Abstract
Merkel cell polyomavirus (MCPyV) is a ubiquitous skin infection that can cause Merkel cell carcinoma (MCC), a highly lethal form of skin cancer with a nearly 50% mortality rate. Since the discovery of MCPyV in 2008, great advances have been made to improve our understanding of how the viral encoded oncoproteins contribute to MCC oncogenesis. However, our knowledge of the MCPyV infectious life cycle and its oncogenic mechanisms are still incomplete. The incidence of MCC has tripled over the past two decades, but effective treatments are lacking. Only recently have there been major victories in combatting metastatic MCC with the application of PD-1 immune checkpoint blockade. Still, these immune-based therapies are not ideal for patients with a medical need to maintain systemic immune suppression. As such, a better understanding of MCPyV's oncogenic mechanisms is needed in order to develop more effective and targeted therapies against virus-associated MCC. In this review, we discuss current areas of interest for MCPyV and MCC research and the progress made in elucidating both the natural host of MCPyV infection and the cell of origin for MCC. We also highlight the remaining gaps in our knowledge on the transcriptional regulation of MCPyV, which may be key to understanding and targeting viral oncogenesis for developing future therapies.
Keywords: Merkel cell polyomavirus, Merkel cell carcinoma, Natural host, Cell of origin for MCC
1. MCPyV and MCC
Merkel cell carcinoma (MCC) is a rare but highly aggressive skin cancer. The diagnosis of MCC relies on analyses of histological features and immunological staining of a skin biopsy, in which the immunohistochemical marker cytokeratin-20 (CK20) is used to distinguish MCC from other neuroendocrine carcinomas [1,2]. Initially described by Cyril Toker in 1972 [3], this cancer metastasizes rapidly and responds relatively poorly to chemotherapy, resulting in low survival rates in its patients [[4], [5], [6], [7], [8], [9], [10]]. MCCs have also demonstrated the intrinsic ability to resist immunological eradication [6,[11], [12], [13], [14], [15]]. Though still rare, the incidence of MCC has tripled in the decades following its discovery [[16], [17], [18]]. Anti-PD1 immune checkpoint therapies against metastatic MCC first demonstrated promising responses in a 2015 study [19]. The following clinical trials exploring the safety and efficacy of anti-PD1 and anti-PDL-1 infusions generated significant response rates with relatively low incidence of adverse events [8,[20], [21], [22]]. Since there are no effective chemotherapeutic treatments available for metastatic MCC, these immune checkpoint blockade therapies greatly benefitted patients [7,17]. However, these immune-based therapies are not ideal for patients with a medical need to maintain systemic immune suppression [23,24]. These shortcomings demonstrated the need for developing more effective and targeted therapies.
The major risk factors for MCC development include excessive UV exposure, advanced age, and immunosuppression, such as that of HIV/AIDS patients and organ transplant recipients [1,25]. The Chang and Moore group at the University of Pittsburgh hypothesized that the increased risk of MCC in immunocompromised individuals was indicative of a possible infectious origin for the cancer. To pursue this idea, they developed the digital transcriptome subtraction technique and used it to identify a novel polyomavirus, subsequently named Merkel cell polyomavirus (MCV or MCPyV), in MCC samples [26]. The MCPyV genome is clonally integrated within the MCC genome, suggesting that viral integration is an event that occurs prior to oncogenesis and that MCPyV is a likely etiological agent for this cancer. Subsequent studies have confirmed that MCPyV DNA is integrated in approximately 80% of MCC cases [[26], [27], [28], [29], [30], [31]].
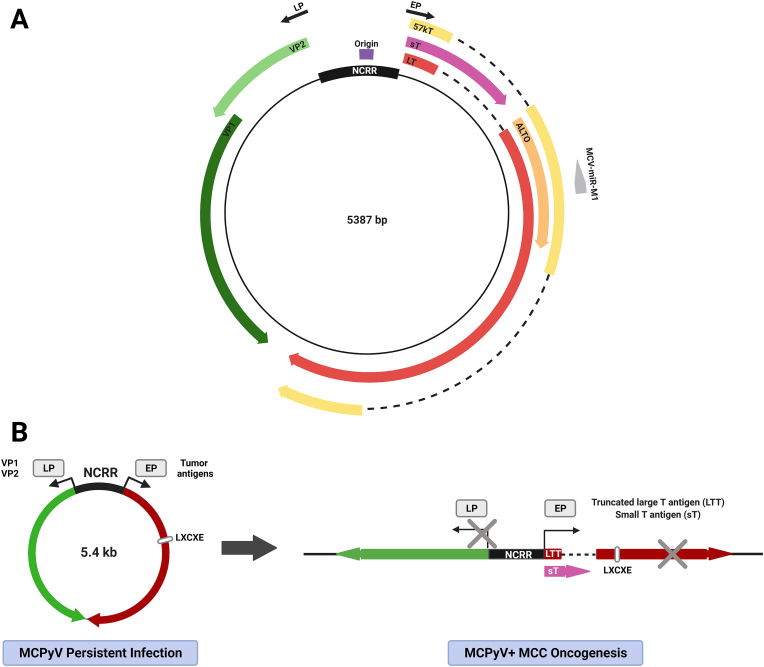
MCPyV is a nonenveloped circular dsDNA virus with a genome of approximately 5.4 kb [32] (Fig. 1A). Transcription of its genes, which are classified as early or late based on the sequential manner in which they are expressed during infection, is controlled by the noncoding regulatory region (NCRR) containing both early and late promoters as well as the origin of replication (Fig. 1A) [33,34]. The early promoter (EP) drives the expression of the large tumor antigen (LT) and small tumor antigen (sT), in addition to the 57 kT antigen and Alternate frame of the LT open reading frame (ALTO) [26,32,35]. The late region encodes the viral capsid proteins VP1 and VP2 [[36], [37], [38]] and a miRNA [39].
Fig. 1.
MCPyV gene expression in infection and MCC. A) MCPyV genomic map. The NCRR contains the origin of replication and bidirectional promoters, which drive the expression of the viral early genes (right) and late genes (left). B) The MCPyV genome is maintained as a replication-competent episome in persistently infected cells, which expresses the early and late viral genes in a temporal manner. In MCPyV-positive MCC, the MCPyV genome is clonally integrated in the host cell genome. Integrated MCPyV continues to express sT and a truncated LT (LTT) that preserve the expression of the N-terminal LXCXE Rb-binding domain but not the C terminal domains needed for regulating viral replication. The LTT truncation mutations and the lack of late gene expression in MCC are denoted by gray Xs. Abbreviations: NCRR, noncoding regulatory region; EP, early promoter; LP, late promoter; LT, large tumor antigen; sT, small tumor antigen; 57 kT, 57 kDa tumor antigen; ALTO, alternate large tumor antigen open reading frame; LTT, truncated large tumor antigen.
MCPyV infection is widespread and commonly asymptomatic in the general population [40,41]. During long-lasting persistent infection, the virus replicates and exists as an episome within infected nonmalignant human cells (Fig. 1B). The viral DNA discovered in MCCs is frequently integrated into the cellular genome. Viral integration occurs at random sites in the genome, though more commonly on chromosome 5 [[42], [43], [44]]; the virus integrates as either a single copy or as a concatemer of multiple copies [42]. The integration event typically results in tumor-specific truncation mutations that preserve the expression of sT and LT truncation (LTT) mutants retaining the N-terminal RB-binding LXCXE motif, which is essential for inhibiting RB's tumor suppressor function (Fig. 1B) [26,[45], [46], [47], [48]]. LTT and sT expressed from the integrated viral genome are the major oncoproteins that support continued tumor cell growth and survival (Fig. 1B) [[47], [48], [49], [50], [51]].
Since the discovery of MCPyV more than a decade ago, much of MCPyV research has been focused on how the MCPyV-encoded oncoproteins stimulate cellular transformation to promote MCC development. These efforts have been summarized in a number of previous reviews [[52], [53], [54], [55], [56]]. On the other hand, the MCPyV life cycle and how it contributes to MCC development remain largely unknown. This set of knowledge is needed for developing more effective treatments for MCC [1,7,57]. In this review, we discuss the gaps in our understanding of the biology driving MCPyV-associated oncogenesis. We examine emerging evidence on the natural host of MCPyV infection and the question of the MCC cell of origin. We will also elaborate on the importance of studying the largely unexplored mechanisms that regulate MCPyV transcription, which drives viral infection and MCC development.
2. MCPyV tropism and infectious host
Merkel cell carcinoma was initially believed to originate from Merkel cells, a rare population of mechanoreceptor cells within the epidermis [3,58]. After the discovery of MCPyV in 2008 [26], evidence quickly arose that MCPyV establishes asymptomatic and persistent infection in a large proportion of the human population [29,40,41,[59], [60], [61], [62]], with initial exposure often occurring during early childhood and a seroprevalence that increases among older age groups [40,59,60,63,64]. As expected based on its association with MCC, MCPyV is detectable within the skin [40,65]. However, a variety of studies have also detected MCPyV in respiratory samples, urine, and blood [[66], [67], [68], [69], [70], [71]]. Additionally, in 2010, Schowalter et al. discovered that MCPyV is chronically shed from the skin, with millions of viral genome equivalents detectable in skin swabs [40]. Schowalter et al. therefore proposed that highly rare Merkel cells may not be abundant enough within the skin to support this level of virion production, and that the infectious host cell was more likely a common skin cell type, such as keratinocytes or melanocytes [40]. It was also suggested that MCPyV infection is intrinsically tied to epidermal differentiation, similar to the biology of human papillomavirus (HPV) infection in keratinocytes [40].
