the routine diagnostic laboratory and the clinician to better define the clinical utilities of these tests. By using four dilution panels of high-titer sera infected with different HCV genotypes, the linearity of COBAS HCM-2 was quite reliable with respect to its dynamic range of 103 to 106 copies/ml, whatever the genotype. For high concentrations, the less linear results may be indicative of a saturation effect in the RT-PCR. However, the relatively poor linearity for high-titer sera reported in previous studies that evaluated the first and second versions of the manual AMPLICOR assay (14, 17) was not observed in the present study.
The mean difference in quantification values between bDNA-2 and COBAS HCM-2 was, on average, 1.00 log for samples infected with genotype 1 and 0.93 log for samples infected with a genotype other than genotype 1, in contrast to previously reported differences of about 2 logs between bDNA-2 and the first version of the AMPLICOR HCV MONITOR assay for non-genotype 1-infected specimens (23, 28). Although a significant linear relationship between the results of COBAS HCM-2 and bDNA-2 was observed in the present study, the Spearman rank correlation coefficient r was only 0.693. Hence, these two assays, based on fundamentally different technologies, should not to be used interchangeably, as observed in previous studies (9, 14, 17, 23, 28).
In the current study, sex, patient age, mode of transmission, liver enzyme levels, and severity of liver disease did not correlate with serum HCV RNA levels, as measured by either COBAS HCM-2 or bDNA-2. This was consistent with the results of previous studies, which used different quantitative HCV technologies (9, 10, 13, 21, 28). Also, consistent with the results of other studies of bDNA-2 and version 2.0 of the manual AMPLICOR assay (9, 17, 23, 28), no genotype-dependent differences in viral load were observed in the present study. Our study demonstrated that, by COBAS HCM-2, the discrepancies in the correlation between HCV genotypes and viral load that have been reported with the first version of the AMPLICOR assay (9, 14, 17, 23, 28) and competitive RT-PCR assays (10) no longer exist. Since underestimation of the viral load for non-genotype 1-infected samples with the first version of the AMPLICOR assay might be due to the genotype-specific differences in the strength of base pairings and in the secondary structure of the 5′ noncoding region in which the PCR primers anneal (22), introduction of 16% dimethyl sulfoxide into the amplification kit to reduce the secondary structure appears to improve the amplification efficiency for non-genotype 1 HCV RNA, resulting in more accurate quantitation. Since genotype and pretreatment viral load have been shown to be two major prognostic markers for chronic hepatitis C patients receiving IFN-α treatment (8, 16, 27, 28, 29), genotype-specific differences in the efficiency of quantitation of HCV RNA may interfere with the roles of these two factors in the outcome of IFN-α treatment. As it is genotype independent, COBAS HCM-2 could provide, prior to and during therapy, a proper assessment of the HCV load and be useful for predicting the outcome of IFN-α treatment in chronic hepatitis C patients. However, further clinical evaluation of COBAS HCM-2 is necessary to document that it is equally efficient for the quantification of HCV genotypes other than genotypes 1 and 2 to extend its usefulness worldwide.
Similar to other reports (8, 16, 28), a correlation between the pretreatment viral load by COBAS HCM-2 and the response to IFN-α was observed. In the 30 patients whose viral loads were monitored during treatment, serum HCV RNA levels 2 weeks after the initiation of IFN-α treatment were shown to be the strongest predictor of a complete response by using multivariate analysis. None of 11 patients with quantifiable viral loads at 4 weeks of therapy achieved a complete response, suggesting that IFN-α treatment may be terminated in this patient group. These results were similar to those in other reports that failure to clear HCV RNA, as determined by qualitative RT-PCR, at 2 or 4 weeks of therapy (7, 26) or an initial decline in serum HCV RNA levels of less than 3 logs in the first 4 weeks of treatment (30) are strongly and independently associated with a very low probability of a complete response to IFN-α. In this limited study, the predictive value of the HCV load at 2 weeks of therapy clearly exceeds the significance of other predictors, such as pretreatment viremia, viral load at 4 weeks of therapy, ratio of viral load reductions at 2 and 4 weeks after therapy, and genotype (8, 16, 28, 29, 30). Further larger studies are needed to elucidate this issue. Initial results indicate that viral load measurements will also provide useful prognostic information for patients receiving the more potent combination therapy (20).
In conclusion, COBAS HCM-2 was found to have greater sensitivity, linearity, and reproducibility than the first version of the AMPLICOR test and quantified HCV genotypes 1 and 2 with equal efficiencies. The ability to automate the test not only improves the assay performance but also allows cost savings in terms of the hands-on time required. Use of COBAS HCM-2 to monitor the HCV load before and during the early phase of treatment appears to be very useful in the planning of IFN-α treatment. On the basis of the present results, the automated COBAS HCM-2 system, which saves hands-on time in the amplification and detection procedures, is a rapid and reliable PCR method for routine quantification of HCV RNA in serum. Further clinical evaluation of COBAS HCM-2 with HCV genotypes other than genotypes 1 and 2 is necessary to extend its usefulness worldwide.