To explore the specific cell types that may act as MCPyV's infectious host, several groups examined the mechanism of viral entry into host cells [37,72]. Ultimately, it was found that glycosaminoglycans such as heparan sulfate are required for the initial attachment of MCPyV virions to the host cell, and that an unknown co-receptor was necessary for full viral entry into the cell; this mechanism was again noted to be similar to the method used by HPV to enter host cells [37]. Immediately post-entry, viral endosome-to-endoplasmic reticulum trafficking appears to be another bottleneck for viral infection in addition to virion attachment and entry, revealing a new obstacle to viral tropism [73].
In the 2010s, there were several attempts to both establish a monolayer cell culture system for MCPyV infection and simultaneously identify the infectious host cell of the virus. Neumann et al. established a transfection system in which a consensus MCPyV genome was capable of replicating in several cell lines, including Human embryonic kidney 293 (HEK293), the lung carcinoma cell line H1299, and the neuroectodermal tumor cell line PFSK-1, achieving detectable viral particle formation [74]. Similarly, Feng et al. designed a replicating MCPyV genome system, which when transfected into HEK293 cells, leads to both early and late gene expression as well as a detectable amount of encapsidated virus [75]. In 2011, Schowalter et al. also established a couple of cell systems that support MCPyV replication, consisting of HEK293-TT (HEK293T cells expressing additional copies of the SV40 T antigen) and HEK293-4T (HEK293-TT cells stably expressing MCPyV LT and sT), both of which are also capable of producing MCPyV virions [37]. Difficulties persisted in discovering a relevant primary cell type that supports infection by native virions, as seen when Schowalter et al. assessed the transducibility of a panel of cell lines by either MCPyV native virus or a pseudovirus harboring a reporter gene. Among the cell types tested, a wide range of cancer-derived cell lines were permissive for MCPyV pseudovirus entry, while only primary human keratinocytes were transducible by pseudovirus among other skin or MCC cell lines. Furthermore, no tested cell types except for HEK293-4T were permissive for viral replication upon infection by native virions [38].
Aside from the U2OS osteosarcoma cell line, which supports low-level MCPyV infection [76], no other permissive cell type was found until 2016, when our group examined the infectability of the entire population of primary cells derived from human foreskin and found that human dermal fibroblasts (HDFs) alone supported the full MCPyV infectious life cycle [77]. Interestingly, Merkel cells do not support MCPyV infection, and consistent with the findings of Schowalter et al., keratinocytes were permissive for pseudovirus entry but not native virus gene expression or replication [77]. MCPyV-infected HDFs show robust replication of the viral genome as well as expression of the early and late genes [77]. The HDF infection model thus affords an exciting platform to investigate the viral and host factors that regulate the MCPyV life cycle.
Interestingly, our analysis of ex vivo skin cultures revealed that MCPyV preferentially infects dermal fibroblasts underlying the basal layer of the epidermis and surrounding hair follicles [77]. Prior to our studies, a number of groups have shown that MCPyV virions are readily detected in eyebrow hair bulbs sampled from healthy human volunteers [[78], [79], [80]]. Together, these observations suggest that MCPyV that have replicated in the dermal cells surrounding hair follicles may subsequently use the follicular space as a path to disseminate to the skin surface and infect new hosts [77]. Physiological processes such as skin wounding and photoaging may allow damaged keratinocytes to release growth factors that promote MCPyV infection [77]. In the event that skin is abraded or irradiated with UV light, proliferation and migration of MCPyV-infected fibroblasts from the dermis to the wounded tissue or shedding layers of sunburned skin could represent another mode of transmission for MCPyV virions to egress from reservoir cells in the deeper layers of the skin and spread to new hosts.
Despite our finding that MCPyV can effectively infect HDFs, the picture of MCPyV infection is far from complete. The identity of the natural host reservoir cells that maintain latent MCPyV infection in the human body remains elusive. As discussed earlier, MCPyV has been detected in a variety of other tissues [66,71,81], raising the possibility that the virus establishes an infectious reservoir at body sites other than the skin, such as the tonsils or within the blood, specifically in inflammatory monocytes [70,71,81]. There are also other additional components that could determine the infectability of the host cell by MCPyV, such as the phosphorylation of viral proteins by host cell kinases to regulate viral replication [82] and the interplay between viral infection and the host immune response [83,84]. The HDF model of MCPyV infection nevertheless presents the opportunity to explore the mechanisms of MCPyV infection within the skin.
We also found that MCPyV's tropism is extremely narrow, as among fibroblasts isolated from common model animals only chimpanzee and human fibroblasts are able to support a significant level of MCPyV infection [85]. This narrow host range of the virus makes it difficult to establish an in vivo infection system [85]. The lack of an animal model for MCPyV infection also presents a major challenge for identifying potential reservoir cells and delineating the complete MCPyV infectious cycle. As suggested by our previous studies, engineering MCPyV chimeras with mammalian polyomaviruses may provide new solution for overcoming the obstacle posed by its narrow host range [85].
3. Cell of origin for MCC
3.1. Merkel cell origin
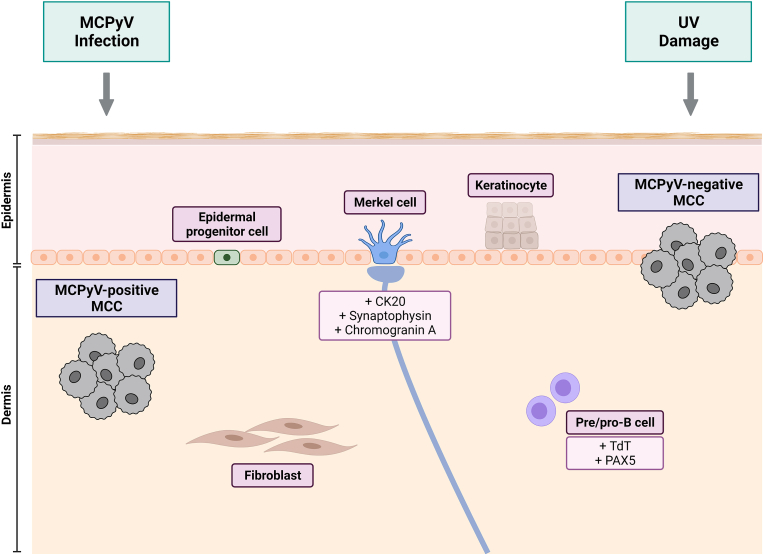
MCC was originally named due to similarities between tumor and Merkel cells observed in ultrastructural studies, such as the presence of distinct neurosecretory granules [58]. Subsequent studies revealed that Merkel cells and MCC also share the expression of specific neuroendocrine markers, including CK20, synaptophysin, and chromogranin A [86,87]. In the years since MCPyV's discovery, the hypothesis that MCC originates from Merkel cells has been called into question. Notably, Merkel cells do not respond to oncogenic stimuli, including that of the MCPyV tumor antigens [88], and are believed to be post-mitotic [89], though recent evidence suggests the existence of proliferative Merkel cells in rare cases of combined MCC and squamous cell carcinoma (SCC) [90]. In addition, the vast majority of MCC cases are found in the dermis or subcutis, while Merkel cells have been suggested to arise through the differentiation of epidermal progenitors during embryonic development [[91], [92], [93]]. These evidence have led to ongoing speculation as to what the origin cell of MCC is, and furthermore, whether it is also the infectious reservoir of MCPyV or a “bystander” cell to MCPyV infection. Additional major hypothesized origin cells of MCC are discussed below and summarized in Fig. 2.
Fig. 2.
Proposed cells of origin for MCC. Several cells of origin have been hypothesized for MCPyV-positive and MCPyV-negative MCC. Merkel cells were historically thought to be the original cell of MCC due to similarities in structure and expression of neuroendocrine marker (CK20, synaptophysin, chromogranin A); however, these cells are post-mitotic and do not respond to oncogenic stimuli [58,[86], [87], [88], [89]]. Epidermal progenitor cells have been proposed as an alternative original cell with proliferative potential [94,95]. Pre- or pro-B cells have also been hypothesized to be the cell of origin, due to the expression of specific B cell markers (including TdT and PAX5) in MCC [96]. Dermal fibroblasts support MCPyV infection and may therefore also act as the site of oncogenic transformation [77]. MCPyV-negative MCC has been specifically proposed to arise from epidermal keratinocytes, which are more prone to UV exposure than dermal cells [97].