REFERENCES
- 1.Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault A M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, et al. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 3.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Colucci G, Gutekunst K. Development of a quantitative PCR assay for monitoring HCV viremia levels in patients with chronic hepatitis C. J Viral Hepat. 1997;4:75–78. doi: 10.1111/j.1365-2893.1997.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 5.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doglio A, Laffont C, Caroli-Bosc F X, Rochet P, Lefebvre J C. Second generation of the automated COBAS Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J Clin Microbiol. 1999;37:1567–1569. doi: 10.1128/jcm.37.5.1567-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavier B, Martunez-Gonzulez M A, Riezu-Boj J I, Lasarte J J, Garcia N, Civeira M P, Prieto J. Viremia after one month of interferon therapy predicts treatment outcome in patients with chronic hepatitis C. Gastroenterology. 1997;113:1647–1653. doi: 10.1053/gast.1997.v113.pm9352868. [DOI] [PubMed] [Google Scholar]
- 8.Hagiwara H, Hayashi N, Mita E, Takehara T, Kasahara A, Fusamoto H, Kamada T. Quantitative analysis of hepatitis C virus RNA in serum during interferon alfa therapy. Gastroenterology. 1993;104:877–883. doi: 10.1016/0016-5085(93)91025-d. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi J, Kishihara Y, Yoshimura E, Tani Y, Yamaji K, Ikematsu H, Ishiko H, Kashiwagi H. Relationship of genotype to level of hepatitis C viremia determined by competitive polymerase chain reaction. J Infect. 1995;30:235–239. doi: 10.1016/s0163-4453(95)90785-8. [DOI] [PubMed] [Google Scholar]
- 11.Kato N, Yokosuka O, Hosoda K, Ito Y, Ohto M, Omata M. Quantification of hepatitis C virus by competitive reverse transcription-polymerase chain reaction: increase of the virus in advanced liver disease. Hepatology. 1993;18:16–20. [PubMed] [Google Scholar]
- 12.Lancet. Acute and chronic hepatitis revisited. Review by an international group. Lancet. 1977;ii:914–919. [PubMed] [Google Scholar]
- 13.Lau J Y N, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 14.Lunel F, Cresta P, Vitour D, Payan C, Dumont B, Frangeul L, Reboul D, Brault C, Piette J C, Huraux J M. Comparative evaluation of hepatitis C virus RNA quantitation by branched DNA, NASBA, and Monitor assays. Hepatology. 1999;29:528–535. doi: 10.1002/hep.510290237. [DOI] [PubMed] [Google Scholar]
- 15.Martell M, Gomez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteba R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, et al. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alpha therapy in chronic hepatitis C. Hepatology. 1995;22(Suppl. 4, part 1):1050–1056. [PubMed] [Google Scholar]
- 17.Mellor J, Hawkins A, Simmonds P. Genotype dependence of hepatitis C virus load measurement in commercial available quantitative assays. J Clin Microbiol. 1999;37:2525–2532. doi: 10.1128/jcm.37.8.2525-2532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto H, Tokita H, Sakamoto M, Horikita M, Kojima M, Iizuka H, Mishiro S. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993;74:2385–2390. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
- 19.Poljak M, Seme K, Koren S. Evaluation of the automated COBAS AMPLICOR hepatitis C virus detection system. J Clin Microbiol. 1997;35:2983–2984. doi: 10.1128/jcm.35.11.2983-2984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poynard T, McHutchison J, Goodman Z, Ling M H, Albrecht J. Is an “a la carte” combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? The ALGOVIRC Project Group. Hepatology. 2000;31:211–218. doi: 10.1002/hep.510310131. [DOI] [PubMed] [Google Scholar]
- 21.Smith D B, Davidson F, Yap P L, Brown H, Kolberg J, Detmer J, Urdea M, Simmonds P. Levels of hepatitis C virus in blood donors infected with different viral genotypes. J Infect Dis. 1996;73:727–730. doi: 10.1093/infdis/173.3.727. [DOI] [PubMed] [Google Scholar]
- 22.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P L, Simmonds P. Variation of hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;6:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 23.Tong C Y W, Hollingsworth R C, Williams H, Irving W L, Gilmore I T. Effect of genotypes on the quantification of hepatitis C virus (HCV) RNA in clinical samples using the Amplicor HCV Monitor Test and the Quantiplex HCV RNA 2.0 Assay (bDNA) J Med Virol. 1998;55:191–196. [PubMed] [Google Scholar]
- 24.Varadaraj K, Skinner D M. Denaturants or cosolvents improve the specificity of PCR amplification of a G+C-rich DNA using genetically engineered DNA polymerases. Gene. 1994;140:1–5. doi: 10.1016/0378-1119(94)90723-4. [DOI] [PubMed] [Google Scholar]
- 25.Winship P R. An improved method for directly sequencing PCR amplified material using dimethylsulfoxide. Nucleic Acids Res. 1989;17:1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaji K, Hayashi J, Kawakami Y, Furusyo N, Sawayama Y, Kishihara Y, Etoh Y, Kashiwagi S. Hepatitis C viral RNA status at two weeks of therapy predicts the eventual response. J Clin Gastroenterol. 1998;26:193–199. doi: 10.1097/00004836-199804000-00009. [DOI] [PubMed] [Google Scholar]
- 27.Yu M L, Chuang W L, Chen S C, Lu S N, Wang J H, Lin Z Y, Hsieh M Y, Wang L Y, Chang W Y. Treatment of chronic hepatitis C with interferon-alpha: a preliminary report. Kaohsiung J Med Sci. 1996;12:581–589. [PubMed] [Google Scholar]
- 28.Yu M L, Chuang W L, Chen S C, Lin Z Y, Hsieh M Y, Wang L Y, Chang W Y. Clinical application of the Quantiplex HCV RNA 2.0 and Amplicor HCV Monitor assays for quantifying serum hepatitis C virus RNA. J Clin Pathol. 1999;52:807–811. doi: 10.1136/jcp.52.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tminaga T, Persing D H. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Collaborative Study Group. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]
- 30.Zeuzem S, Lee J H, Franke A, Roster B, Prommer O, Herrmann G, Roth W K. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology. 1998;27:1149–1156. doi: 10.1002/hep.510270433. [DOI] [PubMed] [Google Scholar]