3.2. Epidermal progenitor cell origin
Cases of mixed tumors consisting of cells from both MCC and another skin cancer have formed the basis of the hypothesis that MCC arises from stem or progenitor cells within the skin. As early as 1998, it was first proposed that a pluripotent stem cell was the origin cell of MCC due to the discovery of several cases of mixed MCC and SCC [94]. Similarly, Kervarrec et al. recently described a mixed tumor consisting of MCPyV-positive MCC and a benign trichoblastoma [95]. While the MCPyV genome was only integrated in the MCC cells, both the MCC and trichoblastoma shared the same somatic mutation profile within the exome, suggesting a common origin in which the viral genome integrated into an existing trichoblastoma cell [95]. This finding ultimately points to a progenitor cell residing in the hair follicle, such as a Merkel cell progenitor, as the cell of origin [95]. Further studies of Atonal homolog 1 (Atoh1), a master regulator of Merkel cell development, support a role of this effector in MCC oncogenesis. Expression of Atoh1 in MCC and fibroblasts induces expression of miR-375, the most common miRNA in MCCs [98]. In vivo studies have also shown that Atoh1 and sT coexpression causes the formation of “MCC-like” aggregates within the mouse epidermis [99]. GLI family zinc finger 1 (Gli1), another essential factor involved in Merkel cell differentiation, also induces a Merkel cell-like phenotype in keratinocytes when coexpressed with LT [100]. However, another interpretation of this evidence is that expression of Merkel cell differentiation factors such as Atoh1 and Gli1 simply contributes to the Merkel cell-like phenotype of MCC, and that the cancer arises from a cell other than the proposed Merkel cell progenitor; the MCPyV tumor antigens may in fact be driving aberrant expression of Merkel cell factors in a skin cell of a different lineage. A few different studies support this possibility. For example, it has been shown that LT inhibits Atoh1 degradation [100], and that LTT expression in fibroblasts induces Atoh1 expression [98]. Similarly, Harold et al. found that the Rb-binding domain of LTT expressed in MCPyV-positive MCC induces the expression of another key Merkel cell differentiation factor, SRY-box transcription factor 2 (Sox2), which in turn upregulates Atoh1 expression [101]. Furthermore, knockdown of LT in MCPyV-positive MCC cells cocultured with keratinocytes causes the cancer cells to undergo neuronal differentiation and display neurite projections and secretory vesicles, suggesting a neuronal origin for the cancer [101]. In addition to direct activity introduced by the viral oncoproteins, epigenetic dysregulation of Merkel cell differentiation factors may also contribute to their aberrant expression in MCC. Sox2 expression and Sonic hedgehog signaling, which is also crucial for Merkel cell specification, are normally restricted to primary hair follicles and touch domes by repressive H3K27me3 marks catalyzed by the Polycomb complex [102,103]. Busam et al. identified a significant reduction in H3K27me3 levels in MCPyV-positive MCC tumor samples, suggesting that dysregulation of Polycomb activity may contribute to ectopic Merkel cell signaling in cancer cells [104]. However, Matsushita et al. later found that H3K27me3 levels are more reduced in combined MCPyV-negative MCC and squamous cell carcinoma (SCC) when compared to pure MCPyV-positive or MCPyV-negative tumors, calling into question the true epigenetic status of the different MCC subsets [105].
3.3. Pre/pro B cell origin
In 2013, it was first proposed by zur Hausen and colleagues that pre/pro B cells are the origin cell of MCC, given that MCC expresses both terminal deoxynucleotidyl transferase (TdT) and paired box protein 5 (PAX5, the coexpression of which is only normally seen in B cells) and immunoglobulin [96]. Subsequent studies confirmed that the presence of B cell markers is positively correlated with MCPyV-positive status in MCC tumors [106,107]. The major questions underlying this hypothesis are whether MCPyV infects pre/pro B cells inside the dermis and whether these cells enter the dermis before or during oncogenesis. Pre/pro B cells are capable of entering the bloodstream under circumstances such as disease [108]. A relatively recent review from the zur Hausen group suggested that pre/pro B cells may circulate in sun-damaged skin [109]. The epidemiological link between MCC and chronic lymphocytic leukemia (CLL) may further support the B cell origin hypothesis. Several studies have established that patients with CLL are predisposed to MCC [[110], [111], [112]]. Though immune suppression caused by CLL has been suggested as the reason for this relationship, the reverse has also been established—namely, that MCC patients are also at a higher risk of subsequent development of CLL [113]. Several groups have also detected MCPyV DNA at low levels in CLL cells [31,[114], [115], [116]]. Though most of these studies discussed the link between CLL and MCC in the context of immune suppression, the fact that the risk for both diseases is reciprocal might suggest some shared etiology.
3.4. Fibroblastic origin
Expanding on our finding that dermal fibroblasts support the full infectious cycle within the human skin, additional models have been proposed to link MCPyV infection to MCC tumorigenesis [77,97,117]. One of the hypotheses is that dermal fibroblasts act as the origin cell for MCC: under yet-to-be-discovered conditions, MCPyV oncogenes expressed in infected HDFs may induce gene expression patterns as observed in MCC to trigger tumor development. In support of this notion, sT's oncogenic properties have been demonstrated in fibroblasts [88], suggesting that dysregulation of normal MCPyV infection may lead to oncogenesis within the infectious host cell. Another theory is that MCPyV actively replicating in the HDFs may accidently enter Merkel cells or their precursor cells residing in the immediate vicinity; the nonpermissive infection environment of Merkel cells may force the replication defective MCPyV genome to integrate into the host cell genome, which may propel Merkel cells to undergo malignant transformation [77].
3.5. Origin for MCPyV-positive and MCPyV-negative MCCs
Adding to the debate over MCC's origin is the additional discussion of whether MCPyV-positive and MCPyV-negative MCC arise from the same origin cell. A number of groups have proposed that there are two different cells of origin for MCC depending on virus status, due in part to the finding that the two cancer subsets have significantly different mutational landscapes: MCPyV-negative MCCs featuring a mutational signature characteristic of UV damage have demonstrated a much higher mutational burden than MCPyV-positive MCCs [118,119]. Hypotheses regarding the origin cell for the two cancer subtypes have evolved over time. In 2014, prior to our group's discovery of dermal fibroblasts as the host cell for MCPyV, Jankowski et al. proposed that MCPyV-positive MCC arises from pre/pro B cells as per zur Hausen's hypothesis, while MCPyV-negative MCC arises from epidermal stem cells [120]. In 2018, Sunshine et al. proposed that MCPyV-positive MCC arises from fibroblasts while MCPyV-negative MCC arises from keratinocytes. This hypothesis is supported by the rationale that epidermal keratinocytes are less protected from UV light than dermal fibroblasts, accounting for the UV-damage mutational signature seen in virus-negative MCC [97].
It has been further suggested that MCPyV-negative MCC may be caused by MCPyV in a hit-and-run mechanism in which cellular transformation is initiated by the virus, which is then lost as tumor development continues to be driven by UV damage-induced mutations [121]. The fact that antibodies against MCPyV VP1 are detectable in the serum and plasma of patients with MCPyV-negative MCC lends support to this possibility [122]. Given the viral oncoprotein addiction of MCPyV-positive MCC [49], a hit-and-run mechanism would only be possible if tumor cells transformed by MCPyV are capable of surviving without viral tumor antigen expression. Indeed, in 2012 Houben et al. established a MCPyV-positive MCC cell line, LoKe, which displays no growth inhibition upon stable knockdown of the MCPyV tumor antigens [121].
3.6. The unique origin of intraepidermal MCC
Further complicating the classification of MCC subtypes are the extremely rare cases of purely intraepidermal MCC, which may be considered another subtype of MCC. Curiously, recorded cases of intraepidermal MCC have all been found to be MCPyV-negative [123,124], with the exception of tumors where the MCPyV status is unknown [125,126]. Narisawa et al. recently described a combined dermal and intraepidermal MCC associated with SCC, in which the dermal and intraepidermal compartments of the MCC had differential expression of the CK20 and CD56 markers, suggesting two separate populations of MCC cells. Furthermore, only the intraepidermal MCC cells expressed the touch dome markers CK15 and CK17, suggesting that this type of MCC may have originated from the touch dome [123]. Therefore, intraepidermal MCC may have its own origin cell distinct from dermal MCC.
4. MCPyV transcription, a key determinant for persistent infection and MCC tumorigenesis
In MCPyV infected cells, EP drives transcription of the early genes to support the viral life cycle during persistent infection (Fig. 1B). In virus-positive MCCs, EP continues to support the expression of the sT and LTT oncogenes [26,[45], [46], [47], [48]] (Fig. 1B). MCPyV-positive MCCs harbor very few genetic mutations [118,127], suggesting that expression of the viral oncogenes is sufficient to drive tumor development. Indeed, LTT and sT have demonstrated robust oncogenic potential to promote tumorigenesis (Fig. 1B) [46,51,128]. MCPyV-positive MCCs are addicted to MCPyV oncoproteins and require their continued expression from the integrated viral genome to survive [46,49,128]. MCPyV EP therefore plays a critical role in both the infectious life cycle of MCPyV and the oncogenic progression of infected cells, representing an attractive anti-cancer therapeutic target for molecular intervention. However, very little is known about the mechanisms that regulate MCPyV EP's transcriptional activity during either MCPyV infection or MCC development. This gap in our knowledge is largely because, until recently, the cellular tropism of MCPyV was unknown and there was a lack of a biologically relevant culture system for studying MCPyV infection and oncogenesis [38].
Additionally, MCPyV differs from other better-studied polyomaviruses to the extent that insights on these related viruses are likely insufficient to bridge the knowledge gap. Compared to other disease-causing polyomaviruses such as BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV), knowledge on MCPyV gene expression mechanisms is relatively sparse [129]. In MCPyV, BKPyV, JCPyV, and simian vacuolating virus 40 (SV40), the NCRR containing the viral origin of replication and bidirectional promoters is considered the major site through which viral gene expression is regulated. However, while the NCRRs of BKPyV, JCPyV, and SV40 all carry tandem repeat elements that function as viral transcription enhancers and binding sites for a number of host cell transcription factors, such elements do not exist in the MCPyV NCRR [129]. Because the MCPyV early and late proximal promoter regions are quite different from those of other human polyomaviruses and SV40, it suggests that distinct cellular factors and molecular mechanisms are involved in controlling MCPyV transcription.
The long search for MCPyV's infectious host cell underscores the virus' highly narrow cellular tropism. When recircularized MCPyV genomes were transfected into a total of 21 cancer cell lines and primary cell cultures, only three were able to support both early and late gene expression [74]. After discovering HDFs as the permissive cells that support productive MCPyV infection in the human skin [77], we showed that while MCPyV can promiscuously enter many types of cells in the human skin, viral early gene expression is detected only in a few cell types such as HDFs and MCC cells, but silenced in nonpermissive cells such as skin keratinocytes [77,85,130]. This finding suggests that MCPyV transcription is the key determinant of the virus' highly restricted host cell tropism and oncogenic potential. Furthermore, the highly narrow host cell tropism of MCPyV suggests that cell-type-specific factors are important for controlling its transcriptional activity [77,85,130]. For example, cell-type-specific epigenetic modifications and regulatory factors may underline MCPyV's highly distinctive transcriptional activity, which in turn defines its unique host cell tropism and oncogenic potential. In addition, cis-acting tissue-specific enhancer(s) and silencer(s) in the MCPyV genome may also control EP-driven transcription.
Epigenetic modifications play an important role in transcriptional regulation [131]. Histone H3 and H4 acetylation and methylation on H3K4/H3K36 are associated with transcriptional activation, whereas tri-methylation of H3K9 and H3K27 is linked to transcriptional repression. After MCPyV genomes are transfected into PFSK-1 cells that support MCPyV gene expression, transactivational histone marks such as H3K4me3 are detected in the NCRR region [132]. In addition, it has been shown that histones are encapsidated into purified MCPyV virions [36]. These studies suggest that MCPyV DNA is packaged into histone-bound nucleosomes likely carrying epigenetic modifications that control its transcription in infected cells. Whether MCPyV EP acquires different epigenetic modifications in permissive and nonpermissive cells to support its distinct transcription activities remains an important question for future studies.
Very little is known about the cellular and viral factors that regulate MCPyV transcription during either viral infection or MCC development. Nevertheless, some studies have begun to uncover both the viral intrinsic components and host cellular factors that regulate MCPyV expression. For example, in 2009, Seo et al. discovered that the MCPyV-encoded miRNA, named mcv-miR-M1, targets the viral early transcripts for degradation [39]. Using an in vitro MCPyV replication system, Theiss et al. subsequently found that mcv-miR-M1 negatively regulates transcription of MCPyV genes encoded by the early region that contains its complementary sequence [132]. While wild-type MCPyV genomes can persist as extrachromosomal episomes for several months in transfected PFSK-1 cells, the MCPyV mcv-miR-M1 knockout mutant is lost at an increased rate, indicating that mcv-miR-M1 may help to establish a state of low-level, long-term episomal persistence [132]. MCPyV EP carries four LT binding sites [34]; it has also been shown that LT can stimulate EP transcription activity in an autoregulatory manner [133]. In addition, LT is enriched at conspicuous viral DNA foci in MCPyV-infected cells [77], further indicating that this viral protein may be recruited to support MCPyV gene expression. Interestingly, MCPyV EP can also be activated in the absence of LT [134]. This LT-independent mode likely supports the first phase of early transcription that occurs immediately after viral entry when there is no LT protein present in the cells. These findings also suggest that, after the first set of LT is expressed in infected cells, they can contribute to EP transcription in the subsequent productive phase of infection, possibly by directing cellular transcription factors or chromatin modifiers to EP. The basal transcriptional activity of MCPyV EP could also be stimulated in permissive cells or repressed in nonpermissive cells by cell type-specific transcription factors uniquely present in these cells.
MCPyV's highly restricted transcription may also result from binding of cis-acting enhancer(s) by transcription activators in permissive cells and/or silencer(s) by transcription repressors in nonpermissive cells. Through in silico analysis of the MCPyV NCRR, Ajuh et al. identified a number of putative NCRR-binding transcription factors, several of which are also known to control the transcription of the related polyomaviruses JCPyV and BKPyV [133]. In JCPyV, the 98-bp tandem repeat demonstrates robust enhancer activity in permissive human fetal glial cells, and functions as a key determinant for JCPyV's highly restricted host cell specificity [135]. Because MCPyV also has a very narrow host range, it is conceivable that an “enhancer-like” element exists in MCPyV genome to support its distinct transcriptional activities in permissive cells. However, unlike other polyomaviruses [[136], [137], [138]], MCPyV does not carry the tandem repeat enhancer elements [129], and its enhancer region remains unknown. It is also unclear whether there is a cis-acting silencer(s) that could inhibit MCPyV early transcription in non-permissive cells. All of these components remain to be investigated in order to fully understand the mechanism of MCPyV transcription.
5. Future perspectives
Decades after the initial description of MCC, there is still no conclusive answer as to what the origin cell of MCC is. Addressing this gap in our knowledge may require advanced technologies. Even before the discovery of MCPyV, gene expression profiling of MCC cell lines by microarray analysis had already distinguished two subtypes (‘Classic’ and ‘Variant’) of MCCs [139]. Since the chromatin accessibility profile of a cancer's origin cell often determines the mutational landscape of the tumor genome [[140], [141], [142], [143]], modern technologies such as ATAC-seq or other epigenetic profiling tools may be crucial in matching the mutational signature of MCC to its cell of origin. Therefore, building a clearer picture of the genomic and epigenomic signature of MCC may be the most effective way to determine the cell from which MCC originates.
Uncovering the origin of MCCs will also be essential for understanding the relationship between MCPyV-positive and MCPyV-negative MCC, and whether they are indeed two subtypes of MCC or two different tumors with similar characteristics. In the clinical context, studies on whether the two categories of MCC are associated with different disease severity and outcome are conflicting [30,144], yet both MCPyV-positive and -negative MCC respond similarly to anti-PD(L)-1 therapy [21,22]. Further evidence suggests that MCPyV-positive and MCPyV-negative MCCs are two tumor subtypes with distinct morphological and immunohistochemical features driven by alternative tumorigenic pathways [1,127,145]. In addition, several groups have revealed the two subsets’ highly different mutational landscapes [118,119,127,146], pointing to the possible existence of multiple cell types of origin for MCC based on virus status. These studies indicate that MCPyV-positive and MCPyV-negative MCC are two different cancers with a convergent phenotype [97,120]. New insight on the origin of MCC will facilitate the development of targeted approaches needed for treating MCPyV-positive and MCPyV-negative MCC.
The infectious tropism of MCPyV identified thus far includes only human and chimpanzee dermal fibroblasts, and viral gene expression rather than viral entry appears to be the major bottleneck for establishing a productive infection [38,77,85]. MCPyV gene expression is therefore likely dependent on cell-type specific factors or mechanisms. Therefore, a more complete understanding of the cell populations that function as an infectious reservoir for MCPyV would be highly beneficial for identifying what these cell-type specific elements may be, and may also be helpful for elucidating the origin cell for MCPyV-positive MCC specifically.
Both the tumor-specific LTT and sT antigens expressed from the integrated MCPyV genome in MCC have demonstrated oncogenic functions in supporting uncontrolled cellular proliferation that ultimately inflict oncogenesis [46,50,88]. Depletion of these tumor antigens from MCPyV-positive MCC cell lines causes cancer cell death [49], making MCPyV oncogene expression an attractive target for developing virus-targeted therapies to treat MCPyV-associated MCCs. Unlike cancers that arise from mutations of endogenous genes, MCPyV-positive tumors carry low mutational burdens and are addicted to the expression of MCPyV T-antigens that drive disease progression. For this reason, virus-targeted therapies would hypothetically be well tolerated and highly desirable. For instance, a natural product identified in our recent study is able to induce killing of MCPyV-positive MCCs at nanomolar concentrations but exhibits little to no toxicity in healthy cells or MCPyV-negative MCCs at much higher concentrations [134]. In the absence of effective targeted therapies for MCC cancer, elucidation of the mechanisms controlling MCPyV transcription is imperative for identifying druggable targets for specific blockage of viral oncogene expression and MCC tumor growth. Decoding the regulatory mechanisms of MCPyV transcription will likely also offer important clues for understanding the origin cell(s) of MCPyV-positive MCC, the highly restricted host cell tropism of MCPyV, as well as the tumorigenic events leading to MCPyV-driven oncogenesis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
Research in the Jianxin You laboratory is supported by the National Institutes of Health (NIH) Grants R01CA187718, R21AR074073, R21AI149761, the NCI Cancer Center Support Grant (NCI P30 CA016520), and the Penn CFAR pilot award (P30 AI 045008).
Acknowledgments
The authors would like to thank all members of the Jianxin You laboratory for their comments and feedback on this manuscript. We apologize to our colleagues whose primary research findings could not be cited due to space constraints. Figures were created with BioRender.com.
References
- 1.Becker J.C., Stang A., DeCaprio J.A., Cerroni L., Lebbé C., Veness M., Nghiem P. Merkel cell carcinoma. Nat. Rev. Dis. Prim. 2017;3:17077. doi: 10.1038/nrdp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott M.P., Helm K.F. Cytokeratin 20: a marker for diagnosing Merkel cell carcinoma. Am. J. Dermatopathol. 1999;21:16–20. doi: 10.1097/00000372-199902000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Toker C. Trabecular carcinoma of the skin. Arch. Dermatol. 1972;105:107–110. [PubMed] [Google Scholar]
- 4.Agelli M., Clegg L.X. Epidemiology of primary Merkel cell carcinoma in the United States. J. Am. Acad. Dermatol. 2003;49:832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- 5.Agelli M., Clegg L.X., Becker J.C., Rollison D.E. The etiology and epidemiology of Merkel cell carcinoma. Curr. Probl. Cancer. 2010;34:14–37. doi: 10.1016/j.currproblcancer.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia S., Afanasiev O., Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Curr. Oncol. Rep. 2011;13:488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colunga A., Pulliam T., Nghiem P. Merkel cell carcinoma in the age of immunotherapy: facts and hopes. Clin. Cancer Res. 2017;24:2035–2043. doi: 10.1158/1078-0432.CCR-17-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer J.G., Blom A., Doumani R., Lewis C., Tarabadkar E.S., Anderson A., Ma C., Bestick A., Parvathaneni U., Bhatia S., et al. Response rates and durability of chemotherapy among 62 patients with metastatic Merkel cell carcinoma. Canc. Med. 2016;5:2294–2301. doi: 10.1002/cam4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nghiem P., Kaufman H.L., Bharmal M., Mahnke L., Phatak H., Becker J.C. Systematic literature review of efficacy, safety and tolerability outcomes of chemotherapy regimens in patients with metastatic Merkel cell carcinoma. Future Oncol. 2017;13:1263–1279. doi: 10.2217/fon-2017-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schadendorf D., Lebbe C., Zur Hausen A., Avril M.F., Hariharan S., Bharmal M., Becker J.C. Merkel cell carcinoma: epidemiology, prognosis, therapy and unmet medical needs. Oxford J. Canc. 2017;1990(71):53–69. doi: 10.1016/j.ejca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Afanasiev O.K., Nagase K., Simonson W., Vandeven N., Blom A., Koelle D.M., Clark R., Nghiem P. Vascular E-selectin expression correlates with CD8 lymphocyte infiltration and improved outcome in Merkel cell carcinoma. J. Invest. Dermatol. 2013;133:2065–2073. doi: 10.1038/jid.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowlatshahi M., Huang V., Gehad A.E., Jiang Y., Calarese A., Teague J.E., Dorosario A.A., Cheng J., Nghiem P., Schanbacher C.F., et al. Tumor-specific T cells in human Merkel cell carcinomas: a possible role for Tregs and T-cell exhaustion in reducing T-cell responses. J. Invest. Dermatol. 2013;133:1879–1889. doi: 10.1038/jid.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer J.G., Afanasiev O.K., McClurkan C., Paulson K., Nagase K., Jing L., Marshak J.O., Dong L., Carter J., Lai I., et al. Merkel cell polyomavirus-specific CD8(+) and CD4(+) T-cell responses identified in Merkel cell carcinomas and blood. Clin. Canc. Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson K.G., Iyer J.G., Simonson W.T., Blom A., Thibodeau R.M., Schmidt M., Pietromonaco S., Sokil M., Warton E.M., Asgari M.M., et al. CD8+ lymphocyte intratumoral infiltration as a stage-independent predictor of Merkel cell carcinoma survival: a population-based study. Am. J. Clin. Pathol. 2014;142:452–458. doi: 10.1309/AJCPIKDZM39CRPNC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulson K.G., Iyer J.G., Tegeder A.R., Thibodeau R., Schelter J., Koba S., Schrama D., Simonson W.T., Lemos B.D., Byrd D.R., et al. Transcriptome-wide studies of Merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J. Clin. Oncol. 2011;29:1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgson N.C. Merkel cell carcinoma: changing incidence trends. J. Surg. Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 17.Paulson K.G., Park S.Y., Vandeven N.A., Lachance K., Thomas H., Chapuis A.G., Harms K.L., Thompson J.A., Bhatia S., Stang A., et al. Merkel cell carcinoma: current United States incidence and projected increases based on changing demographics. J. Am. Acad. Dermatol. 2017;78:457–463. doi: 10.1016/j.jaad.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stang A., Becker J.C., Nghiem P., Ferlay J. The association between geographic location and incidence of Merkel cell carcinoma in comparison to melanoma: an international assessment. Oxford J. Canc. 2018;1990(94):47–60. doi: 10.1016/j.ejca.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantripragada K., Birnbaum A. Response to anti-PD-1 therapy in metastatic Merkel cell carcinoma metastatic to the heart and pancreas. Cureus. 2015;7:e403. doi: 10.7759/cureus.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman H.L., Russell J., Hamid O., Bhatia S., Terheyden P., D'Angelo S.P., Shih K.C., Lebbe C., Linette G.P., Milella M., et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman H.L., Russell J.S., Hamid O., Bhatia S., Terheyden P., D'Angelo S.P., Shih K.C., Lebbé C., Milella M., Brownell I., et al. Updated efficacy of avelumab in patients with previously treated metastatic Merkel cell carcinoma after ≥1 year of follow-up: JAVELIN Merkel 200, a phase 2 clinical trial. J. Immunother. Canc. 2018;6:7. doi: 10.1186/s40425-017-0310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nghiem P., Bhatia S., Lipson E.J., Sharfman W.H., Kudchadkar R.R., Brohl A.S., Friedlander P.A., Daud A., Kluger H.M., Reddy S.A., et al. Durable tumor regression and overall survival in patients with advanced Merkel cell carcinoma receiving pembrolizumab as first-line therapy. J. Clin. Oncol. 2019;37:693–702. doi: 10.1200/JCO.18.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barailler H., Salomon G., Dutriaux C., Prey S., Gérard E., Dousset L., Mertens C., Beylot-Barry M., Meyer N., Pham-Ledard A. Adverse events, need for hospitalization and systemic immunosuppression in very older patients (over 80 years) treated with anti-PD-1 for metastatic melanoma. J. Geriatr. Oncol. 2020;11:1340–1343. doi: 10.1016/j.jgo.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D.B., Sullivan R.J., Menzies A.M. Immune checkpoint inhibitors in challenging populations. Cancer. 2017;123:1904–1911. doi: 10.1002/cncr.30642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engels E.A., Frisch M., Goedert J.J., Biggar R.J., Miller R.W. Merkel cell carcinoma and HIV infection. Lancet. 2002;359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 26.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arora R., Gupta K., Vijaykumar A., Krishna S. DETECTing Merkel cell polyomavirus in Merkel tumors. Front. Mol. Biosci. 2020;7:10. doi: 10.3389/fmolb.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kervarrec T., Tallet A., Miquelestorena-Standley E., Houben R., Schrama D., Gambichler T., Berthon P., Le Corre Y., Hainaut-Wierzbicka E., Aubin F., et al. Diagnostic accuracy of a panel of immunohistochemical and molecular markers to distinguish Merkel cell carcinoma from other neuroendocrine carcinomas. Mod. Pathol. 2019;32:499–510. doi: 10.1038/s41379-018-0155-y. [DOI] [PubMed] [Google Scholar]
- 29.Pastrana D.V., Tolstov Y.L., Becker J.C., Moore P.S., Chang Y., Buck C.B. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5:e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrama D., Peitsch W.K., Zapatka M., Kneitz H., Houben R., Eib S., Haferkamp S., Moore P.S., Shuda M., Thompson J.F., et al. Merkel cell polyomavirus status is not associated with clinical course of Merkel cell carcinoma. J. Invest. Dermatol. 2011;131:1631–1638. doi: 10.1038/jid.2011.115. [DOI] [PubMed] [Google Scholar]
- 31.Shuda M., Arora R., Kwun H.J., Feng H., Sarid R., Fernández-Figueras M.T., Tolstov Y., Gjoerup O., Mansukhani M.M., Swerdlow S.H., et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gjoerup O., Chang Y., George F.V.W., George K. Advances in Cancer Research. Academic Press; 2010. Chapter 1-update on human polyomaviruses and cancer; pp. 1–51. [DOI] [PubMed] [Google Scholar]
- 33.Harrison C.J., Meinke G., Kwun H.J., Rogalin H., Phelan P.J., Bullock P.A., Chang Y., Moore P.S., Bohm A. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J. Mol. Biol. 2011;409:529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwun H.J., Guastafierro A., Shuda M., Meinke G., Bohm A., Moore P.S., Chang Y. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter J.J., Daugherty M.D., Qi X., Bheda-Malge A., Wipf G.C., Robinson K., Roman A., Malik H.S., Galloway D.A. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:12744. doi: 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schowalter R.M., Buck C.B. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 2013;9:e1003558. doi: 10.1371/journal.ppat.1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schowalter R.M., Pastrana D.V., Buck C.B. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7:e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schowalter R.M., Reinhold W.C., Buck C.B. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS One. 2012;7:e42181. doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo G.J., Chen C.J., Sullivan C.S. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Schowalter R.M., Pastrana D.V., Pumphrey K.A., Moyer A.L., Buck C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolstov Y.L., Pastrana D.V., Feng H., Becker J.C., Jenkins F.J., Moschos S., Chang Y., Buck C.B., Moore P.S. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J. Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czech-Sioli M., Günther T., Therre M., Spohn M., Indenbirken D., Theiss J., Riethdorf S., Qi M., Alawi M., Wülbeck C., et al. High-resolution analysis of Merkel Cell Polyomavirus in Merkel Cell Carcinoma reveals distinct integration patterns and suggests NHEJ and MMBIR as underlying mechanisms. PLoS Pathog. 2020;16:e1008562. doi: 10.1371/journal.ppat.1008562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martel-Jantin C., Filippone C., Cassar O., Peter M., Tomasic G., Vielh P., Brière J., Petrella T., Aubriot-Lorton M.H., Mortier L., et al. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. Virology. 2012;426:134–142. doi: 10.1016/j.virol.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 44.Starrett G.J., Thakuria M., Chen T., Marcelus C., Cheng J., Nomburg J., Thorner A.R., Slevin M.K., Powers W., Burns R.T., et al. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med. 2020;12:30. doi: 10.1186/s13073-020-00727-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borchert S., Czech-Sioli M., Neumann F., Schmidt C., Wimmer P., Dobner T., Grundhoff A., Fischer N. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J. Virol. 2014;88:3144–3160. doi: 10.1128/JVI.02916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houben R., Adam C., Baeurle A., Hesbacher S., Grimm J., Angermeyer S., Henzel K., Hauser S., Elling R., Bröcker E.B., et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 47.Sastre-Garau X., Peter M., Avril M.F., Laude H., Couturier J., Rozenberg F., Almeida A., Boitier F., Carlotti A., Couturaud B., et al. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J. Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 48.Shuda M., Feng H., Kwun H.J., Rosen S.T., Gjoerup O., Moore P.S., Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houben R., Shuda M., Weinkam R., Schrama D., Feng H., Chang Y., Moore P.S., Becker J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuda M., Kwun H.J., Feng H., Chang Y., Moore P.S. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaegen M.E., Mangelberger D., Harms P.W., Vozheiko T.D., Weick J.W., Wilbert D.M., Saunders T.L., Ermilov A.N., Bichakjian C.K., Johnson T.M., et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Invest. Dermatol. 2014;135:1415–1424. doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grundhoff A., Fischer N. Merkel cell polyomavirus, a highly prevalent virus with tumorigenic potential. Curr. Opin. Virol. 2015;14:129–137. doi: 10.1016/j.coviro.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Liu W., MacDonald M., You J. Merkel cell polyomavirus infection and Merkel cell carcinoma. Curr. Opin. Virol. 2016;20:20–27. doi: 10.1016/j.coviro.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W., You J. Molecular mechanisms of Merkel cell polyomavirus transformation and replication. Annu. Rev. Virol. 2020;7:289–307. doi: 10.1146/annurev-virology-011720-121757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spurgeon M.E., Lambert P.F. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wendzicki J.A., Moore P.S., Chang Y. Large T and small T antigens of Merkel cell polyomavirus. Curr. Opin. Virol. 2015;11:38–43. doi: 10.1016/j.coviro.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terheyden P., Becker J.C. New developments in the biology and the treatment of metastatic Merkel cell carcinoma. Curr. Opin. Oncol. 2017;29:221–226. doi: 10.1097/CCO.0000000000000363. [DOI] [PubMed] [Google Scholar]
- 58.Tang C.K., Toker C. Trabecular carcinoma of the skin: an ultrastructural study. Cancer. 1978;42:2311–2321. doi: 10.1002/1097-0142(197811)42:5<2311::aid-cncr2820420531>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 59.Chen T., Hedman L., Mattila P.S., Jartti T., Ruuskanen O., Söderlund-Venermo M., Hedman K. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J. Clin. Virol. 2011;50:125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Kean J.M., Rao S., Wang M., Garcea R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tolstov Y.L., Knauer A., Chen J.G., Kensler T.W., Kingsley L.A., Moore P.S., Chang Y. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg. Infect. Dis. 2011;17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Touzé A., Le Bidre E., Laude H., Fleury M.J., Cazal R., Arnold F., Carlotti A., Maubec E., Aubin F., Avril M.F., et al. High levels of antibodies against Merkel cell polyomavirus identify a subset of patients with Merkel cell carcinoma with better clinical outcome. J. Clin. Oncol. 2011;29:1612–1619. doi: 10.1200/JCO.2010.31.1704. [DOI] [PubMed] [Google Scholar]
- 63.Pastrana D.V., Wieland U., Silling S., Buck C.B., Pfister H. Positive correlation between Merkel cell polyomavirus viral load and capsid-specific antibody titer. Med. Microbiol. Immunol. 2012;201:17–23. doi: 10.1007/s00430-011-0200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viscidi R.P., Rollison D.E., Sondak V.K., Silver B., Messina J.L., Giuliano A.R., Fulp W., Ajidahun A., Rivanera D. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin. Vaccine Immunol. 2011;18:1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wieland U., Mauch C., Kreuter A., Krieg T., Pfister H. Merkel cell polyomavirus DNA in persons without Merkel cell carcinoma. Emerg. Infect. Dis. 2009;15:1496–1498. doi: 10.3201/eid1509.081575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abedi Kiasari B., Vallely P.J., Klapper P.E. Merkel cell polyomavirus DNA in immunocompetent and immunocompromised patients with respiratory disease. J. Med. Virol. 2011;83:2220–2224. doi: 10.1002/jmv.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bialasiewicz S., Lambert S.B., Whiley D.M., Nissen M.D., Sloots T.P. Merkel cell polyomavirus DNA in respiratory specimens from children and adults. Emerg. Infect. Dis. 2009;15:492–494. doi: 10.3201/eid1503.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goh S., Lindau C., Tiveljung-Lindell A., Allander T. Merkel cell polyomavirus in respiratory tract secretions. Emerg. Infect. Dis. 2009;15:489–491. doi: 10.3201/eid1503.081206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Husseiny M.I., Anastasi B., Singer J., Lacey S.F. A comparative study of Merkel cell, BK and JC polyomavirus infections in renal transplant recipients and healthy subjects. J. Clin. Virol. 2010;49:137–140. doi: 10.1016/j.jcv.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mertz K.D., Junt T., Schmid M., Pfaltz M., Kempf W. Inflammatory monocytes are a reservoir for Merkel cell polyomavirus. J. Invest. Dermatol. 2010;130:1146–1151. doi: 10.1038/jid.2009.392. [DOI] [PubMed] [Google Scholar]
- 71.Pancaldi C., Corazzari V., Maniero S., Mazzoni E., Comar M., Martini F., Tognon M. Merkel cell polyomavirus DNA sequences in the buffy coats of healthy blood donors. Blood. 2011;117:7099–7101. doi: 10.1182/blood-2010-09-310557. [DOI] [PubMed] [Google Scholar]
- 72.Erickson K.D., Garcea R.L., Tsai B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 2009;83:10275–10279. doi: 10.1128/JVI.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker M., Dominguez M., Greune L., Soria-Martinez L., Pfleiderer M.M., Schowalter R., Buck C.B., Blaum B.S., Schmidt M.A., Schelhaas M. Infectious entry of Merkel cell polyomavirus. J. Virol. 2019;93 doi: 10.1128/JVI.02004-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neumann F., Borchert S., Schmidt C., Reimer R., Hohenberg H., Fischer N., Grundhoff A. Replication, gene expression and particle production by a consensus Merkel cell polyomavirus (MCPyV) genome. PLoS One. 2011;6:e29112. doi: 10.1371/journal.pone.0029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng H., Kwun H.J., Liu X., Gjoerup O., Stolz D.B., Chang Y., Moore P.S. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J., Wang X., Diaz J., Tsang S.H., Buck C.B., You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013;87:9173–9188. doi: 10.1128/JVI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu W., Yang R., Payne A.S., Schowalter R.M., Spurgeon M.E., Lambert P.F., Xu X., Buck C.B., You J. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe. 2016;19:775–787. doi: 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellaud G., Gheit T., Pugin A., Pretet J.L., Tommasino M., Mougin C., Aubin F. Prevalence of human polyomavirus DNA in eyebrow hairs plucked from patients with psoriasis treated with TNF inhibitors. J. Eur. Acad. Dermatol. Venereol. : JEADV. 2015;29:1019–1021. doi: 10.1111/jdv.12439. [DOI] [PubMed] [Google Scholar]
- 79.Hampras S.S., Michel A., Schmitt M., Waterboer T., Kranz L., Gheit T., Fisher K., Sondak V.K., Messina J., Fenske N., et al. Merkel cell polyomavirus (MCV) T-antigen seroreactivity, MCV DNA in eyebrow hairs, and squamous cell carcinoma. Infect. Agents Cancer. 2015;10:35. doi: 10.1186/s13027-015-0030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peretti A., Borgogna C., Rossi D., De Paoli L., Bawadekar M., Zavattaro E., Boldorini R., De Andrea M., Gaidano G., Gariglio M. Analysis of human beta-papillomavirus and Merkel cell polyomavirus infection in skin lesions and eyebrow hair bulbs from a cohort of patients with chronic lymphocytic leukaemia. Br. J. Dermatol. 2014;171:1525–1528. doi: 10.1111/bjd.13215. [DOI] [PubMed] [Google Scholar]
- 81.Saláková M., Košlabová E., Vojtěchová Z., Tachezy R., Šroller V. Detection of human polyomaviruses MCPyV, HPyV6, and HPyV7 in malignant and non-malignant tonsillar tissues. J. Med. Virol. 2016;88:695–702. doi: 10.1002/jmv.24385. [DOI] [PubMed] [Google Scholar]
- 82.Diaz J., Wang X., Tsang S.H., Jiao J., You J. Phosphorylation of large T antigen regulates Merkel cell polyomavirus replication. Cancers. 2014;6 doi: 10.3390/cancers6031464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krump N.A., Wang R., Liu W., Yang J.F., Ma T., You J. Merkel cell polyomavirus infection induces an antiviral innate immune response in human dermal fibroblasts. J. Virol. 2021;95 doi: 10.1128/JVI.02211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shahzad N., Shuda M., Gheit T., Kwun H.J., Cornet I., Saidj D., Zannetti C., Hasan U., Chang Y., Moore P.S., et al. The T antigen locus of Merkel cell polyomavirus downregulates human Toll-like receptor 9 expression. J. Virol. 2013;87:13009–13019. doi: 10.1128/JVI.01786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu W., Krump N.A., MacDonald M., You J. Merkel cell polyomavirus infection of animal dermal fibroblasts. J. Virol. 2018;92 doi: 10.1128/JVI.01610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanly A.J., Elgart G.W., Jorda M., Smith J., Nadji M. Analysis of thyroid transcription factor-1 and cytokeratin 20 separates Merkel cell carcinoma from small cell carcinoma of lung. J. Cutan. Pathol. 2000;27:118–120. doi: 10.1034/j.1600-0560.2000.027003118.x. [DOI] [PubMed] [Google Scholar]
- 87.Moll I., Kuhn C., Moll R. Cytokeratin 20 is a general marker of cutaneous Merkel cells while certain neuronal proteins are absent. J. Invest. Dermatol. 1995;104:910–915. doi: 10.1111/1523-1747.ep12606183. [DOI] [PubMed] [Google Scholar]
- 88.Shuda M., Guastafierro A., Geng X., Shuda Y., Ostrowski S.M., Lukianov S., Jenkins F.J., Honda K., Maricich S.M., Moore P.S., et al. Merkel cell polyomavirus small T antigen induces cancer and embryonic Merkel cell proliferation in a transgenic mouse model. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moll I., Zieger W., Schmelz M. Proliferative Merkel cells were not detected in human skin. Arch. Dermatol. Res. 1996;288:184. doi: 10.1007/BF02505222. [DOI] [PubMed] [Google Scholar]
- 90.Narisawa Y., Inoue T., Nagase K. Evidence of proliferative activity in human Merkel cells: implications in the histogenesis of Merkel cell carcinoma. Arch. Dermatol. Res. 2019;311:37–43. doi: 10.1007/s00403-018-1877-x. [DOI] [PubMed] [Google Scholar]
- 91.Morrison K.M., Miesegaes G.R., Lumpkin E.A., Maricich S.M. Mammalian Merkel cells are descended from the epidermal lineage. Dev. Biol. 2009;336:76–83. doi: 10.1016/j.ydbio.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poulsen M. Merkel-cell carcinoma of the skin. Lancet Oncol. 2004;5:593–599. doi: 10.1016/S1470-2045(04)01593-1. [DOI] [PubMed] [Google Scholar]
- 93.Van Keymeulen A., Mascre G., Youseff K.K., Harel I., Michaux C., De Geest N., Szpalski C., Achouri Y., Bloch W., Hassan B.A., et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iacocca M.V., Abernethy J.L., Stefanato C.M., Allan A.E., Bhawan J. Mixed Merkel cell carcinoma and squamous cell carcinoma of the skin. J. Am. Acad. Dermatol. 1998;39:882–887. doi: 10.1016/s0190-9622(98)70372-x. [DOI] [PubMed] [Google Scholar]
- 95.Kervarrec T., Aljundi M., Appenzeller S., Samimi M., Maubec E., Cribier B., Deschamps L., Sarma B., Sarosi E.-M., Berthon P., et al. Polyomavirus-positive Merkel cell carcinoma derived from a trichoblastoma suggests an epithelial origin of this Merkel cell carcinoma. J. Invest. Dermatol. 2020;140:976–985. doi: 10.1016/j.jid.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 96.zur Hausen A., Rennspiess D., Winnepenninckx V., Speel E.-J., Kurz A.K. Early B-cell differentiation in Merkel cell carcinomas: clues to cellular ancestry. Cancer Res. 2013;73:4982. doi: 10.1158/0008-5472.CAN-13-0616. [DOI] [PubMed] [Google Scholar]
- 97.Sunshine J.C., Jahchan N.S., Sage J., Choi J. Are there multiple cells of origin of Merkel cell carcinoma? Oncogene. 2018;37:1409–1416. doi: 10.1038/s41388-017-0073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fan K., Gravemeyer J., Ritter C., Rasheed K., Gambichler T., Moens U., Shuda M., Schrama D., Becker J.C. MCPyV large T antigen-induced atonal homolog 1 is a lineage-dependency oncogene in Merkel cell carcinoma. J. Invest. Dermatol. 2020;140:56–65. doi: 10.1016/j.jid.2019.06.135. e53. [DOI] [PubMed] [Google Scholar]
- 99.Verhaegen M.E., Mangelberger D., Harms P.W., Eberl M., Wilbert D.M., Meireles J., Bichakjian C.K., Saunders T.L., Wong S.Y., Dlugosz A.A. Merkel cell polyomavirus small T antigen initiates Merkel cell carcinoma-like tumor development in mice. Cancer Res. 2017;77:3151. doi: 10.1158/0008-5472.CAN-17-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kervarrec T., Samimi M., Hesbacher S., Berthon P., Wobser M., Sallot A., Sarma B., Schweinitzer S., Gandon T., Destrieux C., et al. Merkel cell polyomavirus T antigens induce Merkel cell-like differentiation in GLI1-expressing epithelial cells. Cancers. 2020;12:1989. doi: 10.3390/cancers12071989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harold A., Amako Y., Hachisuka J., Bai Y., Li M.Y., Kubat L., Gravemeyer J., Franks J., Gibbs J.R., Park H.J., et al. Conversion of Sox2-dependent Merkel cell carcinoma to a differentiated neuron-like phenotype by T antigen inhibition. Proc. Natl. Acad. Sci. U. S. A. 2019;116:20104–20114. doi: 10.1073/pnas.1907154116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bardot E.S., Valdes V.J., Zhang J., Perdigoto C.N., Nicolis S., Hearn S.A., Silva J.M., Ezhkova E. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J. 2013;32:1990–2000. doi: 10.1038/emboj.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perdigoto C.N., Dauber K.L., Bar C., Tsai P.-C., Valdes V.J., Cohen I., Santoriello F.J., Zhao D., Zheng D., Hsu Y.-C., et al. Polycomb-mediated repression and sonic hedgehog signaling interact to regulate Merkel cell specification during skin development. PLoS Genet. 2016;12:e1006151. doi: 10.1371/journal.pgen.1006151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Busam K.J., Pulitzer M.P., Coit D.C., Arcila M., Leng D., Jungbluth A.A., Wiesner T. Reduced H3K27me3 expression in Merkel cell polyoma virus-positive tumors. Mod. Pathol. 2017;30:877–883. doi: 10.1038/modpathol.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsushita M., Iwasaki T., Wardhani L.O., Kuwamoto S., Nonaka D., Nagata K., Kato M., Kitamura Y., Hayashi K. Decreased H3K27me3 expression is associated with Merkel cell polyomavirus-negative Merkel cell carcinoma, especially combined with cutaneous squamous cell carcinoma. Anticancer Res. 2019;39:5573–5579. doi: 10.21873/anticanres.13751. [DOI] [PubMed] [Google Scholar]
- 106.Hoang M.P., Donizy P., Wu C.L., Kopczynski J., Pieniazek M., Miller D.M., Ryś J. TdT expression is a marker of better survival in Merkel cell carcinoma, and expression of B-cell markers is associated with Merkel cell polyomavirus. Am. J. Clin. Pathol. 2020;154:38–47. doi: 10.1093/ajcp/aqaa017. [DOI] [PubMed] [Google Scholar]
- 107.Johansson B., Sahi H., Koljonen V., Böhling T. The expression of terminal deoxynucleotidyl transferase and paired box gene 5 in Merkel cell carcinomas and its relation to the presence of Merkel cell polyomavirus DNA. J. Cutan. Pathol. 2019;46:26–32. doi: 10.1111/cup.13372. [DOI] [PubMed] [Google Scholar]
- 108.Pilarski L.M., Mant M.J., Ruether B.A. Pre-B cells in peripheral blood of multiple myeloma patients. Blood. 1985;66:416–422. [PubMed] [Google Scholar]
- 109.Sauer C.M., Haugg A.M., Chteinberg E., Rennspiess D., Winnepenninckx V., Speel E.J., Becker J.C., Kurz A.K., zur Hausen A. Reviewing the current evidence supporting early B-cells as the cellular origin of Merkel cell carcinoma. Crit. Rev. Oncol. Hematol. 2017;116:99–105. doi: 10.1016/j.critrevonc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 110.Howard R.A., Dores G.M., Curtis R.E., Anderson W.F., Travis L.B. Merkel cell carcinoma and multiple primary cancers. Cancer epidemiology biomarkers &. Prevention. 2006;15:1545. doi: 10.1158/1055-9965.EPI-05-0895. [DOI] [PubMed] [Google Scholar]
- 111.Kaae J., Hansen A.V., Biggar R.J., Boyd H.A., Moore P.S., Wohlfahrt J., Melbye M. Merkel cell carcinoma: incidence, mortality, and risk of other cancers. J. Natl. Canc. Inst. 2010;102:793–801. doi: 10.1093/jnci/djq120. [DOI] [PubMed] [Google Scholar]
- 112.Koljonen V., Kukko H., Pukkala E., Sankila R., Böhling T., Tukiainen E., Sihto H., Joensuu H. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br. J. Cancer. 2009;101:1444–1447. doi: 10.1038/sj.bjc.6605306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koljonen V., Kukko H., Tukiainen E., Böhling T., Sankila R., Joensuu H., Pukkala E. Second cancers following the diagnosis of Merkel cell carcinoma: a nationwide cohort study. Canc. Epidemiol. 2010;34:62–65. doi: 10.1016/j.canep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 114.Cimino P.J., Jr., Bahler D.W., Duncavage E.J. Detection of Merkel cell polyomavirus in chronic lymphocytic leukemia T-cells. Exp. Mol. Pathol. 2013;94:40–44. doi: 10.1016/j.yexmp.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 115.Haugg A.M., Speel E.J., Pantulu N.D., Pallasch C., Kurz A.K., Kvasnicka H.M., Cathomas G., Wendtner C.M., zur Hausen A. Fluorescence in situ hybridization confirms the presence of Merkel cell polyomavirus in chronic lymphocytic leukemia cells. Blood. 2011;117:5776–5777. doi: 10.1182/blood-2011-03-339895. [DOI] [PubMed] [Google Scholar]
- 116.Teman C.J., Tripp S.R., Perkins S.L., Duncavage E.J. Merkel cell polyomavirus (MCPyV) in chronic lymphocytic leukemia/small lymphocytic lymphoma. Leuk. Res. 2011;35:689–692. doi: 10.1016/j.leukres.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 117.Nirenberg A., Steinman H., Dixon J., Dixon A. Merkel cell carcinoma update: the case for two tumours. J. Eur. Acad. Dermatol. Venereol. 2020;34:1425–1431. doi: 10.1111/jdv.16158. [DOI] [PubMed] [Google Scholar]
- 118.Goh G., Walradt T., Markarov V., Blom A., Riaz N., Doumani R., Stafstrom K., Moshiri A., Yelistratova L., Levinsohn J., et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2016;7:3403–3415. doi: 10.18632/oncotarget.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Starrett G.J., Marcelus C., Cantalupo P.G., Katz J.P., Cheng J., Akagi K., Thakuria M., Rabinowits G., Wang L.C., Symer D.E., et al. Vol. 8. 2017. Merkel Cell Polyomavirus Exhibits Dominant Control of the Tumor Genome and Transcriptome in Virus-Associated Merkel Cell Carcinoma. mBio. e02079–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jankowski M., Kopinski P., Schwartz R., Czajkowski R. Merkel cell carcinoma: is this a true carcinoma? Exp. Dermatol. 2014;23:792–794. doi: 10.1111/exd.12490. [DOI] [PubMed] [Google Scholar]
- 121.Houben R., Grimm J., Willmes C., Weinkam R., Becker J.C., Schrama D. Merkel cell carcinoma and Merkel cell polyomavirus: evidence for hit-and-run oncogenesis. J. Invest. Dermatol. 2012;132:254–256. doi: 10.1038/jid.2011.260. [DOI] [PubMed] [Google Scholar]
- 122.Carter J.J., Paulson K.G., Wipf G.C., Miranda D., Madeleine M.M., Johnson L.G., Lemos B.D., Lee S., Warcola A.H., Iyer J.G., et al. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J. Natl. Canc. Inst. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Narisawa Y., Inoue T., Nagase K. Dermal and intraepidermal Merkel cell carcinoma with squamous cell carcinoma: a report of a rare case with special reference to the touch dome. Am. J. Dermatopathol. 2021;43:15–20. doi: 10.1097/DAD.0000000000001618. [DOI] [PubMed] [Google Scholar]
- 124.Navarrete-Dechent C., Cordova M., Aleissa S., Battle L.R., Ganly I., Pulitzer M., Rossi A.M. Dermoscopy and reflectance confocal microscopy of intraepidermal Merkel cell carcinoma. Australas. J. Dermatol. 2021;62:238–241. doi: 10.1111/ajd.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brown H.A., Sawyer D.M., Woo T. Intraepidermal Merkel cell carcinoma with No dermal involvement. Am. J. Dermatopathol. 2000;22:65–69. doi: 10.1097/00000372-200002000-00013. [DOI] [PubMed] [Google Scholar]
- 126.Song Y., Zheng S., Guo Y. Intraepidermal Merkel cell carcinoma mimicking melanoma in situ: a case report. Am. J. Dermatopathol. 2020;42:977–980. doi: 10.1097/DAD.0000000000001793. [DOI] [PubMed] [Google Scholar]
- 127.Harms P.W., Vats P., Verhaegen M.E., Robinson D.R., Wu Y.M., Dhanasekaran S.M., Palanisamy N., Siddiqui J., Cao X., Su F., et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–3727. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shuda M., Chang Y., Moore P.S. Merkel cell polyomavirus-positive Merkel cell carcinoma requires viral small T-antigen for cell proliferation. J. Invest. Dermatol. 2014;134:1479–1481. doi: 10.1038/jid.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang J.F., You J. Regulation of polyomavirus transcription by viral and cellular factors. Viruses. 2020;12 doi: 10.3390/v12101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu W., Krump N.A., Buck C.B., You J. Merkel cell polyomavirus infection and detection. JoVE : JoVE. 2019;144 doi: 10.3791/58950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 132.Theiss J.M., Günther T., Alawi M., Neumann F., Tessmer U., Fischer N., Grundhoff A. A comprehensive analysis of replicating Merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog. 2015;11:e1004974. doi: 10.1371/journal.ppat.1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ajuh E.T., Wu Z., Kraus E., Weissbach F.H., Bethge T., Gosert R., Fischer N., Hirsch H.H. Novel human polyomavirus noncoding control regions differ in bidirectional gene expression according to host cell, large T-antigen expression, and clinically occurring rearrangements. J. Virol. 2018;92 doi: 10.1128/JVI.02231-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu W., Krump N.A., Herlyn M., You J. Combining DNA damage induction with BCL-2 inhibition to enhance Merkel cell carcinoma cytotoxicity. Biology. 2020;9 doi: 10.3390/biology9020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kenney S., Natarajan V., Strike D., Khoury G., Salzman N.P. JC virus enhancer-promoter active in human brain cells. Science (New York, N.Y.) 1984;226:1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- 136.Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc. Natl. Acad. Sci. U. S. A. 1981;78:943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moreau P., Hen R., Wasylyk B., Everett R., Gaub M.P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.White M.K., Safak M., Khalili K. Regulation of gene expression in primate polyomaviruses. J. Virol. 2009;83:10846–10856. doi: 10.1128/JVI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Van Gele M., Boyle G.M., Cook A.L., Vandesompele J., Boonefaes T., Rottiers P., Van Roy N., De Paepe A., Parsons P.G., Leonard J.H., et al. Gene-expression profiling reveals distinct expression patterns for Classic versus Variant Merkel cell phenotypes and new classifier genes to distinguish Merkel cell from small-cell lung carcinoma. Oncogene. 2004;23:2732–2742. doi: 10.1038/sj.onc.1207421. [DOI] [PubMed] [Google Scholar]
- 140.Alonso-Curbelo D., Ho Y.J., Burdziak C., Maag J.L.V., Morris J.P.t., Chandwani R., Chen H.A., Tsanov K.M., Barriga F.M., Luan W., et al. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature. 2021;590:642–648. doi: 10.1038/s41586-020-03147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362 doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuppe C., Ibrahim M.M., Kranz J., Zhang X., Ziegler S., Perales-Patón J., Jansen J., Reimer K.C., Smith J.R., Dobie R., et al. Decoding myofibroblast origins in human kidney fibrosis. Nature. 2021;589:281–286. doi: 10.1038/s41586-020-2941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Polak P., Karlić R., Koren A., Thurman R., Sandstrom R., Lawrence M., Reynolds A., Rynes E., Vlahoviček K., Stamatoyannopoulos J.A., et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Higaki-Mori H., Kuwamoto S., Iwasaki T., Kato M., Murakami I., Nagata K., Sano H., Horie Y., Yoshida Y., Yamamoto O., et al. Association of Merkel cell polyomavirus infection with clinicopathological differences in Merkel cell carcinoma. Hum. Pathol. 2012;43:2282–2291. doi: 10.1016/j.humpath.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 145.Kervarrec T., Tallet A., Miquelestorena-Standley E., Houben R., Schrama D., Gambichler T., Berthon P., Le Corre Y., Hainaut-Wierzbicka E., Aubin F., et al. Morphologic and immunophenotypical features distinguishing Merkel cell polyomavirus-positive and negative Merkel cell carcinoma. Mod. Pathol. 2019;32:1605–1616. doi: 10.1038/s41379-019-0288-7. [DOI] [PubMed] [Google Scholar]
- 146.Wong S.Q., Waldeck K., Vergara I.A., Schröder J., Madore J., Wilmott J.S., Colebatch A.J., De Paoli-Iseppi R., Li J., Lupat R., et al. UV-associated mutations underlie the etiology of MCV-negative Merkel cell carcinomas. Cancer Res. 2015;75:5228–5234. doi: 10.1158/0008-5472.CAN-15-1877. [DOI] [PubMed] [Google Scholar]